Abstract
Background
Both environmental and genetic factors have been implicated in the induction of autoimmune disease. Therefore, it is important to understand the pathophysiological significance of the gut microbiota and host genetic background that contribute to an autoimmune disease such as inflammatory bowel disease (IBD). We have previously reported that mice deficient for suppressor of cytokine signaling-1 (SOCS1), in which SOCS1 expression was restored in T and B cells on an SOCS1–/– background (SOCS1–/–Tg mice), developed systemic autoimmune diseases accompanied by spontaneous colitis.
Methods
To investigate whether the proinflammatory genetic background affects the gut microbiota, we used SOCS1–/–Tg mice as a model of spontaneous chronic colitis. Fecal samples were collected from SOCS1–/–Tg mice and SOCS1+/+Tg (control) mice at 1 and 6 months of age, and the fecal bacterial 16S ribosomal RNA genes were sequenced using the Illumina MiSeq platform.
Results
Gut microbial diversity was significantly reduced and the intestinal bacterial community composition changed in SOCS1–/–Tg mice in comparison with the control mice. Interestingly, the population of Prevotella species, which is known to be elevated in ulcerative colitis and colorectal cancer patients, was significantly increased in SOCS1–/–Tg mice regardless of age.
Conclusion
Taken together, these results suggest that the proinflammatory genetic background owing to SOCS1 deficiency causes dysbiosis of the gut microbiota, which in turn generates a procolitogenic environment.
Keywords: Colitis, Dysbiosis, Genetic factors, Prevotella, Suppressor of cytokine signaling-1
Introduction
About 1 × 1014 microbes exist in the human gastrointestinal tract [1]. The intestinal mucosa is constantly exposed to an enormous number of microbes, including bacteria, viruses, fungi, and protozoa that raises the risk of infection. Accordingly, the intestinal immune system must strictly control the delicate balance between immune tolerance to commensal microbes and immune response to pathogens. Therefore, immune dysregulation may result in dysbiosis, an imbalance in the composition of bacterial populations in the gut, and intestinal autoimmune diseases, both of which are affected by environmental and genetic factors [2, 3].
Intestinal autoimmune disorders have been implicated in environmental factors such as the Western diet, early childhood antibiotic exposure, and microbial infections. Recent advances in culture-independent sequencing have provided many insights into the relationship between gut microbiota and intestinal autoimmune diseases. It has been reported, for example, that 17 selected strains of intestinal bacteria, including Clostridium clusters IV, XIVa, and XVIII, which are known to be reduced in ulcerative colitis (UC) patients, have a potent capacity to induce colonic Treg cells [4]. Furthermore, dysbiosis is associated not only with inflammatory bowel disease (IBD) but also with other autoimmune diseases. For example, intestinal Prevotella copri has been implicated in the pathogenesis of rheumatoid arthritis (RA) [5]. Moreover, Faecalibacterium, Prevotella, and Anaerostipes were less abundant at the genus levels in the gut microbiota of multiple sclerosis patients [6]. Although there is increasing evidence for an association between dysbiosis and autoimmune diseases, disease causality related to altered microbial communities is still uncertain. In addition to the environmental factors, genetic factors are shown to be critical for the disease process of IBD. Mice lacking interleukin (IL)-1 receptor antagonist, IL-2, IL-10, and other cytokine-related molecules develop autoimmune responses and IBD [7, 8, 9].
Suppressor of cytokine signaling-1 (SOCS1), a negative regulator of interferon (IFN) signaling, has been implicated in the pathogenesis of spontaneous colitis [10, 11, 12]. SOCS1 knockout (SOCS1–/–) mice die within 3 weeks of birth due to severe systemic inflammation caused by excessive IFNγ signaling [13]. In contrast, SOCS1–/–Tg mice, in which SOCS1 is expressed in T and B lymphocytes, but not in nonlymphoid cells, are able to survive for over 6 months; however, these mice develop systemic autoimmune-like disease including spontaneous colitis within 6 months of age [10, 14]. These observations clearly indicate that a genetic factor resulting in excessive cytokine signaling predisposes to spontaneous autoimmune colitis. However, it is not yet fully understood how the proinflammatory genetic background affects the gut microbiota.
In this study, we employed SOCS1–/–Tg mice as an autoimmune colitis model. Analysis of the fecal microbiome using 16S ribosomal RNA (rRNA) gene amplicon sequencing revealed that SOCS1–/–Tg mice showed reduced microbial diversity and altered bacterial composition in the gut. Interestingly, the relative abundance of Prevotella species, which is known to be elevated in UC [15] and colorectal cancer (CRC) [16] patients, was significantly increased in SOCS1–/–Tg mice regardless of age.
We propose that the proinflammatory environment owing to SOCS1 deficiency resulted in dysbiosis of the gut microbiota characterized by increased Prevotella, which is a potential exacerbating factor for autoimmune colitis.
