Abstract
Granulosa cells (GCs) have many functions in the endocrine system. Most notably, they produce progesterone following ovulation. However, it has recently been proven that GCs can change their properties when subjected to long-term culture. In the present study, GCs were collected from hyper-stimulated ovarian follicles during in vitro fertilization procedures. They were grown in vitro, in a long-term manner. RNA was collected following 1, 7, 15 and 30 days of culture. Expression microarrays were used for analysis, which allowed to identify groups of genes characteristic for particular cellular processes. In addition, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to validate the obtained results. Two ontological groups characteristic for processes associated with the development and morphogenesis of the heart were identified during the analyses: ‘Heart development’ and ‘heart morphogenesis’. The results of the microarrays revealed that the highest change in expression was demonstrated by the lysyl Oxidase, oxytocin receptor, nexilin F-actin binding protein, and cysteine-rich protein 3 genes. The lowest change was exhibited by odd-skipped related transcription factor 1, plakophilin 2, transcription growth factor-β receptor 1, and kinesin family member 3A. The direction of changes was confirmed by RT-qPCR results. In the present study, it was suggested that GCs may have the potential to differentiate towards other cell types under long-term in vitro culture conditions. Thus, genes belonging to the presented ontological groups can be considered as novel markers of proliferation and differentiation of GCs towards the heart muscle cells.
Keywords: human granulosa cells, in vitro culture, proliferation, differentiation, heart development
Introduction
The folliculogenesis is a compound process that leads to the formation of the fully grown ovarian follicle (1,2). The developed follicle is composed of several cell layers, including theca cells (TCs) and granulosa cells (GCs). Our recent study has shown that porcine GCs undergo permanent morphological modification when kept in the primary in vitro culture system. After long-term culture, GCs underwent transformation into spindle-like structures, which was accompanied by changes in their gene expression profile. Moreover, the GCs display structural and functional role in the active pre-ovulatory and ovulatory follicle, forming follicular architecture and expressing a number of hormone receptors. Some of that properties seem to prevail ex vivo, as we have observed a time-dependent expression of LHR, FSHR, CYP19, and progesterone receptor genes, in porcine GCs during long-term in vitro culture.
Using a porcine model, we have recently shown that GCs, isolated from pre-ovulatory ovarian follicles, can be successfully cultured in vitro for short and/or long time periods. Moreover, we found that these GCs are characterized by increased in vitro proliferation capability. Although the increase in cells' proliferation is accompanied by cellular ageing and subsequently decreased by a number of passages, the GCs have also displayed significant differentiational capacity. It was also presented that these cells may differentiate even into cell types unassociated with the reproductive process. Kossowska-Tomaszczuk recently determined that after long-term cultivation and under influence of several substances, including LIF and dexamethasone, GCs may differentiate into chondroblasts, osteoblasts and other adult precursor cells (3,4). These experiments have also shown how the huge differentiational plasticity of human GCs during primary in vitro culture can be used in clinical situations. This may open a new gate in the potential application of human granulosa cells, which are nowadays mostly treated as remnant IVF material, mostly in autologous and donor-host grafts, as they exhibit increased stem-like specificity.
The experiments by Kossowska-Tomaszczuk open the possibility of finding new stemness markers in human ovarian granulosa cells. Microarray assays allow for robust and complex transcriptomic analysis, leading to the discovery of new molecular markers of cells function. In this work, we have used the microarrays to describe the transcriptomic profile of human GCs during their long-term primary in vitro culture. We have detected the expression of genes belonging to more than five hundred gene ontology groups, associated with the ‘biology of human GCs’ during their in vitro proliferation and differentiation. In this study, we have investigated the ‘heart development and morphogenesis’ ontological group, as the potential source of new markers of GCs plasticity.
Materials and methods
A large part of the materials and methods section is based on other, published work of the author, presenting results from the same cycle of studies describing human granulosa cells (5).
Patients and collection of granulosa cells
The GCs were derived from patients undergoing in vitro fertilization (IVF) procedures, who had given their informed, written consent to be included in this protocol. The study group consisted of 8 patients, aged 18–40 years, with diagnosed infertility, referred to the Division of Infertility and Reproductive Endocrinology, Poznan University of Medical Sciences, Poznan, Poland. Patients underwent IVF procedure, based on controlled ovarian hyperstimulation protocol, adjusted to the patient's initial infertility workup and ovarian response. Stimulation was performed with human recombinant FSH (Gonal-F, Merck Serono) and highly purified hMG-HP (Menopur, Ferring). The injections with cetrorelix acetate (Cetrotide, Merck Serono) were administered in an adequate dose, to suppress pituitary function. Ovulation triggering was based on the subcutaneous injection of 6,500 U of hCG (Ovitrelle, Merck-Serono). GC containing follicular fluid was collected during transvaginal, ultrasound-guided oocyte pick-up, performed 36 h after human chorionic gonadotropin administration. The content of follicles that were over 16 mm in diameter was immediately passed to an embryologist, who isolated the oocyte, and pooled the follicular fluid from each ovary. The fresh pooled samples were centrifuged for 10 min at 200 g, to separate and collect GCs. Patients with polycystic ovary syndrome (PCOS), endometriosis, and diminished ovarian reserve (serum antimüllerian hormone (AMH) less than 0.7 ng/ml, and/or day 2–3 FSH serum level higher than 15 mU/ml, and/or antral follicle count less than 9) were excluded from the study. This research has been approved by Poznan University of Medical Sciences Bioethical Committee with resolution 558/17.
Primary cell culture
The collected cells were washed twice by centrifugation at 200 × g for 10 min at RT in the culture medium. Medium consisted of Dulbecco's modified Eagle's medium (DMEM, Sigma; Merck KGaA, Darmstadt, Germany), 2% foetal bovine serum FBS (FBS; Sigma; Merck KGaA), 4 mM L-glutamine (stock 200 mM, Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 10 mg/ml gentamicin (Invitrogen; Thermo Fisher Scientific, Inc.), 10,000 U/ml penicillin, and 10,000 µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were cultivated at 37°C under aerobic conditions (5% CO2). Once adherent cells were more than 90% confluent, they were detached with 0.05% trypsin-EDTA (Invitrogen; Thermo Fisher Scientific, Inc.) for 1–2 min and counted using a ‘Neubauer improved’ counting chamber (ISO LAB Laborgerate GmbH, DIN EN ISO Certified 9001). GCs were cultivated for 30 days. The medium was changed twice a week. Finally, total RNA was isolated from GCs after 1, 7, 15 and 30 days.
Total RNA isolation
Total RNA was isolated at 4 time periods, after 1, 7, 15 and 30 days of cultivation. The improved Chomczyński-Sacchi method was used for the RNA isolation (6). The GCs were suspended in 1 ml of a monophase solution of guanidine thiocyanate and phenol (TRI Reagent®, Sigma; Merck KGaA). The chloroform was then added, with the samples centrifuged to obtain three separate phases. RNA was located in the upper, aqueous phase. RNA was then stripped with 2-propanol (Sigma; Merck KGaA, catalogue number I9516), added in an amount adequate for 1 ml of TRI-reagent, and washed with 75% ethanol. Samples prepared in that way were used for further analysis.
