Abstract
Objectives
Forkhead transcription factor, O subgroup, member 1 (FOXO1) is a regulatory protein that plays an essential role in cellular homeostasis. A biological function as a tumor suppressor has been proposed. Here, we examined FOXO1 expression in human pancreatic ductal adenocarcinoma (PDAC) and its precursor lesions.
Methods
We immunohistochemically labeled tissue samples from 47 patients with PDAC for FOXO1 protein. In addition, we extracted data from the Cancer Genome Atlas and the Cancer Cell Line Encyclopedia and studied a potential association with well-established genetic variants. A publicly available microarray dataset of 102 PDAC samples was used to explore the influence of FOXO1 expression on patients' clinical outcome.
Results
Normal ductal epithelium universally expressed nuclear and cytoplasmic FOXO1. Reduced expression was observed in PanIN lesions and PDAC of all cases. Analysis of several datasets showed that the FOXO1 gene transcript levels do not correlate with KRAS, TP53, SMAD4, or CDKN2A mutation status, but positively correlate with patients' outcomes.
Conclusions
Loss of FOXO1 protein is identified as an early event during PDAC development and may be independent of the top 4 mutated cancer genes. Because of its strong expression in normal ductal cells, immunohistochemical detection of FOXO1 can function as a valuable test to establish the diagnosis of transformation and malignancy in pancreatic tissues.
Key Words: Pancreas; Pancreatic intraepithelial neoplasia; Forkhead transcription factor, O subgroup, member 1; Immunohistochemistry; KRAS; Tumor suppressor
Introduction
Pancreatic adenocarcinoma (PAC) remains one of the deadliest forms of cancer with a 5-year survival rate of about 6% [1]. This dismal outcome is mainly due to the lack of methods for early detection and to the fact that it develops early resistance against most forms of conventional cytotoxic therapy [1]. Important progress seen in the past years have, however, improved our understanding of the genetic basis and the biological pathways of this disease and opened up new therapeutic options for targeting tumor metabolism [2, 3]. Yet a better understanding of how metabolic pathways are regulated in cancer is necessary to devise more effective therapies.
Malignant tumors occurring in the pancreas are classified according to the cell of origin and can be divided into endocrine, exocrine and mesenchymal tumors. This sub-classification has strong implications on prognosis and treatment strategy of patients [4]. Exocrine tumors account for more than 95% of all pancreatic cancers. They arise either from (i) pancreatic acinar cells, termed acinar cell carcinoma, or more commonly, from (ii) epithelial cells of the pancreatic duct system. As all solid tumors, pancreatic ductal adenocarcinomas (PDAC) are a mixture of neoplastic and non-neoplastic cells as well as an extracellular matrix. Given this complexity, immunohistochemistry is the most effective technique available to assess protein abundance and localization on the cellular and sub-cellular levels.
The forkhead transcription factor, O subgroup, member 1 (FOXO1) regulates cellular metabolic mechanisms [5, 6]. It is reported to function as a putative tumor suppressor in several types of cancer [7, 8, 9]. Because changes in metabolism are regarded to be critical during malignant transformation [3, 10, 11, 12], we studied the levels of FOXO1 throughout the development of human PDAC.
Materials and Methods
Formalin-fixed, paraffin-embedded tissue samples from PDAC patients (n = 47) were sectioned, deparaffinized, and hydrated. Indigenous peroxidase was inhibited by H2O2, and the sections were incubated with an anti-human FOXO1 antibody (Cat# PA1–39515, Thermo Fisher Scientific, 1: 100 dilution, Rockford, IL, USA) and finally counterstained with hematoxylin. The assay was first established manually according to the manufacturer's protocol. Horseradish peroxidase enzyme and 3-amino-9-ethylcarbazole substrate (cat#3464, Dako, Glostrup, Denmark) were applied. Next, we established an automated FOXO1 staining using the Leica BOND-MAX Autostainer (Leica Biosystems, Melbourne, VIC, Australia) according to the manufacturer's recommendations. Instead of the 3-amino-9-ethylcarbazole, 3,3′-Diaminobenzidine substrate was used (Bond Polymer Refine Detection kit, cat#DS9800, Leica, UK). Microscopic images were captured using a Nikon Eclipse 80i microscope supplied with a Nikon's Digital Sight DS-Fi1camera and the NIS-Elements D software (Nikon, Vienna, Austria). Demographic data of patients included in this study on age and gender are provided as online supplementary Table 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000492433).
