Abstract
Cervical cancer is the second most common cancer in women worldwide. Human papillomavirus (HPV) infection appears to be a necessary factor in the development of almost all cases (>95%) of cervical cancer. HPV E6 induces a change of control of p53 stabilization from Hdm2 to E6/E6AP in HPV-infected cells. It is well known that the LxxLL motif of cellular ubiquitin ligase E6AP binds to the pocket of E6 and causes a conformational change to enable E6 to bind p53 competently. In the ternary complex E6/E6AP/p53, p53 is polyubiquitinated by E6AP and subsequently degraded by a proteasome. Therefore, these cells are deficient in the processes regulated by p53, including apoptosis, damaged DNA repair, and the cell cycle. In the present study, it was demonstrated that quercetin induced G2 phase cell cycle arrest and apoptosis in both HeLa and SiHa cells, accompanied by an increase of p53 and its nuclear signal. It was also observed that quercetin increased the level of the p21 transcript and the pro-apoptotic Bax protein, which are two p53-downstream effectors. However, quercetin did not alter the expression of the HPV E6 protein in cervical cancer cells; therefore, the increase in p53 occurred in an E6 expression-independent manner. Furthermore, molecular docking demonstrated that quercetin binds stably in the central pocket of E6, the binding site of E6AP. These data suggest that quercetin increases the nuclear localization of p53 by interrupting E6/E6AP complex formation in cervical cancer cells.
Keywords: apoptosis, cell cycle, cervical cancer, DNA damage, HPV E6, p53, quercetin, ubiquitin ligase E6AP
Introduction
Cervical cancer is the second most frequent cancer in women with ~530,000 novel cases each year (1). It is well established that persistent infection with high-risk oncogenic human papillomavirus (HPV) is the main etiological agent and initial step in cervical carcinogenesis (2). Although, the best characterized target of the E6 protein of HPV is p53, HPV E6 binds to a diverse range of cellular proteins by altering its stability in a proteasome-dependent manner (3,4). In p53 degradation, the central pocket of E6 directly binds to the LxxLL motif of the ubiquitin ligase E6AP, which induces a conformational change in HPV E6 allowing the formation of a complex with p53 (5,6). In this ternary complex, named E6/E6AP/p53, p53 is polyubiquitinated by E6AP and subsequently degraded by a proteasome (7,8). Therefore, p53 responses such as those resulting from DNA damage, which includes cell cycle arrest, DNA damage repair and apoptosis, are deficient. Hence, the cell is more susceptible to the acquisition of genomic instability and finally cell transformation (9). Therefore, the reactivation of p53 has been suggested to be an effective strategy for cancer therapy in wild-type p53 retaining tumor cells, as its reactivation can result in cell cycle arrest and/or apoptosis by activating or repressing the transcription of apoptotic genes and cell cycle regulatory genes (10).
Flavonoids are widely distributed in plants and are associated with a diverse range of biological activities that include inhibition of phosphoinositide-3-kinase, reactive oxygen species (ROS) generation, DNA damage, cell cycle arrest and apoptosis, as well as p53 restoration (11–13). For instance, the catechin-like flavonoid, epigallocatechin gallate (EGCG) increased the level of the p53 protein accompanied by a reduction of HPV E6 protein in HeLa and CaSki cells (14). Furthermore, the flavonoid luteolin and flavonoid-like synthetic compounds inhibited the binding between HPV E6 and E6AP in vitro and induced an increased expression of the p53 and p21 proteins in cervical cancer cells (15).
Several studies have demonstrated the anticancer activity of quercetin, a polyphenolic flavonoid, against a number of types of cancer cells, such as hepatocellular carcinoma cells where quercetin inhibited the cell proliferation through cell cycle arrest, apoptosis and DNA fragmentation, together with an increase of the total p53 protein and p53 phosphorylation (16). In addition, in melanoma cells, quercetin induced apoptosis by a p53/Bax-dependent mechanism and was correlated with an increase in ROS (17). However, a common mechanism for quercetin-induced p53 restoration has not been well established in HPV-positive cervical cancer cells.
In the present study, it was reported that quercetin arrested the cell cycle in G2 phase and triggered apoptosis in cervical cancer cells. Also, it was observed that quercetin promoted the activation of p53 by an increase of total p53 protein and its nuclear localization, together with the increase of expression of its transcriptional targets including Bax and p21. However, quercetin did not decrease the expression of HPV E6, the agent responsible for the decrease of p53 in these cells. In addition, the molecular docking results predict that quercetin would be able to interrupt the association of E6 with E6AP by binding to the E6 pocket and therefore preventing the formation of the p53 binding cleft on E6 and finally p53 degradation.
Materials and methods
Cell lines and treatments
Human cervical cancer cells expressing HPV-16 (SiHa cells), HPV 18 (HeLa cells) were obtained from the American Type Culture Collection (Manassas, VA, USA) and human foreskin fibroblasts (HFF cells) were kindly provided by Dr. Ramón González (CIDC, UAEM, Cuernavaca, Mor, México). All the cells were maintained in Dulbecco's Modified Eagle's Medium High Glucose (DMEM HG, Caisson Labs, UT, USA) supplemented with 10% (v/v) Fetal Bovine Serum (Biowest LLC, MO, USA) at 37°C in a humidified atmosphere of 5% CO2. Treatment with quercetin or taxol (Sigma aldrich; St. Louis, MO, USA) did not exceed 0.5% of DMSO.
Cell viability
Cell viability was measured using [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] inner salt MTS assay (Promega, Madison WI, USA). Briefly, a total of 4X103 SiHa, HeLa or HFF cells per well were plated in a 96-well plate and allowed to grow during overnight. Cells were exposed to increasing concentrations of quercetin by triplicate for 48 h. Subsequently, 20 µl of MTS reagent was added into each well containing the untreated and treated cells in 100 µl DMEM HG and incubated at 37°C for 3 h. Then the absorbance values were measured at 490 nm in an automatic microplate reader (Promega, Madison, WI, USA). Data were analyzed, and cell viability rate was calculated in GraphPad PRISM version 6.01 statistical program and the IC50 values were determined by regression analysis.
Flow cytometry
HeLa and SiHa cells were treated with quercetin at IC50, whilst HFF cells were exposed to 500 µM quercetin during 48 h. The cells were separately treated with 5 nM taxol (as G2/M control). Control and treated cells were harvested, centrifuged and fixed in cold 70% ethanol. Fixed cells were incubated with 10 µg/ml ribonuclease A and 10 µg/ml propidium iodide during 30 min on ice. Then 10,000 events were acquired in flow cytometer (FACSCalibur; Beckman Coulter, Inc., Brea, CA, USA). Obtained data were analyzed using the FlowJo Software (Tree Star, Inc., Ashland, OR, USA) to generate DNA content frequency histograms. The experiments were conducted by triplicate in three independent experiments and the statistical analysis was performed using ANOVA and subsequent Dunnett test with P<0.05 considered to indicate a statistically significant difference.
