Abstract
Despite the wide application of nanomaterials, toxicity studies of nanoparticles (NP) are often limited to in vitro cell models, and the biological impact of NP exposure in mammals has not been thoroughly investigated. Zinc oxide (ZnO) NPs are commonly used in various consumer products. To evaluate the effects of the inhalation of ZnO NP in mice, we studied splice junction expression in the lungs as a proxy to gene expression changes analysis. Female ICR mice were treated with 6.46 × 104 and 1.93 × 106 NP/cm3 for 3 days and 3 months, respectively. An analysis of differential expression and alternative splicing events in 298 targets (splice junctions) of 68 genes involved in the processes relevant to the biological effects of ZnO NP was conducted using next-generation sequencing. Three days of exposure resulted in the upregulation of IL-6 and downregulation of BID, GSR, NF-kB2, PTGS2, SLC11A2, and TXNRD1 splice junction expression; 3 months of exposure increased the expression of splice junctions in ALDH3A1, APAF1, BID, CASP3, DHCR7, GCLC, GCLM, GSR, GSS, EHHADH, FAS, HMOX-1, IFNγ, NF-kB1, NQO-1, PTGS1, PTGS2, RAD51, RIPK2, SRXN1, TRAF6, and TXNRD1. Alternative splicing of TRAF6 and TXNRD1 was induced after 3 days of exposure to 1.93 × 106 NP/cm3. In summary, we observed changes of splice junction expression in genes involved in oxidative stress, apoptosis, immune response, inflammation, and DNA repair, as well as the induction of alternative splicing in genes associated with oxidative stress and inflammation. Our data indicate the potential negative biological effects of ZnO NP inhalation.
Keywords: zinc oxide nanoparticles, inhalation, splice junction expression, alternative splicing
Zinc oxide nanoparticles (ZnO NPs) are a commonly used material with a yearly production of over 30 000 tons. Because of their antimicrobial properties ZnO NP are applied in medical and cosmetics products, but they are also found in sunscreens, pigments, catalysts, and in electronic devices. Due to the wide application of cosmetics, skin is the main route of exposure of ZnO NP for consumers. Inhalation exposure occurs mostly in industrial production by zinc oxide fumes during activities such as thermal cutting, welding of galvanized steel, or melting. Such exposure may result in zinc fever characterized by throat irritation, cough, and various respiratory and flu-like symptoms. Systemic inflammatory effects of ZnO NP inhalation have recently been demonstrated in human volunteers exposed to ZnO particle doses of 1.0 and 2.5 mg/m3, concentrations below occupational exposure limits of 5.0 mg/m3 set in many countries (Monsé et al., 2018).
In the respiratory system of experimental animals, the administration of ZnO NP caused the induction of processes associated with immune response and oxidative stress, although permanent pathological changes in the organs were usually not found. In C57/Bl6 mice, sub-acute inhalation (2 weeks, 4 h/day, 3.5 mg/m3) resulted in increased number of macrophages in bronchoalveolar lavage (BAL) fluid and production of cytokines IL12(p40) and MIP-1α. However, no significant histopathological changes in the lungs were observed (Adamcakova-Dodd et al., 2014). Inhalation of occupationally relevant doses of ZnO NP (1.4 and 4.9 mg/m3 for 2 weeks) affected total cell, neutrophil, LDH, and total protein levels in Sprague Dawley rats. Inflammatory pathology in the lungs and degeneration and necrosis of the myocardium were also observed (Chuang et al., 2014). In F344 rats, a 4-week exposure (6 h/day, 5 days/week) to 2 and 10 mg/m3 ZnO NP caused transient increase in total cell and neutrophil count and elevated levels of cytokine-induced neutrophil chemoattractant (CINC-1, CINC-2) and heme oxygenase 1 (HO-1) in BAL. However, no persistent lung inflammation or fibrosis was observed (Morimoto et al., 2016).
The pulmonary effects caused by inhalation of various types of nanoparticles differ by the severity of the response and the duration of the effects. In most cases lung inflammation was observed, as eg, after inhalation of metal and metal oxide nanoparticles (ZnO, TiO2, Al2O3, CeO2, Fe2O3.Fe3O4, MnFe2O4, CrOOH, Co, CuO) (Lai et al., 2018; Larsen et al., 2016; Présumé et al., 2016; Wan et al., 2017) or single wall carbon nanotubes (SWCNT) (Mercer et al., 2008). SWNT increased collagen deposition in the lungs but the inflammation was transient, whereas CeO2 inhalation gave rise to a more persistent inflammation. MnFe2O4 and CrOOH caused mild peribronchiolar fibrosis. Co induced extensive lung inflammation, interstitial fibrosis, and proliferation of interstitial cells. CuO promoted collagen accumulation and expression of the progressive fibrosis marker α-SMA in the lungs. In a comparison of several metal oxide NP, ZnO was the only one causing toxic effects in the airways (Larsen et al., 2016). The toxicity of ZnO NP is caused by the presence of particles, as well as by their ability to dissolve in the lungs. It has been shown that the particulate nature of ZnO NP contributes to the systemic negative effects induced by inhalation of these nanoparticles. A comparison of effects associated with ZnO NP and zinc nitrate inhalation revealed acute inflammation at bronchioalveolar junctions of the lungs and cytokine secretion in BAL. However, activity of LDH in BAL and glutamate oxaloacetate transaminase, glutamate pyruvate transaminase, and creatine phosphokinase in blood were induced only by ZnO NP (Chen et al., 2015). The release of Zn2+ ions in cytosol, and their sequestration by mitochondria followed by mitochondrial dysfunction and apoptosis, plays also an important role in ZnO NP toxicity (Kao et al., 2012). The solubility of ZnO NP depends on the chemical properties of the solvent: it is lower in RPMI medium and moderately hard water than in DMEM medium (Reed et al., 2012). After intratracheal instillation into the lungs of rats, ZnO NP remained intact at around neutral pH and rapidly dissolved under acidic conditions in the lysosomes causing lysosomal destabilization and cell death. Interestingly, no dissolution of ZnO NP was observed in the artificial interstitial fluid (Cho et al., 2011).
To investigate the mechanisms of NP toxicity in vivo, gene expression profiling has been used. Although the studies of the pulmonary effects of ZnO NP exposure in experimental animals are limited to intratracheal injection (Fukui et al., 2015) and intranasal instillation (Saptarshi et al., 2015), gene expression changes following the inhalation of other metal-based NP (eg, copper, gold, iron, silver, TiO2, and NiO) were analyzed. Most of the studies focused on global gene expression changes using microarrays, but in some, a targeted analysis was performed (Adamcakova-Dodd et al., 2015; Horie et al., 2016; Liberda et al., 2014; Morimoto et al., 2011). DNA damaging agents not only alter gene expression, but may also induce alternative splicing which preferentially affects the genes implicated in DNA repair, cell-cycle control and apoptosis (Shkreta and Chabot, 2015).
In this study, we aimed to analyze splice junction (SJ) expression changes in mice exposed by the inhalation to two concentrations of ZnO NP for 3 days and 3 months. The analysis of SJ and exon expression is applied to detect alternative splicing events (Li et al., 2015). Here, we used differential SJ expression as a proxy to analyze changes in the expression of genes involved in oxidative stress, immune response, inflammation, apoptosis, DNA damage and repair, and cell cycle regulation as well as to study the alternative splicing events possibly associated with exposure to ZnO NP.
MATERIALS AND METHODS
Animals
Adult female ICR mice (6 weeks old, average weight 24 g) obtained from the Animal facility of Masaryk University (Brno, Czech Republic) were allowed to acclimate to laboratory conditions for at least 1 week before the inhalation experiments. The ICR mice were used because as an outbred strain they exhibit genetic variability comparable with that naturally occurring in human populations (Cui et al., 1993). Commercial diet and water were provided ad libitum. The experiments were approved by the Ethical Board of the Institute of Animal Physiology and Genetics (No. 081/2010; March 29, 2010).