Materials and Methods
Mice
SOCS1–/–Tg mice were described previously [10, 14]. Briefly, littermates of SOCS1–/–Tg mice and SOCS1+/+Tg (control) mice from heterozygous (SOCS1+/–Tg) intercrosses were used in these experiments. Each of the experimental groups (SOCS1–/–Tg and control) were housed in separate cages directly after weaning. The mice were maintained in specific pathogen-free facilities in the Division of Laboratory Animal Science at Oita University (Oita, Japan). All experiments using these mice were approved by and were performed according to the guidelines of the Oita University Animal Ethics Committee. To determine the genotype of each mouse, PCR analysis was performed by using primer sets 10F: AGAGCTTGGGCGACCTCACC and mJAB: TCAGGTAGTCACGGAGTACC for the SOCS1 Tg allele, and JAB P1: CAGGCACCCACTCCTGGCCTT, JAB P2: TGGCCATTCGGCCTGGCCTT, and JAB P3: GCCTTCTTGACGAGTTCTTCTG for the SOCS1 wild-type and deficient allele [13].
Sample Collection
Fecal samples were collected directly from the anus into 1.5-mL microtubes, and these samples were stored at −20°C within 1 h after collection. Fecal samples were collected in the evening (4 p.m. to 9 p.m.) to obtain reproducible data, because it has been shown that the intestinal microbiota oscillates rhythmically over a 24-h period in both mice and humans [17]. Fecal samples were collected from 1-month-old (young) mice and 6-month-old (old) mice. For 16S rRNA sequencing, fecal samples derived from 2–6 mice were pooled in one tube and then subjected to the analysis. The mice were euthanized at 6 months of age. For each mouse, a portion of the proximal colon near the cecum was frozen immediately in liquid nitrogen, and stored at −80°C until analysis by real-time reverse transcription polymerase chain reaction (RT-PCR). The remaining proximal colon and distal colon were fixed in 10% formaldehyde, embedded in paraffin, and stained with hematoxylin-eosin to examine colitis. These experiments were repeated twice independently.
Illumina MiSeq Sequencing
For library preparation, the V3–V4 region of bacterial 16S rRNA genes was amplified by PCR using universal primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) [18]. The 16S rRNA gene amplicon sequencing was performed by Hokkaido System Science Co. Ltd. (Hokkaido, Japan) with an Illumina MiSeq platform (Illumina, San Diego, CA, USA). Briefly, the PCR products were purified, and paired-end DNA sequencing (2 × 300 bp) was carried out on the Illumina MiSeq platform using a MiSeq reagent kit version 3.
Sequence Data Analysis
Barcoded Illumina reads were processed using the microbial genomics module on a CLC Genomics Workbench version 8.5.1 (Qiagen Inc., Hilden, Germany). Index and adapter sequences were removed from the raw fastq files, and quality filtering was performed with the following parameters (ambiguous limit 2, quality limit 0.05, minimum sequence length 200, and minimum number of reads 100). Chimeric reads were detected and filtered from the paired-end reads using the chimera crossover detection algorithm with the following parameters (merge paired-end read: mismatch cost 1, minimum score 40, gap cost 4, and maximum unaligned end mismatches 2; remove chimera read: chimera crossover cost 3 and Kmer size 6). Reference-based operational taxonomic unit (OTU) clustering was achieved at 97% identity with the Greengenes database version 13_8 (similarity percentage 97, minimum occurrences 2, fuzzy match duplicates FALSE, and find best match TRUE). Low-abundance OTUs with combined abundance across all samples of less than 10 were eliminated. A total of 788 OTUs were predicted; these were aligned using the MUSCLE [19] algorithm, and a neighbor-joining tree with a Jukes-Cantor nucleotide substitution model was constructed. Alpha diversity indices (bias-corrected Chao1, Shannon entropy, and phylogenetic diversity) were calculated, and rarefaction curves were plotted with a maximum rarefaction depth of 5,000 sequences per sample. Beta diversity was calculated as follows: principal coordinate analysis based on the generalized UniFrac distance was conducted to compare microbial communities [20]. The generalized UniFrac kernel with α = 0.5 provided the highest power [21].
Statistical Analysis
Generalized UniFrac distances used for microbial community comparison were statistically evaluated by permutational multivariate analysis (PERMANOVA) in the microbial genomics module of the CLC Genomics Workbench (QIAGEN). The number of permutations was defined as 99,999. Comparison of alpha diversity indices and the relative proportion of bacterial genera between the SOCS1–/–Tg and SOCS1+/+Tg groups were evaluated using the Student t test. p values < 0.05 were considered significant. Linear discriminant analysis (LDA) and LDA effect size (LEfSe) were estimated by using LEfSe software with the default settings in the Galaxy platform [22, 23].
P. copri Colonization
Prevotella copri was obtained from RIKEN BRC Microbe division (Japan Collection of Microorganism). P. copri was cultured for 48 h on phenylethyl alcohol brucella blood agar (Kyokuto Pharmaceutical Industrial Co. Ltd, Tokyo, Japan) in an anaerobic condition. The colonies of P. copri were collected and suspended in skimmed milk. The suspension was orally administrated to C57BL/6 mice using a sonde for 12 consecutive days.
