Abstract
To prevent the settlement and/or the growth of fouling organisms (i.e. bacteria, fungi or microalgae), benthic sessile species have developed various defense mechanisms among which the production of chemical molecules. While studies have mostly focused on the release of chemical compounds by single species, there exist limited data on multi-species assemblages. We used an integrative approach to explore the potential interactive effects of distinct assemblages of two corals species and one giant clam species on biofouling appearance and composition. Remarkably, we found distinct biofouling communities suggesting the importance of benthic sessile assemblages in biofouling control. Moreover, the assemblage of 3 species led to an inhibition of biofouling, likely through a complex of secondary metabolites. Our results highlight that through their different effect on their near environment, species assemblages might be of upmost importance for their survival and therefore, should now be taken into account for sustainable management of coral reefs.
Introduction
Benthic sessile communities are under direct and constant pressure of their surrounding environment. Therefore, many species have developed various mechanisms such as direct and indirect biotic interactions to maximize their survival1. Each of these interactions can be either positive or negative and are responsible to a large extent for structuring communities2. Most studies on bioactive products have focused on the competition and defense processes that may lead to the discovery and commercialization of new bioactive metabolites3.
In sessile coral reef organisms, numerous studies have identified a wide variety of secondary metabolites such as terpenoids (e.g. siphonodictine4), saponins (e.g. muricins5), macrolides (e.g. latrunculins6), and steroids (e.g. verumbsteroids7) that can reduce the growth or even lead to the death of neighboring species. The production of such secondary metabolites, mainly non-polar, synthesized constitutively or inducible8, strongly influences sessile organisms’ behaviors in response to local competition or predation pressures. The latter are particularly pervasive in marine biofouling environments where strong developmental and organism-environment interactions take place9.
Biofouling is an ubiquitous phenomenon in the marine environment and a common feature of a wide variety of natural and artificial structures10. Biofouling formation typically starts with the adhesion of dissolved organic matter onto a surface which leads to physiochemical changes11 and to the development of a bacterial mat or early biofilm layer12. Successfully settled bacterial communities may influence the successive settlement of micro-algae (e.g. diatoms, cyanobacteria), fungi and protists, which are all precursors for colonization by larger fouling organisms or macrofouling9.
Biofouling formation onto a substrate, its growth rate and the type of species able to colonize this habitat are modulated by various interactions and involve complex biochemical, behavioral or physical mechanisms9,13. The external environment plays an important role in the biofouling formation, in particular through physical and chemical conditions14,15. For example, the availability of nutrients will play a critical role in the selection of early colonizers and the successive community composition of the biofouling16. Light and temperature increase also play important roles in enhancing the propagation of biofilm and biofouling17,18. Besides these environmental factors, many antifouling agents have been characterized from sessile marine organisms, in particular antifouling molecules produced by sponges, soft corals, and seaweeds12,19,20. Microorganisms associated with marine algae and invertebrates, such as epibionts, also possess antifouling potential12. Despite the growing knowledge that biofouling-associated antifouling compounds are damaging to both larval and adult stages of hard corals and giant clams21–23, there exist very limited data on the actual processes and the specific molecules involved in antifouling activities of multiple interactive coral reef species. There is now growing evidence that hard corals release antifouling active substances3, which can confer different competitive abilities to coral species against algae24.
Studies on Antarctic marine benthos have shown that by bringing together different defense strategies, mainly regulated by chemical interactions such as deterrent or repellent molecules, species assemblages create a complex model of interaction which may help protect Antarctic organisms from competition, for space and fouling pressure notably25. In coral reef ecosystems, the variety of antifouling compounds produced by sessile organisms, even among coral genera, prompted us to study the importance of species biodiversity and interaction on coral reefs’ fitness. Our work investigated their potential cumulative effect on biofouling formation and their ability to minimize the dramatic impact that uncontrolled biofouling expansions (i.e shifts from coral to algal dominated reefs) may have on coral reef ecosystems.
Thus, in this study we combined cytological, metagenomic and metabolomic approaches to explore the interactive effects of multiple benthic species assemblages on the biofouling appearance and composition under normal and thermal stress conditions. Two common Indo-Pacific scleractinian coral species, Pocillopora damicornis and Acropora cytherea, and the giant clam Tridacna maxima were artificially grouped in distinct assemblages, exposed to varying water temperatures, and their influence on algal biofouling formation was investigated.
Material and Methods
Sample collection and preparation
Corals and giant clams collection
Acropora cytherea (n = 4) and Pocillopora damicornis (n = 4) colonies were collected at low tide (∼2 m) in Moorea lagoon, French Polynesia (17°30′S, 149°50′W fringing reef Linareva26). The sampled colonies were then cut into nubbins using side-cutting pliers to produce a minimum of 45 small fragments each. All the samples were grown on tables in a common garden located in the lagoon area of the InterContinental Moorea Resort & Spa during an 8 months period. The giant clams T. maxima were purchased at a French Polynesian distributor on Reao Island (18°28′S, 136°25′W; Company identification number - N°Tahiti: 139 519), and then acclimated in the common garden. The three species were placed on 3 different distant (>3 m) tables in order to avoid inter-species’ interactions. A CITES permit was obtained to allow for samples export (CITES – FR1698700087 – E).
Experimental design
One to three species were placed in aquariums. The different associations of organisms per aquarium are thereafter referred to as “assemblage”. Five distinct assemblages of coral and/or giant clams species were studied: P. damicornis + A. cytherea + T. maxima (PAT); P. damicornis + A. cytherea (PA); A. cytherea + T. maxima (AT); P. damicornis (P) or T. maxima (T) alone. Three nubbins from 4 coral colonies per species (12 nubbins per coral species) and/or 12 giant clams were used for each assemblage. Therefore, the 40 L aquariums were composed of either 12 (assemblages T and P) or 24 (assemblages PA and AT) or 36 (assemblages PAT) organisms (Supplementary S1a). For each experiment (A, B and C), one aquarium without macroorganisms was kept as a control at lagoon temperature (W). Aquariums were randomly chosen to avoid a potential aquarium effect. Each aquarium was open-circuit with seawater pumped directly from Moorea Opunohu’s bay. Two filters of 60 microns each were used to remove sediment, and the water flow in aquariums was 20 L/h. The water temperature within aquariums was controlled with the Biotherm pro system (Hobby, Stukenbrock, Germany). Temperature/Light Data Loggers (P/N U22-001, Onset, Bourne, Massachusetts; or Ruskin, Ottawa, Canada) recorded temperature data every 10 minutes. All aquariums received the same light using LED lamps (PlanetPro ELOS, Verona, Italy), following a diurnal cycle.
The experiment was performed in duplicate (exp. A and B), spaced two weeks apart, except for the AT assemblage which was also studied in duplicate but during the same experiment (exp. C). In each of A and B experiments, 4 distinct assemblages were studied in parallel (assemblages PAT, PA, T and P; 2 aquariums per assemblage; n = 8 aquariums per experiment). After an acclimation period of 12 days, for each set of assemblage, one of the two aquariums was maintained at the ambient lagoon temperature (27 °C; LT; control), and the second one was submitted to a stress temperature (ST). The stress temperature was performed as follows: increased 1 °C per day from day 12 to day 17 up to 32 °C where it was maintained for 3 days (from day 17 to day 19), before being decreased back to 27 °C (day 20). Similarly, in experiment C, after the acclimation period, two aquariums of AT assemblages were placed at lagoon temperature and the remaining two were exposed to the stress temperature (n = 4 aquariums).
Biofilm and nutrient sampling
During the experiments, biofilm deposited on each aquarium wall was scraped at day 12 and 20 using a cover glass slide, and stored in 70% ethanol at −20 °C until further DNA analysis. Three replicates of water samples (40 mL) from aquariums were sampled at the same time and kept at −20 °C until the analysis of nitrite (NO2−), nitrate (NO3−), phosphate (PO42−), ammonium (NH4+) and silicate (Si-OH4; Technicon Autoanalyzer system, Southampton, United Kingdom). We chose to analyze samples from day 12 (T0) during the experiments A, B and C; samples from day 17 (T1) during experiment B and from day 19 (T2) samples during experiment B and C. A synthesis of the tested samples as well as the statistics used are on Supplementary S1b and S1c.
