Abstract
Contactin-associated protein-like 4 (Cntnap4) is a member of the neurexin superfamily of transmembrane molecules that have critical functions in neuronal cell communication. Cntnap4 knockout mice display decreased presynaptic gamma-aminobutyric acid (GABA) and increased dopamine release that is associated with severe, highly penetrant, repetitive, and perseverative movements commonly found in human autism spectrum disorder patients. However, no known function of Cntnap4 has been revealed besides the nervous system. Meanwhile, secretory protein neural EGFL-like 1 (Nell-1) is known to exert potent osteogenic effects in multiple small and large animal models without the off-target effects commonly found with bone morphogenetic protein 2. In this study, while searching for a Nell-1-specific cell surface receptor during osteogenesis, we identified and validated a ligand/receptor-like interaction between Nell-1 and Cntnap4 by demonstrating: 1) Nell-1 and Cntnap4 colocalization on the surface of osteogenic-committed cells; 2) high-affinity interaction between Nell-1 and Cntnap4; 3) abrogation of Nell-1-responsive Wnt and MAPK signaling transduction, as well as osteogenic effects, via Cntnap4 knockdown; and 4) replication of calvarial cleidocranial dysplasiaslike defects observed in Nell-1-deficient mice in Wnt1-Cre-mediated Cntnap4-knockout transgenic mice. In aggregate, these findings indicate that Cntnap4 plays a critical role in Nell-1-responsive osteogenesis. Further, this is the first functional annotation for Cntnap4 in the musculoskeletal system. Intriguingly, Nell-1 and Cntnap4 also colocalize on the surface of human hippocampal interneurons, implicating Nell-1 as a potential novel ligand for Cntnap4 in the nervous system. This unexpected characterization of the ligand/receptor-like interaction between Nell-1 and Cntnap4 indicates a novel biological functional axis for Nell-1 and Cntnap4 in osteogenesis and, potentially, in neural development and function.
Keywords: NEURAL EGFL-LIKE 1 (NELL-1), CONTACTIN-ASSOCIATED PROTEIN-LIKE 4 (CNTNAP4), OSTEOGENESIS, LIGAND/RECEPTOR-LIKE INTERACTION, NELL-1/CNTNAP4 AXIS
Introduction
Neural EGFL-like 1 (Nell-1) is a unique secretory extracellular protein(1) that contains a secretory signal peptide, an N-terminal laminin G (LamG) domain (which overlaps with a thrombospondin-1-like [TSPN] module), a coiled-coil region, several von Willebrand factor-like repeats with five chordin-like, cysteine-rich (CR) domains, and six epidermal growth factor (EGF)-like repeats.(1,2) The combination of the LamG domain, CR domains, and EGF-like repeats in Nell-1 suggests specific ligand-receptor interactions; however, Nell-1 does not interact with known EGF-like receptors.(3–6) Watanabe and colleagues first sequenced Nell-1 from a human fetal brain cDNA library,(7) whereas our group first identified Nell-1’s osteogenic function from its increased expression in active bone formation sites of human craniosynostosis patients.(3) Through gain- and loss-of-function models, we have demonstrated Nell-1’s essential role in craniofacial and appendicular skeletogenesis.(8–10) For example, Nell-1 loss-of-function in newborn mice demonstrates reduced calvarial bone growth, enlarged sagittal sutures, and short body lengths.(1,9,11) Reaffirming the importance of Nell-1 in human development, a similar phenotype of delayed cranial fontanelle and suture closure with a short body stature was recently reported in a 3-year-old Japanese girl with Nell-1 gene locus deletion.(12) Excitingly, the osteogenic potential of Nell-1 has been validated in several small and large animal models consisting of rodents, sheep, and non-human primate.(13–16) Currently, the gold standard for bone graft substitutes is bone morphogenetic protein 2 (BMP2). BMP2, however, has several well-documented, off-target effects including postoperative inflammation, osteoclastogenesis, adipogenesis, and ectopic bone formation that hinder its clinical application. Because these adverse effects have not been observed after Nell-1 administration,(15,17–20) Nell-1 may hold great promise as an alternative therapeutic option for local or systemic bone regeneration.
Meanwhile, contactin-associated protein-like 4 (Cntnap4, also known as Caspr4) is a transmembrane neurexin superfamily receptor with vital functions in neurodevelopment and neurocognition.(21–23) Cntnap4 belongs to a large category of synaptic cell adhesion molecules (SCAMs)(24) that have been implicated in a wide variety of neurodevelopmental and neuropsychiatric disorders. Located on the presynaptic membrane of interneurons, Cntnap4 is known to regulate inhibitory interneuron synapse maturation as well as synaptic transmission in dopaminergic and GABAergic neurons; its loss in transgenic mouse models replicates common behavioral abnormalities observed in individuals with autism spectrum disorders (ASD).(22) Since the initial description of Cntnap4 in 2002(21) and the first Cntnap protein in 1997,(25) there has been, until now, no known extracellular ligand associated with these critical neuronal proteins. Moreover, there is no report for their function beside the nervous system.(26) Here, in an effort to identify the cell surface–specific receptor of Nell-1, we discovered and verified a novel ligand/receptor-like interaction between Nell-1 and Cntnap4 that plays a critical role in Nell-1-responsive osteogenesis. This is the first functional annotation for Cntnap4 in the musculoskeletal system. Furthermore, this unexpected characterization of Nell-1/Cntnap4 axis opens new avenues of further investigations of musculoskeletal development and regeneration, as well as of neural-skeletal cross-talk.
Materials and Methods
Animal breeding, skeletal staining, and micro-CT analysis
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All of the animals were handled according to the Chancellor’s Animal Research Committee (protocols number: 2012–041) of the University of California, Los Angeles (UCLA). All surgery was performed under sodium pentobarbital anesthesia, and every effort was made to minimize suffering.
Cntnap4flox/flox-GFP (mCntnap4-CKO-GFP-CKI) mice were provided by Applied StemCell, Inc. (Milpitas, CA, USA) mated with Wnt1-Cre mice (stock #022501; Jackson Laboratory, Bar Harbor, ME, USA) to obtain Cntnap4flox/+;Wnt1-Cre mice (Supplemental Materials and Methods, subsection 1). Mating was carried out overnight. Animal groups were of mixed sexes unless otherwise stated. All mice were housed in the light- and temperature-controlled UCLA vivarium.
Skeletal staining with Alcian blue and Alizarin red was performed on neonatal mice following the previously described protocols.(11,27) In brief, neonatal mice were euthanized with an overdose of phenobarbital, skinned, and eviscerated. Subsequently, samples were fixed in 95% ethanol for 24 hours at room temperature. Standard staining was performed using Alcian blue for negatively charged proteoglycans and Alizarin red for calcium to provide a gross distinction between cartilage and mineralized tissue, respectively. Parameters of high-resolution micro-CT and 3D reconstruction are detailed in the Supplemental Materials and Methods, subsection 2.
Histological analysis and immunocytochemistry (IHC) staining
Calvarial tissues dissected from six 60-day-old C57BL/6 mice were fixed in 4% ice-cold paraformaldehyde for 24 hours and decalcified with 19% EDTA (MilliporeSigma, Burlington, MA, USA) for 14 days before paraffin embedding. Hematoxylin and eosin (H&E) and IHC staining were performed on 5-μm deparaffinized sections. H&E-stained sections were used for histologic analysis by an experienced pathologist. Primary antibodies used in this study are summarized in Supplemental Table S1. Images were documented by a Keyence BZ-X710 system (Itasca, IL, USA).
Phage display biopanning
HIS-Select Nickel Magnetic Agarose Beads (MilliporeSigma) were selected to immobilize recombinant, full-length, human Nell-1 with polyhistidine-tag (His–tagged Nell-1; Supplemental Materials and Methods, subsection 3). The phage display cDNA library was constructed from the Human Brain cDNA Library using the Novagen T7Select System (MilliporeSigma). An aliquot of the amplified phage display cDNA library was incubated with His-tagged Nell-1–coated beads for four rounds of biopanning screens. Subsequently, the phages bound to the His-tagged Nell-1–coated beads were eluted, and 100 plaques were selected to amplify the phage DNA by PCR with the provided primers and buffers in the Novagen T7Select System. Sequences spanning over 500 bp in size were selected and sequenced by Laragen Inc. (Culver City, CA, USA) (Supplemental Fig. S1). Dissociation constant ELISA was performed to examine the binding affinity with the phages (Supplemental Materials and Methods, subsection 4).