In addition, we extracted data from the Cancer Genome Atlas (TCGA) [13, 14] and the Cancer Cell Line Encyclopedia [15], and studied a potential association with well-established genetic variants. We compared FOXO1 mRNA levels from several PAC samples (n = 168) and pancreatic carcinoma cell lines (n = 43) that harbor multiple mutations in the KRAS, TP53, SMAD4, or CDKN2A genes; the most common mutations in pancreatic carcinoma [16, 17, 18]. Online supplementary Table 2 provides demographic data of patients in the TCGA data set. Names of the 43 cell lines and the reported genetic alterations are provided as online supplementary Table 3. Graphs were generated by GraphPad Prism version 5.
Finally, a publicly available microarray dataset of 102 PDAC samples (by using the R2: Genomics Analysis and Visualization Platform [http://r2.amc.nl]) was used to explore the influence of FOXO1 expression on patients' clinical outcome [19]. The project has been approved by the Ethics Committee of the Medical University of Graz (number: 23-279 ex 10/11).
Results
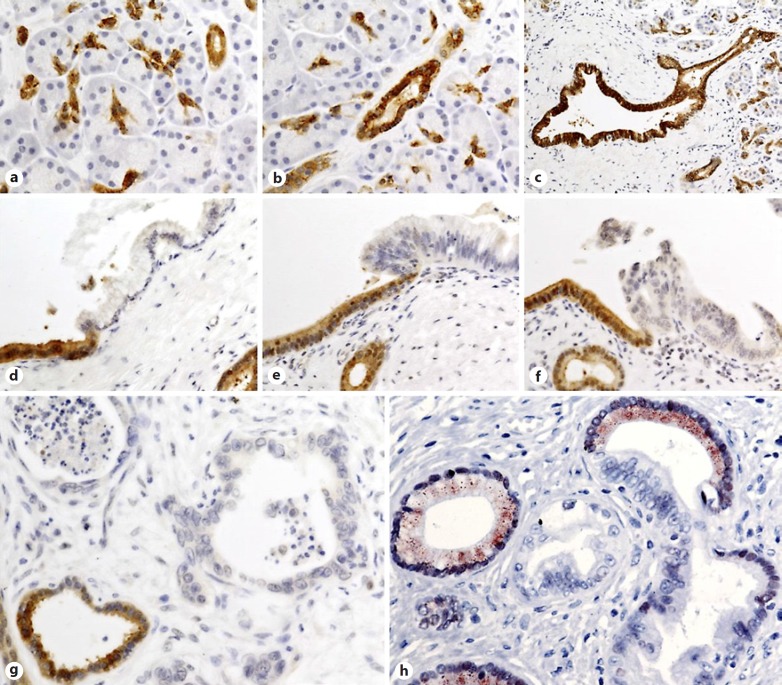
By use of immunohistochemical analysis, FOXO1 positive signal was detected in morphologically normal ductal epithelium but not in acinar cells. We observed an intense signal in all epithelial cells of the pancreatic duct system including centroacinar cells, intra- and interlobular ducts and the main pancreatic duct. These cells showed strong nuclear and cytoplasmic expression. Interestingly, acinar cells were negative for FOXO1, indicating that the staining was specific to ductal cells and that the assay was able to distinguish between acinar and centroacinar cells (Fig. 1a–c). In contrast to the abundant expression in the ductal epithelium, in samples from the 47 patients, FOXO1 signal was reduced or it was undetectable in all neoplastic and malignant lesions arising from these cells including PanIN1, the initial step of the PDAC development (Fig. 1d–h).
Fig. 1.
