Abstract
The present study aimed to investigate the expression of microRNA-495 (miR-495) in non-small cell lung cancer (NSCLC) tissues and cells, as well as its function on the proliferation of lung cancer cells. The expression of miR-495 in 122 pairs of NSCLC tissues and matched paracarcinoma tissues, as well as in human lung cancer cell lines (A549, H460, H1650, H520 and SK-MES-1) and the normal human pulmonary bronchial epithelial cell line 16HBE was determined using reverse transcription quantitative polymerase chain reaction (RT-qPCR). As predicted by bioinformatics analysis, high mobility group A2 (HMGA2) may be a potential target gene of miR-495. In addition, the regulatory function of miR-495 on its target gene HMGA2 was evaluated using a dual-luciferase reporter assay, RT-qPCR and western blotting. Furthermore, the effect of miR-495 on the proliferation of A549 lung cancer cells was investigated using a Cell Counting Kit-8 (CCK-8) assay. The results demonstrated that the expression of miR-495 in NSCLC tissues and cells was significantly downregulated compared with the control. In addition, downregulated expression of miR-495 was associated with tumor differentiation, lymph node metastasis and tumor, node and metastasis staging. Additionally, a dual-luciferase reporter assay revealed that miR-495 could directly associated with the 3′-untranslated region of HMGA2. Upregulated expression of miR-495 significantly downregulated the mRNA and protein expression levels of HMGA2 in A549 cells. Furthermore, the results of CCK-8 assay revealed that upregulated expression of miR-495 significantly suppressed the proliferation of A549 cells; HMGA2 overexpression reversed this inhibition. In summary, the findings of the present study demonstrated that miR-495 was downregulated in NSCLC tissues and cells. In addition, miR-495 suppressed the proliferation of lung cancer cells by directly targeting HMGA2.
Keywords: microRNA-495, lung cancer, high mobility group AT-hook 2, proliferation
Introduction
Lung cancer is one of the most common type of life-threatening malignancies worldwide (1). Non-small cell lung cancer (NSCLC) accounts for 80–85% of cases in lung cancer (2). Surgery-oriented multimodality therapy is the most effective method for the treatment of lung cancer (3); however, there are no symptoms in most patients with lung cancer at early-stage of this disease, and majority of the patients are diagnosed at the moderate to advanced stage, which has deprived them from the opportunity of radical surgery (4). Recently, molecular therapy against the targeting of genes has become a promising treatment of tumors (5). In summary, investigating the mechanisms underlying lung cancer and identifying the potential molecular targets are important in the treatment of this disease.
MicroRNAs (miRNAs) are a class of endogenous and highly conserved single-stranded non-coding small RNAs consisting of 18–22 nucleotides. MiRNAs can bind with the 3′-untranslated region (UTR) of their downstream target gene, consequently degrading mRNA or inhibiting translation at the post-transcription level. Each miRNA regulates numerous target genes; while a specific target mRNA can also be regulated by several miRNAs at the same time (6). They function in a complex regulatory network and act as oncogene or tumor suppressor during the development of tumors (7,8). Recent research has revealed that miR-495 functions as tumor suppressor gene in the majority of solid tumors, including gastric cancer (9), esophageal cancer (10), colorectal cancer (11), renal cell carcinoma (12) and bladder cancer (13); however, the underlying mechanism remains unknown. To investigate the regulatory function of miR-495 in NSCLC, the expression levels of miR-495 in NSCLC and paracarcinoma tissues, as well as in NSCLC cells and normal pulmonary bronchial epithelial cells were compared in the present study. In addition, the association between the expression of miR-495 and the clinicopathological features of patients with NSCLC was evaluated. A549 lung cancer cells were transfected with miR-495 mimics; the effects of miR-495 overexpression on the proliferation of A549 cells were revealed. Additionally, whether high mobility group A2 (HMGA2) is a potential target gene of miR-495 was investigated via bioinformatics analysis in the present study.
Materials and methods
Patient samples
A total of 122 pairs of NSCLC and matched paracarcinoma samples (≥5 cm from tumor margin; 80 males and 42 females; aged 45–77 years old) were obtained via surgery in Weihai Central Hospital (Weihai, China) from June 2014 to June 2016. The tissues were snap-frozen in liquid nitrogen within 30 min following surgery and stored at −80°C until further use. None of the patients received chemotherapy or radiotherapy prior to surgery. In addition, no malignancy in other organs was reported. miR-495 expression was categorized into low and high groups according to the mean value. All pathological sections were examined and verified by two senior pathologists. The clinical features of patients were presented in Table I. TNM staging was carried out according to the 7th edition of the Union for International Cancer Control (UICC) staging Committee TNM staging standards (14). Informed consent was obtained from all patients. The present study was approved by the Medical Ethics Committee of Weihai Central Hospital.
Table I.