Biofouling observation and cytology
Throughout the experiments, photographs of the aquarium were taken (days 15 or 19) and daily observations of the biofouling were recorded.
At beginning of each experimental run, two sterile microscope cover glasses and two sterile glass slides were placed in each aquarium. At the end of the experiment, the slides were observed in order to identify the most predominant genera per assemblage (Supplementary S1c). For the aquarium without macroorganisms (W), two slides for experiment B and C were observed.
DNA extraction, PCR amplification and sequencing
Biofilm DNAs were extracted using the CTAB-based protocol described in Rouzé et al.27 and stored at −20 °C until PCR amplification. Two different genetic markers were selected for metabarcoding analysis. The 16S ribosomal RNA V3/V4 region was amplified with the Bakt_341 forward (5′-CCTACGGGNGGCWGCAG-3′) and Bakt_805 reverse (5′-GACTACHVGGGTATCTAATCC- 3′) primers28. The 18S ribosomal RNA V4 region was amplified with the primer pair D512for18S forward (5′-ATTCCAGCTCCAATAGCG-3′) and the D978rev18S reverse (5′-GACTACGATGGTATCTAATC-3′), specifically designed for characterizing diatom communities29. The primers were modified to include IlluminaTM overhang adaptors as described by Kozich et al.30.
The polymerase chain reactions were performed (Supplementary S1c) and products were visualised on a 2% agarose gel. Samples were purified using magnetic beads (Agencourt Bioscience Corporation, Grand Rapids, Michigan), quantified (Qubit® 2.0 Fluorometer, Invitrogen, Carlsbard, California) and diluted at equimolar concentration to 3 ng/µL. Amplicons from DNA samples previously obtained from two high-latitudinal invasive species (Ciona savignyi and Asterias amurensis) from a previous study31, were used as internal quality controls. Nuclease-free UltraPure™ water (Thermo Fisher Scientific, Waltham, Massachussetts) was used as negative control. The samples were sent to New Zealand Genomics Limited at the University of Auckland for final high-throughput sequencing library preparation. Libraries were sequenced on a MiSeq Illumina™ platform using a 2 × 250 base-pair (bp) paired-end protocol.
Metabolomic sample preparation and Liquid Chromatography-Mass Spectrometry (LC-MS) analysis
Seawater samples from input pipes (n = 5) were collected through a tap placed before the aquariums, and seawater samples from aquariums (n = 23) were individually siphoned and filtered using cartridges Strata-XL 100 u polymeric reversed phase (Phenomenex, Torrance, California). Six liters of seawater per sample were filtered using an extraction chamber.
Samples were extracted from the Strata-XL cartridges and then centrifuged to separate the residual water. The organic phase was transferred in a glass hemolysis tube before drying under nitrogen flow and freeze-dried overnight. Before LC-MS analysis, organic extracts were prepared at a concentration of 1 mg/mL in methanol and filtrated on Uptidisk (PTFE, 0.2 mm, Interchim, San Diego, California).
LC-MS analyses were performed with an LCQ Fleet 2300 (Thermo Scientific) to determine metabolites in the seawater of each assemblages. The samples were analyzed using a positive ion mode with full scan MS window of 200-2000 m/z. A quality control strategy was carried out using a Latin square design for samples, quality control and methanol blank samples in order to reduce the potential error of instrumental drift.
To determine the exact mass of secondary metabolites selected, LC-HRMS analyses were performed on a UHPLC system (Vanquish, Thermo Scientific) interfaced to a QTOF mass spectrometer (MaxisImpact I, Bruker Daltonics, Billerica, Massachussetts). Finally, to strengthen the putative identification of selected metabolites, LC-HRMS and LC-HRMSMS analyses were performed on a UHPLC system (Vanquish, Thermo Scientific) interfaced to an Orbitrap mass spectrometer (Q-exactive Plus Hybrid, Thermo Scientific, Supplementary S1c). Finally, metabolites were assigned according to literature and databases such as LIPID MAPS, Metlin or MarinLit32–34.
Cytology, nutrients, metabarcoding and metabolomics methods are detailed in the Supplementary File S1c.
Experiments
All experiments were performed in accordance with relevant guidelines and regulations.
Results
Significant change of seawater composition between experiments but only slightly between assemblages
The principal component analysis of the seawater nutrients (Supplementary Table S2 and Figure S3) enabled to distinguish each experiment. Nutrients were more abundant in the experiments A and C than in the experiment B, and Si-OH4 were more abundant in the experiment A compared to the two others. For each experiment, slight significant variations of nutrient concentrations were observed between conditions (stress-lagoon temperature) or between assemblages. Whatever the experiments, NH4+ and NO2− concentrations were systematically higher in all assemblages containing giant clams (Tukey test p < 0.05). Moreover, NH4+ concentrations were higher in PAT assemblages than in T assemblages. However, while in the experiment A, the range of NH4+ concentrations in PAT and T assemblages were of [1.493-3.016] µmol/L and of [1.059-1.287] µmol/L, respectively, NH4+ concentrations in the experiment B were less than 1 µmol/L (except for 1 sample) in PAT assemblages, which is lower than the range in T assemblage of the experiment A. Overall, the differences of water composition (relative and absolute concentrations) within experiments remained lower than the differences of water composition between experiments.
Difference of Biofouling appearance according to the type of assemblages
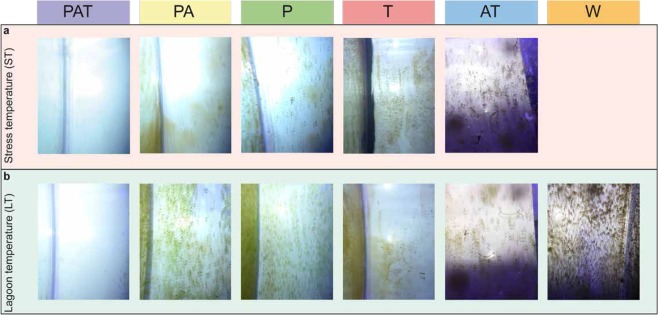
Ten days following the beginning of the acclimation period, the aquarium walls harbored various biofouling aspects that were specifically linked to the type of assemblages under investigation (Fig. 1). This phenomenon was similar between the thermal stress and control conditions (compare ST – LT, Fig. 1), was consistently observed at the end of all experiments and was reproducible. The walls of the aquariums containing PAT assemblage were clean with just a barely perceptible light green biofilm layer. Aquariums containing the T assemblage were covered with a brown biofouling and mucus. Brown and green biofouling were also perceptible on the aquarium walls of the P assemblage, contrasting with only a green layer apparent in the aquariums with either PA or AT assemblages. The most severe biofouling, e.g. murky aquarium walls, appeared in the W aquariums, devoid of macroorganisms. Since no visual difference of biofouling appearance was observed between the two conditions (ST and LT), we chose to focus on the core community by assemblage in order to characterize the different phenotypes observed. Therefore, all data were pooled by assemblage without consideration of treatments.
Figure 1.
Photographs of aquarium’s walls according to the assemblages (a) with or (b) without thermal stress. PAT: P. damicornis, A. cytherea and T. maxima; PA: P. damicornis and A. cytherea; P: P. damicornis; T: T. maxima; AT: A. cytherea and T. maxima; W: no macroorganisms; stress temperature (ST); lagoon temperature (LT, control). Photographs: Isis Guibert.
