Abstract
Increased expression of CCL18 has been observed in various malignancies and in the urine samples of patients with bladder cancer (BC). However, the roles of CCL18 in the development, progression and metastasis of BC remain unclear. The present study demonstrated that CCL18 expression was significantly associated with advanced clinical stages of BC. Furthermore, exogenous CCL18 promoted cell invasion and migration, and induced cell epithelial-mesenchymal transition (EMT) in BC cells. Western blotting demonstrated that E-cadherin, an epithelial marker, was decreased, whereas matrix metalloproteinase (MMP)-2 and vascular endothelial growth factor (VEGF)-C were increased in CCL18-treated cells. Blocking CCR8 via a small molecule inhibitor or short hairpin (sh)RNA mitigated the decrease in E-cadherin, and increase in MMP-2 and VEGF-C, caused by human recombinant (r)CCL18. CCR8 knockdown by shRNA reversed rCCL18-induced cancer cell invasion, migration and EMT. In conclusion, these data suggested that CCL18 may promote migration, invasion and EMT by binding CCR8 in BC cells. Inhibition of CCL18 activity by blocking CCR8 could be a potential therapeutic strategy for preventing the progression of BC.
Keywords: bladder cancer, CCL18, CCR8
Introduction
Bladder cancer (BC) is the fourth most common type of cancer in men in the United States and is responsible for ~150,000 cases of mortality worldwide; therefore, it is considered an important health problem (1,2). Approximately 75% of patients have non-muscle-invasive BC (NMIBC) (Ta, T1) and 25% have muscle-invasive BC (≥T2). An important clinical issue is the high rate of recurrence associated with NMIBC. Although radical cystectomy and pelvic lymphadenectomy can be performed, a high percentage of patients progress to advanced invasive tumors. Tumors at a high risk for recurrence and progression require neoadjuvant treatment (3).
Chemokine (C-C motif) ligand (CCL)18 is a C-C chemokine that is highly expressed in human lung tissues and plasma; CCL18 serves numerous functions in immune modulation and cancer progression (4). Recent studies have reported that increased expression levels of CCL18 are observed in various malignancies, including ovarian cancer, lung cancer, oral squamous cell cancer and breast cancer (5–8). It has also been suggested that CCL18 acts as a potential urinary biomarker in BC (9,10); however, the molecular mechanisms underlying the effects of CCL18 on BC remain unknown. C-C motif receptor 8 (CCR8) is a G protein-coupled receptor, which is known to be expressed by immune cells, including T-helper 2 lymphocytes, natural killer cells, and monocytes (11). In humans, CCR8 is selectively activated by CCL1/CC chemokine I-309 (12). Recently, Islam et al reported that CCR8 is a functional receptor for CCL18 (13).
The present study revealed that CCL18 promoted the migration and invasion of BC cells. However, the underlying molecular mechanisms by which CCL18 induces epithelial-mesenchymal transition (EMT) in BC cells and contributes to cancer cell invasion have not been identified. In addition, the present study demonstrated that excessive expression of CCL18 in tumor tissues was associated with tumor stage and poor prognosis in patients with BC. Furthermore, the present data indicated that CCL18 may promote migration, invasion and EMT through the G protein-coupled receptor chemokine CCR8.
Materials and methods
Gene expression profiling
Expression of CCL18 in BC (BLCA dataset) was checked using The Cancer Genome Atlas (TCGA) data. TCGA, launched by the National Institutes of Health (NIH, Bethesda, MD, USA), is a publicly funded project that comprises a comprehensive ‘atlas’ of cancer genomic profiles. The gene expression levels of CCL18 in 404 patients with BC compared with 28 normal samples from TCGA database and GTEx were analyzed using the GEPIA web server (14). The Pathology Atlas from the Human Protein Atlas (www.proteinatlas.org/pathology) was used to perform analyses based on the mRNA expression levels of CCL18 in BC tissue and the clinical outcomes (survival). The data in the Pathology Atlas are based on the integration of publicly available data from TCGA. In addition, gene expression profiling studies involving several clinical samples were performed to analyze the expression of CCL18 in a dataset available through the Gene Expression Omnibus (GSE31684; http://www.ncbi.nlm.nih.gov/geo/) (15).
Patients and tissue samples
The present study was approved by the Research Ethics Committee of the First Affiliated Hospital, Nanchang University (Nanchang, China). Informed consent was obtained from all patients. All specimens were handled and anonymized according to ethical and legal standards. Immunohistochemical analysis of CCL18 was conducted using 64 primary BC tissues and 30 adjacent noncancerous bladder tissues. Detailed information on the clinical features of all patients in this study is presented in Table I.
Table I.
Association of CCL18 expression with the clinicopathological characteristics of bladder cancer.
| CCL18 expression levels in tumor tissues | |||
|---|---|---|---|
| Parameter | High (Grade ≥2) (n=37) | Low (Grade <1) (n=27) | P-value |
| Agea, mean ± SD | 62.7±9.1 | 64.6±8.8 | 0.403 |
| Sex, n (%)b | |||
| Female | 6 (54.5) | 5 (45.5) | 0.809 |
| Male | 31 (58.5) | 22 (41.5) | |
| Tumor stage, n (%)c | 0.018 | ||
| T1 | 10 (47.6) | 11 (52.4) | |
| T2 | 10 (43.5) | 13 (56.5) | |
| T3 | 9 (75.0) | 3 (25.0) | |
| T4 | 8 (100.0) | 0 (0.0) | |
| Tumor grade, n (%)a | 0.447 | ||
| Low grade | 12 (48.0) | 13 (52.0) | |
| High grade | 25 (64.1) | 14 (35.9) | |
Age data were analyzed by a
Student's t-test. Other data were analyzed using.
