Abstract
Discovery of new catalysts for demanding aqueous reactions is challenging. Here, we describe methodology for selection of catalytic phages by taking advantage of localized assembly of the product of the catalytic reaction that is screened for. A phage display library covering 109 unique dodecapeptide sequences is incubated with nonassembling precursors. Phages which are able to catalyze formation of the self-assembling reaction product (via amide condensation) acquire an aggregate of reaction product, enabling separation by centrifugation. The thus selected phages can be amplified by infection of Escherichia coli. These phages are shown to catalyze amide condensation and hydrolysis. Kinetic analysis shows a minor role for substrate binding. The approach enables discovery and mass-production of biocatalytic phages.
Despite advances,1–3 robust design and discovery approaches for catalysis of demanding chemical reactions, especially in aqueous media, remain elusive. In contrast, effective methods are available for selection of selective binders based on peptides and oligonucleotides.4–8 Screening for binders is relatively straightforward, involving exposure of a combinatorial library to surface immobilized ligands, followed by selection through elution of bound molecules. Similarly, screening for catalysts is possible by exposing sequence libraries to surface immobilized transition state analogues, using libraries of antibodies,9,10 phages11 or proteins.12 In addition to catalyst discovery by screening, de novo design approaches are being developed, usually by combining a synthetic substrate binding pocket or nanoparticle surface with catalytic residues, e.g., using peptides with strategically positioned catalytic groups, such as imidazole or proline.3,13–18 Remarkably, the dipeptide seryl-histidine (SH) was shown to catalyze hydrolytic reactions,19 suggesting that a surface or binding pocket may not be essential for catalysis.
Here, we describe direct selection for catalysis rather than transition state binding, by creating a physical link between catalyst and reaction product through its aggretation at the site of catalytic formation. Library screening by co-localization of reaction products and catalysts was previously explored using solid-phase combinatorial peptide libraries by detection of a localized colorimetric reaction product or by co-immobilization of catalyst candidates with the reaction product.20 Biocatalytic self-assembly has not been used for catalyst screening and provides options for a wide range of reactions.21–24
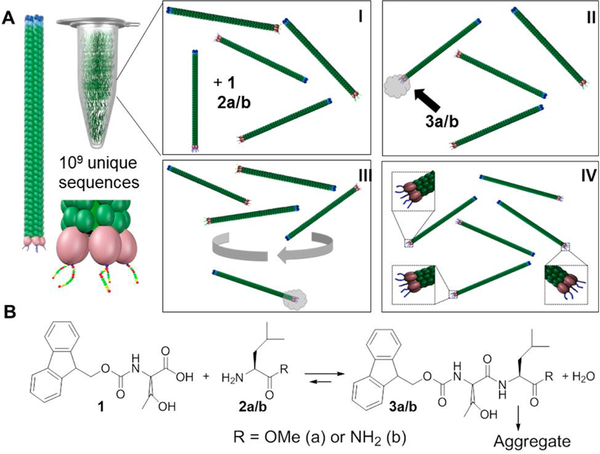
We use display libraries of M13 filamentous phage viruses, consisting of 2.7 × 109 unique phages which display five copies of a dodecapeptide sequence at their tip (Figure 1A), which is encoded by DNA embedded within the phage backbone.20 By combining phage display with catalytic self-assembly21–24 co-localization of catalyst and supramolecular aggregation25–27 of the reaction product may be expected. Thus, catalytic viruses remain associated with self-assembled reaction products,25–27 enabling isolation by centrifugation (Figure 1A). As proof of concept, we herein search for catalysts for amide condensation/hydrolysis in water (Figure 1B), an extremely challenging reaction with an estimated uncatalyzed half-life in the range of 300 years.28
Figure 1.