Associations between miR-495 expression and the clinicopathological characteristics of patients with non-small cell lung cancer.
| miR-495 [n (%)] | ||||
|---|---|---|---|---|
| Clinicopathological features | n | Low expression | High expression | P-value |
| Gender | 0.316 | |||
| Male | 80 | 40 (50.00) | 40 (50.00) | |
| Female | 42 | 25 (59.52) | 17 (40.48) | |
| Age (year) | 0.179 | |||
| >60 | 72 | 42 (58.33) | 30 (41.67) | |
| ≤60 | 50 | 23 (46.00) | 27 (54.00) | |
| Tumor size | 0.312 | |||
| >5 cm | 53 | 31 (58.49) | 22 (41.51) | |
| ≤5 cm | 69 | 34 (49.28) | 35 (50.72) | |
| Tumor differentiation | 0.008a | |||
| Well + Moderate | 77 | 34 (44.16) | 43 (55.84) | |
| Poorly | 45 | 31 (68.89) | 14 (31.11) | |
| TNM stage | 0.024a | |||
| I + II | 84 | 39 (46.43) | 45 (53.57) | |
| III | 38 | 26 (68.42) | 12 (31.58) | |
| Smoking | 0.298 | |||
| Yes | 51 | 30 (58.82) | 21 (41.18) | |
| No | 71 | 35 (49.30) | 36 (50.70) | |
| Lymphatic metastasis | 0.005a | |||
| Yes | 72 | 46 (63.89) | 26 (36.11) | |
| No | 50 | 19 (38.00) | 31 (62.00) | |
| Pathology classification | 0.081 | |||
| Squamous cell carcinoma | 54 | 24 (44.44) | 30 (55.56) | |
| Adenocarcinoma | 68 | 41 (60.29) | 27 (39.71) | |
P<0.05. miR, microRNA; TNM, tumor, node and metastasis.
Cell culture
Human NSCLC cell lines A549, H460, H1650, H520 and SK-MES-1, as well as the normal human pulmonary bronchial epithelial cell line 16HBE were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Roswell Park Memorial Institute 1640 medium (RPMI-1640) (HyClone; GE Healthcare, Chicago, IL, USA) containing 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified atmosphere of 5% CO2. Cell lines with the highest relative expression of miR-495 were selected for subsequent experiments.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from tissues or cells was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocols. The purity of the extracted RNA was determined using an ultraviolet spectrophotometer. cDNA was synthesized using a PrimeScript RT reagent kit (Promega Corporation, Madison, WI, USA) according to the manufacturer's protocols. The primers of miR-495, HMGA2 and the internal reference U6, GAPDH were obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The primer sequences used were as follows: miR-495 forward, 5′-GGAGCTTGAGCGGATGGCGA-3′ and reverse, 5′-TTAGCGGAGCGGGAGGGCGA-3′; U6 forward, 5′-CATCACCATCAGGAGAGTCG-3′ and reverse, 5′-TGACGCTTGCCCACAGCCTT3′; HMGA2 forward, 5′-CCAACCGGTGAGCCCTCT-3′ and reverse, 5′-TTGAGCTGCTTTAGAGGGAC-3′; GAPDH forward, 5′-CGCTGAGTACGTCGTGGAGT-3′ and reverse, 5′-GTCGCTGTTGAAGTCAGAGGAG-3′. qPCR was performed using a GoTaq® RT-qPCR system (Promega Corporation) according to the manufacturer's protocols. The reaction conditions were as follows: 2 min at 94°C, 20 sec at 94°C and 30 sec at 60°C for 40 cycles. The relative expression of miR-495 and HMGA2 mRNA were calculated using the 2−ΔΔCq method (15). All assays were performed in triplicate.
Cell transfection
A549 cells at the logarithmic phase were seeded on the 6-well plate (1.5×106 cells/well) and cultured overnight in RPMI-1640 without antibiotics. Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for transfection according to the manufacturer's protocols. Following transfection for 24 h, the cells were collected for subsequent experiments. The cells were divided into 3 groups: i) Negative control (NC) group, transfection with mimics-NC plasmid (50 nM, sense: 5′-UUCUCCGAACGUGUCACGUTT-3′; anti-sense: 5′-ACGUGACACGUUCGGAATT-3′); ii) miR-495 mimics group, transfection with miR-495 mimics plasmid (50 nM, sense: 5′-AAACAAACAUGGUGCACUUCUU-3′; anti-sense: 5′-GAAGUGCACCAUGUUUGUUUUU-3′) and iii) miR-495 mimics + HMGA2 group, co-transfection with miR-495 mimics and HMGA2-expressing plasmids. The miR-495 mimics, mimics-NC and HMGA2 expression plasmids were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China).
Cell counting kit-8 (CCK-8) assay
Cells were harvested following transfection and inoculated with 10 µl CCK-8 solution (Blue Skies Biotech. Inc.; LakePharma., Inc., Worcester, MA, USA) for 24, 48, 72 and 96 h according to the manufacturer's protocols. Following an incubation at 37°C for an additional 2 h in the dark, the absorbance at 450 nm was measured using a plate reader.
