Abstract
Background
In the recent years, genomic and pan-genomic studies have become increasingly important. Culturomics allows to study human microbiota through the use of different culture conditions, coupled with a method of rapid identification by MALDI-TOF, or 16S rRNA. Bacterial taxonomy is undergoing many changes as a consequence. With the help of pan-genomic analyses, species can be redefined, and new species definitions generated.
Results
Genomics, coupled with culturomics, has led to the discovery of many novel bacterial species or genera, including Akkermansia muciniphila and Microvirga massiliensis. Using the genome to define species has been applied within the genus Klebsiella. A discontinuity or an abrupt break in the core/pan-genome ratio can uncover novel species.
Conclusions
Applying genomic and pan-genomic analyses to the reclassification of other bacterial species or genera will be important in the future of medical microbiology. The pan-genome is one of many new innovative tools in bacterial taxonomy.
Reviewers
This article was reviewed by William Martin, Eric Bapteste and James Mcinerney.
Open peer review
Reviewed by William Martin, Eric Bapteste and James Mcinerney.
Keywords: Genomic, Novel species, Pan-genome, Taxonomy
Background
The study of digestive bacterial ecosystems was initially explored by microbial culture in the 1970s [1–4]. The birth of genomics, followed by the development of Next Generation Sequencing (NGS) methods in 2004, made it possible to discover the uncultivable, such as Akkermansia muciniphila [5] thanks to metagenomics. Since the emergence of metagenomics, microbial culture has gradually been replaced by molecular tools for complex microbiota study [6]. In 2015, a new approach called “culturomics” was developed and intensive culture assays were carried out to select the 18 best culture conditions to cultivate the largest number of isolates [7]. Culturomics has also allowed the identification of a large number of prokaryotic species, as it allows the simultaneous combination of different culture conditions, using 16S rRNA gene amplification and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) [8]. As a result, by increasing the number of species of bacteria being discovered, a new polyphasic method for bacterial species description has emerged; the “taxonogenomics” [9]. In order to describe a new bacterium, taxonogenomics complements classic features with the description of the whole genome sequence with the proteomic information obtained by MALDI-TOF MS and has permitted to describe, for example, Microvirga massiliensis [10].
The current classification of bacterial species relies on a combination of phenotypic and genotypic properties [11–13]. The genotypic criteria used for bacterial taxonomy was the genomic G + C content composition, DNA-DNA hybridization, and, later, the 16S rRNA gene [14, 15]. However, these genotypic criteria were limited since they required the use of restrictive genetic tools. For instance, DNA-DNA hybridization uses a 70% threshold for species discrimination. However, it cannot be used for all prokaryote genera, as described for Rickettsia species [16, 17]. Furthermore, the comparison of the single gene 16S rRNA [18–20], as well as the low conventional divergence between two 16S rRNA genes [21] of two organisms, causes a slight and limited bacterial description [22]. Indeed, the experiments of Acinas et al. involving 76 whole genomes shows an extreme diversity (11.6%) of the identity of 16S rRNA genes in the bacteria Thermoanaerobacter tengcongensis [23]. Except for Thermoanaerobacter tengcongensis, 16S rRNA gene is generally more conserved, and therefore not universally reliable for determining taxonomic relationships at the species level. Moreover, the variation of nucleotides observed in several copies of rRNA genes in single organisms, as well as the possibility that 16S rRNA genes are derived from horizontal gene transfer, can lead to well-established relationships between taxa, into phylogenetic trees [18]. Nevertheless, 16S rRNA gene is currently the gold standard tool for prokaryotic taxonomy [13]. With the emergence of first-generation sequencing in 1975–77 [24, 25], followed by high-throughput sequencing in 2004 [26], access to complete genetic information was deeply revolutionized. Thanks to these modern high-throughput sequencing technologies, a considerable amount of data is generated, enabling studies based on pan-genomic analyses (Fig. 1). The first definition of the pan-genome was proposed by Tettelin et al. [27] in 2005, just after the beginning of the era of high-throughput sequencing. A pan-genome can be defined as being the entire gene content belonging to a study group [28–30]. The applications are multiple, including the study of pathogenicity [31, 32], the mobilome [33], resistome [34], prediction of the lifestyle of bacteria [35], and also for taxonomy. Indeed, pan-genome study allows a reclassification of the species [36], thus clarifying and improving the traditional criteria previously presented.
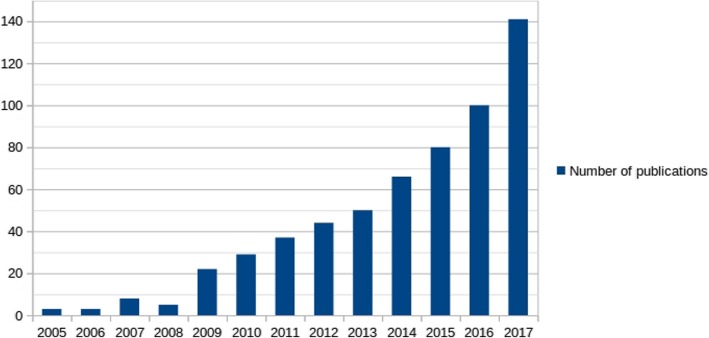
Fig. 1.