Fisher's exact test (two-sided) or
χ2 Pearson (two-sided) test.
Cell culture and treatment
The 5637 human BC cell line was purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences, Shanghai Institute of Cell Biology (Shanghai, China) and was maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Media were supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in a humidified incubator containing 5% CO2. The small molecule CCR8 inhibitor R243 (cat. no. AOB2014) was purchased from AOBIOUS, Inc. (Gloucester, MA, USA). For chemokine treatment, 5637 cells pretreated with or without R243 (5 µM) for 6 h at 37°C/5% carbon dioxide (16), were exposed to 50 or 100 ng/ml CCL18 (PeproTech, Inc., Rockville, MD, USA) for 36 h.
Short hairpin (sh)RNA transfection
For transfection, 5637 cells were plated in 6-well plates at 2×105/well. The human CCR8 shRNA plasmid and non-target shRNA (NT shRNA) were obtained from Invitrogen; Thermo Fisher Scientific, Inc. The targeted sequences were: Human CCR8 (5′-CCGGGGATTATACACTTGACCTCAGTGTCGACACTGAGGTCAAGTGTATAATCCTTTTTTG-3′) and non-target (NT) shRNA (5′-CCGGCAACAAGATGAAGAGCACAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT-3′). Once the 5637 cells reached 50–60% confluence, 1.25 µg/ml CCR8 shRNA and NT shRNA were transfected using Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and Opti-MEM medium (Gibco; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. A total of 6 h post-transfection, the medium was replaced with fresh medium containing 10% FBS. NT shRNA was used as the negative control.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
A total of 24, 48 and 72 h post-transfection, total RNA was extracted from 5637 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was synthesized using a Takara PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan) according to manufacturer's protocols. qPCR was performed using SYBR Premix Ex Taq kit (Takara Bio, Inc., Otsu, Japan) in the ABI PRISM® 7500 real-time PCR system (Thermo Fisher Scientific, Inc.). The thermocycling conditions were initial denaturation (1 cycle, 95°C, 30 sec), PCR reaction (40 cycles, 95°C, 5 sec, and 60°C, 30 sec for annealing and elongation). The data of RT-qPCR were normalized against an internal control β-actin and relative expression levels were evaluated using the 2−∆∆Cq method and then expressed as fold changes (17). The oligonucleotide sequences of the RT-qPCR primers are listed in Table II.
Table II.
Oligonucleotide sequences for polymerase chain reaction amplification.
| Gene | NCBI no. | Sequence (5′-3′) | Product size (bp) |
|---|---|---|---|
| CCR8 | NM_005201.3 | F: GTGTGACAACAGTGACCGACT | 173 |
| R: CTTCTTGCAGACCACAAGGAC | |||
| E-cadherin | NM_001317186.1 | F: AGCTGCCCAGAAAATGAAAAAGG | 203 |
| R: GTGTATGTGGCAATGCGTTCTC | |||
| MMP-2 | NM_001302510.1 | F: CCTCTCCACTGCCTTCGATA | 129 |
| R: TGGGAGGAGTACAGTCAGCA | |||
| VEGF-C | NM_005429.2 | F: GGCTGGCAACATAACAGAGAA | 159 |
| R: CCCCACATCTATACACACCTCC | |||
| β-actin | NM_001101.3 | F: CATGTACGTTGCTATCCAGGC | 250 |
| R: CTCCTTAATGTCACGCACGAT |
CCR8, chemokine (C-C motif) receptor 8; F, forward; MMP-2, matrix metalloproteinase-2; NCBI, National Center for Biotechnology Information; R, reverse; VEGF-C, vascular endothelial growth factor-C.
Western blot analysis
Total proteins were extracted from cells using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China). Proteins were quantified using a Pierce bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.), followed by western blot analysis. 40 µg proteins from cell lysates were subjected to 12% SDS-PAGE. Once proteins were transferred to nitrocellulose membranes, they were incubated with antibodies targeting β-actin (cat. no. 58169, 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), CCR8 (cat. no. ab32131, 1:500; Abcam, Cambridge, UK), E-cadherin (cat. no. 3195, 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), matrix metalloproteinase (MMP)-2 (cat. no. 40994, 1:1,000; Cell Signaling Technology, Inc.) and vascular endothelial growth factor (VEGF)-C (cat. no. 2445, 1:1,000; Cell Signaling Technology, Inc.), followed by incubation with a horseradish peroxidase-labeled goat anti-rabbit secondary antibody (cat. no. ab6721, 1:3,000; Abcam) for 1.5 h at room temperature. Protein bands were visualized by Millipore enhanced chemiluminescence (cat. no. WBKLS0500; EMD Millipore, Billerica, MA, USA). ImageJ 1.45 software (NIH) was used to perform densitometric analysis of each band.