Microarray expression analysis and statistics
Total RNA (100 ng) from each pooled sample was subjected to two rounds of sense cDNA amplification (Ambion® WT Expression kit). The obtained cDNA was used for biotin labelling and fragmentation, with the use of GeneChip® WT Terminal Labeling and Hybridization (Affymetrix). Biotin-labelled fragments of cDNA (5.5 µg) were hybridized to the Affymetrix® Human Genome U219 Array (48°C/20 h). Microarrays were then washed and stained according to the technical protocol using the Affymetrix GeneAtlas Fluidics Station. The array strips were scanned employing Imaging Station of the GeneAtlas System. Preliminary analysis of the scanned chips was performed using Affymetrix GeneAtlas™ Operating Software. The quality of gene expression data was confirmed according to the quality control criteria provided by the software. The obtained CEL files were imported into downstream data analysis software.
All of the presented analyses and graphs were performed using Bioconductor and R programming languages. Each CEL file was merged with a description file. The Robust Multiarray Averaging (RMA) algorithm was used to correct background, normalize, and summarize the results. To determine the statistical significance of the analyzed genes, moderated t-statistics from the empirical Bayes method were performed. The obtained P-value was corrected for multiple comparisons using Benjamini and Hochberg's false discovery rate. The selection of significantly altered genes was based on a P-value beneath 0.05 and expression higher than two-fold.
Differentially expressed genes were subjected to selection by examination of genes involved in heart development and morphogenesis. The differentially expressed gene list (separated for up- and down-regulated genes) was uploaded to the DAVID software (Database for Annotation, Visualization and Integrated Discovery) (7).
Subsequently, the relationship between the genes belonging to chosen GO terms was analysed with the use of GOplot package (8). The GoPlot package had calculated the Z-score: The number of upregulated genes minus the number of downregulated genes divided by the square root of the count. This information allowed to estimate the direction of changes in each gene-ontology term.
Moreover, interactions between differentially expressed genes/proteins belonging to the chosen GO terms were investigated by STRING10 software (Search Tool for the Retrieval of Interacting Genes) (9). The list of gene names was used as a query for the interaction prediction. The search criteria were based on co-occurrences of genes/proteins in scientific texts (text mining), co-expression, and experimentally observed interactions. The results of these analyses generated a gene/protein interaction network where the intensity of the edges reflected the strength of the interaction score.
Finally, the functional interactions between the genes belonging to the chosen GO BP terms were investigated using REACTOME FIViz application from the Cytoscape 3.6.0 software. This app accesses the pathways stored in the Reactome database, allowing for pathway enrichment analysis of a set of genes, visualization of hit pathways using manually laid-out pathway diagrams directly in Cytoscape, and investigation of functional relationships among genes in hit pathways. The app can also access the Reactome Functional Interaction (FI) network, a highly reliable, manually curated pathway-based protein functional interaction network covering over 60% of human proteins.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
The RT-qPCR method was performed to confirm the results obtained in the analysis of expression microarrays. Three genes were selected from each heatmap: The ones showing highest, lowest, and intermediate-level of expression. Changes in the level of expression of those genes were then examined. Three biological samples of each gene were used for the analysis. Each test was performed in 3 replicates. Reverse transcription was based on the protocols and reagents of SABiosciences (RT2 First Stand kit-330401), using a Veritimer 96-well Thermal Cycler. One microgram of each gene's RNA transcript was used for reverse transcription. Real-time PCR was performed using the 7900HT Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.), RT2 SYBR® Green ROX™ qPCR Master Mix (Qiagen Sciences, Gaithersburg, MD, USA) and sequence-specific primers (Table I). Glyceraldehyde-3-phosphate dehydrogenase (GADPH), β-actin (ACTB), hypoxanthine phosphoribosyltransferase 1 (HRPT1) were used as reference genes. Gene expression was analyzed using the relative quantification (RQ) method. The q-PCR starters were designed using Primer3Plus software (primer3plus.com/cgi-bin/dev/primer3plus.cgi).
Table I.
Oligonucleotide sequences of primers used for reverse transcription-quantitative polymerase chain reaction analysis.
| Gene | Primer sequences (5′-3′) |
|---|---|
| PCNA | F: GGCGTGAACCTCACCAGTAT |
| R: TCTCGGCATATACGTGCAAA | |
| LOX | F: CAGAGGAGAGTGGCTGAAGG |
| R: CCAGGTAGCTGGGGTTTACA | |
| OXTR | F: TTCTTCGTGCAGATGTGGAG |
| R: GGACGAGTTGCTCTTTTTGC | |
| BMP4 | F: AAGCGTAGCCCTAAGCATCA |
| R: TGGTTGAGTTGAGGTGGTCA | |
| TPM1 | F: CTCTCAAAGATGCCCAGGAG |
| R: TCTCATCTGCTGCCTTCTCA | |
| TNNC1 | F: ACCTCTTCCGCATGTTTGAC |
| R: CCACACCCTTCATGAACTCC | |
| TTN | F: GGCATCCCCAAACCTAAAAT |
| R: TTTTGCCACTGCTGATTCTG | |
| TGFBR1 | F: TGTTGGTACCCAAGGAAAGC |
| R: CACTCTGTGGTTTGGAGCAA | |
| SMARCD3 | F: CTCTGAAGAGGCCCATGAAG |
| R: GAACTTCCGCTTCTGTTTGC | |
| NDST1 | F: TCACCTTCAACCTGGGCTAC |
| R: ACGGACTGGTTGTGGAAAAG | |
| GAPDH | F: TCAGCCGCATCTTCTTTTGC |
| R: ACGACCAAATCCGTTGACTC | |
| ACTB | F: AAAGACCTGTACGCCAACAC |
| R: CTCAGGAGGAGCAATGATCTTG | |
| HPRT | F: TGGCGTCGTGATTAGTGATG |
| R: ACATCTCGAGCAAGACGTTC |
F, forward; R, reverse; PCNA, proliferating cell nuclear antigen; LOX, Lysyl Oxidase; OXTR, oxytocin receptor; BMP4, bone morphogenetic protein 4; TPM1, tropomyosin 1; TNNC1, troponin 1; TTN, titin; TGFBR1, transcription growth factor-β receptor 1; SMARCD3, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3; NDST1, N-deacetylase and N-sulfotransferase 1; ACTB, β-actin; HPRT, hypoxanthine phosphoribosyltransferase 1.