Bioinformatic prediction and dual-luciferase reporter assay
HMGA2 was identified as the potential direct target of miR-495 via the bioinformatics database miRanda version August 2010 Release (available at: www.microrna.org). The wild-type (wt) pmirGLO-HMGA2-wtUTR and mutant (mut) pmirGLO-HMGA2-mtUTR plasmids were synthesized by Shanghai GenePharma Co., Ltd. A549 cells (5×105 cells/well) were plated onto 6-well plates. Then, 100 ng pmirGLO-HMGA2-wtUTR or pmirGLO-HMGA2-mtUTR were used to co-transfect A549 cells with 50 nM miR-495 mimics or mimics-NC using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were harvested 24 h post-transfection. Luciferase activity was evaluated 48 h post-transfection using a Dual-Luciferase® Reporter Assay (Promega Corporation), and the levels of firefly luciferase activity were normalized to that of Renilla luciferase.
Western blotting
Transfected A549 cells were collected and washed with PBS for three times. The total protein of cells was extracted with the protein lysis buffer (Cell Signaling Technology Inc., Danvers, MA, USA), and the concentration of protein was determined using a bicinchoninic acid assay. Protein samples (40 µg) were separated on a 10% SDS-PAGE gel for 30 min and transferred to polyvinylidene difluoride membranes. Then, the membranes were blocked in tris-buffered-saline-Tween-20 (TBST; containing 0.1% Tween-20) with 5% skimmed milk for 2 h at room temperature and incubated with primary antibodies against HMGA2 (1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA; cat. no. sc-130024) and β-actin (1:2,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA; cat. no. sc-130065) at 4°C overnight with agitation. Following washing, the membranes were incubated with goat anti-mouse horseradish peroxidase-conjugated secondary antibody (1:2,000; Santa Cruz Biotechnology, Inc.; sc-516102) at room temperature for 2 h. Signals were visualized using an enhanced chemiluminescence luminescent reagent (cat. no. sw2030; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) and were developed using a gel imaging system. β-actin was used as the internal reference. Bands were analyzed with Image J software (version 1.51j8; National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
SPSS 19.0 software (IBM Corp., Armonk, NY, USA) was employed to perform statistical analysis. Data were presented as the mean ± standard deviation and analyzed using a Student's t-test or analysis of variance (ANOVA). A Student-Newman-Keuls test was used as a post-hoc test following ANOVA. Enumeration data were compared using a χ2 test. All experiments were repeated three times. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-495 is downregulated in NSCLC tissues and cells
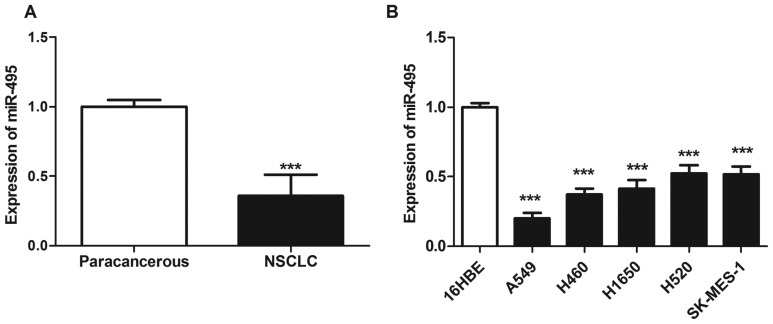
The expression levels of miR-495 were significantly downregulated in NSCLC tissues compared with in matched paracarcinoma tissues (P<0.05; Fig. 1A). Furthermore, the expression of miR-495 was also significantly reduced in NSCLC cell lines A549, H460, H1650, H520 and SK-MES-1 compared with in the normal pulmonary bronchial epithelial cell line 16HBE (P<0.05; Fig. 1B). In addition, the relative expression levels of miR-495 in A549 cells were the lowest. Therefore, A549 was selected for further cytological study to ectopically overexpress miR-495.
Figure 1.
Expression of miR-495 is downregulated in NSCLC tissues and cells. (A) Reverse transcription-quantitative polymerase chain reaction was used to detect the expression levels of miR-145 in NSCLC and paracarcinoma tissues. The expression of miR-495 was significantly downregulated in NSCLC tissues compared with matched paracarcinoma tissues. ***P<0.001 vs. paracarcinoma tissues. (B) Expression levels of miR-495 were also reduced in NSCLC cell lines A549, H460, H1650, H520 and SK-MES-1 compared with the normal pulmonary bronchial epithelial cell line 16HBE. ***P<0.001 vs. 16HBE cells. MiR, microRNA; NSCLC, non-small cell lung cancer.
Expression of miR-495 is associated with the clinicopathological parameters of patients with NSCLC
The mean miR-495 expression levels in all patients were used as the threshold to divide the patients into miR-495 high and low expression groups. Downregulated expression of miR-495 was associated with tumor differentiation, lymph node metastasis and clinical stage (P<0.05; Table I), but was not associated with age, sex, tumor size, smoking and pathological classification (Table I).