Gene expression profiling
Based on previously published functional investigations of Nell-1, 12 different cell lines and primary cells isolated from bone or relevant tissues were selected for use in the current study (Supplemental Table S2). Total RNA was isolated from subconfluent cells by TRIzol Reagent, followed by DNase treatment. An amount of 1 mg RNA was injected for reverse transcription with the iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad Laboratories Inc., Hercules, CA, USA). 1 μL reverse transcription product was used for real-time PCR with SsoAdvanced Universals Probes Supermix (Bio-Rad) and TaqMan primers/probe sets (listed in Supplemental Table S3) on a QuantStudio3 system (Thermo Fisher Scientific, Canoga Park, CA, USA) in triplicate.
Nell-1/Cntnap4 physical binding analysis
Serum-starved MC3T3-E1 pre-osteoblasts (Subclone 4, ATCC CRL-2593; ATCC, Manassas, VA, USA) and primary newborn mouse calvaria cells (NMCC)(10) were used to confirm the interaction between Nell-1 and Cntnap4 by immunocytochemistry (ICC) staining, pull-down, co-immunoprecipitation (Co-IP), and Duolink proximity ligation assay (PLA) analysis (Supplemental Materials and Methods, subsections 5–8). Primary antibodies used in this study are summarized in Supplemental Table S1. The binding properties of Nell-1 and Cntanp4 (Supplemental Materials and Methods, subsection 9) were analyzed on a Biacore 3000 instrument (Biacore AB, Uppsala, Sweden) by the UCLA Surface Plasmon Resonance (SPR) Core. More details are provided in the Supplemental Materials and Methods, subsection 10.
RNA interference (RNAi) and in vitro osteogenic differentiation assays
MC3T3-E1 cells were transfected with Cntnap4 shRNA Lentiviral particles (Santa Cruz Biotechnology, Dallas, TX, USA) or non-target control shRNA by Lipofectamine 3000. The positive transfected colonies were selected by Puromycin (MilliporeSigma).
Control and stable Cntnap4-knockdown (Cntnap4-KD) MC3T3-E1 cells were seeded on 24-well plates for alkaline phosphatase (ALP), Alizarin red, and IHC staining. In addition, cells were seeded on 6-well plates for osteogenic genes expression assay. Both cell types were cultured in osteogenic differentiation medium (α-MEM, 10% fetal bovine serum [FBS; Thermo Fisher Scientific], 50 μg/mL ascorbic acid, and 10 mM β-glycerophosphate [MilliporeSigma]) with or without 500 ng/mL Nell-1 or 100 ng/mL BMP2 (Medtronic, Minneapolis, MN, USA). Protein isolation and Western blot were performed as previously described.(28)
Mouse calvarial bone explant cultivation
Calvarial vaults of neonatal C57BL/6 mice were harvested and cultured as previously described.(28) Subsequently, the calvarial vaults were randomly assigned to experimental groups (8 calvarial vaults/group). Lentiviral particles of Cntnap4 shRNA (Santa Cruz Biotechnology) and CMV-Nell-1 (GenTarget Inc., San Diego, CA, USA) were added to the culture medium (serum-free BGJb medium [Biggers, Gwatkin, Judah] supplemented with 1× L-glutamine, 100 unit/mL of penicillin, 100 μg/mL of streptomycin, 2.5 μg/mL of amphotericin B, 100 μg/mL of L-ascorbic acid, and 10 mM glycerophosphate) at day 0 and day 1, respectively. Alizarin Complexone (2 μg/mL, MilliporeSigma) was added to the culture medium at day 4, and the medium, containing Alizarin Complexone, was changed every 3 days thereafter. The explants were fixed in 4% paraformaldehyde at day 10. The Alizarin Complexone deposition on explants was observed with an Olympus SZX12 fluorescent microscope. Overlap width of the frontal and parietal bones, as well as the area of unclosed fontanel, were quantified using Image-Pro Plus (Media Cybernetics, Warrendale, PA, USA). To avoid surveyor bias, three experienced pathologists recognized and measured the overlapped widths in a double-blinded fashion. Each sample was measured three times, and the means of the values were plotted.
Statistical analysis
All statistical analyses were conducted in consultation with the UCLA Statistical Biomathematical Consulting Clinic. Initial sample numbers were determined using a power analysis to give α = 0.05 and power = 0.8. No data were excluded from the analyses. Statistical significance was performed with OriginPro 8 (Origin Lab Corp., Northampton, MA, USA) and included the one-way ANOVA and two-sample t test. Individual comparisons between two groups were determined by the Mann-Whitney test for nonparametric data. Statistical significance was determined at the p < 0.05 level.
Results
T7 phage particles with 1st and 2nd LamG extracellular domains of Cntnap4 exhibit high binding affinity to Nell-1
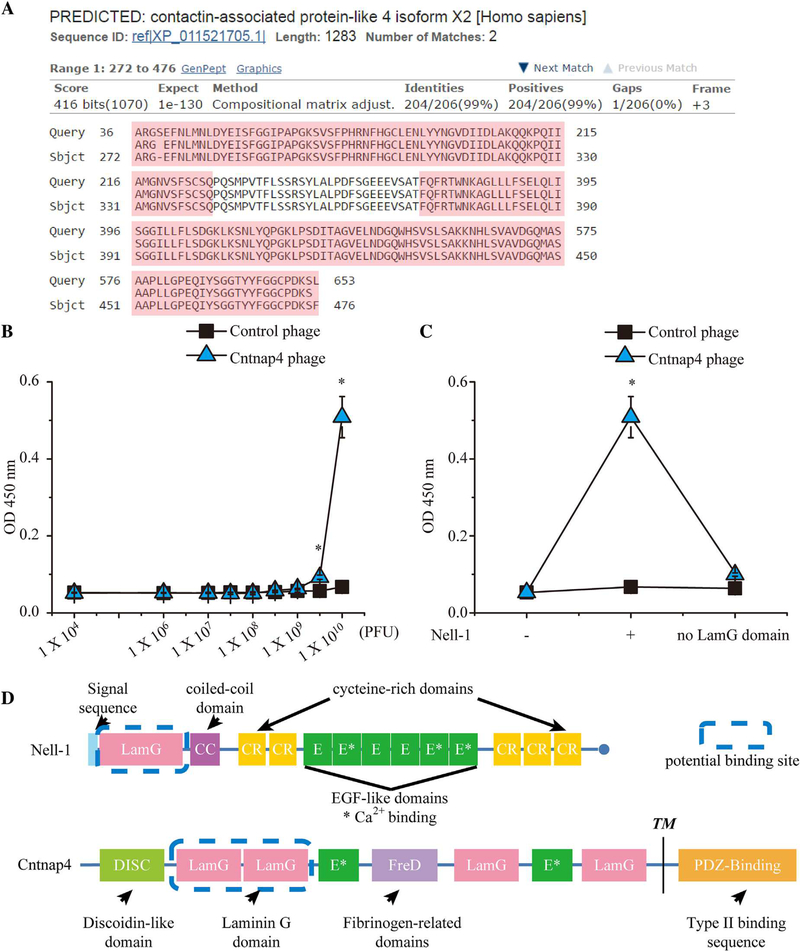
To identify the native binding protein(s) of Nell-1, we used a human brain cDNA library to construct a T7 phage display cDNA library for biopanning. After four rounds of biopanning, the phages bound to the His-tagged Nell-1–coated magnetic beads were eluted. We selected 100 plaques for phage DNA amplification by PCR and sequenced DNA fragments over 500 bp in size (Supplemental Fig. S1). Twenty-three Nell-1-binding candidates were identified, nine of which were transmembrane proteins (Supplemental Table S4). We repeatedly detected phage particles harboring a sequence that aligned with the first and second LamG extracellular domains of Cntnap4 (Fig. 1A), which exhibited high binding affinity to the full-length Nell-1 protein (Fig. 1B, C). Further studies indicated that the N-terminal LamG domain is essential for the Nell-1/Cntnap4 interaction as deletion of the Nell-1 LamG domain nearly eliminated Cntnap4 phage binding to Nell-1 (Fig. 1C, D).
Fig. 1.