Number of publications per year for all pan-genome studies in the genomic database on the PubMed website (https://www.ncbi.nlm.nih.gov/pubmed)
Mapping strategy
NGS application in genomics and metagenomics
This technical revolution offers new fields of application, such as genomics (whole-genome sequencing, WGS), single-cell sequencing (SCS) and metagenomics. In particular, complete genome sequencing allows a better understanding of the genetic basis of phenotypic variability, but also to analyze biodiversity and estimate the diversity of single nucleotide polymorphism (SNP) [37]. Single-cell genomics allows the sequencing of a single isolated cell by capturing and amplifying its DNA. Single-cell sequencing is able to produce partial [38] and complete [39] genomes. SCS DNA allows the identification and assembly of genomes from uncultivated microorganisms [40–43] but it is estimated that only 1% of bacterial species have been cultivated in laboratory [44]. Nevertheless, SCS has its own limits along with inherent biases [40, 43], especially during the amplification step, including false-positive and false-negative errors, allelic dropout events and coverage non-uniformity [45]. However, single-cell genomics and metagenomics are two complementary approaches to analyze bacterial communities [46]. The sequencing of metagenomes has made it possible to inventory the diversity of microbial ecosystems [47] and to determine the interactions between microorganisms in ecosystems [48]. This technique, which allows DNA sequencing of bacteria present in specific environments, offers many advantages. Indeed, it opens a window on a world of great wealth that was hitherto totally unknown.
Assembly, finishing and annotations
Organism sequencing has recently become accessible in many laboratories where specific consortium sequencing projects are proposed, and currently the number of projects is exponentially growing. However, NGS technologies have higher error rates (~ 0.1–15%) and smaller read lengths (35–1000 bp) than those obtained from Sanger sequencing platforms [49]. After sequencing, the data produced (reads) are computationally reconstructed into longer continuous sequences (contigs), a step called assembly [50]. This process consists of an overlap of reads aimed at reconstructing the initial sequence of the genome. It’s called de novo when no reference is available. The assembly with Sanger data is based on two-by-two comparison of reads, looking for overlapping sequences of minimal length with associated identity percentages (CAP assembler, for example [51]). The change of scale due to the huge volume of data, the short-read lengths and the non-uniform confidence in base calling, excluded this assembly strategy. The most commonly used approach for the assembly of short-read data is based on treating k-mers. All k-mers length substrings of all sequencing reads appear as nodes in a graph. The nodes are joined by edges if the nodes share a k-1 length substring. This graph is known as the de Bruijn graph [52]. Short read de Bruijn graph assemblers determine optimal paths through this graph to form the contigs of an assembly. Each contig ends when there is no outgoing edge from a visited node, or when the branching in the graph becomes too complex to be resolved. Frequently, the raw de Bruijn graph is reduced by collapsing any linear chains of nodes and edges into a single metanode. An assembly quality can then be assessed using a set of metrics. The usual ones are the total count of contigs, scaffolds, their total length, N50 (length of the smallest contig in the set of contigs that represent at least 50% of the genome), and their average length [53, 54]. Bilen et al., removed scaffolds less than 800 bp in size or less than 25% of the median depth (identified as possible contaminants) [55]. A good metric is also the proportion of reads mapped back, or not, to the contigs [53]. The assembly quality is an important criterion for ulterior analysis such as rate of lateral transfer or annotation [56]. The next important step required is genome annotation. To make genomic data valuable, a reliable and correct annotation is essential [57]. It is used to identify, locate and distinguish gene function using similarities when studying protein databases by BLAST [58], and can provide a basis for many genome analyses [59]. Organisms whose genome is now fully sequenced have revealed that nearly 40% of the genes identified have no assigned function; either because they do not resemble any known genes, or because (for half of them) they resemble other genes with unknown function [60]. The first step in annotation is to predict gene function, which is usually done individually for each gene using computational tools. However, the identification of gene function requires the combination of several complementary experimental approaches, whether by computer (in silico analysis), biochemical, or genetic (in vivo and in vitro analysis). The second step in annotation is to identify relationships between genes, proteins and regulatory elements. These relationships can be of very varied nature: physical interactions between proteins/DNA, proteins/RNA and proteins/proteins, networks regulating gene expression, metabolic pathways or others.
Example of a mapping study: Akkermansia muciniphila
Powerful approaches based on mapping short-read sequences to a reference genome are used to analyze WGS data from closely related isolates [5, 61–64]. Several bioinformatics tools are used in a mapping study, such as the CLC genomics Workbench (CLC bio, Aarhus, Denmark).
By using metagenomics data from a human stool sample, the genome of Akkermansia muciniphila was successfully assembled after mapping the short-read sequences [5]. The stool sample was from a patient admitted to the intensive care unit and treated with a 10-day course of imipenem [65]. High-level colonization by the Verrucomicrobia phylum was reported in this patient. Indeed, the reads from pyrosequencing are classified in the phylum range and up to 84% of these reads belong to Verrucomicrobia phyla. Reads were generated from a SOLiD sequencer, whereas short-reads shotgun and paired-end runs were generated on a 454 sequencer. Both technologies generated 1.4 G bases of metagenomic sequence data from the sample. The several mapping sequences from SOLiD and 454 data against the A. muciniphila type strain ATCC BAA-835 allowed to obtain the genome of A. muciniphila strain Urmite with 1 scaffold and 58 contigs.
The presence of a range of putative antibiotic resistance genes (ARGs) from different antibiotic classes has been demonstrated in a recent study of A. muciniphila strain Urmite [5]. The putative ARG were beta-lactamase, macrolides, vancomycin, chloramphenicol, sulfonamide, tetracycline and trimethoprim.
The short-read sequence mapping approach of a reference genome has been successful in assembling a genome directly from a human stool sample. However, this approach remains limited since very divergent sequences, not present in the reference, could not be detected [64]. Indeed, if the draft genome obtain contains highly divergent additional genes with respect to the reference genome, we may lose this information and not be able to completely reconstruct the original genetic content. Consequently, mapping method cannot be used on very divergent genomes [5]. These limitations will be progressively lessened by the exponential growth of data and subsequent genomes available in generalist databases.
