Abstract
(−)-Epigallocatechin-3-gallate (EGCG), a major constituent of green tea, is a potential anticancer agent, but the molecular mechanisms of its effects are not well-understood. The present study was conducted to examine the mechanism of EGCG in lung cancer cells. Alterations in long non-coding RNAs (lncRNAs) and mRNAs were investigated in lung cancer cells treated with EGCG by lncRNA microarray analysis. Furthermore, the functions and signaling pathways regulated by EGCG were predicted by bioinformatics analysis. A total of 960 lncRNAs and 1,434 mRNAs were significantly altered following EGCG treatment. These lncRNAs were distributed across nearly all human chromosomes and the mRNAs were involved in the cell cycle and the mitotic cell cycle process. Through a combination of microarray and bioinformatics analysis, 20 mRNAs predicted to serve a key role in the EGCG regulation were identified, and certain regulatory networks involving EGCG-regulated lncRNAs were predicted. In conclusion, EGCG affects the expression of various lncRNAs and mRNAs in the cells, therefore affecting cell functions. The results of the present study provide an insight into the mechanism of EGCG, which may be useful for therapeutic development.
Keywords: (−)-epigallocatechin-3-gallate, long non-coding RNAs, microarray, lung cancer, bioinformatics analysis
Introduction
Lung cancer is one of the most common solid tumors and has very high global incidence and mortality rates (1). Although therapeutic treatment of lung cancer has made great progress, the prognosis of lung cancer patients remains unsatisfactory and treatment causes a number of side effects (2). Natural dietary molecules are promising candidates for novel therapeutic agents in cancer. Green tea is one of the most widely consumed beverages, worldwide (3). Furthermore, (−)-epigallocatechin-3-gallate (EGCG), the main component of green tea polyphenols, was demonstrated to have various biological activities and demonstrates potential as a chemical and therapeutic agent in several diseases, including various cancers (3–8) and atherosclerosis (9).
Long non-coding RNAs (lncRNAs) are defined as a class of transcripts longer than 200 nucleotides (nt) in the cytoplasm and nucleus (10–13). LncRNAs, characterized by the complexity and diversity of their sequences, have been implicated in a wide spectrum of cellular activities and diseases (14,15). Emerging studies have demonstrated that certain lncRNAs are frequently abnormally regulated in several types of cancer and serve important roles in the occurrence and development of tumors (16–19). Accumulating evidence has confirmed the activity of EGCG in lung cancer therapy and many lncRNAs and mRNAs are involved in regulating tumorigenesis (10,20–22). However, microarray analysis of the differential expression profiles of lncRNAs and mRNAs in lung cancer cells treated with EGCG has not been reported.
In the present study, the EGCG-regulated lncRNAs and mRNAs were investigated in lung cancer cells by bioinformatics analysis. Based on the microarray analysis, a set of lncRNAs and mRNAs was identified whose expression levels were significantly modulated by EGCG in lung cancer cells, some of which are novel. However, the extent to which these genes are associated with lung cancer cells through EGCG remains unclear and needs to be addressed in the near future.
Materials and methods
Cell culture and cell proliferation assay
The human lung cancer cell lines A549 (lung adenocarcinoma) and NCI-H460 (large cell lung carcinoma) were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cells were plated in 96-well plates (5,000 cells/well) and were incubated overnight (37°C, 5% CO2), followed by treatment with EGCG (20, 40, 80, 160, 320 µM) and 0 µM EGCG (control group) for 24 or 48 h respectively. The MTT-based assay (Sangon Biotech Co., Ltd., Shanghai, China) was performed to determine viable cell numbers at 37°C and DMSO was used to dissolve the formazan. Absorbance at 570 nm was measured and the 50% maximal inhibitory concentration (IC50) of EGCG for A549 and NCI-H460 cells were calculated by Graphpad Prism version 6.0 (GraphPad Software, Inc., CA, USA).
RNA extraction and quality control
Total RNA was extracted from 1×106 A549 or NCI-H460 cells treated with different concentration of EGCG and control group respectively, using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), purified with a mirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and was quantified using a spectrophotometer (NanoDrop ND-1000; NanoDrop, Wilmington, DE, USA). The RNA integrity of each sample was assessed by capillary electrophoresis using the RNA 6000 Nano Lab-on-a-Chip kit and Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Only RNA extracts with RNA integrity values >6 were subjected to further analysis.
RNA amplification, labeling, and hybridization
complementary (c)DNA labeled with a fluorescent dye (Cy5 or Cy3-deoxycytidine triphosphate; CapitalBio Technology Co., Ltd., Beijing, China) was produced by Eberwine's linear RNA amplification method as previously described (23), and subsequently RNase H enzymatic reaction (37°C for 45 min, and followed by 95°C for 5 min). The labeled cDNAs were purified using a Capital Bioc RNA Amplification and Labeling kit (CapitalBio Corporation, Beijing, China) and then hybridized with specific probes (CapitalBio Technology Co., Ltd.) in a hybridization oven (Xinghua Analytical Instrument Factory, Jiangsu, China) overnight at 45°C.
Microarray analysis
The expression levels of lncRNAs and mRNAs were determined using Gene Spring software V13.0 (Agilent Technologies). The differentially expressed lncRNAs and mRNAs between the EGCG-treated and control groups were identified based on the threshold values of ≥2 and ≤-2 fold-change and paired Student's t-test P<0.05. Data were log2-transformed and median-centered using genes and Adjust Data function of CLUSTER 3.0 software (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) and analyzed by hierarchical clustering with average linkages. Tree visualization was performed using the Java TreeView (Stanford University School of Medicine, Stanford, CA, USA; http://sourceforge.net/projects/jtreeview/files). Gene-lung cancer associations were investigated by using the DisGeNET database (http://www.disgenet.org/web/DisGeNET/menu), which records disease-associated genes and provides literature support, as in the previous reference (24).
Gene Ontology (GO), pathway and disease analysis
GO analysis was performed using an online database (www geneontology.org). The GO database provides a network of three structured definition terms that describe the properties of a gene product (25). Pathway analysis of the differentially expressed genes was performed according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (www.genome.jp/kegg/). Specifically, Fisher's exact test and χ2 tests were performed to classify the GO category and select the significant pathway. The false discovery rate (FDR) was calculated and the threshold of significance was defined as P<0.05. FunDO database (http://fundo.nubic.northwestern.edu/) was used for disease analysis.
Network analysis of EGCG target genes
Search Tool for Interactions of Chemicals (STITCH; http://stitch.embl.de/) is a database of interactions and correlations between compounds and genes. The EGCG direct target genes were searched and analyzed by using STITCH according to the reference (26). Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org/) is a database of protein-protein interactions (27). The interaction of EGCG direct target gene and differential expression oncogene affected by EGCG was determined in the STRING database, and only the interactions with a combined score > 0.4 were considered as significant. Finally, the network of compound and gene interactions was mapped.