Production of ZnO nanoparticles and their characterization
ZnO NPs were generated continuously using an evaporation-condensation-oxidation technique. A ceramic crucible containing a small amount of granular zinc was placed inside the ceramic reactor tube of a vertically oriented furnace (Carbolite TZF 15/50/610; Carbolite, Hope Valley, UK). Zinc was evaporated at the center of the furnace at a temperature of 465°C; metal vapor was carried out of the furnace with an inert nitrogen gas stream at a flow rate of 1.15 L/min. In the furnace output zinc vapor was diluted with a U-HEPA filtrated air stream at a flow rate of 1.80 L/min resulting in the oxidation of zinc to zinc oxide and the formation of ZnO NP by a chemical vapor condensation process. The stream with ZnO NP was then split into two streams at a ratio of approximately 1:30. Before entering the inhalation chamber, both streams were further diluted with a stream of purified humidified air (10 L/min) and used for whole-body inhalation experiments.
The morphology of ZnO NP was studied using scanning transmission electron microscopy (STEM). The particles were collected on copper S160-4 TEM grids (3 mm in diameter, 400 mesh grids; Agar Scientific, Electron Technology, Stansted, Essex, UK) by electrostatic precipitation using a Nanometer Aerosol Sampler (model 3089; TSI, Shoreview, Minnesota). The size and shape of ZnO NPs were then analyzed using the Magellan 400 L XHR microscope (FEI Company, Hillsboro, Oregon) operating in the scanning transmission electron microscope (STEM) mode.
The particle number concentration and the size distribution of ZnO NP in the inhalation chambers were measured continuously in the size range of 7.64–229.6 nm using a Scanning Mobility Particle Sizer Spectrometer (SMPS Spectrometer; model 3936L72; DMA model 3081, CPC model 3772; TSI) at 5-min intervals.
Exposure to ZnO NP
Adult mice were exposed to ZnO NPs in a whole-body inhalation chamber described in detail in our previous study (Vecera et al., 2011). The mice (5 animals/group) in two exposure groups were exposed continuously for 3 days, or 3 months, respectively (24 h/day, 7 days/week). The control animals were exposed to clean, filtered air without nanoparticles in the same inhalation chamber. The inhalation experiment started at the same time for both exposure groups and the controls and was carried out in parallel with two concentrations of ZnO NPs of ca 6.46 × 104 and 1.93 × 106 particles/cm3, respectively, corresponding to mass concentrations of 20 µg/m3 and 625 µg/m3, respectively. The estimated deposited dose of ZnO for lower NP concentration was 0.009 and 0.269 µg of ZnO/g of mouse body weight for 3 days and 3 months exposure period, respectively (Bide et al., 2000, Mitchell et al., 2007). For higher NPs concentration, the estimated deposited dose of ZnO was 0.275 and 8.332 µg of ZnO/g of mouse body weight for 3 days and 3 months period, respectively. Because no maximum human inhalation residue limit has been established for zinc, the inhalation doses for our experiment were set up based on acute and intermediate oral exposure, 300 µg Zn/kg/day (https://www.atsdr.cdc.gov/mrls/mrllist.asp; last accessed January 1, 2019), and the assumption that the pulmonary deposition fraction of ZnO is 100%. In this case, the inhalation deposited doses of 3 µg/kg/day for acute, and 917 µg/kg/day for subchronic exposure correspond to mass concentration of 20 µg/m3 and 625 µg Zn/m3 and nanoparticle number concentration of 6.46 × 104 and 1.93 × 106 P/cm3, respectively. The exposure doses were within the range on previous reports of inhalation exposure to ZnO NP (Adamcakova-Dodd et al., 2014; Chuang et al., 2014; Morimoto et al., 2016). To prevent the adsorption of nanoparticles on feeding, a special dispenser has been used. In a 10.5 cm × 17 cm × 2 cm polyvinyl chloride block five openings of a 1.5 cm diameter were drilled longitudinally. The openings were filled with feed and the dispenser was placed, in a nearly vertical position, in a wire feeding compartment of the polycarbonate box in the inhalation chamber. At the end of exposure (after 3 days, or 3 months, respectively) the mice were sacrificed by cervical dislocation. Lungs were collected for biochemical, electron microscopic, and SJ expression analyses.
Chemical analysis of Zn in the lungs
For the analysis of Zn content, lung tissue was collected from five animals from each exposure group (a ZnO NP concentration of 6.46 × 104 and 1.93 × 106 NP/cm3) and five control animals. Samples were rinsed in high purity water and stored in vials at −25°C until further processing. The samples were decomposed by microwave assisted digestion in 3 ml of concentrated sub-boil grade nitric acid using Quartz Distillation System (model MSBQ 2; Maasen, Eningen, Germany). The samples, including blanks, were treated in precleaned quartz tubes of closed pressurized autoclave system (UltraWave, Milestone Srl, Italy). The decomposition program consisted of four steps with the following characteristics: step 1: 10 min with a temperature ramp between 100 and 120°C; step 2: 5 min with a temperature ramp between 120 and 200°C; step 3: 3 min with a temperature ramp between 200 and 250°C; step 4: 5 min at 250°C. After cooling down (10 min), digests were quantitatively transferred to vials and diluted with ultrapure water to the final mass of 10 g/vial.
Zinc content in digests was determined by flame and electrothermal atomic absorption spectrometry (ET AAS) using AA 3110 and AAnalyst 600 (Perkin-Elmer, Inc., Shelton, Connecticut). For quantification, a method of standard addition calibration was applied.
Transmission electron microscopy of ZnO NP in the lungs
The samples were fixed in 3% glutaraldehyde in cacodylate buffer, postfixed in 2% OsO4 solution in phosphate buffer, dehydrated in 50%, 70%, 90%, and 100% acetone and embedded in the Epon-Durcupan epoxy resin mixture (Epon 812 Serva, Germany; Durcupan, ACM Fluka, Switzerland). Thin sections were cut at 60 nm on an ultramicrotome Leica EM UC7 (Leica Mikrosysteme GmbH, Vienna, Austria) and placed on 50 mesh formvar-coated nickel grids. The sections were contrasted using 2% uranyl acetate and 2% lead citrate. The samples were observed at 80 kV using a Philips EM 208 transmission electron microscope (FEI Company, Eindhoven, The Netherlands).
Histopathological examination of lung tissue
Lung tissue samples were fixed overnight in 10% buffered neutral formaldehyde at 4°C. The samples were then dehydrated in an increasing series of ethanol, treated with xylene and embedded in paraffin. Serial histological sections of 5 μm thickness were prepared and selected slides were stained in hematoxylin-eosin and Green Trichrome using standard histological techniques. Sections were examined by light microscopy in a blinded fashion.
RNA extraction
For RNA extraction lung tissue samples were flash-frozen in liquid nitrogen and kept at −80°C until further processing. To extract RNA, the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Hilden, Germany) was used. The tissue (200–600 mg) was homogenized under liquid nitrogen using a mortar and pestle; homogenates were transferred to tubes containing the lysis buffer provided with the kit. RNA was extracted from the homogenates according to the manufacturer’s instructions and quantified using a Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts). To achieve equal RNA concentration, the samples were precipitated by sodium acetate and ethanol, and redissolved in an appropriate volume of RNase-free water. The integrity of RNA was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc, Santa Clara, California). An RNA Integrity Number (RIN) ranged from 5.3 to 7.9. Isolated RNA was stored at −80°C until further processing.
Differential splice junction expression analysis by next-generation sequencing
TruSeq targeted RNA custom-made assays were prepared using DesignStudio (https://designstudio.illumina.com/; last accessed January 1, 2019). The assays consisted of 298 targets (SJs) in 68 selected genes including those playing a role in oxidative stress, immune response, inflammation, apoptosis, DNA damage and repair, and cell cycle regulation. For 16 of the genes the available assays covered all SJs; this allowed for the discovery of potentially novel transcript variants. For the other genes, the analysis of alternative splicing was only possible for known transcript variants provided that the assays targeted SJs that would allow distinguishing between these variants (13 genes). In summary, among the selected 68 genes our custom made assay allowed us to analyze transcript variants in 29 (42.6%) genes. For a complete list of selected genes and targeted SJs, see Supplementary Table 1.
