Abstract
The present study aimed to evaluate the antitumor effect of Scutellaria barbata polysaccharides (SBPS) in a hepatoma mouse model and examine the serum proteins involved in the tumorigenesis and SBPS treatment. A hepatoma model was established by the subcutaneous inoculation of murine hepatocellular carcinoma into Kunming mice. The treatment (once a day) lasted until the tumor weight in the model group was ~1 g (~7-10 days after inoculation). The sera proteins from each group were then collected and subjected to two-dimensional gel electrophoresis. Differentially expressed proteins were screened out and representatives were identified using matrix-assisted laser desorption ionization time-of-flight mass spectrometry. SBPS treatment at different doses significantly inhibited hepatoma growth (all P<0.01 vs. model group). The comparative serum proteomics showed that pseudouridine synthase 1 and chain A of the signal recognition particle Alu RNA-binding heterodimer (Srp9/14) were increased in the serum of the H22 hepatoma-bearing mice, and both were reduced by SBPS treatment. Mitochondrial ribosomal protein L24 was absent from the serum of H22 hepatoma-bearing mice, and was restored by SBPS treatment to approximately the normal level. Taken together, SBPS inhibited the growth of hepatic carcinoma in mice and affected serum proteomic profiling.
Keywords: hepatoma, Scutellaria barbata polysaccharides, serum proteomics, two-dimensional gel electrophoresis, matrix-assisted laser desorption ionization time-of-flight mass spectrometry
Introduction
Scutellaria barbata D. Don (S. barbata; Ban zhi lian in Chinese) is used as an immunomodulatory and antitumor agent in traditional Chinese medicine (1). Extracts of S. barbata have exhibited growth-inhibitory effects in a number of types of cancer in vitro and/or in vivo, including liver cancer (2,3). A recent study revealed that the immunomodulatory function of S. barbata extracts on Th1 and Th17 immune responses was involved in its antitumor effect when treating hepatoma H22-bearing mice (4). The active ingredients in S. barbata extracts were investigated regarding their different inhibitory effects on hepatocarcinoma and underlying mechanisms, and it was found that total flavonoids from S. barbata impaired the viability (5) and metastatic capability (6) of MHCC97H human hepatocarcinoma cells in vitro, which were associated with regulation of the mitochondrial apoptotic pathway (5) and matrix metalloproteinases (6), respectively.
S. barbata polysaccharides (SBPSs) are another category of main active ingredients extracted from S. barbata (7). SBPS have been reported to not only inhibit the proliferation of lung carcinoma cells in vitro and in a subcutaneous xenograft model in a dose-dependent manner, but also downregulate phosphorylated (p)-c-Met and its downstream signaling molecules, including p-extracellular signal-regulated kinase (p-ERK) and p-protein kinase B (p-AKT) (8). Yang et al showed that SBPS potently inhibited cell proliferation and human epidermal growth factor receptor (HER)2 phosphorylation in HER2-mutated non-small cell lung cancer in vitro and in xenograft models (9). In addition, a previous study revealed that SBPS inhibited the proliferation and elevated the apoptosis of human colon cancer HT29 cells. SBPS also inhibited the epithelial-mesenchymal transition of HT29 cells by upregulating the mRNA expression of E-cadherin and downregulating that of N-cadherin and vimentin (10). However, there has been limited investigation of the effects of SBPS on liver cancer so far. In addition, comprehensive analyses of the molecular mechanisms underlying the antitumor effect of SBPS using high-throughput omics technologies are required.
Serum contains an abundance of proteins that are important indicators of physiological or pathological states. Therefore, comprehensive determination of the proteome of human serum with high accuracy and availability has the potential to identify disease biomarkers, monitor disease development, and identify the mechanisms underlying disease development (11). Matrix-assisted laser (induced) desorption ionization (MALDI)-based profiling of serum proteomics has been approved for routine diagnostics (12). Examining serum proteomics with the assistance of MALDI time-of-flight mass spectrometry (MALDI-TOF MS) analysis has identified several potential targets for treating hepatocellular carcinoma (13,14).
In the present study, an experimental model of mice bearing H22 hepatic carcinoma was established and treated with different dosages of SBPS. The antitumor effect of SBPS on hepatic carcinoma was investigated. In addition, two-dimensional gel electrophoresis (2-DE) was applied to establish the serum protein patterns of normal mice, tumor-bearing mice and SBPS-treated tumor-bearing mice. The differential protein spots were analyzed using MALDI-TOF MS and the resulting protein sequences were used to search the National Center for Biotechnology Information (NCBI) database for validation. Six serum proteins were identified between the tumor-bearing mice and the tumor-bearing mice treated with SBPS. This study may provide a novel perspective for treating hepatocellular carcinoma with S. barbata extracts.
Materials and methods
Establishment of the hepatic carcinoma mouse model, treatment, and sample collection
Kunming mice (23±1°C,12 h light/dark cycle, with free access to food and water), weighing 20±2.0 g (males and females in equal numbers), were provided by Heilongjiang University of Chinese Medicine (Heilongjiang, China). The murine hepatocellular carcinoma was purchased from the Cancer Institute and Hospital (Chinese Academy of Medical Sciences, Beijing, China). The study was approved by the Ethics Committee of Mudanjiang Medical University (Mudanjiang, China).
Under sterile conditions, the ascites from mice bearing H22 hepatoma (established previously) for 6–7 days were extracted, diluted at a ratio of 1:3 with sterile saline, and centrifuged at 4°C and 300 × g for 5 min, as previously described (15). The cell concentration was adjusted to ~2×l07/ml, with cell viability ≥95%. Mice at the age of 6–8 weeks were subcutaneously inoculated with 0.2 ml of cell suspension in the right forelimb axillary region to establish the solid tumor model.
