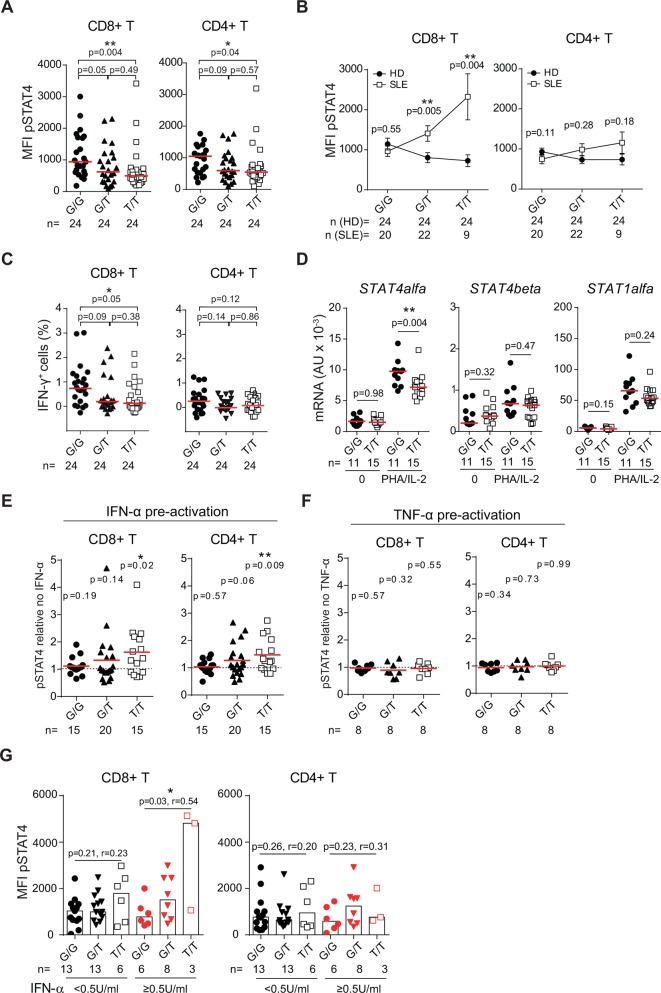
Intronic single nucleotide polymorphisms (SNP) in STAT4, tagged by rs7574865[T], are associated with increased susceptibility to develop systemic lupus erythematosus (SLE) and a more severe disease phenotype with nephritis and stroke.1 2 Recently, we demonstrated that T cells from patients with SLE carrying the STAT4 risk allele rs7574865[T] have an enhanced induction of STAT4 protein following PHA/interleukin (IL)-2 activation, which results in increased IL-12-induced phosphorylation of STAT4 (pSTAT4) and interferon (IFN)-γ production.3 As the majority of STAT4 risk allele carriers do not develop disease, we asked whether the STAT4 risk allele exerts the same effect in healthy individuals. In studies of IL-12-stimulated cells from 72 healthy women (online supplementary materials and methods) we unexpectedly found that STAT4 risk allele carriership was associated with a decreased pSTAT4 in CD8+ and CD4+ T cells from healthy individuals (figure 1A). Notably, in the absence of STAT4 risk alleles healthy individuals and patients with SLE phosphorylated STAT4 to the same extent (figure 1B). However, with increasing numbers of STAT4 risk alleles, patients with SLE responded stronger to IL-12 stimulation. The decreased IL-12 response in healthy STAT4 risk allele carriers resulted in a decreased production of IFN-γ in CD8+ T cells and a slight, but non-significant, decrease in CD4+ T cells (figure 1C). Demonstrating the specificity for IL-12, phorbol 12-myristate 13-acetate-induced IFN-γ production was not affected (online supplementary figure 1). Thus, the STAT4 risk allele rs7574865[T] associates with opposite directional effects in cells from healthy donors compared with patients with SLE.
annrheumdis-2018-213836supp001.docx (14.9KB, docx)
annrheumdis-2018-213836supp002.eps (1.5MB, eps)
Figure 1.
IFN-α enhances the IL-12 response in T cells from STAT4 risk allele carriers. (A–C) PHA/IL-2 preactivated peripheral blood mononuclear cells (PBMC) from healthy donors (HD) were restimulated with IL-12. (A, B) Phosphorylation of STAT4 (pSTAT4) (50 ng/mL IL-12, 20 min) and (C) frequency of IFN-γ+ cells (5 ng/mL IL-12, 15 hours) were determined using flow cytometry. (B) Mean with error bars depicting the SEM. Data from patients with SLE are previously published.3 (D) mRNA expression levels of STAT4alpha, STAT4beta and STAT1alpha in CD8+ memory T cells from healthy individuals before and after 6 hours of stimulation with PHA/IL-2 were determined with qRT-PCR. (A–D) Statistical comparisons were made using two-tailed Mann-Whitney U tests. (E, F) PBMCs from healthy individuals were preactivated with (E) 100 U/mL IFNαb2, (F) 1 ng/mL TNF-α, or mock treated for 24 hours before PHA/IL-2 stimulation. IL-12-induced pSTAT4 (100 ng/mL, 20 min) in cytokine pretreated cells was divided by IL-12-induced pSTAT4 in mock-treated cells and two-tailed one-sample t-tests were used to assess whether the means differ from 1. (G) IL-12-induced pSTAT4 in patients with SLE stratified for plasma levels of IFN-α below (black) or above (red) the detection limit of 0.5 U/mL. P values for two-tailed Spearman’s correlation tests and the Spearman’s correlation coefficient (r) are denoted. (A–G) G/G, homozygous protective; G/T, heterozygous; T/T, homozygous risk for rs7574865. Red horizontal lines depict the (A–D) median or (E, F) mean values. *P<0.05; ** P<0.01. IFN, interferon; IL, interleukin; n, number of individuals in each group; SLE, systemic lupus erythematosus; TNF, tumour necrosis factor.
