Abstract
Chronic physiological stress impacts animal fitness by catabolizing metabolic stores and suppressing reproduction. This can be especially deleterious for capital breeding carnivores such as marine mammals, with potential for ecosystem-wide effects. However, the impacts and indicators of chronic stress in animals are currently poorly understood. To identify downstream mediators of repeated stress responses in marine mammals, we administered adrenocorticotropic hormone (ACTH) once daily for four days to free-ranging juvenile northern elephant seals (Mirounga angustirostris) to stimulate endogenous corticosteroid release, and compared blubber tissue transcriptome responses to the first and fourth ACTH administrations. Gene expression profiles were distinct between blubber responses to single and repeated ACTH administration, despite similarities in circulating cortisol profiles. We identified 61 and 12 genes that were differentially expressed (DEGs) in response to the first ACTH and fourth administrations, respectively, 24 DEGs between the first and fourth pre-ACTH samples, and 12 DEGs between ACTH response samples from the first and fourth days. Annotated DEGs were associated with functions in redox and lipid homeostasis, suggesting potential negative impacts of repeated stress on capital breeding, diving mammals. DEGs identified in this study are potential markers of repeated stress in marine mammals, which may not be detectable by endocrine profiles alone.
Introduction
Increasing anthropogenic activity has been correlated with loss of biodiversity in many ecosystems1. Consequences of anthropogenic disturbance, such as declines in apex predator populations, are causing detrimental shifts in food web dynamics2. For example, the collapse of large marine carnivore populations has been correlated with altered trophic structure and decline of commercially valuable fish stocks3. Marine mammal populations are threatened by noise pollution, ship strikes, fisheries competition and bycatch, prey declines, habitat loss, and climate change4–7. In addition to direct effects on survival, anthropogenic disturbance may impact marine mammal health, fitness, and population persistence by causing physiological stress.
Stress response pathways maintain homeostasis and mediate the relationship between an animal and its external environment8. The mammalian stress response is mediated by the hypothalamic-pituitary-adrenal (HPA) axis9. In response to psychological or physiological stress, corticotropin-releasing hormone (CRH) is released by the hypothalamus, triggering the release of adrenocorticotropic hormone (ACTH) by the pituitary gland. This stimulates synthesis of corticosteroids (glucocorticoids and mineralocorticoids) by the adrenal glands. Stress studies in terrestrial mammals have primarly focused on downstream effects of glucocorticoids (e.g. cortisol) rather than mineralocorticoids, but a number of studies have shown that the mineralocorticoid aldosterone is an important component of stress responses in marine mammals10–12. Corticosteroids circulate in blood complexed with binding globulins (e.g. corticosteroid binding globulin); free hormones exert biological effects by binding to cell surface and intracellular receptors – glucocorticoid receptor, GR, and mineralocorticoid receptor, MR – in target tissues (e.g. adipose, muscle, and liver). Cell surface receptors alter signaling pathways, while intracellular hormone-receptor complexes translocate to the nucleus, bind glucocorticoid and mineralocoid response elements of target genes, and impact gene transcription13. Altered expression of target genes, such as those encoding metabolic enzymes and hormones, promotes the physiological adjustments required to overcome the stressor. These adjustments include mobilization of glucose and lipid stores to meet increased energy demands and transient suppression of energetically expensive processes, such as reproduction, growth, and immune responses9. While short-term (acute) stress responses are adaptive14–16, repeated (chronic) stress exposure may deplete energy stores and impair immunity and reproduction, reducing fitness. This could be detrimental to threatened animal populations that already experience nutritional stress and low fecundity, such as some marine mammals17. However, little is known about the indicators and downstream effects of repeated stress in wildlife, hindering the ability of conservation biologists to identify stressed animals and predict the effects of repeated stress on population stability18,19.
Marine mammals rely on endogenous energy reserves to support life history stages characterized by high energy expenditure and nutrient limitation. For example, semiaquatic marine mammals (e.g. pinnipeds – seals and sea lions) reproduce on land using a capital breeding strategy, in which animals rely solely on energy stores to fuel metabolic demands of reproduction20,21. Many fully aquatic marine mammals (e.g. cetaceans) are also capital breeders that migrate long distances between foraging and breeding grounds22. Both examples necessitate energy accumulation and storage during feeding and energy mobilization during fasting, a process that occurs for several weeks to several months as part of marine mammal life history. The main energy depot in cetaceans and pinnipeds is blubber, a specialized type of subcutaneous adipose tissue that plays significant roles in metabolism (similarly to white adipose tissue, WAT, in other mammals) and thermoregulation. Blubber is vertically stratified into at least two layers: an outer layer used for thermoregulation, and an inner, metabolically active layer23. WAT is a major target of stress hormones in mammals24 and studies in marine mammals have shown that acute HPA axis activation mobilizes stored lipids10,25. It is plausible that chronic or repeated stress would deregulate fasting metabolism and deplete lipid stores, among other impacts, limiting the ability of capital breeders to meet the energetic demands of reproduction and leading to declines in fitness.
In this study, we evaluated the effects of repeated HPA axis activation on blubber gene expression in a fasting-adapted marine mammal, the northern elephant seal (Mirounga angustirostris). Elephant seals are an established marine mammal study system in which variation in baseline hormone levels26, responses to acute ACTH administration14,25,27,28, and metabolic adjustments during fasting29,30 have been described. In addition, anesthesia procedures have been shown to minimize handling stress in this species, enabling measurements of true baseline hormone and gene expression levels25. We simulated repeated physiological stress exposure by administering synthetic ACTH to juvenile elephant seals once daily for 4 days, inducing significant elevations in corticosteroid levels that we described in a previous publication12. To evaluate downstream mechanisms by which repeated HPA axis activation may impact metabolic homeostasis, we compared transcriptome responses of elephant seal blubber tissue to the first and last ACTH administration by RNA sequencing (RNAseq). We identified 27 genes that were differentially expressed in blubber during the response to the fourth versus the first ACTH administration, many of which were associated with lipid storage and mobilization. These genes are potential biomarkers for discriminating between acute and repeated ACTH responses in marine mammals, which otherwise cannot be distinguished by cortisol profiles alone.
Materials and Methods
Study animals and experimental design
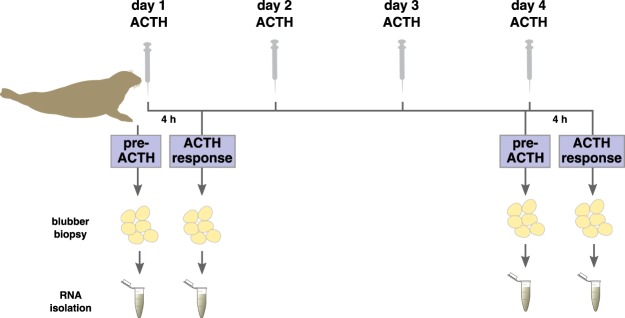
Juvenile (0.8-year old) northern elephant seals (Mirounga angustirostris) were sampled at Año Nuevo State Reserve (San Mateo County, CA) in Aug.–Nov. 2016. All animal handling protocols were approved by Sonoma State University and University of the Pacific Institutional Animal Care and Use Committees (IACUC), Department of the Navy Bureau of Medicine and Surgery (BUMED), and were conducted under National Oceanic and Atmospheric Administration (NOAA) Fisheries Permit No. 19108. All procedures involving animals were conducted in accordance with the relevant guidelines and regulations of IACUC and BUMED protocols and the NOAA permit. Repeated stress was simulated by administering ACTH to each study animal once every 24 hours for 4 consecutive days. Blood and blubber samples were collected before ACTH administration (“pre-ACTH”) and 4 hours after ACTH administration (“ACTH response”) on the first and fourth days (Fig. 1). Details of the experimental manipulation and hormone response have been reported previously12. In summary, ACTH administration induced significant elevation in circulating cortisol and aldosterone levels and decline in total triiodothyronine (T3) levels. Cortisol levels recovered to baseline within 24 hours of each ACTH administration and cortisol responses to ACTH did not vary in magnitude between the first and fourth administrations. However, aldosterone responses to ACTH were higher on day 4 than on day 1 and did not recover to baseline between administrations (for details, see McCormley et al.12). Four of the 7 animals that participated in the repeated ACTH administration experiment described in McCormley et al.12 (seals 2, 4, 6, and 7) were selected for the current study based on amounts of sequencing quality RNA that could be obtained from blubber samples.
Figure 1.