Confirmation of binding affinity between Cntnap4 phages and Nell-1 using a binding dissociation constant ELISA assay. (A) The Basic Local Alignment Search Tool (BLAST) result of the amino acid sequence displayed by the T7 phage constructed with human brain cDNA matched the human Cntnap4 partial protein sequence. Query = the amino acid sequence enclosed by the T7 phage DNA; Sbjct = the matched amino acid sequence Cntnap4. The LamG domains are highlighted in pink. (B) By increasing the number of phages incubated with Nell-1 precoated ELISA plates, the Cntnap4 phage demonstrated significantly higher binding affinity than the control phage. (C) The Cntnap4 phage revealed high binding affinity only to full-length Nell-1 and not to LamG domain-deleted Nell-1. (D) Structures of Nell-1 and Cntnap4 and their potential interaction domains. Nell-1 is a secreted protein composed of 810 amino acids with a molecular weight of ~90 kDa before N-glycosylation and oligomerization. It contains several structural motifs including a laminin G (LamG) domain, a coiled-coil (CC) domain, five cysteine-rich (CR) domains, and six epidermal growth factor (E)-like domains. Cntnap4 is a transmembrane protein of 1310 amino acids consisting of a large extracellular domain, a single membrane-spanning domain, and a short cytoplasmic region at the carboxy-terminus. The extracellular region is composed of a discoidin-like domain (DISC), a fibrinogen-related domain (FreD), two E repeats, and four LamG domains. The cytoplasmic region contains a binding site for PDZ domains. The potential binding domain of Nell-1 and Cntnap4 is highlighted by the blue dashed line. TM= transmembrane. Mean ± SEM of six independent experiments performed in triplicate is shown. *p < 0.05 when compared with control phage.
Nell-1 and Cntnap4 bind on the plasma membrane of osteogenic-committed cells
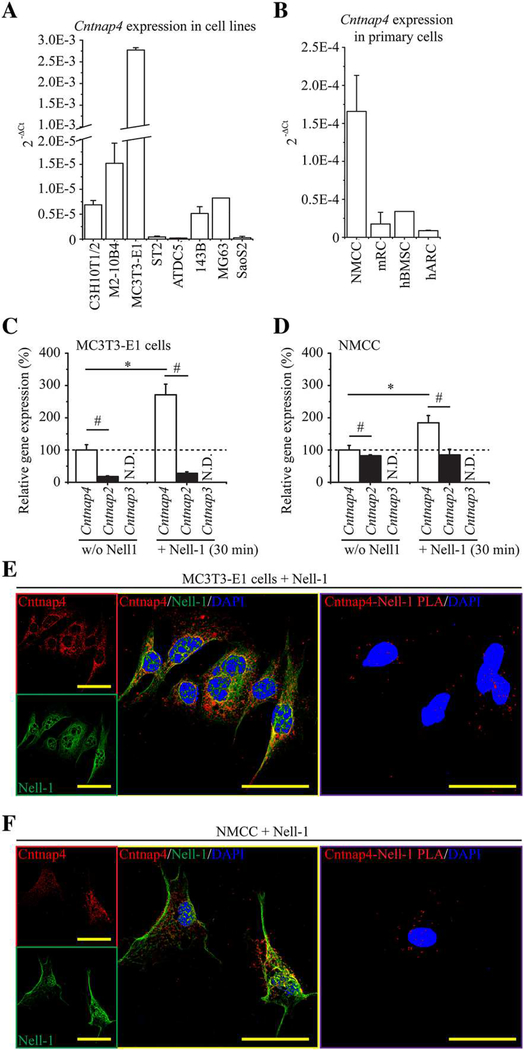
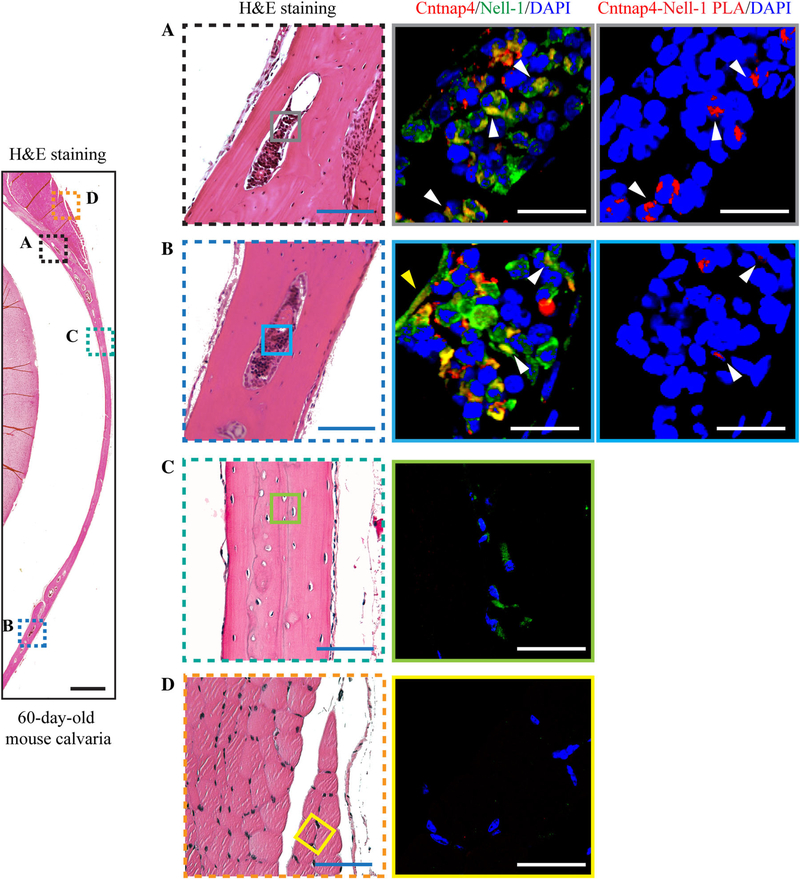
Of the 12 Nell-1-responsive cell lines capable of osteogenic and/or chondrogenic differentiation (Supplemental Table S2), Cntnap4 expression was highest in the osteogenic-committed MC3T3-E1 cell line(29,30) (Fig. 2A), as opposed to less differentiated cell lines (C3H10T1/2, ST-2, 143B, MG63, and SaoS2(29,31–34)), predominantly adipogenic-committed cell line M2–10B4,(35) or chondrogenic-committed cell line ATDC5.(36) Meanwhile, NMCC, which are known to be highly osteogenic,(10) exhibited significantly higher levels of Cntnap4 expression when compared with primary mouse rib chondrocytes, human articular chondrocytes, or human bone marrow stem cells (Fig. 2B). Further, exogenously administered Nell-1 protein significantly upregulated Cntnap4 expression in both MC3T3-E1 pre-osteoblasts and primary NMCC (Fig. 2C, D). Using confocal laser scanning microscopy (CLSM), we confirmed the colocalization of Nell-1 and Cntnap4 on the plasma membrane of MC3T3-E1 and NMCC in the samples incubated with recombinant human Nell-1 (Fig. 2E, F). We further validated these results utilizing an in situ Duolink PLA (Fig. 2E, F). Additionally, we observed the most prominent Nell-1/Cntnap4 binding in the marrow cavity rather than in the calcified regions of 60-day-old mouse calvarial bone or the surrounding muscle (Fig. 3).
Fig. 2.
Cntnap4 and Nell-1 colocalization in osteogenic-committed cells. (A) Of the eight types of tested cell lines, MC3T3-E1 pre-osteoblasts expressed the highest levels of Cntnap4. (B) Of the four types of tested primary cells, NMCC exhibited the highest expression levels of Cntnap4. NMCC = newborn mouse calvarial cells. mRC = mouse rib chondrocytes. hBMSC = human bone marrow stem cells. hARC = human articular chondrocytes. Mean ± SEM of six independent experiments performed in triplicate is shown (A, B). In addition, Nell-1 significantly increased the levels of Cntnap4 in both MC3T3-E1 pre-osteoblasts (C) and NMCC (D). On the contrary, expression of Cntnap2, which was markedly lower than that of Cntnap4, was not responsive to Nell-1 simulation. Mean ± SEM of three independent experiments performed in duplicate is shown (C, D). *p < 0.05 when compared with the group without Nell-1 treatment; #p < 0.05 when compared with the Cntnap4 expression. Moreover, CLSM revealed the colocalization of Nell-1 and Cntnap4 in MC3T3-E1 pre-osteoblasts after 30 minutes of incubation with exogenous recombinant human Nell-1 (E). Colocalization was predominantly found on the plasma membrane. The direct Nell-1/Cntnap4 interaction was validated by Duolink PLA. Similar Nell-1 and Cntnap4 colocalization and protein interactions were also observed in the plasma membrane of NMCC with 30 minutes of Nell-1 treatment (F). Scale bar = 50 μm.
Fig. 3.
Cntnap4 and Nell-1 colocalization in mouse calvarial bone marrow cavities. (A, B) Calvarial bones of 60-day-old mice showed high-intensity double staining of Nell-1 and Cntnap4 inside the bone marrow cavities. White arrows = marrow cavity cells with both Nell-1/Cntnap4 colocalization staining and PLA signaling; yellow arrow = bone lining cells with Nell-1/Cntnap4 colocalization staining. Nell-1 was detected in the calcified bone of 60-day-old mouse calvaria (C) but not in the surrounding muscle (D). However, unlike the marrow cavity where Nell-1 and Cntnap4 colocalize, Cntnap4 was barely detected in the calcified areas of mouse calvarial bone and the surrounding muscle. Scale bar = 500 μm (black), 100 μm (blue), and 20 μm (white).