Induction of DSS Colitis
One day after the administration of P. copri, the mice were given 2.5% DSS (dextran sulfate sodium) in drinking water for 8 days followed by water alone for an additional 3 days. During the course of induction of DSS colitis, the severity of colitis was measured by body weight change. Mice were eventually sacrificed and colonic mucosal damage was evaluated by microscopic observation of HE-stained colon sections.
Quantification of TNFα by Real-Time PCR
Total RNA from the colons of DSS colitis-induced mice was isolated using TRI Reagent (Molecular Research Center Inc., Cincinnati, OH, USA); reverse-transcription into cDNA was carried out using a Verso cDNA synthesis kit (Thermo Fisher Scientific Inc., Waltham, MA, USA). Real-time PCR was performed using the SYBR green kit (KAPA SYBR FAST qPCR kit, Kapa Biosystem Inc., Wilmington, MA, USA) with a LightCycler 96 (Roche, Merck, Darmstadt, Germany). Each mRNA level was normalized to β-actin mRNA expression. The amplification conditions were: 45 cycles of 95°C (5 s) and 60°C (30 s). The sequences of the primers were as follows: TNFα FW: 5′-CCACCACGCTCTTCTGTCTA-3′; TNFα RV: 5′-TCCTCCACTTGGTGGTTTGT-3′; β-actin FW: 5′-CTTCCTCCCTGGAGAAGAGCTATGAGC-3′; β-actin RV: 5′-GCCTAGAAGCACTTGCGGTGCACG-3′.
Results
Levels of Cytokine Gene Expression Assessed by Real-Time RT-PCR
We used SOCS1–/–Tg mice as a model of spontaneous autoimmune colitis. As shown in a previous report, these mice exhibited inflammation not only in the colon but also in various organs [14]. We analyzed the colonic mRNA expression levels of proinflammatory cytokines such as TNFα, IL-1β, IL-6, IL-12p40, IFNγ, and MIP-1α by real-time RT-PCR. Consistent with our previous report [10], colonic TNFα expression was significantly upregulated in SOCS1–/–Tg mice compared to control mice, while no change was observed in the expression of IL-1β, IL-6, IFNγ, IL-12p40, and MIP-1α (data not shown). Furthermore, we confirmed the elevation of TNFα expression by Western blot analysis (data not shown). The colitis in SOCS1–/–Tg mice was also apparent in the histological analysis, which exhibited a hyperplasia on the mucosa (data not shown). The number of goblet cells determined by alcian blue staining was significantly reduced in SOCS1–/–Tg mice (data not shown).
Gut Microbial Composition in SOCS1–/–Tg and SOCS1+/+Tg mice
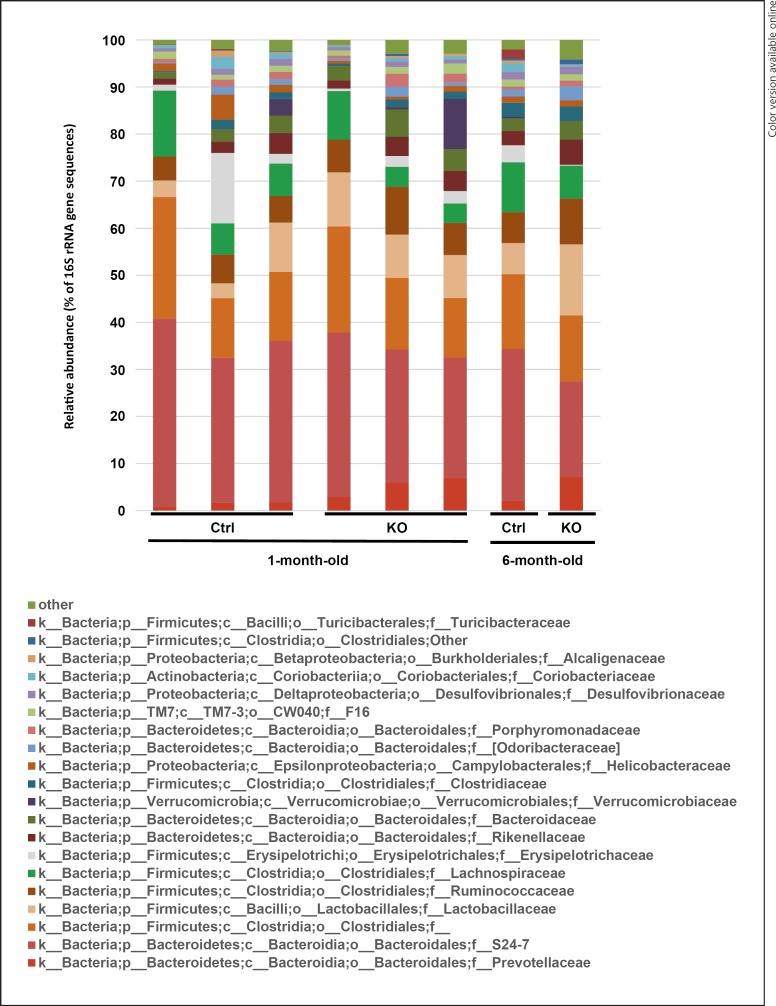
To clarify whether the proinflammatory background of SOCS1–/–Tg mice affects the gut microbiota, 16S metagenome was performed with the fecal samples collected from young and old mice. The bacterial 16S rRNA genes in the samples were sequenced for phylogenetic analysis using the Illumina MiSeq platform, allowing us to investigate total 788 OTUs belonging to 9 phyla, 18 classes, 24 orders, 42 families, and 62 genera. The phyla of Bacteroidetes and Firmicutes were abundant in both SOCS1–/–Tg and control mice (data not shown). The relative abundance of taxa at the family level was compared between SOCS1–/–Tg and control mice at 1 and 6 months of age (Fig. 1). S24–7, Clostridiaceae, Lactobacillaceae, Ruminococcaceae, and Lachnospiraceae were abundant in both groups; however, Prevotellaceae was more abundant in SOCS1–/–Tg mice than in control mice, regardless of age.