Repeated ACTH administration experiment (described in McCormley et al.12) and blubber tissue sampling design. ACTH was administered to juvenile elephant seals once every 24 hours for 4 days. Blubber biopsies were collected immediately before ACTH administration (“pre-ACTH”) and 4 hours after ACTH administration (“ACTH response”) on the first and fourth days. RNA isolated from blubber tissue was used for transcriptome sequencing. Seal (color modified, from https://pixabay.com), syringe (from https://pixabay.com), adipose (enhanced contrast, from https://openclipart.org), and microcentrifuge tube (from http://www.clker.com) images were obtained under CC0 1.0 Universal: CC0 1.0 Public Domain Dedication. License: https://creativecommons.org/publicdomain/zero/1.0.
ACTH administration and blubber sampling
Study animals were chemically immobilized and blood samples were collected as previously described12. Blubber samples (“day 1 pre-ACTH”) were collected from the posterior flank region of each animal using a 6.0 mm diameter biopsy punch (Miltex, USA). The inner half (closest to musculature) of each biopsy sample was isolated, minced, and placed in cryovials containing RNAlater™ Stabilization Solution (1.5 mL per ~300 mg tissue; Invitrogen, USA). Samples were incubated for 24 hours at 4 °C, after which RNAlater™ solution was removed and samples were stored at −80 °C until RNA isolation. After baseline sampling, animals received 20 U of corticotropin LA gel (Wedgewood Pharmacy, USA) via intramuscular injection (mass specific dose: 0.17 ± 0.02 U/kg). A paired set of blubber and blood samples was collected 4 hours after ACTH administration (“day 1 ACTH response”); biopsies were collected from the contralateral side of the animal to the ACTH injection. On the second and third days of the experiment, ACTH was administered approximately 24 hours after the injection from the previous day, but no post-ACTH sampling was conducted. On the fourth day, pre-ACTH sampling (“day 4 pre-ACTH”), ACTH administration, and post-ACTH sampling (“day 4 ACTH response”) were conducted as described for the first day.
RNA isolation
Blubber samples were homogenized by bead beating in Qiazol (Qiagen, USA; ~100 mg tissue per 0.5 mL) using Bullet Blender Storm 24 (Next Advance, USA; Speed 12, two 2-minute cycles). After bead beating, another 0.5 mL of Qiazol was added to each sample and incubated for 5 minutes at room temperature with occasional vortexing. Lysates were further disrupted using QiaShredder tubes (Qiagen, USA) to shear genomic DNA and centrifuged for 15 minutes to separate lipids and cellular components. RNA was isolated from homogenates using the Lipid RNeasy Tissue Kit (Qiagen, USA) after chloroform (VWR Life Sciences, USA) phase extraction according to manufacturer’s protocol. Genomic DNA was removed using a 20-minute on-column DNase I digest (Qiagen, USA). RNA concentration was quantified using Qubit 3.0 Fluorometer Broad Range RNA Assay (Life Technologies, USA). RNA integrity was evaluated using the Total RNA 6000 Pico kit on the 2100 Bioanalyzer (Agilent Technologies, USA). Mean (±SD) RNA integrity number (RIN) and 28S/18S rRNA abundance ratios were 6.81 ± 0.32 and 1.03 ± 0.27, respectively. We used Qiagen RNAlater™ for preservation of blubber samples and isolation of RNA with RIN ≥ 7 in a previous study14. However, Ambion RNAlater™ reagent used in this study did not adequately prevent RNA degradation. Due to the logistical challenges of the experiment, blubber sampling could not be repeated and we proceeded with RNA sequencing, using ribosomal RNA depletion instead of polyA mRNA enrichment during library preparation to reduce 3′ degradation bias31. Adequate amounts sequencing-quality RNA were obtained from 14 samples: the entire sample set (day 1 pre-ACTH, day 1 ACTH response, day 4 pre-ACTH, day 4 ACTH response) from seals 4, 6 and 7, and two samples (day 1 ACTH response, day 4 ACTH response) from seal 2.
Library preparation and RNA sequencing
cDNA library preparation and Illumina HiSeq 4000 sequencing were conducted at the University of California, Berkeley QB3 Vincent J. Coates Genomics Sequencing Laboratory. Ribosomal RNA (rRNA) was depleted using Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat; Illumina, Inc., USA). Yield and quality of mRNA were assessed using fluorometric methods and an Agilent 2100 Bioanalyzer, respectively (Agilent Technologies, USA). PrepX directional RNAseq library kits (WaferGen Bio-systems Inc., USA) were used for library preparation and libraries were quantified with KAPA Library Quantification kits for Illumina platforms (Kapa Biosystems, USA) using a Bio-Rad CFX Connect (Bio-Rad Laboratories, USA). Thirteen cycles of indexing PCR using KAPA High Fidelity Hotstart Amplification Kits (Kapa Biosystems, USA) were used before library quantification and validation. Individual libraries were barcoded, indexed as TruSeq single index per library, pooled in equimolar concentrations (400 ng), and sequenced as 100 base-pair paired-end reads on the Illumina HiSeq 4000. This generated a total of 150,055 sequenced megabases and a mean (±SD) of 26.9 ± 4.2 million paired-end 100-bp reads per sample. Sequencing data were demultiplexed and converted from bclfile to fastq file with Illumina’s bcl2Fastq software v2.18 (Illumina, USA). Raw data were uploaded to NCBI Sequence Read Archive (SRA accession: SRP157071).
De novo transcriptome assembly
All computational analyses were conducted using the Extreme Science and Engineering Discovery Environment (XSEDE)32 Bridges Large High Performance Computing Cluster at the Pittsburg Supercomputing Center through allocation TG-IBN150010. Transcriptome assembly was conducted using Trinity v2.4.033. Sequencing adapters and poor quality bases were trimmed from reads using Trimmomatic run in Trinity with default settings. Quality of raw and trimmed reads was evaluated using FastQC v0.11.334. De novo assembly was conducted after in silico read normalization (50X coverage, kmer size 25 bp) using Trinity with all default settings (paired-end, strand-specific FR mode). Read mapping metrics were obtained using Bowtie2 v2.2.735 and transcriptome completeness was analyzed using BUSCO v1.2236 (metazoan BUSCO dataset downloaded on 8/24/2017).
Transcriptome annotation
The assembly was annotated by aligning translated DNA query sequences against the UniProtKB/SwissProt protein sequence database (downloaded on 9/29/2017; “BLASTX”) using DIAMOND v.0.8.3137 with e-value threshold for significant matches of 1e-3 and the “more-sensitive” option. Putative protein sequences encoded in the transcriptome were predicted using TransDecoder v3.0.138 and annotated against the UniProtKB/SwissProt protein sequence database (“BLASTP”) using DIAMOND with e-value cutoff of 1e-3. Differentially expressed genes that did not have hits to the SwissProt database were manually annotated against the NCBI RefSeq database using NCBI BLASTX v2.7.1 with an e-value cutoff of 1e-5. Functional annotation of KEGG categories and GO enrichment was performed using DAVID Bioinformatics Resources v6.839 with the human genome as background. Categories were considered significantly enriched at p < 0.05 (adjusted for multiple comparisons using Benjamini-Hochberg correction40).
Gene expression analyses
All gene expression analyses were conducted using the Trinity pipeline. Transcript abundance was estimated using Kallisto v0.43.041. Differential expression analysis was performed at the gene level using the DESeq2 package in Bioconductor v3.542 in R v3.4.1 with false discovery rate cutoff of 0.05 and log2 fold-change cutoff of 1.0. Pairwise comparisons between sampling conditions are shown in Table 3. Libraries from seal 2 were included only in the ACTH response comparison (day 4 ACTH response vs day 1 ACTH response). All other comparisons were conducted using libraries from seals 4, 6, and 7 only. Protein-protein interaction network prediction was conducted using STRING v10.543. Network data were imported into Cytoscape v3.5.044 and filtered by experimental interactions using the edge weighted spring embedded layout. Unconnected nodes were removed from the analysis. Cytoskape network statistics were calculated using the network analyzer tool with an undirected analysis.
Table 3.
Number of DEGs identified in each pairwise comparison using DESeq 2.
| Comparison name | Pairwise comparison | total DEGs |
upreg. DEGs | downreg. DEGs |
annotated unique DEGs |
|---|---|---|---|---|---|
| First ACTH response | Day 1 ACTH response/Day 1 pre-ACTH | 315 | 302 | 13 | 61 |
| Fourth ACTH response | Day 4 ACTH response/Day 4 pre-ACTH | 29 | 21 | 8 | 12 |
| Overall ACTH response | Day 4 ACTH response/Day 1 pre-ACTH | 624 | 479 | 145 | 99 |
| Pre-ACTH comparison | Day 4 pre-ACTH/Day 1 pre-ACTH | 66 | 39 | 27 | 24 |
| ACTH response comparison | Day 4 ACTH response/Day 1 ACTH response | 27 | 23 | 4 | 12 |
DEGs: differentially expressed genes. Annotated unique DEGs: differentially expressed elephant seal homologs of vertebrate proteins with known function, with multiple transcript isoforms collapsed to a single gene.