Mitotic nuclei staining
HeLa and SiHa cells were grown on slide and treated with quercetin and taxol as described in flow cytometry assay. The cells were fixed with 4% paraformaldehyde in PEM buffer (PIPES 100 mM pH 6.9, EGTA 5 mM, MgCl2 2 mM) for 15 min followed 4% paraformaldehyde in NaHCO3 for 45 min. The cells were permeabilized with 0.1% Triton in PBS 1X for 10 min. Nuclei were stained with DAPI 0.2 µg/ml, at 37°C for 10 min. Cell images were observed using a fluorescence microscope and acquired by NIS-Elements software. At least 200 nuclei were counted from different fields acquired in distinct days to determine the percentage of mitotic nuclei.
Epifluorescence microscopy
HeLa or SiHa cells were exposed to their respective quercetin IC50. HFF cells were exposed at 140 µM quercetin, the highest concentration employed in cervical cancer cells. After 72 h, attached cells on slide were stained with acridine orange/ethidium bromide (AO/EB, 1:1 molecular relation). The images were immediately captured using an epifluorescence microscope to determinate quercetin-induced apoptotic traits. Different controls were included in the experiment: Cells not treated, as negative control; cell treated at 1 mM H2O2 for 3 h, as apoptosis control; and cells immersed in 80–90°C water for 15 sec, as necrosis control. It is well known that AO can pass through cell membrane, but EB cannot. Necrotic cells stain red but have a nuclear morphology resembling that of viable cells. Apoptotic cells appear green, and morphological changes such as formation of apoptotic bodies are observed. The criteria for identification are as follows: Viable cells appear to have green nucleus with intact structure; early apoptosis cells exhibit a bright green nucleus showing condensation of chromatin; late apoptosis appears as dense orange areas of chromatin condensation; and orange intact nucleus depicts secondary necrosis (18,19). The experiments were conducted by triplicate in three independent experiments.
Immunofluorescence of p53 protein
50,000 HeLa or SiHa cells were grown on glass coverslips in a 24-well cell culture plate and allowed to attach during overnight. Upon quercetin IC50 treatment, both HeLa and SiHa cells were fixed and permeabilized as mentioned in the Mitotic nuclei staining section. The cells were then incubated with the primary antibody mouse anti-p53 (1:250; DO-1, Santa Cruz Biotechnology, Dallas, TX, USA) overnight at 4°C. The secondary antibody anti-mouse Alexa 488 (1:1,000) was added and incubated for two h at 37°C to visualize the p53 localization. The nuclei were stained with DAPI (0.2 µg/ml). The images were acquired in confocal microscopy. The acquisitions for DAPI and Alexa488 were made sequentially to avoid crossover of fluorescence emission. These experiments were conducted by duplicated in two different days.
Western blot analysis
Following quercetin treatment, HeLa and SiHa cells were lysed with RIPA buffer (Tris-HCl 50 mM, NaCl 150 mM, NP40 1%, sodium deoxycholate 0.5%, SDS 0.1%) containing protease inhibitor cocktail (Sigma, MO, USA). Protein concentration was determined by bicinchoninic acid method using Pierce™ BCA Protein Assay kit (Pierce biotechnology, IL, USA). Equal protein amounts from all samples were loaded and resolved in SDS-PAGE 12% and transferred to PVDF membrane. The membranes were blocked with 5% milk in TBS-Tween-20 for 1 h, and then incubated overnight at 4°C with the following primary antibodies: p53 (DO-1, 1:1,000 dilution), HPV18 E6 (G-7, 1:200 dilution), HPV16/18 E6 (C1P5, 1:100 dilution), Bax (2D2, 1:500 dilution), Bcl-2 (C-2, 1:500), from Santa Cruz Biotechnology. The proteins were visualized using the ChemiDoc-XRS Imaging System (Bio-Rad Laboratories, Inc. CA, USA), using supersignal west femto maximum sensitivity substrate (Thermo Fischer Scientific, IL, USA) as a developer. GAPDH antibody primary (GA1R) from Santa Cruz Biotechnology was used as the loading control. The experiment was executed in triplicate and quantification was performed via densitometric analysis using ImageJ TM software. The densitometric data were normalized with the loading control. The level of the expression of the proteins was determined in function to the respective control.
RT-PCR
To determinate the level of transcript of p21 gene, total RNA from SiHa and HeLa cells were obtained following 48 h of quercetin IC50 exposure. 50 ng of DNase I-treated total RNAs were retro-transcribed and PCR amplified in one step as follow: cDNA synthesis, 50°C for 30 min; retrotranscriptase inactivation, 95°C for 15 min; 35 cycles of DNA amplification at 95°C for 30 sec, 52°C for 30 sec, 72°C for 1 min; final extension, 72°C for 5 min, in Mastercycler gradient Serial No. 5331 (Eppendorf, Hamburg, Germany) by using One-Step RT-PCR Hot-Start kit (Thermo Fisher, CA, USA). Specific oligonucleotides of p21 (GenBank reference sequence: NM_001291549) (forward sequence, 5′→3′TTAGCAGCGGAACAAGGAGT and reverse sequence, 5′→3′AGAAACGGGAACCAGGACAC; product size, 252 bp) and of the loading control, GAPDH (GenBank reference sequence, NM_002046.6) (forward sequence, 5′→3′CAACGACCACTTTGTCAAGC and reverse sequence, 5′-GGTGGTCCAGGGGTCTTACT-3′; product size, 115 bp). RT-PCR reactions were resolved in agarose gel 1.2% and visualized in ChemiDoc XRS Imaging System (Bio-Rad Laboratories, Inc, CA, USA). The densitometric analysis was performed as mentioned in the Western blot section.
Caspase 3/7 activity assay
4×103 HeLa or SiHa cells were plated in 96-well plate and allowed overnight. Then, cells were treated at IC50 of quercetin for 72 h. After treatment, the caspasas 3/7 activity was tested by Caspase-Glo 3/7 Assay from Promega, following manufacturer's instructions. The results were represented as relative units of luminescence and represented in graphs. The statistical analysis was performed by One-way ANOVA with P<0.05 considered to indicate a statistically significant difference.