(A) M13 phage and library: I, 1 and 2a/b are added; II, 3a/b is formed as localized aggregates at the tip of catalytic phages; III, phage/aggregates are separated by centrifugation; IV, amplification in E. coli host.; (B) Catalytic condensation and assembly of Fmoc-T and L-NH2/OMe.32
The library of M13 phages is incubated in the presence of the soluble precursors 9-fluorenylmethoxycarbonyl-threonine (Fmoc-T, 1) and leucine-methyl ester (L-OMe, 2a) or leucine-amide (L-NH2, 2b). Upon catalytic condensation, Fmoc-peptides 3a/b are formed that aggregate upon formation (Figure 1B). While peptide hydrolysis (rather than condensation) is normally favored in aqueous media, the free energy reduction associated with the self-assembly of the reaction product shifts the reaction toward amide condensation, as shown previously using enzymatic catalysis.29 The low aqueous solubility of 3a/b prevents dissolution, enabling localized assembly at the site of catalysis (i.e., at the phage tip) (Figure 1A, II). Similar spatiotemporal co-localization of catalytic formation and self-assembly was previously demonstrated24 including in cellular expressed enzymes30 and using catalytic micropatterned surfaces.31 Formation of localized aggregates facilitates the separation and isolation of catalytic phage by gentle centrifugation (Figure 1A, III). This panning process is similar to that used in the identification of phages that catalyze nucleation and growth of zinc oxide (ZnO).32 For amplification, thus isolated phage particles require to be removed from 3a aggregates, which was achieved using subtilisin (from Bacillus licheniformis) to hydrolyze the terminal methyl ester (Supporting Information (SI) 1). Phages were amplified in Escherichia coli. After two subsequent rounds of panning, this process revealed approximately 700 colonies, from which 20 were picked for sequencing (SI 2, Table S1).
To demonstrate that selection occurs through the proposed catalytic assembly at the peptide-displaying domains, amplified phages were mixed with the 1/2a precursors (Figure 2A, left), and these phages were subsequently imaged by transmission electron microscopy (TEM) (Figure 2C). TEM images show that aggregates form at the tips of CP4 phages within 30 min (see also SI Figures S3.3–3.5) while no such aggregates were observed on non-selected phages (SI Figures 3.1–3.2). It is possible that besides 3a, unreacted Fmoc-T (1) also contributes by selective binding to the phage tip, a process which may contribute to catalysis through locally enhanced concentration of 1. Further work is required to explore the relative importance of substrate and product aggregation at the phage terminus. When using 3a in the panning process, there is the possibility that the selected phages catalyze hydrolysis of the terminal ester bond (as discussed later). Ester bonds are more labile than amides and hydrolysis of the methyl ester in 3a causes a change in amphiphilic balance, which results in disassembly.33 Therefore, we also tested the amide terminated 1/2b to avoid the competing ester hydrolysis reaction. The panning procedure was identical to that for the 1/2a-based system, except for that aggregates were removed by washing with an acetonitrile/water mixture (SI 1). After one round of panning 18 plaques were obtained, with three dominant sequences identified (CPN1 (7 times), CPN2 (2), and CPN3 (2); additional sequences occurred once (SI 2, Table S2)).
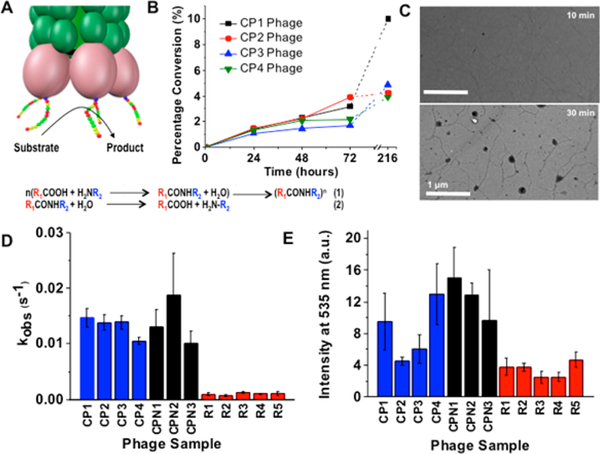
Figure 2.