Results
Blubber transcriptome assembly
Blubber biopsies were collected from 4 juvenile northern elephant seals immediately prior to ACTH administration (“pre-ACTH”) and 4 hours following ACTH administration (“ACTH response”) on the first and fourth days of the experiment (Fig. 1). Morphometric data, ACTH doses, and corticosteroid concentrations measured at the four sampling points are presented in Table 1 (from McCormley et al.12). Fourteen cDNA libraries from day 1 pre-ACTH (seals 4, 6, and 7), day 1 ACTH response (seals 2, 4, 6, and 7), day 4 pre-ACTH (seals 4, 6, and 7), and day 4 ACTH response (seals 2, 4, 6, and 7) were sequenced using two Illumina HiSeq 4000 lanes. The libraries from seal 2 were used for assembly but were included only in the ACTH response comparison to increase power for detection of differentially expressed genes (see Methods). Raw sequencing reads were uploaded to NCBI Short Read Archive (BioProject ID: PRJNA485363, SRA accession: SRP157071). A single reference transcriptome was assembled de novo using Trinity software. We assembled 1.36 billion bases into 2,031,456 contigs (or “transcripts”) in 1,216,779 gene clusters (Table 2). The high number of transcripts is common for de novo assemblies45 and is likely a result of sequence polymorphism, alternative splicing, and variability in individuals’ responses to ACTH. The transcriptome assembly is available at figshare: (https://figshare.com/s/37cba07b2e9877f25b50).
Table 1.
Experimental data for juvenile elephant seals used in the RNAseq study, including cortisol and aldosterone concentrations measured immediately before and 4 hours following administration of ACTH (pre-ACTH and ACTH response, respectively; from McCormley et al.12).
| Subject | Sex | Mass (kg) | ACTH (U/kg) | Day | pre-ACTH cortisol (nM) | ACTH response cortisol (nM) | pre-ACTH aldosterone (pM) | ACTH response aldosterone (pM) |
|---|---|---|---|---|---|---|---|---|
| Seal 2 | M | 119 | 0.17 | 1 4 |
198.7 202.2 |
760.2 2626.1 |
812.6 1204.2 |
1465.8 2110.5 |
| Seal 4 | M | 125 | 0.16 | 1 4 |
429.1 227.7 |
1298.5 1704.1 |
828.4 1476.5 |
2000.1 2608.5 |
| Seal 6 | F | 103 | 0.19 | 1 4 |
357.9 494.3 |
1542.1 4454.3 |
670.9 1610.1 |
1251.1 2747.1 |
| Seal 7 | F | 99 | 0.20 | 1 4 |
394.6 355.5 |
2598.6 2319.2 |
792.2 1291.3 |
1415.0 2123.0 |
ACTH: adrenocorticotropic hormone.
Aldosterone responses to the fourth ACTH administration were significantly enhanced relative to the first (p < 0.05), while cortisol responses to the first and fourth ACTH administrations did not vary in magnitude12.
Table 2.
Transcriptome assembly metrics.
| Metric | Value |
|---|---|
| Mean (±SD) reads per sample | 26.9 ± 4.2 million |
| Assembled bases | 1,361,692,561 |
| Assembled contigs (transcripts) | 2,031,456 |
| Assembled transcript families | 1,216,779 |
| Annotated transcripts | 372,783 |
| Mean transcript length | 670.3 bp |
| Read mapping rate | 92.40% |
| Complete metazoan BUSCOs | 85.8% |
| Complete and single-copy metazoan BUSCOs | 51.6% |
| Complete and duplicated metazoan BUSCOs | 34.1% |
| Fragmented metazoan BUSCOs | 13.8% |
| Missing metazoan BUSCOs | 0.4% |
BUSCOs: Benchmarking Universal Single-Copy Orthologs. Read mapping rate: percent of sequenced reads mapping as proper pairs back to the assembly.
To evaluate read representation in the reference transcriptome, we mapped sequenced reads back to the assembly. Ninety-two percent of reads mapped back as proper pairs to the assembly, suggesting that it accurately represents sequenced reads (Table 2). To evaluate assembly completeness, we performed a search for metazoan Benchmarking Universal Single-Copy Orthologs (BUSCOs). Out of 978 metazoan BUSCOs in the OrthoDB catalog, 85.8% were present as complete orthologs in the seal assembly, 51.6% had matches to a single seal transcript, 34.1% were duplicated, 13.8% were fragmented, and only 0.4% were missing from the assembly (Table 2). This suggests that the blubber assembly is fairly complete as it contains the majority of orthologs predicted to be expressed in all metazoans. Due to the large number of short and potentially redundant transcripts in the assembly, we filtered the transcriptome using Transdecoder to retain only transcripts with putative protein-coding regions. The in silico-translated proteome contained 266,916 predicted proteins, some of which were predicted from multiple open reading frames of the same transcripts.
To identify seal homologs of vertebrate genes with known functions, the raw transcriptome assembly was annotated by translated nucleotide search (BLASTX) and the Transdecoder-predicted peptides were annotated by protein search (BLASTP) against the UniProt SwissProt protein database (e-value threshold 1e-3). BLASTX homologs were detected for 372,783 elephant seal transcripts and BLASTP homologs were detected for 164,557 Transdecoder-predicted elephant seal proteins. Transcriptome annotation data are available at figshare (https://figshare.com/s/37cba07b2e9877f25b50). To identify gene functional categories (KEGG pathways) overrepresented in the blubber transcriptome, we used the DAVID functional annotation tool. Fifty-two KEGG pathways were enriched in the assembly relative to the human genome background (adjusted p < 0.05). The largest category was metabolic pathways, which contained 882 elephant seal homologs. Other pathways of interest included the child categories purine metabolism (135 genes), insulin signaling pathway (112 genes), Wnt signaling pathway (102 genes), AMPK signaling pathway (101 genes), insulin resistance (96 genes), thyroid hormone signaling pathway (94 genes), carbon metabolism (91 genes), adipocytokine signaling pathway (58 genes), fatty acid metabolism (44 genes), and fatty acid degradation (39 genes), among others. These KEGG enrichment data were similar to those from other elephant seal blubber transcriptomes14,46.
Differential gene expression
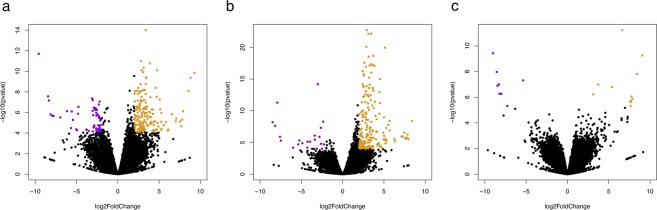
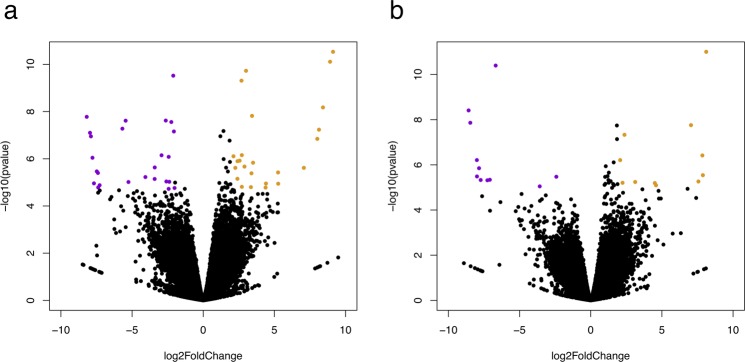
Differences in transcript abundance between sampling conditions were identified using DESeq2. The numbers of differentially expressed (DEG) and annotated genes (adjusted p-value < 0.05 and FDR < 0.05) identified in each pairwise comparison between conditions are shown in Table 3. The magnitude and significance of gene expression differences in each comparison are presented in Figs 2 and 3. We first examined changes in gene expression in response to the first ACTH administration, fourth ACTH administration, and over the course of the entire experiment (day 1 pre-ACTH vs. day 4 ACTH response; “overall ACTH response;” Fig. 2). We then compared expression between day 1 and day 4 pre-ACTH conditions (“pre-ACTH comparison”) and between day 1 and day 4 ACTH responses (“ACTH response comparison,” Fig. 3).