Molecular docking
The HPV16 E6 crystal structure bound to LxxLL E6AP motif and fusioned to Maltose Binding Protein (MBP) was taken from PDB (ID: 4GIZ) (20). Missing hydrogens from E6 were added using charmm-gui server. The energy from E6 structure was minimized to avoid any bad contacts with 100 steepest descent steps using charmm version 42 with charmm36 parameters. Marvin was used for drawing, displaying and characterizing chemical structures of quercetin, luteolin, CAF24 and C170. Minimized E6 structure was used as target to conduct 1,000 independent blind docking runs. The initial pdbqt files and the configuration files were set using Pymol and Autodock Vina Plugin for Pymol. The grid center was set on (3.76, 60.0, 30.29) with a size of 50×40×60Å using 1Å of spacing size. The analysis was made using local unix scripts. Final structure images were made using VMD molecular software. Protein-ligand interactions maps were made with Maestro.
Statistical analysis
Data were analyzed using the GraphPad Prism version 6.01 statistical program (GraphPad Software, Inc.). Statistical differences between groups were evaluated using one-way analysis of variance followed by the Dunnett's test or t-test. P<0.05 was considered to indicate a statistical significance.
Results
Cytotoxic effect of quercetin in cervical cancer cells
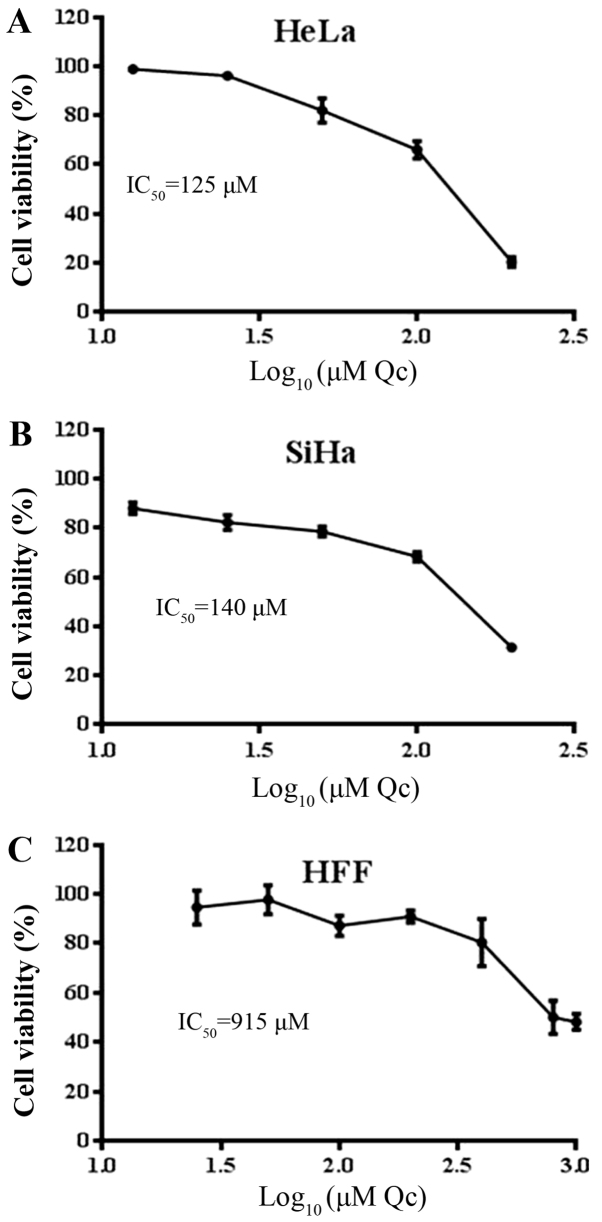
To determine the cytotoxic effect of quercetin on HeLa and SiHa cells, a cell viability assay was performed following 48 h of quercetin exposure. The results demonstrated that quercetin decreased the viability of the HeLa and SiHa cells at a similar concentration, with an IC50 of 125 and 140 µM, respectively (Fig. 1A and B). Nevertheless, in HFF cells quercetin required ~1,000 µM to reach the cytotoxic effects demonstrated in cervical cancer cells (Fig. 1C). This result demonstrates that quercetin exhibits selective toxicity for the cervical cancer cells, HeLa and SiHa, compared with the HFF cells.
Figure 1.
Effect of quercetin on HeLa, SiHa and HFF cell viability. Increasing concentrations of quercetin were employed to assess cell viability at 48 h. In cervical cancer cells, HeLa (A) and SiHa (B) were 0–200 µM and in HFF cells (C) were 0–1,000 µM. Viability data represent the mean ± SD to each concentration of quercetin. Qc IC50 was calculated and indicated into each graph. HFF, human foreskin fibroblasts; Qc, Quercetin.
Induction of cell cycle arrest by quercetin in cervical cancer cells
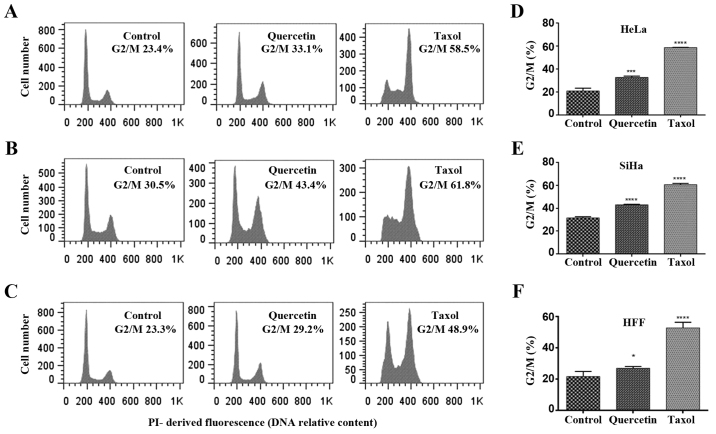
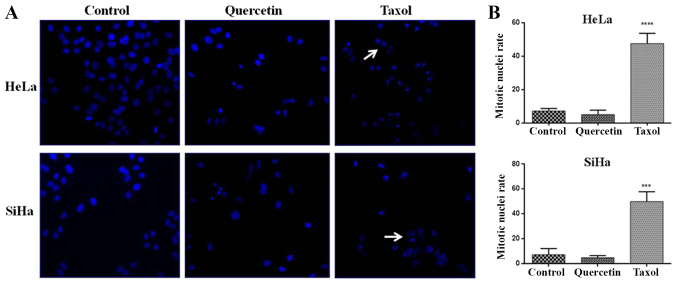
Quercetin has demonstrated multiple biological activities, including cell cycle arrest. To investigate whether the quercetin-induced toxicity is due to its effect on cell proliferation, cell cycle analysis was performed by flow cytometry (Fig. 2). The results revealed that quercetin did not induce significant changes in HFF cells at 140 µM (data not shown), whilst at a high concentration (500 µM) a barely significant increase in the cell population of G2/M phase (with 4n DNA content) was reached (Fig. 2C and D). Furthermore, quercetin at IC50 induced a significant increase of G2/M phases in HeLa and SiHa cells (Fig. 2A-F), as did treatment with taxol (used as a positive control). Taxol is a known mitosis blocker due to its ability to bind tubulin, resulting in stabilization of microtubules, which blocks metaphase/anaphase progression (21). However, quercetin does not have a unique mechanism of cell cycle arrest. To discriminate between quercetin-induced G2 and M phase arrest in HeLa and SiHa cells, DAPI-stained nuclei were analyzed by epifluorescence microscopy following 48 h of quercetin treatment. Following exposure to taxol, a significant increase of mitotic nuclei in HeLa and SiHa cells was observed, whilst significant changes were not detected when HeLa and SiHa cells were challenged with quercetin (Fig. 3A and B). These results indicate that quercetin arrested in the G2 phase rather than the M phase of the cell cycle in cervical cancer cells.