Analysis of lncRNA binding transcription factors
We downloaded the transcriptional factors for each of the 20 differential expression oncogenes most affected by EGCG treatment from the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgTracks?hgsid=698152999_7PLacukNAMsjS2GSMIFtIxAic9jW). Then, the lncRNA-transcriptional factor-oncogene regulatory network was constructed by using the LncPro database (http://bioinfo.bjmu.edu.cn/lncpro/) as previously described (28).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The Total RNA of 1×106 A549 cells treated with 80 µM EGCG for 48 h and the control group cells was extracted as described above respectively. The PrimeScript™ RT Reagent kit (Takara Bio, Inc., Otsu, Japan) was used for cDNA synthesis and genomic DNA removal. qPCR was performed using SYBR® Premix Ex Taq™ (Takara Bio, Inc., Otsu, Japan) and in an ABI 7000 instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.) for 40 cycles (95°C for 15 sec, 60°C for 1 min) after an initial 3 min degeneration at 95°C, and β-actin was used as an internal control (primers for qPCR listed in Table I). The relative gene expression data was analyzed by 2−ΔΔCq method as previously described (29).
Table I.
The primers for RT-qPCR.
| Gene | Name | Sequence |
|---|---|---|
| ENSG00000272796.1 | Sense | CATTGGACTGGGTGAGGC |
| Antisense | TTGAGCACGGTAGAGGAGAC | |
| ENSG00000254054.2 | Sense | GATTCCACTGATATTTACTGAA |
| Antisense | GGACCGCTCCTTTGATGC | |
| ENSG00000260630.2 | Sense | GCCGTGAGTGAAGGGCAGAG |
| Antisense | TTCCAGAGCCGCCAGTAGGG | |
| ENSG00000235142.2 | Sense | AGCTGGAATGCAGATGGG |
| Antisense | AGCACAGGCTCAAGGGAC | |
| ENSG00000224063.1 | Sense | AGAACTGATTTTAGAATGCCA |
| Antisense | CCAATGTATTTGCCAAGA | |
| ENSG00000251018.2 | Sense | AGACCTATTGGAACTGACT |
| Antisense | AGAACCTGAGTGCCTTGT | |
| ENSG00000226403.1 | Sense | CTGGTCCTCGCAGTCCGC |
| Antisense | CTCTTTCCCAAAGGGCAC | |
| PSMC3IP | Sense | GCGGATCAGGACCAGTTT |
| Antisense | TTTGACCAGGACTAGGCG | |
| ENSG00000230109.1 | Sense | AAAACTATGAGAAAACTGGGTC |
| Antisense | ATGTAAGTTTCTGATTGGTCC | |
| ENSG00000130600.10 | Sense | GAGGAGCTGAGTGGGACC |
| Antisense | TTGATGTTGGGCTGATGAG | |
| ALDOC | Sense | GGCATCAAGGTTGACAAGGG |
| Antisense | GCTGGCAGATACTGGCATAA | |
| HABP2 | Sense | AATAAGTGTCAGAAAGTGCAAAA |
| Antisense | CAGCGGTAGTAGGGAGGA | |
| CFB | Sense | TATGAAGACCACAAGTTGAAGT |
| Antisense | GTATAGCAAGTCCCGGATC | |
| IL11 | Sense | TGCACAGCTGAGGGACAA |
| Antisense | CCGCAGGTAGGACAGTAGG | |
| SMOC1 | Sense | AGATGACGGGTCTAAGCC |
| Antisense | ATCACCAAGTGTTTAATCCATA | |
| AREG | Sense | CTCGGCTCAGGCCATTAT |
| Antisense | AGCCAGGTATTTGTGGTTCG | |
| EREG | Sense | CTGGGTTTCCATCTTCTA |
| Antisense | TGTTATTGACACTTGAGCC | |
| MTRNR2L6 | Sense | TAGGGACTTGTATGAATGAC |
| Antisense | ATAGGTTGCTCGGAGGTT | |
| POLR3G | Sense | CACCTGAAGAAAGACAAG |
| Antisense | TCAGTATTAGTGAGTGGTGT | |
| LYAR | Sense | TTTCTGGGGCGATGACTA |
| Antisense | TTGGGGCTGACATTGGGT | |
| β-actin | Sense | AGGGGCCGGACTCGTCATACT |
| Antisense | GGCGGCACCACCATGTACCCT |
Statistical analysis
The experiments were performed three times. All results are expressed as the mean ± standard deviation. A paired Student's t test was performed to compare the two groups in the microarray analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Analysis of cell proliferation
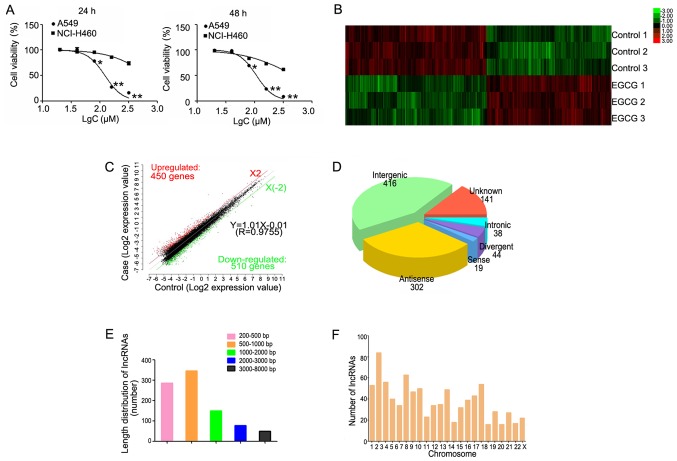
Treatment with EGCG inhibited lung cancer cell proliferation in dose-dependent manner (Fig. 1A). EGCG inhibited the growth of A549 cells, with IC50 values of 122.4 and 108.2 µM at 24 and 48 h, respectively. Additionally, the IC50 values of EGCG in NCI-H460 cells were 693.2 and 485.1 µM at 24 and 48 h, respectively. These results demonstrated that A549 cells are more sensitive to EGCG than NCI-H460 cells. Therefore, A549 cells treated with 80 µM (≈IC30) for 48 h were used for microarray analysis.
Figure 1.
Hierarchical clustering of differentially expressed lncRNAs. (A) The survival rate of lung cancer cells following treatment with EGCG detected by MTT assay. (B) Hierarchical clustering of 3 EGCG-treated samples (EGCG1-3) versus 3 untreated samples (CON1-3) was based on the 960 dysregulated lncRNAs. Red represents upregulated lncRNAs, while green represents downregulated lncRNAs. (C) Red, black, and green dots represent lncRNAs with expression that was increased, equivalent, or decreased between the EGCG-treated and control groups. (D) Length distribution of 960 differentially expressed lncRNAs. (E) Class distribution of dysregulated lncRNAs, including 416 intergenic, 302 antisense, 141 unknown, 44 divergent, 38 intronic, and 19 sense lncRNAs. (F) Chromosome enrichment analysis of 960 differentially expressed lncRNAs on chromosomes (DNA). *P<0.05, **P<0.01 vs. NCI-H460. EGCG, (−)-epigallocatechin-3-gallate; lncRNA, long non-coding RNA.
Analysis of differentially expressed lncRNAs
A total of 19,446 lncRNAs were detected by Gene Spring software V13.0. Based on the results of the lncRNA expression profile, differentially expressed lncRNAs between the EGCG-treated and control groups were identified. According to their expression levels in the samples, the clusters were hierarchically clustered (Fig. 1B). The threshold was set to a fold-change ≥2, P<0.05, and FDR <0.05 and a total of 960 differential expressed lncRNAs, including 450 and 510 up- and downregulated lncRNAs, respectively were identified (Fig. 1C).