Finding novel species
Novel species identification
- Culturomics (MALDI-TOF-MS and 16S rRNA)
In recent years, with the introduction of a rapid and inexpensive identification method using MALDI-TOF mass spectrometry, the volume of microbial culture has considerably increased, which now allows to more readily detect pathogenic bacterial species. This technique of reference for bacterial identification in clinical microbiology laboratories has also developed a new concept to study human microbiota also called “microbial culturomics” [7, 66]. Culturomics was developed in a study of a complex microbiota and allows a large number of isolates to be grown through the selection of the 18 best culture conditions [7, 9]. This technique is based on the diversification of culture conditions by varying the time and temperature of incubation but also culture medium composition and atmosphere [66]. In a preliminary work, this approach allowed the cultivation of 340 bacterial species, including 31 novel species, as well as species belonging to rare phyla (Synergistetes and Deinococcus-Thermus), using 212 different cultivation conditions [66]. A detailed analysis made it possible to select successively the 70 then the 18 most appropriate culture conditions in order to explore the greatest possible diversity for each sample. Another work has permitted to cultivated more than 50% of the known species of the human digestive tract, including 247 novel species [7, 8]. Novel species are identified by MALDI-TOF, and 16S rRNA gene sequencing for non-identified spectra in MALDI-TOF.
- other methods
Novel species were also identified with multilocus sequence analysis (MLSA) of several concatenated housekeeping gene sequences (rrs, recA, gyrB, dnaK, glnII and rpoD) [67, 68]. The housekeeping genes are usually involved in the expression and maintenance of genetic information at the transcription or translation level. The recA gene is essential for the maintenance and repair of DNA and is a good resolutive tool for predicting lineage and genus among rhizobial strains [69]. Phylogenetic analysis to discover new species also uses DNA sequences of the internal transcribed spacer 2 (ITS2) region [70].
The nucleotide sequence or the peptide sequence can also be used. In general, the peptide sequence is preferred, since it can avoid some biases inherent to the G + C content of the organism studied, as well as the degeneration on the third nucleotide of the codon [71].
Taxonogenomics strategy and the example of a novel species study: Microvirga massiliensis
This new polyphasic strategy, called “taxonogenomics”, systematically combines phenotypic and genomic criteria [9, 72]. Using this strategy, 15 novel bacteria have officially been considered as new species and/or new genera in official validation lists No. 153 and No. 155 by the International Taxonomy Committee of the International Journal of Systematic and Evolutionary Microbiology [66, 73–83]. Lagier et al., identified a novel bacterial species, Microvirga massiliensis, from a human stool sample using culturomics and metagenomics approaches [66]. Recently, the description of the genome of Microvirga massiliensis sp. nov. strain JC119T was realised via the taxonogenomics approach [10]. The draft genome of this bacterium is 9,207,211 bp long, which is the largest bacterial genome of a human isolate. Of the 8762 predicted genes, 8685 were protein-coding genes, 77 were rRNA genes, including 21 rRNA genes, and the genome exhibits a G + C content of 63.28%.
The bacterial pan-genome
History of taxonomy and the concept of bacterial species
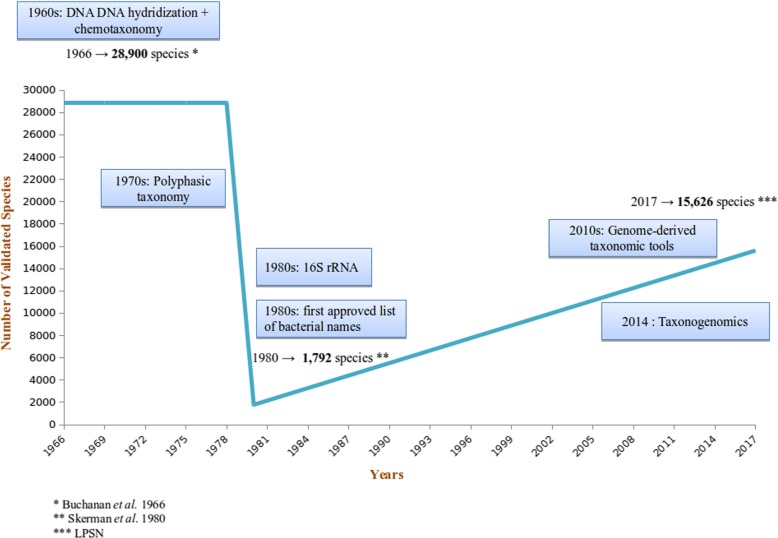
The number of bacterial species has constantly changed according to the classification criteria used. Indeed, in 1966, Buchanan et al. [84] published a census of 28,900 bacterial species, manifesting an urgent need for a better classification. The situation was improved in 1980 by Skerman, McGowan and Sneath [85], who, thanks to long and tedious efforts, managed to reduce the number of valid bacterial species to 1792. As an example, the taxonomy of Salmonella species was thoroughly modified, with the creation of subspecies and serovars instead of sub-genera and species, respectively, and the distribution of strains into two distinct species: Salmonella enterica and Salmonella bongori, on the basis of DNA-DNA hybridization [86, 87]. To date (May, 2017), this number has risen up to 15,626 species (listed in List of Prokaryotic names with Standing in Nomenclature (LPSN), www.bacterio.net) (Fig. 2).
Fig. 2.
Validated number of bacterial names over the years. The blue box indicates the major advances in molecular biology
Since the early 1990s, because of advances in molecular biology, bacterial classification has been in perpetual upheaval. Taxonomic information is essential regarding the identification, nomenclature and classification of microbial strains [88] and to better understanding the biodiversity and relationships among living microorganisms [89].
Currently, prokaryote taxonomy relies on polyphasic combinations of phenotypic properties (pathogenesis, morphology, environmental and culture conditions), chemotaxonomic properties (chemical composition of cellular components) and genotypic properties [11, 90], including DNA-DNA hybridization (DDH), DNA G + C content [91] and 16S rRNA sequence similarity [12, 92] The application of molecular hybridization methods provides a genomic definition of the bacterial species, taking into account the similarity rate and the thermal stability of the hybrids obtained by the DNAs of two bacterial isolates. Isolates belonging to the same species are characterized by homologies of their DNA, which result in hybridization percentages greater than or equal to 70%, and stability of the hybrids formed below 5 °C [93–95]. This definition is still recognized by the international bacterial taxonomy committees.