For the library preparation, TruSeq Targeted RNA Expression kit (Illumina, San Diego, California) was used. The library was prepared according to the manufacturer’s instructions. RNA samples (200–600 ng, depending on the RIN) were used to synthesize cDNA, that was further hybridized to the Oligo pool (targeted SJs) and PCR amplified in the presence of adapter indexes provided by Illumina. A single-end sequencing reaction was performed using the MiSeq system (Illumina) and the MiSeq Reagent kit v3 (150-cycle).
Analysis of sequencing data—splice junction counts
The primary data (FastQ files) were used for subsequent analysis. The sequencing was performed as 150 base long single-end reads, but the real length between the designed primers was 50 bases, so the primary data were trimmed to this length. Quality control of the primary data before and after trimming was done by FastQC. Ensembl reference mouse genome GRCm38 (mm10) was downloaded from Illumina iGenomes (https://support.illumina.com/sequencing/sequencing_software/igenome.html; last accessed January 1, 2019). The appropriate Refseq annotation of mouse genes (in a gtf format) was downloaded from UCSC Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables; last accessed January 1, 2019). A reference gtf file for junction counting was made according to the experimental design to correspond to the control group. Primary data were aligned to the reference mm10 genome using Tophat2 (ver. 2.1.0). Counts for individual SJs were obtained by HTSeq-count (ver. 0.6.1); data were annotated using biomaRt (ver. 2.26.1). The differences in counts between the control and exposed groups were analyzed by the Student’s t test (SPSS v20.0; IBM, Armonk, New York); correction for multiple testing was performed using the Storey Tibshirani FDR method (Storey and Tibshirani, 2003). Changes in relative SJ expression, expressed as fold change, were calculated by the DESeq2 method (Love et al., 2014).
Analysis of sequencing data—alternative splicing events
To analyze alternative splicing events, the Multivariate Analysis of Transcript Splicing (MATS; http://rnaseq-mats.sourceforge.net; last accessed January 1, 2019; ver. 3.2.5; [Shen et al., 2014]) and the Spliced Transcripts Alignment to a Reference (STAR; https://github.com/alexdobin/STAR; last accessed January 1, 2019; ver. 2.5; [Dobin et al., 2013]) tools were used. As an annotation file, mouse mm10 genome in a gtf format (generated at http://genome.ucsc.edu/cgi-bin/hgTables; last accessed January 1, 2019) was used. Files containing sequencing data (in a bam format) were compared between the exposed and control animals, for both tested concentrations and time intervals. The following alternative splicing events were analyzed: skipped exons, alternative 5′ and 3′ splice sites, mutually exclusive exons, and retained introns. Pooled differential splicing tests were performed using rMATS-STAT (https://github.com/Xinglab/rMATS-STAT; last accessed January 1, 2019) to identify the events modulated by ZnO NP. A graphical presentation of the results was conducted using the Sashimi plots (https://github.com/Xinglab/rmats2sashimiplot; last accessed January 1, 2019).
Real-time quantitative PCR (RT-qPCR) verification of the sequencing data
One microgram of RNA from each sample was used for complementary DNA (cDNA) synthesis using the Transcriptor High Fidelity cDNA synthesis Kit (Roche, Basel, Switzerland). The original protocol was modified by using 2.5 μM oligo(dT) and 10 μM random hexamers for priming in a 20 μl reaction volume. cDNA synthesis was run using the following conditions: 30 min at 55°C and 5 min at 85°C. RT-qPCR was performed using the 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, California). Each RT-qPCR reaction was carried out in a final volume of 14 μl containing 2.5 μl of diluted cDNA, 3.8 μl of water and 7 μl of master mix. To determine the level of each target gene, 0.7 μl of a specifically designed assay (Custom designed real-time PCR assay with Double-Dye probe; Primerdesign, Eastleigh, UK) was added to the reaction mixture. Cycling conditions were: 2 min at 95°C followed by 40 cycles of amplification (10 s at 95°C and 60 s at 60°C). The baseline and threshold values of RT-qPCR experiments raw data were assessed with SDS Relative Quantification Software version 2.3 (Applied Biosystems, Waltham, Massachusetts) to determine Ct values. The expression levels of target genes were normalized to the reference genes (EIF4A2 and CANX). The reference genes were selected according to the stability of gene expression during experimental conditions using the geNorm Reference Gene Selection Kit (Primerdesign, Eastleigh, UK). Relative changes in normalized gene levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The sequences of primers used in RT-qPCR are shown in Supplementary Table 2.
Statistical analysis
To compare the Zn content in the lungs of exposed and control animals, compare RT-qPCR results, number of SJs and to calculate correlations between data, SPSS v20.0 (IBM) was used. The normality of distribution was checked by the Kolmogorov-Smirnov test. As the data were distributed normally, the Student’s t test was used for the comparison of individual groups. The correlation between RT-qPCR and SJ expression results was calculated using the Pearson test. The mean values ± standard deviations of the analyzed parameters are reported in the figures. Venn diagrams were prepared in Bioinformatics & Evolutionary Genomics tool (http://bioinformatics.psb.ugent.be/webtools/Venn/; last accessed January 1, 2019).
RESULTS
ZnO NP Characterization
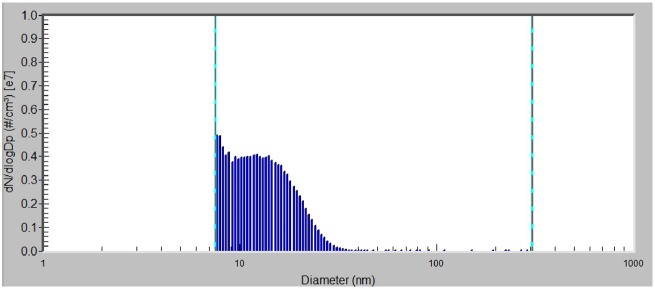
The size distribution of ZnO NP in the inhalation chamber was analyzed separately for the chamber with the concentration of 6.46 × 104 and 1.93 × 106 ZnO NP/cm3, respectively. In the chamber with the higher concentration of ZnO NP, mode diameter, geometric mean diameter, and average total number concentration of nanoparticles was: 7.64 nm, 12.7 nm, and 1.93 × 106 particles/cm3, respectively; average mass concentration was 625 µg/m3. The size distribution of nanoparticles expressed in number concentration is shown in Figure 1. In the chamber with the lower concentration of NPs, mode diameter, geometric mean diameter, and average total number concentration of ZnO NP was 7.64 nm, 12.6 nm, and 6.46 × 104 particles/cm3, respectively; average mass concentration was 20.2 µg/m3. The micrographs from STEM analysis showed that the nanoparticles are formed by agglomerates in the size range of about 6–21 nm (Figure 2).
Figure 1.
Size distribution of ZnO NP expressed in number concentration of nanoparticles in the inhalation chamber, with a concentration of 1.93 × 106 ZnO NP/cm3 (mode diameter 7.64 nm, geometric mean diameter 12.7 nm).
Figure 2.
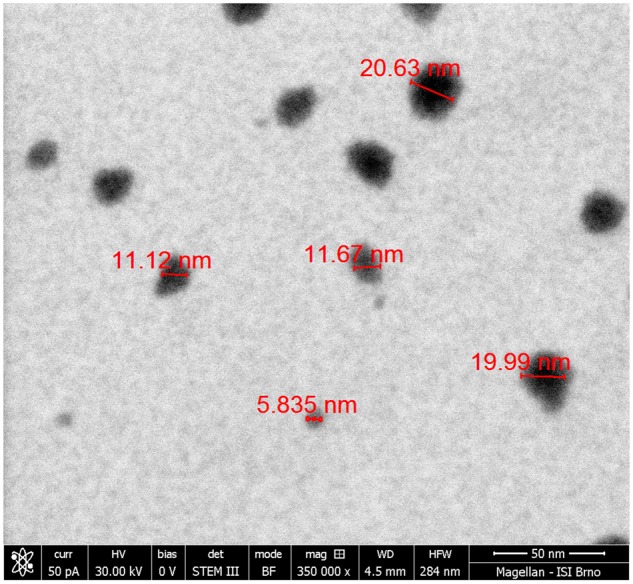
The scanning transmission electron micrograph showing the agglomerates of primary ZnO NP.