(A) Catalytic activity of selected phages showing (1) amide condensation and (2) hydrolysis. (B) Conversion % of amide condensation 1/2b by HPLC. (C) TEM analysis of 1/2a after 10 and 30 min incubation with CP4 phage. (D) Hydrolysis of FRET peptide (based on 72 h time course, SI 5, Figure S5.1,2) with 0.0167 μM phage, 2 μM FRET peptide. (E). Activity of phages in FITC-Casein hydrolysis by fluorescence.
There is no obvious sequence similarity between the “hits” of both panning experiments (Table 1, SI Tables S1 and S2). It is apparent that the majority of peptides selected contain amino acids that are typically associated with nucleophilic catalysis (e.g., H, S, D, E, C). It should be noted that this is not unexpected based on random selection (for example, a chance of 12/20 = 0.6 for the presence of histidine in a dodecapeptide sequence and a chance of 0.6 × 0.6 = 0.36 for both a serine and histidine). Among the 18 peptides identified in 1/2a panning, 4 peptides containing both serine (S) and histidine (H) were initially selected based on the reported catalytic activity of SH.19 These 4, in addition to the 3 identified using 1/2b (CPN1 and 2 have S and H, while CPN3 has no H) plus 5 random controls were assessed further (Table 1).
Table 1.
Peptide Sequences Selected by Catalytic Self-Assemblya
| Name | Sequence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CP1 | T | D | H | T | H | N | K | G | Y | A | N | K |
| CP2 | T | S | H | P | S | Y | Y | L | T | G | S | N |
| CP3 | S | H | Q | A | L | Q | E | M | K | L | P | M |
| CP4 | S | M | E | S | L | S | K | T | H | H | Y | R |
| CPN1 | A | M | H | S | L | V | G | P | A | F | N | R |
| CPN2 | H | D | T | S | E | Q | L | L | V | A | P | S |
| CPN3 | D | L | R | S | C | T | A | C | A | V | N | A |
| Random 1 | N | Y | A | D | L | L | E | L | T | G | R | D |
| Random 2 | K | Y | E | L | N | Q | S | V | Y | H | P | V |
| Random 3 | R | L | H | Q | A | E | I | N | V | Q | F | A |
| Random 4 | S | A | C | S | A | A | N | H | Y | L | L | N |
| Random 5 | E | G | S | T | G | A | L | R | H | S | A | N |
CP1–4 were selected using 1/2a; CPN1–3 were selected using 1/2b. In addition, five random phages were selected as controls. Histidines are italicized.
In order to demonstrate catalytic activity of the selected phages, they were amplified and transferred from LB medium to buffer (SI 6). First, we tested the ability of the amplified phages to catalyze amide condensation using 1/2b (an initial attempt to quantify the condensation using 1/2a gave irreproducible results, likely due to aforementioned competing ester hydrolysis catalyzed by these phages). Gratifyingly, four selected phages, CP1–4, showed significant amide condensation over time (CP1 gave 3.2%, CP2 3.9%, CP3 1.7%, and CP4 2.2% condensation product at 72 h which increased further after 216 h, Figure 2B). The condensation rates of these phages are compatible with the time scale used for the panning experiments completed in 3 days. CPN1–3 gave rise to low levels of measurable condensation, with values only marginally better than the background (the condensation product is 0.98% for CPN1 and 1.15% for CPN2 and CPN3 after 72 h of incubation). We hypothesize that the formation of aggregates at the tip of phages is self-limiting, likely related to prevention of diffusion to the catalytic site once it is blocked with aggregated material, thus preventing further condensation and assembly. Unexpectedly, randomly selected phages (Table 1) showed a low level of amide condensation by HPLC (up to 1% after 72 h), although no aggregation was observed in TEM images suggesting the formed 3b were not localized and aggregated at the phage tip. The spontaneous formation of these low levels of 3b is likely an autocatalytic process, which we are currently investigating.