The mice bearing hepatic carcinoma H22 were randomly divided into five groups (n=10 in each group): Model (injected with normal saline), CTX [injected with CTX at 30 mg/(kg·day)], SBPS high-dose [injected with SBPS at 200 mg/(kg·day)], SBPS moderate-dose [injected with SBPS at 100 mg/(kg·day)] and SBPS low-dose groups [injected with SBPS at 50 mg/(kg·day)]. The doses were determined based on previous literature (16–19). The injection volume was 10 ml/(kg·day) for mice in all five groups. From 6 h post-inoculation, saline, CTX, and SBPS were intraperitoneally injected once a day for 7 days. SBPS was purchased from Ningbo Dekang Biochem Co., Ltd. (Ningbo, China) and cyclophosphamide (CTX; cat. no. 12060625) was purchased from Heng Rui Medicine Co., Ltd. (Jiangsu, China).
The mice were sacrificed 24 h after the final drug administration by cervical dislocation, and blood samples were collected from the orbital sinus. The blood samples were maintained at 4°C for 30 min, and then centrifuged at 3,000 × g and 4°C for 20–30 min to obtain serum. Sera samples from the same group were pooled together and stored at −80°C.
The tumors were removed and weighed to calculate the tumor inhibition rate as follows: Inhibition rate = (1 - average tumor weight of treatment group/average tumor weight of model group) ×100%.
2-DE and image analysis
The serum samples were preprocessed using the Calbiochem® ProteoExtract™ Albumin/IgG Removal kit (EMD Millipore; Merck KGaA, Darmstadt, Germany), and then precipitated with cold acetone. The precipitated samples were resolved in protein extraction buffer (200 µl per 10 mg samples) containing 1 mM PMSF, 2 mM EDTA and 10 mM DTT.
The protein concentration was determined using the 2-D Quant kit from Amersham Biosciences; GE Healthcare Life Sciences, Little Chalfont, UK). Subsequently, 50 or 200 µg of proteins were loaded in each lane of immobilized pH gradient (IPG) strips. The first dimension of 2-DE was performed in IPG isoelectric focusing. The strips were balanced and then transferred onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels for the second dimension, which was followed by silver staining (0.25% AgNO3, 0.015% formaldehyde) for 20 min. The 2-DE gels were scanned using the PowerLook 2100 XL scanner system (Umax Company, Fremont, CA, USA) and processed using ImageMaster 5.0 software (GE Healthcare Life Sciences).
MALDI-TOF MS
The protein spots were excised from the gel manually, digested with trypsin and desalinated. The peptide mixtures were identified via Ultraflex TOF/TOF MS (Bruker Daltonics, Bremen, Germany) in the reflection mode with a 20-kV accelerating voltage and 22.4-kV reflecting voltage. A 1-ns pulsed laser light was produced by a nitrogen laser at 337 nm, with an ion extraction delay of 0 ns. Subsequently, 100 shots were accumulated per spectrum, and trypsin autolysis peaks were used as internal calibrant and adrenocorticotropic hormone as external calibrant to obtain peptide mass fingerprints (PMFs).
Protein identification
The signal peaks of single isotopes were acquired from the PMF images using Flexanalysis 3.0 software, and PMFs were searched against the NCBI database (https://www.ncbi.nlm.nih.gov/protein) using the Mascot program (Matrix Science, Ltd., London, UK, http://www.matrixscience.com/search_form_select.html). The search parameters were: Mus musculus; trypsin digestion; ion species: Monoisotopic and MH+; carbamidomethyl modification of cysteine as a fixed modification; oxidation of methionine as a variable modification; and mass error of peptide fragments: ±0.1%.
Statistical analysis
The data were analyzed using SPSS 13.0 statistical analysis software (SPSS, Inc., Chicago, IL, USA). The experimental results are expressed as the mean ± standard deviation. The differences among groups were compared using one-way analysis of variance and Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Antitumor effect of SBPS on H22 hepatoma
To evaluate the effect of SBPS on hepatoma growth, the tumor-bearing mice were injected with SBPS, CTX, or saline every day for 7 days (~7-10 days after H22 cell inoculation). The maximum tumor volume was ~10-15 mm3. Compared with the model group, the tumor weights of the CTX and SBPS high-, moderate-, and low-dose groups were significantly decreased (all P<0.01; Table I). Compared with the CTX group, the low- and high-dose SBPS groups were significantly different (both P<0.05; Table I); the moderate-dose SBPS group exhibited comparable antitumor effects (P>0.05; Table I). Therefore, SBPS efficiently inhibited the growth of hepatoma in mice and the inhibitory rate of the moderate-dose SBPS was the highest (49.68%; Table I).
Table I.
Antitumor effect of SBPS on H22 hepatoma.
| Group | Dose [mg/(kg · day)] | Tumor weight (g) | Inhibition rate (%) |
|---|---|---|---|
| Model | – | 1.083±0.236 | – |
| CTX | 30 | 0.390±0.119a | 63.99 |
| High-dose SBPS | 200 | 0.627±0.163a,b | 43.03 |
| Moderate-dose SBPS | 100 | 0.505±0.185a | 49.68 |
| Low-dose SBPS | 50 | 0.680±0.157a,b | 37.21 |
Mice bearing H22 hepatic carcinoma were randomly divided into five groups: Model, CTX, and SBPS high-dose/middle-dose/low-dose groups. From 6 h post-inoculation, saline, CTX and SBPS were intraperitoneally injected once a day for 7 days. The mice were sacrificed 24 h after the final drug administration. The results are expressed as the mean ± standard deviation and were analyzed using analysis of variance and Tukey's post hoc test.