Among the differentially expressed lncRNAs, there were 416 intergenic, 302 antisense, 141 unknown, 44 divergent, 38 intronic, and 19 sense lncRNAs (Fig. 1D). The length of the differentially expressed lncRNAs ranged from 200 base pairs (bp) to 8 kb, with the lengths of 660 lncRNAs (68.75%) ranging between 200–1,000 bp (Fig. 1E). Additionally, these differentially expressed lncRNAs were distributed in nearly all human chromosomes (Fig. 1F).
Analysis of differentially expressed mRNAs
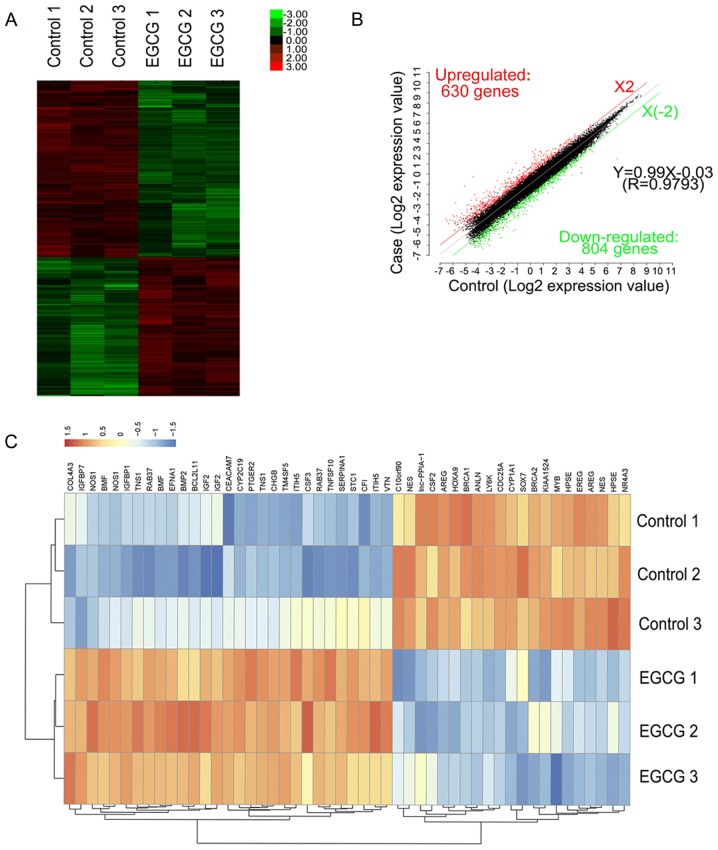
A total of 1,434 mRNAs were detected whose expression significantly differed between lung cancer cells treated with EGCG and the control group (fold-change ≥2.0, P<0.05, and FDR <0.05). Of these, 804 mRNAs were downregulated and 630 were upregulated. Through hierarchical clustering analysis, different expression patterns were predicted (Fig. 2A and B). The top 10 upregulated and 10 downregulated mRNAs between the two groups are listed in Table II. The top 5 upregulated and 5 downregulated mRNAs associated with lung cancer are listed in Table III. Genes associated with lung cancer in the DisGeNET database were investigated. By comparing the results of the database with the results obtained upon EGCG treatment, 168 known lung cancer genes in the EGCG-treated groups were identified, and the top 50- changed genes were plotted as a heat map (Fig. 2C).
Figure 2.
Hierarchical clustering of the differentially expressed mRNAs. (A) Hierarchical clustering of 3 EGCG treatment samples (EGCG1-3) versus 3 untreated samples (CON1-3) was based on the 1,434 dysregulated mRNAs. Red represents upregulated mRNAs, while green represents downregulated mRNAs. (B) Red, black, and green dots represent mRNAs with expression that was increased, equivalent, or decreased between the EGCG-treated and control groups. (C) Known lung cancer oncogenes in the heat map were significantly differentially expressed following EGCG treatment. EGCG, (−)-epigallocatechin-3-gallate; lncRNA, long non-coding RNA.
Table II.
Top 10 up and downregulated mRNAs following EGCG treatment.
| Upregulated mRNAs | Downregulated mRNAs | |||
|---|---|---|---|---|
| mRNA | FC(abs) | mRNA | FC(abs) | |
| CEACAM7 | 36.10 | MTRNR2L6 | 18.14 | |
| AGT | 30.47 | CYP1A1 | 10.22 | |
| TF | 15.28 | ID4 | 9.65 | |
| TNS1 | 10.05 | CORO1A | 8.27 | |
| SMOC1 | 9.04 | HMGCS2 | 7.21 | |
| IL11 | 8.17 | MTRNR2L8 | 6.22 | |
| ALDOC | 8.15 | SP5 | 6.14 | |
| NRAP | 8.06 | HPDL | 5.72 | |
| KCNJ16 | 7.61 | GREB1 | 5.58 | |
| BMF | 7.59 | CEMIP | 5.53 | |
abs, absolute; EGCG, (−)-epigallocatechin-3-gallate; FC, fold-change.
Table III.
Top 5 up- and downregulated mRNAs associated with lung cancer.
| Author, year | Genesymbol | FC(abs) | Regulation | Reported function in lung cancer | (Refs.) |
|---|---|---|---|---|---|
| Pan et al, 2016 | TTR | 6.91 | Up | Reduced transthyretin expression in sera of lung cancer | (21) |
| Liu et al, 2007 | SLC5A10 | 5.21 | Up | DNA methylation in SLC5A8 expression in lung cancer cell lines, Reduced or lost expression of SLC5A8 was observed in tumor tissues | (50) |
| Park et al, 2013 and Tsai et al, 2014 | RAB37 | 4.73 | Up | Lung cancer patients with metastasis and poor survival show low hRAB37 protein expression | (51) |
| Knockdown of hRAB37 in low metastasis cell line led to a significant increase in cell migration | (52) | ||||
| Wu et al, 2009 | ITIH5 | 3.46 | Up | ITIH5 mRNA expression was significantly decreased in NSCLC compared to normal lung tissue, ITIH5 may be a novel putative tumor suppressor gene in NSCLC | (53) |
| Dötsch et al, 2015 | IGFBP7 | 3.08 | Up | (IGFBP7) was considered a tumor suppressor gene in lung cancer. | (54) |
| Chen et al, 2011 | GREB1 | 5.58 | Down | The expression of GREB1 in lung cancers is significantly higher than that in normal lung tissues, and closely related to smoking | (55) |
| Zhang et al, 2012 and McLemore et al, 1990 | CYP1A1 | 10.22 | Down | A biomarker for lung cancer risk, and closely related to smoking | (56,57) |
| Taioli et al, 1998 | CLSPN | 5.24 | Down | Claspin contribute to the radioresistance of lung cancer brain metastases | (58) |
| Choi et al, 2014 and Taniguchi et al, 2016 | AREG | 4.72 | Down | AREG can cause lung tumorigenesis and resistance to crizotinib | (59,60) |
| Taron et al, 2004 | BRCA1 | 3.08 | Down | BRCA1 as an indicator of chemoresistance in lung cancer | (61) |
abs, absolute; AREG, amphiregulin; BRCA1, breast cancer 1; CLSPN, claspin; CYP1A1, cytochrome P450 family 1 subfamily A member 1; FC, fold-change; IGFBP7, insulin like growth factor binding protein 7; ITIH5, inter-α-trypsin inhibitor heavy chain family member 5; SLC5A10, solute carrier family 5 member 10; TTR, transthyretin.