The forkhead transcription factor, O subgroup, member 1 (FOXO1) expression in normal and neoplastic pancreatic tissues. Strong nuclear and cytoplasmic expression in centroacinar cells (a), intralobular duct (b) and interlobular duct (c). Compared to its prominent signal in non-transformed ductal cells, FOXO1 is absent in transformed cells of pancreatic intraepithelial neoplasia (PanIN) PanIN1 (d), PanIN2 (e), and PanIN3 (f) lesions. Invasive pancreatic ductal adenocarcinoma (PDAC) cells in (g) also show no detectable FOXO1 expression, whereas the adjacent non-neoplastic duct (lower left) is positive. The (h) micrograph demonstrates the potential application of FOXO1 immunohistochemistry (IHC) to aid in the histopathological evaluation of pancreas. Visualization of the antibody-antigen binding is performed using horseradish peroxidase (HRP)/3,3′-Diaminobenzidine (DAB; brown signal in micrographs a–g), or HRP/3-amino-9-ethylcarbazole (AEC; red signal in micrograph h).
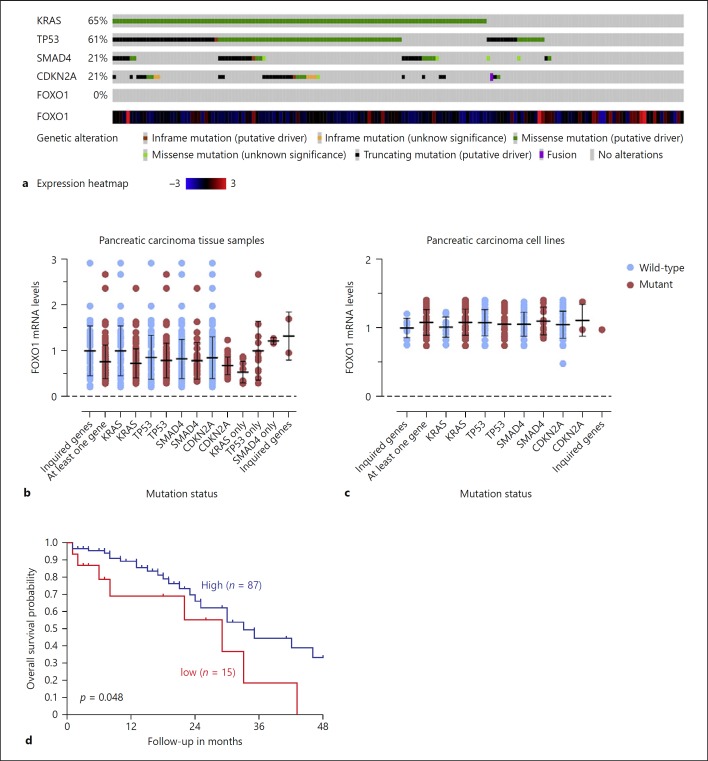
Data available from a public database showed that the FOXO1 gene harbors no mutations in 168 tumor samples from patients diagnosed with PAC (TCGA, PanCancerAtlas) (Fig. 2a). Interestingly, FOXO1 mRNA levels were similar in the 168 PAC tumor samples and the 43 pancreatic carcinoma cancer cell lines. There was no correlation between FOXO1 mRNA levels and mutations in KRAS, TP53, SMAD4, or CDKN2A, the top 4 genes mutated in pancreatic carcinoma (Fig. 2a–c).
Fig. 2.
Analysis of the forkhead transcription factor, O subgroup, member 1 (FOXO1) gene mutation and transcript levels. None of the tumors in the Cancer Genome Atlas (TCGA) data set has a mutation in FOXO1 gene (a; n = 168). FOXO1 mRNA levels in pancreatic adenocarcinoma (PAC) tissue samples (b; n = 168) or in pancreatic carcinoma cell lines (c; n = 43). The data were normalized to mRNA levels in tumor samples or cell lines with wild-type KRAS, TP53, SMAD4 and CDKN2A. Data are presented as means and error bars represent the SD of the mean. d Kaplan scan for FOXO1 derived from clinically annotated human pancreatic cancer gene expression set (Yey: 4hm44K, http://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Patients with low FOXO1 expression in their tumor tissue had significantly shorter overall survival after 48 months than patients with high FOXO1 levels (p = 0.048, log-rank test).