Statistical analysis
The analysis of the presented results was conducted using Bioconductor (www.bioconductor.org) and R programming languages (R version 3.5.1; www.r-project.org). To determine the statistical significance of the analysed genes, moderated t-statistics from the empirical Bayes method were performed. The P-value was corrected for multiple comparisons using Benjamini and Hochberg's false discovery rate. P<0.05 was considered to indicate a statistically significant difference. The statistical significance of enriched GO terms and KEGG pathways was performed by DAVID database software (v.6.8; david.ncifcrf.gov). Statistical significance was calculated using Benjamini method. Each GO term and KEGG pathway were considered significantly enriched if they contained at least 5 differently expressed genes and showed P<0.05. Statistical analysis of RT-qPCR results was conducted using Real Statistics Resource Pack add-on for MS Excel 2016 (Microsoft Corporation, Redmond, WA, USA).
Results
Whole transcriptome profiling by Affymetrix microarray allowed for the analysis of expression of gene transcripts in human ovarian granulosa cells during long-term in vitro culture. Using Affymetrix® Human HgU 219 Array we have examined the expression of 22480 transcripts. Genes with a fold change higher then abs (2) and with a corrected P-value lower than 0.05 were considered differentially expressed. This set of genes consisted of 2278 different transcripts.
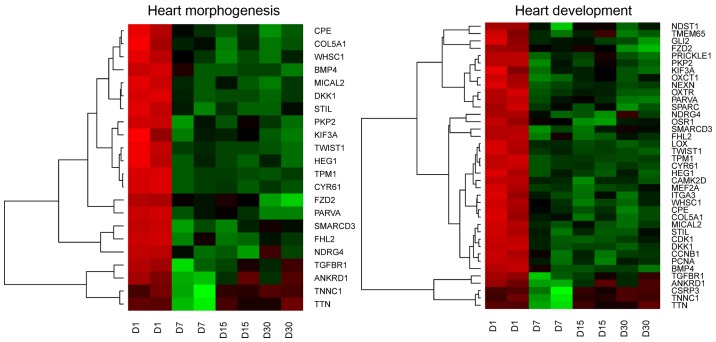
DAVID (Database for Annotation, Visualization and Integrated Discovery) software was used for extraction of gene ontology biological process terms (GO BP). Up- and down-regulated gene sets were subjected to DAVID search separately, and only gene sets where adj. P-value was lower than 0.05 were selected. The DAVID software analysis showed that differentially expressed genes belong to 582 Gene ontology groups and 45 KEGG pathways. In this study, we focused on ‘heart morphogenesis’ and ‘heart development’ GO BP terms. These sets of genes were subjected to hierarchical clusterization procedure and presented as heatmaps (Fig. 1). The gene symbols, fold changes in expression, Entrez gene IDs and corrected P-values of those genes were shown in Table II.
Figure 1.
Heat map representation of differentially expressed genes belonging to the ‘heart morphogenesis’ and ‘heart development’ Gene Ontology Biological Process terms. Arbitrary signal intensity acquired from microarray analysis is represented by colours (green indicates higher and red indicates lower expression). Log2 signal intensity values for any single gene were resized to Row Z-Score scale (from −2, the lowest expression to +2, the highest expression for a single gene).
Table II.
Gene symbols, fold changes in expression, Entrez gene IDs and corrected P-values of studied genes.
| Nazwa | foldD7_D1 | foldD15_D1 | foldD30_D1 | adj.P.Val.D7_D1 | adj.P.Val.D15_D1 | adj.P.Val.D30_D1 | Entrez.Gene ID |
|---|---|---|---|---|---|---|---|
| LOX | 53,155,736 | 80,241,544 | 102,191,488 | 0.0008508 | 0.0005524 | 0.0004038 | 4,015 |
| OXTR | 38,386,169 | 2,260,881 | 54,854,523 | 0.0006796 | 0.001012 | 0.0003993 | 5,021 |
| NEXN | 35,206,319 | 19,134,152 | 29,664,545 | 0.0012665 | 0.0019093 | 0.0010376 | 91,624 |
| CSRP3 | 27,412,789 | 1,585,293 | 1,057,341 | 0.0191031 | 0.6877273 | 0.9672829 | 8,048 |
| TPM1 | 17,883,412 | 16,942,974 | 18,049,628 | 0.0004891 | 0.0004529 | 0.0003078 | 7,168 |
| ITGA3 | 16,927,809 | 18,842,178 | 22,470.094 | 0.0277094 | 0.0222552 | 0.016508 | 3,675 |
| CDK1 | 13,880,751 | 18,568,254 | 12,196,334 | 0.002233 | 0.0014905 | 0.0018488 | 983 |
| TWIST1 | 13,410,656 | 16,061,334 | 20,741,443 | 0.0013489 | 0.0010638 | 0.000668 | 7,291 |
| ANKRD1 | 11,887,373 | 2,508,097 | 2,617,912 | 0.0247916 | 0.2787596 | 0.2411828 | 27,063 |
| MICAL2 | 1,054,092 | 9,421,967 | 12,712,885 | 0.0068803 | 0.0072788 | 0.0042113 | 9,645 |