The development of DNA sequencing has led to the determination of a threshold in order to define the species based on the similarity of gene sequences, initially based on the 16S rRNA gene. These data were later compared to those obtained by DDH [96, 97]. As for the classification of prokaryotes, the 16S rRNA gene is an effective molecular marker due to its functional stability, conservation and universal presence [98]. However, for bacterial taxonomy, this gene has several limitations; notably, the presence of SNPs in the rRNA operon in a single genome [23, 99]; the use of a single gene that may not reflect the evolution history of the genome (~ 0.07% of a genome) [19, 100] or the high degree of conservation in a same genus, like Brucella or Rickettsia [101]. The divergence of 1.3% accepted from two sequences, corresponding to 50 million years of divergence [17, 22] also brings a limitation such that the presence in multiple of the 16 s rRNA genes and sometimes variable copies [102, 103]. VanBerkum et al., showed that a small portion of the 16S rRNA gene sequence of Bradyrhizobium elkanii is originated from a Mesorhizobium spp. genome by lateral transfer [100, 104].
What happens when two species have a 98.6% similarity percentage? Are they two distinct species? Establishing a threshold is not biological and cannot be based essentially on this criterion, especially when different thresholds are used by different biologists.
With the advent of whole genomes sequencing, phylogeny has entered a new era: the era of phylogenomics. Many studies have already demonstrated the importance of genomes in bacterial taxonomy by suggesting a focus on the presence or absence of genes within genomes [105–107]; the gene content [108]; the presence of SNPs or indels in conserved genes [109]; the comparison of orthologous genes [110]; the study of metabolic pathways and chromosome gene order [111, 112]; or by sequence similarity at the genome level, estimated by parameters such as “digital DDH”, Average Nucleotide Identity (ANI) or AGIOS using the Genome-To-Genome Distance Calculator (GGDC), ANI calculator and in-lab pipeline named Marseille Average Genomic identity (MAGi) softwares [113–116].
Pan-genome study for taxonomic purposes: The Klebsiella genus
The taxonomic classification of Klebsiella species has been the subject of a long controversy. Klebsiella species are part of the large Enterobacteriaceae family, which are Gram-negative bacteria. Originally, the Klebsiella genus was divided into pathovars linked to the diseases they caused: Klebsiella pneumoniae, Klebsiella ozaenae and Klebsiella rhinoscleromatis [117]. With the development of new tools such as G + C content composition, DNA-DNA hybridization and 16S rRNA sequencing [11, 95], classification of Klebsiella species has been continuously revised [118, 119]. K. ozaenae and K. rhinoscleromatis were notably reclassified as K. pneumoniae subspecies [70, 120, 121].
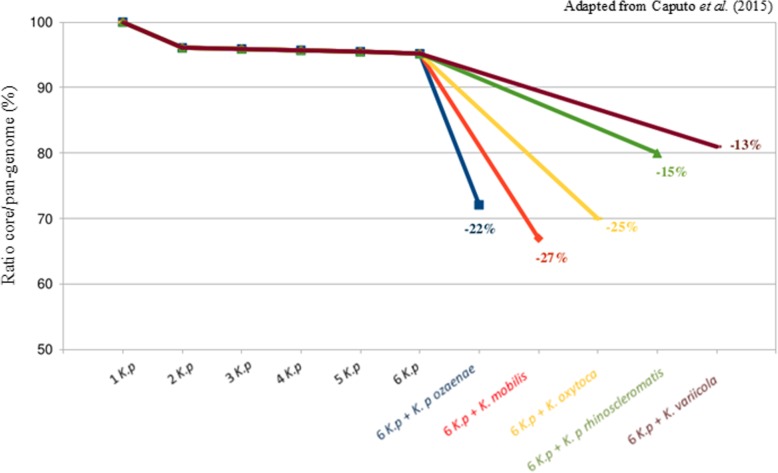
PNan-genome analyses were performed for different strains and subspecies of K. pneumoniae, K. oxytoca, K. variicola and K. mobilis [36]. We determined that the core/pan-genome ratio for six K. pneumoniae subsp. pneumoniae strains was 94%. Then, we determined this ratio by comparing successively K. pneumoniae subsp. pneumoniae to K. mobilis, K. variicola, K. oxytoca, K. pneumoniae subsp. ozaenae or K. pneumoniae subsp. rhinoscleromatis genomes (Fig. 3). The ratios obtained were 67, 81, 69, 72 and 79% respectively. Therefore, we observed a discontinuity variation in the ratio for each of these species/subspecies, with a difference ranging from 13 to 27% with the bona fide Klebsiella pneumoniae species. We estimate that this ratio break is greater than 10% with no transition zone and reflects individual biological species. Accordingly, the authors said that K. pneumoniae subsp. ozaenae or K. pneumoniae subsp. rhinoscleromatis can be considered as species. A recent study has demonstrated the same break in the core/pan-genome ratio of E. coli strains, after the additions of Shigella strains [122]. The COG analysis and the KEGG analysis showed large differences which once again highlighted the very distinct genomic content of K. pneumoniae subsp. ozaenae or K. pneumoniae subsp. rhinoscleromatis.
Fig. 3.
The core/pan-genome ratio for all strains of Klebsiella studied
This pan-genomic analysis enabled to conlude that K. pneumoniae subsp. ozaenae or K. pneumoniae subsp. rhinoscleromatis, which exhibit as many differences between them as with other Klebsiella species, may be distinct Klebsiella species [36] or, following Ereshefsky’s eliminative pluralism on species philosophy [123], distinct pan-genome-derived species.