Figure 2.
Volcano plots showing log2 fold change (x-axis) and significance (-log10 * adjusted p-value; y-axis) of genes differentially expressed during the overall ACTH response (a), first ACTH response (b), and fourth ACTH response (c). Significantly upregulated and downregulated genes are shown in orange and purple, respectively. Log2 fold-change and significance cutoffs for differential expression were |1| and 0.05, respectively.
Figure 3.
Volcano plots showing log2 fold change (x-axis) and significance (-log10 * adjusted p-value; y-axis) of genes differentially expressed in the pre-ACTH (a) and ACTH response (b) comparisons. Significantly upregulated and downregulated genes are shown in orange and purple, respectively. Log2 fold-change and significance cutoffs for differential expression were |1| and 0.05, respectively.
We identified 61 annotated, unique (i.e. all transcript isoforms collapsed into a single gene) DEGs in response to the first ACTH administration (Supplementary File 1), 12 DEGs in response to the fourth ACTH administration (Supplementary File 2), 99 DEGs in the overall ACTH response (Supplemetary File 3), 24 DEGs in the pre-ACTH comparison (Table 4, Supplementary File 4), and 12 DEGs in the ACTH response comparison (Table 5, Supplementary File 5). Several DEGs had hits to poorly characterized genomic regions of other mammals (first ACTH response: 31, fourth ACTH response: 7, overall ACTH response: 34, pre-ACTH comparison: 8, ACTH response comparison: 6), and thus could not provide functional insights. Functional annotation of DEG lists identified 2 Gene Ontology (GO) biological process categories enriched in first ACTH response (positive regulation of fat cell differentiation, negative regulation of transcription from RNA polymerase II promoter), 18 GO categories enriched in overall ACTH response (including fatty acid transport, long-chain fatty acid metabolic process, long-chain fatty acid-CoA ligase activity, positive regulation of fat cell differentiation, response to hypoxia, transcription factor binding) and 1 GO category enriched in the ACTH response comparison (zinc ion binding). KEGG categories were significantly enriched only in the overall ACTH response dataset (ECM-receptor interaction, focal adhesion, pathways in cancer, and small cell lung cancer).
Table 4.
Genes differentially expressed in the pre-ACTH comparison (between pre-ACTH samples from day 1 and pre-ACTH samples from day 4).
| Transcript | Log2 FC | Uniprot or NCBI Accession | Gene Name | Pathway/Process |
|---|---|---|---|---|
| TRINITY_DN590927_c3_g12 | 8.41 | GCNT7_PIG | Beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase 7 (GCNT7) | protein glycosylation |
| TRINITY_DN560952_c14_g1 | 5.27 | XP_020935075.1 | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 1 isoform X1 [Sus scrofa] (PFKFB1) | glycolysis and gluconeogenesis, response to GCs |
| TRINITY_DN566526_c4_g1 | 3.00 | XM_008712174.1 | PREDICTED: Ursus maritimus THAP domain containing 8 (THAP8), transcript variant X2, mRNA (THAP8) | transcription factor |
| TRINITY_DN590504_c4_g4 | 2.90 | ACSM1_BOVIN | Acyl-coenzyme A synthetase ACSM1, mitochondrial precursor (ACSM1) |
fatty acid activation and metabolism |
| TRINITY_DN560507_c9_g1 | 2.69 | HMCS2_HUMAN | Hydroxymethylglutaryl-CoA synthase, mitochondrial precursor (HMGCS2) | ketogenesis |
| TRINITY_DN592323_c1_g1 | 2.41 | TGO1_BOVIN | Transport and Golgi organization protein 1 homolog precursor (MIA3) |
vesicular transport, collagen secretion |
| TRINITY_DN578525_c2_g2 | 2.13 | 209L2_MACMU | CD209 antigen-like protein 2 (CD209L2) | immune response |
| TRINITY_DN568360_c2_g1 | 1.91 | KCRU_HUMAN | Creatine kinase U-type, mitochondrial precursor (CKMT1A) |
phosphocreatine synthesis |
| TRINITY_DN591927_c4_g2 | 1.43 | XM_006736873.1 | cysteine dioxygenase type 1 (CDO1) | adipogenesis, taurine biosynthesis, cysteine degradation |
| TRINITY_DN571555_c4_g2 | 1.42 | GPX3_RAT | Glutathione peroxidase 3 precursor (GPX3) | antioxidant response |
| TRINITY_DN572974_c28_g1 | 1.33 | XM_021692420.1 | adiponectin, C1Q and collagen domain containing (ADIPOQ) | fatty acid oxidation, insulin sensitivity |
| TRINITY_DN566880_c2_g2 | 1.20 | HUTH_BOVIN | Histidine ammonia-lyase (HAL) | amino acid degradation |
| TRINITY_DN592891_c7_g1 | 1.13 | MGST1_PIG | Microsomal glutathione S-transferase 1 (MGST1) | antioxidant response |
| TRINITY_DN575636_c4_g1 | −1.30 | ZBT46_MOUSE | Zinc finger and BTB domain-containing protein 46 (Zbtb46) |
transcriptional repression, immune cell development |
| TRINITY_DN576776_c0_g1 | −2.02 | ITA2_HUMAN | Integrin alpha-2 precursor (ITGA2) |
ECM organization, cell-matrix adhesion |
| TRINITY_DN545745_c4_g5 | −2.06 | FMOD_HUMAN | Fibromodulin precursor (FMOD) | collagen fibril formation |
| TRINITY_DN57258 _c0_g6 | −2.42 | HBA_ODORO | Hemoglobin subunit alpha (HBA) |
oxygen transport |
FC: fold change (day 4 pre-ACTH/day 1 pre-ACTH); adjusted p < 0.05.
Table 5.
Genes differentially expressed in the ACTH response comparison (between ACTH response samples from day 1 and ACTH response samples from day 4).
| Transcript ID | Log2 FC | Uniprot or NCBI Accession | Gene Name | Pathway/Process |
|---|---|---|---|---|
| TRINITY_DN551341_c9_g1 | 1.94 | GLRA2_RAT | Glycine receptor subunit alpha-2 (GLRA2) | glycine receptor |
| TRINITY_DN570625_c1_g4 | 1.81 | ATS16_HUMAN | A disintegrin and metalloproteinase with thrombospondin motifs 16 (ADAMTS16) | blood pressure regulation |
| TRINITY_DN587322_c7_g1 | 1.23 | PIAS4_HUMAN | E3 SUMO-protein ligase PIAS4 (PIAS4) | protein sumoylation, cell stress response, metabolic homeostasis |
| TRINITY_DN592677_c7_g3 | 1.18 | XM_022511147.1 | Perilipin 1 (PLIN1) | lipolysis regulation, insulin sensitivity |
| TRINITY_DN549777_c0_g1 | 1.09 | PLIN4_HUMAN | Perilipin-4 (PLIN4) | triacylglyceride packaging |
| TRINITY_DN587150_c2_g2 | 1.06 | ZA2G_BOVIN | Zinc-alpha-2-glycoprotein (AZGP1) | lipid mobilization |
| TRINITY_DN591317_c4_g2 | 1.04 | ACSL1_MOUSE | Long-chain-fatty-acid–CoA ligase 1 (ACSL1) | fatty acid catabolism, fatty acid synthesis |
| TRINITY_DN587426_c4_g1 | 1.02 | CIDEA_MOUSE | Cell death activator CIDE-A (CIDEA) | lipolysis regulation, insulin sensitivity |
| TRINITY_DN591034_c2_g1 | −1.96 | BGH3_PIG | Transforming growth factor-beta-induced protein ig-h3 (TGFBI) | cell adhesion, adipogenesis |
| TRINITY_DN589692_c4_g2 | −2.82 | XM_002919434.3 | PREDICTED: Ailuropoda melanoleuca C1q and tumor necrosis factor related protein 3, mRNA (C1QTNF3) | inhibition of gluconeogenesis and inflammation |
FC: fold change (day 4 ACTH response/day 1 ACTH response); adjusted p < 0.05.