Amide hydrolysis, the direct reversal of amide condensation as used in the phage screening, was tested by monitoring degradation of a Förster resonance energy transfer (FRET)-peptide, E(EDANS)-GTLGK(DABCYL). This substrate contains the TL sequence as used in the panning reaction, flanked with a fluorophore/quencher pair separated by glycine (G) spacers. Upon hydrolysis, emission is enhanced (Figure 2D; see also SI 4), which indicates that the FRET peptide is degraded. All CP1–4 and CPN1–3 phage samples showed hydrolytic activity and low levels of background hydrolysis were observed for phages with random peptide sequences (R1–5). The kinetics of hydrolysis were measured through the increase of fluorescence at 493 nm over time and converted to concentrations using a calibration curve. A rate contstant was determined as kobs = substrate converted [mol]/catalyst [mol]/time [s]. These values are expressed on the basis of concentration of the selected peptides (five per phage). The thus obtained rate constants, kobs = (1–2) × 10−2 s−1, are an order of magnitude higher than levels of hydrolysis detected for the five control phages, kobs = 1 × 10−3 s−1 (Figure 2D). The best performing catalyst, CPN2 showing a 20-fold rate enhancement compared to the control phages (for comparison, enzymatic hydrolysis of amide bonds typically has kcat values in the range of 102−103 s−1).28 It is worth noting that CPN2 does not contain the SH sequence previously associated with the general hydrolysis action of amide bonds.17 Moreover, CPN3 shows amidase activity while it does not contain histidine, suggesting that a different catalytic mechanism is responsible. Amide hydrolysis catalyzed by phages was further assessed in the hydrolysis of fluorescein labeled casein (Figure 2E). Significant hydrolytic activity was observed for CP1, CP4, and CPN1–3, while no activity above background was observed for control phages, CP2 and CP3.
Typically, biocatalysts that display amide hydrolysis activity (such as serine proteases) also show esterase activity at higher rates. Therefore, in order to gain further insight in the catalytic rates of selected catalytic phages for the comparison with other examples of designed peptide-based catalysts, ester hydrolysis was investigated using p-nitrophenyl acetate (pNPA), a standard assay for the comparative assessment of hydrolase activities. CPN3 showed the highest activity, approximately 2.5 times higher than that of the background, followed by CP4 = CPN1,2 > CP1 (Figure 3A). The catalytic phages of CP2 and CP3 showed the similar activity to controls. The observed activity of the control phages is most likely related to well-documented esterase activity of imidazole residues34 contained in two residues present at the pIII protein domain of the phage. Interestingly, these results demonstrate that amidase and esterase activities do not directly correlate and depend on the peptide sequences. The results also demonstrate that histidine is not crucial for ester hydrolysis, with CPN3 showing highest activity.
Figure 3.
Hydrolysis of para-nitrophenyl acetate. The phage concentration was 0.0167 μM, and the concentration of pNPA was 6 mM. (A) Kobs values of the ester hydrolysis by various catalytic phages. (B) Catalytic activity of the CPN3 phage at substrate concentrations of 2–10 mM.
In order to explore the role of the phage itself in catalysis, the hydrolytic activity of the chemically synthesized peptides was tested. It was found that while hydrolytic rates in pNPA hydrolysis were 3 orders of magnitude lower, a similar trend of relative rates compared to that of phages was observed (Figures 3A and S5), demonstrating that the phage protein, but also the peptide sequence influences activity. A remarkable observation is the (relatively) enhanced activity of free peptide CPN3 in ester hydrolysis. When compared to literature values, the rate observed for CPN3 of kobs = 3.5 × 10−3 s−1 (Figure S4.3b, 10 mM) is an order of magnitude lower compared to the best value observed for supramolecular peptide systems described by Korendovych,3 which benefits from substrate binding, multi-valency, and which is strictly zinc dependent. Attempts to quantify amide condensation by free peptides yielded inconclusive results, due to the aforementioned background condensation observed for 1/2b.