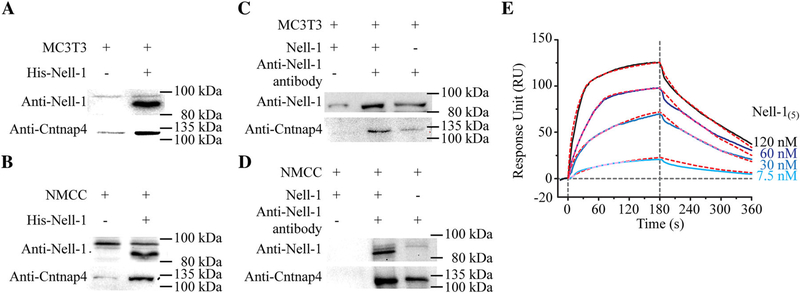
We next used His-tagged Nell-1 to successfully pull down Cntnap4 from the lysate of MC3T3-E1 pre-osteoblasts and primary NMCC without the use of cross-linking reagents (Fig. 4A, B), which demonstrates a strong physical interaction between Cntnap4 and Nell-1. Subsequently, we performed a Co-IP assay using an antibody against Nell-1 and thus confirmed Cntnap4 and Nell-1 binding in both MC3T3-E1 pre-osteoblasts and primary NMCC (Fig. 4C, D). Moreover, SPR analysis revealed the high binding affinity between Nell-1 and the immobilized extracellular portion of Cntnap4 (Cntnap4extra): KD = 32.8 ± 0.9 nM, ka = (2.39 ± 0.05) × 105 1/Ms, and kd = 0.0078 ± 0.0002 1/s (Fig. 4E). These data indicate a ligand/receptor-like interaction(37) between Nell-1 and Cntnap4.
Fig. 4.
Physical interaction between Nell-1 and Cntnap4. Pull-down assays were performed with MC3T3-E1 pre-osteoblasts (A) and NMCC (B). Increased Cntnap4 was detected when beads were coated with His-tagged Nell-1. Co-immunoprecipitation assay with MC3T3-E1 pre-osteoblasts (C) and NMCC (D) demonstrated an increase in Cntnap4 when cells were incubated with Nell-1. To confirm specificity, no Cntnap4 was detected when the agarose beads were not coated with anti-Nell-1 antibody. (E) SPR assay was performed to assess the binding affinity and dynamic relationship between pentameric Nell-1 (Nell-1(5)) and the immobilized Cntnap4extra, which demonstrated classical ligand-receptor binding. Red dashed lines present the kinetic projection performed by Scrubber 2.0 (BioLogic Software Pty Ltd., Campbell, Australia).
Although extracellular ligands for protein members of the Cntnap family have not been previously described, we tested whether Nell-1, as the first identified extracellular ligand of Cntnap4, may also bind Cntnap2 and Cntnap3 in osteogenic-committed cells. Overall, we did not detect any Cntnap3 expression in cultured MC3T3-E1 pre-osteoblasts, primary NMCC, or in mouse calvarial bone cells in vivo (Fig. 2C, D and Supplemental Fig. S2A, B). We did detect Cntnap2 expression in both MC3T3-E1 pre-osteoblasts and NMCC; however, Cntnap2 expression was significantly lower than that of Cntnap4. Further, unlike Cntnap4, Cntnap2 expression was not markedly induced by Nell-1 stimulation (Fig. 2C, D) and only a few bone marrow cells exhibited binding between Nell-1 and Cntnap2 (Supplemental Fig. S2C, D). These data suggest that Cntnap4 is more associated with osteogenesis when compared with Cntnap2 or Cntnap3.
Depleting Cntnap4 expression diminishes Nell-1-responsive osteogenesis
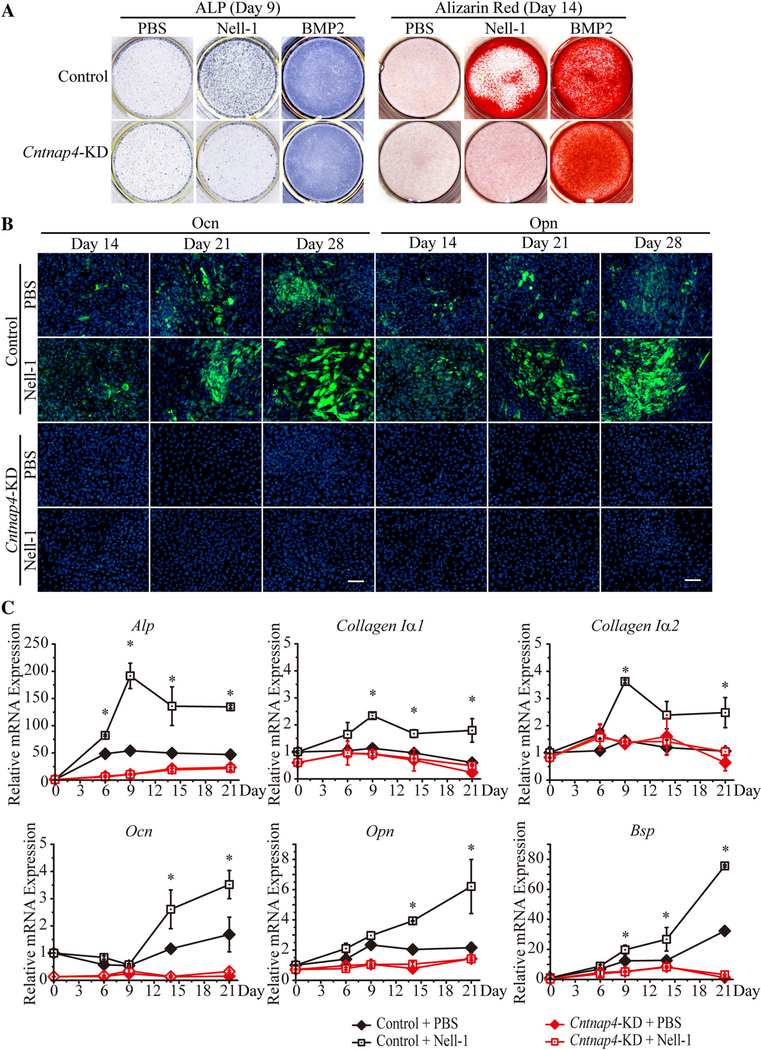
To determine the necessity of Cntnap4 for Nell-1-mediated osteogenesis, we established a stable Cntnap4-KD MC3T3-E1 cell line whose Cntnap4 expression was approximately 85% lower than control scramble-shRNA transfected (Control) MC3T3-E1 cells (Supplemental Fig. S3). In Control MC3T3-E1 cells, Nell-1 protein treatment markedly increased ALP and Alizarin red staining (Fig. 5A). However, in Cntnap4-KD MC3T3-E1 cells, only negligible staining was observed subsequent to Nell-1 treatment (Fig. 5A). Interestingly, both control MC3T3-E1 cells and Cntnap4-KD MC3T3-E1 cells exhibited similar robust osteogenic responses to BMP2, an osteogenic protein with a signaling pathway distinct from Nell-1.(1) These data indicate that Cntnap4-KD specifically targeted Nell-1 but not BMP2-mediated MC3T3-E1 cell osteogenic differentiation. Additionally, Nell-1 protein significantly enhanced osteocalcin (Ocn) and osteopontin (Opn) expression in control MC3T3-E1 pre-osteoblasts, but this effect was abrogated in Nell-1-treated Cntnap4-KD MC3T3-E1 cells (Fig. 5B). Concomitant gene expression profiling of osteogenic markers Alp, Collagen Iα1, Collagen Iα2, Ocn, Opn, and bone sialoprotein (Bsp) (Fig. 5C) revealed inhibited osteogenic differentiation in Cntnap4-KD MC3T3-E1 cells and a consequential lack of response to Nell-1. These data show that Cntnap4 is indispensable for the osteogenic bioactivity of Nell-1.
Fig. 5.