We next manually categorized unique annotated DEGs based on known function using literature search to understand their potential roles in repeated stress responses and metabolic homeostasis. Genes that were upregulated (n = 58) during the first ACTH response included many known GC targets and genes of interest involved in oxidative stress defenses (GPX3), fatty acid oxidation (ACADM), immune function (TLR4), adipogenesis (DKK1), lipid and monocarboxylate transport (SLC16A9), ketogenesis (HMGCS2), insulin resistance and obesity (ZBTB16), NADP biosynthesis and saturated fatty acid metabolism (NADK2), circadian rhythm (PER1), and inactive steroid receptor complexes (FKBP5). Other differentially expressed genes were associated with sphingolipid metabolism, DNA replication and repair, cytoskeleton and ECM remodeling, protein synthesis and posttranslational modification, protein transport, and embryonic development and cell differentiation. Genes downregulated (n = 3) during the first ACTH response were associated with ECM remodeling and insulin resistance (ADAMTS9), cell growth and migration (ABI2), and negative regulation of tyrosine kinase activity (PTPN12) (Supplementary File 1).
Genes upregulated (n = 10) during the fourth ACTH response included those associated with protein degradation (CACYBP), inhibition of apoptosis (DYRK3), and transcriptional regulation (NCOR1), as well as those involved in cell adhesion and migration, sphingolipid biosynthesis, and nucleotide metabolism. Genes downregulated (n = 2) during the fourth ACTH response included an activin receptor associated with adipogenesis (ACVR2A) and a protein glycosylation enzyme (Supplementary File 2). Only two DEGs (ABI2 and SPTSSB) were common between day 1 and day 4 ACTH responses.
Genes of interest that were upregulated (n = 75) during the overall ACTH response included those associated with lipolysis (CES1D), ketogenesis (HMGCS2), adipogenesis (DKK1), oxidative stress (FMO2), regulation of energy balance (LEP), fatty acid oxidation (ACSL1), fatty acid biosynthesis (ACSM1, ACSM4, SCD2, GPD1), lipid binding and transport (APOD, SLC27A6, ABCA6, ABCA10), steroid hormone transport (SLC10A6), sphingolipid metabolism (ACER2), and amino acid transport (SLC1A3). Upregulated genes also included an enzyme that links glycolytic and triglyceride synthesis pathways (GPD1), angiotensinogen (AGT) precursor, and a transcriptional activator that increases sensitivity to low GC concentrations (GMEB1). Genes of interest that were downregulated (n = 24) over the course of the entire experiment included those associated with inflammation and insulin resistance (CD44), stress and hormone responses (DRG1), local production of triiodothyronine (DIO2), regulation of glycolysis (PFKFB1), gene transcription and adipocyte differentiation (JUNB), as well as those involved in ECM remodeling and cell adhesion (Supplementary File 3).
Annotated genes upregulated (n = 13) in the pre-ACTH comparison were associated with processes of interest including glycolysis (PFKFB1), ketogenesis (HMGCS2), fatty acid activation and metabolism (ACSM1), regulation of fatty acid oxidation and insulin sensitivity (ADIPOQ), adipogenesis (CDO1), protection from oxidative damage (GPX3, MGST1), immune response (CD209L2), and amino acid degradation (HAL). Other upregulated genes were associated with phosphocreatine synthesis, collagen secretion, gene transcription, and protein glycosylation. Genes downregulated (n = 11) in the pre-ACTH comparison were associated with transcription (ZBTB46), suppression of inflammation (SETD6), oxidative stress protection (GDAP1) and transcriptional repression by nuclear receptors and inhibition of adipogenesis (NCOR1). Other downregulated genes were associated with ECM organization, cell adhesion, and motility, hematopoiesis and oxygen transport (Table 4, Supplementary File 4).
Annotated genes that were upregulated (n = 9) in the ACTH response comparison included those involved in lipid droplet formation (PLIN1, PLIN4, CIDEA), lipolysis (AZGP1), fatty acid metabolism (ACSL1), as well as cell adhesion and ECM remodeling, protein sumoylation, and glycine binding. Genes downregulated in the ACTH response comparison (n = 3) included those involved in adipogenesis, adipokine signaling, and fatty acid uptake (THBS1), lipid oxidation (C1QTNF3), and cell adhesion (TGFBI)(Table 5, Supplementary File 5).
Lastly, we examined the potential for interaction between proteins encoded by annotated DEGs by conducting protein-protein interaction (PPI) network analysis using the overall ACTH response dataset. The predicted PPI network contained 20 nodes, 25 edges, network density of 0.132, network heterogeneity of 0.687, network centralization of 0.263, average local clustering coefficient of 0.135, and PPI network enrichment p-value of 0.00386 (Fig. 4), suggesting that DEGs are likely functioning as a coordinated network to regulate cellular and metabolic responses to repeated ACTH administration.
Figure 4.
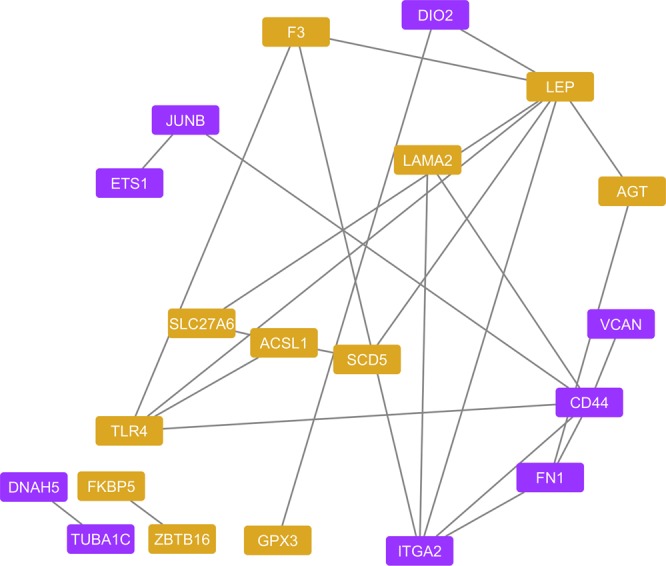
Predicted protein-protein interaction (PPI) network (p < 0.05) for genes differentially expressed over the course of the experiment (overall ACTH response). Upregulated and downregulated genes are shown in orange and purple, respectively.
Discussion
Overall, this study shows that gene expression differences between single and repeated ACTH responses in marine mammal blubber are detectable despite similarities in cortisol responses, providing more sensitive indication of stress response states than endocrine profiles alone. We administered ACTH once daily for four days to juvenile northern elephant seals and previously reported significant elevation in corticosteroid levels in response to the first and fourth administrations12. However, while the aldosterone response was facilitated by repeated ACTH administration, the cortisol response was not, highlighting the unreliability of glucocorticoid measurements alone for discriminating between acute and repeated HPA axis activation. Therefore, we used RNAseq to examine downstream changes in blubber gene expression in response to repeated ACTH administration. Such changes in blubber gene activity reflect activation of genomic and cell-surface hormone receptors, including glucocorticoid receptor (GR), mineralocorticoid receptor (MR), and the adrenocorticotropic hormone receptor (MC2R) in multiple cell types, capturing the complexity of target tissue responses47–49. We identified DEGs during the responses to both the first and fourth ACTH administration, although the number of DEGs identified in response to the latter was much smaller than the former. We also identified transcriptional differences between pre-ACTH and post-ACTH response states from the first and fourth days. Annotated DEGs were associated with functions such as lipid and carbohydrate metabolism, adipogenesis, adipokine regulation of energy balance, insulin resistance, oxidative stress, transcriptional regulation, and ECM remodeling in other mammals and provide insights into mechanisms by which repeated HPA axis activation may affect marine mammal physiology.
Genes upregulated in response to the first ACTH challenge were associated with protection from oxidative damage, fatty acid oxidation, immune response, adipogenesis, nutrient transport, sphingolipid and ketone metabolism, cell differentiation, cytoskeletal and ECM remodeling, and insulin resistance and obesity in other animals, while downregulated genes were associated with ECM remodeling and cell migration. Several identified DEGs are primarily known for their roles in other biological processes (e.g. embryonic development) and have only recently been implicated in lipid metabolism and adipogenesis. These include GDF3 and DKK1, which promote adipogenesis and metabolic homeostasis50,51, and the transcriptional regulator ZBTB16, which plays a role in obesity and brown adipose energetics52. This is consistent with pro-adipogenic effects of GCs in other mammals24. Other genes upregulated in response to the first ACTH adminstration included those associated with immune response, which is consistent with studies in other mammals in which acute stress enhanced immune capacity, while chronic stress had immunosuppressive effects53. DKK1 and similar functional classes of genes associated with adipogenesis, immune response, and extracellular matrix function were also upregulated in blubber 2 hours after a single ACTH administration14. This suggests that transcriptional changes measured 2 and 4 hours after ACTH administration are capturing the same biological response in blubber. Upregulation of genes involved in lipid metabolism is consistent with known lipolytic effects of corticosteroids and the increase in circulating free fatty acid levels that has been measured in response to ACTH in this species25,28,54. Interestingly, one upregulated gene, FKBP5, is a GR antagonist55, suggesting a potential negative feedback mechanism to limit acute responses to ACTH. Genes downregulated in response to the first ACTH administration were assocated with ECM remodeling and cell migration, suggesting suppression of some energy-intensive functions during the ACTH response. However, many upregulated genes were also involved in ECM remodeling, highlighting the complexity of this specialized connective tissue.