Due to the observed sequence diversity, it will be a major endeavor to establish the mechanisms underlying the hydrolytic activities. In order to assess a possible role of the substrate binding, catalytic rates were compared at varying substrate concentrations for both phage and free peptide (Figures 3B and S5b). The catalytic profiles do not suggest Michaelis–Menten kinetics, rather a close to linear relationship within the concentration range studied (solubility of pNPA becomes limiting over 10 mM), indicating that the Km value is not yet approached under these conditions and therefore a kcat value cannot be determined. This result implies a minor role of substrate binding in catalysis, which differentiates our systems from previously reported self-assembled catalytic systems,3,13,15,16 and suggesting operation more akin to small molecule catalysts. One approach for improvement of catalytic activity is introduction of binding pockets.3
In summary, the strategy demonstrated in this work, combining the catalytic self-assembly of product with the phage display library, was successful in the identification of seven phages displaying dodecapeptides that enable the hydrolysis of ester and amide bonds under physiological conditions. The approach allows for non-biased discovery as we make no assumptions about the origins of catalysis. There is no apparent sequence homology in the peptide sequences, further demonstrating that function can be found in random, unevolved peptide sequences.35
Supplementary Material
ACKNOWLEDGMENTS
Development of modified phage display library methodology for catalytic peptide discovery and selection of catalytic peptides through this approach were supported by the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering under Award No. DE-FG-02- 01ER45935. Hunter College infrastructure is supported by Grant No. MD007599 from the National Institute on Minority Health and Health Disparities (NIMHD) of the National Institutes of Health (NIH). R.V.U. acknowledges funding from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/EMERgE/ERC grant agreement no. 258775. Y.M. thanks the Japan Society for the Promotion of Science, and the International Training Program provided through Tokyo University of Agriculture and Technology. We thank Prof. Y. Xu for the assistance in biological experiments, Dr. L. Yang for mass spectrometry, Prof. P. J. Halling and Dr. A. Mulholland for helpful discussions, and Vivomotion for phage images.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
Optimization of phage elution, selected sequences, additional TEM analysis, amide condensation, FRET peptide hydrolysis, and detailed methods. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Nanda V; Koder RL Nat. Chem 2010, 2, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Golynskiy MV; Seelig B Trends Biotechnol. 2010, 28, 340. [DOI] [PubMed] [Google Scholar]
- (3).Rufo CM; Moroz YS; Moroz OV; Stöhr J; Smith TA; Hu X; DeGrado WF; Korendovych IV Nat. Chem 2014, 1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Whaley SR; English DS; Hu EL; Barbara PF; Belcher AM Nature 2000, 405, 665. [DOI] [PubMed] [Google Scholar]
- (5).Naik RR; Stringer SJ; Agarwal G; Jones SE; Stone MO Nat. Mater 2002, 1, 169. [DOI] [PubMed] [Google Scholar]