Figure 2.
Cell cycle arrest by quercetin. At 48 h of the treatment with quercetin (A) 125 µM for HeLa cells, (B) 140 µM for SiHa cells and (C) 500 µM for HFF cells, cells were stained with propidium iodide to determine cell cycle distribution by FACS flow cytometry. No treated cells were used as negative control. A total of 5 nM taxol was used as positive control. (D-F) Statistical analysis was carried out in GraphPad Prism software using one-way analysis of variance followed Dunnett's multiple comparisons test. *P<0.05, ***P<0.001 and ****P<0.0001 vs. control group (D-F). HFF, human foreskin fibroblasts.
Figure 3.
Mitotic nuclei rate in cervical cancer cells. (A) Upon incubation for 48 h with quercetin IC50 or 5 nM taxol in HeLa or SiHa cells, the nuclei were stained with 4′,6-diamidino-2-phenylindole and visualized by epifluorescence microscopy. White arrow indicates mitotic nuclei in metaphase. (B) Statistical analysis was carried out in GraphPad Prism software by one-way analysis of variance followed Dunnett's multiple comparisons test. ***P<0.001 vs. control group. HFF, human foreskin fibroblasts.
Effect of quercetin on p53 expression and its cellular localization in cervical cancer cells
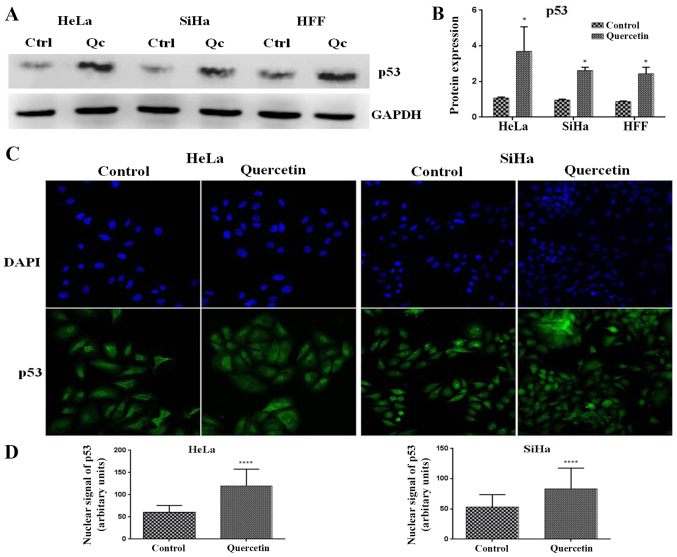
p53 is a master cell cycle regulator, which is able to induce the expression of genes involved in different checkpoints of the cell cycle. To investigate whether quercetin modified the expression of p53 in cervical cancer cells, western blotting was performed. It was observed that quercetin significantly increased the level of p53 in HeLa, SiHa and HFF cells (Fig. 4A and B). Furthermore, it is well known that p53 nuclear translocation is necessary for its transcriptional activity and cell cycle regulation effects. Notably, it was observed that quercetin significantly increased the p53 nuclear signal in HeLa and SiHa cells (Fig. 4C and D). These results suggest that quercetin promotes the nuclear activities of p53 in HeLa and SiHa cells.
Figure 4.
Analysis of p53 expression and nuclear localization of p53 in cervical cancer cells induced by Qc. (A) Whole cell extracts from HeLa and SiHa cells (IC50 Qc) or HFF cells (140 µM of Qc) were obtained and analyzed by western blot to determine changes of expression of p53. GAPDH was used as a loading control. The quantification was performed via densitometric analysis and the data were normalized with the loading control. (B) Statistical analysis was performed by one-way analysis of variance followed Dunnett's multiple comparisons test. *P<0.05. The HeLa or SiHa cells were treated with quercetin at IC50. At 24 h of treatment the cells were fixed and immunostained with anti-p53 (DO-1) and anti-mouse Alexa 488. 4′,6-diamidino-2-phenylindole was used to localize nuclei. (C) The images were acquired using a confocal microscope. (D) Nuclear signals of p53 was quantified and compared between control and quercetin groups. The statistical significance was determined by T-test analysis. ****P<0.0001. HFF, human foreskin fibroblasts; Ctrl, control; Qc, Quercetin.
Upregulation of target genes of p53
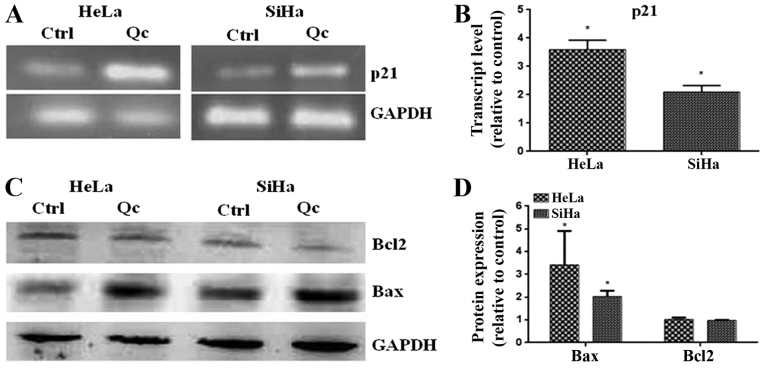
To demonstrate the activity of p53 induced by quercetin, the level of transcript of p21 and the levels of pro-apoptotic Bax protein, two well-known target genes of p53, were evaluated. The results demonstrated that quercetin increased the level of the transcript of p21 in HeLa and SiHa cells (Fig. 5A and B). additionally, quercetin significantly increased the level of the pro-apoptotic Bax protein (Fig. 5C and D). These results suggest that quercetin promotes the transcriptional activity of p53.