| BMP4 | 10,489,539 | 17,221,545 | 21,243,468 | 0.0204669 | 0.0100715 | 0.0070848 | 652 |
| DKK1 | 8,954,961 | 9,635,247 | 10.055,325 | 0.0024355 | 0.0020428 | 0.0015034 | 22,943 |
| COL5A1 | 8,845,619 | 15,695,652 | 18,51,511 | 0.016744 | 0.0071873 | 0.005229 | 1,289 |
| PRICKLE1 | 7,301,909 | 4,726,273 | 9,353,689 | 0.0106664 | 0.0202006 | 0.0056925 | 144,165 |
| GLI2 | 6,93,552 | 7,103,782 | 14,112,758 | 0.0191991 | 0.0163042 | 0.0056787 | 2,736 |
| CCNB1 | 6,641,253 | 8,124,624 | 7,003,387 | 0.0318394 | 0.0211764 | 0.0237061 | 891 |
| STIL | 56,288 | 540,042 | 4,900,424 | 0.0141252 | 0.0134497 | 0.0143661 | 6,491 |
| TTN | 4,908,741 | 1,271,044 | 1,144,088 | 0.0047259 | 0.4647411 | 0.6871576 | 7,273 |
| NDRG4 | 4,817,497 | 7,098,036 | 3,324,764 | 0.0375828 | 0.0171188 | 0.0672424 | 65,009 |
| CPE | 4,531,794 | 6,031,618 | 8,102,031 | 0.0083776 | 0.0043779 | 0.0024281 | 1,363 |
| CYR61 | 4,318,043 | 424,915 | 4,154,475 | 0.0011941 | 0.0011841 | 0.0009317 | 3,491 |
| HEG1 | 4,138,551 | 4,604,733 | 4,311,688 | 0.0083402 | 0.0059934 | 0.0059049 | 57,493 |
| FZD2 | 3,767,679 | 3,278,544 | 13,843,888 | 0.0057158 | 0.0072683 | 0.0006712 | 2,535 |
| TNNC1 | 3,349,751 | 1,217,197 | 1,066,087 | 0.0053652 | 0,4460103 | 0.8215354 | 7,134 |
| FHL2 | 3,073,142 | 3,898,276 | 2,849,228 | 0.0282662 | 0.0139549 | 0.0283799 | 2,274 |
| MEF2A | 3,019,241 | 3,647,594 | 2,798,678 | 0.0041095 | 0.002502 | 0.0039542 | 4,205 |
| WHSC1 | 2,810,461 | 3,935,288 | 3,454,839 | 0.017237 | 0.0063714 | 0.0075821 | 7,468 |
| TMEM65 | 2,790,505 | 19,071 | 3,094,836 | 0.0273694 | 0.0922242 | 0.0162561 | 157,378 |
| SMARCD3 | 2,528,987 | 2,267,934 | 175,636 | 0.0106455 | 0.0135429 | 0.0390055 | 6,604 |
| NDST1 | 2,511,612 | 1,661,929 | 2,158,918 | 0.0456627 | 0.0897835 | 0.0622311 | 3,340 |
| OXCT1 | 2,382,383 | 180,451 | 1,980,136 | 0.0045742 | 0.0135215 | 0.0075821 | 5,019 |
| PARVA | 2,313,442 | 2,094,473 | 3,017,466 | 0.0060141 | 0.0080188 | 0.0021427 | 55,742 |
| PCNA | 2,300,166 | 2,407,186 | 2,057,527 | 0.0170402 | 0.0125668 | 0.0210382 | 5,111 |
| CAMK2D | 2,259,943 | 2,712,418 | 2,455,167 | 0.0133284 | 0.0063681 | 0.0076605 | 817 |
| SPARC | 2,230,562 | 2,335,873 | 2,990,594 | 0.0204267 | 0.0152481 | 0.0060567 | 6,678 |
| KIF3A | 219,478 | 1,850,005 | 2,377,716 | 0.0242452 | 0.04543 | 0.0142647 | 11,127 |
| TGFBR1 | 2,188,996 | 1,585,418 | 146,801 | 0.026412 | 0.0065456 | 0.0556766 | 7,046 |
| PKP2 | 2,108,275 | 183,479 | 2,092,256 | 0.0277094 | 0.0459483 | 0.0231586 | 5,318 |
| OSR1 | 2,071,946 | 4,175,767 | 2,370,344 | 0.0060753 | 0.0009141 | 0.0029158 | 130,497 |
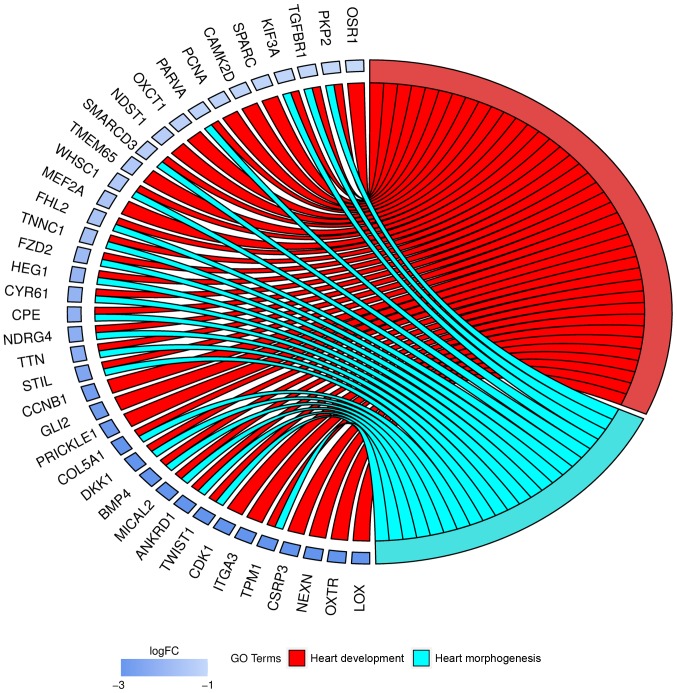
Moreover, genes that formed one particular GO group can also belong to other different GO term categories. Because of that, we explored the gene intersections between selected GO BP terms. The relation between those GO BP terms was presented as circle plot (Fig. 2).
Figure 2.
Representation of the mutual association between ‘heart morphogenesis’ and ‘heart development’ GO Biological Process terms. The ribbons indicate which gene belongs to which categories. The genes were sorted by logFC from most to least changed gene. FC, fold change; GO, Gene Ontology.
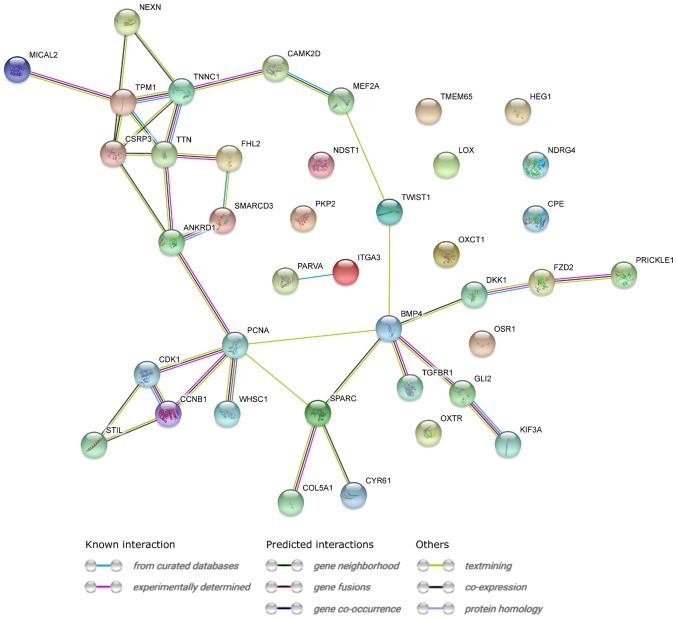
STRING interaction network was generated among differentially expressed genes belonging to each of selected GO BP terms. Using this prediction method yielded a molecular interaction network formed between protein products of studied genes (Fig. 3).
Figure 3.
STRING-generated interaction network among differentially expressed genes belonging to the ‘heart morphogenesis’ and ‘heart development’ Gene Ontology Biological Process terms. The intensity of the edges reflects the strength of the interaction score.
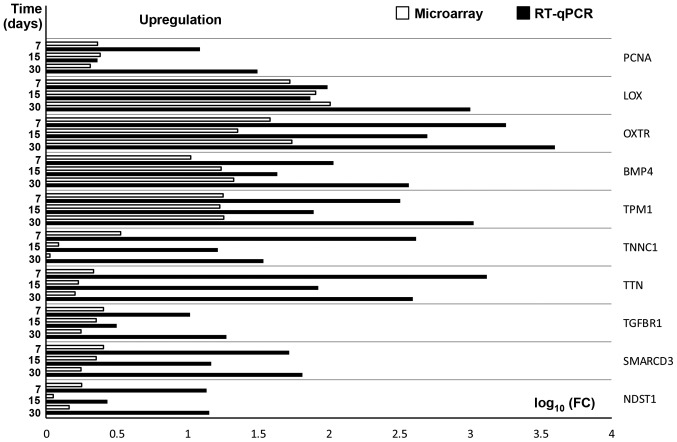
RT-qPCR was conducted to validate the results obtained during microarray analysis. The outcomes were presented and compared in a form of a bar graph (Fig. 4).