P<0.01, compared with the model group
P<0.05, compared with the CTX group. N=10/group. CTX, cyclophosphamide; SBPS, Scutellaria barbata polysaccharides.
2-DE mapping of serum proteins in the normal, tumor and SBPS groups
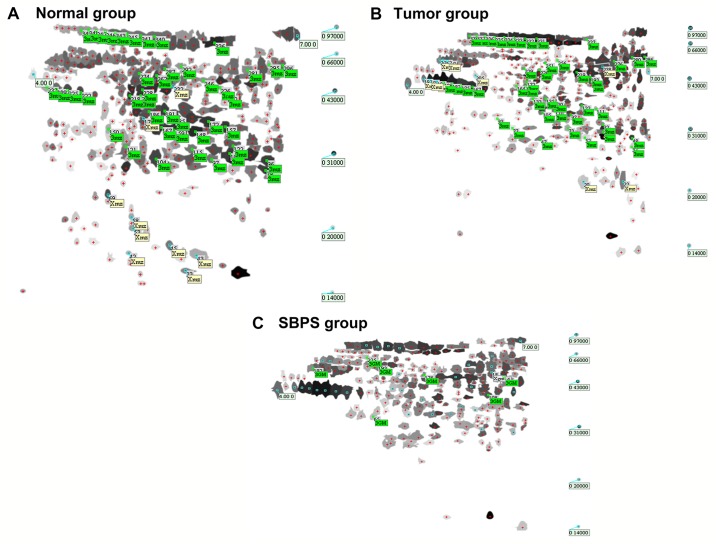
The total numbers of serum protein spots in the normal, tumor, and SBPS groups were 347±5, 335±3 and 252±7, respectively. Software analysis showed 62 protein spots with a change of >3-fold between the normal and tumor groups; 18 spots had a change of >3-fold between the tumor and SBPS groups (Fig. 1A-C).
Figure 1.
Representative two-dimensional gel electrophoresis images showing the protein spots from the sera of different groups. Protein spots in (A) normal group, (B) tumor group and (C) SBPS group. SBPS, Scutellaria barbata polysaccharides.
Analysis of the differences in the 2-DE map
The clear, large and heavily stained protein spots were selected as differentially expressed proteins from the three independent 2-DE images. The normal group vs. model group and the SBPS group vs. model group were compared to identify the differentially expressed serum proteins. The spots with fold-changes of >3 or <0.3 (tumor/normal or SBPS/tumor) were selected (Tables II and III). Table IV shows the serum proteins which were differentially expressed between the normal and tumor-bearing mice but rescued by SBPS treatment. Among them, spots 33, 176, 232 and 257 were expressed in the normal group, absent in the tumor group, and to a certain extent, restored in the SBPS group; whereas spots 20, 22, 205 and 238 exclusively appeared in the tumor group and disappeared upon SBPS treatment. In addition, spots 150, 196, 198, 283 and 310 in the tumor group were lower than those in the normal group; however, following SBPS treatment, the expression levels of these proteins were either restored to an extent (spots 150 and 196) or even exceeded the normal level (spots 198, 283 and 310). By contrast, spots 234, 296 and 349 in the tumor group were higher than those in the normal group; following SBPS treatment, the expression of these proteins were either restored to an extent (spot 234) or were below the normal level (spots 296 and 349).
Table II.
Differentially expressed proteins between the normal and tumor-bearing mice with a fold-change of >3 or <0.3.
| ID | Protein isoelectric point | MW (Da) | Fold-change (tumor vs. normal) |
|---|---|---|---|
| 33a | 5.73 | 15,799 | – |
| 42a | 5.09 | 17,393 | – |
| 43a | 5.86 | 17,211 | – |
| 45a | 5.55 | 18,195 | – |
| 53a | 5.13 | 19,925 | – |
| 58a | 5.12 | 21,334 | – |
| 69a | 4.85 | 24,353 | – |
| 176a | 5.25 | 37,256 | – |
| 232a | 5.58 | 45,714 | – |
| 157 | 6.18 | 35,678 | 0.140 |
| 79 | 6.64 | 28,093 | 0.144 |
| 218 | 5.10 | 41,945 | 0.159 |
| 97 | 6.02 | 30,039 | 0.198 |
| 228 | 5.24 | 43,818 | 0.202 |
| 262 | 5.40 | 51,344 | 0.206 |
| 287 | 5.49 | 55,973 | 0.209 |
| 336 | 6.06 | 77,358 | 0.217 |
| 172 | 5.99 | 36,731 | 0.219 |
| 159 | 5.60 | 35,150 | 0.235 |
| 104 | 5.39 | 30,112 | 0.239 |
| 167 | 5.46 | 35,627 | 0.242 |
| 150 | 4.85 | 35,300 | 0.244 |
| 186 | 5.31 | 38,657 | 0.256 |
| 109 | 6.23 | 31,088 | 0.270 |
| 122 | 6.27 | 32,303 | 0.273 |
| 121 | 5.05 | 32,280 | 0.279 |
| 215 | 5.22 | 42,005 | 0.280 |
| 292 | 5.68 | 57,306 | 0.282 |
| 274 | 5.20 | 52,815 | 0.292 |
| 185 | 5.60 | 37,389 | 0.294 |
| 148 | 5.84 | 34,827 | 0.304 |
| 191 | 5.49 | 38,767 | 0.306 |
| 115 | 5.80 | 31,780 | 0.311 |
| 96 | 6.66 | 29,846 | 0.329 |
| 348 | 4.63 | 93,782 | 3.16 |
| 340 | 5.37 | 85,259 | 3.32 |
| 296 | 6.83 | 57,848 | 3.33 |
| 349 | 4.54 | 93,040 | 3.48 |
| 281 | 6.43 | 54,928 | 3.61 |
| 295 | 6.68 | 58,487 | 3.61 |
| 226 | 6.12 | 44,861 | 3.72 |