HMGA2 is a direct target gene of miR-495
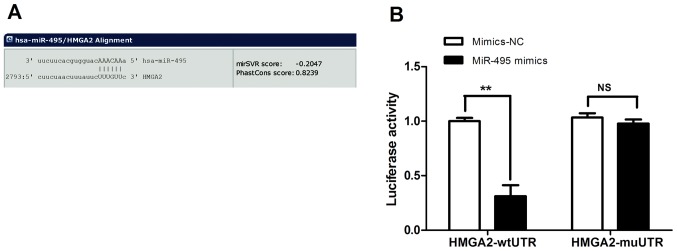
HMGA2 was predicted to be a potential direct target gene of miR-495 using the target gene prediction database miRanda and gene functional analysis. In addition, there were complementary binding sites to the seed sequence of miR-495 in the 3′-UTR of HMGA2 (Fig. 2A). To further investigate whether HMGA2 was the direct regulatory target gene of miR-495, A549 cells were co-transfected with miR-495 mimics or mimics-NC, and pmirGLO-HMGA2-wtUTR or pmirGLO-HMGA2-mtUTR. The results of the dual-luciferase reporter assay revealed that the luciferase activity of cells in the miR-495mimics + pmirGLO-HMGA2-wtUTR group was significantly reduced compared with that of the mimics-NC + pmirGLO-HMGA2-wtUTR group (Fig. 2B). Additionally, no significant difference was observed between cells in miR-495mimics + pmirGLO-HMGA2-mt group and the mimics-NC + pmirGLO-HMGA2-mtUTR group (P>0.05; Fig. 2B). These results indicated that miR-495 could specifically bind to the 3′-UTR of HMGA2; however, this effect was not detected in the presence of 3′-UTR-mutated HMGA2, suggesting that HMGA2 may be the direct target of miR-495.
Figure 2.
HMGA2 is a direct target gene of miR-495. (A) MiR-495 and HMGA2 binding sequence. (B) Dual-luciferase assay was used to determine whether HMGA2 was directly targeted by miR-495. The luciferase activity of cells in miR-495-mimics + pmirGLO-HMGA2-wtUTR group was significantly downregulated compared with in the mimics NC + pmirGLO-HMGA2-wtUTR group. **P<0.01 vs. mimic-NC + pmirGLO-HMGA2-wtUTR. No significant difference between cells in miR-495 mimics + pmirGLO-HMGA2-mtUTR group and the mimics-NC + pmirGLO-HMGA2-mtUTR group was observed. P>0.05. HMGA2, high mobility group A2; miR, microRNA; NS, not significant; NC, negative control; mt, mutant; UTR, untranslated region; wt, wild-type.
MiR-495 inhibits the proliferation of A549 cells in a HMGA2-dependant manner
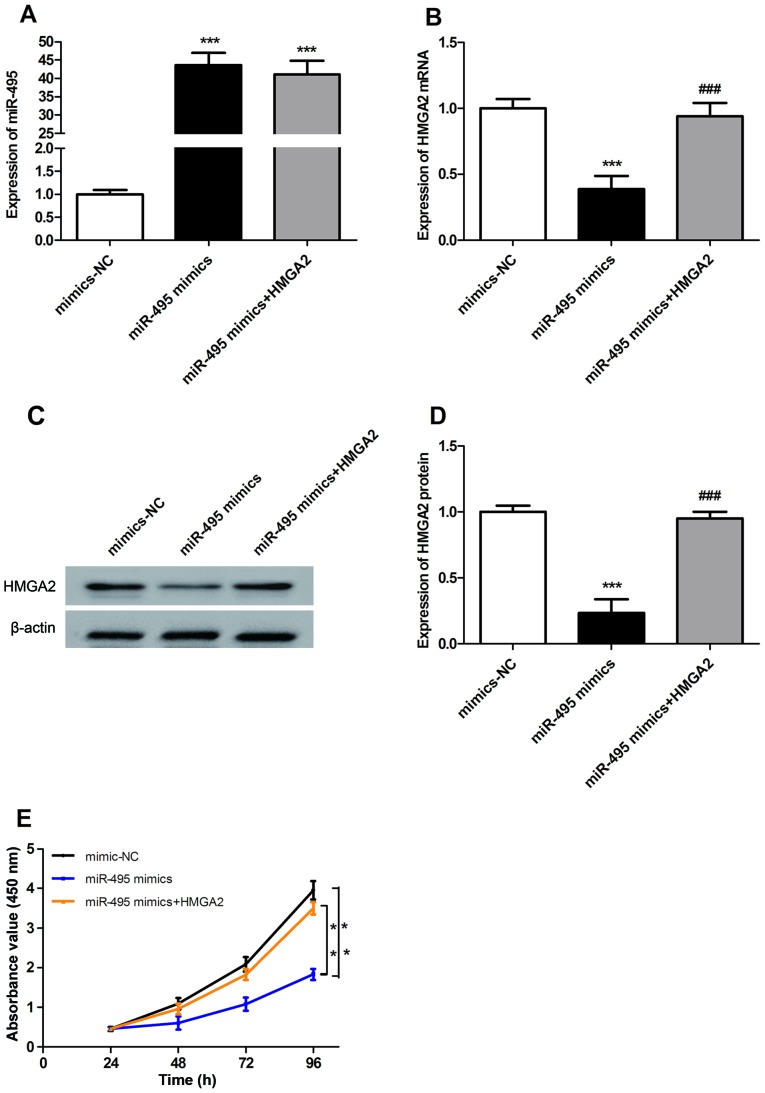
The expression levels of miR-495 in A549 cells in the miR-495 mimics and miR-495 mimics + HMGA2 groups were significantly upregulated compared with in the mimic-NC group (P<0.001; Fig. 3A); however, there was no significant difference in miR-495 expression between miR-495 mimics and miR-495 mimics + HMGA2 groups (P>0.05; Fig. 3A). The mRNA and protein expression levels of HMGA2 in transfected A549 cells were evaluated using RT-qPCR and western blotting. There were no significant differences in the mRNA and protein expression levels of HMGA2 between mimic-NC and miR-495 mimics + HMGA2 groups (P>0.05; Fig. 3B-D). However, the mRNA and protein expression levels of HMGA2 in the miR-495 mimics group were significantly downregulated compared with those in the mimic-NC group (P<0.001; Fig. 3B-D). Furthermore, the expression of HMGA2 in the miR-495 mimics + HMGA2 group was significantly upregulated compared with in the miR-495 mimics group (P<0.001; Fig. 3B-D). In addition, the results of the CCK-8 assay revealed that upregulated expression of miR-495 could significantly inhibit the proliferation of A549 cells compared with the control (P<0.01), while HMGA2 overexpression could significantly reversed this inhibition (P<0.01; Fig. 3E). The above results show that miR-495 inhibits the proliferation of A549 cells in a HMGA2-dependant manner.