Distinct taxonomic groups according to the assemblages
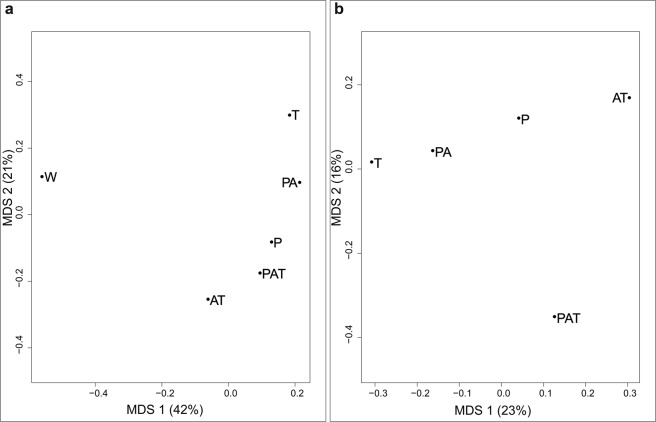
Seven taxonomic groups were defined using cytological observations: Chlorophyta, diatoms (Bacillariophyta), cyanobacteria (Cyanophyta), dinoflagellates (Dinoflagellata), Ochrophyta, Rhodophyta and undetermined. Within these, identifications were assigned at the lowest possible taxonomic level (family or genus). We characterized 201 distinct associated taxa across all assemblages. Classical multidimensional scaling analysis of the 6 assemblages based on the Jaccard’s Index (Fig. 2a) showed a strong dissimilarity (separation on the first axis) between aquariums without (W) and with (PAT, PA, AT, P and T) macroorganisms. The second axis separated the remaining five assemblages from T to AT. After exclusion of the control in the analysis, a similar distribution than the second axis of the first classical multidimensional scaling (from T to AT) was directly observed on the first axis and the second axis mainly separated PAT from all others (Fig. 2b).
Figure 2.
Classical multidimensional scaling from the Jaccard’s index of assemblages with (a) and without (b) control. PAT: P. damicornis, A. cytherea and T. maxima; PA: P. damicornis and A. cytherea; AT: A. cytherea and T. maxima; P: P. damicornis; T: T. maxima and W: no macroorganisms (control).
Different family and genus composition of the three main taxonomic groups according to the assemblages
We analysed the biofouling of PAT, PA, P and T assemblages using metabarcoding. After data processing of 1 202 179 paired-end reads, 711 313 contigs (16 505–83 708 per assemblage) with an average length of 420 bp (18S analysis) and 340 076 contigs (3 862–38 158 per assemblage) with an average length of 428 bp (16S analysis) were selected for taxonomic and functional analysis.
As the three main steps of biofouling are successively the development of bacteria, diatoms and algae, we focused on these three groups. The bacterial group composed of 13 847 unique contigs was divided into 44 phyla and 856 genera (Supplementary Table S4). Of the 8 330 unique contigs identified from the 18 S analysis, 6 369 were assigned to the diatoms group (Ochrophyta phylum), which divided into 63 genera (Supplementary Table S5). For the algae analysis, 19 990 unique contigs were assigned into 4 phyla (Chlorophyta, Euglenida, Eukaryota, Phaeophyceae) according to Silva database. From those phyla, 29 genera were identified (Supplementary Table S6).
In order to characterize the biofouling for each assemblage, we first compared the presence of families and genera for the three groups. Amongst the 28 characterized families of bacteria (Supplementary Figure S7), 13 families were found in all assemblages and 3 were found exclusively in the T assemblage (Methylocystaceae, Legionellaceae and Hyphomicrobiaceae). We also noticed that 12 bacterial families were present in 2 or 3 assemblages sharing or not a common species. For example, 3 families (Oceanopirillaceae, Halomonadaceae and Granulosinoccaceae) were found in all assemblages containing P. damicornis (PAT, PA and P). Other families were found in 2 assemblages such as Pseudoalteromonadaceae in PA and T assemblages but not in the PAT assemblage. Amongst the 17 identified families of diatoms, 12 were common to all assemblages, 2 (undetermined Ochromonadales and undetermined Coscinodiscophytina) were common to PA and P assemblages and 3 (Rhizosolenids, Dictyochophyceae and undetermined Pedinellales) were found exclusively in the PA assemblage. For the algae group, 28 families were identified and 15 were common to all assemblages. No specific families were determined for the PAT assemblage but 2 were specific to PA (Fucaceae and undetermined Eutroptiales) and to T (Ulvophyceae and Ceramiaceae) assemblages. Furthermore, Peridiniaceae, undetermined Rotaliida and Florideophyceae families were identified in all assemblages except for the PAT assemblage (Supplementary Table S4-S6).
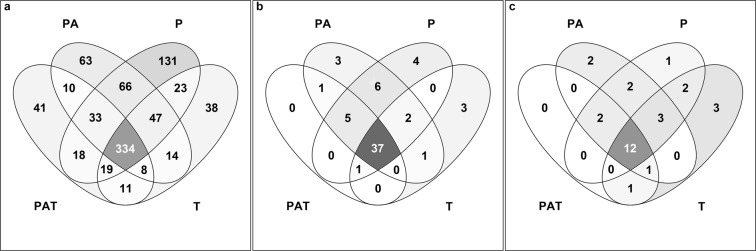
At genus level, a qualitative analysis revealed which genera of bacteria, diatom, and algae were common or specific to different assemblages (Fig. 3). Out of the 856 bacterial genera assigned, 474, 576, 672 and 494 were observed in PAT, PA, P and T assemblages, respectively, among which 334 were common. However, 41, 63, 131 and 38 genera were specific to PAT, PA, P and T assemblages, respectively. Interestingly, 47 genera were common to all assemblages except PAT (Fig. 3a). Few diatoms genera were found (63), but similarly to the bacterial group, 37 genera common to all assemblages were found (Fig. 3b). No specific diatoms were observed in the PAT assemblage, but 3 genera in PA, 4 in P, and 3 in T assemblages were specifically observed. Noteworthy, 2 diatom genera (Melosira sp. and Psammoneis sp.) were common to all assemblages except PAT. Concerning the algae group, only 29 genera were assigned and 12 were common to all assemblages (Fig. 3c). Similar to diatoms, no algal genera were specific to the PAT assemblage, and 3 (undetermined Peridiniceae, undetermined Florideophyceae and undetermined Rotaliida) were common to all the assemblages except this one.
Figure 3.
Venn diagrams of genera detected in the assemblages. (a) bacteria, (b) diatoms (c) algae. PAT: P. damicornis, A. cytherea and T. maxima; PA: P. damicornis and A. cytherea; P: P. damicornis and T: T. maxima.
Characterization of biofouling from each assemblage by specific species abundance and metabolic function of bacteria
To complement the qualitative analysis, we analyzed the relative abundance of bacteria. All assemblages were dominated by Rhodobacteraceae (~25-45%). Flavobacteriaceae were also found in large quantities (~5-15%) in all assemblages (Supplementary Figure S7). Nevertheless, the bacterial community composition yielded a specific “barcode community fingerprint” for each assemblage (Fig. 4). As shown, the barcode fingerprint can be clustered into 8 sub-communities. These clusters were rarely found in the same abundance level between assemblages. In fact, high abundances (red color, Fig. 4) of clusters 1, 2 and 5 are specific to PAT, T and P assemblages, respectively.
Figure 4.