Opposing directional effects have been described for SNPs in terms of neurotransmitter receptor binding in patients with psychiatric diseases and healthy individuals,4 5 but to the best of our knowledge this is the first time such an effect is described in patients with SLE. Another example of this phenomenon is the differential regulation of mRNA expression in different cell types, and in in vitro stimulated cells compared with unstimulated cells.6 The underlying mechanisms for these observations are not completely known, but may include cell-type-specific expression or context-dependent induction of transcription factors.
When exploring these possibilities, we observed that basal mRNA levels of STAT4alpha, STAT4beta and STAT1alpha did not correlate with STAT4 genotype in healthy donor CD8+ memory T cells. However, following PHA/IL-2 stimulation, homozygous risk individuals displayed a defective induction of STAT4alpha mRNA expression, which is essential for IL-12-induced IFN-γ production,7 whereas STAT4beta and STAT1alpha mRNA were normally induced (figure 1D).
Given the important role of type I IFN in SLE, we hypothesised that IFN-α may be an environmental modulator of the STAT4 risk allele. Indeed, preactivation of healthy donor cells with IFN-α for 24 hours before PHA/IL-2 stimulation augmented the IL-12-induced pSTAT4 in CD8+ and CD4+ T cells from risk allele carriers, but not in non-risk allele carriers (figure 1E). In contrast, preactivation of cells with tumour necrosis factor-α did not affect IL-12-induced pSTAT4 (figure 1F). In support of an interaction with IFN-α and the STAT4 risk allele in patients with SLE in vivo, stratification of the patients from our previous study3 into those with plasma levels of IFN-α above or below the detection limit 0.5 U/mL revealed that the increased IL-12-induced pSTAT4 in CD8+ T cells from risk allele carriers was specific to the IFN-α+ patients (figure 1G).
In conclusion, the STAT4 risk allele rs7574865[T] has opposite effects in healthy individuals compared with patients with SLE and we identify IFN-α as an environmental modifier of the STAT4 risk allele. This finding may have implications for why the majority of risk allele carriers do not develop disease, and may also suggest a mechanism where STAT4 risk allele carriers are at risk to develop SLE when IFN is produced, for instance, during a viral infection.
Acknowledgments
We thank Martin Joelsson, Lisbeth Fuxler and Karolina Tandre for assisting in collecting the PBMCs. Genotyping was performed by the SNP&SEQ Technology Platform, which is part of Science for Life Laboratory and the National Genomics Infrastructure at Uppsala University, supported by the Swedish Research Council (VR-RFI) and the Knut and Alice Wallenberg Foundation.
Footnotes
Handling editor: Josef S Smolen
Contributors: NH performed the experiments and analysed the data. NH and LR designed the study, interpreted the data and wrote the manuscript.
Funding: This work was supported by grants from the Swedish Research Council for Medicine and Health (D0283001), the Swedish Rheumatism Association, King Gustav V’s 80-Year Foundation, the Knut and Alice Wallenberg Foundation (2011.0073), the Swedish Society of Medicine and the Ingegerd Johansson donation, and Erik, Karin and Gösta Selander’s Foundation.
Competing interests: None declared.
Patient consent: Not required.
Ethics approval: The study was approved by the Regional Ethics Board in Uppsala (2009/013).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Bolin K, Sandling JK, Zickert A, et al. . Association of STAT4 polymorphism with severe renal insufficiency in lupus nephritis. PLoS One 2013;8:e84450 10.1371/journal.pone.0084450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Svenungsson E, Gustafsson J, Leonard D, et al. . A STAT4 risk allele is associated with ischaemic cerebrovascular events and anti-phospholipid antibodies in systemic lupus erythematosus. Ann Rheum Dis 2010;69:834–40. 10.1136/ard.2009.115535 [DOI] [PubMed] [Google Scholar]
- 3. Hagberg N, Joelsson M, Leonard D, et al. . The STAT4 SLE risk allele rs7574865[T] is associated with increased IL-12-induced IFN-γ production in T cells from patients with SLE. Ann Rheum Dis 2018;77:1070–7. 10.1136/annrheumdis-2017-212794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Savitz J, Hodgkinson CA, Martin-Soelch C, et al. . DRD2/ANKK1 Taq1A polymorphism (rs1800497) has opposing effects on D2/3 receptor binding in healthy controls and patients with major depressive disorder. Int J Neuropsychopharmacol 2013;16:2095–101. 10.1017/S146114571300045X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cannon DM, Klaver JK, Gandhi SK, et al. . Genetic variation in cholinergic muscarinic-2 receptor gene modulates M2 receptor binding in vivo and accounts for reduced binding in bipolar disorder. Mol Psychiatry 2011;16:407–18. 10.1038/mp.2010.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fairfax BP, Knight JC. Genetics of gene expression in immunity to infection. Curr Opin Immunol 2014;30:63–71. 10.1016/j.coi.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoey T, Zhang S, Schmidt N, et al. . Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. Embo J 2003;22:4237–48. 10.1093/emboj/cdg393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2018-213836supp001.docx (14.9KB, docx)
annrheumdis-2018-213836supp002.eps (1.5MB, eps)