Figure 3.
MiR-495 inhibits the proliferation of A549 cells in a HMGA2-dependant manner. (A) Reverse transcription-quantitative polymerase chain reaction was used to determine the expression levels of miR-495 following transfection. The expression of miR-495 in the miR-495 mimics and miR-495 mimics + HMGA2 groups was significantly upregulated compared with in the mimic-NC group. ***P<0.001 vs. mimic-NC. (B-D) Expression of HMGA2 in A549 cells in the miR-495 mimics group was significantly downregulated compared with in the mimic-NC group. The protein expression levels of HMGA2 in the miR-495 mimics + HMGA2 groups were significantly upregulated compared with those in miR-495 mimics group. ###P<0.001 vs. miR-495 mimics; ***P<0.001 vs. mimic-NC. (E) Cell proliferation was evaluated by a Cell Counting Kit-8 assay. MiR-495 inhibited the proliferation of A549 cells, whereas HMGA2 overexpression could reverse this inhibition. **P<0.01. MiR, microRNA; HMGA2, high mobility group A2; NC, negative control.
Discussion
Lung cancer is a frequently-occurring malignancy, and its initiation and development are complex processes involving numerous genetic alterations (16); however, the molecular mechanisms underlying this disease remain unclear. Recently, the roles of miRNA in numerous diseases have attracted increasing attention with advances in of epigenetic research (17). Aberrant expression of miRNA in the majority of malignancies has been revealed; miRNAs serve an indispensable role in the initiation and development of malignancies (18,19).
MiR-495 is a novel miRNA that is located in chromosome 14q32.31 (20). It serves an important role in cell proliferation, apoptosis, and the immune and inflammatory responses (21). It has been revealed that aberrant expression of miR-495 was associated with tumor cell proliferation, apoptosis and chemoresistance (22). In addition, miR-495 functioned as tumor suppressor in the majority of solid tumors (9–13); however, the expression of miR-495 was upregulated in some solid tumors, including hepatocellular carcinoma (23) and breast cancer (24), and thus may be considered as an oncogene. In the present study, the expression levels of miR-495 in NSCLC and matched paracarcinoma tissues, as well as in human lung cancer and normal human pulmonary bronchial epithelial cells were compared. The results revealed that miR-495 expression in NSCLC tissues and cells was significantly downregulated. In addition, to investigate the effects of miR-495 on the proliferation of lung cancer cells, miR-495 mimics were successfully transfected into A549 cells. Thus, A549 cells overexpressing miR-495 were established; overexpression of miR-495 significantly inhibited the proliferation of A549 cells, indicating that miR-495 could function as a tumor suppressor gene in NSCLC.
MiRNAs may regulate numerous target genes; however, each target gene can also be regulated by several miRNAs. The role of miRNA in tumors depends on its regulatory functions on downstream target genes (25). Li et al (9) reported that miR-495 could inhibit the invasion and metastasis of gastric cancer cells in a phosphatase of regenerating liver 3-dependent manner. Mao et al (10) revealed that miR-495 could suppress the invasion of esophageal cancer by targeting protein kinase. Chu et al (26) reported that miR-495 could inhibit the proliferation and metastasis of NSCLC cells in a metastasis associated 1 family member 3-dependent manner. In the present study, the potential target gene of miR-495 was predicted to be HMGA2 using target gene prediction database miRanda and gene functional analysis; there were complementary binding sites to the seed sequence of miR-495 in the 3′-UTR of HMGA2. Additionally, a dual-luciferase reporter gene assay revealed that miR-495 could specifically bind to the 3′-UTR of HMGA2. Furthermore, the expression levels of HMGA2 were further determined using RT-qPCR and western blotting; upregulation of miR-495 significantly downregulated HMGA2 expression. The findings of the present study demonstrated that miR-495 inhibited the expression of HMGA2, thus HMGA2 may be one of the direct regulatory target genes of miR-495.