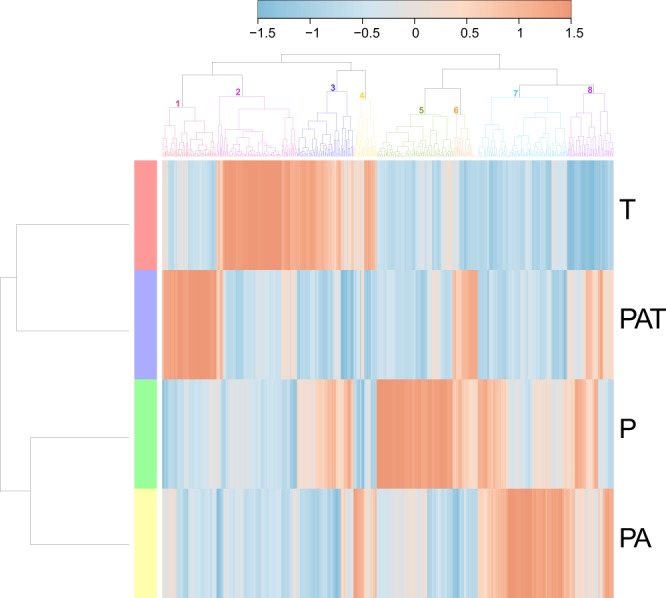
Heatmap based on bacterial genera and showing specific ‘barcode community fingerprints’ for each assemblage. Communities’ differences are showed on a relative scale, with enrichment (red) and depletion (blue) of sequence abundance calculated using the Ward’s clustering method and a Pearson distance measure. T: T. maxima; PAT: P. damicornis, A. cytherea and T. maxima; P: P. damicornis; PA: P. damicornis and A. cytherea.
We further compared the metabolic functions of bacterial communities within each assemblage (Fig. 5). Similar to the qualitative analysis, bacterial functions were clustered according to the differential abundance of bacterial species and their putative associated functions. Amongst the 5 clusters, 3 were linked to specific assemblages. Cluster 1 was specific to PAT with an increase of bacteria with “iron oxidizer” and “syntrophic” functions. Cluster 3, where 9 bacterial functions increased, was specific to T, and cluster 4 where “sulfur metabolizing”, “stores polyhydroxybutyrat” and “cellobiose degrading” functions increased was specific to PA. Interestingly, the two other clusters shared similar increase in bacterial functions between two assemblages. Cluster 2 showed an increase in “atrazine metabolism” and “nitrite reducer” functional responses in PAT and T assemblages. Increase of 8 bacterial functions in cluster 5 was common to P and PA assemblages amongst which “selenite reducer”, “chitin degradation”, “alkane degrader” and “streptomycin producer” were strongly represented. Consequently, PAT assemblage was closer to T assemblage than to PA and P assemblages.
Figure 5.
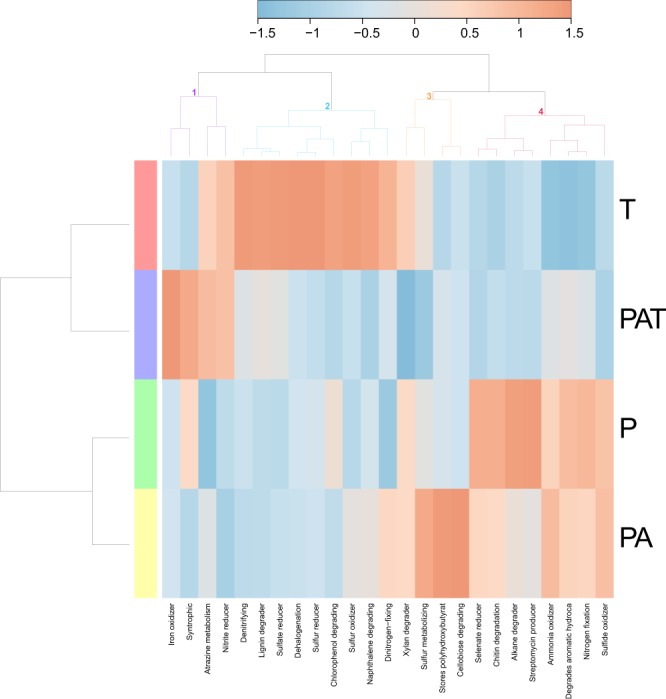
Heatmap of bacterial “metabolism by phenotype”. Data show functional differences derived from each assemblage on a relative scale with enrichment (red) and depletion (blue) of gene functions using a Pearson distance measure and the Ward’s clustering method. T: T. maxima; PAT: P. damicornis, A. cytherea and T. maxima; P: P. damicornis; PA: P. damicornis and A. cytherea.
Secondary metabolites released in seawater are related to each assemblage
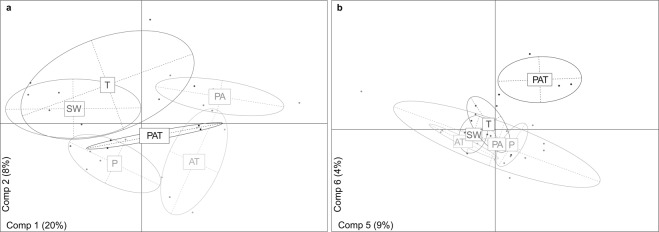
To determine if secondary metabolites might account for the difference of biofouling communities, a chemical analysis of the tanks’ seawater was performed. The overall LC-MS analysis allowed for the detection of 1 488 distinct metabolites. A large majority of them (1 370), were detected in both assemblages and incoming seawater. Particularly, no specific metabolites were detected in PAT. A quantitative Partial least Square Discriminant Analysis on metabolites was then conducted to discriminate each assemblage. The first components discriminated every assemblage (Fig. 6a), with PAT at the center of the plot, indicating an average mix of all metabolites rather than the emergence of new metabolites. In order to determine the Variable Importance in the Projection (VIP) potentially contributing to the inhibition of biofouling effect, we focused on the components which are clearly responsible for the separation of PAT from other assemblages (5th and 6th components, Fig. 6b). Thereby, 55 metabolites were selected among which the most abundant were kept as VIPs (n = 28). A similar approach was performed for the less abundant metabolites (68). Nevertheless, none of them were selected because none were specifically less abundant in PAT. The exact mass of VIPs was determined using LC-HRMS analyses with a High Resolutive QTOF mass spectrometer (Supplementary Figure S8). From these masses, the molecular formulas were estimated and putative identifications were assessed for 5 VIPs on 28 most abundant VIPs (Table 1). The 5 VIPs were all detected in each PAT assemblage regardless of the treatment (PAT.L and PAT.S). Identifications were strengthened by full and fragmentation spectra issued from MS analyses with a Qexactive mass spectrometer. According to available literature and databases, VIPs were assigned to carotenoid (VIP 630) and lipids (VIPs 399, 425, 979 and 981). Using the absorption spectra, the carotenoid was identified as peridinin which exhibited a maximum wavelength at 473-475 nm35,36. For the 4 other VIPs, spectrum and proposed mechanisms of fragmentation in the literature leads to the putative identification of 3 VIPs among which two digalactosyldiacyglycerols (DGDG 18:5/20:5 and DGDG 20:5/18:4)37,38 and the brassicasterol39. Based on mass and calculated formula, the last VIP might be a sterol.
Figure 6.
Partial least Square Discriminant Analysis of organic extracts metabolites from seawater’s aquariums. (a) Distribution of the metabolites on the first two components (Comp1 and Comp2) allowing the separation according to the assemblages. (b) Distribution on the fifth and six components (Comp5 and Comp6) providing the separation of PAT from other assemblages. PAT: P. damicornis, A. cytherea and T. maxima; PA: P. damicornis and A. cytherea; P: P. damicornis; T: T. maxima; AT: A. cytherea and T. maxima and SW: incoming seawater.
Table 1.
Accurate mass measurements, molecular formulas, and putative identification of the 5 determined VIPs.
| Ionic form | Experimental m/z | Calculated molecular formula | Calculated m/z | Mass error [ppm] | MS/MS fragmentation | Putative identification |
|---|---|---|---|---|---|---|
| [M + H]+ | 399.3624 | C28H47O | 399.3621 | 0.6 | 281.3509a, 297.2574, 255.2104, 135.1167 | Brassicasterol |
| [M + H]+ | 425.3413 | C29H45O2 | 425.3414 | −0.3 | 407.3306a, 191.1064, 173.0959 | Sterol |
| M°+ | 630.3539 | C39H50O7 | 630.3551 | −1.9 | 603.7519, 553.3241, 520.0495, 487.2805 | Peridinin |
| [M + Na+] | 979.5394 | C53H80O15Na | 979.5389 | 0.5 | 705.3445b, 677.3134c | DGDG 18:5/20:5 |
| [M + Na+] | 981.5551 | C53H82O15Na | 981.5546 | 0.6 | 679.3299b, 705.3453c | DGDG 20:5/18:4 |
a[M + H - H2O]+, b[M + Na - FA1]+, c[M + Na - FA2]+, FA = fatty acid.