Cntnap4 is indispensable for Nell-1 osteogenic bioactivity in vitro. (A) ALP staining on day 9 and Alizarin red staining on day 14 revealed increased staining in the Nell-1 and BMP2 groups of control shRNA-transfected MC3T3-E1 cells. In Cntnap4-KD MC3T3-E1 cells, high staining intensities of ALP and Alizarin red were only present in the BMP2 group. (B) A time-dependent, steady increase in Ocn and Opn staining was observed in both PBS and recombinant human Nell-1-treated control MC3T3-E1 cells. At each time point, the Nell-1-treated group demonstrated increased staining intensity when compared with the PBS-treated group. In Cntnap4-KD MC3T3-E1 cells, neither PBS nor Nell-1 treatment resulted in detectable positive staining of Ocn or Opn. (C) In Control MC3T3-E1 cells, Alp, Collagen Iα1, and Collagen Iα2 reached peak expression levels 9 days after stimulation, whereas Ocn, Opn, and Bsp displayed time-dependent patterns of increased expression. Notably, higher expression levels were detected for each gene in the Nell-1 treatment group when compared with the PBS group. However, Cntnap4-KD MC3T3-E1 cells did not exhibit significant changes in any osteogenic markers nor any differences between the PBS group and the Nell-1 group. Mean ± SEM of six independent experiments performed in triplicate is shown. *p < 0.05 when compared with the Control + PBS group. Scale bar = 100 μm.
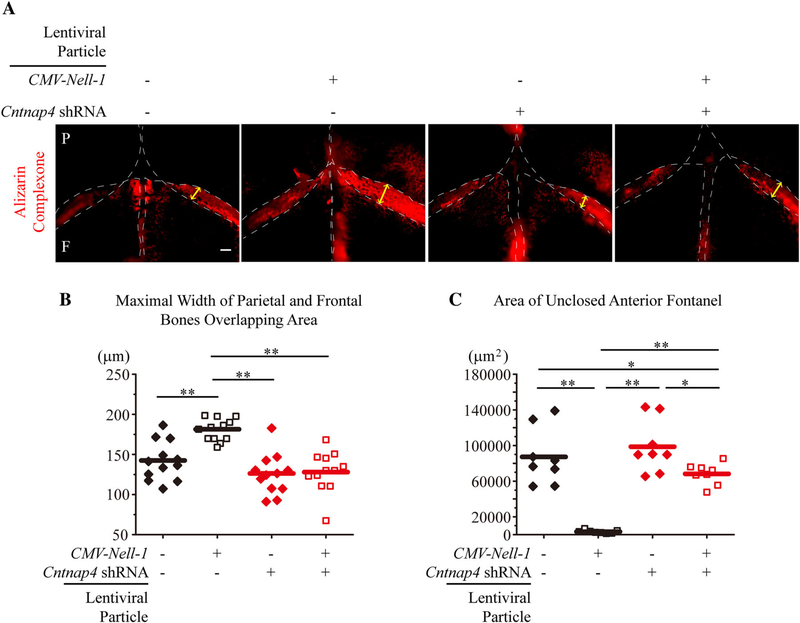
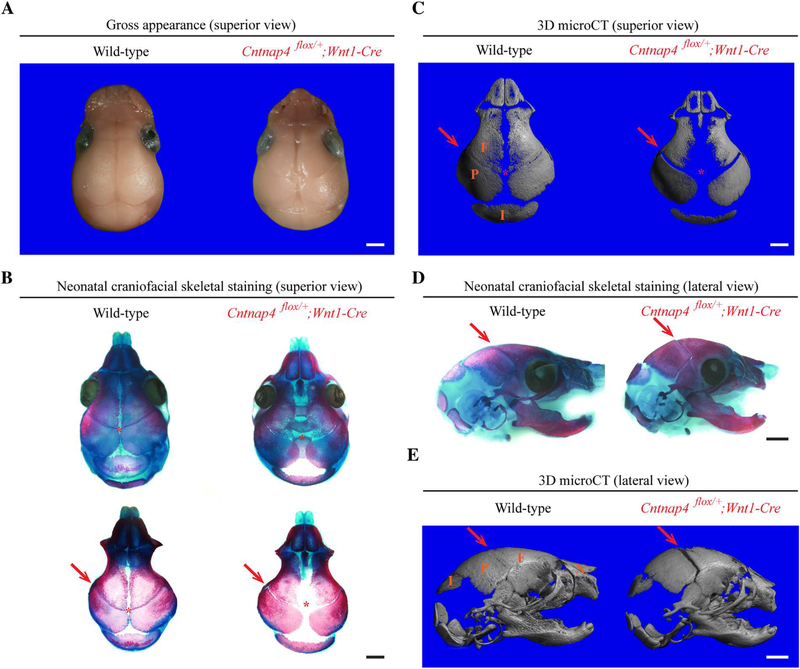
To further confirm the role of Cntnap4 in Nell-1-mediated osteogenesis, we transduced neonatal mouse calvarial explants with control (empty), CMV-Nell-1, Cntnap4 shRNA, or CMV-Nell-1 + Cntnap4 shRNA lentiviral particles, and cultured them ex vivo. After 10 days of culture, the explants transfected with CMV-Nell-1 exhibited increased bone formation (delineated by increased mineral apposition in Alizarin Complexone) (Fig. 6A), increased bony overlaps between the parietal and frontal bones (in the coronal sutures) (Fig. 6B), and narrowed anterior fontanels (Fig. 6C) relative to the explants transfected with control lentiviral particles. Moreover, Cntnap4-KD completely ablated the osteogenic effects of Nell-1 overexpression in the calvarial explants transfected with CMV-Nell-1 (Fig. 6). In agreeance with the ex vivo calvarial explant cultivation, defective mineralization and bone formation were also observed in the coronal suture of neonatal mice with Wnt1-Cre-mediated deletion of Cntnap4 (Cntnap4flox/+;Wnt1-Cre) (Fig. 7). It is worth noting that Wnt1-Cre-mediated Cntnap4-knockout also replicated the effects of Nell-1 deficiency on craniofacial skeletal developmental anomalies in neonatal mice.(11) These data further demonstrate the essential role of Cntnap4 in Nell-1-mediated osteogenesis and calvarial bone development.
Fig. 6.
Cntnap4-KD blocks the osteogenic effects of Nell-1 ex vivo. (A) Mineral deposition in the mouse calvarial explants was revealed by Alizarin Complexone during the culture period. Lentiviral overexpression of Nell-1 increased the density of Alizarin Complexone; however, when Nell-1 was overexpressed in Cntnap4-KD samples, the Alizarin Complexone staining was comparable to the control (without Cntnap4-KD or Nell-1 overexpression). (B) Quantification of the maximal width of the frontal and parietal bone overlapping area. Nell-1 overexpression alone increased the overlapping area, whereas Cntnap4-KD alone slightly reduced the overlapping area. When the Cntnap4-KD samples were treated with Nell-1 lentiviral overexpression, the maximal width of the overlapping area remained unchanged (similar to that of the control group). (C) Quantification of the unclosed anterior fontanel area. The calvarial explants in the Nell-1 overexpression group demonstrated completely closed fontanels, whereas anterior fontanels in the control group remained open. Cntnap4-KD alone slightly inhibited the closure of the anterior fontanels. The Cntnap4-KD + Nell-1 overexpression group showed a largely open fontanel area. The edge of each calvarial bone is outlined by a white dotted line; yellow arrows represent the maximal width of the frontal and parietal bone overlapping area (in the coronal suture). P=parietal; F=frontal. Eight calvaria explants were used for each group. For B and C, the means were used as center values. The Mann–Whitney test was used for statistical analysis. p < 0.05, p < 0.01. Scale bar = 100 μm.
Fig. 7.
Wnt1-Cre-mediated Cntnap4-knockout (Cntnap4flox/+;Wnt1-Cre) neonatal mice demonstrate decreased craniofacial development. Representative superior view of gross appearance (A), skeletal staining (B), and 3D micro-CT reconstruction (C) images of the craniofacial skeleton of neonatal Cntnap4flox/+;Wnt1-Cre mouse and its wild-type littermate demonstrate the significant difference in the cranial bone formation and sutural patency. In comparison with its wild-type littermate, the neonatal Cntnap4flox/+;Wnt1-Cre mouse has significantly less bone in the anterior fontanel of calvaria (red asterisk). In addition, defective mineralization and bone formation are also observed in the coronal suture (red arrows) of the Cntnap4flox/+;Wnt1-Cre mouse skull, which can also be appreciated from the lateral view (D) and 3D micro-CT reconstructions (E). Six pairs of neonatal Cntnap4flox/+;Wnt1-Cre mice and their wild-type littermates were compared. F = frontal bone; P = parietal bone; I = interparietal bone; N = nasal bone. Scale bar = 1 mm.