Conclusions
Since the introduction of DNA sequencing by Sanger and Coulson in 1977, great progress has been made. A growing amount of data is being generated, requiring continuously advanced computer processing. Numerous studies illustrating different methods have been published in different fields, such as genome assembly and annotation, as well as research on new bacterial species and the taxonomic classification of bacteria.
Regarding genome analysis, a complete genome of Akkermansia muciniphila was obtained directly from human stool samples by an original approach. Based on taxonogenomics, the creation of Microvirga massiliensis sp. nov. containing the strain JC119T could be done.
Taxonomy traditionally operates with the principle of discontinuous variation. A break in the ratio of the core/pan-genome means that there is no transition from one species to another, leading to a definition of different species. This leap in the ratio represents a major difference between genomes. These irreconcilable differences cannot exist within a single species. The great discontinuity variation in the core/pan-genome ratio observed in Klebsiella species may help to redefine these species. Thus, we believe that pan-genome studies can help to better visualize gene content differences, inform species (re)definitions or classify species on their own according to discontinuous genomic content. However, this strategy will have to be empirically and systematically tested in the currently proposed genera.
Genomics challenges taxonomy. We are at the very beginning of the interpretation of the genome for taxonomic purposes.
Acknowledgments
This study was supported by Mediterranée Infection and the National Research Agency under the program “Investissements d’avenir”, reference ANR-10-IAHU-03.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Abbreviations
- ANI
Average nucleotide identity
- ARG
Antibiotic resistance gene
- BSR
Blast score ratio
- COG
Cluster of orthologous group
- DDH
DNA-DNA hybridization
- GGDC
Genome-to-genome distance calculator
- GOLD
Genomes online database
- ITS
Internal transcribed spacer
- KEGG
Kyoto encyclopedia of genes and genomes
- LPSN
List of prokaryotic names with standing in nomenclature
- MALDI-TOF
Matrix-assisted laser desorption/ionization time-of-flight
- MLSA
Multilocus sequence analysis
- NGS
Next-generation sequencing
- SCS
Single-cell sequencing
- SNP
Single nucleotide polymorphism
- WGS
Whole-genome sequencing
Authors’ contributions
DR designed the research project. AC performed analysis and wrote the manuscript. PEF provided support. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Aurélia Caputo, Email: aurelia.caputo@univ-amu.fr.
Pierre-Edouard Fournier, Email: pierre-edouard.fournier@univ-amu.fr.
Didier Raoult, Phone: +33 4 13 73 20 01, Email: didier.raoult@gmail.com.
References
- 1.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 2.Finegold SM, Attebery HR, Sutter VL. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am J Clin Nutr. 1974;27:1456–1469. doi: 10.1093/ajcn/27.12.1456. [DOI] [PubMed] [Google Scholar]
- 3.Moore WE, Holdeman LV. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol. 1974;27:961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagier J-C, Edouard S, Pagnier I, Mediannikov O, Drancourt M, Raoult D. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev. 2015;28:208-36. [DOI] [PMC free article] [PubMed]
- 5.Caputo A, Dubourg G, Croce O, Gupta S, Robert C, Papazian L, et al. Whole-genome assembly of Akkermansia muciniphila sequenced directly from human stool. Biol Direct. 2015;10:5. doi: 10.1186/s13062-015-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagier J-C, Million M, Hugon P, Armougom F, Raoult D. Human gut microbiota: repertoire and variations. Front Cell Infect Microbiol. 2012;2:136. doi: 10.3389/fcimb.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagier J-C, Hugon P, Khelaifia S, Fournier P-E, La Scola B, Raoult D. The rebirth of culture in microbiology through the example of Culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237-64. [DOI] [PMC free article] [PubMed]
- 8.Lagier J-C, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Fournier P-E, Lagier J-C, Dubourg G, Raoult D. From culturomics to taxonomogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. 2015;36:73–78. doi: 10.1016/j.anaerobe.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Caputo A, Lagier J-C, Azza S, Robert C, Mouelhi D, Fournier P-E, et al. Microvirga massiliensis sp. nov., the human commensal with the largest genome. MicrobiologyOpen. 2016;5:307–322. doi: 10.1002/mbo3.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staley JT. The bacterial species dilemma and the genomic-phylogenetic species concept. Philos Trans R Soc B Biol Sci. 2006;361:1899–1909. doi: 10.1098/rstb.2006.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tindall BJ, Rosselló-Móra R, Busse H-J, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010;60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 13.Kämpfer P, Glaeser SP. Prokaryotic taxonomy in the sequencing era--the polyphasic approach revisited. Environ Microbiol. 2012;14:291–317. doi: 10.1111/j.1462-2920.2011.02615.x. [DOI] [PubMed] [Google Scholar]
- 14.Rosselló-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 15.Drancourt M, Berger P, Raoult D. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J Clin Microbiol. 2004;42:2197–2202. doi: 10.1128/JCM.42.5.2197-2202.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drancourt M, Raoult D. Taxonomic position of the Rickettsiae: current knowledge. FEMS Microbiol Rev. 1994;13:13–24. doi: 10.1111/j.1574-6976.1994.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 17.Ogata H, Audic S, Renesto-Audiffren P, Fournier PE, Barbe V, Samson D, et al. Mechanisms of evolution in rickettsia conorii and R. Prowazekii. Science. 2001;293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- 18.Sentausa E, Fournier P-E. Advantages and limitations of genomics in prokaryotic taxonomy. Clin Microbiol Infect. 2013;19:790–795. doi: 10.1111/1469-0691.12181. [DOI] [PubMed] [Google Scholar]