Figure 4.
RT-qPCR validation of microarray results, presented in a form of a bar graph. FC was presented in its logarithmic form to provide clear comparability of the results. PCNA, Proliferating cell nuclear antigen; LOX, Lysyl Oxidase; OXTR, oxytocin receptor; BMP4, Bone morphogenetic protein 4; TPM1, Tropomyosin 1; TNNC1, Troponin 1; TTN, Titin; TGFBR1, Transcription growth factor-β receptor 1; SMARCD3, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3; NDST1, N-deacetylase and N-sulfotransferase 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; FC, fold change.
As can be seen, the direction of changes in expression was confirmed in all examples. However, the scale of differences in transcript levels varied between both of the methods analysed.
The GCs have undergone extensive morphological modifications through the time of the culture. They changed their shape from star-like structures to broader, fibroblast-like form. These observable changes were presented as photographs, taken under an inverted microscope, using a relief contrast (Fig. 5).
Figure 5.
Pictures of human ovarian granulosa cell cultures used for the experiment, presenting changes in their morphology following 24 h, and 7, 15 and 30 days of in vitro culture. All of the photographs were taken using an inverted microscope, under relief phase contrast (magnification, ×10, 20 and 40, as indicated).
Discussion
Ovarian follicles are formed during the process of folliculogenesis and oogenesis. The follicle consists of the oocyte, follicular fluid and oocyte-associated cells-granulosa cells (GCs) (10). In physiological conditions, GCs largely influence the growth and development of the ovarian follicle. In addition, they produce gap junctions that allow for their bi-directional communication with the oocyte (11,12). During this dialogue, nutrients and metabolites are delivered to the oocyte (13). In addition, the key role of GCs in physiological conditions is the production of estradiol during follicular growth and the secretion of progesterone after ovulation (14).
The process of folliculogenesis begins with the change of GCs' shape from flattened to cuboidal through numerous mitotic divisions of cells. A characteristic feature is the production of follicle stimulating hormone receptor (FSHR) by the GCs. In preovulatory follicle, four types of GCs can be distinguished: The outermost layer is the granulosa membrane, the periantral granulosa is located inside the follicle, the intermediate layer forms the cumulus oophorus, with corona radiata being the final layer. Granulosa cells forming the corona radiata directly surround the oocyte (15). The individual types of GCs forming the four layers are functionally different, secreting different molecules and expressing different receptors (16).
Recent studies indicate that there is a pool of stem cells in mature mouse and human ovaries that support oogenesis and folliculogenesis. Thanks to these stem cells, the primary follicular resource can be produced during the reproductive period. Previously, it was thought that the number of primary follicles is determined and established shortly before the birth of the female (17,18). In recent years, however, the mentioned stem cells have become the major focus of scientists. It is proven that every tissue or organ (including the ovary) contains a population of stem cells capable of differentiating (19–22).
Many authors point to new possibilities of using GCs in regenerative medicine and broadly understood transplantology. GC containing follicular fluid, transferred for utilization during routine in vitro fertilization procedures, may be the source of stem cells. Several authors clearly indicate these previously unnoticed properties of GCs. Many articles follow a different lead, suggesting that GCs are subject to the transdifferentiation process. Transdifferentiation is the transformation of already differentiated cells into another cell type, which consists of two stages: cells dedifferentiation, followed by the natural developmental path towards a specific cell lineage. During this process, the mature cell is reprogrammed and changes into another cell type under the influence of the appropriate factors (23–25). Kossowska-Tomaszczuk et al (3,4) conducted experiments in which they indicated that GCs have stem-like properties. Luteinized GC subpopulations express the OCT-4 gene, escalation of which is characteristic for mesenchymal stem cells. In addition, the research team carried out the differentiation of GCs cells towards osteoblasts, neuroblasts and chondroblasts. They succeeded to obtain populations of differentiated cells (3,4). The results of our research indicate that GCs have stem cell properties and can differentiate, in the long-term primary in vitro culture, towards e.g.: osteoblasts or muscle cells (unpublished results). The results of our research have been confirmed by the literature. Oki et al (26) stated that pig-derived GCs may transdifferentiate towards osteoblasts both in vitro and in vivo. They conducted a microarray analysis clearly indicating that GCs show the GC-specific marker genes, together with osteogenic markers, during transdifferentiation (26). Brevini et al (27) proved that GCs, under the influence of external factors (Vascular endothelial growth factor-VEGF and 5-azacytidine-5-aza-CR), can differentiate towards muscle cells. Their research confirms the change in the phenotype of GCs. In addition, the differentiated GCs expressed the genes characteristic for muscle cells (including Desmin and Myosin heavy chain).
Following up on Brevini et al (27), we have analyzed the results of microarrays in terms of the possibility of GC differentiation into cardiomyocytes. It turned out that, in long-term in vitro culture, GCs express some genes involved in the process of heart morphogenesis and development.
In our research, we identified 39 genes responsible for heart development and morphogenesis processes. All of these genes are presented on heatmaps (Fig. 1). Among all genes, 22 are involved in both: Heart development and heart morphogenesis ontology groups. The remaining 17 genes are characteristic for the heart development process. The expression of these genes could confirm the theory that GCs can differentiate into cells proprietary to the heart tissue, during in vitro cell culture.
It is known that the heart is formed from the mesoderm that arises during the process of embryo gastrulation. In humans, the mesoderm begins to form at the third week of embryo development (28). At this stage, the mesodermal cells are still precardiac cells. Mesodermal cells differentiate into cardiac cells in response to endodermal induction signals-primarily bone morphogenetic proteins (BMPs) (29). Five transcription factors that play a primary role in cardiac development in various organisms have been identified as NKX2.5, Mef2, GATA, Tbx and Hand (30). Precardiac cells are multipotent and differentiate in myocardial, endothelial and smooth muscle cells through a process called progressive lineage restriction (31). The last stage of cardiac development is the differentiation of the atrial and ventricular specific myocytes and heart conduction cells (32). In our studies, we showed the expression of the Mef2A gene, and therefore suggest that GCs, in long-term in vitro culture, show features of mesenchymal stem cells that can differentiate into heart cells.