Discussion
This is the first study demonstrating that biofouling formation is strongly influenced by surrounding benthic sessile species. In particular, we show that in its most severe form, the resulting effect of a species-specific assemblage is biofouling inhibition. Biofouling is a complex sequential process, which can be modulated by many abiotic and biotic factors, ranging from external factors to intrinsic influence of the marine taxa participating in biofouling formation itself.
Among these factors, nutrients are known to influence growth and productivity of various organisms such as bacteria and algae. In our study, nutrient contents in the seawater of each aquarium were compatible with those observed in previous nutrient analyses from the Moorea lagoon26,40. An increase of silicates was notably observed during the wet season as well as after strong rains41, as was the case for all aquariums of the A experiment. This link with external parameters can be explained by the design of our experiment, which consisted of an open-circuit with a water flow of 20 L/h per aquarium of 40 L. Therefore, the lagoon seawater composition mainly accounted for the significant variation of nutrient composition between the 3 experiments. As the effect of species assemblage on biofouling development was reproducible, this nutrient discrepancy between experiments could not account for the observed differences in biofouling development.
Studies have shown that bacteria, plankton and phytoplankton as well as dissolved organic and inorganic molecules are filtered from seawater and ingested by corals and bivalves42,43. This feeding can lead to a depletion of 30 to 45% of total chlorophyll a and picoplankton above a scleractinian coral dominated reef when compared with adjacent waters42. Tridacna species are able to filter up to 58% of algal cells from their surrounding water44. Moreover, a recent study has shown that corals are able to feed on diatoms such as Thalassiosira sp.43. In light of these results, and taking into account that giant clams filter large amounts of seawater (e.g. water filtration rate of one Tridacna crocea is 2-3 L/h)44, filtering activities could reduce the number of species participating in biofouling development and so could disrupt some steps of biofouling formation. Therefore, whenever another heterotrophic species is added to an assemblage, the community of microorganisms within the aquariums’ seawater could be modified (e.g. additive depletion of microorganisms). Such modifications could account for the differences in biofouling composition and development observed between assemblages, the stronger impact of these additive depletion effects leading to the inhibition of biofouling (e.g. lack of key species required for biofouling progression). Biofouling development was strongly inhibited in the three species assemblages, PAT, of which aquarium water was depleted of 47 bacteria, 2 diatoms and 3 algae genera when compared to the other assemblages. Among these lacking organisms, some bacterial taxa e.g. Rhodobacteraceae that are important biofilm precursors45, could be required for effective biofouling formation. Further studies of giant clam and coral filter-feeding species interactions will be helpful to better understanding their respective putative roles in depleting the microorganisms involved in biofouling progression. However, our experimental conditions with high seawater renewal and successive samplings led to four organisms per species at the end of the experiments. Therefore, corals and giant clams’ nutrition alone cannot be responsible for the disappearance of biofouling key species. Even though relevant, this hypothesis is not sufficient to account for the observed difference in biofouling development.
In contrast to ingested filtered organisms, those associated with invertebrate species, bacteria and microalgae, have been well characterized. Bacteria have been found living on the surface, in the tissues and in the mucus of coral species46–48. Because mucus contains a lot of inorganic phosphate and dissolved organic carbon, it is suspected as an effective inducer of bacterial growth49. Moreover, the diversity and quantity of the hosted bacteria differs not only between species but also according to their environment50. Dobrestov and collaborators12 have shown that numerous bacteria associated with corals possess an anti-bacterial activity, and antibacterial compounds in coral mucus have notably been found to select specific bacterial population leading to an antifouling effect51. Interestingly, 7 bacterial species were specifically found in PAT assemblages. Among them, bacteria belonging to the genus Lysobacter which are able to degrade biofilm and could contribute to the observed antifouling effect52. Corals and giant clams are known to produce bioactive compounds such as sterols which are described as chemical defensive substances53,54. Indeed, living corals are free of fouling and this particularity come from the production of secondary metabolites, especially sterols playing key roles in allelopathy with the ability to inhibit the growth of organisms around them54. Thereby, a fouling-resistant composition, non-toxic for the environment and applicable on immersed equipment, was made with sterols55. Even though no specific metabolite to the PAT assemblage was detected, suggesting that no new compound is produced in this context, 5 VIPs characterizing the PAT assemblage were highlighted. Among them, one was assigned to carotenoids and 4 to lipids. The carotenoid, identified as peridinin, is a specific pigment found in Symbiodinium dinoflagellates from corals and giant clams35. Interestingly, like most of the carotenoids, peridinin exhibits an inhibitory activity even at low concentration. As peridinin can inhibit cell proliferation and can exhibit a cytotoxic activity56,57, it might contribute to the biofouling inhibition observed in PAT assemblage. Moreover, among the lipids identified, one was assigned to brassicasterol and two to DGDG. Brassicasterol is found in various marine organisms such as cnidarians, sponges or diatoms58–60. In addition to having significant anti-inflammatory properties60, this sterol exhibits activity against organisms such as protozoans61. Furthermore, DGDG are also found in marine organisms, especially in Symbiodinium from both corals and giant clams62,63. Digalactosyldiacylglycerols possess strong nitric oxide inhibitory properties leading to antiviral activity64,65. Moreover, DGDG have been identified as antifouling agent demonstrating repellent activity notably of blue mussels66. These data suggest that a synergism of secondary metabolites might lead to antifouling activity as it has already been assumed in a previous study67. Therefore, in the present study, either due to their abundance as VIPs or to their synergism, carotenoid and lipids might be responsible for the biofouling inhibition specifically observed in PAT assemblage. In this regard, as mucus is continuously released by both corals and giant clams68, we cannot exclude that mucus flocs and/or secreted molecules such as antibiotics could adhere to the aquarium’s walls, playing a role in biofouling production. This effect will be, as discussed for nutrition, dependent on the different species present in the aquarium, with possible positive or negative interactions.
Regardless of the mechanisms involved in this inhibition, our experimental results suggest that biofouling develops differently, depending on the type and complexity of the sessile species assemblages and this may have important implications in coral reef ecology. Moreover, these phenotypes were maintained throughout experimental trials, even during the five days short thermal stress, suggesting that occasional increases in seawater temperature are not sufficient to induce differences of biofouling appearances. Global warming, ocean acidification, and local human impacts are considered as the main causes of coral reefs deterioration globally69–71. The combination of these factors usually prevent resilience of coral reefs, and favour the shift from coral-dominated to algal-dominated reef ecosystems72–74. Our discovery of an assemblage dependent antifouling activity is all the more important that biofouling inhibition occurs even in brief increased temperature conditions. Another major point of our results is that the loss of one species in a three species assemblage leads to a change in the algal inhibition spectrum, the two species assemblage being less active against or more prone to algal development. Despite the timeliness of understanding ecological shifts towards algal-dominated tropical habitats, a gap of knowledge still exists about the mechanisms governing these shifts75. It is well known that coral reefs taxa, e.g. scleractinian corals species, are differentially sensitive to environmental stressors76–78 and that, among sessile species, giant clams are notably more resistant than corals to sea water warming79,80. Therefore, environmental stress raises sequential and differential loss of species. This selective loss of species may impact the structuration of coral reefs benthic communities, leading to a change in the species interaction network and therefore, as exemplified by our work, might strongly impact their defence mechanisms against prerequisite steps of algal development. On the contrary, maintaining the diversity may temper this coral-algal shift. Hence, our study pinpoints that sessile coral reef species diversity, distribution and fitness might strongly influence this coral-algal shift, underlining the putative dramatic consequences of the decrease in biodiversity and the health of coral reefs.