Cntnap4 mediates Nell-1-stimulated MAPK and Wnt signaling in osteogenic-committed cells
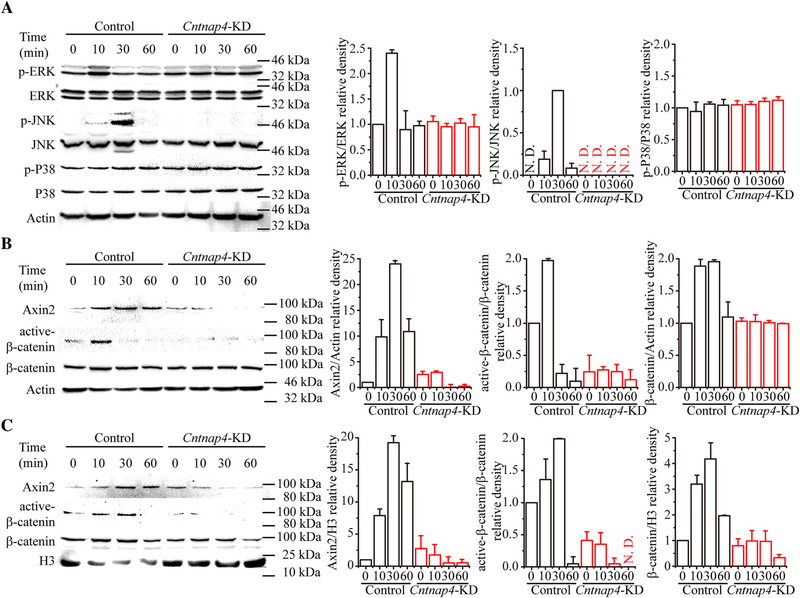
Because Cntnap4-KD specifically diminished the MC3T3-E1 cells’ robust osteogenic response to Nell-1 but not BMP2 (Fig. 5A), it was hypothesized that Cntnap4 is specifically involved in osteogenesis regulated by Nell-1. Because Nell-1 is known to activate Wnt and MAPK transduction during osteogenesis,(16,28,38,39) we next examined the effect of Cntnap4-KD on Wnt and MAPK signaling pathways during osteogenic differentiation. In agreement with our previous studies, Nell-1 induced high ERK and JNK phosphorylation/activation levels in control MC3T3-E1 cells. Conversely, Nell-1-responsive ERK or JNK activation was markedly diminished in Cntnap4-KD MC3T3-E1 cells (Fig. 8A). Similarly, although Nell-1 treatment significantly elevated intracellular and nuclear levels of Axin2 and active β-catenin in control MC3T3-E1 pre-osteoblasts, Cntnap4-KD completely abolished Nell-1-responsive Wnt signaling (Fig. 8B, C). These findings demonstrate that Cntnap4 is critical and specific for Nell-1-mediated activation of MAPK and Wnt signaling in osteogenic-committed cells (Fig. 9).
Fig. 8.
Cntnap4 is indispensable for the Nell-1-responsive activation of MAPK and Wnt signaling pathways in vitro. (A) Activation of MAPK signaling in Control and Cntnap4-KD MC3T3-E1 pre-osteoblasts stimulated with Nell-1. In Control MC3T3-E1 cells, significantly higher levels of pERK and pJNK were detected 10 minutes and 30 minutes after Nell-1 stimulation, respectively. There was no change detected in the phosphorylation level of P38. In Cntnap4-KD MC3T3-E1 cells, Nell-1 stimulation did not alter the expression levels of pERK and pJNK. Expression of Wnt signaling molecules in the whole cell lysate (B) and cell nuclear lysate (C) of Control and Cntnap4-KD MC3T3-E1 cells treated with Nell-1. In Control MC3T3-E1 cells, Nell-1 significantly increased the expression levels of Axin2 and active β-catenin, whereas no effect was observed on these markers in Cntnap4-KD cells treated with Nell-1. The charts demonstrate mean relative band intensity (normalized to control MC3T3-E1 at 0 minutes) ± SEM for three individual experiments. p < 0.05 when compared with the Control group at 0 minutes, #p < 0.05 when compared with the Cntnap4-KD group at 0 minutes.
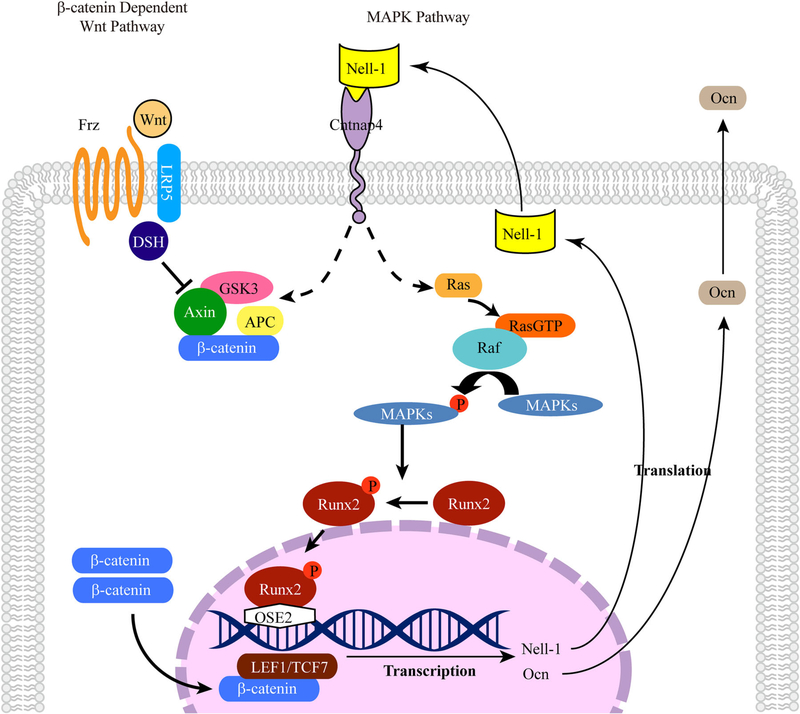
Fig. 9.
Schematic diagram of Nell-1 signaling pathways during osteogenesis. As a secreted molecule, Nell-1 initiates cellular signaling through binding to its specific receptor, Cntnap4, on the cell surface. The MAPK and Wnt signaling pathways play critical roles in Nell-1-mediated osteogenesis. Nell-1 preferentially activates ERK and JNK in MAPK signaling and also promotes the phosphorylation/activation of Runx2, which stimulates the expression of Nell-1 and Ocn by directly binding to the OSE2 region of their promoters. In addition, Nell-1 promotes the expression of Axin2 and active β-catenin and increases the nuclear translocation of active β-catenin.
Discussion
Our past studies have detected all-trans retinoic acid-induced differentiation factor (ATRAID, also known as apoptosis-related protein 3 [APR3]) as an intracellular Nell-1 binding protein in osteosarcoma- and kidney-derived cell lines.(39) Because ATRAID is a protein mainly present on the lysosomal membrane,(40) it is more likely to be involved in endocytosis and/or transportation of Nell-1 rather than extracellular Nell-1 binding and signal transduction. This may also explain our previous observation that ATRAID can bind with multiple truncated Nell-1 constructs with or without the N-terminal LamG domain.(39) In addition, we also reported integrin-β1 binding to the C-terminus of Nell-1.(41,42) Integrin-β1, however, is not Nell-1-specific because integrin-β1 binds promiscuously with a broad range of molecules.(41,43,44)
During the search for a specific cell surface receptor for Nell-1-mediated osteogenesis, we unexpectedly identified a high-binding affinity, ligand/receptor-like interaction between Nell-1 and Cntnap4. Transmembrane receptor Cntnap4 has known roles in connecting pre- and post-synapses critical to synapse development and cortical interneuron function.(21–24) Until present, however, Cntnap4 had no known connection to osteogenesis and, importantly, no known ligand. The unexpected discovery of Cntnap4 as a cell membrane binding protein for Nell-1 in osteogenic-committed cells, and Nell-1 as a ligand of Cntnap4, provides a foundational framework for the broader investigation of the Nell-1/Cntnap4 functional axis. From an osseous standpoint, Nell-1-overexpressing mice exhibit a spectrum of anomalies ranging from craniosynostosis to acrania,(41) whereas N-ethyl-N-nitrosourea (ENU)-induced Nell-1-deficient mice exhibit calvarial defects similar to human cleidocranial dysplasia (CCD) patients.(11) In this study, when we focused on calvarial skeletal development, Cntnap4 deficiency on osteogenesis was evidenced at the onset of Wnt1-Cre expression (Fig. 7). As found in the ENU-induced Nell-1-deficient mice,(11) neonatal Wnt1-CreCntnap4/+ mice have severe underdevelopment of craniofacial bone accompanied with widened sutures, which highly resembles the CCD-like calvarial abnormalities.(42) Considering the incomplete penetrance perinatally of the calvarial disorders found in Nell-1 transgenic mice, without specific focus in the calvarial bone at the neonatal stage, the skeletal abnormalities could possibly be overlooked in the previous studies using a Cntnap4-eGFP knock-in/knockout mouse model for assessing the Cntanp4’s function in synaptic transmission without reporting the grossly apparent calvarial defects.(22) Therefore, to our best knowledge, the current study is the first report that functionally annotates Cntnap4 in a system other than the central nervous system.(26)
Clearly, the further systematical investigation is necessary to elucidate the distribution and functions of Cntnap4 in the entire musculoskeletal system among different development stages. Interestingly, integrin-β1 binds to the C-terminus of Nell-1,(43) while Cntnap4 binds to the N-terminus of Nell-1. From the standpoint of the mechanism of action, this raises the possibility for the potential interplay between integrin-β1- and Cntnap4-mediated Nell-1-responsive signal transduction, particularly Wnt signal transduction.(16) Meanwhile, Nell-1/Cntnap4 binding is predominately detected in the marrow cavity instead of the calcified region of mouse calvarial bones (Fig. 3), suggesting that Nell-1/Cntnap4 may function in a specific stage of osteogenic differentiation. Moreover, our preliminary studies revealed that chondrogenic committed ATDC5 cells exhibited notably lower Cntnap4 expression (Fig. 2A), whereas Cntnap4-KD did not affect Nell-1-stimulated increases of nuclear factor of activated T cells 2 (Nfatc2), a known Nell-1 primary response gene in ATDC5 cells(44) (Supplemental Fig. S4). Thus, the influence of Cntnap4 on Nell-1-responsive signal transduction in chondrogenic-committed cells is distinct from it in osteogenic-committed cells, which suggests that Cntnap4 may mediate Nell-1’s function in a cell type-dependent manner. Considering the remarkable differences between intramembranous ossification that primarily occurs in the skull and endochondral ossification that occurs in other skeletal areas, the involvement of Cntnap4 in osteogenesis may also differ among different types of bones. Therefore, future studies investigating the temporospatial distribution and function of Cntnap4 in musculoskeletal development/regeneration are warranted. Such studies will likely require multiple transgenic mouse models, including but not limited to the currently used Wnt1-Cre-mediated Cntnap4-knockout transgenic mice.