In addition, this is confirmed by the number of studies that describe the transition of multipotent mesodermal precursor cells into myoblasts, and then into mature myocytes. The myocyte-specific enhancer factor 2 (MEF2) (33) is responsible for the above process. Xu et al (34), in their studies, proved that overexpression of Mef2A causes cardiomyopathy and hypertension. Liu et al (35) demonstrated that Mef2 becomes active during heart formation in the early stages of embryogenesis and is responsible for the differentiation of atria and ventricles of the heart. Other studies demonstrate the key role of Mef2A in maintaining the appropriate number of mitochondria and the proper architecture of the heart's cells. Mef2a-deficient mice showed high mortality within one week after birth and marked dilation of the ventricle. These mice have been proven to be susceptible to sudden death (36). The above studies indicate the key role played by Mef2a in the formation of the heart. This, together with the findings of our research that shows that GCs can give rise to heart cells, opens up a way for further research into the application of human granulosa in heart regeneration medicine. As it has already been mentioned, inducers of the endoderm, such as BMPs, are an important group of factors involved in the development of the heart. In long-term in vitro culture, GCs express the BMP4 gene. According to the literature, this protein is involved in the development of endocardial epithelium and the correct formation of particular chambers of the heart. Deficiency of BMP4 in mice causes an abnormal number of cardiac cells, defective valve structure, defects in the septum of the heart, and abnormal heart valve formation (37,38).
Another gene characteristic for both morphogenesis and heart development is LOX (Lysyl oxidase). This gene shows the biggest change in expression in relation to the initial condition during long-term in vitro culture. Li et al (39) demonstrated that lysyl oxidase is present in the smooth muscle cells of the blood vessels of rats. In addition, Kaneda et al (40) noticed that LOX may be a suppressor of ovarian or stomach tumours. Our research indicates that this gene is expressed in granulosa cells and thus they could possibly differentiate into the blood vessels of the heart and be a suppressor of pathological changes in the ovary.
The gene responsible for the formation of the oxytocin receptor (oxytocin receptor-OXTR) showed prominently increased expression. To date, the expression of this gene has been associated with fertility, lactation and reproductive behaviour. Recently, mice research has suggested that OXTR plays a large role in the regulation and development of social behaviour (41). In addition, in recent years, the role of the above gene in the formation of the cardiovascular system has been indicated (42).
Another gene characteristic for the development and morphogenesis of the heart is NEXN (Nexilin F-actin binding protein) and Cysteine-rich protein 3 (CSRP3) also known as Muscle LIM protein (MLP). These genes show activity in the fetal and adult heart, as well as skeletal muscle adhesion and migration during embryogenesis, and are responsible for hyperthyroid cardiomyopathy (43–45).
During the long-term in vitro cultivation of GCs, it was noted that these cells show expression of cyclin-dependent kinases (CDKs), specifically CDK1. These compounds are responsible for the regulation of the cell cycle of cardiomyocytes (46). This serves as further evidence that in vitro, GCs may be differentiated into cardiomyocytes. The above genes showed clearly increased expression during long-term in vitro GC culture.
Among the genes characteristic for the development and morphogenesis of the heart, none showed a drastic decline during long-term in vitro culture. Some of these genes showed only a smaller change in expression. These include OSR1, PKP2, TGFBR1, KIF3A.
The OSR1 gene is expressed in GCs under physiological conditions but also participates in the formation of cardiac septa (47,48). We can assume that even a small change in the expression of this gene may indicate the possibility of GC differentiation. Similar observations were found in the case of the PKP2 gene (plakophilin). Plakophilin is a component of desmosomes and is necessary for the proper functioning and distribution of connexins, a type gap junction protein. It was noted that, during the silencing of the PKP2 gene in rat cardiac myocytes, there was a reduction in the number of gap junctions between the heart cells. This led to changes within the right ventricle (49). Other studies indicate that connexin is also present in GCs and is necessary for bi-directional communication between the oocyte and granulosa cells (50).
Another gene that seems to be characteristic for the processes related to the development of heart is TNNC1 (troponin c). This gene is responsible for the proper functioning of the muscle fibres of the heart, with all of its known mutations leading to cardiomyopathy (51). Another very important gene, demonstrating that GCs in long-term in vitro culture can undergo differentiation/transdifferentiation into cardiac muscle, is SMARCD3 (actin-dependent regulator of chromatin, subfamily D). Lickert et al (52) noticed that this gene is expressed in the hearts of early mouse embryos. Deficiency of this gene caused defects in heart morphogenesis, as well as abnormal differentiation of myocardium and skeletal muscles. In humans, the impairment of this gene is associated with the occurrence of many congenital heart defects (52). Other studies show that, in general, mesodermal cells may undergo transdifferentiation towards cardiac myocytes. SMARCD3, among others, is deeply involved in this process. It is noted that the presence of this gene can cause the transdifferentiation of mesoderm cells into cardiomyocytes (53). It can, therefore, be suggested that GCs, in long-term in vitro culture, show stem-like potential and may undergo differentiation or transdifferentiation towards cardiomyocytes.
It may be concluded that GCs have many characteristics of stem cells in long-term in vitro culture. The presented results suggest that it is possible to differentiate GCs into cells that build the heart and, at the same time, it may become a new possibility in the treatment of heart diseases and regenerative medicine of blood vessels. However, the conclusions were based only on large-scale transcriptomic analysis. While validated by more focused RT-qPCR tests in terms of the direction of changes, the scale of differences in transcript levels varied between the methods. This may be caused by many factors, mainly associated with the huge array of processes occurring from the time of transcription to expression of protein products. Therefore, the conclusions of this study require further analyses on the protein level, followed by practical trials of obtaining a stable, differentiated population of cardiac cells coming from GC cultures, which could later be applied in possible clinical situations.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Polish National Science Centre (grant nos. 2014/15/B/NZ7/00999 and UMO-2016/21/B/NZ9/03535) and from Poznan University of Medical Sciences (grant no. 502-01-02227367-08414).