Zinc Concentration, ZnO NP Detection, and Histopathological Changes in the Lungs
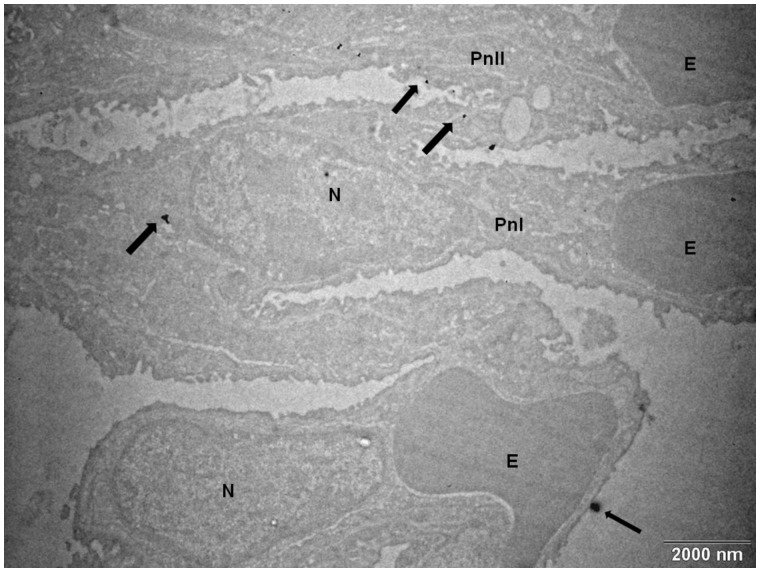
The concentration of zinc in the lungs differed significantly between the controls, and animals exposed for 3 days to 1.93 × 106 ZnO NP/cm3 (Zn content [µg/g tissue]: 13.1 ± 0.10 vs 20.6 ± 0.38, p < .05). However, no such difference was observed for the 3-month exposure period (Figure 3). The chemical analysis of the Zn content in the lungs of mice exposed to 6.46 × 104 ZnO NP/cm3 did not show a significant difference between the exposed and control animals. Scanning transmission electron microscopy showed the presence of ZnO NP in pneumocytes after 3 months of exposure to 1.93 × 106 ZnO NP/cm3 (Figure 4); however, for a lower concentration and/or shorter inhalation period no ZnO NP were detected. Histopathological evaluation revealed a goblet cell hyperplasia at the terminal bronchioli of the lungs of animals exposed to 1.93 × 106 ZnO NP/cm3 for 3 months (Figure 5).
Figure 3.
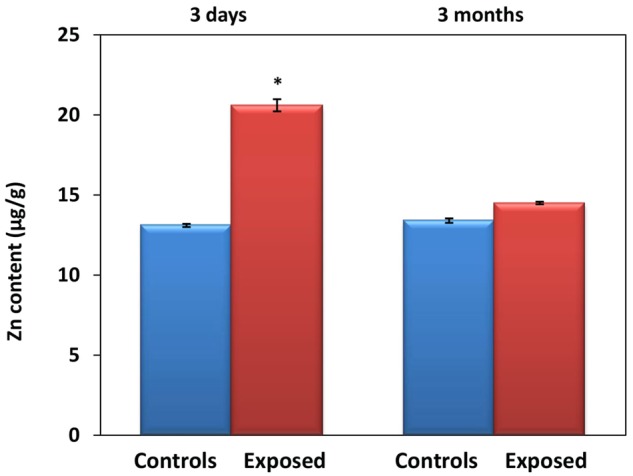
Mean Zn content (± SD) in the lungs of animals exposed to 1.93 × 106 ZnO NP/cm3 for 3 days and 3 months, respectively.
Figure 4.
The transmission electron micrograph of ZnO NP (arrows) in pneumocytes after 3 months exposure to 1.93 × 106 ZnO NP/cm3. Abbreviations: E, erythrocyte; N, nucleus; Pn, pneumocyte.
Figure 5.
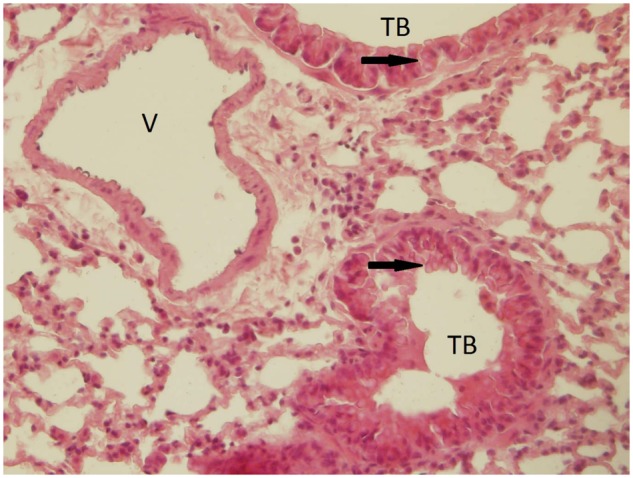
A goblet cell hyperplasia at the terminal bronchioli of the lungs (indicated by the arrow) of animals after 3 months exposure to 1.93 × 106 ZnO NP/cm3. This finding, not observed in the control group, may be related to irritation as a consequence of ZnO NP inhalation. Abbreviations: TB, terminal bronchioli; V, vessel.
Differential Splice Junction Expression Analysis
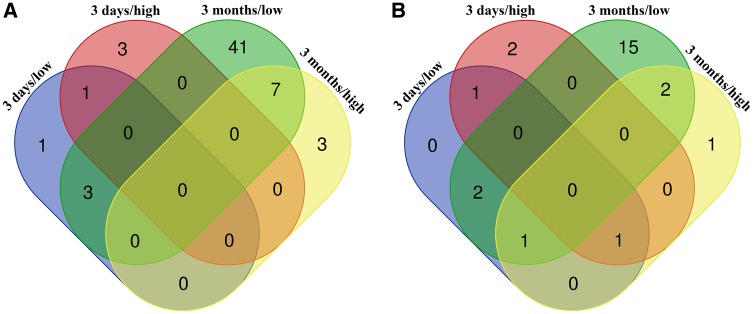
To analyze the effect of ZnO NP exposure on the expression of the studied genes, we assessed the differences in the abundances (read counts) of individual SJs within the selected genes. Three days of exposure resulted in significant changes in the expression of 8 SJs in 7 genes (Supplementary Table 3, Figs. 6A and 6B), whereas 3 months of exposure significantly affected 54 SJs in 22 genes (Supplementary Table 4, Figs. 6A and 6B). For the shorter treatment period, the expression was mostly downregulated; after the longer inhalation of ZnO NP all significant changes were associated with the upregulation of SJ expression. Interestingly, the significant results obtained for 3 months exposure were mostly those induced by 6.46 × 104 ZnO NP/cm3, a lower tested concentration; this was not confirmed for 3 days of exposure. There was very little overlap of deregulated SJs and respective genes between both exposure periods and tested doses (Figs. 6A and 6B). Significant effects for both concentrations of ZnO NP were found only for the expression of SJs in BID (3 days exposure), FAS, NQO1, and TXNRD1 (3 months exposure). Overall, the affected genes included those participating in apoptotic (APAF1, BID, CASP3, FAS), oxidative stress (ALDH1A3, DHCR7, EHHADH, GCLC, GCLM, GSR, GSS, HMOX1, NQO1, PTGS1, PTGS2, SRXN1, TXNRD1) and immunological response (IFN-γ, IL-6, NF-κB1, NF-κB2, RIPK2, TRAF6), ion transport (SLC11A2), and DNA repair (RAD51). A complete list of deregulated SJs with at least one significant result for the given gene is provided in Supplementary Tables 5A and 5B.