Fig. 1.
Relative abundance of bacterial family composition in the pooled feces. SOCS1–/–Tg (KO) and SOCS1+/+Tg (control) mice were analyzed at 1 month and 6 months of age. Stacked columns represent the abundance of a given family as a percentage of the total bacterial sequences in the pooled sample derived from 2–6 mice. Each stacked column represents pooled fecal samples from 2–6 mice per group. The 20 most representative families are shown.
Species Richness of the Gut Microbiota Was Reduced in SOCS1–/–Tg Mice
Next, we calculated alpha diversity and beta diversity to reveal the species diversity. The filtered reads containing 290,451 reads (median 33,745 reads per group, SD ± 5,138) was used for further alpha diversity and beta diversity analysis.
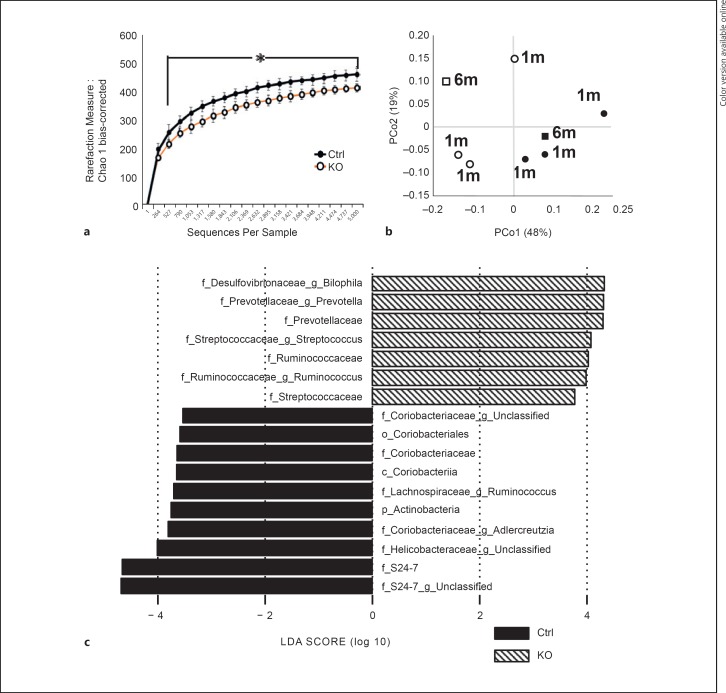
Alpha diversity was compared using the following indices: bias-corrected Chao1, Shannon entropy, and phylogenetic diversity. Each analysis revealed that the diversity of the microbiota in the young SOCS1–/–Tg mice was decreased compared to that in young control mice (data not shown). The same was true for the diversity of microbiota in old mice (data not shown). Alpha diversity estimated by bias-corrected Chao1 using combined data from young and old mice showed a significant reduction of species richness in SOCS1–/–Tg mice (Fig. 2a).
Fig. 2.
Comparison of bacterial taxa between SOCS1–/–Tg and control mice. a Analysis of alpha diversity in SOCS1–/–Tg mice (KO) and SOCS1+/+Tg mice (control). Diversity indices in KO mice versus control mice were predicted by bias-corrected Chao1. The data are presented as the mean ± SD. Statistical differences were analyzed by Student t test. * p < 0.05 was considered statistically significant. b Principal coordinate analysis of generalized UniFrac distance analysis for SOCS1–/–Tg mice (KO) and SOCS1+/+Tg mice (control). Each dot represents a microbial community from KO (white) and control (black) mice, 1-month-old mice (circles) and 6-month-old mice (squares), 2–6 mice per dot. p < 0.03, calculated by PERMANOVA. c LEfSe shows differentially abundant OTUs between SOCS1–/–Tg mice and SOCS1+/+Tg mice. LDA scores (log10) for the most prevalent taxa in KO mice are represented on the positive scale, whereas negative LDA scores indicate enriched taxa in control mice.