Immunohistochemistry
Tissue sections from paraffin- embedded bladder cancer were dewaxed in xylene and rehydrated in a graded alcohol series. Following microwave-induced antigen retrieval (Tris-EDTA pH 9.0), the slides were washed with PBS and incubated with primary antibody against CCL18 (cat. no. ab104867, 1:200; Abcam) overnight at 4°C. The sections were incubated with secondary antibody at room temperature. for 30 min after washing with PBST. Color detection was performed by liquid DAB+ substrate chromogen system (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) according to the manufacturer's protocols. Slides were counterstained with hematoxylin. Hemalum was used to stain nuclei of cells blue at room temperature for 20 sec. Immunohistochemical images of CCL18 expression were analyzed according to a previous study (18). Representative areas with 5 or 20 CCL18 cells per ×400 high-power field were chosen and images were captured randomly as references for grading. The densities of CCL18+ cells were graded as follows: Grade 1, <5 positive cells/microscopic field; grade 2, 5–20 cells/field; and grade 3, >20 cells/field. Grade 1 was considered to be low-level CCL18+, and grades 2 and 3 were considered to be high-level CCL18+.
Cell migration and Transwell assays
Cell migration was determined using a wound-healing assay. Briefly, 5637 cells were seeded in six-well culture dishes and cultured at 37°C to form a confluent monolayer. After transfection with or without CCR8 shRNA for 24 h, and a wound was made by scratching the monolayer with a 10 µl pipette tip. The wounded monolayer was then washed three times with PBS to remove cell debris and treated with CCL18 for 24 h at 37°C. After scratching, the area of the cell-free scratch was imaged under an Olympus CKX41 inverted microscope (Olympus Corporation, Tokyo, Japan) at 0 and 24 h. Cell invasive capacities were measured using a Transwell assay (Corning Incorporated, Corning, NY, USA). Briefly, 5637 cells were treated with CCL18 and/or transfected with CCR8 shRNA for 24 h, and were then seeded in the upper chamber of the Transwell system. The upper chamber of the insert was precoated with 0.1 ml (300 µg/ml) Matrigel matrix (Corning Incorporated) for the invasion assay. The invaded cells on the underside of the membrane were counted. In this assay, prepared cells were seeded in the upper chamber with serum-free medium and the medium of the lower chamber was supplemented with 10% FBS as a chemoattractant. Following incubation for 24 h, the cells were fixed with 4% formaldehyde at 37°C for 30 min. Cells that did not invade through the pores were removed with a cotton swab. Cells that had invaded to the lower surface of the membrane were stained with crystal violet at 37°C for 15 min. Finally, images of five representative fields at ×100 magnification were randomly captured using a light microscope (Carl Zeiss AG, Oberkochen, Germany) and the number of cells in each well was semi-quantified.
Statistical analysis
All data are presented as the means ± standard deviation in at least three replicates per group. Statistical analysis was performed to determine the significance of the differences between groups using one-way analysis of variance (with post hoc Turkey's honest significant difference test) or Student's t-test. χ2 analysis and Fisher's exact test were used to examine the association between CCL18 expression and the clinicopathological features of patients with BC. The effects of CCL18 expression levels on survival were estimated using the Kaplan-Meier method and were compared by the log-rank test. Wilcoxon-signed rank test was used to compare CCL18 expression between 30 paired cancerous and noncancerous tissues of patients with BC. All statistical analyses were performed using GraphPad Prism 7.00 software for Windows (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Association of CCL18 expression with the clinicopathological characteristics of BC
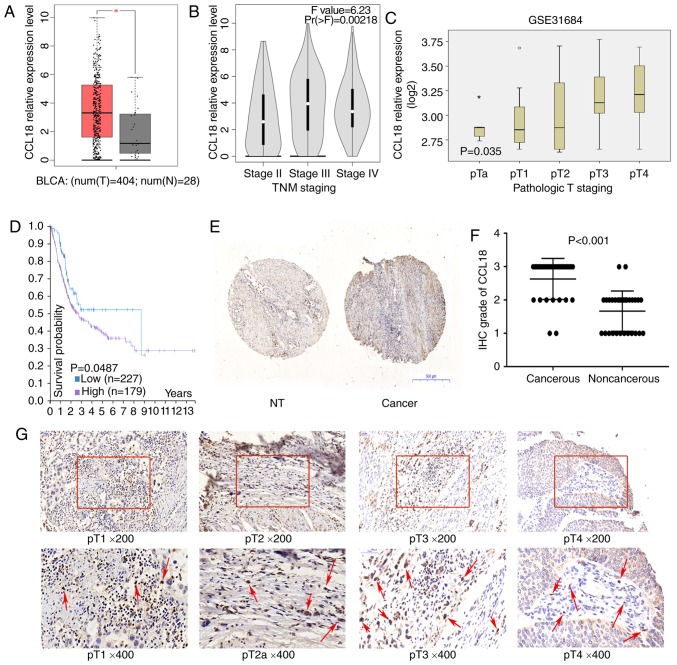
To assess whether CCL18 may be involved in the development of BC, this study first investigated the mRNA expression levels of CCL18 using TCGA dataset. As illustrated in Fig. 1A, the mRNA expression levels of CCL18 were significantly higher in BC tissues compared with in nontumor tissues. In addition, CCL18 mRNA expression was higher in BC tissues at stages III and IV compared with at stage II (Fig. 1B and C). Kaplan-Meier analysis indicated that higher CCL18 expression was associated with worse patient survival (log-rank test, P=0.0487; Fig. 1D). Subsequently, the expression of CCL18 was investigated in BC tissues using immunohistochemistry. As reported in a previous study (10), there was no epithelial staining for CCL18; however, inflammatory cells in the stroma were CCL18-positive. It was revealed that CCL18 expression was higher in the majority of cancerous samples compared with in noncancerous tissues (Fig. 1E). Furthermore, the protein expression levels of CCL18 were detected in tissues obtained from 30 patients with BC by immunohistochemical staining. The results demonstrated that CCL18 expression was significantly stronger in the majority of BC tissues compared with in paired normal tissues (Fig. 1F). As shown in Fig. 1G, CCL18 was weakly detected in stage I and II BC tissues, but was strongly detected in stage III and IV BC tissues in tissue samples employed in the present study. Subsequently, the association between CCL18 expression in BC cases and various clinical outcomes was analyzed. As shown in Table I, higher CCL18 expression was associated with higher pathological stages (P<0.05). These data suggested that CCL18 was elevated in BC tissues and may be associated with the progression of BC.