Figure 6.
Venn diagrams reporting numbers of unique and common deregulated splice junctions (A) and respective genes (B) in the lungs after inhalation exposure to ZnO NP. Three days/low—exposure to 6.46 × 104 NP/cm3 for 3 days; 3 days/high—exposure to 1.93 × 106 NP/cm3 for 3 days; 3 months/low—exposure to 6.46 × 104 NP/cm3 for 3 months; 3 days/high—exposure to 1.93 × 106 NP/cm3 for 3 months.
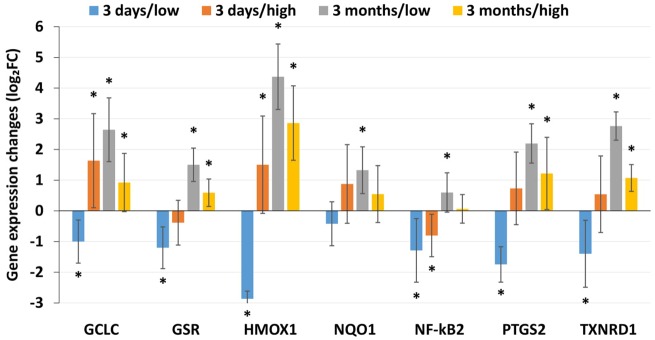
To validate our approach of using SJ expression as a proxy for gene expression changes analysis, the differential SJ expression data obtained for GCLC, GSR, HMOX, NQO1, NF-κB2, PTGS2, and TXNRD1 after exposure to both concentrations of ZnO NP in both time intervals were correlated with mRNA expression assessed using RT-qPCR (Figure 7). The results showed a very good agreement between the methods: they significantly correlated (R = 0.961, p < .001) and for 96.4% of the gene/ZnO NP concentration/exposure time combinations the data were comparable for both approaches. The difference was observed only for the NF-κB2 expression after 3 months of exposure to the higher concentration of ZnO NP (a nonsignificant upregulation detected by RT-qPCR in contrast with a downregulation observed by the next-generation sequencing [NGS] approach; data not shown).
Figure 7.
mRNA expression levels assessed by RT-qPCR for selected genes. The expression significantly correlated with differential SJ expression data obtained for these genes (R = 0.961, p < .001) indicating that SJ expression could be used as a valid approach to study differential gene expression. Three days/low—exposure to 6.46 × 104 NP/cm3 for 3 days; 3 days/high—exposure to 1.93 × 106 NP/cm3 for 3 days; 3 months/low—exposure to 6.46 × 104 NP/cm3 for 3 months; 3 days/high—exposure to 1.93 × 106 NP/cm3 for 3 months.
Alternative Splicing Events Analysis
Alternative splicing (AS) events were analyzed based on the counts of SJs in the individual genes following inhalation exposure to ZnO and in the controls. Overall, we found changes in AS events associated with 3 days of exposure to ZnO NP, whereas 3 months of exposure had no effects on the process. Exon skipping, but no other AS event, was affected after exposure to 1.93 × 106 ZnO NP/cm3; we observed no effect of the lower ZnO NP concentration. We detected an increased frequency of AS of TRAF6 and TXNRD1, genes participating in the inflammatory and oxidative stress response, respectively, following exposure to ZnO NP. For TRAF6, two transcript variants (a longer [NM_009424.3; 6188 bp] and a shorter [NM_001303273.1; 5985 bp] one) have been described. The exposure to ZnO NP increased the activity of AS and thus the frequency of the shorter NM_001303273.1 variant. The average inclusion level for the skipped exon was 37.3% for the exposed and 59.7% for the control animals (p < .001) (Table 1). Four transcript variants of TXNRD1 have been reported: NM_001042523.1, NM_001042513.1, NM_015762.2, and NM_001042514.1. In our study, we identified two of these variants: a longer variant (NM_001042513.1; 3417 bp) and a shorter variant (NM_015762.2; 3310 bp). We found significant differences between the frequencies of these variants: the exposure to 1.93 × 106 ZnO NP/cm3 was associated with an increased frequency of the shorter variant when compared with the controls indicating the induction of AS. The average inclusion level for the skipped exon in the exposed animals was 9.9%, whereas in the controls it reached 31.1% (p < .001) (Table 1).
Table 1.
Alternative Splicing Events in Lung Tissues of Mice Following 3 Days Exposure to 1.93 × 106 ZnO NP/cm3
| Gene | RefSeq ID (Longer/Shorter Transcript Variant) | Exposed (N = 5) |
Controls (N = 5) |
Inclusion level difference (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Number of junctions included | Number of junctions skipped | Mean inclusion level (%) | Number of junctions included | Number of junctions skipped | Mean inclusion level (%) | |||
| TRAF6 | NM_009424.3/NM_001303273.1 | 62 ± 38a | 54 ± 33b | 37.3e | 136 ± 94 | 42 ± 20 | 59.6 | −22.3 |
| TXNRD-1 | NM_001042513.1/NM_015762.2 | 20 ± 13c | 89 ± 60d | 9.9e | 33 ± 20 | 42 ± 31 | 31.1 | −21.2 |
“Number of junctions included” reports the mean number of reads (± SD)/animal mapped to the splice junctions included in the longer transcript variant; “number of junctions skipped” reports the mean number of reads (± SD)/animal mapped to the skipped splice junctions; “mean inclusion level” is the normalized mean percentage of splice junctions included in the longer transcript variant; “inclusion level difference” reports the difference between the mean inclusion level in exposed and control animals, a negative value indicates a higher proportion of transcript variants with skipped exons and activity of alternative splicing in the exposed animals; details of the calculation are provided in Shen et al. (2014).
p = .145, bp = .508, cp = .252, dp = .161 for a comparison of number of junctions included or number of junctions skipped between the exposed and the control animals. The mean inclusion levels for the skipped exons statistically differed (ep < .001) between the studied groups for both genes.
DISCUSSION
As ZnO NPs are widely used in numerous consumer products and industrial applications, it is important to obtain sufficient information about the biological impact of ZnO NP exposure to identify potential health risks. In general, exposure to nanoparticles depends on the method of their production and their real-life applications. Thus, the same type of NP can enter the body by different routes and the respiratory tract is the main and typical route of entry to human organism for many NPs (Cho et al., 2012; Kendall et al., 2011; Li et al., 2013). Therefore, the lungs often represent an organ with maximum exposure to nanoparticles and increased susceptibility to their toxic effects.
Although zinc is an essential mineral that plays an important role in cellular metabolism, and is present in active sites of enzymes and acts as an antioxidant (Cooper, 2008), it has numerous toxic effects at higher concentrations. In the case of airway exposure of experimental animals to ZnO NP, these effects are mostly associated with inflammatory responses and necrosis caused by the ability of ZnO NP to generate reactive oxygen species (Cho et al., 2010, 2011; Chuang et al., 2014; Luyts et al., 2014; Vandebriel and De Jong, 2012; Xu et al., 2014).
In this study, we aimed to evaluate the effects of ZnO NP inhalation in mice. Particularly, we focused on SJ expression changes in the lungs as a primary target organ after the inhalation of two concentrations of ZnO NP (6.46 × 104 and 1.93 × 106 ZnO NP/cm3) for 3 days and 3 months to mimic acute and subchronic exposure. As alternative splicing is a mechanism that contributes to transcriptomic and proteomic diversity in higher eukaryotes after eg, DNA damage, we further concentrated on the induction of AS in the lungs of exposed animals.