RT-qPCR verification of differentially expressed lncRNAs and mRNAs
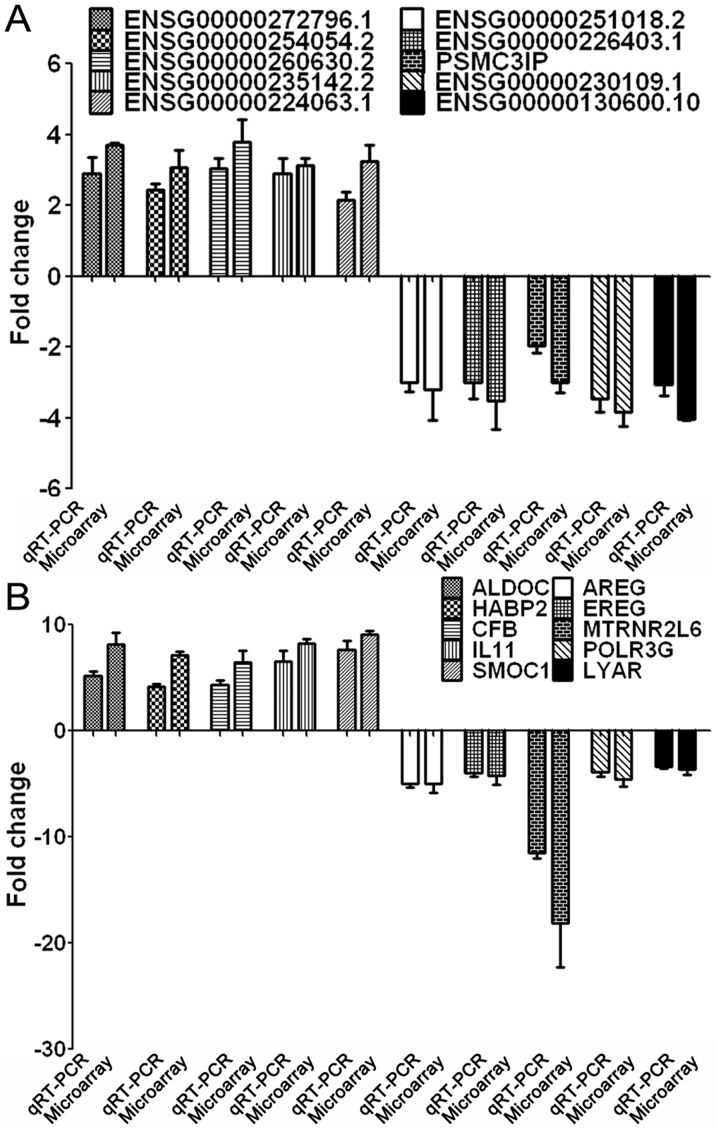
A total of 10 differentially expressed lncRNAs were randomly selected, including 5 upregulated (ENSG00000272796.1, ENSG00000254054.2, ENSG00000260630.2, ENSG00000235142.2 and ENSG00000224063.1) and 5 downregulated lncRNAs (ENSG00000251018.2, ENSG00000226403.1, PSMC3IP, ENSG00000230109.1 and SG00000130600.10), for verification in these lung cancer cells. The RT-qPCR results of these 10 selected lncRNAs were consistent with those from the microarray analysis (Fig. 3A). Additionally, 10 differentially expressed mRNAs were also randomly selected, including 5 upregulated (aldolase C, hyaluronan-binding protein 2, complement factor B, interleukin (IL) 11, and secreted modular calcium-binding protein 1) and 5 downregulated mRNAs (amphiregulin, epiregulin, humanin-like protein 6, polymerase (RNA) III (DNA Directed) Polypeptide G, and Ly1 antibody reactive). The results for all 10 selected mRNAs were consistent with those from the microarray analysis (Fig. 3B).
Figure 3.
RT-qPCR validation of the microarray analysis results. (A) RT-qPCR analysis of 10 dysregulated lncRNAs, including 5 upregulated and 5 downregulated lncRNAs. (B) RT-qPCR analysis of 10 dysregulated mRNAs, consisting of 5 upregulated and 5 downregulated mRNAs. ALDOC, Aldolase C; AREG, Amphiregulin; CFB, Complement factor B; EREG, Epiregulin; HABP, Hyaluronan-binding protein; IL, Interleukin; lncRNA, long non-coding RNA; LYAR, Ly1 antibody reactive; MTRNR2L6, Humanin-like protein; POLR3G, Polymerase (RNA) III (DNA Directed) Polypeptide G; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SMOC, Secreted modular calcium-binding protein.
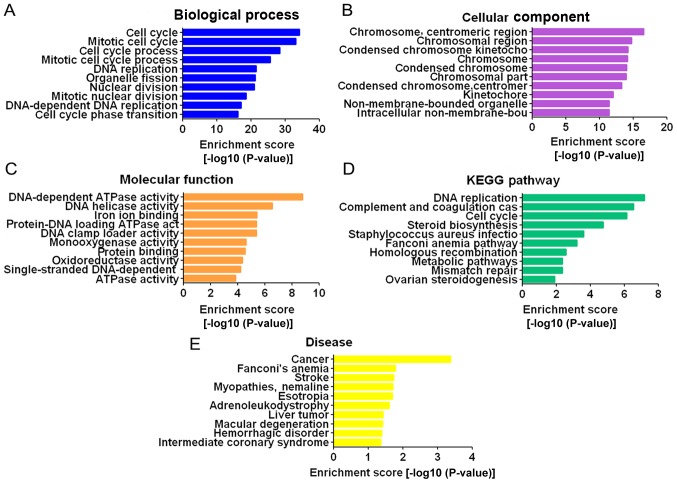
GO and pathway analysis for differentially expressed mRNAs
GO analysis consists of three parts: Biological processes, cellular components, and molecular functions. Through GO analysis, it was demonstrated that the differentially expressed mRNAs were mainly enriched in the GO terms ‘cell cycle’, ‘mitotic cell cycle’, ‘cell cycle process’ and ‘mitotic cell cycle process’ for biological processes (Fig. 4A); ‘chromosome’, ‘centromeric region’, ‘chromosomal region’, ‘condensed chromosome kinetochore’ and ‘chromosome’ for cellular components (Fig. 4B); and DNA-dependent ATPase activity’, ‘DNA helicase activity’ and ‘iron ion binding’ for molecular functions (Fig. 4C). Pathway analysis was based on the KEGG database. The differentially expressed mRNAs were associated with DNA replication, complement and coagulation cascades, cell cycle and other pathways (Fig. 4D). The dysregulated mRNAs were associated with a number of diseases, of which cancer was the most relevant (Fig. 4E).
Figure 4.
Pathways and GO terms for differentially expressed mRNAs. (A-C) GO analysis. (D) KEGG pathway analysis. (E) FunDO disease analysis. GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes.