Figure 5.
(A-D) Evaluation of the expression of p21 and Bax in cervical cancer cells. (A) After treatment of HeLa or SiHa cells with quercetin at IC50 for 48 h, total RNA was obtained to determine the level of p21 transcript. (C) From whole cell extracts of HeLa or SiHa cells treated during 48 h to quercetin at IC50, western blots were carried out to determine the expression of Bax and Bcl-2. (B and D) The quantification was performed via densitometric analysis and the data were normalized with the loading control (GAPDH). The relative transcript level of p21 in the control group was normalized to 1. Statistical significance was determined by t-test analysis. *P<0.05 vs. control in each respective cell line. Bcl2, B-cell lymphoma 2; Bax, Bcl2-associated × protein; Ctrl, control; Qc, Quercetin.
Induction of apoptosis by quercetin in cervical cancer cells
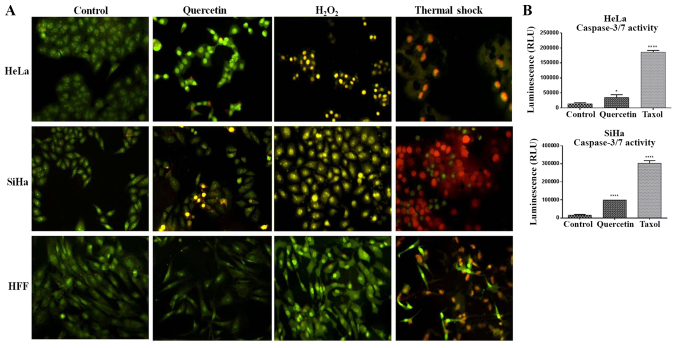
Due to quercetin increasing the expression of p53 and Bax proteins, two known apoptotic genes, the effect of quercetin in the induction of apoptosis in cervical cancer cells was evaluated. Therefore, apoptotic morphological traits were analyzed using AO/EB staining, as well as measuring the activity of caspases 3/7. As presented in Fig. 6A, the nuclei of the untreated cells were homogenously stained with AO in comparison to quercetin-treated cells or the apoptosis positive control (H2O2), which displayed intensely nuclei stained, presumably due to the chromatin condensation. Furthermore, cells treated with quercetin, as in the apoptosis control, exhibited membrane blebbing and apoptotic bodies, which were more evident in the SiHa cells compared with the HeLa cells. In addition, quercetin significantly induced the activation of caspases 3/7 in HeLa and SiHa cells (Fig. 6B). These results together with the augmentation of the Bax/Bcl-2 ratio in HeLa and SiHa cells (Fig. 5C and D), an early event of mitochondrial outer membrane perforation and subsequent apoptosis, indicate that quercetin triggers apoptosis in these cells.
Figure 6.
Analysis of apoptosis in cervical cancer cells. (A) The acridine orange/ethidium bromide double-staining was performed to evaluate morphological changes caused by the induction of apoptosis at 72 h of quercetin exposure in HeLa or SiHa cells. Control represents cells no treated; Qc indicates treatment with quercetin at IC50 in HeLa and SiHa cells or 140 µM in HFF cells; H2O2 indicates apoptotic control by treatment with 1 mM H2O2 for 3 h; and thermal shock represents necrosis control by exposure of the cells to boiled water for 20 sec. Images were acquired at 20× magnification. (B) The activity of caspases 3/7 was evaluated by triplicate using caspase-GLO Assay at 72 h of quercetin treatment in both HeLa and SiHa cells. Taxol at 5 nM was used as a positive control. Statistical significance was determined by one-way analysis of variance followed Dunnett's multiple comparisons test. *P<0.05 and ****P<0.0001 vs. control group. HFF, human foreskin fibroblasts; Bcl2, B-cell lymphoma 2; Bax, Bcl2-associated × protein; Ctrl, control; Qc, Quercetin.
Quercetin-induced p53 activation is E6 HPV protein expression-independent
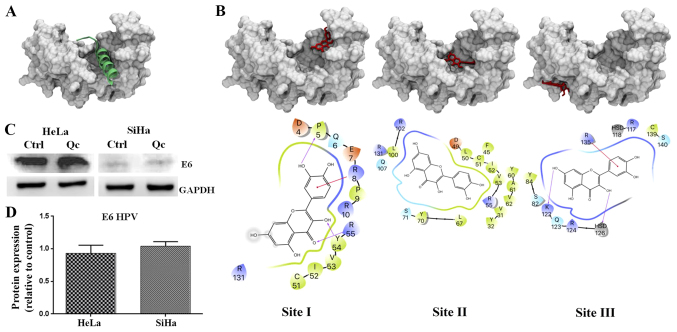
In HPV positive cervical cancer cells, the stability of p53 is totally E6 dependent (22). Therefore quercetin, which caused an activation of p53, would also be expected to induce a decrease in E6 expression. However, western blotting demonstrated that HPV E6 expression remains constant despite the increase of p53 in HeLa and SiHa cells (Fig. 7C). An explanation for the activation of p53 by quercetin in cervical cancer cells could be associated with the disruption of the E6/E6AP complex assembly rather than downregulation of E6 expression. This assumption is supported by the report that flavonoids structurally similar to quercetin were capable of interrupting E6-E6AP association in vitro (15).
Figure 7.
Molecular docking of quercetin on HPV16 E6 protein structure and analysis of expression of HPV E6 in cervical cancer cells after treatment with quercetin. (A) E6 protein in complex with the helix of E6AP containing LxxLL peptide is shown. (B) From left to right, quercetin in site I, II and III and its respective contacts (below) with E6 residues are displayed. (C) Western blot analysis was carried out from whole cell extracts of HeLa or SiHa cells exposed to quercetin at IC50 for 24 h to determine the HPV E6 expression. (D) The quantification was performed via densitometric analysis and the data were normalized with the loading control (GAPDH). Statistical significance was determined by t-test analysis.