Considering the physicochemical properties of ZnO NP, their toxicity may be associated with both the presence of particles and the dissolution of ZnO followed by the release of Zn2+ in the organism. Currently, it is believed that the negative biological effects of ZnO NP are mostly associated with Zn2+ release (Reed et al., 2012). A study by Gilbert et al. (2012) suggests that in the human bronchial epithelial cells (BEAS-2B) ZnO NP are first accumulated intracellularly and then completely dissolved generating Zn2+ that is complexed by molecular ligands. The data obtained in Wistar rats indicate that following dissolution in the lungs Zn2+ translocate to the blood (Wang et al., 2010). In our study, we observed an increased content of Zn in the lungs of the animals exposed to 1.93 × 106 NP/cm3 for 3 days when compared with the controls; after 3 months of exposure, there was no difference between the groups. At the same time, STEM detected the presence of ZnO NP in the lungs after 3 months, but not after 3 days of exposure. This observation suggests that a longer inhalation period may have resulted in the saturation of mechanisms responsible for ZnO NP dissolution in lysosomes, causing the shift of balance between the particulate and dissolved form of ZnO NP toward the particulate form. This fact may have contributed to the differences in the SJ expression and AS events induced after acute and subchronic exposure to ZnO NP.
Gene expression changes in the lungs following ZnO exposure have been investigated in rats and mice in several studies. The inhalation of ZnO fumes induced mRNA levels of metallothionein, a protein responsible for binding heavy metals, thus providing protection against their toxicity (Cosma et al., 1992). The increase in expression levels was observed at ZnO concentrations of 1–5 mg/m3 and returned to the control levels 24 h after exposure. The intratracheal instillation of ZnO NP increased mRNA levels of heme oxygenase-1 (HMOX1), interleukin-6 (IL-6), cytokine-induced neutrophil chemoattractant (CINC)-1 and -3, and metallothionein-1 24 h after exposure. The elevated levels were not detected in the samples collected 1 week after instillation (Fukui et al., 2015). In another study, intranasal instillation of ZnO NP resulted in the increased expression of eotaxin mRNA 24 h after exposure; the expression of monocyte chemo attractant protein (MCP)-1 and tumor necrosis factor alpha (TNF-α) mRNA was not affected (Saptarshi et al., 2015). Although the data are limited and routes of ZnO delivery differ, the results indicate that in the pulmonary system ZnO induces the expression of genes associated with immune response and oxidative stress.
In our study, we investigated the changes of expression of a larger set of genes participating in oxidative stress, immune response, inflammation, apoptosis, DNA damage and repair, and cell cycle regulation. We used targeted RNA sequencing that allowed us not only to assess changes in the expression of selected SJs but also to identify the induction of AS following exposure to ZnO NP. The analysis of SJ expression revealed the following notable results: (1) The number of deregulated SJs was lower after 3 days exposure than after the longer inhalation period. (2) The expression of most of the SJs was downregulated after 3 days of exposure to ZnO NP, whereas the 3 months of exposure caused an upregulation of SJ expression. (3) Most of the significant results obtained after the longer ZnO NP treatment were observed for the lower concentration (6.46 × 104 NP/cm3); the concentration of 1.93 × 106 NP/cm3 had mostly no significant effect.
The acute exposure caused a significant decrease in SJs expression levels in genes associated with apoptosis (BID, BH3 interacting-domain death agonist), oxidative stress response (GSR, glutathione reductase; PTGS2, prostaglandin synthase 2; TXNRD1, thioredoxin reductase 1), metal transportation (SLC11A2, solute carrier family 11 member 2), and inflammation/immunity (NF-κB2). Interleukin 6, an important cytokine secreted by T cells and macrophages to modulate immune response, was the only protein whose SJ expression was induced after 3 days of ZnO NP inhalation. In contrast to this data, the subchronic 3 months of ZnO NP inhalation induced the SJ expression in genes encoding proteins involved in the execution of the apoptotic response (APAF1, a protein responsible for the initiation of apoptosis; CASP3, a member of caspase cascade that activates caspase 6 and 7; FAS, a receptor playing a central role in the initiation of apoptosis) and oxidative stress including HMOX1 (an enzyme responsible for conversion of heme to biliverdin, that also possesses anti-inflammatory properties), ALDH3A1 (a protein that plays a role in response to lipid peroxidation), NQO1 (a quinone reductase participating in detoxification reactions), EHHADH (a protein involved in the peroxisomal oxidation of fatty acids), enzymes associated with glutathione metabolism (GSS, GSR, GCLM, GCLC), SRXN1 (an enzyme that reduces cysteine-sulfinic acid formed as a result of exposure to oxidants), PTGS1 (a key protein in prostaglandin biosynthesis that is expressed as a result of inflammation), and DHCR7 (a protein participating in cholesterol biosynthesis that reduces oxidative stress by dehydrocholesterol reduction). Other deregulated SJs included those in genes participating in immune response and inflammation (NF-κB1 and TRAF6, a signal transducer in the NF-κB pathway; RIPK2 and IFN-γ that play a role in innate and adaptive immune response) and DNA repair (Rad51 that plays a role in homologous recombination). Thus, our results suggest that there is a stark difference in the response of the organism to acute and subchronic exposure to ZnO NP. Whereas short-term inhalation seemed to fail to activate protective mechanisms in the lungs, long-term exposure was associated with the induction of such processes. Indeed, some reports indicate development of clinical tolerance after repeated inhalation exposure to ZnO fumes and ZnO NP (Adamcakova-Dodd et al., 2014). Thus, our observations may represent a molecular manifestation of such processes. Whereas it is difficult to explain this data, we should still bear in mind that the SJ expression was assessed and the protein levels of the respective markers may have been different. Such analyses, however, were out of the scope of our study. Overall, most of the deregulated SJs were detected in the genes associated with oxidative stress response and/or inflammation. Both processes, characteristic for NP exposure, are known to play an important role in induction of neurodegenerative and cardiovascular diseases, as well as in cancer (Madl et al., 2014). We may thus assume, that acute inhalation of ZnO NP may increase the risk of these diseases, whereas a long-term, chronic exposure may induce protective mechanisms that decrease the risk of ZnO NP exposure.
DNA damage is one of the factors affecting alternative splicing, particularly in the genes involved in DNA repair, cell cycle control and apoptosis (Shkreta and Chabot, 2015). Various DNA damaging agents have been shown to induce AS (reviewed in [Shkreta and Chabot, 2015]), but the effect of nanoparticles exposure has not yet been studied. A recent study revealed that ROS production induced by paraquat affected the AS of DNA repair genes including APAF-1, H-ras, ERCC1, SKP2, and BIN1 (Vivarelli et al., 2013). The generation of ROS is a mechanism also implicated in the negative effects of NP exposure. To investigate AS, methods based on RT-PCR or microarray techniques have been used. However, these approaches only allow for the detection of known, reported transcript variants. A recent development of NGS brought the opportunity to discover novel transcript variants provided that the whole-genome transcriptome was analyzed. This approach has recently been used to detect AS events in HepG2 cells following exposure to benzo[a]pyrene, a human carcinogen (van Delft et al., 2012). In this study, we used the MiSeq system and assays provided in Illumina DesignStudio to detect SJs in the selected genes.