The surprising identification of Nell-1 as a ligand for Cntnap4 will facilitate future studies not only in bone but also potentially in neuronal function. Previous Nell-1 studies have focused primarily on the skeletal system, although Nell-1 was first identified in both the fetal and adult human brain(7) with abundant Nell-1 expression detected in developing and adult nervous systems.(2) Nell-1 and Cntnap4 display largely overlapping spatial expression patterns in the hippocampus, inferior olivary nucleus, and spinal cord,(2,21,45) and our ongoing studies have confirmed colocalization of Nell-1 and Cntnap4 in the human hippocampus (Supplemental Fig. S5) as well as in osteogenic-committed cells. Interestingly, transcriptome analyses implicate both Nell-1 and Cntnap4 in several neurodegenerative and neuropsychiatric disorders such as ASD and Alzheimer’s disease.(46–48) This raises intriguing possibilities of whether a functional Nell-1/Cntnap4 axis also is involved in neural development. Besides osseous anomalies, we have also observed neurologic anomalies in Nell-1 gain- and loss-of-function mice. For example, our preliminary studies revealed that, like Cntnap4-mutant mice,(22) Nell-1-mutant mice also have aberrant behaviors that can be rescued by specific neuropsychiatric pharmaceutical intervention (unpublished data). Although the influence of the central nervous system on bone development and morphogenesis is widely recognized, recent studies suggest that the skeleton also orchestrates the development and cognitive functions of the nervous system.(49–52) It is worth noting that some young CCD patients have delayed development of motor skills such as crawling and walking,(53) which are developmental skill milestones associated with multiple brain areas. Beyond physical development delays, some CCD patients also exhibit cognitive disorders.(54) Determining whether the described cognitive disorders in CCD patients(52) can be replicated in Nell-1/Cntnap4-deficient models is of great interest. Therefore, further investigations are warranted to demonstrate whether the functional involvement of the Nell-1/Cntnap4 axis in neural tissues is as active as it is in bone.
Taken together, the identification and further detailed investigation of this new functional Nell-1/Cntnap4 axis will facilitate understanding of bioactivity of Nell-1, as well as Cntnap4, in bone development and regeneration. Moreover, these studies will offer a critical insight on signaling between the nervous system and skeleton during conditions of health and disease.
Supplementary Material
Acknowledgments
We thank Dr David Myszka from the University of Utah for his assistance in SPR data analysis, Dr Karen Lyons from University of California, Los Angeles, for her insightful scientific input and critical reading of the manuscript, and Richard Song from University of California, Los Angeles, for his critical editing of the manuscript.
This work was supported by the NIAMS (R01 AR066782-01, AR068835-01A1, AR061399-01A1), NIH/National Center for Advancing Translational Science (NCATS) UCLA CTSI Grant (No. UL1TR001881), the National Aeronautics and Space Administration GA-2014-154, the International S&T Cooperation Program of China (No. 2013DFB30360), and the National Natural Science Foundation of China (No. 81200762). CLSM was performed at the Center for NanoScience Institute Advanced Light Microscopy/Spectroscopy Shared Resource Facility at UCLA, which was supported by funding from the NIH-NCRR shared resources (CJX1-443835-WS-29646) and the NSF Major Research Instrumentation (CHE-0722519).
Authors’ roles: CL, ZZ, XZ, KT, and CS designed the study, analyzed data, and wrote the manuscript. CL performed all the experiments and analysis except for the following: ZZ performed the confocal imaging and analysis; ZZ, XC, WJ, and FC performed the expression plasmid construction and protein purification; PH and JKK performed the Cntnap4 knockout mice maintenance and micro CT scanning; ZZ and SS analyzed the SPR data; and SP produced and validated the Nell-1 lentiviral particles. FC, KT, and CS provided the financial support. EAB, GA, and ECC assisted in writing and revising the manuscript.
Footnotes
Disclosures
XZ, KT, and CS are inventors of Nell-1-related patents. They are founders and/or past board members of Bone Biologics Inc./Bone Biologics Corp., which sublicenses Nell-1 patents from the UC Regents, who also hold equity in the company. All other authors state that they have no conflicts of interest.
References
- 1.Zhang X, Zara J, Siu RK, Ting K, Soo C. The role of NELL-1, a growth factor associated with craniosynostosis, in promoting bone regeneration. J Dent Res. 2010;89(9):865–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuroda S, Oyasu M, Kawakami M, et al. Biochemical characterization and expression analysis of neural thrombospondin-1-like proteins NELL1 and NELL2. Biochem Biophys Research Commun. 1999;265(1): 79–86. [DOI] [PubMed] [Google Scholar]
- 3.Ting K, Vastardis H, Mulliken JB, et al. Human NELL-1 expressed in unilateral coronal synostosis. J Bone Miner Res. 1999;14(1):80–9. [DOI] [PubMed] [Google Scholar]
- 4.Campbell ID, Baron M. The structure and function of protein modules. Philos T Roy Soc B. 1991;332(1263):165–70. [DOI] [PubMed] [Google Scholar]
- 5.Abreu JG, Coffinier C, Larrain J, Oelgeschlager M, De Robertis EM. Chordin-like CR domains and the regulation of evolutionarily conserved extracellular signaling systems. Gene. 2002; 287(1–2): 39–47. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Kuroda S, Carpenter D, et al. Craniosynostosis in transgenic mice overexpressing Nell-1. J Clin Invest. 2002;110(6):861–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe TK, Katagiri T, Suzuki M, et al. Cloning and characterization of two novel human cDNAs (NELL1 and NELL2) encoding proteins with six EGF-like repeats. Genomics. 1996;38(3):273–6. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Carpenter D, Bokui N, et al. Overexpression of Nell-1, a craniosynostosis-associated gene, induces apoptosis in osteoblasts duringcraniofacialdevelopment.JBoneMinerRes.2003;18(12):2126–34. [DOI] [PubMed] [Google Scholar]
- 9.Desai J, Shannon ME, Johnson MD, et al. Nell1-deficient mice have reduced expression of extracellular matrix proteins causing cranial and vertebral defects. Hum Mol Genet. 2006;15(8): 1329–41. [DOI] [PubMed] [Google Scholar]
- 10.Truong T, Zhang X, Pathmanathan D, Soo C, Ting K. Craniosynostosis-associated gene nell-1 is regulated by runx2. J Bone Miner Res. 2007;22(1):7–18. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Ting K, Pathmanathan D, et al. Calvarial cleidocraniodysplasia-like defects with ENU-induced Nell-1 deficiency. J Craniofac Surg. 2012;23(1):61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dateki S, Watanabe S, Kinoshita F, Yoshiura K, Moriuchi H. Identification of 11p14.1-p15.3 deletion probably associated with short stature, relative macrocephaly, and delayed closure of the fontanelles. Am J Med Genet A. 2017;173(1):217–20. [DOI] [PubMed] [Google Scholar]
- 13.Aghaloo T, Cowan CM, Chou YF, et al. Nell-1-induced bone regeneration in calvarial defects. Am J Pathol. 2006;169(3):903–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Zara JN, Siu RK, et al. Nell-1 enhances bone regeneration in a rat critical-sized femoral segmental defect model. Plast Reconstr Surg. 2011;127(2):580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siu RK, Lu SS, Li W, et al. Nell-1 protein promotes bone formation in a sheep spinal fusion model. Tissue Eng Part A. 2011; 17(7–8): 1123–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James AW, Shen J, Zhang X, et al. NELL-1 in the treatment of osteoporotic bone loss. Nat Commun. 2015;6:7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.James AW, Pan A, Chiang M, et al. A new function of Nell-1 protein in repressing adipogenic differentiation. Biochem Biophys Res Commun. 2011;411(1):126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue J, Peng J, Yuan M, et al. NELL1 promotes high-quality bone regeneration in rat femoral distraction osteogenesis model. Bone. 2011;48(3):485–95. [DOI] [PubMed] [Google Scholar]