Figure 1.
CCL18 expression in bladder cancer tissues. (A) Expression of CCL18 in bladder cancer samples (red box) and adjacent tissues (grey box), *P<0.001. (B) Expression of CCL18 in TNM stage II, III and IV bladder cancer samples in The Cancer Genome Atlas BLCA dataset. (C) Expression of CCL18 in bladder cancer samples, according to the pathological stage, in the GSE31684 dataset. (D) Results of overall survival analysis in patients stratified according to CCL18 mRNA expression (Kaplan-Meier test) in the Human Protein Atlas. (E and F) IHC staining of CCL18 in 30 paired noncancerous and cancerous tissues obtained from patients with bladder cancer (magnification, ×40). (G) Protein expression levels of CCL18 in bladder tumor tissues, as observed by IHC staining. Red arrows indicate CCL18-positive cells in the stroma. Hemalum was used to stain nuclei of cells blue. CCL18 immunoreactivity in stromal cells were located in cytoplasm; no CCL18 staining was detected in the nuclei of cells. BLCA, bladder cancer; CCL18, chemokine (C-C motif) ligand 18; IHC, immunohistochemistry; NT, nontumorous.
CCL18 regulates the expression of E-cadherin, MMP2 and VEGF-C in 5637 BC cells
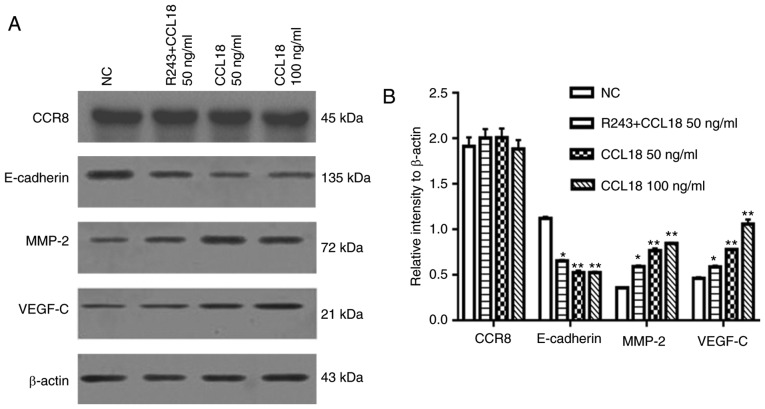
EMT is thought to serve a key role in the invasion and metastasis of numerous types of tumor (19). EMT is an evolutionarily conserved developmental program, which is believed to have a critical role in carcinogenesis and tumor metastasis by enhancing mobility and invasion. Our previous study revealed that VEGF-C, MMP-2 and MMP-9 are immunosuppressive factors, which serve important roles in BC invasion and metastasis (20). To determine whether CCL18 enhanced BC invasion and metastasis through EMT, and by altering MMP-2 and VEGF-C expression, 5637 BC cells were treated with rCCL18. Following treatment of 5637 BC cells with 50 or 100 ng/ml CCL18 for 36 h, the expression levels of E-cadherin were significantly lower, and the expression levels of MMP-2 and VEGF-C were higher compared with in the control group (Fig. 2).
Figure 2.
CCL18 affects the expression levels of E-cadherin, MMP-2 and VEGF-C. (A and B) 5637 bladder cancer cells were treated with PBS or CCL18 (50 or 100 ng/ml) for 36 h, with or without pretreatment with the CCR8 inhibitor R243. Protein expression levels of E-cadherin, MMP-2 and VEGF-C were detected in 5637 cells by western blot analysis. *P<0.05 compared with the 50 ng/ml CCL18 and NC groups; **P<0.05 compared with the NC group. CCL18, chemokine (C-C motif) ligand 18; MMP-2, matrix metalloproteinase-2; NC, normal control; VEGF-C, vascular endothelial growth factor-C.