We found AS events (specifically exon skipping) to be induced after 3 days of exposure to 1.93 × 106 NP/cm3. Longer treatment and/or lower concentration of ZnO NP had no impact on the process. For both affected genes (TRAF6 and TXNRD1), the frequency of the shorter splicing variant increased following the treatment. TRAF6 encodes tumor necrosis factor receptor-associated factor 6, a protein that serves as a downstream actor of multiple receptor families with immunoregulatory functions (Walsh et al., 2015), as well as in NF-κB activation following genotoxic stress, resulting in double strand DNA damage (Hadian and Krappmann, 2011). The NCBI database reports two transcript variants of TRAF6, both detected in our study, differing in one exon but encoding the same protein (NM_009424.3, 6188 bp; NM_001303273.1, 5985 bp; https://www.ncbi.nlm.nih.gov/gene/22034; last accessed January 1, 2019). However, no information on the possible functional impact of the presence/absence of the exon is available. TXNRD1 encodes thioredoxin reductase 1, an enzyme that uses electrons from NADPH to reduce oxidized thioredoxin and other protein and nonprotein substrates, and thus helps to protect the organism against oxidative stress (Arnér 2009; Cebula et al., 2015). Four transcript variants of TXNRD1 are reported at the NCBI database (https://www.ncbi.nlm.nih.gov/gene/50493; last accessed January 1, 2019): variant 1 encoding the longer cytosolic isoform 1 (NM_001042523.1); and variant 2 (NM_001042513), variant 3 (NM_015762.2), and variant 4 (NM_001042514.1), all encoding the same cytosolic isoform 2. Although no information on the functional differences of both isoforms in mice is available, in human cells stably overexpressing individual isoforms, the alteration of genes associated with differentiation has been observed (Nalvarte et al., 2015). In our study, we were able to distinguish between two transcript variants differing in one exon (NM_001042513 and NM_015762.2). Similarly to TRAF6, the exposure to ZnO NP resulted in higher frequency of the shorter variant (NM_015762.2). Thus, although we showed that inhalation of ZnO NP affects AS in the exposed mice, the functional impact of such changes, if any, could not be determined based on our data.
LIMITATIONS OF THE STUDY
Although we used NGS to detect differential SJ expression, due to technical reasons (the capacity of the MiSeq system) we had to limit the number of genes for which this analysis has been conducted. Due to budget restrictions, protein expression analyses in the lungs have not been conducted; these would potentially shed more light on the mechanisms of response to ZnO NP inhalation. Finally, our results might be affected by the fact that due to logistics reasons (the size of the inhalation chamber and the need for separate cages in case of both genders) only female mice were exposed. As in general, gender differences in gene expression of mice have been observed (Shvedova et al., 2015), a study on both genders would provide more complete information on the biological effects on ZnO NP inhalation.
CONCLUSIONS
In our study, we aimed to evaluate the biological consequences of acute and subchronic exposure of mice to ZnO NP. To the best of our knowledge, this is the first study analyzing the effects of inhalation exposure to ZnO NP using NGS technology, allowing not only the analysis of differential SJ expression, but also the detection of alternative splicing. Our data showed SJ expression changes in the lungs, concerning particularly the processes associated with oxidative stress, immune response, and inflammation after subchronic exposure to ZnO NP. We further detected the induction of alternative splicing following acute exposure. In summary, exposure to ZnO NP by inhalation caused potentially negative biological response in the experimental animals.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the assistance provided by the Research Infrastructure NanoEnviCZ, supported by the Ministry of Education, Youth, and Sports of the Czech Republic under Project No. LM2015073. The authors report no conflict of interest.
FUNDING
This study was supported by Czech Science Foundation (P503/12/G147, 18-02079S), the Ministry of Youth, Education, and Sports of the Czech Republic (LO1508, CZ.02.1.01/0.0/0.0/16_013/00 01821), and Operational Program Prague—Competitiveness (CZ.2.16/3.1.00/21528).
REFERENCES
- Adamcakova-Dodd A., Monick M. M., Powers L. S., Gibson-Corley K. N., Thorne P. S. (2015). Effects of prenatal inhalation exposure to copper nanoparticles on murine dams and offspring. Part. Fibre Toxicol. 12, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamcakova-Dodd A., Stebounova L. V., Kim J. S., Vorrink S. U., Ault A. P., O’Shaughnessy P. T., Grassian V. H., Thorne P. S. (2014). Toxicity assessment of zinc oxide nanoparticles using sub-acute and sub-chronic murine inhalation models. Part. Fibre Toxicol. 11, 15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnér E. S. J. (2009). Focus on mammalian thioredoxin reductases—Important selenoproteins with versatile functions. Biochim. Biophys. Acta 1790, 495–526. [DOI] [PubMed] [Google Scholar]
- Bide R. W., Armour S. J., Yee E. (2000). Allometric respiration/body mass data for animals to be used for estimates of inhalation toxicity to young adult humans. J. Appl. Toxicol. 20, 273–290. [DOI] [PubMed] [Google Scholar]
- Cebula M., Schmidt E. E., Arnér E. S. J. (2015). TrxR1 as a potent regulator of the Nrf2-Keap1 response system. Antioxid. Redox Signal. 23, 823–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-K., Ho C.-C., Chang H., Lin J.-F., Yang C. S., Tsai M.-H., Tsai H.-T., Lin P. (2015). Particulate nature of inhaled zinc oxide nanoparticles determines systemic effects and mechanisms of pulmonary inflammation in mice. Nanotoxicology 9, 43–53. [DOI] [PubMed] [Google Scholar]
- Cho W.-S., Duffin R., Howie S. E. M., Scotton C. J., Wallace W. A. H., Macnee W., Bradley M., Megson I. L., Donaldson K. (2011). Progressive severe lung injury by zinc oxide nanoparticles; the role of Zn2+ dissolution inside lysosomes. Part. Fibre Toxicol. 8, 27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.-S., Duffin R., Poland C. A., Howie S. E. M., MacNee W., Bradley M., Megson I. L., Donaldson K. (2010). Metal oxide nanoparticles induce unique inflammatory footprints in the lung: Important implications for nanoparticle testing. Environ. Health Perspect. 118, 1699–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.-S., Duffin R., Thielbeer F., Bradley M., Megson I. L., MacNee W., Poland C. A., Tran C. L., Donaldson K. (2012). Zeta potential and solubility to toxic ions as mechanisms of lung inflammation caused by metal/metal oxide nanoparticles. Toxicol. Sci. 126, 469–477. [DOI] [PubMed] [Google Scholar]
- Chuang H.-C., Juan H.-T., Chang C.-N., Yan Y.-H., Yuan T.-H., Wang J.-S., Chen H.-C., Hwang Y.-H., Lee C.-H., Cheng T.-J. (2014). Cardiopulmonary toxicity of pulmonary exposure to occupationally relevant zinc oxide nanoparticles. Nanotoxicology 8, 593–604. [DOI] [PubMed] [Google Scholar]
- Cooper R. (2008). Zinc toxicology following particulate inhalation. Indian J. Occup. Environ. Med. 12, 10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma G., Fulton H., DeFeo T., Gordon T. (1992). Rat lung metallothionein and heme oxygenase gene expression following ozone and zinc oxide exposure. Toxicol. Appl. Pharmacol. 117, 75–80. [DOI] [PubMed] [Google Scholar]
- Cui S., Chesson C., Hope R. (1993). Genetic variation within and between strains of outbred Swiss mice. Lab. Anim. 27, 116–123. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H., Iwahashi H., Endoh S., Nishio K., Yoshida Y., Hagihara Y., Horie M. (2015). Ascorbic acid attenuates acute pulmonary oxidative stress and inflammation caused by zinc oxide nanoparticles. J. Occup. Health 57, 118–125. [DOI] [PubMed] [Google Scholar]
- Gilbert B., Fakra S. C., Xia T., Pokhrel S., Mädler L., Nel A. E. (2012). The fate of ZnO nanoparticles administered to human bronchial epithelial cells. ACS Nano 6, 4921–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadian K., Krappmann D. (2011). Signals from the nucleus: Activation of NF-B by cytosolic ATM in the DNA damage response. Sci. Signal. 4, pe2. [DOI] [PubMed] [Google Scholar]