| 222 | 4.54 | 42,726 | 4.11 |
| 343 | 4.93 | 89,953 | 4.28 |
| 345 | 5.07 | 87,836 | 4.28 |
| 246 | 5.93 | 48,676 | 4.30 |
| 347 | 4.71 | 92,488 | 4.34 |
| 341 | 5.22 | 86,281 | 4.61 |
| 346 | 4.82 | 90,850 | 4.62 |
| 225 | 4.39 | 43,407 | 5.15 |
| 210 | 6.20 | 41,975 | 5.54 |
| 229 | 4.26 | 44,024 | 7.16 |
| 233 | 4.15 | 45,002 | 8.41 |
| 20b | 6.19 | 21,225 | – |
| 22b | 6.68 | 21,243 | – |
| 192b | 4.21 | 42,905 | – |
| 195b | 4.37 | 42,243 | – |
| 197b | 4.28 | 42,557 | – |
| 205b | 4.85 | 44,630 | – |
| 238b | 6.43 | 49,488 | – |
| 262b | 4.45 | 53,826 | – |
| 266b | 4.41 | 53,975 | – |
| 269b | 4.50 | 53,310 | – |
Proteins found in normal mice but not in tumor-bearing mice, sorted according to ID number in ascending order.
Proteins found in tumor-bearing mice but not in normal mice, sorted according to ID number in ascending order. All other proteins are sorted according to the fold-change of protein levels (tumor/normal) in ascending order. MW, molecular weight.
Table III.
Differentially expressed proteins between the SBPS-treated and tumor-bearing mice with a fold-change of >3 or <0.3.
| ID | Protein isoelectric point | MW (Da) | Fold-change (SPBS vs. tumor) |
|---|---|---|---|
| 12a | 4.66 | 15,103 | – |
| 16a | 4.64 | 19,473 | – |
| 20a | 6.19 | 21,225 | – |
| 22a | 6.68 | 21,243 | – |
| 100a | 4.56 | 34,106 | – |
| 106a | 4.49 | 34,385 | – |
| 138a | 5.92 | 37,999 | – |
| 205a | 4.85 | 44,630 | – |
| 238a | 6.43 | 49,488 | – |
| 260a | 6.10 | 53,604 | – |
| 213 | 6.69 | 46,770 | 0.271 |
| 266 | 4.41 | 53,975 | 3.490 |
| 268 | 5.20 | 54,874 | 3.690 |
| 93 | 5.12 | 33,111 | 3.740 |
| 305 | 5.07 | 61,268 | 3.760 |
| 144 | 6.52 | 38,538 | 4.190 |
| 239 | 5.76 | 50,451 | 4.510 |
| 18b | 6.64 | 49,338 | – |
Proteins found in tumor-bearing mice but not in SBPS-treated mice, sorted according to the ID no. in ascending order.
Proteins found in SBPS-treated mice but not in tumor-bearing mice. All other proteins are sorted according to the fold-change of protein levels (tumor/normal) in ascending order. SBPS, Scutellaria barbata polysaccharides; MW, molecular weight.
Table IV.
Serum proteins differentially expressed between the normal and tumor-bearing mice but rescued by SBPS treatment.
| Fold-change | |||||||
|---|---|---|---|---|---|---|---|
| ID | PI | MW (Da) | Normal | Tumor | SBPS | Expression level (tumor vs. normal) | Fold-change (SPBS vs. normal) |
| 257a | 6.42 | 50,071 | 1,354.16 | – | 318.46 | – | 0.235 |
| 232a | 5.58 | 45,714 | 675.51 | – | 559.23 | – | 0.828 |
| 33a | 5.73 | 15,799 | 623.64 | – | 137.48 | – | 0.220 |
| 176a | 5.25 | 37,256 | 417.18 | – | 567.86 | – | 1.360 |
| 310 | 4.25 | 61,307 | 1,109.01 | 85.52 | 2,060.76 | 0.0771 | 1.860 |
| 218 | 5.10 | 41,945 | 847.48 | 145.76 | 444.41 | 0.172 | 0.524 |
| 150 | 4.85 | 35,300 | 435.68 | 81.82 | 307.92 | 0.188 | 0.707 |
| 283 | 6.13 | 55,710 | 668.29 | 126.31 | 1,830.14 | 0.189 | 2.740 |
| 196 | 6.38 | 39,969 | 1,319.16 | 299.31 | 1,261.29 | 0.227 | 0.956 |
| 198 | 6.70 | 40,396 | 306.08 | 116.53 | 1,047.45 | 0.381 | 3.420 |
| 325 | 4.90 | 71,029 | 187.60 | 94.61 | 357.99 | 0.504 | 1.910 |
| 296 | 6.83 | 57,848 | 761.96 | 1,954.87 | 583.35 | 2.57 | 0.766 |
| 349 | 4.54 | 93,040 | 539.96 | 1,444.62 | 187.35 | 2.68 | 0.347 |
| 234 | 6.65 | 46,292 | 263.39 | 1,485.30 | 404.69 | 5.64 | 1.540 |
| 205b | 4.85 | 44,630 | – | 138.75 | – | – | – |
| 20b | 6.19 | 21,225 | – | 696.90 | – | – | – |
| 22b | 6.68 | 21,243 | – | 1,382.00 | – | – | – |
| 238b | 6.43 | 49,488 | – | 2,139.15 | – | – | – |
Proteins found in normal mice but not in tumor-bearing mice, sorted in descending order of protein levels in the serum of normal mice.