HMGA2 is a non-histone chromosomal protein and member of the high mobility group (27). The role of HMGA2 in tumors has been widely studied, and HMGA2 was reported to be a potential oncogene (28). HMGA2 is abundantly expressed in solid tumor tissues, including thyroid cancer (29), ovarian cancer (30), prostate cancer (31), gallbladder adenocarcinoma (32), esophageal squamous carcinoma (33), bladder cancer (34), NSCLC (35) and gastric cancer (36). In addition, HMGA2 binds to the promoter of the cyclin gene, upregulating the expression of cyclin and accelerating the G2/M stage of the cell cycle, and consequently promoting tumorigenesis (37). E2F transcription factor 1 (E2F1) is a member of the transcription factor E2F family and regulates the progression of the cell cycle from G1 to S phase (38). Fedele et al (39) revealed that overexpressed HMGA2 could bind to phosphorylated retinoblastoma protein, consequently enhancing the activity of E2F1 and resulting in the impaired proliferation of tumor cells. Cyclin A serves an important role in promoting the mitosis and proliferation of cells; it is an important regulatory protein of the cell cycle (40). HMGA2 activates the promoter of the cyclin A gene to induce its expression, thereby disrupting the cell cycle and promoting the occurrence of tumors (41). A recent study also demonstrated that in human pituitary adenoma, HMGA2 directly associated with the promoter of the cyclin B2 gene and upregulate its expression, consequently accelerating the transition of G2 to M phase (42). Additionally, Morishita et al (43) revealed that HMGA2 promoted epithelial mesenchymal transition, activated transcription growth factor β (TGF-β) receptor II, and activated the TGF-β signaling pathway, which facilitated the invasion and metastasis of tumor cells. It was reported that downregulated expression level of HMGA2 in NSCLC cells notably suppressed the proliferation of lung cancer cells (44). In the present study, A549 cells were co-transfected with miR-495 mimics and HMGA2-overexpressing plasmids. The results revealed that the expression levels of HMGA2 in the miR-495 mimics + HMGA2 group were significantly upregulated compared with the control, indicating that the inhibitory effects of miR-495 on HMGA2 expression was successfully reversed. Furthermore, the results of CCK-8 assay suggested that HMGA2 overexpression significantly reversed the inhibition of miR-495 mimics on the proliferation of A549 cells, indicating that miR-495 suppressed the proliferation of A549 cells in a HMGA2-dependent manner.
In summary, the present study revealed that miR-495 expression was downregulated in NSCLC tissues and cells. Therefore, HMGA2 may be one of the direct target genes of miR-495. In addition, miR-495 could inhibit the expression of HMGA2, consequently suppressing the proliferation of lung cancer cells. The results of the present study may provide insight into the mechanisms underlying the inhibition of lung cancer cell proliferation. In conclusion, miR-495 may be considered as a potential target in the treatment of lung cancer via gene therapy in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HW designed the experiments. JS, YQ and TS performed the experiments. JS and HW analyzed the data. JS wrote the manuscript. All authors have read and approved the manuscript.
Ethics approval and consent to participate
The present study was approved by the Medical Ethics Committee of Weihai Central Hospital (Weihai, China). Informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Ridge CA, Mcerlean AM, Ginsberg MS. Epidemiology of lung cancer. Semin Intervent Radiol. 2013;30:93–98. doi: 10.1055/s-0033-1342949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakelee H, Kelly K, Edelman MJ. 50 Years of progress in the systemic therapy of non-small cell lung cancer. Am Soc Clin Oncol Educ Book. 2014:177–189. doi: 10.14694/EdBook_AM.2014.34.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhan P, Qian Q, Yu LK. Prognostic value of COX-2 expression in patients with non-small cell lung cancer: A systematic review and meta-analysis. J Thorac Dis. 2013;5:40–47. doi: 10.3978/j.issn.2072-1439.2013.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheng M, Zhao Y, Wang F, Li S, Wang X, Shou T, Luo Y, Tang W. Targeted drugs for unselected patients with advanced non-small-cell lung cancer: A network meta-analysis. J Thorac Dis. 2016;8:98–115. doi: 10.3978/j.issn.2072-1439.2016.01.