Genes upregulated in response to the fourth ACTH administration included those associated with regulation of transcription, protein degradation, inhibition of apoptosis, and cell migration, while downregulated genes included those associated with adipogenesis and post-translational protein modification. Significantly fewer DEGs were identified in response to the fourth ACTH administration compared to the first, potentially because many cellular responses were already engaged by the previous ACTH responses. Upregulated genes included two that may protect blubber tissue from deleterious effects of repeated corticosteroid exposure: DYRK3, a kinase that phosphorylates and activates the sirtuin SIRT1 and promotes cell survival during stress56 and NCOR1, a nuclear corepressor that mediates ligand-induced downregulation of the GR gene57. DYRK3 was also upregulated in response to a single ACTH administration in a previous study14. Surprisingly, the type II BMP receptor ACVR2A, which promotes adipogenesis by GDF6 signaling58, was downregulated during the fourth ACTH response, whereas pro-adipogenic factors were upregulated in response to a single ACTH administration in this and a previous study14. These results suggest that the transcriptional response to a fourth sequential ACTH administration may serve to promote cell survival and limit adipogenesis and GC sensitivity. However, it is unclear whether this potential negative feedback is sufficient to prevent deleterious consequences of repeated HPA axis activation, as there were dramatic gene expression differences between the baseline state on day 1 and the ACTH response state on day 4.
The largest number of DEGs was identified in the overall ACTH response comparison, as this comparison captured all gene expression differences between pre-ACTH samples on day 1 and post-ACTH samples on day 4. Upregulated genes of interest suggest that repeated ACTH administration impacted lipid and ketone metabolism, adipogenesis, redox homeostasis, and thyroid hormone signaling. Upregulation of genes encoding leptin and the ketogenesis enzyme HMGCS2 suggests a potential increase in energy expenditure and ketone production to support metabolic demands of repeated HPA axis activation. However, leptin is also involved in modulating immune responses59, and its role in elephant seal metabolism is currently unclear60. Genes associated with lipolysis and fatty acid oxidation were upregulated concomitant with those involved in fatty acid and triglyceride synthesis (e.g. ACSM1). ACSM1 was also upregulated 24 hours after a single ACTH administration in a previous study14. Glucorticoid-associated lipogenesis has been described in obese humans24,61, and may be mediated by MR62. Upregulation of GPD1, which promotes triglyceride synthesis in response to excess glucose, and AGT, a component of the renin-angiotensin (RAS) system associated with obesity, suggest that repeated ACTH administration may potentially increase lipogenesis in a naturally obese and insulin-resistant mammal30. Downregulation of the iodothyronine deiodinase DIO2, which converts thyroxine to bioactive triiodothyronine (T3), is consistent with suppression of circulating T3 levels after repeated ACTH administration that was shown in the same animals12. Therefore, these data suggest that fasting-adapted mammals may simultaneously metabolize and maintain lipid stores during repeated HPA axis perturbation. However, suppression of T3 and increased lipogenesis may decrease energy expenditure and impair the ability of fasting-adapted marine mammals to participate in energetically demanding life history stages.
Our previous studies have shown that while cortisol levels returned to baseline within 24 hours of each ACTH administration12, changes in gene expression in response to ACTH could be detected even when cortisol was no longer elevated14. To identify whether transcriptional changes alone can be used to detect repeated ACTH exposure, we compared gene expression profiles in blubber samples collected immediately prior to the first and fourth ACTH administrations. Genes upregulated in pre-ACTH samples from day 4 relative to day 1 were associated with regulation of glycolysis, ketogenesis, fatty acid biosynthesis, adipokine signaling, oxidative stress, and adipogenesis. Downregulated genes were associated with genetic and epigenetic regulation of transcription, mitochondrial dynamics, and immune cell differentiation. These results suggest that recovery from repeated ACTH administration may involve metabolic and cellular adjustments such as increased glycolysis and fatty acid oxidation, synthesis of fatty acids and ketones, antioxidant responses, and increased adipogenesis. Upregulation of antioxidant enzymes GPX3 and MGST1 may serve to protect seal tissues from oxidative damage associated with exposure to GCs63,64. Two genes upregulated in day 4 pre-ACTH samples (24 hours after the third ACTH administration) – ACSM1 and CDO1 – were also upregulated 24 hours after a single ACTH administration in a previous study14. However, genes associated with ketogenesis and antioxidant defenses were not altered in response to a single ACTH challenge and may therefore serve as markers of recently experienced repeated HPA axis activation.
To identify transcriptional differences between blubber responses to single and repeated ACTH administrations, we compared the ACTH response samples from days 1 and 4. We identified nine DEGs that were upregulated and three that were downregulated in day 4 ACTH response samples compared to those from day 1, and suggest them as potential markers of repeated HPA axis stimulation in marine mammal blubber. Upregulated genes were associated with lipid droplet formation, energy homeostasis, and lipolysis, while downregulated genes were associated with adipogenesis and lipid uptake and oxidation. Three upregulated genes encoded the lipid droplet-associated proteins CIDEA and the perilipins PLIN1 and PLIN4, which are involved in regulation of lipolysis and lipid droplet expansion65. While GCs have been shown to upregulate genes encoding some lipid droplet proteins66,67, they mainly affect perilipins by inhibitory phosphorylation that facilitates lipolysis68. Transcriptional upregulation of perilipins in response to repeated ACTH administration may function to restrict lipolysis in an attempt to conserve lipid stores in a fasting-adapted mammal. Other genes upregulated during the fourth ACTH response compared to the first included the glycoprotein AZGP1 and glycine receptor GLRA2, which promote lipolysis69, ADAMTS16, which is associated with regulation of blood pressure70, PIAS4, which inhibits AMPK and SIRT1 (and thus derepresses lipid and protein synthesis71,72), and CNTNAP3, a cell adhesion protein that is a marker of brown adipocytes73. Downregulated genes included THBS1, which is associated with adipocyte proliferation and fatty acid uptake74, C1QTNF3, an adipokine that increases hepatic lipid oxidation and decreases lipid synthesis, gluconeogenesis and inflammation75, and TGFBI, a TGF beta family member associated with type 2 diabetes and adipogenesis76. While interpretation of DEG roles in ACTH responses is complicated by their pleiotropic (or poorly described) functions and the heterogeneity of cell types in blubber tissue, these data suggest that repeated ACTH administration primarily increases lipid catabolism and dysregulates lipid storage. This is consistent with significant declines in circulating triglyceride levels detected after 4 ACTH administrations77. Therefore, repeated HPA axis activation may impact the ability of marine mammals to maintain prolonged fasts and high levels of energy expenditure required to participate in key life history stages. For example, depletion of energy reserves by repeated stress would impact reproductive expenditure (e.g. milk production, male-male competition), decreasing fitness78.
To our knowledge, this study is the first to examine changes in gene expression in wild marine mammals in response to repeated ACTH administration. Despite the limitations of small sample size, high degree of individual variability in ACTH responses, and small number of ACTH administrations, we identified dozens of DEGs that were differentially expressed in response to repeated ACTH administration. Most significally, we identified gene expression differences between single and repeated ACTH responses in blubber despite similarities in cortisol responses, providing more sensitive indication of response states than endocrine profiles alone. Further work is necessary to correlate transcriptional changes with metabolic and redox profiles and validate the ability of these biomarkers to discriminate baseline stress states in marine mammals. Nevertheless, these findings provide some of the first insights into the physiological impacts of repeated HPA axis stimulation in free-ranging marine mammals, and suggest potential molecular markers of repeated stress exposure that can complement other approaches used by conservation biologists to evaluate the effects of anthropogenic activity on threatened wildlife populations.