Principal coordinate analysis was conducted based on generalized UniFrac distances (Fig. 2b). The principal component scores accounted for 48% (PCo1) and 19% (PCo2) of the total variance. The microbial communities of SOCS1–/–Tg mice and control mice were clearly separated from each other when compared on the basis of the host genotype, but not when compared by age. Taken together, these results clearly indicated that the bacterial communities in SOCS1–/–Tg mice were significantly reduced and separated from those in control mice.
Gut Microbiome Composition Was Altered in SOCS1–/–Tg Mice
We next analyzed the abundance of bacterial species in the pooled data of SOCS1–/–Tg mice and control mice using LEfSe software (LDA score [log10] > 3.0; p < 0.05; Fig. 2c). Positive LDA scores indicated an increased relative abundance in SOCS1–/–Tg mice, while negative LDA scores indicated an increased relative abundance in control mice. SOCS1–/–Tg mice had a higher relative abundance of Prevotella, Bilophila, Ruminococcus, and Streptococcus (LDA score [log10] > 3.0), whereas S24–7 was the most enriched family in control mice (Fig. 2c).
P. copri Did Not Alter the Disease Phenotype of DSS-Induced Colitis in Mice
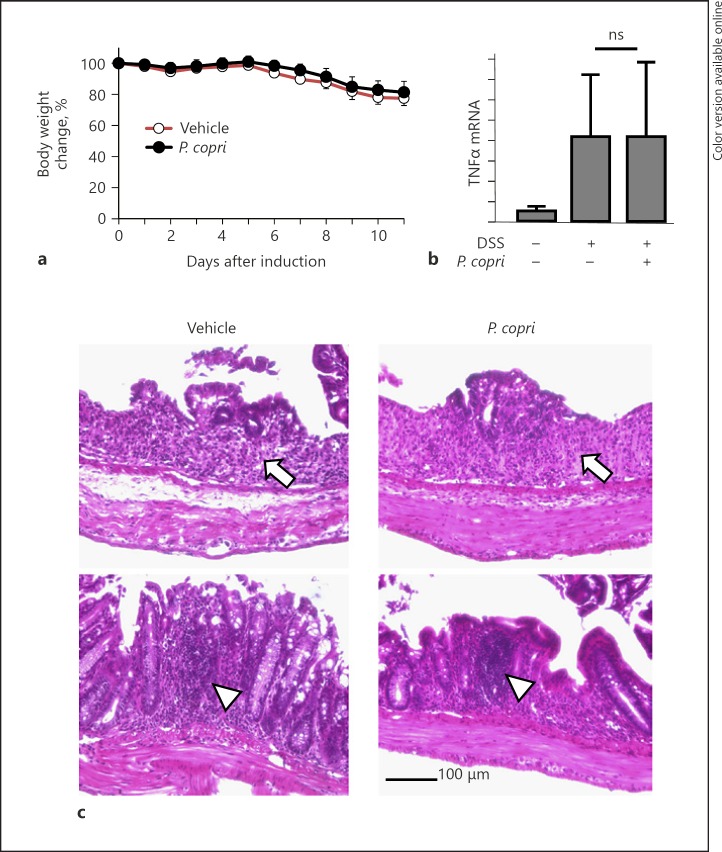
Since we observed the increased abundance of Prevotella, which is known as the pathogenesis of RA [5] in SOCS1–/–Tg mice, we examined the effect of transfer of feces from SOCS1–/–Tg mice to colonize the microbiota in WT mice. However, we failed to observe a change in the gut microbiota in SOCS1+/+Tg mice (no colitis) cohousing with SOCS1–/–Tg mice. We then examined the effect of inoculation of P. copri from human feces in the DSS-induced colitis model. One day prior to the administration of DSS, wild-type mice were given P. copri using a stomach sonde. As shown in Figure 3a, the body weight was comparably reduced in both P. copri-inoculated and vehicle-treated mice. TNFα mRNA expression levels were also increased to a similar extent by DSS treatment in both groups (Fig. 3b). In addition, histological damage of colonic mucosa was seen in both groups of mice (Fig. 3c).
Fig. 3.
P. copri colonization did not affect the phenotype of DSS colitis. a Body weight change in vehicle-treated (n = 5) and P. copri-inoculated mice (n = 6) receiving 2.5% DSS in drinking water. The data are presented as the mean ± SD. b Relative mRNA expression levels of TNFα in the colon of the DSS-induced mice. c Representative images of HE-stained colon samples from the DSS-induced mice. Arrows indicate mucosal tissue destruction and loss of goblet cells; arrowheads show lymphocyte accumulation in lamina propria.