Bioinformatic analysis of potential mechanisms of EGCG regulating lncRNAs/mRNAs
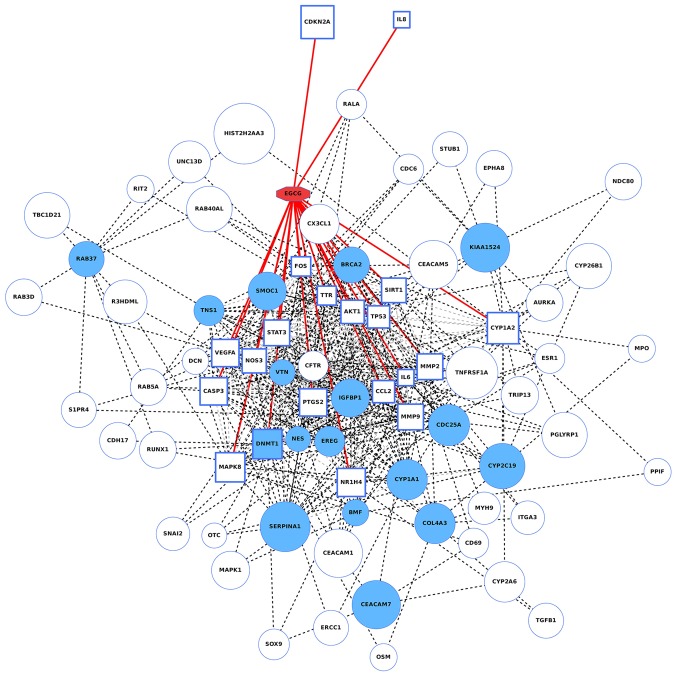
A total of 20 EGCG target genes were detected using the STITCH database; these genes were AKT1, Caspase 3, IL-6, prostaglandin-endoperoxide synthase 2 (PTGS2), tumor protein 53 (TP53), vascular endothelial growth factor A (VEGFA), FOS, mitogen-activated protein kinase 8 (MAPK8), nitric oxide synthase 3 (NOS3), signal transducer and activator of transcription 3 (STAT3), matrix metalloproteinase 9 (MMP9), C-C motif chemokine ligand 2 (CCL2), transthyretin (TTR), MMP2, DNA (cytosine-5)-methyltransferase 1 (DNMT1), sirtuin 1 (SIRT1), nuclear receptor subfamily 1 group H member 4 (NR1H4), cytochrome P450 family 1 subfamily A member 2 (CYP1A2), cyclin dependent kinase inhibitor 2A (CDKN2A), and IL-8. The genes are annotated as being directly targeted by EGCG (26). According to the lung cancer tissues’ gene expression levels, obtained from The Cancer Genome Atlas database, it was demonstrated that the expression levels of these 20 genes were closely associated to the lncRNAs regulated by EGCG, among which DNMT1 is also a well-known lung cancer-associated gene. All the 20 EGCG direct target genes and 20 oncogenes mostly affected by EGCG were entered into the STRING database for network construction. The network of gene interactions as well as compound-gene interactions were illustrated in Fig. 5. A broad linkage between EGCG target genes and known lung cancer-associated genes suggested that EGCG was very likely to affect NSCLC by acting on these genes.
Figure 5.
Compound and gene interaction network diagram. Red represents EGCG, blue borders represent the target genes for direct action of EGCG, blue circles or squares represent known lung cancer genes, red solid lines indicate that EGCG may directly interact with the genes, and light gray dashed lines indicate gene-gene interactions. The black dotted lines indicate the shortest path from the target genes of EGCG to known oncogenes. EGCG, (−)-epigallocatechin-3-gallate.
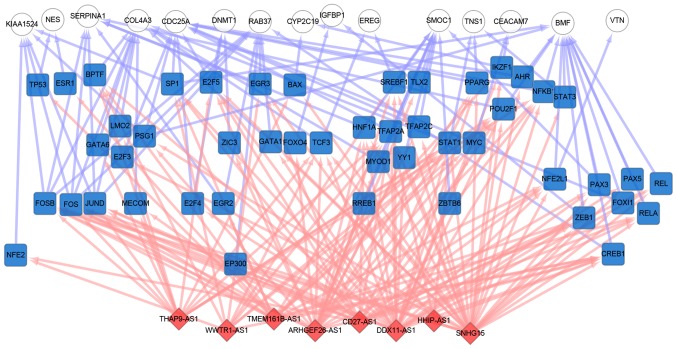
LncPro was used to predict the interaction of the top 10 lncRNAs and top 20 oncogenes affected by EGCG. A total of 8 of the 10 lncRNAs were demonstrated to bind the transcriptional factors (TF) of relevant oncogenes with relatively high binding scores. The network of lncRNA-TF-oncogenes is demonstrated in Fig. 6, where the lncRNAs can bind to nearly all oncogene-associated TFs, including B-cell lymphoma-2-modifying factor and collagen type IV α3 chain, which are regulated by additional lncRNA-bound transcriptional factors.
Figure 6.
lncRNA-TF-oncogene regulatory network diagram. Red represents lncRNAs, blue represents transcriptional factors, and white represents functional lung cancer genes. lncRNA, long non-coding RNA; TF, transcriptional factor.
Discussion
EGCG, the main active and water-soluble component of green tea polyphenols, accounting for 9–13% of the green weight of green tea, has been demonstrated to possess anticancer activity (30,31). The IC50 value (in A549 cells) conversion for green tea was about 0.4–2 g, while this is less than a daily intake (15–30 g) of people who have tea drinking habits. The golden ratio of brewing tea is 1:50, that is, 50 ml of boiling water poured into 1 g of tea. However, there are differences between in vitro and in vivo experiments, which need further verification.
As a powerful cancer chemopreventive agent, the anti-tumor effect of EGCG has been recognized in numerous studies conducted in various countries (6,32,33); however, it is difficult to determine the molecular mechanisms of the anticancer effects of EGCG, particularly in vivo. Therefore, in the present study, the expression levels of lncRNAs and mRNAs in EGCG-treated and control groups were analyzed in genomic studies combined with bioinformatic analysis. Based on the bioinformatic analysis, the general characteristics, functional comments, and pathways of the differentially expressed lncRNAs and mRNAs were identified. The present results provide a comprehensive understanding of the lncRNAs and mRNAs in lung cancer cells regulated by EGCG, and may help to reveal the molecular mechanisms of EGCG in lung cancer, but these need to be validated further.
Previous studies demonstrated that ~18% of encoded protein genes that produce lncRNAs are associated with cancer, while only ~9% of all encoded protein-coding genes are associated with cancer (34). Increasing evidence has revealed the importance of lncRNAs in lung cancer. For example, a previous study demonstrated that Oct4 regulates the expression of the lncRNAs nuclear paraspeckle assembly transcript 1 and metastasis associated lung adenocarcinoma transcript 1 promoting lung cancer progression (35). The lncRNA HOTAIR was demonstrated to regulate lung cancer cell growth and metastasis (36). The lncRNA XLOC008466 was reported as an oncogene in human non-small cell lung cancer by virtue of its targeting miR-874 (37). However, studies on lncRNAs regulated by EGCG in lung cancer are limited. The present study aimed to identify significantly down- or upregulated lncRNAs and mRNAs in lung cancer cells treated with EGCG. First, a lung cancer cell line demonstrating sensitivity to EGCG was selected by MTT analysis and was treated with 80 µΜ EGCG for 48 h, followed by microarray analysis.