Quercetin is predicted to bind to the E6AP binding pocket in HPV E6
To predict the binding between quercetin and E6 protein, the quercetin ligand was subjected to 1,000 independent molecular blind docking assays against the E6 protein structure (PDB ID: 4GIZ) (20). The same operation was performed for flavonoids luteolin, CAF24 and C170 (reference ligands), which were demonstrated by Cherry et al in 2013 to be capable of binding specifically to HPV E6 protein to disrupt the association with E6AP (Fig. 7A) (15). The results predicted that all ligands, including quercetin, were able to interact with the E6 pocket, necessary for the binding with E6AP. Docking predicted that quercetin could bind in three sites on the E6 protein (Fig. 7B). One of them (known as site II) was the E6 pocket site where quercetin bound with the lowest energy (−7.08±-0.18; Fig. 7B, middle panel). Notably, quercetin in site I (Fig. 7B, left panel) made contact with the Pro5, Gln6, Glu7, Arg8, Pro9 and Arg10 residues of E6 (Table I) that were previously described to participate in E6-p53 interactions (20). Quercetin in site II made contact with Asp49 of E6 which establishes polar interactions with His115 in the L1 loop of p53. Also, in site II quercetin contacted residues Leu50, L100 and Arg102 of E6 that impact on the architecture of E6 necessary for competent interaction with p53, which are discussed below. These results predict that quercetin would be able to interrupt the association of E6 with E6AP by binding to the E6 pocket and therefore preventing the formation of the p53 binding cleft on E6 and finally p53 degradation. Taken together, these results suggest that quercetin reactivates p53, preventing the formation of the E6/E6AP complex, leading to cell cycle arrest in the G2 phase and apoptosis.
Table I.
Summary of the contacts of each ligand with the residues of the HPV E6 protein (PDB ID: 4GIZ).
| Ligand | Site 1 | Site 2 | Site 3 | Energy (kcal/mol) in site 2 |
|---|---|---|---|---|
| Quercetin | D4, P5, Q6, E7, R8, P9, R10, C51, I52, V53, Y54, R55, R131. | V31, Y32, F45, D49, L50, C51, I52, V53, R55, Y60, A61, V62, L67, Y70, S71, L100, R102, Q107, R131. | S82, Y84, R117, H118, K122, Q123, R124, H126, R135, C139, S140. | −7.08±-0.18 |
| CAF24 | P5, Q6, E7, R8, P9, R10, C51, I52, V53, Y54, R55. | R10, K11, V31, Y32, F45, D49, L50, C51, V53, R55, A61, V62, L67, Y70, S71, I73, S74, R77, H78, R102, Q107, I128, R131, T133. | K72, I73, E75, Y76, Y79, S82, R124, H126, R135. | −7.99±-0.18 |
| C170 | * | P5, R8, P9, R10, K11, V31, Y32, F45, D49, L50, C51, I52, V53, Y54, R55, Y60, A61, V62, C66, L67, F69, Y70, S71, I73, S74, E75, R77, H78, L100, R102, C103, I104, Q107, I128, R129, R131, T133. | * | −6.8±-0.17 |
| Luteolin | D4, P5, Q6, E7, R8, P9, R10 C51, I52, V53, Y54, R55, R131. | R10, K11, V31, Y32, F45, R48, D49, L50, C51, I52, V53, Y60, A61, V62, C66, L67, Y70, S71, S74, Q75, H78, L100, R102, G107, R131. | S82, Y84, R117, H118, K122, G123, R124, H126, R135, S140. | −6.82±-0.19 |
not observed.
Discussion
Currently vaccination against most prevalent oncogenic HPVs as well as cervical cancer screening employed mainly in developed countries, has reduced the incidence and the mortality associated with this malignancy (23,24). However, cervical cancer is still one of the most common types of cancer in women worldwide. Aside from chemotherapy there are no specific treatments for HPV. The development of safe and effective drugs with minimal side effects is required for cervical cancer. In fact, the results of the present study demonstrated that quercetin possesses a selective toxicity towards cervical cancer derived cells infected with HPV16 (SiHa cells) and HPV18 (HeLa cells), [which are responsible for ~70% of all cervical cancer cases worldwide (1)] compared with HFF cells (considered normal cells), which required around a 7–8 fold higher concentration to exhibit the cytotoxic effects presented in the cervical cancer cell lines. In general, the quercetin IC50 in cervical cancer cells lines resulted high, which is a common feature for flavonoids (13,25). Indeed, further in vivo studies need to be performed to verify the pharmacokinetic of quercetin, as well as the synthesis of analogs to enhance its potency in future works.
Quercetin at high concentrations (>40 µM) is able to act as a prooxidant molecule causing DNA damage and resulting in cell cycle arrest and/or p53-dependent or independent mitochondrial apoptosis (12,25). Flow cytometry assays demonstrated that in SiHa and HeLa cells quercetin induced an increase in the G2/M phase cell population, whilst in HFF cells, the increase in the G2/M phase was barely significant and required a ~3-fold higher concentration of quercetin, which indicated a selectivity to cervical cancer cells in comparison with HFF cells. Furthermore, this arrest in G2/M phase did not correlate with an increase of the mitotic nuclei rate in HeLa and SiHa cells following quercetin treatment, suggesting that the arrest occurred in the G2 phase. Furthermore, the G2 phase cell cycle arrest was accompanied by the restoration of p53, by the increase of total p53 protein and its nuclear localization in HeLa and SiHa cells following quercetin treatment. In fact, a common feature between all these cell types is the status of the wild type p53, whose restoration could activate the p53 responses, such as cell cycle arrest and apoptosis, which has been widely studied (23).
The best-characterized p53 target gene is p21, for which the promoter contains two highly conserved p53-responsive elements on which p53 directly binds to activate p21 transcription (26). In response to DNA damage, p53 induces p21 transcription and then the p21 protein can associate and inhibit the mitosis-promoting factor, cyclin dependent kinase1-Cyclin B1 to arrest in the G2 phase (27). In the present study, an accumulation of the p21 transcript in HeLa and SiHa cells was observed following exposure to quercetin, demonstrating the reactivation of p53 as a transcriptional factor. Therefore, the upregulation of p21 together with the reactivation of p53 strongly suggest its participation in the G2 phase cell cycle arrest.