Proteins found in tumor-bearing mice but not in normal mice, sorted in ascending order of protein levels in the serum of tumor-bearing mice. All the other proteins are sorted according to the fold-change of protein levels (tumor/normal) in ascending order. SBPS, Scutellaria barbata polysaccharides; MW, molecular weight.
MALDI-TOF MS analysis
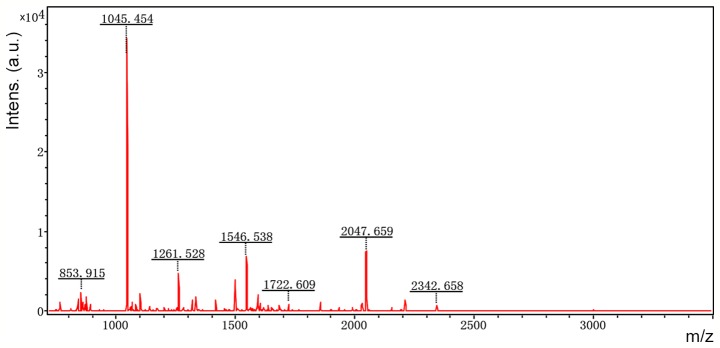
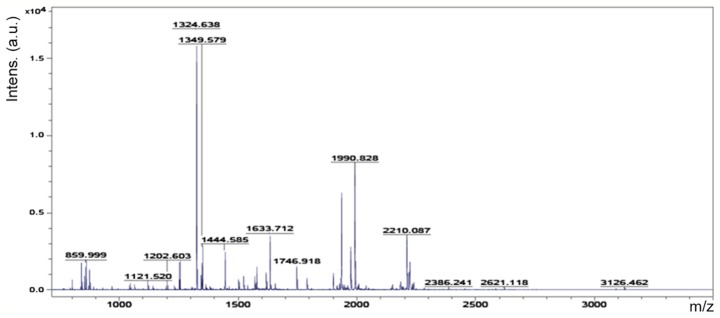
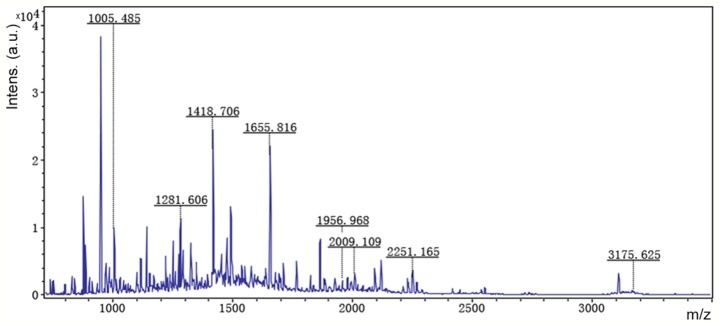
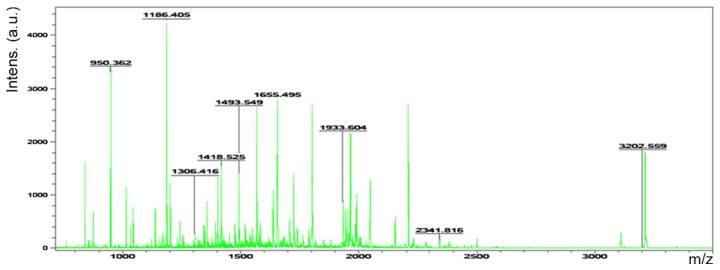
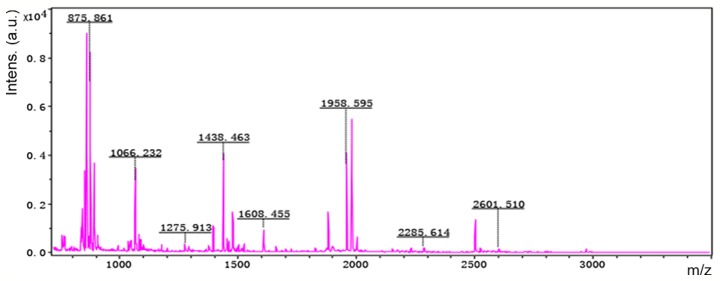
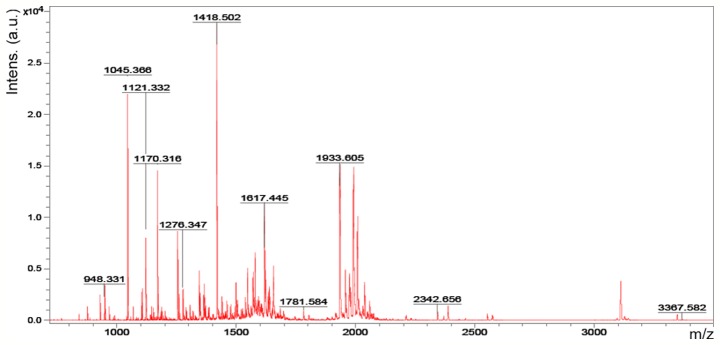
Accordingly, six representative protein spots (150, 196, 234, 296, 232 and 238) were selected for further analysis. Through MALDI-TOF MS analysis, the respective PMFs of spots 150 (Fig. 2), 196 (Fig. 3), 234 (Fig. 4), 296 (Fig. 5), 232 (Fig. 6) and 238 (Fig. 7) were obtained. The six protein spots were identified in the NCBI database (Table V). Taken together, the results showed that pseudouridine synthase 1 (Pus1p; spot 234) and chain A of the signal recognition particle Alu RNA-binding heterodimer (Srp9/14; spot 296) were increased in the serum of H22 hepatoma-bearing mice, and both were reduced by SBPS treatment. In addition, mitochondrial ribosomal protein L24 (MRPL24; spot 232) was absent from the serum of H22 hepatoma-bearing mice, and this was restored approximately to the normal level by SBPS treatment.