CCR8 is required for migration and invasion of BC cells via CCL18
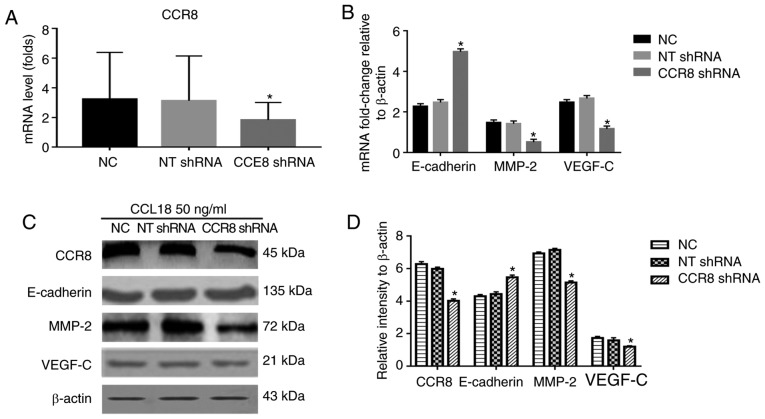
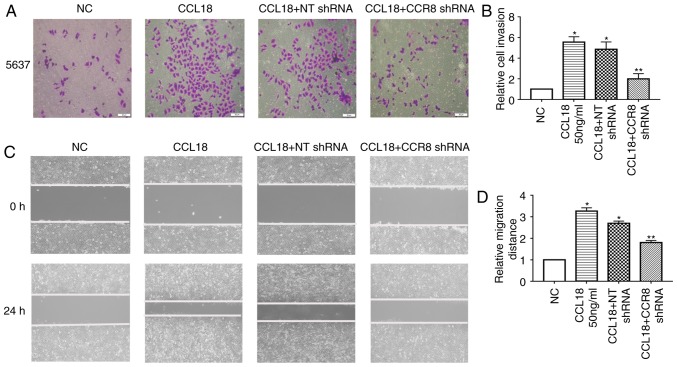
CCL18 has numerous receptors, including PITPNM family member 3 (PITPNM3) and G protein-coupled estrogen receptor 1 (GPR30), which mediate CCL18-induced migration of cancer cells, and promote tumor invasion and metastasis (8,21). Recently, Islam et al reported that CCR8 is a functional receptor for CCL18 (13). To identify whether CCR8 was associated with CCL18-induced tumor invasion and EMT, this study examined the expression levels of E-cadherin, MMP-2 and VEGF-C in 5637 BC cells treated with a small molecule CCR8 inhibitor, R243, or transfected with CCR8 shRNA, followed by CCL18 treatment (Figs. 2 and 3). As shown in Fig. 2, R243 reversed the decreased levels of E-cadherin and increased levels of MMP-2 and VEGF-C caused by CCL18, without affecting CCR8 levels. The results of RT-qPCR confirmed that the mRNA expression levels of CCR8 were downregulated by CCR8 shRNA (Fig. 3A); CCR8 protein expression was also reduced in response to CCR8 shRNA (Fig. 3D). RT-qPCR analysis revealed that the mRNA expression levels of E-cadherin were increased, whereas those of MMP-2 and VEGF-C were decreased compared with in cells transfected with NT shRNA (Fig. 3B). Similar results were revealed by western blot analysis for the protein expression levels of E-cadherin, MMP-2 and VEGF-C (Fig. 3C and D). Subsequently, the migratory and invasive abilities of BC cells were reduced in CCR8-knockdown cells compared with in control cells (Fig. 4A-D).
Figure 3.
Effects of CCR8 shRNA on CCL18-induced expression of E-cadherin, MMP-2 and VEGF-C. 5637 bladder cancer cells were treated with CCL18 (50 ng/ml), CCL18 + NT shRNA or CCL18 + CCR8 shRNA for 36 h. (A) mRNA expression levels of CCR8 after being transfected with CCR8 shRNA was examined by reverse transcription-quantitative polymerase chain reaction. β-actin was used as an internal control. (B) mRNA expression levels of CCR8, E-cadherin, MMP-2 and VEGF-C were examined by reverse transcription-quantitative polymerase chain reaction. β-actin was used as an internal control. (C and D) Protein expression levels of CCR8, E-cadherin, MMP-2 and VEGF-C in 5637 cells were detected by western blotting. β-actin was used as an internal control. *P<0.05 compared with the NC and NT shRNA groups. CCL18, chemokine (C-C motif) ligand 18; CCR8, chemokine (C-C motif) receptor 8; MMP-2, matrix metalloproteinase-2; NC, normal control; NT, non-target; shRNA, short hairpin RNA; VEGF-C, vascular endothelial growth factor-C.
Figure 4.
5637 bladder cancer cells were treated with CCL18 (50 ng/ml), CCL18 + NT shRNA or CCL18 + CCR8 shRNA for 24 h. Invasion and migration were detected by (A and B) Transwell assays and (C and D) wound healing assays, respectively (magnification, ×100). *P<0.05 compared with the NC group. **P<0.05 compared with the CCL18 + NT shRNA group. CCL18, chemokine (C-C motif) ligand 18; CCR8, chemokine (C-C motif) receptor 8; NC, normal control; NT, non-target; shRNA, short hairpin RNA.