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acunzo M, Croce CM. MicroRNA in cancer and cachexia-a mini-review. J Infect Dis. 2015;212(Suppl 1):S74–S77. doi: 10.1093/infdis/jiv197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorel O, Dewals BG. MicroRNAs in large herpesvirus DNA genomes: Recent advances. Biomol Concepts. 2016;7:229–239. doi: 10.1515/bmc-2016-0017. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Zhang G, Li D, Jie Z, Chen H, Xiong J, Liu Y, Cao Y, Jiang M, Le Z, Tan S. Methylation-associated silencing of miR-495 inhibit the migration and invasion of human gastric cancer cells by directly targeting PRL-3. Biochem Biophys Res Commun. 2015;456:344–350. doi: 10.1016/j.bbrc.2014.11.083. [DOI] [PubMed] [Google Scholar]
- 10.Mao Y, Li L, Liu J, Wang L, Zhou Y. MiR-495 inhibits esophageal squamous cell carcinoma progression by targeting Akt1. Oncotarget. 2016;7:51223–51236. doi: 10.18632/oncotarget.9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan L, Yao J, Qiu J. miRNA-495 suppresses proliferation and migration of colorectal cancer cells by targeting FAM83D. Biomed Pharmacother. 2017;96:974–981. doi: 10.1016/j.biopha.2017.11.138. [DOI] [PubMed] [Google Scholar]
- 12.Lu Y, Liu WG, Lu JH, Liu ZJ, Li HB, Liu GJ, She HY, Li GY, Shi XH. LncRNA UCA1 promotes renal cell carcinoma proliferation through epigenetically repressing p21 expression and negatively regulating miR-495. Tumour Biol. 2017;39:1010428317701632. doi: 10.1177/1010428317701632. [DOI] [PubMed] [Google Scholar]
- 13.Tan M, Mu X, Liu Z, Tao L, Wang J, Ge J, Qiu J. microRNA-495 promotes bladder cancer cell growth and invasion by targeting phosphatase and tensin homolog. Biochem Biophys Res Commun. 2017;483:867–873. doi: 10.1016/j.bbrc.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Wu N, Zheng Q, Feng Y, Yan S, Lv C, Li S, Wang Y, Yang Y. Evaluation of the 7th edition of the TNM classification for lung cancer at a single institution. J Cancer Res Clin Oncol. 2014;140:1189–1195. doi: 10.1007/s00432-014-1636-0. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schimittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Mehta A, Dobersch S, Romero-Olmedo AJ, Barreto G. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015;34:229–241. doi: 10.1007/s10555-015-9563-3. [DOI] [PubMed] [Google Scholar]
- 17.Hesse M, Arenz C. MicroRNA maturation and human disease. Methods Mol Biol 1095. 2014:11–25. doi: 10.1007/978-1-62703-703-7_2. [DOI] [PubMed] [Google Scholar]
- 18.Salido-Guadarrama I, Romero-Cordoba S, Peralta-Zaragoza O, Hidalgo-Miranda A, Rodríguez-Dorantes M. MicroRNAs transported by exosomes in body fluids as mediators of intercelluar communication in cancer. Onco Targets Ther. 2014;7:1327–1338. doi: 10.2147/OTT.S61562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Yao H, Lin S, Zhu X, Shen Z, Lu G, Poon WS, Xie D, Lin MC, Kung HF. Transcriptional and epigenetic regulation of human microRNAs. Cancer Lett. 2013;331:1–10. doi: 10.1016/j.canlet.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Song Y, Liu D, Zhao J, Xu J, Ren J, Hu Y, Wang Z, Hou Y, Zhao G. MiR-495 promotes senescence of mesenchymal stem cells by targeting Bmi-1. Cell Physiol Biochem. 2017;42:780–796. doi: 10.1159/000478069. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Yang Y, Feng Z. Suppression of microRNA-495 alleviates high-glucose-induced retinal ganglion cell apoptosis by regulating Notch/PTEN/Akt signaling. Biomed Pharmacother. 2018;106:923–929. doi: 10.1016/j.biopha.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 22.Bai Z, Wang J, Wang T, Li Y, Zhao X, Wu G, Yang Y, Deng W, Zhang Z. The MiR-495/Annexin A3/P53 axis inhibits the invasion and EMT of colorectal cancer cells. Cell Physiol Biochem. 2017;44:1882–1895. doi: 10.1159/000485877. [DOI] [PubMed] [Google Scholar]
- 23.Yang H, Cho ME, Li TW, Peng H, Ko KS, Mato JM, Lu SC. MicroRNAs regulate methionine adenosyltransferase 1A expression in hepatocellular carcinoma. J Clin Invest. 2013;123:285–298. doi: 10.1172/JCI63861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang-Verslues WW, Chang PH, Wei PC, Yang CY, Huang CK, Kuo WH, Shew JY, Chang KJ, Lee EY, Lee WH. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30:2463–2474. doi: 10.1038/onc.2010.618. [DOI] [PubMed] [Google Scholar]
- 25.Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;15:1–19. doi: 10.1093/bib/bbs075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu H, Chen X, Wang H, Du Y, Wang Y, Zang W, Li P, Li J, Chang J, Zhao G, Zhang G. MiR-495 regulates proliferation and migration in NSCLC by targeting MTA3. Tumour Biol. 2014;35:3487–3494. doi: 10.1007/s13277-013-1460-1. [DOI] [PubMed] [Google Scholar]