Figure 2.
Peptide mass fingerprint of protein spot 150.
Figure 3.
Peptide mass fingerprint of protein spot 196.
Figure 4.
Peptide mass fingerprint protein spot 234.
Figure 5.
Peptide mass fingerprint of protein spot 296.
Figure 6.
Peptide mass fingerprint of protein spot 232.
Figure 7.
Peptide mass fingerprint of protein spot 238.
Table V.
Mass spectrometry results of differentially expressed protein spots.
| ID | Protein name | gi | MS score | Peptides matched (n) | Predicted MW (Da) | Protein isoelectric point | Sequence coverage (%) |
|---|---|---|---|---|---|---|---|
| 150 | Unnamed | gi|26352025 | 85 | 24 | 40,885 | 9.22 | 52 |
| 196 | LOC101056021 | gi|407261771 | 87 | 8 | 10,789 | 11.87 | 89 |
| 234 | Pseudouridine synthase 1, isoform CRA_b | gi|148688057 | 82 | 21 | 51,127 | 9.10 | 44 |
| 296 | Chain A, signal recognition particle Alu RNA binding heterodimer, Srp9/14 | gi|157829621 | 73 | 21 | 26,560 | 9.64 | 70 |
| 232 | Mitochondrial ribosomal protein L24, isoform CRA_b | gi|148683388 | 66 | 11 | 27,036 | 9.49 | 49 |
| 238 | mCG16645, isoform CRA_b | gi|148703223 | 77 | 27 | 51,801 | 5.47 | 43 |
gi, GenInfo identifier; MW, molecular weight.
Discussion
In the present study, a hepatoma mouse model was established to evaluate whether SBPS had an antitumor effect on H22 hepatoma in vivo and to determine how SBPS treatment affected the serum proteome of tumor-bearing mice. The study found that SBPS at different dosages inhibited hepatoma growth. Differentially expressed proteins were identified on comparing the serum proteome of the healthy control mice, tumor-bearing mice and SBPS-treated tumor-bearing mice. Accordingly, several abnormally expressed serum proteins in the tumor-bearing mice were rescued by SBPS administration, indicating the antitumor effect of SBPS. In the present study, the results showed that the inhibitory effects of SBPS on the tumors were not dose-dependent. The tumor inhibition rate was lower in the high-dose group than that in the moderate-dose group. It may be that excessive doses of drugs increase the burden on the liver and kidney, and may have a negative impact on the anticancer mechanisms in the body, such as immune function. Even if the dose-limiting toxicity has been examined in a previous study, additional toxicity testing of SBPS is required in order to determine the exact mechanisms of action.
Other than the direct inhibition of cancer cell proliferation, SBPS also showed antioxidant effects in vitro and anti-angiogenic and immune-regulation effects in vivo, which may systematically regulate the metabolism in patients with cancer or tumor-bearing animals. Therefore, proteomic analysis is suitable for the comprehensive evaluation of serum proteins, which can reflect the multifaceted effects of SBPS on hepatoma mice. SBPS inhibits tumor growth via different molecular pathways in various types of cancers, indicating the cancer type-dependent effects of SBPS. Seven differentially expressed proteins were identified between healthy control mice, tumor-bearing mice and SBPS-treated tumor-bearing mice were in the literature, including three functional proteins relevant to solid tumors, three proteins with unknown functions and an unnamed protein.
PUS enzymes catalyze the site-specific isomerization of uridine to generate pseudouridine (PU) (20). Elevated PU excretion has been detected in different diseases, particularly in the urine of patients with advanced cancer (21,22). Previously, the positivity of urinary and serum PU in patients with hepatoma was reported to be 71.3% and 70.0%, respectively, and PU levels were reduced to normal levels following tumor resection (23). The present study showed that the serum level of Pus1p was low in normal mice, increased in tumor-bearing mice, but restored to a low level in SBPS-treated tumor-bearing mice. This may partially explain why PU is abnormally increased in patients with hepatoma, as Pus1p catalyzes the generation of PU. Serum Pus1p may be of clinical significance in the diagnosis and monitoring of primary liver cancer. However, the way in which SBPS inhibits the expression or secretion of Pus1p requires investigation in the future.
The heterodimeric protein complex SRP9/SRP14, as a component of the SRP, binds to 7SL RNA or cytoplasmic Alu RNA to form a complex known as Alu RNP (24). In these two forms, the SRP9/14 dimer mainly delays peptide elongation and possibly inhibits the initiation of protein synthesis (25). Reports have indicated the involvement of SRP9/14 in cancer development. For example, Rho et al combined 2-D PAGE and tandem mass spectrometry to identify five differentially expressed proteins between normal colon and colon cancer tissues. SRP9 was one of the upregulated proteins in cancer tissues, which was also confirmed by western blot analysis and immunohistochemistry, suggesting the upregulation of SRP9 is a candidate biomarker of colon cancer (26). An early study revealed that SRP9 was one of the upregulated genes in Chinese patients with hepatocellular carcinoma (27). Consistently, the expression level of SRP9 was found to be increased in tumor tissues. Regulation of the expression of SRP9/14 in liver cancer remains to be elucidated; however, alterations in the expression levels of SRP9/14 indicate that SBPS regulates specific protein synthesis within the tumor or in the whole body of tumor-bearing mice.