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WK conducted the experiments, chose the models and wrote the manuscript. MBrą designed the experiments and wrote the manuscript. PC performed data analysis and figure preparation, and wrote the manuscript. ArB conducted the experiments, performed data analysis and prepared the figures. MJN wrote the manuscript and performed data analysis. KO conducted the experiments, revised the medical methodology, and performed material extraction and preparation. MaJ conducted the experiments, and performed data analysis and language corrections. MiJ designed the experiments and performed models analysis. LP designed the study and revised the medical methodology (particularly that of in vitro fertilization procedures). AnB designed the study, and revised the methodology and the manuscript. DR designed the study. MTS designed the study, revised the methodology and critically revised the manuscript for important intellectual content. MBru designed the experiments, supervised the project, was involved in the medical procedure design and approved the final draft of the manuscript. MZ revised the methodology, performed data analysis, provided writing assistance and approved the final draft of the manuscript. MN supervised the project, designed the experiments (particularly the primary cell culture procedure), provided editorial supervision and approved the final draft of the manuscript. BK supervised the project, designed the study, revised the methodology, provided editorial supervision, and wrote the manuscript. All authors read and approved the final manuscript
Ethics approval and consent to participate
The present study has been approved by Poznan University of Medical Sciences Bioethical Committee with resolution 558/17. All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rybska M, Knap S, Jankowski M, Jeseta M, Bukowska D, Antosik P, Nowicki M, Zabel M, Kempisty B, Jaśkowski JM. Cytoplasmic and nuclear maturation of oocytes in mammals-living in the shadow of cells developmental capability. Med J Cell Biol. 2018;6:13–17. doi: 10.2478/acb-2018-0003. [DOI] [Google Scholar]
- 2.Rybska M, Knap S, Jankowski M, Jeseta M, Bukowska D, Antosik P, Nowicki M, Zabel M, Kempisty B, Jaśkowski JM. Characteristic of factors influencing the proper course of folliculogenesis in mammals. Med J Cell Biol. 2018;6:33–38. doi: 10.2478/acb-2018-0006. [DOI] [Google Scholar]
- 3.Kossowska-Tomaszczuk K, De Geyter C, De Geyter M, Martin I, Holzgreve W, Scherberich A, Zhang H. The multipotency of luteinizing granulosa cells collected from mature ovarian follicles. Stem Cells. 2009;27:210–219. doi: 10.1634/stemcells.2008-0233. [DOI] [PubMed] [Google Scholar]
- 4.Kossowska-Tomaszczuk K, De Geyter C. Cells with stem cell characteristics in somatic compartments of the ovary. Biomed Res Int 2013. 2013:310859. doi: 10.1155/2013/310859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kranc W, Brązert M, Ożegowska K, Nawrocki MJ, Budna J, Celichowski P, Dyszkiewicz-Konwińska M, Jankowski M, Jeseta M, Pawelczyk L, et al. Expression profile of genes regulating steroid biosynthesis and metabolism in human ovarian granulosa cells-a primary culture approach. Int J Mol Sci. 2017;18(pii):E2673. doi: 10.3390/ijms18122673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 7.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter W, Sánchez-Cabo F, Ricote M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 9.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2004;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kranc W, Budna J, Kahan R, Chachuła A, Bryja A, Ciesiółka S, Borys S, Antosik MP, Bukowska D, Brussow KP, et al. Molecular basis of growth, proliferation, and differentiation of mammalian follicular granulosa cells. J Biol Regul Homeost Agents. 2017;31:1–8. [PubMed] [Google Scholar]
- 11.Mora JM, Fenwick MA, Castle L, Baithun M, Ryder TA, Mobberley M, Carzaniga R, Franks S, Hardy K. Characterization and significance of adhesion and junction-related proteins in mouse ovarian follicles. Biol Reprod. 2012;86(153):1–14. doi: 10.1095/biolreprod.111.096156. [DOI] [PubMed] [Google Scholar]
- 12.Kempisty B, Ziółkowska A, Piotrowska H, Zawierucha P, Antosik P, Bukowska D, Ciesiółka S, Jaśkowski JM, Brüssow KP, Nowicki M, Zabel M. Real-time proliferation of porcine cumulus cells is related to the protein levels and cellular distribution of Cdk4 and Cx43. Theriogenology. 2013;80:411–420. doi: 10.1016/j.theriogenology.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Su YQ, Sugiura K, Eppig JJ. Mouse oocyte control of granulosa cell development and function: Paracrine regulation of cumulus cell metabolism. Semin Reprod Med. 2009;27:32–42. doi: 10.1055/s-0028-1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeppesen JV, Kristensen SG, Nielsen ME, Humaidan P, Dal Canto M, Fadini R, Schmidt KT, Ernst E, Yding Andersen C. LH-receptor gene expression in human granulosa and cumulus cells from antral and preovulatory follicles. J Clin Endocrinol Metab. 2012;97:E1524–E1531. doi: 10.1210/jc.2012-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Findlay JK, Kerr JB, Britt K, Liew SH, Simpson ER, Rosairo D, Drummond AE. Ovarian physiology: Follicle development, oocyte and hormone relationships. Anim Reprod. 2009;6:16–19. [Google Scholar]
- 16.Nguyen T, Lee S, Hatzirodos N, Hummitzsch K, Sullivan TR, Rodgers RJ, Irving-Rodgers HF. Spatial differences within the membrana granulosa in the expression of focimatrix and steroidogenic capacity. Mol Cell Endocrinol. 2012;363:62–73. doi: 10.1016/j.mce.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Guigon CJ, Magre S. Contribution of germ cells to the differentiation and maturation of the ovary: Insights from models of germ cell depletion. Biol Reprod. 2006;74:450–458. doi: 10.1095/biolreprod.105.047134. [DOI] [PubMed] [Google Scholar]
- 18.Johnson J, Canning J, Kaneko T, Pru JK, Tilly JL. Germline stem cells and follicular renewal in the postnatal mammalian ovary. Nature. 2004;428:145–150. doi: 10.1038/nature02316. [DOI] [PubMed] [Google Scholar]
- 19.Parte S, Bhartiya D, Telang J, Daithankar V, Salvi V, Zaveri K, Hinduja I. Detection, characterization, and spontaneous differentiation in vitro of very small embryonic-like putative stem cells in adult mammalian ovary. Stem Cells Dev. 2011;20:1451–1464. doi: 10.1089/scd.2010.0461. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Virant-Klun I, Rozman P, Cvjeticanin B, Vrtacnik-Bokal E, Novakovic S, Rülicke T, Dovc P, Meden-Vrtovec H. Parthenogenetic embryo-like structures in the human ovarian surface epithelium cell culture in postmenopausal women with no naturally present follicles and oocytes. Stem Cells Dev. 2009;18:137–149. doi: 10.1089/scd.2007.0238. [DOI] [PubMed] [Google Scholar]
- 21.Virant-Klun I, Zech N, Rozman P, Vogler A, Cvjeticanin B, Klemenc P, Malicev E, Meden-Vrtovec H. Putative stem cells with an embryonic character isolated from the ovarian surface epithelium of women with no naturally present follicles and oocytes. Differentiation. 2008;76:843–856. doi: 10.1111/j.1432-0436.2008.00268.x. [DOI] [PubMed] [Google Scholar]