Discussion
SOCS1 is an intracellular protein involved in the negative regulation of the cytokine-JAK-STAT pathway [24]. We have previously reported that deficiency of SOCS1 results in the hyperactivation of STAT1, STAT3, and NF-κB, which in turn leads to spontaneous colitis in SOCS1–/–Tg mice and SOCS1–/–TCRα–/– mice [10, 11]. These signaling pathways are also known to be activated in human IBD [25, 26, 27, 28]. Hence, we employed SOCS1–/–Tg mice as an autoimmune colitis model in this study.
Activation of Toll-like receptor (TLR) signaling by commensal bacteria results in the production of inflammatory cytokines such as TNFα, and consequently the development of spontaneous colitis [29]. Indeed, it is reported that removing commensal bacteria by the administration of antibiotics is effective to a certain degree for inducing remission in IBD patients [30] as well as for the prevention of colitis in SOCS1–/–Rag2–/– mice [12]. As reported previously [10], we have observed that only TNFα expression, but not IL-1β or IL-6, was upregulated in SOCS1–/–Tg mice (data not shown). Proinflammatory cytokines such as TNFα, IL-1β, and IL-6 are known to be induced by the activation of NF-κB, which is a transcription factor downstream of TLR signaling. Mansell et al. [31] demonstrated that removal of SOCS1 regulation potentiates Mal-dependent phosphorylation and transactivation of NF-κB subunit p65, leading to amplified inflammatory responses. Hence, SOCS1 deficiency in nonlymphoid cells leads to activate NF-κB, which may increase proinflammatory cytokine production. There are two main signal pathways downstream of TLR4, mediated by MyD88 and TRIF [32]. It is known that the early phase NF-κB activation is controlled by the MyD88-dependent pathway [33], while the late phase NF-κB activation is controlled by the TRIF-dependent pathway. Upon activation of TLR signaling, TNFα is induced in the relatively early phase, while IL-1β and IL-6 are induced in the late phase. In addition, it has been revealed that TNFα is induced in the early phase in an IκBζ-independent manner, while IL-6 is induced in the late phase in an IκBζ-dependent manner [34]. These results suggest that TLR-mediated IL-6 induction is regulated in a gene expression process of at least two steps, involving inducible IκBζ that is not required for the TNFα induction. However, it was unclear how the proinflammatory cytokine milieu affects gut microbiota. Therefore, in this study we conducted 16S rRNA gene amplicon sequencing of the fecal microbiota in SOCS1–/–Tg mice. The data revealed intestinal microbial dysbiosis including a decrease in species diversity characterized by a predominant population of the Prevotellaceae family in the mutant mice.
Recent advances in metagenomic 16S analysis have enabled us to better understand the association between the microbiota and pathophysiological conditions in the host. Several studies have indicated that IBD patients have dysbiosis, in which taxa in the phylum Firmicutes are consistently decreased and taxa in phylum Proteobacteria are increased [35, 36, 37]. In addition, some immunocompromised mice show a high correlation between colitis and dysbiosis. For example, IL-10-deficient mice, a widely used IBD model, show reduced bacterial diversity and an increase in taxa in the phylum Proteobacteria [38]. Moreover, mice doubly deficient in T-bet and Rag2 (T-bet–/–Rag2–/– mice), which are also known to develop UC, exhibit a higher proportional representation of taxa in the order Bacteroidales [39]. Mice deficient in NLRP6 (NOD-like receptor family pyrin domain containing 6), which is highly expressed in colonic epithelial cells, show an increased representation of the phylum Bacteroidetes (Prevotellaceae) and candidate phylum TM7 [40].
In this study, we found that the proportions of taxa in the genera Prevotella, Bilophila, and Streptococcus were significantly increased in SOCS1–/–Tg mice. Prevotella, which is a genus of obligate anaerobic Gram-negative bacilli, exists in the oral cavity and gastrointestinal tract in healthy humans and predominates in the mucosal tissue of UC [15], CRC [16], and RA patients [5]. Furthermore, it has been demonstrated that P. copri exacerbates DSS-induced colitis [5], suggesting that Prevotella is a potential risk factor in the aggravation of colitis. However, in our experiments, inoculation of P. copri from humans in mice did not worsen the phenotype of DSS-induced colitis, probably due to the failure to colonize the mice with P. copri. It is known that Prevotellaceae produce sulfatases which degrade mucus oligosaccharides leading to disruption of the mucosal barrier function [41]. Impairment of the barrier function of the epithelial cell layer results in chronic inflammation such as IBD, and indeed sulfatases are increased in the intestinal mucosa in IBD patients [42]. Interestingly, Prevotella is increased in both CRC patients and SOCS1–/–Tg mice in which colorectal carcinomas develop spontaneously; however, the pathophysiological relevance of Prevotella should be solved.