- (6).Ellington AD; Szostak JW Nature 1990, 346, 818. [DOI] [PubMed] [Google Scholar]
- (7).Tuerk C; Gold L Science 1990, 249, 505. [DOI] [PubMed] [Google Scholar]
- (8).Bastian AA; Marcozzi A; Herrmann A Nat. Chem 2012, 4, 789. [DOI] [PubMed] [Google Scholar]
- (9).Pollack SJ; Jacobs JW; Schultz PG Science 1986, 234, 1570. [DOI] [PubMed] [Google Scholar]
- (10).Tramontano A; Janda KD; Lerner RA Science 1986, 234, 1566. [DOI] [PubMed] [Google Scholar]
- (11).Forrer P; Jung S; Pluckthun A Curr. Opin. Struct. Biol 1999, 9, 514. [DOI] [PubMed] [Google Scholar]
- (12).Arnold FH Acc. Chem. Res 1998, 31, 125. [Google Scholar]
- (13).Guler MO; Stupp SI J. Am. Chem. Soc 2007, 129, 12082. [DOI] [PubMed] [Google Scholar]
- (14).Gao Y; Zhao F; Wang Q; Zhang Y; Xu B Chem. Soc. Rev 2010, 39, 3425. [DOI] [PubMed] [Google Scholar]
- (15).Zupeng H; Huang G; Guan S; Wang Y; Shi G; et al. J. Mater. Chem. B 2013, 1, 2297. [DOI] [PubMed] [Google Scholar]
- (16).Zaramella D; Scrimin P; Prins LJ J. Am. Chem. Soc 2012, 134, 8396. [DOI] [PubMed] [Google Scholar]
- (17).Berdugo C; Miravet JF; Escuder B Chem. Commun 2013, 49, 10608. [DOI] [PubMed] [Google Scholar]
- (18).Coppage R; Slocik JM; Ramezani-Dakhel H; Bedford NM; Heinz H; Naik R; Knecht MR J. Am. Chem. Soc 2013, 135, 11048. [DOI] [PubMed] [Google Scholar]
- (19).Li Y; et al. Bioorg. Med. Chem 2000, 8, 2675. [DOI] [PubMed] [Google Scholar]
- (20).(a) Krattiger P; McCarthy C; Pfaltz A Angew. Chem., Int. Ed 2003, 42, 1722. [DOI] [PubMed] [Google Scholar]; (b) Krattiger P; Kovasy R; Revell JD; Ivan S; Wennemers H Org. Lett 2005, 7, 1101. [DOI] [PubMed] [Google Scholar]
- (21).Winkler S; Wilson D; Kaplan DL Biochemistry 2000, 39, 12739. [DOI] [PubMed] [Google Scholar]
- (22).Collier JH; Messersmith PB Bioconjugate Chem. 2003, 14, 748. [DOI] [PubMed] [Google Scholar]
- (23).Yang ZM; et al. Adv. Mater 2004, 16, 1440. [Google Scholar]
- (24).Williams RJ; Smith AM; Collins R; Hodson N; Das AK; Ulijn RV Nat. Nanotechnol 2009, 4, 19. [DOI] [PubMed] [Google Scholar]
- (25).Kiyonaka S; et al. Nat, Mater. 2004, 3, 58. [DOI] [PubMed] [Google Scholar]
- (26).Estroff LA; Hamilton AD Chem. Rev 2004, 104, 1201. [DOI] [PubMed] [Google Scholar]
- (27).Hirst AR; Escuder B; Miravet JF; Smith DK Angew. Chem., Int. Ed 2008, 47, 8002. [DOI] [PubMed] [Google Scholar]
- (28).Wolfenden R; Snider MJ Acc. Chem. Res 2001, 34, 938. [DOI] [PubMed] [Google Scholar]
- (29).Toledano S; Williams RJ; Jayawarna V; Ulijn RV J. Am. Chem. Soc 2006, 128, 1070. [DOI] [PubMed] [Google Scholar]
- (30).Zhou J; Du XW; Gao Y; Shi JF; Xu BJ Am. Chem. Soc 2014, 136, 2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Olive AGL; Abdullah NH; Ziemecka I; Mendes E; Eelkema R; van Esch JH Angew.Chem., Int. Ed 2014, 53, 4132. [DOI] [PubMed] [Google Scholar]
- (32).Wei Z; Maeda Y; Matsui H Angew. Chem., Int. Ed 2001, 50, 10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Das AK; Collins R; Ulijn RV Small 2008, 4, 279. [DOI] [PubMed] [Google Scholar]
- (34).Broo KS; Brive L; Ahlberg P; Baltzer LJ Am. Chem. Soc 1997, 119, 11362. [Google Scholar]
- (35).Cherny I; Korolev M; Koehler AN; Hecht MH ACS Synth. Biol 2012, 1, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.