GO analysis revealed that these differentially expressed mRNAs are associated with different biological processes, including cell cycle, mitotic cell cycle, DNA replication, organelle fission, nuclear division, mitotic nuclear division, and cell cycle phase transition. Thus, the present results indicate that EGCG may exert its anticancer function by regulating lung cancer cell proliferation, including suppression of the cell cycle and mitotic cell cycle. Furthermore, a previous mechanistic study demonstrated that EGCG inhibits lung cancer-associated processes, including anchorage-independent growth and the cell cycle, by directly targeting the epidermal growth factor receptor (EGFR) signaling pathway (38).
Additionally, a study on salivary adenoid cystic carcinoma revealed that the EGCG anticancer effect is mediated via the EGFR/extracellular regulated kinase (ERK) signaling transduction and mitochondrial apoptosis pathways (34). Differential proteomic analysis demonstrated that EGCG inhibited hepatoma-derived growth factor and activated apoptosis to increase the chemosensitivity of non-small cell lung cancer (39). Based on the present results, it may be hypothesized that the various lncRNAs, mRNAs, or proteins regulated by EGCG identified in these studies constitute or regulate important cellular components, including the chromosome centromeric region and chromosome region, but this needs to be investigated further. Several studies have confirmed that EGCG significantly inhibits the expression of telomerase and increases the chemosensitivity of lung cancer cells (21,40,41).
GO analysis indicated that the mRNAs induced by EGCG are involved in various molecular functions, including DNA-dependent ATPase activity, DNA helicase activity and iron ion binding. A previous study reported that EGCG regulates the cross-talk between JWA (also known as ADP Ribosylation Factor Like GTPase 6 Interacting Protein 5) and topoisomerase Iiα regulating lung cancer cell invasion (42). Antioxidant supplementation is known to increase the risk of lung cancer, and EGCG as an exemplary antioxidant reported to induce significant cell death and DNA damage in human lung cells through a reductive mechanism (43). KEGG pathway enrichment analysis indicated that EGCG inhibited lung cancer by affecting DNA replication, complement and coagulation cascades, cell cycle, and other signaling pathways, including the Wnt/β-catenin, unclear respirator factor2/Kelch-like ECH-associated protein 1, mitogen-activated kinase/ERK, EGFR, STAT3, and AKT signaling pathways (38,44–47). However, the precise co-regulatory functions and mechanisms of lncRNA-mRNA require further investigation.
Additionally, combined analysis of the microarray chip sequencing and bioinformatics results obtained from databases revealed a number of key factors in the EGCG process, including TP53 and DNMT1, which are well-known cancer-associated genes (48,49). These factors also demonstrate high variability in cancer patients; therefore, the levels of variation in these factors may affect the anticancer effects of EGCG in patients, but additional studies are necessary to determine the effects of these factors.
In conclusion, the present study provided the lncRNA and RNA expression profile in vitro. In the present study, 960 lncRNAs and 1,434 mRNAs were identified to be differentially expressed in lung cancer cells treated with EGCG compared with those without treatment. Furthermore, through cluster analysis and GO function and KEGG pathway analysis, it was demonstrated that EGCG is associated with possible regulation of the expression of several lncRNA and mRNAs, but this needs to be investigated further.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- EGCG
(−)-epigallocatechin-3-gallate
- lncRNAs
long non-coding RNAs
- nt
nucleotides
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- FDR
false discovery rate
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- TF
transcription factor
Funding
The present study was supported by the Science and Technology Project of Hunan Province, China (grant no. 2013FJ3036).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
D-LH and GW conceived and designed the study. D-LH and L-HZ performed the experiments. GW provided the financial aid and approved the final version. JY acquired data. Y-FH, L-HZ and DW analyzed the data. H-HZ contributed to the interpretation of the results. Y-FH, DW and H-HZ drafted and revised the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Nørskov MS, Dahl M, Tybjærghansen A. Genetic variation in GSTP1, lung function, risk of lung cancer and mortality. J Thorac Oncol. 2017;12:1664–1672. doi: 10.1016/j.jtho.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Yang JC, Mok T, Han B, Orlando M, Puri T, Park K. A review of regimens combining pemetrexed with an epidermal growth factor receptor tyrosine kinase inhibitor in the treatment of advanced nonsquamous non-small-cell lung cancer. Clin Lung Cancer. 2018;19:27–34. doi: 10.1016/j.cllc.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Cao J, Han J, Xiao H, Qiao J, Han M. Effect of tea polyphenol compounds on anticancer drugs in terms of anti-tumor activity, toxicology, and pharmacokinetics. Nutrients. 2016;8:E762. doi: 10.3390/nu8120762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oya Y, Mondal A, Rawangkan A, Umsumarng S, Iida K, Watanabe T, Kanno M, Suzuki K, Li Z, Kagechika H, et al. Down-regulation of histone deacetylase 4, −5 and −6 as a mechanism of synergistic enhancement of apoptosis in human lung cancer cells treated with the combination of a synthetic retinoid, Am80 and green tea catechin. J Nutr Biochem. 2017;42:7–16. doi: 10.1016/j.jnutbio.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Hong OY, Noh EM, Jang HY, Lee YR, Lee BK, Jung SH, Kim JS, Youn HJ. Epigallocatechin gallate inhibits the growth of MDA-MB-231 breast cancer cells via inactivation of the β-catenin signaling pathway. Oncol Lett. 2017;14:441–446. doi: 10.3892/ol.2017.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M, Li JJ, Gu QH, An J, Cao LM, Yang HP, Hu CP. EGCG induces lung cancer A549 cell apoptosis by regulating Ku70 acetylation. Oncol Rep. 2016;35:2339–2347. doi: 10.3892/or.2016.4587. [DOI] [PubMed] [Google Scholar]
- 7.Jiang L, Tao C, He A, He X. Overexpression of miR-126 sensitizes osteosarcoma cells to apoptosis induced by epigallocatechin-3-gallate. World J Surg Oncol. 2014;12:383. doi: 10.1186/1477-7819-12-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng PB, Hu CP, Xiong Z, Yang HP, Li YY. Treatment with EGCG in NSCLC leads to decreasing interstitial fluid pressure and hypoxia to improve chemotherapy efficacy through rebalance of Ang-1 and Ang-2. Chin J Nat Med. 2013;11:245–253. doi: 10.3724/SP.J.1009.2013.00245. [DOI] [PubMed] [Google Scholar]