Beyond the biofouling phenotypes, with its extreme form translating into biofouling inhibition, the present study also highlighted the functional interactions resulting from sessile species assemblages. Our data showed that specific communities of bacteria, diatoms and algae characterized the biofouling composition of each assemblage. The bacterial functional profiling revealed an increase of “stores polyhydroxybutyrat” in PA assemblage that could be linked to a need of energy storage molecule or to a stress81. In line with this interpretation, microbiomes in PA assemblages included species of Pseudoalteromonaceae that are able to degrade a form of polyhydroxybutyrat referred to as poly-3-hydroxybutyrate82. Other functions such as sulfur reducer, iron oxidizer, streptomycin producer and cellobiose degrading were, respectively, strictly found in T, PAT, P and PA. Additionally, some specific functions such as nitrite reducer and atrazine metabolism for T and PAT or degrade aromatic hydrocarbon for PAT, P and PA, are shared by some but not all of the assemblages. Therefore, the presence of nitrite reducer activities in both PAT and T assemblages could account for the systematically higher level of NH4+ in these assemblages. Such common features between some specific assemblages highlight the potential cumulative functional effect of the addition of one species in an assemblage, such as degrading aromatic hydrocarbons produced by P in PAT and PA as well as some emergent functions such as iron oxidizer in PAT. Therefore, all these combinations of functions provide a specific capacity to an assemblage for responding to changing environmental conditions. Such a trait can be related to the beneficial effect of plant assemblages for the growth, productivity and protection against pathogens and others, such as in permaculture where combinations of plants are used to enhance resistance and productivity83.
Considering the resulting functional aspects of various species assemblages, with some positive and/or negative interactions, together with their effect on biofouling development, our data support the notion that marine species assemblages should be integrally part of future coral reefs restoration plans as is already the case for terrestrial restoration.
Supplementary information
Acknowledgements
This work was supported by LabexCorail and CNRS fundings. Isis Guibert PhD was supported by Pierre and Marie Curie University – Doctoral school 129. We are grateful to the Academie Française for their financial support through the Walter-Zellidja Grant. The authors thank Dr. Claude Payri and Dr. Catherine Gobin for her precious help on diatoms identification and Delphine Raviglione for his help with the metabolomics analysis. We acknowledge the InterContinental Resort & Spa Moorea and the Moorea Dolphin Center for providing coral garden protected area. We also thank Franck Lerouvreur, Pascal Ung, Ewen Morin and all the students from CRIOBE who helped us on the field.This study was funded by Labex Corail and the Walter Zellidja Grant from the Academie Française.
Author Contributions
I.G., V.B. and G.L. designed the study, I.G., C.S., I.B., V.B., conducted the investigation of the study, I.G., X.P., M.Z., I.B., V.B., did the data curation, I.G., G.L., M.L., I.B., V.B., realized the formal analysis, V.B. supervised the project, I.G. and V.B. acquired the funds. All authors wrote and reviewed the manuscript.
Data Availability
Supplemental information for this article can be found at http://www.nature.com/scientificreports. All data are available upon request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39268-1.
References
- 1.Núñez-Pons L, Avila C. Natural products mediating ecological interactions in Antarctic benthic communities: a mini-review of the known molecules. Nat. Prod. Rep. 2015;32:1114–1130. doi: 10.1039/C4NP00150H. [DOI] [PubMed] [Google Scholar]
- 2.Stachowicz JJ. Mutualism, Facilitation, and the Structure of Ecological Communities. Bioscience. 2001;51:235–246. doi: 10.1641/0006-3568(2001)051[0235:MFATSO]2.0.CO;2. [DOI] [Google Scholar]
- 3.Puglisi MP, Sneed JM, Sharp KH, Ritson-Williams R, Paul VJ. Marine chemical ecology in benthic environments. Nat. Prod. Rep. 2014;31:1510–1553. doi: 10.1039/C4NP00017J. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan B, Faulkner DJ, Webb L. Siphonodictidine, a metabolite of the burrowing sponge Siphonodictyon sp. that inhibits coral growth. Science. 1983;221:1175–6. doi: 10.1126/science.221.4616.1175. [DOI] [PubMed] [Google Scholar]
- 5.Bandurraga MM, Fenical W. Isolation of the muricins. Evidence of a chemical adaptation against fouling in the marine octocoral Muricea fruticosa (gorgonacea) Tetrahedron. 1985;41:1057–1065. doi: 10.1016/S0040-4020(01)96473-7. [DOI] [Google Scholar]
- 6.Kashman Y, Groweiss A, Lidor R, Blasberger D, Carmely S. Latrunculins: NMR study, two new toxins and a synthetic approach. Tetrahedron. 1985;41:1905–1914. doi: 10.1016/S0040-4020(01)96553-6. [DOI] [Google Scholar]
- 7.Cao F, et al. Polyhydroxylated sterols from the south China sea gorgonian Verrucella umbraculum. Helv. Chim. Acta. 2014;97:900–908. doi: 10.1002/hlca.201300397. [DOI] [Google Scholar]
- 8.Quinn RA, et al. Metabolomics of reef benthic interactions reveals a bioactive lipid involved in coral defence. Proc. Biol. Sci. 2016;283:1–10. doi: 10.1098/rspb.2016.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Railkin, A. I. Marine Biofouling Colonization Processes and Defenses. (2004).
- 10.Allison DG. The Biofilm Matrix. Biofouling. 2003;19:139–150. doi: 10.1080/0892701031000072190. [DOI] [PubMed] [Google Scholar]
- 11.Guo W, Ngo HH, Li J. A mini-review on membrane fouling. Bioresour. Technol. 2012;122:27–34. doi: 10.1016/j.biortech.2012.04.089. [DOI] [PubMed] [Google Scholar]
- 12.Dobretsov S, Abed RMM, Teplitski M. Mini-review: Inhibition of biofouling by marine microorganisms. Biofouling. 2013;29:423–441. doi: 10.1080/08927014.2013.776042. [DOI] [PubMed] [Google Scholar]
- 13.von Ammon U, et al. The impact of artificial surfaces on marine bacterial and eukaryotic biofouling assemblages: A high-throughput sequencing analysis. Mar. Environ. Res. 2017;133:57–66. doi: 10.1016/j.marenvres.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Pereira Masi B, Zalmon I, Coutinho R. Effects of structural factors on upwelling fouling community, Southeast Brazil. Brazilian J. Oceanogr. 2016;64:387–400. doi: 10.1590/s1679-87592016128706404. [DOI] [Google Scholar]
- 15.Leonard K, Hewitt CL, Campbell ML, Primo C, Miller SD. Epibiotic pressure contributes to biofouling invader success. Sci. Rep. 2017;7:1–7. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, et al. Impact of reverse nutrient diffusion on membrane biofouling in fertilizer-drawn forward osmosis. J. Memb. Sci. 2017;539:108–115. doi: 10.1016/j.memsci.2017.05.074. [DOI] [Google Scholar]
- 17.Farhat NM, Vrouwenvelder JS, Van Loosdrecht MCM, Bucs SS, Staal M. Effect of water temperature on biofouling development in reverse osmosis membrane systems. Water Res. 2016;103:149–159. doi: 10.1016/j.watres.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Scandura, G. et al. Advanced protection against marine biofouling using solar light. bioRxiv 1–21, 10.1101/106195 (2017).