In addition to the loss of FOXO1 protein expression, we found that low levels of FOXO1 are associated with poor survival in a cohort of 102 patients with PDAC (p < 0.05, log-rank test) (Fig. 2d).
Discussion
In this report, we evaluated FOXO1 protein levels in normal pancreatic tissue, neoplastic, noninvasive lesions known as pancreatic intraepithelial neoplasia, and PDAC. We observed that the expression level of this transcription factor is reduced both in pre-invasive lesions and in PADC.
Based on our observation, we propose that FOXO1 is promising as a histopathological marker to aid the diagnosis of pancreatic cancer. The strong signal of FOXO1 protein in normal ductal epithelium – but not in neoplastic cells – is easily detectable by immunohistochemistry performed on archival formalin-fixed, paraffin-embedded tissue samples. This assay may be particularly relevant when microscopically evaluating (i) surgical resection margins [20] and (ii) small biopsy specimens. Recent research presented an opportunity for early detection of pancreatic cancer by examining exosomes, small particles released from cancer cells into the blood [21]. Reliable histopathological markers are especially valuable to confirm the diagnosis of cancer in samples obtained by core needle biopsy. Further studies are required to confirm our observation and to validate the usefulness of FOXO1 in routine diagnostic surgical pathology.
The fact that FOXO1 is already lost in PanIN1 indicates that it is downregulated at an early stage of transformation to neoplastic cells. It is reported that less than 40% of PanIN1 lesions have a KRAS mutation and far less a TP53 mutation [22, 23] arguing that the reduced levels of this transcription factor, at least in part, are not mediated by the KRAS or TP53 mutations. This assumption was further supported by the bioinformatics data, which showed no correlation between FOXO1 gene expression and any of the tested mutations.
The involvement of FOXO1 as a putative tumor suppressor in pancreatic carcinogenesis is further substantiated by the survival analysis of 102 external patients, which indicates that PDAC patients with low levels of FOXO1 mRNA have a poor prognosis.
Investigating the molecular mechanisms behind the downregulation of FOXO1 protein levels is clearly imperative but beyond the scope of this work. Nevertheless, our findings are bound to add to our current understanding of the biology of pancreatic cancer and should help to confirm the diagnosis of PDAC as well as precursor lesions [24, 25].
Disclosure Statement
The authors declare that they have no conflicts of interest to disclose.
Supplementary Material
Supplementary data
Acknowledgment
This project was supported by the Austrian Science Fund (FWF) project W1226 DK “Metabolic and cardiovascular disease” and the “Oesterreichische Nationalbank” Anniversary Fund: grant 14320. W.A.-Z. is also supported by the Austrian Research Promotion Agency (FFG) funded CBmed project: ADX models for the evaluation of cancer treatments. Formalin-fixed, paraffin-embedded tissue samples from PDAC patients were retrieved from the Biobank of the Medical University of Graz, Austria.
References
- 1.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 3.Perera RM, Bardeesy N. Pancreatic cancer metabolism: breaking it down to build it back up. Cancer Discov. 2015;5:1247–1261. doi: 10.1158/2159-8290.CD-15-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stotz M, Eisner F, Szkandera J, Absenger G, Kornprat P, Lackner C, Samonigg H, Gerger A, Pichler M. Clinico-pathological characteristics and clinical outcome of different histological types of pancreatic cancer in a large middle European series. J Clin Pathol. 2013;66:753–757. doi: 10.1136/jclinpath-2012-201394. [DOI] [PubMed] [Google Scholar]
- 5.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puthanveetil P, Wan A, Rodrigues B. FoxO1 is crucial for sustaining cardiomyocyte metabolism and cell survival. Cardiovasc Res. 2013;97:393–403. doi: 10.1093/cvr/cvs426. [DOI] [PubMed] [Google Scholar]