MRPL24 is one of the proteins encoded by nuclear genes (28). It facilitates the specific requirements of protein synthesis in mitochondria (29). In the present study, MRPL24 was expressed in healthy control mice but absent in tumor-bearing mice, suggesting that this protein was important in the process of tumorigenesis. This suggests that MRPL24 may be a novel target for the diagnosis of liver cancer.
In conclusion, the antitumor effect of SBPS was confirmed in the hepatoma mouse model and seven differentially expressed serum proteins were identified using proteomic analysis in mice with/without SBPS treatment. These differentially expressed proteins supported the antitumor mechanism of SBPS. They may provide valuable clues for the pathogenesis, diagnosis and treatment of liver cancer.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- SBPS
Scutellaria barbata polysaccharides
- MALDI
matrix-assisted laser (induced) desorption ionization
- NCBI
National Center for Biotechnology Information
- IPG
immobilized pH gradient
- PMF
peptide mass fingerprints
- PUS
pseudouridine synthases
- PU
pseudouridine
- SRP9
signal recognition particle 9
- MRPL24
mitochondrial ribosomal protein L24
Funding
This study was supported by grants from the National Natural Science Foundation Emergency Management Project (grant no. 81441113), the Study on the Effect of PAHs on the Leydig Cells of Offspring Rats in Pregnancy and its Mechanism (grant no. 2015/01-2015/12), the Research Project of the Health and Family Planning Commission of Heilongjiang Province (grant no. 2017–582) and the Regulative Effect of MicroRNA on Gene Expression of k-ras in Colorectal Cancer (grant no. 2017/1-2018/12).
Availability of data and materials
The dataset supporting the results of this study are included within the article.
Authors' contributions
GS and LL conceived and supervised the study; GS and LL designed experiments; LL, XX and LW performed experiments; HZ completed the animal experiments and blood sample collection; ZH developed new software and performed simulation experiments; BZ and YC analyzed data; LL, XX and LW wrote the manuscript; GS and LL made manuscript revisions. All authors reviewed the results and approved the final version of the manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Mudanjiang Medical University (Mudanjiang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Yu J, Lei J, Yu H, Cai X, Zou G. Chemical composition and antimicrobial activity of the essential oil of Scutellaria barbata. Phytochemistry. 2004;65:881–884. doi: 10.1016/j.phytochem.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Dai ZJ, Wang XJ, Li ZF, Ji ZZ, Ren HT, Tang W, Liu XX, Kang HF, Guan HT, Song LQ. Scutellaria barbate extract induces apoptosis of hepatoma H22 cells via the mitochondrial pathway involving caspase-3. World J Gastroenterol. 2008;14:7321–7328. doi: 10.3748/wjg.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai ZJ, Wu WY, Kang HF, Ma XB, Zhang SQ, Min WL, Kang WF, Ma XB, Zhang SQ, Min WL, et al. Protective effects of Scutellaria barbata against rat liver tumorigenesis. Asian Pac J Cancer Prev. 2013;14:261–265. doi: 10.7314/APJCP.2013.14.1.261. [DOI] [PubMed] [Google Scholar]
- 4.Kan X, Zhang W, You R, Niu Y, Guo J, Xue J. Scutellaria barbata D. Don extract inhibits the tumor growth through down-regulating of Treg cells and manipulating Th1/Th17 immune response in hepatoma H22-bearing mice. BMC Complement Altern Med. 2017;17:41. doi: 10.1186/s12906-016-1551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao J, Lu WF, Dai ZJ, Lin S, Zhao Y, Li S, Zhao NN, Wang XJ, Kang HF, Ma XB, Zhang WG. Induction of apoptosis by total flavonoids from Scutellaria barbata D. Don in human hepatocarcinoma MHCC97-H cells via the mitochondrial pathway. Tumour Biol. 2014;35:2549–2559. doi: 10.1007/s13277-013-1336-4. [DOI] [PubMed] [Google Scholar]
- 6.Dai ZJ, Wang BF, Lu WF, Wang ZD, Ma XB, Min WL, Kang HF, Wang XJ, Wu WY. Total flavonoids of Scutellaria barbata inhibit invasion of hepatocarcinoma via MMP/TIMP in vitro. Molecules. 2013;18:934–950. doi: 10.3390/molecules18010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye CL, Huang Q. Extraction of polysaccharides from herbal Scutellaria barbata D. Don (Ban-Zhi-Lian) and their antioxidant activity. Carbohydr Polym. 2012;89:1131–1137. doi: 10.1016/j.carbpol.2012.03.084. [DOI] [PubMed] [Google Scholar]
- 8.Yang X, Yang Y, Tang S, Tang H, Yang G, Xu Q, Wu J. Anti-tumor effect of polysaccharides from Scutellaria barbata D. Don on the 95-D xenograft model via inhibition of the C-met pathway. J Pharmacol Sci. 2014;125:255–263. doi: 10.1254/jphs.13276FP. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Yang G, Hou G, Liu Q, Hu W, Zhao PU, He YI. Scutellaria barbata D. Don polysaccharides inhibit the growth of Calu-3 ×enograft tumors via suppression of the HER2 pathway and angiogenesis. Oncol Lett. 2015;9:2721–2725. doi: 10.3892/ol.2015.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun P, Sun D, Wang X. Effects of Scutellaria barbata polysaccharide on the proliferation, apoptosis and EMT of human colon cancer HT29 cells. Carbohydr Polym. 2017;167:90–96. doi: 10.1016/j.carbpol.2017.03.022. [DOI] [PubMed] [Google Scholar]