- 19.Qian P-Y, Li Z, Xu Y, Li Y, Fusetani N. Mini-review: Marine natural products and their synthetic analogs as antifouling compounds: 2009–2014. Biofouling. 2015;31:101–122. doi: 10.1080/08927014.2014.997226. [DOI] [PubMed] [Google Scholar]
- 20.Dobretsov S, Teplitski M, Paul V. Mini-review: Quorum sensing in the marine environment and its relationship to biofouling. Biofouling. 2009;25:413–427. doi: 10.1080/08927010902853516. [DOI] [PubMed] [Google Scholar]
- 21.Gomez ED, Mingoa-Licuanan SS. Achievements and lessons learned in restocking giant clams in the Philippines. Fish. Res. 2006;80:46–52. doi: 10.1016/j.fishres.2006.03.017. [DOI] [Google Scholar]
- 22.Diaz-Pulido, G., Harii, S., McCook, L. J. & Hoegh-Guldberg, O. The impact of benthic algae on the settlement of a reef-building coral. Coral Reefs 274–279 (2010).
- 23.Vieira C, et al. Allelopathic interactions between the brown algal genus Lobophora (Dictyotales, Phaeophyceae) and scleractinian corals. Sci. Rep. 2016;6:1–11. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nugues MM, Bak RPM. Differential competitive abilities between Caribbean coral species and a brown alga: A year of experiments and a long-term perspective. Mar. Ecol. Prog. Ser. 2006;315:75–86. doi: 10.3354/meps315075. [DOI] [Google Scholar]
- 25.Figuerola, B. et al. In Marine Ecosystems(ed. Cruzado, D. A.) 105–126 at www.intechopen.com (2012).
- 26.Rouzé H, Lecellier G, Langlade M, Planes S, Berteaux-Lecellier V. Fringing reefs exposed to different levels of eutrophication and sedimentation can support the same benthic communities. Mar. Pollut. Bull. 2015;92:212–221. doi: 10.1016/j.marpolbul.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Rouzé H, et al. An updated assessment of Symbiodinium spp. that associate with common scleractinian corals from Moorea (French Polynesia) reveals high diversity among background symbionts and a novel finding of clade B. PeerJ. 2017;5:1–22. doi: 10.7717/peerj.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcelino, V. R. & Verbruggen, H. Multi-marker metabarcoding of coral skeletons reveals a rich microbiome and diverse evolutionary origins of endolithic algae. Sci. Rep. 1–9, 10.1038/srep31508 (2016). [DOI] [PMC free article] [PubMed]
- 29.Blaxter ML. The promise of a DNA taxonomy. Philos. Trans. R. Soc. B Biol. Sci. 2004;359:669–679. doi: 10.1098/rstb.2003.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pochon, X., Bott, N. J., Smith, K. F. & Wood, S. A. Evaluating detection limits of next-generation sequencing for the surveillance and monitoring of international marine pests. PLoS One8, 1-12 (2013). [DOI] [PMC free article] [PubMed]
- 32.Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:606–612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blunt, J. & Munro, M. MarinLit: A database of the marine natural products literature. http://pubs.rsc.org/marinlit/ (2017).
- 34.Guijas C, et al. METLIN: A technology platform for identifying knowns and unknowns. Anal. Chem. 2018;90:3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeffrey SW, Haxo FT. Photosynthetic pigments of symbiotic dinoflagellates (zooxanthellae) from corals and clams. Biol. Bull. 1968;135:149–165. doi: 10.2307/1539622. [DOI] [Google Scholar]
- 36.Zigmantas D, Hiller RG, Sundstrom V, Polivka T. Carotenoid to chlorophyll energy transfer in the peridinin-chlorophyll-a-protein complex involves an intramolecular charge transfer state. Proc. Natl. Acad. Sci. 2002;99:16760–16765. doi: 10.1073/pnas.262537599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imbs AB, et al. Polyunsaturated molecular species of galactolipids: Markers of zooxanthellae in a symbiotic association of the soft coral Capnella sp. (Anthozoa: Alcyonacea) Russ. J. Mar. Biol. 2015;41:461–467. doi: 10.1134/S1063074015060048. [DOI] [Google Scholar]
- 38.Cutignano A, et al. Profiling of complex lipids in marine microalgae by UHPLC/tandem mass spectrometry. Algal Res. 2016;17:348–358. doi: 10.1016/j.algal.2016.05.016. [DOI] [Google Scholar]
- 39.Mo S, Dong L, Hurst WJ, Breemen RBvan. Quantitative analysis of phytosterols in edible oils using APCI liquid chromatography-tandem mass spectrometry. NIH Public Access. 2013;48:949–956. doi: 10.1007/s11745-013-3813-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nahon S, et al. Spatial and temporal variations in stable carbon (δ13C) and nitrogen (δ15N) isotopic composition of symbiotic scleractinian corals. PLoS One. 2013;8:1–17. doi: 10.1371/journal.pone.0081247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrera-Silveira JA. Spatial heterogeneity and seasonal patterns in a tropical coastal lagoon. J. Coast. Res. 1994;10:738–746. [Google Scholar]
- 42.Houlbrèque F, Delesalle B, Blanchot J, Montel Y, Ferrier-Pagès C. Picoplankton removal by the coral reef community of La Prévoyante, Mayotte Island. Aquat. Microb. Ecol. 2006;44:59–70. doi: 10.3354/ame044059. [DOI] [Google Scholar]
- 43.Piccinetti CC, et al. Herbivory in the soft coral Sinularia flexibilis (Alcyoniidae) Sci. Rep. 2016;6:22679. doi: 10.1038/srep22679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neo ML, Eckman W, Vicentuan K, Teo SLM, Todd PA. The ecological significance of giant clams in coral reef ecosystems. Biol. Conserv. 2015;181:111–123. doi: 10.1016/j.biocon.2014.11.004. [DOI] [Google Scholar]
- 45.Elifantz H, Horn G, Ayon M, Cohen Y, Minz D. Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 2013;85:348–357. doi: 10.1111/1574-6941.12122. [DOI] [PubMed] [Google Scholar]
- 46.Kooperman N, Ben-Dov E, Kramarsky-Winter E, Barak Z, Kushmaro A. Coral mucus-associated bacterial communities from natural and aquarium environments. FEMS Microbiol. Lett. 2007;276:106–113. doi: 10.1111/j.1574-6968.2007.00921.x. [DOI] [PubMed] [Google Scholar]
- 47.Sharp KH, Distel D, Paul VJ. Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 2012;6:790–801. doi: 10.1038/ismej.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee STM, Davy SK, Tang SL, Kench PS. Mucus sugar content shapes the bacterial community structure in thermally stressed Acropora muricata. Front. Microbiol. 2016;7:1–11. doi: 10.3389/fmicb.2016.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakajima R, et al. In situ release of coral mucus by Acropora and its influence on the heterotrophic bacteria. Aquat. Ecol. 2009;43:815–823. doi: 10.1007/s10452-008-9210-y. [DOI] [Google Scholar]
- 50.Meistertzheim AL, et al. Patterns of bacteria-host associations suggest different ecological strategies between two reef building cold-water coral species. Deep. Res. Part I Oceanogr. Res. Pap. 2016;114:12–22. doi: 10.1016/j.dsr.2016.04.013. [DOI] [Google Scholar]
- 51.Ralston E, Swain G. Bioinspiration—the solution for biofouling control? Bioinspir. Biomim. 2009;4:1–10. doi: 10.1088/1748-3182/4/1/015007. [DOI] [PubMed] [Google Scholar]
- 52.Gökçen A, Vilcinskas A, Wiesner J. Biofilm-degrading enzymes from Lysobacter gummosus. Virulence. 2014;5:378–387. doi: 10.4161/viru.27919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Withers NW, Kokke WC, Fenical W, Djerassi C. Sterol patterns of cultured zooxanthellae isolated from marine invertebrates: Synthesis of gorgosterol and 23-desmethylgorgosterol by aposymbiotic algae. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3764–3768. doi: 10.1073/pnas.79.12.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang CY, et al. Chemical defensive substances of soft corals and Gorgonians. Acta Ecol. Sin. 2008;28:2320–2328. doi: 10.1016/S1872-2032(08)60031-1. [DOI] [Google Scholar]
- 55.Tyson, B. V. & Reynolds, K. J. Fouling-resistant composition comprising sterols and/or dervatives thereof. 1, 1–10 (2011).