Supplementary information
Acknowledgements
The authors are grateful to Drs. D. Costa and P. Robinson, the rangers at Año Nuevo State Park, and UC Santa Cruz’s Año Nuevo State Reserve for facilitating access to the animals. This is scientific contribution #226 of the National Marine Mammal Foundation. This work was supported by Office of Naval Research Award number N00014-15-1-2773 to C.D. Champagne, J. I. Khudyakov, D.E. Crocker, and D.S. Houser. This work used the Extreme Science and Engineering Discovery Environment (XSEDE; allocation TG-IBN150010 to J.I. Khudyakov), which is supported by National Science Foundation grant number ACI-1548562.
Author Contributions
J.I.K., C.D.C., D.E.C. and D.S.H. conceived the study. C.D.C., J.I.K., D.E.C., J.S.D., A.P.S. and M.C.M. participated in sample collection and sample processing. J.S.D. and J.I.K. isolated RNA for sequencing. J.S.D., L.P.B. and J.I.K. analyzed the data. J.S.D. and J.I.K. drafted the manuscript. C.D.C., D.E.C., M.C.M., L.P.B., A.P.S. and D.S.H. edited, revised, and approved the final version of the manuscript.
Data Availability
Raw sequencing reads were uploaded to NCBI Short Read Archive (BioProject ID: PRJNA485363, SRA accession: SRP157071). The transcriptome assembly and annotation data are available at figshare (https://figshare.com/s/37cba07b2e9877f25b50).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39089-2.
References
- 1.Ceballos G, et al. Accelerated modern human-induced species losses: Entering the sixth mass extinction. Sci Adv. 2015;1:e1400253. doi: 10.1126/sciadv.1400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ripple WJ, et al. Status and ecological effects of the world’s largest carnivores. Science. 2014;343:1241484. doi: 10.1126/science.1241484. [DOI] [PubMed] [Google Scholar]
- 3.Springer AM, et al. Sequential megafaunal collapse in the North Pacific Ocean: an ongoing legacy of industrial whaling? Proc Natl Acad Sci USA. 2003;100:12223–12228. doi: 10.1073/pnas.1635156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maxwell SM, et al. Cumulative human impacts on marine predators. Nat Commun. 2013;4:2688. doi: 10.1038/ncomms3688. [DOI] [PubMed] [Google Scholar]
- 5.Davidson AD, et al. Drivers and hotspots of extinction risk in marine mammals. Proc Natl Acad Sci USA. 2012;109:3395–3400. doi: 10.1073/pnas.1121469109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkinson S, Demaster DP, Calkins DG. Anthropogenic causes of the western steller sea lion eumetopias jubatus population decline and their threat to recovery. Mammal Review. 2008;38:1–18. doi: 10.1111/j.1365-2907.2008.00128.x. [DOI] [Google Scholar]
- 7.Kunc, H. P., McLaughlin, K. E. & Schmidt, R. Aquatic noise pollution: implications for individuals, populations, and ecosystems. Proc Biol Sci283, 10.1098/rspb.2016.0839 (2016). [DOI] [PMC free article] [PubMed]
- 8.Boonstra R, Fox C. Reality as the leading cause of stress: rethinking the impact of chronic stress in nature. Functional Ecology. 2013;27:11–23. doi: 10.1111/1365-2435.12008. [DOI] [Google Scholar]
- 9.Sapolsky RM, Romero LM, Munck AU. How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr Rev. 2000;21:55–89. doi: 10.1210/edrv.21.1.0389. [DOI] [PubMed] [Google Scholar]
- 10.Champagne, C. D. et al. Comprehensive endocrine response to acute stress in the bottlenose dolphin from serum, blubber, and feces. Gen Comp Endocrinol, 10.1016/j.ygcen.2018.05.015 (2018). [DOI] [PubMed]
- 11.Atkinson S, Crocker D, Houser D, Mashburn K. Stress physiology in marine mammals: how well do they fit the terrestrial model? J Comp Physiol B. 2015;185:463–486. doi: 10.1007/s00360-015-0901-0. [DOI] [PubMed] [Google Scholar]
- 12.McCormley MC, et al. Repeated adrenocorticotropic hormone administration alters adrenal and thyroid hormones in free-ranging elephant seals. Conserv Physiol. 2018;6:coy040. doi: 10.1093/conphys/coy040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadmiel M, Cidlowski JA. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci. 2013;34:518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khudyakov JI, Champagne CD, Meneghetti LM, Crocker DE. Blubber transcriptome response to acute stress axis activation involves transient changes in adipogenesis and lipolysis in a fasting-adapted marine mammal. Sci Rep. 2017;7:42110. doi: 10.1038/srep42110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breuner CW, Patterson SH, Hahn TP. In search of relationships between the acute adrenocortical response and fitness. Gen Comp Endocrinol. 2008;157:288–295. doi: 10.1016/j.ygcen.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 16.Christiansen JJ, et al. Effects of cortisol on carbohydrate, lipid, and protein metabolism: studies of acute cortisol withdrawal in adrenocortical failure. J Clin Endocrinol Metab. 2007;92:3553–3559. doi: 10.1210/jc.2007-0445. [DOI] [PubMed] [Google Scholar]
- 17.Fujiwara M, Caswell H. Demography of the endangered North Atlantic right whale. Nature. 2001;414:537–541. doi: 10.1038/35107054. [DOI] [PubMed] [Google Scholar]
- 18.Pirotta, E. et al. Understanding the population consequences of disturbance. Ecology and Evolution, 10.1002/ece3.4458 (2018). [DOI] [PMC free article] [PubMed]
- 19.Breuner CW, Delehanty B, Boonstra R. Evaluating stress in natural populations of vertebrates: total CORT is not good enough. Functional Ecology. 2013;27:24–36. doi: 10.1111/1365-2435.12016. [DOI] [Google Scholar]
- 20.Champagne, C., Crocker, D., Fowler, M. A. & Houser, D. Fasting Physiology of the Pinnipeds: The Challenges of Fasting While Maintaining High Energy Expediture and Nutrient Delivery for Lactation 309–336 (Springer, Berlin, Heidelberg, 2012).
- 21.Boyd IL. State-dependent fertility in pinnipeds: contrasting capital and income breeders. Functional Ecology. 2000;14:623–630. doi: 10.1046/j.1365-2435.2000.t01-1-00463.x. [DOI] [Google Scholar]
- 22.Braithwaite JE, Meeuwig JJ, Hipsey MR. Optimal migration energetics of humpback whales and the implications of disturbance. Conserv Physiol. 2015;3:cov001. doi: 10.1093/conphys/cov001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strandberg U, et al. Stratification, composition, and function of marine mammal blubber: the ecology of fatty acids in marine mammals. Physiol Biochem Zool. 2008;81:473–485. doi: 10.1086/589108. [DOI] [PubMed] [Google Scholar]
- 24.Lee MJ, Pramyothin P, Karastergiou K, Fried SK. Deconstructing the roles of glucocorticoids in adipose tissue biology and the development of central obesity. Biochim Biophys Acta. 2014;1842:473–481. doi: 10.1016/j.bbadis.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Champagne C, Tift M, Houser D, Crocker D. Adrenal sensitivity to stress is maintained despite variation in baseline glucocorticoids in moulting seals. Conserv Physiol. 2015;3:cov004. doi: 10.1093/conphys/cov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jelincic JA, Tift MS, Houser DS, Crocker DE. Variation in adrenal and thyroid hormones with life-history stage in juvenile northern elephant seals (Mirounga angustirostris) Gen Comp Endocrinol. 2017;252:111–118. doi: 10.1016/j.ygcen.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Khudyakov JI, Preeyanon L, Champagne CD, Ortiz RM, Crocker DE. Transcriptome analysis of northern elephant seal (Mirounga angustirostris) muscle tissue provides a novel molecular resource and physiological insights. BMC Genomics. 2015;16:64. doi: 10.1186/s12864-015-1253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khudyakov JI, Champagne CD, Preeyanon L, Ortiz RM, Crocker DE. Muscle transcriptome response to ACTH administration in a free-ranging marine mammal. Physiol Genomics. 2015;47:318–330. doi: 10.1152/physiolgenomics.00030.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crocker DE, Champagne CD, Fowler MA, Houser DS. Adiposity and fat metabolism in lactating and fasting northern elephant seals. Adv Nutr. 2014;5:57–64. doi: 10.3945/an.113.004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houser DS, Champagne CD, Crocker DE. A non-traditional model of the metabolic syndrome: the adaptive significance of insulin resistance in fasting-adapted seals. Front Endocrinol (Lausanne) 2013;4:164. doi: 10.3389/fendo.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen EA, et al. Effect of RNA integrity on uniquely mapped reads in RNA-Seq. BMC Res Notes. 2014;7:753. doi: 10.1186/1756-0500-7-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Towns J, et al. XSEDE: Accelerating Scientific Discovery. Comput Sci Eng. 2014;16:62–74. doi: 10.1109/Mcse.2014.80. [DOI] [Google Scholar]
- 33.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leggett RM, Ramirez-Gonzalez RH, Clavijo BJ, Waite D, Davey RP. Sequencing quality assessment tools to enable data-driven informatics for high throughput genomics. Front Genet. 2013;4:288. doi: 10.3389/fgene.2013.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 37.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 38.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini, Y. & Hochberg, Y. Controlling The False Discovery Rate - A Practical And Powerful Approach To Multiple Testing. J. Royal Statist. Soc., Series B57, 10.2307/2346101 (1995).