- 19.O’Malley MA, Koonin EV. How stands the tree of life a century and a half after the origin? Biol Direct. 2011;6:32. doi: 10.1186/1745-6150-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dagan T, Roettger M, Stucken K, Landan G, Koch R, Major P, et al. Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol Evol. 2013;5:31–44. doi: 10.1093/gbe/evs117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pei AY, Oberdorf WE, Nossa CW, Agarwal A, Chokshi P, Gerz EA, et al. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl Environ Microbiol. 2010;76:3886–3897. doi: 10.1128/AEM.02953-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochman H, Elwyn S, Moran NA. Calibrating bacterial evolution. Proc Natl Acad Sci U S A. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol. 2004;186:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambardar S, Gupta R, Trakroo D, Lal R, Vakhlu J. High throughput sequencing: an overview of sequencing chemistry. Indian J Microbiol. 2016;56:394–404. doi: 10.1007/s12088-016-0606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome.”. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Mira A, Martín-Cuadrado AB, D’Auria G, Rodríguez-Valera F. The bacterial pan-genome:a new paradigm in microbiology. Int Microbiol Off J Span Soc Microbiol. 2010;13:45–57. doi: 10.2436/20.1501.01.110. [DOI] [PubMed] [Google Scholar]
- 31.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, et al. Origins of the E. Coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A. 2009;106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.den Bakker HC, Cummings CA, Ferreira V, Vatta P, Orsi RH, Degoricija L, et al. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics. 2010;11:688. doi: 10.1186/1471-2164-11-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olivares J, Bernardini A, Garcia-Leon G, Corona F, B Sanchez M, Martinez JL. The intrinsic resistome of bacterial pathogens. Front Microbiol. 2013;4:103. doi: 10.3389/fmicb.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diene SM, Merhej V, Henry M, Filali AE, Roux V, Robert C, et al. The Rhizome of the Multidrug-Resistant Enterobacter aerogenes Genome Reveals How New “Killer Bugs” Are Created because of a Sympatric Lifestyle. Mol Biol Evol. 2012;30:mss236. [DOI] [PubMed]
- 36.Caputo A, Merhej V, Georgiades K, Fournier P-E, Croce O, Robert C, et al. Pan-genomic analysis to redefine species and subspecies based on quantum discontinuous variation: the Klebsiella paradigm. Biol Direct. 2015;10:55. doi: 10.1186/s13062-015-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camiolo S, Sablok G, Porceddu A. Altools: a user friendly NGS data analyser. Biol Direct. 2016;11:8. doi: 10.1186/s13062-016-0110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, et al. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 39.Woyke T, Tighe D, Mavromatis K, Clum A, Copeland A, Schackwitz W, et al. One bacterial cell, one complete genome. PLoS One. 2010;5:e10314. doi: 10.1371/journal.pone.0010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 41.Marcy Y, Ouverney C, Bik EM, Lösekann T, Ivanova N, Martin HG, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLean JS, Lombardo M-J, Badger JH, Edlund A, Novotny M, Yee-Greenbaum J, et al. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci. 2013;110:E2390–E2399. doi: 10.1073/pnas.1219809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lasken RS. Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol. 2012;10:631–640. doi: 10.1038/nrmicro2857. [DOI] [PubMed] [Google Scholar]
- 44.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Navin NE. Advances and applications of single cell sequencing technologies. Mol Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doud DFR, Woyke T. Novel approaches in function-driven single-cell genomics. FEMS Microbiol Rev. 2017;41:538–548. doi: 10.1093/femsre/fux009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nasheri N, Petronella N, Ronholm J, Bidawid S, Corneau N. Characterization of the genomic diversity of norovirus in linked patients using a metagenomic deep sequencing approach. Front Microbiol. 2017;8:73. doi: 10.3389/fmicb.2017.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng FC-H, Shaw GT-W, Weng C-Y, Yang Y-J, Wang D. Inferring microbial interactions in the gut of the Hong Kong whipping frog (Polypedates megacephalus) and a validation using probiotics. Front Microbiol. 2017;8:525. doi: 10.3389/fmicb.2017.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ekblom R, Wolf JBW. A field guide to whole-genome sequencing, assembly and annotation. Evol Appl. 2014;7:1026–1042. doi: 10.1111/eva.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Idury RM, Waterman MS. A new algorithm for DNA sequence assembly. J Comput Biol J Comput Mol Cell Biol. 1995;2:291–306. doi: 10.1089/cmb.1995.2.291. [DOI] [PubMed] [Google Scholar]
- 53.Cabau C, Escudié F, Djari A, Guiguen Y, Bobe J, Klopp C. Compacting and correcting trinity and oases RNA-Seq de novo assemblies. PeerJ. 2017;5:e2988. doi: 10.7717/peerj.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao MD, Nguyen SH, Ganesamoorthy D, Elliott AG, Cooper MA, Coin LJM. Scaffolding and completing genome assemblies in real-time with nanopore sequencing. Nat Commun. 2017;8:14515. doi: 10.1038/ncomms14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bilen M, Beye M, Fonkou MDM, Khelaifia S, Cadoret F, Armstrong N, et al. Genomic and phenotypic description of the newly isolated human species Collinsella bouchesdurhonensis sp. nov. MicrobiologyOpen. 2018;7(5):e00580. doi: 10.1002/mbo3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu D, Hunt M, Tsai IJ. Inferring synteny between genome assemblies: a systematic evaluation. BMC Bioinformatics. 2018;19:26. doi: 10.1186/s12859-018-2026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett. 2016;363. [DOI] [PubMed]