- 56.Tsushima M, et al. Inhibitory effect of natural carotenoids on Epstein-Barr virus activation activity of a tumor promoter in raji cells. A screening study for anti-tumor promoters. Biol. Pharm. Bull. 1995;18:227–233. doi: 10.1248/bpb.18.227. [DOI] [PubMed] [Google Scholar]
- 57.Ishikawa C, Jomori T, Tanaka J, Senba M, Mori N. Peridinin, a carotenoid, inhibits proliferation and survival of HTLV-1-infected T-cell lines. Int. J. Oncol. 2016;49:1713–1721. doi: 10.3892/ijo.2016.3648. [DOI] [PubMed] [Google Scholar]
- 58.Abou-Hussein DR, Badr JM, Youssef DTA. Dragmacidoside: A new nucleoside from the red sea sponge Dragmacidon coccinea. Nat. Prod. Res. 2014;28:1134–1141. doi: 10.1080/14786419.2014.915828. [DOI] [PubMed] [Google Scholar]
- 59.Nawi, N. M., Simoh, S., Zainal, A. & Rahman, Z. A. GC-MS analysis of phytochemical contents of Curcuma aeruginosa and Curcuma caesia and their antioxidant potential. In Proceeding of the 30th Annual Seminar of The Malaysian Natural Products Soceity Moving Translational Research In Natural Products Forward International Conference of Natural Products 2014 (ICNP2014) 60–66 (2014).
- 60.Samorì C, Costantini F, Galletti P, Tagliavini E, Abbiati M. Inter- and intraspecific variability of nitrogenated compounds in gorgonian corals via application of a fast one-step analytical protocol. Chem. Biodivers. 2018;15:1–11. doi: 10.1002/cbdv.201700449. [DOI] [PubMed] [Google Scholar]
- 61.Carreira de Paula J, et al. Trypanocidal activity of organic extracts from the Brazilian and Spanish marine sponges. Rev. Bras. Farmacogn. 2015;25:651–656. doi: 10.1016/j.bjp.2015.08.011. [DOI] [Google Scholar]
- 62.Kneeland, J. M. Lipid biomarkers of coral stress: calibration and exploration. at http://mit.dspace.org/handle/1721.1/33935 (2011).
- 63.Awai K, Matsuoka R, Shioi Y. Lipid and fatty acid compositions of Symbiodinium strains. Proc. 12th Int. Coral Reef Symp. 2012;1:9–12. [Google Scholar]
- 64.Banskota AH, Stefanova R, Gallant P, McGinn PJ. Mono- and digalactosyldiacylglycerols: Potent nitric oxide inhibitors from the marine microalga Nannochloropsis granulata. J. Appl. Phycol. 2013;25:349–357. doi: 10.1007/s10811-012-9869-2. [DOI] [Google Scholar]
- 65.Ríos-Ibarra CP, et al. Downregulation of inducible nitric oxide synthase (iNOS) expression is implicated in the antiviral activity of acetylsalicylic acid in HCV-expressing cells. Arch. Virol. 2014;159:3321–3328. doi: 10.1007/s00705-014-2201-5. [DOI] [PubMed] [Google Scholar]
- 66.Katsuoka M, Ogura C, Etoh H, Sakata K, Ina K. Galactosyl- and sulfoquinovosyldiacyl-glycerols isolated from the brown algae, Undaria pinnatifida and Costaria costata as repellents of the blue mussel, Mytilus edulis. Agric. Biol. Chem. 1990;54:3043–3044. [Google Scholar]
- 67.Ramasamy M, Balasubramanian U. Identification of bioactive compounds and antimicrobial activity of marine clam Anadara granosa (Linn.) Int. J. Sci. Nat. 2012;3:263–266. [Google Scholar]
- 68.Soo P, Todd PA. The behaviour of giant clams (Bivalvia: Cardiidae: Tridacninae) Mar. Biol. 2014;161:2699–2717. doi: 10.1007/s00227-014-2545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meltvedt A, Jadot C. Progression of the coral-algal phase shift in the Caribbean: A case study in Bonaire, Dutch Caribbean. Mar. Technol. Soc. J. 2014;48:33–41. doi: 10.4031/MTSJ.48.6.4. [DOI] [Google Scholar]
- 70.Arias-González, J. E. et al. A coral-algal phase shift in Mesoamerica not driven by changes in herbivorous fish abundance. PLoS One 1–17. 10.1371/journal.pone.0174855 (2017). [DOI] [PMC free article] [PubMed]
- 71.Hoegh-Guldberg O, et al. Coral reefs under rapidclimate change and ocean acidification. Science. 2007;318:1737–1742. doi: 10.1126/science.1152509. [DOI] [PubMed] [Google Scholar]
- 72.McManus JW, Polsenberg JF. Coral-algal phase shifts on coral reefs: Ecological and environmental aspects. Prog. Oceanogr. 2004;60:263–279. doi: 10.1016/j.pocean.2004.02.014. [DOI] [Google Scholar]
- 73.Diaz-Pulido G, McCook LJ. The fate of bleached corals: Patterns and dynamics of algal recruitment. Mar. Ecol. Prog. Ser. 2002;232:115–128. doi: 10.3354/meps232115. [DOI] [Google Scholar]
- 74.Hauri C, Fabricius KE, Schaffelke B, Humphrey C. Chemical and physical environmental conditions underneath mat- and canopy-forming macroalgae, and their effects on understorey corals. PLoS One. 2010;5:1–9. doi: 10.1371/journal.pone.0012685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jorissen H, Skinner C, Osinga R, De Beer D, Nugues MM. Evidence for water-mediated mechanisms in coral–algal interactions. Proc. R. Soc. B Biol. Sci. 2016;283:1–10. doi: 10.1098/rspb.2016.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loya, et al. Coral bleaching: The winners and the losers. Ecol. Lett. 2001;4:122–131. doi: 10.1046/j.1461-0248.2001.00203.x. [DOI] [Google Scholar]
- 77.Ries JB, Cohen AL, McCorkle DC. Marine calcifiers exhibit mixed responses to CO2-induced ocean acidification. Geology. 2009;37:1131–1134. doi: 10.1130/G30210A.1. [DOI] [Google Scholar]
- 78.Rodolfo-Metalpa R, et al. Coral and mollusc resistance to ocean acidification adversely affected by warming. Nat. Clim. Chang. 2011;1:308–312. doi: 10.1038/nclimate1200. [DOI] [Google Scholar]
- 79.Andréfouët S, et al. Climate variability and massive mortalities challenge giant clam conservation and management efforts in French Polynesia atolls. Biol. Conserv. 2013;160:190–199. doi: 10.1016/j.biocon.2013.01.017. [DOI] [Google Scholar]
- 80.Buck BH, Rosenthal H, Ulrich S-P. Effect of increased irradiance and thermal stress on the symbiosis of Symbiodinium microadriaticum and Tridacna gigas. Aquat. Living Resour. 2002;15:107–117. doi: 10.1016/S0990-7440(02)01159-2. [DOI] [Google Scholar]
- 81.Ali AA, Shaban KA, Tantawy EA. Effect of Poly-β-Hydroxybutyrate (PHB) and glycogen producing endophytic bacteria on yield, growth and nutrient contents in rice cultivated in saline soil. Appl. Sci. Reports. 2014;8:134–142. [Google Scholar]
- 82.Mergaert J, Swings J. Biodiversity of microorganisms that degrade bacterial and synthetic polyesters. J. Ind. Microbiol. 1996;17:463–469. [Google Scholar]
- 83.Bane, P. Earth care manual: A permaculture handbook for britain and other temperate climates. (2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplemental information for this article can be found at http://www.nature.com/scientificreports. All data are available upon request.