- 19.Shen J, James AW, Zara JN, et al. BMP2-induced inflammation can be suppressed by the osteoinductive growth factor NELL-1. Tissue Eng Part A. 2013; 19(21–22): 2390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen J, James AW, Zhang X, et al. Novel Wnt regulator NEL-like molecule-1 antagonizes adipogenesis and augments osteogenesis induced by bone morphogenetic protein 2. Am J Pathol. 2016;186(2):419–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spiegel I, Salomon D, Erne B, Schaeren-Wiemers N, Peles E. Caspr3 and caspr4, two novel members of the caspr family, are expressed in the nervous system and interact with PDZ domains. Mol Cell Neurosci. 2002;20(2):283–97. [DOI] [PubMed] [Google Scholar]
- 22.Karayannis T, Au E, Patel JC, et al. Cntnap4 differentially contributes to GABAergic and dopaminergic synaptic transmission. Nature. 2014;511(7508):236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin FT, Futagawa T, Li D, et al. Caspr4 interaction with LNX2 modulates the proliferation and neuronal differentiation of mouse neural progenitor cells. Stem Cells Dev. 2015;24(5):640–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon J, Amini S, White MK. General overview of neuronal cell culture. Methods Mol Biol. 2013;1078:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou Y, Zhang WF, Liu HY, et al. Structure and function of the contactin-associated protein family in myelinated axons and their relationship with nerve diseases. Neural Regen Res. 2017;12(9):1551–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cowan CM, Cheng S, Ting K, et al. Nell-1 induced bone formation within the distracted intermaxillary suture. Bone. 2006;38(1):48–58. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Ting K, Bessette CM, et al. Nell-1, a key functional mediator of Runx2, partially rescues calvarial defects in Runx2(+/−) mice. J Bone Miner Res. 2011;26(4):777–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnsdorf EJ, Jones LM, Cater DR, Jacobs CR. Multipotent characteristics of periosteal cells and fibroblasts In: Arnsdorf EJ, editor. Guiding osteogenic lineage commitment: the role of mesenchymal stem cell biology and the mechanical microenvironment. Ann Arbor, MI: ProQuest LLC; 2008. [Google Scholar]
- 30.Yazid MD, Ariffin SHZ, Senafi S, Razak MA, Wahab RMA. Determination of the differentiation capacities of murines’ primary mononucleated cells and MC3T3-E1 cells. Cancer Cell Int. 2010;10:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohseny AB, Machado I, Cai YP, et al. Functional characterization of osteosarcoma cell lines provides representative models to study the human disease. Lab Invest. 2011;91(8):1195–205. [DOI] [PubMed] [Google Scholar]
- 32.Tsai SW, Liou HM, Lin CJ, et al. MG63 osteoblast-like cells exhibit different behavior when grown on electrospun collagen matrix versus electrospun gelatin matrix. PloS One. 2012;7(2): e 31200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson L, Wang SS, Tawfik O, et al. Effect of 25-hydroxyvitamin D3 and 1 alpha,25 dihydroxyvitamin D-3 on differentiation and apoptosis of human osteosarcoma cell lines. J Orthop Res. 2012;30(5):831–44. [DOI] [PubMed] [Google Scholar]
- 34.Taylor SM, Jones PA. Multiple new phenotypes induced in 10t1/2-cells and 3t3-cells treated with 5-azacytidine. Cell. 1979;17(4):771–9. [DOI] [PubMed] [Google Scholar]
- 35.Rohilla M, Bal A, Singh G, Joshil K. Phenotypic and functional characterization of ductal carcinoma in situ-associated myoepithelial cells. Clin Breast Cancer. 2015;15(5):335–42. [DOI] [PubMed] [Google Scholar]
- 36.Yao Y, Wang Y. ATDC5: an excellent in vitro model cell line for skeletal development. J Cell Biochem. 2013;114(6):1223–9. [DOI] [PubMed] [Google Scholar]
- 37.Pattnaik P Surface plasmon resonance: applications in understanding receptor-ligand interaction. Appl Biochem Biotechnol. 2005;126(2):79–92. [DOI] [PubMed] [Google Scholar]
- 38.Bokui N, Otani T, Igarashi K, et al. Involvement of MAPK signaling molecules and Runx2 in the NELL1-induced osteoblastic differentiation. FEBS Lett. 2008;582(2):365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen F, Walder B, James AW, et al. NELL-1-dependent mineralisation of Saos-2 human osteosarcoma cells is mediated via c-Jun N-terminal kinase pathway activation. Int Orthop. 2012;36(10): 2181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding X, Chen Y, Han L, Qiu W, Gu X, Zhang H. Apoptosis related protein 3 is a lysosomal membrane protein. Biochem Biophys Res Commun. 2015;460(4):915–22. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Cowan CM, Jiang X, et al. Nell-1 induces acrania-like cranioskeletal deformities during mouse embryonic development. Lab Invest. 2006;86(7):633–44. [DOI] [PubMed] [Google Scholar]
- 42.Otto F, Thornell AP, Crompton T, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–71. [DOI] [PubMed] [Google Scholar]
- 43.Hasebe A, Nakamura Y, Tashima H, et al. The C-terminal region of NELL1 mediates osteoblastic cell adhesion through integrin alpha3beta1. FEBS Lett. 2012;586(16):2500–6. [DOI] [PubMed] [Google Scholar]
- 44.Chen W, Zhang X, Siu RK, et al. Nfatc2 is a primary response gene of Nell-1 regulating chondrogenesis in ATDC5 cells. J Bone Miner Res. 2011;26(6):1230–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura R, Nakamoto C, Obama H, Durward E, Nakamoto M. Structure-function analysis of Nel, a thrombospondin-1-like glycoprotein involved in neural development and functions. J Biol Chem. 2012;287(5):3282–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Connolly JJ, Glessner JT, Hakonarson H. A genome-wide association study of autism incorporating autism diagnostic interview-revised, autism diagnostic observation schedule, and social responsiveness scale. Child Dev. 2013; 84(1): 17–33. [DOI] [PubMed] [Google Scholar]
- 47.Butler MG, Rafi SK, Manzardo AM. High-resolution chromosome ideogram representation of currently recognized genes for autism spectrum disorders. Int J Mol Sci. 2015;16(3):6464–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Guo XQ, Chu JF, Zhang X, Yan ZR, Li YZ. Potential hippocampal genes and pathways involved in Alzheimer’s disease: a bioinformatic analysis. Genet Mol Res. 2015;14(2):7218–32. [DOI] [PubMed] [Google Scholar]
- 49.Masi L Crosstalk between the brain and bone. Clin Cases Miner Bone Metab. 2012;9(1):13–16. [PMC free article] [PubMed] [Google Scholar]
- 50.Chamouni A, Schreiweis C, Oury F. Bone, brain & beyond. Rev Endocr Metab Dis. 2015;16(2):99–113. [DOI] [PubMed] [Google Scholar]
- 51.Karsenty G, Ferron M. The contribution of bone to whole-organism physiology. Nature. 2012;481(7381):314–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rousseaud A, Moriceau S, Ramos-Brossier M, Oury F. Bone-brain crosstalk and potential associated diseases. Horm Mol Biol Clin I. 2016;28(2):69–83. [DOI] [PubMed] [Google Scholar]
- 53.Hassan NMM, Dhillon A, Huang BY. Cleidocranial dysplasia: clinical overview and genetic considerations. Pediatr Dent J. 2016;26(2):45–50. [Google Scholar]
- 54.Takenouchi T, Sato W, Torii C, Kosaki K. Progressive cognitive decline in an adult patient with cleidocranial dysplasia. Eur J Med Genet. 2014;57(7):319–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.