- Horie M., Yoshiura Y., Izumi H., Oyabu T., Tomonaga T., Okada T., Lee B.-W., Myojo T., Kubo M., Shimada M., et al. (2016). Comparison of the pulmonary oxidative stress caused by intratracheal instillation and inhalation of NiO nanoparticles when equivalent amounts of NiO are retained in the lung. Antioxidants (Basel, Switzerland) 5, pii: E4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y.-Y., Chen Y.-C., Cheng T.-J., Chiung Y.-M., Liu P.-S. (2012). Zinc oxide nanoparticles interfere with zinc ion homeostasis to cause cytotoxicity. Toxicol. Sci. 125, 462–472. [DOI] [PubMed] [Google Scholar]
- Kendall M., Ding P., Kendall K. (2011). Particle and nanoparticle interactions with fibrinogen: The importance of aggregation in nanotoxicology. Nanotoxicology 5, 55–65. [DOI] [PubMed] [Google Scholar]
- Lai X., Zhao H., Zhang Y., Guo K., Xu Y., Chen S., Zhang J. (2018). Intranasal delivery of copper oxide nanoparticles induces pulmonary toxicity and fibrosis in C57BL/6 mice. Sci. Rep. 8, 4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen S. T., Jackson P., Poulsen S. S., Levin M., Jensen K. A., Wallin H., Nielsen G. D., Koponen I. K. (2016). Airway irritation, inflammation, and toxicity in mice following inhalation of metal oxide nanoparticles. Nanotoxicology 10, 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Hu X., Bai Y., Alattar M., Ma D., Cao Y., Hao Y., Wang L., Jiang C. (2013). The oxidative damage and inflammatory response induced by lead sulfide nanoparticles in rat lung. Food Chem. Toxicol. 60, 213–217. [DOI] [PubMed] [Google Scholar]
- Li Y., Rao X., Mattox W. W., Amos C. I., Liu B. (2015). RNA-seq analysis of differential splice junction usage and intron retentions by DEXSeq. PLoS One 10, e0136653.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberda E. N., Cuevas A. K., Qu Q., Chen L. C. (2014). The acute exposure effects of inhaled nickel nanoparticles on murine endothelial progenitor cells. Inhal. Toxicol. 26, 588–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyts K., Smulders S., Napierska D., Van Kerckhoven S., Poels K., Scheers H., Hemmeryckx B., Nemery B., Hoylaerts M. F., Hoet P. H. M. (2014). Pulmonary and hemostatic toxicity of multi-walled carbon nanotubes and zinc oxide nanoparticles after pulmonary exposure in Bmal1 knockout mice. Part. Fibre Toxicol. 11, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl A. K., Plummer L. E., Carosino C., Pinkerton K. E. (2014). Nanoparticles, lung injury, and the role of oxidant stress. Annu. Rev. Physiol. 76, 447–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer R. R., Scabilloni J., Wang L., Kisin E., Murray A. R., Schwegler-Berry D., Shvedova A. A., Castranova V. (2008). Alteration of deposition pattern and pulmonary response as a result of improved dispersion of aspirated single-walled carbon nanotubes in a mouse model. Am. J. Physiol. Lung Cell. Mol. Physiol. 294, L87–L97. [DOI] [PubMed] [Google Scholar]
- Mitchell L. A., Gao J., Wal R. V., Gigliotti A., Burchiel S. W., McDonald J. D. (2007). Pulmonary and systemic immune response to inhaled multiwalled carbon nanotubes. Toxicol. Sci. 100, 203–214. [DOI] [PubMed] [Google Scholar]
- Monsé C., Hagemeyer O., Raulf M., Jettkant B., van Kampen V., Kendzia B., Gering V., Kappert G., Weiss T., Ulrich N., et al. (2018). Concentration-dependent systemic response after inhalation of nano-sized zinc oxide particles in human volunteers. Part. Fibre Toxicol. 15, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto Y., Izumi H., Yoshiura Y., Tomonaga T., Oyabu T., Myojo T., Kawai K., Yatera K., Shimada M., Kubo M., et al. (2016). Evaluation of pulmonary toxicity of zinc oxide nanoparticles following inhalation and intratracheal instillation. Int. J. Mol. Sci. 17, 1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto Y., Oyabu T., Ogami A., Myojo T., Kuroda E., Hirohashi M., Shimada M., Lenggoro W., Okuyama K., Tanaka I. (2011). Investigation of gene expression of MMP-2 and TIMP-2 mRNA in rat lung in inhaled nickel oxide and titanium dioxide nanoparticles. Ind. Health 49, 344–352. [DOI] [PubMed] [Google Scholar]
- Nalvarte I., Damdimopoulos A. E., Ruegg J., Spyrou G. (2015). The expression and activity of thioredoxin reductase 1 splice variants v1 and v2 regulate the expression of genes associated with differentiation and adhesion. Biosci. Rep. 35, e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Présumé M., Simon-Deckers A., Tomkiewicz-Raulet C., Le Grand B., Tran Van Nhieu J., Beaune G., Duruphty O., Doucet J., Coumoul X., Pairon J.-C., et al. (2016). Exposure to metal oxide nanoparticles administered at occupationally relevant doses induces pulmonary effects in mice. Nanotoxicology 10, 1535–1544. [DOI] [PubMed] [Google Scholar]
- Reed R. B., Ladner D. A., Higgins C. P., Westerhoff P., Ranville J. F. (2012). Solubility of nano-zinc oxide in environmentally and biologically important matrices. Environ. Toxicol. Chem. 31, 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saptarshi S. R., Feltis B. N., Wright P. F., Lopata A. L. (2015). Investigating the immunomodulatory nature of zinc oxide nanoparticles at sub-cytotoxic levels in vitro and after intranasal instillation in vivo. J. Nanobiotechnol. 13, 6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Park J. W., Lu Z., Lin L., Henry M. D., Wu Y. N., Zhou Q., Xing Y. (2014). rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl. Acad. Sci. 111, E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkreta L., Chabot B. (2015). The RNA splicing response to DNA damage. Biomolecules 5, 2935–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvedova A. A., Kisin E. R., Yanamala N., Farcas M. T., Menas A. L., Williams A., Fournier P. M., Reynolds J. S., Gutkin D. W., Star A., et al. (2015). Gender differences in murine pulmonary responses elicited by cellulose nanocrystals. Part. Fibre Toxicol. 13, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. D., Tibshirani R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U S A 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandebriel R., De Jong W. (2012). A review of mammalian toxicity of ZnO nanoparticles. Nanotechnol. Sci. Appl. 5, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Delft J., Gaj S., Lienhard M., Albrecht M., Kirpiy A., Brauers K., Claessen S., Lizarraga D., Lehrach H., Herwig R., et al. (2012). RNA-seq provides new insights in the transcriptome responses induced by the carcinogen benzo[a]pyrene. Toxicol. Sci. 130, 427–439. [DOI] [PubMed] [Google Scholar]
- Vecera Z., Mikuska P., Moravec P., Smolik J.. 2011. Unique exposure system for the whole body inhalation experiments with small animals. Presented at the NANOCON, pp. 652–654. Tanger Ltd., Brno, Czech Republic.
- Vivarelli S., Lenzken S. C., Ruepp M.-D., Ranzini F., Maffioletti A., Alvarez R., Mühlemann O., Barabino S. M. L. (2013). Paraquat modulates alternative pre-mRNA splicing by modifying the intracellular distribution of SRPK2. PLoS One 8, e61980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M. C., Lee J., Choi Y. (2015). Tumor necrosis factor receptor-associated factor 6 (TRAF6) regulation of development, function, and homeostasis of the immune system. Immunol. Rev. 266, 72–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan R., Mo Y., Zhang Z., Jiang M., Tang S., Zhang Q. (2017). Cobalt nanoparticles induce lung injury, DNA damage and mutations in mice. Part. Fibre Toxicol. 14, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang L., Ding W., Zhang F. (2010). Acute toxicity of ferric oxide and zinc oxide nanoparticles in rats. J. Nanosci. Nanotechnol. 10, 8617–8624. [DOI] [PubMed] [Google Scholar]
- Xu J., Futakuchi M., Alexander D. B., Fukamachi K., Numano T., Suzui M., Shimizu H., Omori T., Kanno J., Hirose A., et al. (2014). Nanosized zinc oxide particles do not promote DHPN-induced lung carcinogenesis but cause reversible epithelial hyperplasia of terminal bronchioles. Arch. Toxicol. 88, 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.