Discussion
To the best of our knowledge, the present study is the first to demonstrate that CCL18 may be frequently overexpressed in BC. Furthermore, it was revealed that CCL18 enhanced bladder cancer cell invasion, migration and EMT. CCL18 has numerous receptors, including PITPNM3 (8) and GPR30 (21), which mediate CCL18-induced migration of cancer cells, and promote tumor invasion and metastasis. CCL18 activates proline-rich tyrosine kinase 2 and the Src kinase PITPNM3 by binding to PITPNM3, which is expressed in breast cancer cells, thereby promoting metastasis of breast cancer (8). However, the functional receptor for CCL18 that mediates BC cell invasion and migration remains unknown. Recently, Islam et al reported that CCR8 is a functional receptor for CCL18 (13). When CCL18 binds to CCR8, CCR8 is activated through G-protein signaling, inducing CCR8 internalization and activating cell chemotaxis and calcium flux in human CCR8-transfected cells. In the present study, it was further deduced that the G-protein coupled receptor CCR8 may mediate signaling by CCL18, and a potential mechanism underlying tumor regulation in BC was determined.
E-cadherin is an epithelial marker, the functional loss of which is a hallmark of EMT, which is thought to promote BC progression and metastasis (22). MMP-2 is considered a mesenchymal marker of EMT (23). In the present study, CCL18 was revealed to mediate EMT through MMP-2-dependent pathways.
MMPs are best known for their profound role in malignant transformation and metastasis via extracellular matrix disruption (24). MMP-2, which is a member of the MMP family, is associated with tumor invasion through degradation of the extracellular matrix and basement membrane. In BC, high MMP2 expression is strongly correlated with decreased survival (25). In addition, MMP2 is overexpressed at the invasive front and is associated with a higher tumor grade in BC (26).
Previous studies have reported that upregulation of the lymphangiogenic growth factor VEGF-C is positively correlated with regional lymph node metastasis and poor survival in BC (27–32). VEGF-C is a protein precursor that must be activated by a converting enzyme (proprotein convertases). VEGF-receptor 3 (VEGF-R3) is one of the receptors of VEGF-C, which promotes lymphangiogenesis, tumor cell migration, invasion and metastasis. Blocking VEGF-C signaling via either small interfering RNA or a VEGF-R3 antibody inhibits lymphatic-based metastatic spread of human malignancies (33).
Immunosuppressive factors can be secreted into the extracellular environment by tumor cells in an autocrine or paracrine fashion, resulting in deep immunosuppressive regions that are formed locally in tumor cells. Our previous study revealed that VEGF-C, MMP-2 and MMP-9, as immunosuppressive factors, serve important roles in BC invasion and metastasis (20). In the present study, it was demonstrated that the expression levels of VEGF-C and MMP-2 were elevated by CCL18 treatment. Furthermore, the mRNA expression levels of CCL18 were assessed in BC tissues according to TCGA data, and protein levels were detected by immunohistochemistry. The mRNA and protein expression levels of CCL18 were higher compared with in noncancerous bladder tissues. In the present cohort, CCL18 upregulation was associated with a high pathological grade. Furthermore, it was revealed that rCCL18 stimulation significantly enhanced the invasive potential of 5637 cells, and the CCL18 receptor, CCR8, mediated this activity in 5637 cells. Conversely, a CCR8 inhibitor or CCR8 shRNA abrogated the decreased levels of E-cadherin, and increased levels of MMP-2 and VEGF-C caused by CCL18. These data indicated that CCR8 may be associated with the oncogenic role of CCL18 in 5637 BC cells.
The CCR8 axis is associated with cancer progression in BC, renal carcinoma (34) and pancreatic cancer (35). R243 is a novel small molecule CCR8 inhibitor, which inhibits the effects of CCR8 in vivo and in vitro (16). CCR8 inhibition by R243 is able to counteract the phenotypes induced by extracellular vesicles decorated with CCL18 in glioblastoma cells (36). The present study demonstrated that the CCR8 inhibitor, R243, abrogated the effects of CCL18 on BC cells, thus suggesting a potential role for R243 in blocking the CCL18/CCR8 axis during BC progression. Although R243 is able to inhibit the effects of CCR8, the expression of CCR8 were unaffected in present study. Additionally, due to very low signal levels in a previous Biacore analysis, determination of intact R243 binding to immobilized CCR8 failed (16). Binding to the receptor or the site of action for R243 remains unclear. Therefore, in this study, CCR8 shRNA was selected, rather than R243, for use in cell migration and invasion experiments.