- 58.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 59.Stein L. Genome annotation: from sequence to biology. Nat Rev Genet. 2001;2:493–503. doi: 10.1038/35080529. [DOI] [PubMed] [Google Scholar]
- 60.Médigue C, Bocs S, Labarre L, Mathé C, Vallenet D. L’annotation in silico des séquences génomiques. médecine/sciences. 2002;18:237–250. [Google Scholar]
- 61.Hernandez D, François P, Farinelli L, Osterås M, Schrenzel J. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishito Y, Osana Y, Hachiya T, Popendorf K, Toyoda A, Fujiyama A, et al. Whole genome assembly of a natto production strain Bacillus subtilis natto from very short read data. BMC Genomics. 2010;11:243. doi: 10.1186/1471-2164-11-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bratcher HB, Corton C, Jolley KA, Parkhill J, Maiden MC. A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics. 2014;15:1138. doi: 10.1186/1471-2164-15-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dubourg G, Lagier J-C, Armougom F, Robert C, Audoly G, Papazian L, et al. High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment. Int J Antimicrob Agents. 2013;41:149–155. doi: 10.1016/j.ijantimicag.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 66.Lagier J-C, Armougom F, Million M, Hugon P, Pagnier I, Robert C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 67.Msaddak A, Rejili M, Durán D, Rey L, Imperial J, Palacios JM, et al. Members of microvirga and Bradyrhizobium genera are native endosymbiotic bacteria nodulating Lupinus luteus in northern Tunisian soils. FEMS Microbiol Ecol. 2017;93. [DOI] [PubMed]
- 68.Busquets A, Gomila M, Beiki F, Mulet M, Rahimian H, García-Valdés E, et al. Pseudomonas caspiana sp. nov., a citrus pathogen in the Pseudomonas syringae phylogenetic group. Syst Appl Microbiol. [cited 2017 Jun 13]; Available from: http://www.sciencedirect.com/science/article/pii/S0723202017300450 [DOI] [PubMed]
- 69.Vinuesa P, León-Barrios M, Silva C, Willems A, Jarabo-Lorenzo A, Pérez-Galdona R, et al. Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates endemic genistoid legumes (Papilionoideae: Genisteae) from the Canary Islands, along with Bradyrhizobium japonicum bv. Genistearum, Bradyrhizobium genospecies alpha and Bradyrhizobium genospecies beta. Int J Syst Evol Microbiol. 2005;55:569–575. doi: 10.1099/ijs.0.63292-0. [DOI] [PubMed] [Google Scholar]
- 70.Wang M, Cao B, Yu Q, Liu L, Gao Q, Wang L, et al. Analysis of the 16S–23S rRNA gene internal transcribed spacer region in Klebsiella species. J Clin Microbiol. 2008;46:3555–3563. doi: 10.1128/JCM.00927-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gupta RS. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev MMBR. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramasamy D, Mishra AK, Lagier J-C, Padhmanabhan R, Rossi M, Sentausa E, et al. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 73.Ramasamy D, Kokcha S, Lagier J-C, Nguyen T-T, Raoult D, Fournier P-E. Genome sequence and description of Aeromicrobium massiliense sp. nov. Stand Genomic Sci. 2012;7:246–257. doi: 10.4056/sigs.3306717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roux V, Lagier J-C, Gorlas A, Robert C, Raoult D. Non-contiguous finished genome sequence and description of Kurthia senegalensis sp. nov. Stand Genomic Sci. 2014;9:1319–1330. doi: 10.4056/sigs.5078947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hugon P, Mishra AK, Lagier J-C, Nguyen TT, Couderc C, Raoult D, et al. Non-contiguous finished genome sequence and description of Brevibacillus massiliensis sp. nov. Stand Genomic Sci. 2013;8:1–14. doi: 10.4056/sigs.3466975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kokcha S, Ramasamy D, Lagier J-C, Robert C, Raoult D, Fournier P-E. Non-contiguous finished genome sequence and description of Brevibacterium senegalense sp. nov. Stand Genomic Sci. 2012;7:233–245. doi: 10.4056/sigs.3256677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oren A, Garrity GM. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol. 2014;66:1–5. [DOI] [PubMed]
- 78.Lagier J-C, Armougom F, Mishra AK, Nguyen T-T, Raoult D, Fournier P-E. Non-contiguous finished genome sequence and description of Alistipes timonensis sp. nov. Stand Genomic Sci. 2012;6:315–324. doi: 10.4056/sigs.2685971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lagier J-C, El Karkouri K, Nguyen T-T, Armougom F, Raoult D, Fournier P-E. Non-contiguous finished genome sequence and description of Anaerococcus senegalensis sp. nov. Stand Genomic Sci. 2012;6:116–125. doi: 10.4056/sigs.2415480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lagier J-C, Gimenez G, Robert C, Raoult D, Fournier P-E. Non-contiguous finished genome sequence and description of Herbaspirillum massiliense sp. nov. Stand Genomic Sci. 2012;7:200–209. doi: 10.4056/sigs.3086474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lagier J-C, El Karkouri K, Mishra AK, Robert C, Raoult D, Fournier P-E. Non contiguous-finished genome sequence and description of Enterobacter massiliensis sp. nov. Stand Genomic Sci. 2013;7:399–412. doi: 10.4056/sigs.3396830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lagier J-C, Elkarkouri K, Rivet R, Couderc C, Raoult D, Fournier P-E. Non contiguous-finished genome sequence and description of Senegalemassilia anaerobia gen. Nov., sp. nov. Stand Genomic Sci. 2013;7:343–356. doi: 10.4056/sigs.3246665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pei J, Chu C, Li X, Lu B, Wu Y. CLADES: A classification-based machine learning method for species delimitation from population genetic data. Mol Ecol Resour. 2018;18(5):1144–1156. doi: 10.1111/1755-0998.12887. [DOI] [PubMed] [Google Scholar]
- 84.Paolozzi L, Liébart J-C, Sansonetti P. Microbiologie: biologie des procaryotes et de leurs virus. Paris: Dunod; 2015. [Google Scholar]
- 85.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Evol Microbiol. 1980;30:225–420. [Google Scholar]
- 86.Reeves MW, Evins GM, Heiba AA, Plikaytis BD, Farmer JJ. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989;27:313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le Minor L, Popoff MY. Designation of Salmonella enterica sp. nov., nom. Rev., as the type and only species of the Genus Salmonella: request for an opinion. Int J Syst Evol Microbiol. 1987;37:465–468. [Google Scholar]