- 41.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 42.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conesa A, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez, B. et al. Adipose Transcriptome Analysis Provides Novel Insights into Molecular Regulation of Prolonged Fasting in Northern Elephant Seal Pups. Physiol Genomics, 10.1152/physiolgenomics.00002.2018 (2018). [DOI] [PMC free article] [PubMed]
- 47.Oakley RH, Cidlowski JA. The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. J Allergy Clin Immunol. 2013;132:1033–1044. doi: 10.1016/j.jaci.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomez-Sanchez E, Gomez-Sanchez CE. The Multifaceted MineralocorticoidReceptor. Compr Physiol. 2014;4:965–994. doi: 10.1002/cphy.c130044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iwen KAH, et al. Melanocortin crosstalk with adipose functions: ACTH directly induces insulin resistance, promotes a pro-inflammatory adipokine profile and stimulates UCP-1 in adipocytes. Journal of Endocrinology. 2008;196:465–472. doi: 10.1677/Joe-07-0299. [DOI] [PubMed] [Google Scholar]
- 50.Zamani N, Brown CW. Emerging roles for the transforming growth factor-{beta} superfamily in regulating adiposity and energy expenditure. Endocr Rev. 2011;32:387–403. doi: 10.1210/er.2010-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christodoulides C, Lagathu C, Sethi JK, Vidal-Puig A. Adipogenesis and WNT signalling. Trends Endocrinol Metab. 2009;20:16–24. doi: 10.1016/j.tem.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plaisier CL, et al. Zbtb16 has a role in brown adipocyte bioenergetics. Nutr Diabetes. 2012;2:e46. doi: 10.1038/nutd.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morey JN, Boggero IA, Scott AB, Segerstrom SC. Current Directions in Stress and Human Immune Function. Curr Opin Psychol. 2015;5:13–17. doi: 10.1016/j.copsyc.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ensminger DC, Somo DA, Houser DS, Crocker DE. Metabolic responses to adrenocorticotropic hormone (ACTH) vary with life-history stage in adult male northern elephant seals. Gen Comp Endocrinol. 2014;204:150–157. doi: 10.1016/j.ygcen.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 55.Bali U, Phillips T, Hunt H, Unitt J. FKBP5 mRNA Expression Is a Biomarker for GR Antagonism. J Clin Endocr Metab. 2016;101:4305–4312. doi: 10.1210/jc.2016-1624. [DOI] [PubMed] [Google Scholar]
- 56.Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem. 2010;285:13223–13232. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramamoorthy S, Cidlowski JA. Ligand-induced repression of the glucocorticoid receptor gene is mediated by an NCoR1 repression complex formed by long-range chromatin interactions with intragenic glucocorticoid response elements. Mol Cell Biol. 2013;33:1711–1722. doi: 10.1128/MCB.01151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang SS, et al. Gdf6 induces commitment of pluripotent mesenchymal C3H10T1/2 cells to the adipocyte lineage. FEBS J. 2013;280:2644–2651. doi: 10.1111/febs.12256. [DOI] [PubMed] [Google Scholar]
- 59.Francisco V, et al. Obesity, Fat Mass and Immune System: Role for Leptin. Front Physiol. 2018;9:640. doi: 10.3389/fphys.2018.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crocker DE, Ortiz RM, Houser DS, Webb PM, Costa DP. Hormone and metabolite changes associated with extended breeding fasts in male northern elephant seals (Mirounga angustirostris) Comp Biochem Physiol A Mol Integr Physiol. 2012;161:388–394. doi: 10.1016/j.cbpa.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 61.Macfarlane DP, Forbes S, Walker BR. Glucocorticoids and fatty acid metabolism in humans: fuelling fat redistribution in the metabolic syndrome. J Endocrinol. 2008;197:189–204. doi: 10.1677/JOE-08-0054. [DOI] [PubMed] [Google Scholar]
- 62.John K, Marino JS, Sanchez ER, Hinds TD., Jr. The glucocorticoid receptor: cause of or cure for obesity? Am J Physiol Endocrinol Metab. 2016;310:E249–257. doi: 10.1152/ajpendo.00478.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spiers JG, Chen HJ, Sernia C, Lavidis NA. Activation of the hypothalamic-pituitary-adrenal stress axis induces cellular oxidative stress. Front Neurosci. 2014;8:456. doi: 10.3389/fnins.2014.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crocker DE, Khudyakov JI, Champagne CD. Oxidative stress in northern elephant seals: Integration of omics approaches with ecological and experimental studies. Comp Biochem Physiol A Mol Integr Physiol. 2016;200:94–103. doi: 10.1016/j.cbpa.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 65.Konige M, Wang H, Sztalryd C. Role of adipose specific lipid droplet proteins in maintaining whole body energy homeostasis. Biochim Biophys Acta. 2014;1842:393–401. doi: 10.1016/j.bbadis.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hochberg I, et al. Gene expression changes in subcutaneous adipose tissue due to Cushing’s disease. J Mol Endocrinol. 2015;55:81–94. doi: 10.1530/Jme-15-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barclay JL, et al. Effects of glucocorticoids on human brown adipocytes. Journal of Endocrinology. 2015;224:139–147. doi: 10.1530/Joe-14-0538. [DOI] [PubMed] [Google Scholar]
- 68.Brasaemle DL, Wolins NE. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J Biol Chem. 2012;287:2273–2279. doi: 10.1074/jbc.R111.309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez YR, et al. Effect of Glycine on Adipocyte Hypertrophy in a Metabolic Syndrome Rat Model. Curr Drug Deliv. 2016;13:158–169. doi: 10.2174/156720181301160314151554. [DOI] [PubMed] [Google Scholar]
- 70.Gopalakrishnan K, et al. Targeted disruption of Adamts16 gene in a rat genetic model of hypertension. P Natl Acad Sci USA. 2012;109:20555–20559. doi: 10.1073/pnas.1211290109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun L, et al. Protein inhibitor of activated STAT 4 (PIAS4) regulates pro-inflammatory transcription in hepatocytes by repressing SIRT1. Oncotarget. 2016;7:42892–42903. doi: 10.18632/oncotarget.9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan Y, et al. SUMOylation of AMPKalpha1 by PIAS4 specifically regulates mTORC1 signalling. Nat Commun. 2015;6:8979. doi: 10.1038/ncomms9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tews D, et al. Comparative gene array analysis of progenitor cells from human paired deep neck and subcutaneous adipose tissue. Mol Cell Endocrinol. 2014;395:41–50. doi: 10.1016/j.mce.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 74.Kong P, et al. Thrombospondin-1 regulates adiposity and metabolic dysfunction in diet-induced obesity enhancing adipose inflammation and stimulating adipocyte proliferation. Am J Physiol Endocrinol Metab. 2013;305:E439–450. doi: 10.1152/ajpendo.00006.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y, Wright GL, Peterson JM. C1q/TNF-Related Protein 3 (CTRP3) Function and Regulation. Compr Physiol. 2017;7:863–878. doi: 10.1002/cphy.c160044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ehrlund A, et al. Transcriptional Dynamics During Human Adipogenesis and Its Link to Adipose Morphology and Distribution. Diabetes. 2017;66:218–230. doi: 10.2337/db16-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Champagne, C. et al. In Society for Marine Mammalogy (2017).
- 78.Crocker, D. E., Williams, J. D., Costa, D. P. & Le Boeuf, B. J. Maternal traits and reproductive effort in northern elephant seals. Ecology82, 3541–3555, doi:10.1890/0012-9658(2001)082[3541:Mtarei]2.0.Co;2 (2001).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads were uploaded to NCBI Short Read Archive (BioProject ID: PRJNA485363, SRA accession: SRP157071). The transcriptome assembly and annotation data are available at figshare (https://figshare.com/s/37cba07b2e9877f25b50).