In addition to the increase of the p21 transcript by p53, the activation of p53 by quercetin also induced high levels of the pro-apoptotic protein Bax. Notably, mitochondrial apoptosis regulated by p53 can occur by activating or repressing the transcription of pro-apoptotic (Bax, Bid, and Noxa) or survival (Bcl-2 and Bcl-xL) genes, respectively (28–30). In addition, p53 can also trigger mitochondrial apoptosis through activation of Bak or inhibition of Bcl-2 by direct association. The increase of the Bax/Bcl-2 ratio results in the formation of the mitochondria outer membrane pore, cytochrome release and the subsequent activation of caspases 3/7 (31,32). Furthermore, quercetin induced caspases 3/7 activity, as well as morphological cell traits associated to apoptosis in HeLa and SiHa cervical cancer cells. This result matches with a previous study in which quercetin induced mitochondrial apoptosis (33).
p53 inactivation by HPV E6 is a general feature in cervical cancer. Therefore, identifying the inhibition of HPV E6 in cervical cancer is of interest in order to reactivate p53 and sensitize cervical cancer cells to apoptosis. Notably, the increase of the p53 protein in HeLa and SiHa cells did not alter the expression of the HPV E6 protein. This result contrasts with the finding reported by Qiao et al (14) in which they demonstrated that treatment with the flavonoid EGCG in HeLa and CaSki cells caused an increase of p53 accompanied by a decrease of HPV E6 protein. Furthermore, it corresponds to the published data by Malecka et al (34) in which the flavonoid gossypetin with only an additional hydroxyl in the C8 position compared with quercetin, disrupted the E6/E6AP association. Similar results were previously described by Cherry et al (15), which clearly demonstrated that in vitro, luteolin (lacking a hydroxyl in the C3 position with respect to quercetin) and synthetic flavonoids disrupted the binding of HPV 16 E6 and the core 70 amino acids containing the E6 binding region (LxxLL) of E6AP and inhibited the E6-induced p53 degradation in rabbit reticulocyte lysate.
The crystallographic structure of the HPV-16 E6 protein (PDB ID: 4GIZ) in complex with the LxxLL peptide of E6AP and p53 core was solved. It was demonstrated that the LxxLL motif E6AP binds to a conserved pocket of the E6 protein causing a conformational change of E6 to allow it to competently interact with p53 (20), since neither E6 nor E6AP are separately able to associate with p53 (8). Based on the solved structure of E6/E6AP/p53 and E6/E6AP, the critical E6 residues to interact with E6AP and p53 were revealed and the functionality in p53 degradation was demonstrated by mutagenesis (6,20). In the present study it was demonstrated that quercetin in site I from the E6 structure established contact with Pro5, Gln6, Arg8 and, Arg10 residues that bring together the N-terminal arm and α1 helix of E6 and N-terminal arm, β1 and β10 of p53. Moreover, E6 Glu7 residue establishes direct contacts with p53, while E6 Pro5, Arg8 and Pro9 residues modulate the conformation of the N-terminal region of E6. In fact, mutations in Pro5, Arg8 or Pro9 impair ternary complex assembly and p53 degradation (35,36). In addition, in site II corresponding to a hydrophobic pocket of E6 where the LxxLL motif E6AP binds, quercetin binding was predicted to be more energetically favorable. In site II quercetin established contacts with Asp49 residue whose substitution to alanine impairs p53 ternary complex assembly and p53 degradation suggesting its essential role in this process. Also, in site II, quercetin made contact with Leu50, L100, Arg102 and Arg131 of E6. Point mutations in the hydrophobic residue Leu50 have been demonstrated to decrease LxxLL peptide binding and p53 degradation. Hydrophobic interactions were established between the E6 residue Leu100 and p53 core residues Leu114 and Trp146. Surrounding the hydrophobic pocket, the charged residue Arg102 holds together the E6 N domain, E6C domain, and the LxxLL motif of E6AP. Arg131 provides structurally equivalent contacts with the E6AP peptide. Point mutations of these arginine residues lead to markedly decreased LxxLL peptide binding and p53 degradation (20). These multiple contacts that quercetin was predicted to establish with critical residues of E6, which are established interactions with E6AP and p53, suggest that quercetin could disrupt the E6AP/E6/p53 complex formation and prevent p53 degradation. However, further in vitro or in vivo binding assays need to be performed to confirm the separation of p53 and E6/E6AP.
Finally, the present study proposes that quercetin can reactivate p53 and induce G2 phase cell cycle arrest and apoptosis in HPV-positive human cervical cancer-derived cells, by inhibiting the interaction of E6/E6AP.
Acknowledgements
The authors thank Dr Ramón González from CIDC and Dr Patricia García López INCan for the relevant suggestions and criticisms to the project.
Funding
Aldo F. Clemente Soto acknowledges fellowship 379417 from CONACYT. Partial support from UAEM (grants nos. SI-DGDI-UAEM/13/289) and PAPIIT-UNAM (IN210316) is acknowledged.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
AFC-S and LG-M conceived the original idea and planned the experiments of the project. AFC-S participated in the experiments. AFC-S and ES-V acquired and analyzed the confocal microscopy images. CM-P carried out the molecular docking. JNS-C supervised the cell viability and flow cytometry assay. All authors analyzed and interpreted the data. OP-Z supervised the project. AFC-S wrote the manuscript with the support of LG-M. All authors contributed and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Marth C, Landoni F, Mahner S, McCormack M, Gonzalez-Martin A, Colombo N, ESMO Guidelines Committee Cervical cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28(Suppl_4):iv72–iv83. doi: 10.1093/annonc/mdx220. [DOI] [PubMed] [Google Scholar]
- 2.Dürst M, Gissmann L, Ikenberg H, zur Hausen H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci USA. 1983;80:3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardiol D, Kühne C, Glaunsinger B, Lee SS, Javier R, Banks L. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene. 1999;18:5487–5496. doi: 10.1038/sj.onc.1202920. [DOI] [PubMed] [Google Scholar]
- 4.Nakagawa S, Huibregtse J. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol Cell Biol. 2000;20:8244–8253. doi: 10.1128/MCB.20.21.8244-8253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ansari T, Brimer N, Vande Pol SB. Peptide interactions stabilize and restructure human papillomavirus type 16 E6 to interact with p53. J Virol. 2012;86:11386–11391. doi: 10.1128/JVI.01236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Zapien D, Ruiz FX, Poirson J, Mitschler A, Ramirez J, Forster A, Cousido-Siah A, Masson M, Vande Pol S, Podjarny A, et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529:541–545. doi: 10.1038/nature16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as an ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 8.Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurlin PJ, Kaur P, Smith PP, Perez-Reyes N, Blanton RA, McDougall JK. Progression of human papillomavirus type 18-immortalized human keratinocytes to a malignant phenotype. Proc Natl Acad Sci USA. 1991;88:570–574. doi: 10.1073/pnas.88.2.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb Perspect Med. 2016;6:a026104. doi: 10.1101/cshperspect.a026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulati N, Laudet B, Zohrabian VM, Murali R, Jhanwar-Uniyal M. The antiproliferative effect of quercetin in cancer cells is mediated via inhibition of the PI3K-Akt/PKB pathway. Anticancer Res. 2006;26:1177–1181. [PubMed] [Google Scholar]