In conclusion, the present data offered convincing evidence to suggest that upregulation of CCL18 may be involved in the aggressive progression of BC partially through the G-protein-coupled receptor CCR8. These results provided information regarding the mechanisms underlying prevention of the progression of BC by inhibiting CCL18 activity via blocking CCR8. However, further studies are required to explore the detailed mechanisms underlying the effects of CCL18 and CCR8 on the regulation of BC progression.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of P.R. China (grant no. 81560419), the Natural Science Foundation of Jiangxi (grant no. 20151BAB205047) and the Jiangxi Province Infrastructure Facilities for Scientific Research Institutes (grant no. 20142BBA13038).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
XL, XX, WD, MH, YWu, ZZ and KZ performed the experiments and generated data. XL, ZZ, YWang, XC, XZ, LC and YL analyzed the data. GW and BF made substantial contributions to the design of the experiments. XL, XX and BF wrote the manuscript. All authors reviewed and approved the manuscript.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Committee of the First Affiliated Hospital, Nanchang University (Nanchang, China). Informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Prasad SM, Decastro GJ, Steinberg GD, Medscape Urothelial carcinoma of the bladder: Definition, treatment and future efforts. Nat Rev Urol. 2011;8:631–642. doi: 10.1038/nrurol.2011.144. [DOI] [PubMed] [Google Scholar]
- 4.Chenivesse C, Tsicopoulos A. CCL18-Beyond chemotaxis. Cytokine. 2018;109:52–56. doi: 10.1016/j.cyto.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Tang Y, Yu H, Yin Q, Li M, Shi L, Zhang W, Li D, Li L. CCL18 from tumor-cells promotes epithelial ovarian cancer metastasis via mTOR signaling pathway. Mol Carcinog. 2016;55:1688–1699. doi: 10.1002/mc.22419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi L, Zhang B, Sun X, Zhang X, Lv S, Li H, Wang X, Zhao C, Zhang H, Xie X, et al. CC chemokine ligand 18(CCL18) promotes migration and invasion of lung cancer cells by binding to Nir1 through Nir1-ELMO1/DOC180 signaling pathway. Mol Carcinog. 2016;55:2051–2062. doi: 10.1002/mc.22450. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Liang X, Li M, Tao X, Tai S, Fan Z, Wang Z, Cheng B, Xia J. Chemokine (CC motif) ligand 18 upregulates Slug expression to promote stem-cell like features by activating the mammalian target of rapamycin pathway in oral squamous cell carcinoma. Cancer Sci. 2017;108:1584–1593. doi: 10.1111/cas.13289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Yao Y, Gong C, Yu F, Su S, Chen J, Liu B, Deng H, Wang F, Lin L, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011;19:541–555. doi: 10.1016/j.ccr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urquidi V, Kim J, Chang M, Dai Y, Rosser CJ, Goodison S. CCL18 in a multiplex urine-based assay for the detection of bladder cancer. PLoS One. 2012;7:e37797. doi: 10.1371/journal.pone.0037797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyake M, Ross S, Lawton A, Chang M, Dai Y, Mengual L, Alcaraz A, Giacoia EG, Goodison S, Rosser CJ. Investigation of CCL18 and A1AT as potential urinary biomarkers for bladder cancer detection. BMC Urol. 2013;13:42. doi: 10.1186/1471-2490-13-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Ambrosio D, Iellem A, Bonecchi R, Mazzeo D, Sozzani S, Mantovani A, Sinigaglia F. Selective up-regulation of chemokine receptors CCR4 and CCR8 upon activation of polarized human type 2 Th cells. J Immunol. 1998;161:5111–5115. [PubMed] [Google Scholar]
- 12.Roos RS, Loetscher M, Legler DF, Clark-Lewis I, Baggiolini M, Moser B. Identification of CCR8, the receptor for the human CC chemokine I-309. J Biol Chem. 1997;272:17251–17254. doi: 10.1074/jbc.272.28.17251. [DOI] [PubMed] [Google Scholar]
- 13.Islam SA, Ling MF, Leung J, Shreffler WG, Luster AD. Identification of human CCR8 as a CCL18 receptor. J Exp Med. 2013;210:1889–1898. doi: 10.1084/jem.20130240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riester M, Taylor JM, Feifer A, Koppie T, Rosenberg JE, Downey RJ, Bochner BH, Michor F. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin Cancer Res. 2012;18:1323–1333. doi: 10.1158/1078-0432.CCR-11-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oshio T, Kawashima R, Kawamura YI, Hagiwara T, Mizutani N, Okada T, Otsubo T, Inagaki-Ohara K, Matsukawa A, Haga T, et al. Chemokine receptor CCR8 is required for lipopolysaccharide-triggered cytokine production in mouse peritoneal macrophages. PLoS One. 2014;9:e94445. doi: 10.1371/journal.pone.0094445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Leung SY, Yuen ST, Chu KM, Mathy JA, Li R, Chan AS, Law S, Wong J, Chen X, So S. Expression profiling identifies chemokine (C-C motif) ligand 18 as an independent prognostic indicator in gastric cancer. Gastroenterology. 2004;127:457–469. doi: 10.1053/j.gastro.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 19.Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol. 2018;13:395–412. doi: 10.1146/annurev-pathol-020117-043854. [DOI] [PubMed] [Google Scholar]