- 11.Zhang AH, Sun H, Yan GL, Han Y, Wang XJ. Serum proteomics in biomedical research: A systematic review. Appl Biochem Biotechnol. 2013;170:774–786. doi: 10.1007/s12010-013-0238-7. [DOI] [PubMed] [Google Scholar]
- 12.Ng EW, Wong MY, Poon TC. Advances in MALDI mass spectrometry in clinical diagnostic applications. Top Curr Chem. 2014;336:139–175. doi: 10.1007/128_2012_413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao XL, Li H, Yu XL, Liang P, Dong BW, Fan J, Li M, Liu FY. Predicting early intrahepatic recurrence of hepatocellular carcinoma after microwave ablation using SELDI-TOF proteomic signature. PLoS One. 2013;8:e82448. doi: 10.1371/journal.pone.0082448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen S, Peng H, Wang Y, Xu M, Lin M, Xie X, Peng B, Kuang M. Screening for immune-potentiating antigens from hepatocellular carcinoma patients after radiofrequency ablation by serum proteomic analysis. BMC Cancer. 2018;18:117. doi: 10.1186/s12885-018-4011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Gu L, Zhong Y, Chen Y, Zhang L, Zhang AR, Sobol RW, Chen T, Li J. Administration of polysaccharide from Panax notoginseng prolonged the survival of H22 tumor-bearing mice. Onco Targets Ther. 2016;9:3433–3441. doi: 10.2147/OTT.S79427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song G, Wang G, Liu H. Experimental study of Scutellaria barbata polysaccharide on synergistic and attenuation effects of on cyclophosphamide. Tradit Chinese Med Inf. 2010;27:107–109. (In Chinese) [Google Scholar]
- 17.Song G, Yu Y, Wang X. Experiments on antitumor activity of Scutellaria barbata polysaccharides and its immunological mechanisms. World Sci Technol Mod Trad Chinese Med. 2011;13:641–643. (In Chinese) [Google Scholar]
- 18.Yu Y, Zhou Q, Yun K. Effects of Scutellaria barbata polysaccharide combined with adjuvant chemotherapy drugs on serum TNF-α and VEGF expression in mice. Tradit Chinese Med Inf. 2010;27:29–31. (In Chinese) [Google Scholar]
- 19.Xu X, Wu L, Li L, et al. Effects of Scutellaria barbata polysaccharide on tissue protein in H22 tumor bearing mice. J Chinese Gerontol. 2016;36:4709–4710. (In Chinese) [Google Scholar]
- 20.Hamma T, Ferre-D'Amare AR. Pseudouridine synthases. Chem Biol. 2006;13:1125–1135. doi: 10.1016/j.chembiol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Waalkes TP, Gehrke CW, Zumwalt RW, Chang SY, Lakings DB, Tormey DC, Ahmann DL, Moertel CG. The urinary excretion of nucleosides of ribonucleic acid by patients with advanced cancer. Cancer. 1975;36:390–398. doi: 10.1002/1097-0142(197508)36:2<390::AID-CNCR2820360214>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 22.Rasmuson T, Bjork GR. Urinary excretion of pseudouridine and prognosis of patients with malignant lymphoma. Acta Oncol. 1995;34:61–67. doi: 10.3109/02841869509093640. [DOI] [PubMed] [Google Scholar]
- 23.Tu Z, Xu S, Wu M. Clinical value of urinary and serum pseudouridine in diagnosis and monitoring of primary liver cancer. Chin Med J (Engl) 1995;108:204–208. [PubMed] [Google Scholar]
- 24.Chang DY, Hsu K, Maraia RJ. Monomeric scAlu and nascent dimeric Alu RNAs induced by adenovirus are assembled into SRP9/14-containing RNPs in HeLa cells. Nucleic Acids Res. 1996;24:4165–4170. doi: 10.1093/nar/24.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger A, Ivanova E, Gareau C, Scherrer A, Mazroui R, Strub K. Direct binding of the Alu binding protein dimer SRP9/14 to 40S ribosomal subunits promotes stress granule formation and is regulated by Alu RNA. Nucleic Acids Res. 2014;42:11203–11217. doi: 10.1093/nar/gku822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rho JH, Qin S, Wang JY, Roehrl MH. Proteomic expression analysis of surgical human colorectal cancer tissues: UP-regulation of PSB7, PRDX1, and SRP9 and hypoxic adaptation in cancer. J Proteome Res. 2008;7:2959–2972. doi: 10.1021/pr8000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Zhu X, Zhu J, Liao S, Tang Q, Liu K, Guan X, Zhang J, Feng Z. Identification of differential expression of genes in hepatocellular carcinoma by suppression subtractive hybridization combined cDNA microarray. Oncol Rep. 2007;18:943–951. [PubMed] [Google Scholar]
- 28.Sylvester JE, Fischel-Ghodsian N, Mougey EB, O'Brien TW. Mitochondrial ribosomal proteins: Candidate genes for mitochondrial disease. Genet Med. 2004;6:73–80. doi: 10.1097/01.GIM.0000117333.21213.17. [DOI] [PubMed] [Google Scholar]
- 29.Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V. Ribosome. The structure of the human mitochondrial ribosome. Science. 2015;348:95–98. doi: 10.1126/science.aaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset supporting the results of this study are included within the article.