- 9.Yin J, Huang F, Yi Y, Yin L, Peng D. EGCG attenuates atherosclerosis through the Jagged-1/Notch pathway. Int J Mol Med. 2016;37:398–406. doi: 10.3892/ijmm.2015.2422. [DOI] [PubMed] [Google Scholar]
- 10.Ma S, Deng X, Yang Y, Zhang Q, Zhou T, Liu Z. The lncRNA LINC00675 regulates cell proliferation, migration, and invasion by affecting Wnt/beta-catenin signaling in cervical cancer. Biomed Pharmacother. 2018;108:1686–1693. doi: 10.1016/j.biopha.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Xi Z, Si J, Nan J. LncRNA MALAT1 potentiates autophagyassociated cisplatin resistance by regulating the microRNA30b/autophagyrelated gene 5 axis in gastric cancer. Int J Oncol. 2019;54:239–248. doi: 10.3892/ijo.2018.4609. [DOI] [PubMed] [Google Scholar]
- 12.Liang H, Zhang C, Guan H, Liu J, Cui Y. LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p. J Cell Physiol. 2018 Oct 26; doi: 10.1002/jcp.27484. Doi: 10.1002/jcp.27484. [DOI] [PubMed] [Google Scholar]
- 13.Kong X, Duan Y, Sang Y, Li Y, Zhang H, Liang Y, Liu Y, Zhang N, Yang Q. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2018 Oct 26; doi: 10.1002/jcp.27587. Doi: 10.1002/jcp.27587. [DOI] [PubMed] [Google Scholar]
- 14.Delás MJ, Hannon GJ. lncRNAs in development and disease: From functions to mechanisms. Open Biol. 2017;7:170121. doi: 10.1098/rsob.170121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng L, Yuan X, Jiang B, Tang Z, Li GC. LncRNAs: Key players and novel insights into cervical cancer. Tumour Biol. 2016;37:2779–2788. doi: 10.1007/s13277-015-4663-9. [DOI] [PubMed] [Google Scholar]
- 16.Yang ZY, Yang F, Zhang YL, Liu B, Wang M, Hong X, Yu Y, Zhou YH, Zeng H. LncRNA-ANCR down-regulation suppresses invasion and migration of colorectal cancer cells by regulating EZH2 expression. Cancer Biomark. 2017;18:95–104. doi: 10.3233/CBM-161715. [DOI] [PubMed] [Google Scholar]
- 17.Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, Shan B, Zheng D, Shi J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2016;31:893–903. doi: 10.1096/fj.201600994R. [DOI] [PubMed] [Google Scholar]
- 18.Zhu H, Zhao H, Zhang L, Xu J, Zhu C, Zhao H, Lv G. Dandelion root extract suppressed gastric cancer cells proliferation and migration through targeting lncRNA-CCAT1. Biomed Pharmacother. 2017;93:1010–1017. doi: 10.1016/j.biopha.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Sun H, Wang G, Peng Y, Zeng Y, Zhu QN, Li TL, Cai JQ, Zhou HH, Zhu YS. H19 lncRNA mediates 17β-estradiol-induced cell proliferation in MCF-7 breast cancer cells. Oncol Rep. 2015;33:3045–3052. doi: 10.3892/or.2015.3899. [DOI] [PubMed] [Google Scholar]
- 20.Li N, Yu J, Luo A, Tang Y, Liu W, Wang S, Liu Y, Song Y, Fang H, Chen B, et al. LncRNA and mRNA signatures associated with neoadjuvant chemoradiotherapy downstaging effects in rectal cancer. J Cell Biochem. 2018 Oct 15; doi: 10.1002/jcb.27796. Doi: 10.1002/jcb.27796. [DOI] [PubMed] [Google Scholar]
- 21.Pan J, Wu X, Wang X, Huang W, Feng Q. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget. 2016;7:43337–43351. doi: 10.18632/oncotarget.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Fang L, Jiang J, Kuang Y, Wang B, Shang X, Han P, Li Y, Liu M, Zhang Z, Li P. The cisplatin-induced lncRNA PANDAR dictates the chemoresistance of ovarian cancer via regulating SFRS2-mediated p53 phosphorylation. Cell Death Dis. 2018;9:1103. doi: 10.1038/s41419-018-1148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips J, Eberwine JH. Antisense RNA amplification: A linear amplification method for analyzing the mRNA population from single living cells. Methods. 1996;10:283–288. doi: 10.1006/meth.1996.0104. [DOI] [PubMed] [Google Scholar]
- 24.Gene Ontology Consortium: Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piñero J, Queralt-Rosinach N, Bravo À, Deu-Pons J, Bauer-Mehren A, Baron M, Sanz F, Furlong LI. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015. 2015:bav028. doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn M, Szklarczyk D, Franceschini A, Campillos M, von Mering C, Jensen LJ, Beyer A, Bork P. STITCH 2: An interaction network database for small molecules and proteins. Nucleic Acids Res. 2010;38:D552–D556. doi: 10.1093/nar/gkp937. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Q, Ren S, Lu M, Zhang Y, Zhu D, Zhang X, Li T. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genomics. 2013;14:651. doi: 10.1186/1471-2164-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Wei R, Mao L, Xu P, Zheng X, Hackman RM, Mackenzie GG, Wang Y. Suppressing glucose metabolism with epigallocatechin-3-gallate (EGCG) reduces breast cancer cell growth in preclinical models. Food Funct. 2018;9:5682–5696. doi: 10.1039/C8FO01397G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker KM, Bauer AC. Green tea catechin, EGCG, suppresses PCB 102-induced proliferation in estrogen-sensitive breast cancer cells. Int J Breast Cancer 2015. 2015:163591. doi: 10.1155/2015/163591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang YQ, Lu JL, Liang YR, Li QS. Suppressive effects of EGCG on cervical cancer. Molecules. 2018;23:E2334. doi: 10.3390/molecules23092334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo KW, Wei Chen, Lung WY, Wei XY, Cheng BH, Cai ZM, Huang WR. EGCG inhibited bladder cancer SW780 cell proliferation and migration both in vitro and in vivo via down-regulation of NF-κB and MMP-9. J Nutr Biochem. 2017;41:56–64. doi: 10.1016/j.jnutbio.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Weng LX, Wang GH, Yao H, Yu MF, Lin J. Epigallocatechin gallate inhibits the growth of salivary adenoid cystic carcinoma cells via the EGFR/Erk signal transduction pathway and the mitochondria apoptosis pathway. Neoplasma. 2017;64:563–570. doi: 10.4149/neo_2017_410. [DOI] [PubMed] [Google Scholar]
- 35.Jen J, Tang YA, Lu YH, Lin CC, Lai WW, Wang YC. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer. 2017;16:104. doi: 10.1186/s12943-017-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Chen WJ, Gan TQ, Zhang XL, Xie ZC, Ye ZH, Deng Y, Wang ZF, Cai KT, Li SK, et al. Clinical significance and effect of lncRNA HOXA11-AS in NSCLC: A study based on bioinformatics, in vitro and in vivo verification. Sci Rep. 2017;7:5567. doi: 10.1038/s41598-017-05856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang R, Li P, Zhang G, Lu C, Wang H, Zhao G. Long non-coding RNA XLOC_008466 functions as an oncogene in human non-small cell lung cancer by targeting miR-874. Cell Physiol Biochem. 2017;42:126–136. doi: 10.1159/000477121. [DOI] [PubMed] [Google Scholar]