- 22.Virant-Klun I. Postnatal oogenesis in humans: A review of recent findings. Stem Cells Cloning. 2015;8:49–60. doi: 10.2147/SCCAA.S32650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buganim Y, Jaenisch R. Transdifferentiation by defined factors as a powerful research tool to address basic biological questions. Cell Cycle. 2012;11:4485–4486. doi: 10.4161/cc.22665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen CN, Burke ZD, Tosh D. Transdifferentiation, metaplasia and tissue regeneration. Organogenesis. 2004;1:36–44. doi: 10.4161/org.1.2.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dzafic E, Stimpfel M, Virant-Klun I. Plasticity of granulosa cells: On the crossroad of stemness and transdifferentiation potential. J Assist Reprod Genet. 2013;30:1255–1261. doi: 10.1007/s10815-013-0068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oki Y, Ono H, Motohashi T, Sugiura N, Nobusue H, Kano K. Dedifferentiated follicular granulosa cells derived from pig ovary can transdifferentiate into osteoblasts. Biochem J. 2012;447:239–248. doi: 10.1042/BJ20120172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brevini TAL, Pennarossa G, Rahman MM, Paffoni A, Antonini S, Ragni G, deEguileor M, Tettamanti G, Gandolfi F. Morphological and molecular changes of human granulosa cells exposed to 5-azacytidine and addressed toward muscular differentiation. Stem Cell Rev. 2014;10:633–642. doi: 10.1007/s12015-014-9521-4. [DOI] [PubMed] [Google Scholar]
- 28.Moorman A, Webb S, Brown NA, Lamers W, Anderson RH. Development of the heart: (1) formation of the cardiac chambers and arterial trunks. Heart. 2003;89:806–814. doi: 10.1136/heart.89.7.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meilhac SM, Lescroart F, Blanpain C, Buckingham ME. Cardiac cell lineages that form the heart. Cold Spring Harb Perspect Med. 2014;4:a013888. doi: 10.1101/cshperspect.a013888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epstein JA. Franklin H. Epstein Lecture. Cardiac development and implications for heart disease. N Engl J Med. 2010;363:1638–1647. doi: 10.1056/NEJMra1003941. [DOI] [PubMed] [Google Scholar]
- 32.Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 33.Breitbart RE, Liang CS, Smoot LB, Laheru DA, Mahdavi V, Nadal-Ginard B. A fourth human MEF2 transcription factor, hMEF2D, is an early marker of the myogenic lineage. Development. 1993;118:1095–1106. doi: 10.1242/dev.118.4.1095. [DOI] [PubMed] [Google Scholar]
- 34.Xu J, Gong NL, Bodi I, Aronow BJ, Backx PH, Molkentin JD. Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J Biol Chem. 2006;281:9152–9162. doi: 10.1074/jbc.M510217200. [DOI] [PubMed] [Google Scholar]
- 35.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci USA. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA, Olson EN. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med. 2002;8:1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 37.McCulley DJ, Kang JO, Martin JF, Black BL. BMP4 is required in the anterior heart field and its derivatives for endocardial cushion remodeling, outflow tract septation, and semilunar valve development. Dev Dyn. 2008;237:3200–3209. doi: 10.1002/dvdy.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdelwahid E, Rice D, Pelliniemi LJ, Jokinen E. Overlapping and differential localization of Bmp-2, Bmp-4, Msx-2 and apoptosis in the endocardial cushion and adjacent tissues of the developing mouse heart. Cell Tissue Res. 2001;305:67–78. doi: 10.1007/s004410100399. [DOI] [PubMed] [Google Scholar]
- 39.Li W, Nellaiappan K, Strassmaier T, Graham L, Thomas KM, Kagan HM. Localization and activity of lysyl oxidase within nuclei of fibrogenic cells. Proc Natl Acad Sci USA. 1997;94:12817–12822. doi: 10.1073/pnas.94.24.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaneda A, Wakazono K, Tsukamoto T, Watanabe N, Yagi Y, Tatematsu M, Kaminishi M, Sugimura T, Ushijima T. Lysyl oxidase is a tumor suppressor gene inactivated by methylation and loss of heterozygosity in human gastric cancers. Cancer Res. 2004;64:6410–6415. doi: 10.1158/0008-5472.CAN-04-1543. [DOI] [PubMed] [Google Scholar]
- 41.Takayanagi Y, Yoshida M, Bielsky IF, Ross HE, Kawamata M, Onaka T, Yanagisawa T, Kimura T, Matzuk MM, Young LJ, Nishimori K. Pervasive social deficits, but normal parturition, in oxytocin receptor-deficient mice. Proc Natl Acad Sci USA. 2005;102:16096–16101. doi: 10.1073/pnas.0505312102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutkowska J, Jankowski M. Oxytocin revisited: Its role in cardiovascular regulation. J Neuroendocrinol. 2012;24:599–608. doi: 10.1111/j.1365-2826.2011.02235.x. [DOI] [PubMed] [Google Scholar]
- 43.Hassel D, Dahme T, Erdmann J, Meder B, Huge A, Stoll M, Just S, Hess A, Ehlermann P, Weichenhan D, et al. Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat Med. 2009;15:1281–1288. doi: 10.1038/nm.2037. [DOI] [PubMed] [Google Scholar]
- 44.Zhao Y, Wei YJ, Cao HQ, Ding JF. Molecular cloning of NELIN, a putative human cytoskeleton regulation gene. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 2001;33:19–24. [PubMed] [Google Scholar]
- 45.Geier C, Gehmlich K, Ehler E, Hassfeld S, Perrot A, Hayess K, Cardim N, Wenzel K, Erdmann B, Krackhardt F, et al. Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum Mol Genet. 2008;17:2753–2765. doi: 10.1093/hmg/ddn259. [DOI] [PubMed] [Google Scholar]
- 46.Ikenishi A, Okayama H, Iwamoto N, Yoshitome S, Tane S, Nakamura K, Obayashi T, Hayashi T, Takeuchi T. Cell cycle regulation in mouse heart during embryonic and postnatal stages. Dev Growth Differ. 2012;54:731–738. doi: 10.1111/j.1440-169X.2012.01373.x. [DOI] [PubMed] [Google Scholar]
- 47.Lan CW, Chen MJ, Jan PS, Chen HF, Ho HN. Differentiation of human embryonic stem cells into functional ovarian granulosa-like cells. J Clin Endocrinol Metab. 2013;98:3713–3723. doi: 10.1210/jc.2012-4302. [DOI] [PubMed] [Google Scholar]
- 48.Zhou L, Liu J, Olson P, Zhang K, Wynne J, Xie L. Tbx5 and Osr1 interact to regulate posterior second heart field cell cycle progression for cardiac septation. J Mol Cell Cardiol. 2015;85:1–12. doi: 10.1016/j.yjmcc.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oxford EM, Musa H, Maass K, Coombs W, Taffet SM, Delmar M. Connexin43 remodeling caused by inhibition of plakophilin-2 expression in cardiac cells. Circ Res. 2007;101:703–711. doi: 10.1161/CIRCRESAHA.107.154252. [DOI] [PubMed] [Google Scholar]
- 50.Kempisty B, Ziółkowska A, Ciesiółka S, Piotrowska H, Antosik P, Bukowska D, Nowicki M, Brüssow KP, Zabel M. Study on connexin gene and protein expression and cellular distribution in relation to real-time proliferation of porcine granulosa cells. J Biol Regul Homeost Agents. 2014;28:625–635. [PubMed] [Google Scholar]
- 51.Parvatiyar MS, Landstrom AP, Figueiredo-Freitas C, Potter JD, Ackerman MJ, Pinto JR. A mutation in TNNC1-encoded cardiac troponin C, TNNC1-A31S, predisposes to hypertrophic cardiomyopathy and ventricular fibrillation. J Biol Chem. 2012;287:31845–31855. doi: 10.1074/jbc.M112.377713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 53.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.