In this study, we examined both young mice and old mice. In humans, it is known that the microbiota of the gut alters with aging, and although the association between gut microbiota and aging is not yet fully understood, diet is one of the most influential factors for altering the composition of the gut microbiota [43]. However, although humans consume a variety of foods during their lifetimes, the mice in our experiments were fed the same food throughout their lifetimes. Therefore, the host genetic background in which the SOCS1 gene is deleted in nonlymphoid cells could be the most influential factor, rather than the aging process, in SOCS1–/–Tg mice.
We have shown that the proinflammatory genetic background affects the gut microbiota potentially leading to dysbiosis. However, it remains to be determined whether upregulation of TNFα results in the perturbation of the gut microbiota. To address this issue, further studies are required to examine whether treatment of SOCS1–/–Tg mice with recombinant TNFα disturbs the gut microbiota (increases in Prevotella). It would be interesting to examine whether the administration of anti-TNFα antibody to SOCS1–/–Tg mice or intercrossing SOCS1–/–Tg mice with TNFα–/– mice can improve dysbiosis. On the other hand, Lankelma et al. [44] reported that gut microbiota disruption by broad-spectrum antibiotics decreases TNFα production by mononuclear cells. Further studies are still needed to verify the cause-effect relationship between the upregulation of TNFα and the loss of diversity of the gut microbiota.
In conclusion, the proinflammatory genetic background due to SOCS1 deficiency leads to dysbiosis accompanied by a reduced diversity of gut microbiota and increased abundance of Prevotella, which may exacerbate autoimmune colitis.
Statement of Ethics
All animal experiments were performed in accordance with the guidelines of the Oita University Animal Ethics Committee (OUAEC) and were approved by the OUAEC.
Disclosure Statement
The authors have no competing financial interests to declare.
Acknowledgements
We thank Ms. Chiharu Aoki for her excellent secretarial assistance, Dr. Naoki Hijiya, Ms. Mami Kimoto, and Ms. Aiko Yasuda for providing technical assistance, and members of Dr. Kobayashi's laboratory for valuable discussions. This work was supported by Grants-in-Aid from the Japan Society for the Promotion of Science (grant No. 15K08953 [K. Mizukami], 26460942 [M.K.], 26460420 [K. Murakami], 26305014 [T.K.], 15K19577 [T. Ozaki], 15H06512 [N.K.], and 16H05191 [Y.Y.]), Suzuken Memorial Foundation (N.K.), GSK Japan Research Grant (S.H.), Kurozumi Medical Foundation (N.K.), and Lotte Research Promotion Grant (S.H.).
verified
References
- 1.Ahmed I, Roy BC, Khan SA, Septer S, Umar S. Microbiome, Metabolome and Inflammatory Bowel Disease. Microorganisms. 2016 Jun;4((2)):20. doi: 10.3390/microorganisms4020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klareskog L, Padyukov L, Lorentzen J, Alfredsson L. Mechanisms of disease: genetic susceptibility and environmental triggers in the development of rheumatoid arthritis. Nat Clin Pract Rheumatol. 2006 Aug;2((8)):425–33. doi: 10.1038/ncprheum0249. [DOI] [PubMed] [Google Scholar]
- 3.Hewagama A, Richardson B. The genetics and epigenetics of autoimmune diseases. J Autoimmun. 2009 Aug;33((1)):3–11. doi: 10.1016/j.jaut.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013 Aug;500((7461)):232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 5.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013 Nov;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T, et al. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to clostridia XIVa and IV clusters. PLoS One. 2015 Sep;10((9)):e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdollahi-Roodsaz S, Joosten LA, Koenders MI, Devesa I, Roelofs MF, Radstake TR, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest. 2008 Jan;118((1)):205–16. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993 Oct;75((2)):253–61. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 9.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998 Nov;66((11)):5224–31. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanada T, Kobayashi T, Chinen T, Saeki K, Takaki H, Koga K, et al. IFNgamma-dependent, spontaneous development of colorectal carcinomas in SOCS1-deficient mice. J Exp Med. 2006 Jun;203((6)):1391–7. doi: 10.1084/jem.20060436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chinen T, Kobayashi T, Ogata H, Takaesu G, Takaki H, Hashimoto M, et al. Suppressor of cytokine signaling-1 regulates inflammatory bowel disease in which both IFNgamma and IL-4 are involved. Gastroenterology. 2006 Feb;130((2)):373–88. doi: 10.1053/j.gastro.2005.10.051. [DOI] [PubMed] [Google Scholar]
- 12.Chinen T, Komai K, Muto G, Morita R, Inoue N, Yoshida H, et al. Prostaglandin E2 and SOCS1 have a role in intestinal immune tolerance. Nat Commun. 2011 Feb;2((1)):190. doi: 10.1038/ncomms1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marine JC, Topham DJ, McKay C, Wang D, Parganas E, Stravopodis D, et al. SOCS1 deficiency causes a lymphocyte-dependent perinatal lethality. Cell. 1999 Sep;98((5)):609–16. doi: 10.1016/s0092-8674(00)80048-3. [DOI] [PubMed] [Google Scholar]