- 20.Fu B, Wang Y, Zhang X, Lang B, Zhou X, Xu X, Zeng T, Liu W, Zhang X, Guo J, Wang G. miR-221-induced PUMA silencing mediates immune evasion of bladder cancer cells. Int J Oncol. 2015;46:1169–1180. doi: 10.3892/ijo.2015.2837. [DOI] [PubMed] [Google Scholar]
- 21.Catusse J, Wollner S, Leick M, Schröttner P, Schraufstätter I, Burger M. Attenuation of CXCR4 responses by CCL18 in acute lymphocytic leukemia B cells. J Cell Physiol. 2010;225:792–800. doi: 10.1002/jcp.22284. [DOI] [PubMed] [Google Scholar]
- 22.McConkey DJ, Choi W, Marquis L, Martin F, Williams MB, Shah J, Svatek R, Das A, Adam L, Kamat A, et al. Role of epithelial-to-mesenchymal transition (EMT) in drug sensitivity and metastasis in bladder cancer. Cancer Metastasis Rev. 2009;28:335–344. doi: 10.1007/s10555-009-9194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orlichenko LS, Radisky DC. Matrix metalloproteinases stimulate epithelial-mesenchymal transition during tumor development. Clin Exp Metastasis. 2008;25:593–600. doi: 10.1007/s10585-008-9143-9. [DOI] [PubMed] [Google Scholar]
- 24.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 25.Davies B, Waxman J, Wasan H, Abel P, Williams G, Krausz T, Neal D, Thomas D, Hanby A, Balkwill F. Levels of matrix metalloproteases in bladder cancer correlate with tumor grade and invasion. Cancer Res. 1993;53:5365–5369. [PubMed] [Google Scholar]
- 26.Kanayama HO, Yokota KY, Kurokawa Y, Murakami Y, Nishitani M, Kagawa S. Prognostic values of matrix metalloproteinase-2 and tissue inhibitor of metalloproteinase-2 expression in bladder cancer. Cancer. 1998;82:1359–1366. doi: 10.1002/(SICI)1097-0142(19980401)82:7<1359::AID-CNCR20>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 27.He W, Zhong G, Jiang N, Wang B, Fan X, Chen C, Chen X, Huang J, Lin T. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J Clin Invest. 2018;128:861–875. doi: 10.1172/JCI96218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandriota SJ, Jussila L, Jeltsch M, Compagni A, Baetens D, Prevo R, Banerji S, Huarte J, Montesano R, Jackson DG, et al. Vascular endothelial growth factor-C-mediated lymphangiogenesis promotes tumour metastasis. EMBO J. 2001;20:672–682. doi: 10.1093/emboj/20.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki K, Morita T, Tokue A. Vascular endothelial growth factor-C (VEGF-C) expression predicts lymph node metastasis of transitional cell carcinoma of the bladder. Int J Urol. 2005;12:152–158. doi: 10.1111/j.1442-2042.2005.01010.x. [DOI] [PubMed] [Google Scholar]
- 30.Miyata Y, Kanda S, Ohba K, Nomata K, Hayashida Y, Eguchi J, Hayashi T, Kanetake H. Lymphangiogenesis and angiogenesis in bladder cancer: Prognostic implications and regulation by vascular endothelial growth factors-A, -C, and -D. Clin Cancer Res. 2006;12:800–806. doi: 10.1158/1078-0432.CCR-05-1284. [DOI] [PubMed] [Google Scholar]
- 31.Chen JC, Chang YW, Hong CC, Yu YH, Su JL. The role of the VEGF-C/VEGFRs axis in tumor progression and therapy. Int J Mol Sci. 2012;14:88–107. doi: 10.3390/ijms14010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keck B, Wach S, Taubert H, Zeiler S, Ott OJ, Kunath F, Hartmann A, Bertz S, Weiss C, Hönscheid P, et al. Neuropilin-2 and its ligand VEGF-C predict treatment response after transurethral resection and radiochemotherapy in bladder cancer patients. Int J Cancer. 2015;136:443–451. doi: 10.1002/ijc.28987. [DOI] [PubMed] [Google Scholar]
- 33.Rinderknecht M, Villa A, Ballmer-Hofer K, Neri D, Detmar M. Phage-derived fully human monoclonal antibody fragments to human vascular endothelial growth factor-c block its interaction with VEGF receptor-2 and 3. PLoS One. 2010;5:e11941. doi: 10.1371/journal.pone.0011941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eruslanov E, Stoffs T, Kim WJ, Daurkin I, Gilbert SM, Su LM, Vieweg J, Daaka Y, Kusmartsev S. Expansion of CCR8(+) inflammatory myeloid cells in cancer patients with urothelial and renal carcinomas. Clin Cancer Res. 2013;19:1670–1680. doi: 10.1158/1078-0432.CCR-12-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Duell EJ, Yu K, Risch HA, Olson SH, Kooperberg C, Wolpin BM, Jiao L, Dong X, Wheeler B, et al. Pathway analysis of genome-wide association study data highlights pancreatic development genes as susceptibility factors for pancreatic cancer. Carcinogenesis. 2012;33:1384–1390. doi: 10.1093/carcin/bgs151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berenguer J, Lagerweij T, Zhao XW, Dusoswa S, van der Stoop P, Westerman B, de Gooijer MC, Zoetemelk M, Zomer A, Crommentuijn MHW, et al. Glycosylated extracellular vesicles released by glioblastoma cells are decorated by CCL18 allowing for cellular uptake via chemokine receptor CCR8. J Extracell Vesicles. 2018;7:1446660. doi: 10.1080/20013078.2018.1446660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.