- 27.Yang F, Zhao L, Mei D, Jiang L, Geng C, Li Q, Yao X, Liu Y, Kong Y, Cao J. HMGA2 plays an important role in Cr (VI)-induced autophagy. Int J Cancer. 2017;141:986–997. doi: 10.1002/ijc.30789. [DOI] [PubMed] [Google Scholar]
- 28.Busch B, Bley N, Müller S, Glaß M, Misiak D, Lederer M, Vetter M, Strauß HG, Thomssen C, Hüttelmaier S. The oncogenic triangle of HMGA2, LIN28B and IGF2BP1 antagonizes tumor-suppressive actions of the let-7 family. Nucleic Acids Res. 2016;44:3845–3864. doi: 10.1093/nar/gkw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin L, Lloyd RV, Henry MR, Erickson LA, Sebo TJ, Rumilla KM, Zhang J. The diagnostic utility of combination of HMGA2 and IMP3 qRT-PCR testing in thyroid neoplasms. Appl Immunohistochem Mol Morphol. 2015;23:36–43. doi: 10.1097/PAI.0000000000000031. [DOI] [PubMed] [Google Scholar]
- 30.Yang J, Zhang Q, Dong JQ, Chang XH, He XJ. Overexpression of high mobility group A2 and its correlation with microRNA let-7 family in serous ovarian cancers. Beijing Da Xue Xue Bao Yi Xue Ban. 2012;44:749–754. (In Chinese) [PubMed] [Google Scholar]
- 31.Zhu C, Li J, Cheng G, Zhou H, Tao L, Cai H, Li P, Cao Q, Ju X, Meng X, et al. miR-154 inhibits EMT by targeting HMGA2 in prostate cancer cells. Mol Cell Biochem. 2013;379:69–75. doi: 10.1007/s11010-013-1628-4. [DOI] [PubMed] [Google Scholar]
- 32.Zou Q, Xiong L, Yang Z, Lv F, Yang L, Miao X. Expression levels of HMGA2 and CD9 and its clinicopathological significances in the benign and malignant lesions of the gallbladder. World J Surg Oncol. 2012;10:92. doi: 10.1186/1477-7819-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q, Lv GD, Qin X, Gen YH, Zheng ST, Liu T, Lu XM. Role of microRNA let-7 and effect to HMGA2 in esophageal squamous cell carcinoma. Mol Biol Rep. 2012;39:1239–1246. doi: 10.1007/s11033-011-0854-7. [DOI] [PubMed] [Google Scholar]
- 34.Ding X, Wang Y, Ma X, Guo H, Yan X, Chi Q, Li J, Hou Y, Wang C. Expression of HMGA2 in bladder cancer and its association with epithelial-to-mesenchymal transition. Cell Prolif. 2014;47:146–151. doi: 10.1111/cpr.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer B, Loeschke S, Schultze A, Weigel T, Sandkamp M, Goldmann T, Vollmer E, Bullerdiek J. HMGA2 overexpression in non-small cell lung cancer. Mol Carcinog. 2007;46:503–511. doi: 10.1002/mc.20235. [DOI] [PubMed] [Google Scholar]
- 36.Kong D, Su G, Zha L, Zhang H, Xiang J, Xu W, Tang Y, Wang Z. Coexpression of HMGA2 and Oct4 predicts an unfavorable prognosis in human gastric cancer. Med Oncol. 2014;31:130. doi: 10.1007/s12032-014-0130-5. [DOI] [PubMed] [Google Scholar]
- 37.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 38.Denechaud PD, Fajas L, Giralt A. E2F1, a novel regulator of metabolism. Front Endocrinol (Lausanne) 2017;8:311. doi: 10.3389/fendo.2017.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fedele M, Pierantoni GM, Visone R, Fusco A. E2F1 activation is responsible for pituitary adenomas induced by HMGA2 gene overexpression. Cell Div. 2006;1:17. doi: 10.1186/1747-1028-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loukil A, Cheung CT, Bendris N, Lemmers B, Peter M, Blanchard JM. Cyclin A2: At the crossroads of cell cycle and cell invasion. World J Biol Chem. 2015;6:346–350. doi: 10.4331/wjbc.v6.i4.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tessari MA, Gostissa M, Altamura S, Sgarra R, Rustighi A, Salvagno C, Caretti G, Imbriano C, Mantovani R, Del Sal G, et al. Transcriptional activation of the cyclin A gene by the architectural transcription factor HMGA2. Mol Cell Biol. 2003;23:9104–9116. doi: 10.1128/MCB.23.24.9104-9116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Angelo D, Esposito F, Fusco A. Epigenetic mechanisms leading to overexpression of HMGA proteins in human pituitary adenomas. Front Med (Lausanne) 2015;2:39. doi: 10.3389/fmed.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morishita A, Zaidi MR, Mitoro A, Sankarasharma D, Szabolcs M, Okada Y, D'Armiento J, Chada K. HMGA2 is a driver of tumor metastasis. Cancer Res. 2013;73:4289–4299. doi: 10.1158/0008-5472.CAN-12-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Cello F, Hillion J, Hristov A, Wood LJ, Mukherjee M, Schuldenfrei A, Kowalski J, Bhattacharya R, Ashfaq R, Resar LM. HMGA2 participates in transformation in human lung cancer. Mol Cancer Res. 2008;6:743–750. doi: 10.1158/1541-7786.MCR-07-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.