- 14.Hanada T, Yoshida H, Kato S, Tanaka K, Masutani K, Tsukada J, et al. Suppressor of cytokine signaling-1 is essential for suppressing dendritic cell activation and systemic autoimmunity. Immunity. 2003 Sep;19((3)):437–50. doi: 10.1016/s1074-7613(03)00240-1. [DOI] [PubMed] [Google Scholar]
- 15.Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol. 2006 May;55((Pt 5)):617–24. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 16.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011 Jan;6((1)):e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014 Oct;159((3)):514–29. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 18.Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011 Oct;5((10)):1571–9. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004 Aug;5((1)):113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012 Aug;28((16)):2106–13. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plantinga A, Zhan X, Zhao N, Chen J, Jenq RR, Wu MC. MiRKAT-S: a community-level test of association between the microbiota and survival times. Microbiome. 2017 Feb;5((1)):17. doi: 10.1186/s40168-017-0239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goecks J, Nekrutenko A, Taylor J, Galaxy Team T, Galaxy Team Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11((8)):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19: Unit 19.10.1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007 Jun;7((6)):454–65. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki A, Hanada T, Mitsuyama K, Yoshida T, Kamizono S, Hoshino T, et al. CIS3/SOCS3/SSI3 plays a negative regulatory role in STAT3 activation and intestinal inflammation. J Exp Med. 2001 Feb;193((4)):471–81. doi: 10.1084/jem.193.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovato P, Brender C, Agnholt J, Kelsen J, Kaltoft K, Svejgaard A, et al. Constitutive STAT3 activation in intestinal T cells from patients with Crohn's disease. J Biol Chem. 2003 May;278((19)):16777–81. doi: 10.1074/jbc.M207999200. [DOI] [PubMed] [Google Scholar]
- 27.Mudter J, Weigmann B, Bartsch B, Kiesslich R, Strand D, Galle PR, et al. Activation pattern of signal transducers and activators of transcription (STAT) factors in inflammatory bowel diseases. Am J Gastroenterol. 2005 Jan;100((1)):64–72. doi: 10.1111/j.1572-0241.2005.40615.x. [DOI] [PubMed] [Google Scholar]
- 28.Neurath MF, Fuss I, Schürmann G, Pettersson S, Arnold K, Müller-Lobeck H, et al. Cytokine gene transcription by NF-kappa B family members in patients with inflammatory bowel disease. Ann N Y Acad Sci. 1998 Nov;859(1 INTESTINAL PL):149–59. doi: 10.1111/j.1749-6632.1998.tb11119.x. [DOI] [PubMed] [Google Scholar]
- 29.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004 Aug;118((3)):285–96. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Khan KJ, Ullman TA, Ford AC, Abreu MT, Abadir A, Marshall JK, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2011 Apr;106((4)):661–73. doi: 10.1038/ajg.2011.72. [DOI] [PubMed] [Google Scholar]
- 31.Mansell A, Smith R, Doyle SL, Gray P, Fenner JE, Crack PJ, et al. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat Immunol. 2006 Feb;7((2)):148–55. doi: 10.1038/ni1299. [DOI] [PubMed] [Google Scholar]
- 32.Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007 Nov;13((11)):460–9. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003 Aug;301((5633)):640–3. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto M, Yamazaki S, Uematsu S, Sato S, Hemmi H, Hoshino K, et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 2004 Jul;430((6996)):218–22. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- 35.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007 Aug;104((34)):13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010 Dec;139((6)):1844–1854.e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 37.Tong M, Li X, Wegener Parfrey L, Roth B, Ippoliti A, Wei B, et al. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS One. 2013 Nov;8((11)):e80702. doi: 10.1371/journal.pone.0080702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maharshak N, Packey CD, Ellermann M, Manick S, Siddle JP, Huh EY, et al. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes. 2013 Jul-Aug;4((4)):316–24. doi: 10.4161/gmic.25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010 Sep;8((3)):292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011 May;145((5)):745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright DP, Rosendale DI, Robertson AM. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol Lett. 2000 Sep;190((1)):73–9. doi: 10.1111/j.1574-6968.2000.tb09265.x. [DOI] [PubMed] [Google Scholar]
- 42.Tsai HH, Dwarakanath AD, Hart CA, Milton JD, Rhodes JM. Increased faecal mucin sulphatase activity in ulcerative colitis: a potential target for treatment. Gut. 1995 Apr;36((4)):570–6. doi: 10.1136/gut.36.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar M, Babaei P, Ji B, Nielsen J. Human gut microbiota and healthy aging: recent developments and future prospective. Nutr Healthy Aging. 2016 Oct;4((1)):3–16. doi: 10.3233/NHA-150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lankelma JM, Belzer C, Hoogendijk AJ, de Vos AF, de Vos WM, van der Poll T, et al. Antibiotic-Induced Gut Microbiota Disruption Decreases TNF-α Release by Mononuclear Cells in Healthy Adults. Clin Transl Gastroenterol. 2016 Aug;7((8)):e186. doi: 10.1038/ctg.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]