- 7.Dansen TB, Burgering BM. Unravelling the tumor-suppressive functions of FOXO proteins. Trends in cell biology. 2008;18:421–429. doi: 10.1016/j.tcb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Xie L, Ushmorov A, Leithauser F, Guan H, Steidl C, Farbinger J, Pelzer C, Vogel MJ, Maier HJ, Gascoyne RD, Moller P, Wirth T. FOXO1 is a tumor suppressor in classical hodgkin lymphoma. Blood. 2012;119:3503–3511. doi: 10.1182/blood-2011-09-381905. [DOI] [PubMed] [Google Scholar]
- 9.Zhang B, Gui LS, Zhao XL, Zhu LL, Li QW. FOXO1 is a tumor suppressor in cervical cancer. Genet Mol Res. 2015;14:6605–6616. doi: 10.4238/2015.June.18.3. [DOI] [PubMed] [Google Scholar]
- 10.Al-Zoughbi W, Huang J, Paramasivan GS, Till H, Pichler M, Guertl-Lackner B, Hoefler G. Tumor macroenvironment and metabolism. Semin Oncol. 2014;41:281–295. doi: 10.1053/j.seminoncol.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sousa CM, Kimmelman AC. The complex landscape of pancreatic cancer metabolism. Carcinogenesis. 2014;35:1441–1450. doi: 10.1093/carcin/bgu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Zoughbi W, Pichler M, Gorkiewicz G, Guertl-Lackner B, Haybaeck J, Jahn SW, Lackner C, Liegl-Atzwanger B, Popper H, Schauer S, Nusshold E, Kindt AS, Trajanoski Z, Speicher MR, Haemmerle G, Zimmermann R, Zechner R, Vesely PW, Hoefler G. Loss of adipose triglyceride lipase is associated with human cancer and induces mouse pulmonary neoplasia. Oncotarget. 2016;7:33832–33840. doi: 10.18632/oncotarget.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cbio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sipos B, Moser S, Kalthoff H, Torok V, Lohr M, Kloppel G. A comprehensive characterization of pancreatic ductal carcinoma cell lines: Towards the establishment of an in vitro research platform. Virchows Arch. 2003;442:444–452. doi: 10.1007/s00428-003-0784-4. [DOI] [PubMed] [Google Scholar]
- 17.Deer EL, Gonzalez-Hernandez J, Coursen JD, Shea JE, Ngatia J, Scaife CL, Firpo MA, Mulvihill SJ. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A, Cicenas J. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer. Cancers. 2017;9 doi: 10.3390/cancers9050042. pii: E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stratford JK, Bentrem DJ, Anderson JM, Fan C, Volmar KA, Marron JS, Routh ED, Caskey LS, Samuel JC, Der CJ, Thorne LB, Calvo BF, Kim HJ, Talamonti MS, Iacobuzio-Donahue CA, Hollingsworth MA, Perou CM, Yeh JJ. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med. 2010;7:e1000307. doi: 10.1371/journal.pmed.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luttges J, Zamboni G, Kloppel G, Recommendation for the examination of pancreaticoduodenectomy specimens removed from patients with carcinoma of the exocrine pancreas A proposal for a standardized pathological staging of pancreaticoduodenectomy specimens including a checklist. Dig Surg. 1999;16:291–296. doi: 10.1159/000018738. [DOI] [PubMed] [Google Scholar]
- 21.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider G, Schmid RM. Genetic alterations in pancreatic carcinoma. Mol Cancer. 2003;2:15. doi: 10.1186/1476-4598-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohr M, Kloppel G, Maisonneuve P, Lowenfels AB, Luttges J. Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: a meta-analysis. Neoplasia. 2005;7:17–23. doi: 10.1593/neo.04445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vyberg M, Torlakovic E, Seidal T, Risberg B, Helin H, Nielsen S. Nordic immunohistochemical quality control. Croat Med J. 2005;46:368–371. [PubMed] [Google Scholar]
- 25.Fitzgibbons PL, Bradley LA, Fatheree LA, Alsabeh R, Fulton RS, Goldsmith JD, Haas TS, Karabakhtsian RG, Loykasek PA, Marolt MJ, Shen SS, Smith AT, Swanson PE. Principles of analytic validation of immunohistochemical assays: guideline from the college of american pathologists pathology and laboratory quality center. Arch Pathol Lab Med. 2014;138:1432–1443. doi: 10.5858/arpa.2013-0610-CP. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data