- 12.Metodiewa D, Jaiswal AK, Cenas N, Dickancaité E, Segura-Aguilar J. Quercetin may act as a cytotoxic prooxidant after its metabolic activation to semiquinone and quinoidal product. Free Radic Biol Med. 1999;26:107–116. doi: 10.1016/S0891-5849(98)00167-1. [DOI] [PubMed] [Google Scholar]
- 13.Srivastava S, Somasagara RR, Hegde M, Nishana M, Tadi SK, Srivastava M, Choudhary B, Raghavan SC. Quercetin, a natural flavonoid interacts with DNA, arrests cell cycle and causes tumor regression by activating mitochondrial pathway of apoptosis. Sci Rep. 2016;6:24049. doi: 10.1038/srep24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiao Y, Cao J, Xie L, Shi X. Cell growth inhibition and gene expression regulation by (−)-epigallocatechin-3-gallate in human cervical cancer cells. Arch Pharm Res. 2009;32:1309–1315. doi: 10.1007/s12272-009-1917-3. [DOI] [PubMed] [Google Scholar]
- 15.Cherry JJ, Rietz A, Malinkevich A, Liu Y, Xie M, Bartolowits M, Davisson VJ, Baleja J, Androphy EJ. Structure based identification and characterization of flavonoids that disrupt human papillomavirus-16 E6 function. PLoS One. 2013;8:e84506. doi: 10.1371/journal.pone.0084506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanigawa S, Fujii M, Hou DX. Stabilization of p53 is involved in quercetin-induced cell cycle arrest and apoptosis in HepG2 cells. Biosci Biotechnol Biochem. 2008;72:797–804. doi: 10.1271/bbb.70680. [DOI] [PubMed] [Google Scholar]
- 17.Thangasamy T, Sittadjody S, Lanza-Jacoby S, Wachsberger PR, Limesand KH, Burd R. Quercetin selectively inhibits bioreduction and enhances apoptosis in melanoma cells that overexpress tyrosinase. Nutr Cancer. 2007;59:258–268. doi: 10.1080/01635580701499545. [DOI] [PubMed] [Google Scholar]
- 18.Yang GY, Liao J, Kim K, Yurkow EJ, Yang CS. Inhibition of growth and induction of apoptosis in human cancer cell lines by tea polyphenols. Carcinogenesis. 1998;19:611–616. doi: 10.1093/carcin/19.4.611. [DOI] [PubMed] [Google Scholar]
- 19.Mohan S, Bustamam A, Ibrahim S, Al-Zubairi AS, Aspollah M, Abdullah R, Elhassan MM. In vitro ultramorphological assessment of apoptosis on CEMss induced by linoleic acid-rich fraction from Typhonium flagelliforme tuber. Evid Based Complement Alternat Med 2011. 2011:421894. doi: 10.1093/ecam/neq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanier K, Charbonnier S, Sidi AO, McEwen AG, Ferrario MG, Poussin-Courmontagne P, Cura V, Brimer N, Babah KO, Ansari T, et al. Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins. Science. 2013;339:694–698. doi: 10.1126/science.1229934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan MA, Toso RJ, Thrower D, Wilson L. Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc Natl Acad Sci USA. 1993;90:9552–9556. doi: 10.1073/pnas.90.20.9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengstermann A, Linares LK, Ciechanover A, Whitaker NJ, Scheffner M. Complete switch from Mdm2 to human papillomavirus E6-mediated degradation of p53 in cervical cancer cells. Proc Natl Acad Sci USA. 2001;98:1218–1223. doi: 10.1073/pnas.98.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Kriekinge G, Castellsagué X, Cibula D, Demarteau N. Estimation of the potential overall impact of human papillomavirus vaccination on cervical cancer cases and deaths. Vaccine. 2014;32:733–739. doi: 10.1016/j.vaccine.2013.11.049. [DOI] [PubMed] [Google Scholar]
- 24.Peirson L, Fitzpatrick-Lewis D, Ciliska D, Warren R. Screening for cervical cancer: A systematic review and meta-analysis. Syst Rev. 2013;2:35. doi: 10.1186/2046-4053-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wätjen W, Michels G, Steffan B, Niering P, Chovolou Y, Kampkötter A, Tran-Thi QH, Proksch P, Kahl R. Low concentrations of flavonoids are protective in rat H4IIE cells whereas high concentrations cause DNA damage and apoptosis. J Nutr. 2005;135:525–531. doi: 10.1093/jn/135.3.525. [DOI] [PubMed] [Google Scholar]
- 26.Laptenko O, Beckerman R, Freulich E, Prives C. p53 binding to nucleosomes within the p21 promoter in vivo leads to nucleosome loss and transcriptional activation. Proc Natl Acad Sci USA. 2011;108:10385–10390. doi: 10.1073/pnas.1105680108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charrier-Savournin FB, Château MT, Gire V, Sedivy J, Piette J, Dulic V. p21-mediated nuclear retention of cyclin B1-Cdk1 in response to genotoxic stress. Moll Biol Cell. 2004;15:3965–3976. doi: 10.1091/mbc.e03-12-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA, Hoffman B, Reed JC. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene. 1994;9:1799–1805. [PubMed] [Google Scholar]
- 29.Sax JK, Fei P, Murphy ME, Bernhard E, Korsmeyer SJ, El-Deiry WS. BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol. 2002;4:842–849. doi: 10.1038/ncb866. [DOI] [PubMed] [Google Scholar]
- 30.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/S1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 31.Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. J Biol Chem. 2004;24:6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomita Y, Marchenko N, Erster S, Nemajerova A, Dehner A, Klein C, Pan H, Kessler H, Pancoska P, Moll UM. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem. 2006;281:8600–8606. doi: 10.1074/jbc.M507611200. [DOI] [PubMed] [Google Scholar]
- 33.Vidya Priyadarsini R, Senthil Murugan R, Maitreyi S, Ramalingam K, Karunagaran D, Nagini S. The flavonoid quercetin induces cell cycle arrest and mitochondria-mediated apoptosis in human cervical cancer (HeLa) cells through p53 induction and NF-κB inhibition. Eur J Pharmacol. 2010;649:84–91. doi: 10.1016/j.ejphar.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 34.Malecka KA, Fera D, Schultz DC, Hodawadekar S, Reichman M, Donover PS, Murphy ME, Marmorstein R. Identification and characterization of small molecule human papillomavirus E6 inhibitors. ACS Chem Biol. 2014;9:1603–1612. doi: 10.1021/cb500229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mietz JA, Unger T, Huibregtse JM, Howley PM. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper B, Schneider S, Bohl J, Jiang Yh, Beaudet A, Vande Pol S. Requirement of E6AP and the features of human papillomavirus E6 necessary to support degradation of p53. Virology. 2003;306:87–99. doi: 10.1016/S0042-6822(02)00012-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.