- 88.Moore ERB, Mihaylova SA, Vandamme P, Krichevsky MI, Dijkshoorn L. Microbial systematics and taxonomy: relevance for a microbial commons. Res Microbiol. 2010;161:430–438. doi: 10.1016/j.resmic.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 89.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, et al. Opinion: re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 90.Hugon P, Dufour J-C, Colson P, Fournier P-E, Sallah K, Raoult D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect Dis. 2015;15:1211–1219. doi: 10.1016/S1473-3099(15)00293-5. [DOI] [PubMed] [Google Scholar]
- 91.Zakhia F, de Lajudie P. Modern bacterial taxonomy: techniques review--application to bacteria that nodulate leguminous plants (BNL) Can J Microbiol. 2006;52:169–181. doi: 10.1139/w05-092. [DOI] [PubMed] [Google Scholar]
- 92.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 93.Krawiec S. Concept of a bacterial species. Int J Syst Bacteriol. 1985;35:217–220. [Google Scholar]
- 94.Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 96.Keswani J, Whitman WB. Relationship of 16S rRNA sequence similarity to DNA hybridization in prokaryotes. Int J Syst Evol Microbiol. 2001;51:667–678. doi: 10.1099/00207713-51-2-667. [DOI] [PubMed] [Google Scholar]
- 97.Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA Reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 98.Ritari J, Salojärvi J, Lahti L, de Vos WM. Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database. BMC Genomics. 2015;16:1056. doi: 10.1186/s12864-015-2265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rainey FA, Ward-Rainey NL, Janssen PH, Hippe H, Stackebrandt E. Clostridium paradoxum DSM 7308T contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiol Read Engl. 1996;142(Pt 8):2087–2095. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 100.Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gándara B, Merino AL, Rogel MA, Martínez-Romero E. Limited Genetic Diversity ofBrucella spp. J Clin Microbiol. 2001;39:235–240. doi: 10.1128/JCM.39.1.235-240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One. 2013;8:e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marchandin H, Teyssier C, Siméon De Buochberg M, Jean-Pierre H, Carriere C, Jumas-Bilak E. Intra-chromosomal heterogeneity between the four 16S rRNA gene copies in the genus Veillonella: implications for phylogeny and taxonomy. Microbiol Read Engl. 2003;149:1493–1501. doi: 10.1099/mic.0.26132-0. [DOI] [PubMed] [Google Scholar]
- 104.van Berkum P, Terefework Z, Paulin L, Suomalainen S, Lindström K, Eardly BD. Discordant phylogenies within the rrn loci of rhizobia. J Bacteriol. 2003;185:2988–2998. doi: 10.1128/JB.185.10.2988-2998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fitz-Gibbon ST, House CH. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 1999;27:4218–4222. doi: 10.1093/nar/27.21.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tekaia F, Lazcano A, Dujon B. The genomic tree as revealed from whole proteome comparisons. Genome Res. 1999;9:550–557. [PMC free article] [PubMed] [Google Scholar]
- 107.Rivera MC, Lake JA. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature. 2004;431:152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- 108.Montague MG, Hutchison CA. Gene content phylogeny of herpesviruses. Proc Natl Acad Sci U S A. 2000;97:5334–5339. doi: 10.1073/pnas.97.10.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gupta RS. The branching order and phylogenetic placement of species from completed bacterial genomes, based on conserved indels found in various proteins. Int Microbiol Off J Span Soc Microbiol. 2001;4:187–202. doi: 10.1007/s10123-001-0037-9. [DOI] [PubMed] [Google Scholar]
- 110.Coenye T, Vandamme P. Extracting phylogenetic information from whole-genome sequencing projects: the lactic acid bacteria as a test case. Microbiol Read Engl. 2003;149:3507–3517. doi: 10.1099/mic.0.26515-0. [DOI] [PubMed] [Google Scholar]
- 111.Huson DH, Steel M. Phylogenetic trees based on gene content. Bioinforma Oxf Engl. 2004;20:2044–2049. doi: 10.1093/bioinformatics/bth198. [DOI] [PubMed] [Google Scholar]
- 112.Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 113.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 114.Auch AF, von Jan M, Klenk H-P, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Auch AF, Klenk H-P, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci. 2010;2:142–148. doi: 10.4056/sigs.541628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, Thompson FL. Microbial genomic taxonomy. BMC Genomics. 2013;14:913. doi: 10.1186/1471-2164-14-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bascomb S, Lapage SP, Willcox WR, Curtis MA. Numerical classification of the tribe Klebsielleae. J Gen Microbiol. 1971;66:279–295. doi: 10.1099/00221287-66-3-279. [DOI] [PubMed] [Google Scholar]
- 118.Cowan ST, Steel M, Shaw C, Duguid JP. A classification of the Klebsiella group. J Gen Microbiol. 1960;23:601–612. doi: 10.1099/00221287-23-3-601. [DOI] [PubMed] [Google Scholar]
- 119.Brenner DJ, Farmer JJ, Hickman FW, Asbury MA, Steigerwalt AG. Taxonomic and Nomenclature Changes in Enterobacteriaceae. [cited 2017 May 31]. Available from: https://www.abebooks.fr/Taxonomic-Nomenclature-Enterobacteriaceae-Don-Brenner-Farmer/4158251114/bd
- 120.Klebsiella OI. Bergey’s man Syst Bacteriol. 1974. [Google Scholar]
- 121.Orskov I, Genus V. Klebsiella. Bergey’s manual of systematic bacteriology. 1984:461–5.
- 122.Rouli L, Merhej V, Fournier P-E, Raoult D. The bacterial pangenome as a new tool for analyzing pathogenic bacteria. New Microbes New Infect. [cited 2015 Jul 20]; Available from: http://www.sciencedirect.com/science/article/pii/S2052297515000529 [DOI] [PMC free article] [PubMed]
- 123.Ereshefsky M. Eliminative Pluralism. Philos Sci. 1992;59:671–690. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.