- 38.Ma YC, Li C, Gao F, Xu Y, Jiang ZB, Liu JX, Jin LY. Epigallocatechin gallate inhibits the growth of human lung cancer by directly targeting the EGFR signaling pathway. Oncol Rep. 2014;31:1343–1349. doi: 10.3892/or.2013.2933. [DOI] [PubMed] [Google Scholar]
- 39.Florespérez A, Marchat LA, Sánchez LL, Romero-Zamora D, Arechaga-Ocampo E, Ramírez-Torres N, Chávez JD, Carlos-Reyes Á, Astudillo-de la Vega H, Ruiz-García E, et al. Differential proteomic analysis reveals that EGCG inhibits HDGF and activates apoptosis to increase the sensitivity of non-small cells lung cancer to chemotherapy. Proteomics Clin Appl. 2016;10:172–182. doi: 10.1002/prca.201500008. [DOI] [PubMed] [Google Scholar]
- 40.Sadava D, Whitlock E, Kane SE. The green tea polyphenol, epigallocatechin-3-gallate inhibits telomerase and induces apoptosis in drug-resistant lung cancer cells. Biochem Biophys Res Commun. 2007;360:233–237. doi: 10.1016/j.bbrc.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 41.Shervington A, Pawar V, Menon S, Thakkar D, Patel R. The sensitization of glioma cells to cisplatin and tamoxifen by the use of catechin. Mol Biol Rep. 2009;36:1181–1186. doi: 10.1007/s11033-008-9295-3. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Shen X, Wang X, Li A, Wang P, Jiang P, Zhou J, Feng Q. EGCG regulates the cross-talk between JWA and topoisomerase IIα in non-small-cell lung cancer (NSCLC) cells. Sci Rep. 2015;5:11009. doi: 10.1038/srep11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu LY, Ou N, Lu QB. Antioxidant induces DNA damage, cell death and mutagenicity in human lung and skin normal cells. Sci Rep. 2013;3:3169. doi: 10.1038/srep03169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou Y, Zhu Q, Li Z, Peng Y, Yu X, Yuan B, Liu Y, Liu Y, Yin L, Peng Y, et al. The FOXM1-ABCC5 axis contributes to paclitaxel resistance in nasopharyngeal carcinoma cells. Cell Death Dis. 2017;8:e2659. doi: 10.1038/cddis.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shanmugam T, Selvaraj M, Poomalai S. Epigallocatechin gallate potentially abrogates fluoride induced lung oxidative stress, inflammation via Nrf2/Keap1 signaling pathway in rats: An in-vivo and in-silico study. Int Immunopharmacol. 2016;39:128–139. doi: 10.1016/j.intimp.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 46.Yun M, Lee D, Park MN, Kim EO, Sohn EJ, Kwon BM, Kim SH. Cinnamaldehyde derivative (CB-PIC) sensitizes chemo-resistant cancer cells to drug-induced apoptosis via suppression of MDR1 and its upstream STAT3 and AKT signalling. Cell Physiol Biochem. 2015;35:1821–1830. doi: 10.1159/000373993. [DOI] [PubMed] [Google Scholar]
- 47.Tang G, Zhang Z, Qian H, Chen J, Wang Y, Chen X, Chen B, Chen Y. (−)-Epigallocatechin-3-gallate inhibits osteosarcoma cell invasiveness by inhibiting the MEK/ERK signaling pathway in human osteosarcoma cells. J Environ Pathol Toxicol Oncol. 2015;34:85–93. doi: 10.1615/JEnvironPatholToxicolOncol.2015011925. [DOI] [PubMed] [Google Scholar]
- 48.Ciccarese C, Massari F, Blanca A, Tortora G, Montironi R, Cheng L, Scarpelli M, Raspollini MR, Vau N, Fonseca J, Lopez-Beltran A. Tp53 and its potential therapeutic role as a target in bladder cancer. Expert Opin Ther Targets. 2017;21:401–414. doi: 10.1080/14728222.2017.1297798. [DOI] [PubMed] [Google Scholar]
- 49.Nuñez NN, Manlove AH, David SS. DNMT1 and cancer: An electrifying link. Chem Biol. 2015;22:810–811. doi: 10.1016/j.chembiol.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 50.Liu L, Liu J, Dai S, Wang X, Wu S, Wang J, Huang L, Xiao X, He D. Reduced transthyretin expression in sera of lung cancer. Cancer Sci. 2007;98:1617–1624. doi: 10.1111/j.1349-7006.2007.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park JY, Kim D, Yang M, Park HY, Lee SH, Rincon M, Kreahling J, Plass C, Smiraglia DJ, Tockman MS, Kim SJ. Gene silencing of SLC5A8 identified by genome-wide methylation profiling in lung cancer. Lung Cancer. 2013;79:198–204. doi: 10.1016/j.lungcan.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai CH, Cheng HC, Wang YS, Lin P, Jen J, Kuo IY, Chang YH, Liao PC, Chen RH, Yuan WC, et al. Small GTPase Rab37 targets tissue inhibitor of metalloproteinase 1 for exocytosis and thus suppresses tumour metastasis. Nat Commun. 2014;5:4804. doi: 10.1038/ncomms5804. [DOI] [PubMed] [Google Scholar]
- 53.Wu CY, Tseng RC, Hsu HS, Wang YC, Hsu MT. Frequent down-regulation of hRAB37 in metastatic tumor by genetic and epigenetic mechanisms in lung cancer. Lung Cancer. 2009;63:360–367. doi: 10.1016/j.lungcan.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 54.Dötsch MM, Kloten V, Schlensog M, Heide T, Braunschweig T, Veeck J, Petersen I, Knüchel R, Dahl E. Low expression of ITIH5 in adenocarcinoma of the lung is associated with unfavorable patients' outcome. Epigenetics. 2015;10:903–912. doi: 10.1080/15592294.2015.1078049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Cui T, Knösel T, Yang L, Zöller K, Petersen I. IGFBP7 is a p53 target gene inactivated in human lung cancer by DNA hypermethylation. Lung Cancer. 2011;73:38–44. doi: 10.1016/j.lungcan.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 56.Zhang L, Xiao H, Zhou H, Santiago S, Lee JM, Garon EB, Yang J, Brinkmann O, Yan X, Akin D, et al. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol Life Sci. 2012;69:3341–3350. doi: 10.1007/s00018-012-1027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mclemore TL, Adelberg S, Liu MC, McMahon NA, Yu SJ, Hubbard WC, Czerwinski M, Wood TG, Storeng R, Lubet RA, et al. Expression of CYP1A1 gene in patients with lung cancer: Evidence for cigarette smoke-induced gene expression in normal lung tissue and for altered gene regulation in primary pulmonary carcinomas. J Natl Cancer Inst. 1990;82:1333–1339. doi: 10.1093/jnci/82.16.1333. [DOI] [PubMed] [Google Scholar]
- 58.Taioli E, Ford J, Trachman J, Li Y, Demopoulos R, Garte S. Lung cancer risk and CYP1A1 genotype in African Americans. Carcinogenesis. 1998;19:813–817. doi: 10.1093/carcin/19.5.813. [DOI] [PubMed] [Google Scholar]
- 59.Choi SH, Yang H, Lee SH, Ki JH, Nam DH, Yoo HY. TopBP1 and Claspin contribute to the radioresistance of lung cancer brain metastases. Mol Cancer. 2014;13:211. doi: 10.1186/1476-4598-13-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taniguchi H, Takeuchi S, Fukuda K, Nakagawa T, Arai S, Nanjo S, Yamada T, Yamaguchi H, Mukae H, Yano S. AREG triggered EGFR activation confers in vivo crizotinib-resistance of EML4-ALK lung cancer and circumvention by EGFR inhibitors. Cancer Sci. 2016;108:53–60. doi: 10.1111/cas.13111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taron M, Rosell R, Felip E, Mendez P, Souglakos J, Ronco MS, Queralt C, Majo J, Sanchez JM, Sanchez JJ, Maestre J. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet. 2004;13:2443–2449. doi: 10.1093/hmg/ddh260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.