Summary
A variety of genetic cardiovascular diseases may one day be curable using gene editing technology. Germline genome editing and correction promises to permanently remove monogenic cardiovascular disorders from the offspring and subsequent generations of affected families. Although technically feasible and likely to be ready for implementation in humans in the near future, this approach remains ethically controversial. Although currently beset by several technical challenges, and not yet past small animal models, somatic genome editing may also be useful for a variety of cardiovascular disorders. It potentially avoids ethical concerns about permanent editing of the germline and allows treatment of already diseased individuals. If technical challenges of Cas9-gRNA delivery (viral vector immune response, nonviral vector delivery) can be worked out, then CRISPR-Cas9 may have a significant place in the treatment of a wide variety of disorders in which partial or complete gene knockout is desired. However, CRISPR may not work for gene correction in the human heart because of low rates of homology directed repair. Off-target effects also remain a concern, although, thus far, small animal studies have been reassuring. Some of the therapies mentioned in this review may be ready for small clinical trials in the near future.
Key Words: CRISPR, gene editing, germline gene correction
Abbreviations and Acronyms: DMD, Duchenne muscular dystrophy; DSB, double-stranded break; HCM, hypertrophic cardiomyopathy; NHEJ, nonhomologous end-joining
Central Illustration
Since the successful sequencing of the human genome more than 15 years ago, there has been an explosion of knowledge regarding the genetic contributions to common cardiovascular diseases, as well as advancement of the understanding of monogenic cardiovascular disorders. Although this knowledge has allowed for the development of potent pharmaceuticals and better risk stratification for cardiovascular diseases, the rapid development of CRISPR-Cas9 techniques in the past 5 years may dramatically change the outlook for novel therapies in cardiovascular disorders, including those previously thought untreatable.
CRISPR, which stands for “clustered regularly interspersed short palindromic repeats,” refers to a mechanism that evolved in bacteria to identify and remove foreign DNA from their genomes, using an RNA guide and CRISPR-associated (Cas) nuclease (1). It was quickly recognized that such systems have the incredible ability to facilitate precise editing of genes in both mature and developing organisms. Although the introduction of CRISR-Cas9 has the potential to revolutionize the mechanistic understanding of cardiovascular diseases and aide in the development of novel pharmacological therapies, it is the prospect of genome modification in humans that would transform how cardiovascular diseases are treated.
Before the latest development of gene editing techniques, a few approaches that allowed gene targeting in animals existed (e.g., the generation of “knockout” mice). However, these techniques were time consuming, technically challenging, and inefficient. CRISPR has revolutionized gene targeting because it allows for production of double-stranded DNA breaks at precise, user-directed locations within the genome, thus facilitating editing of a chosen segment of DNA in an efficient manner (1).
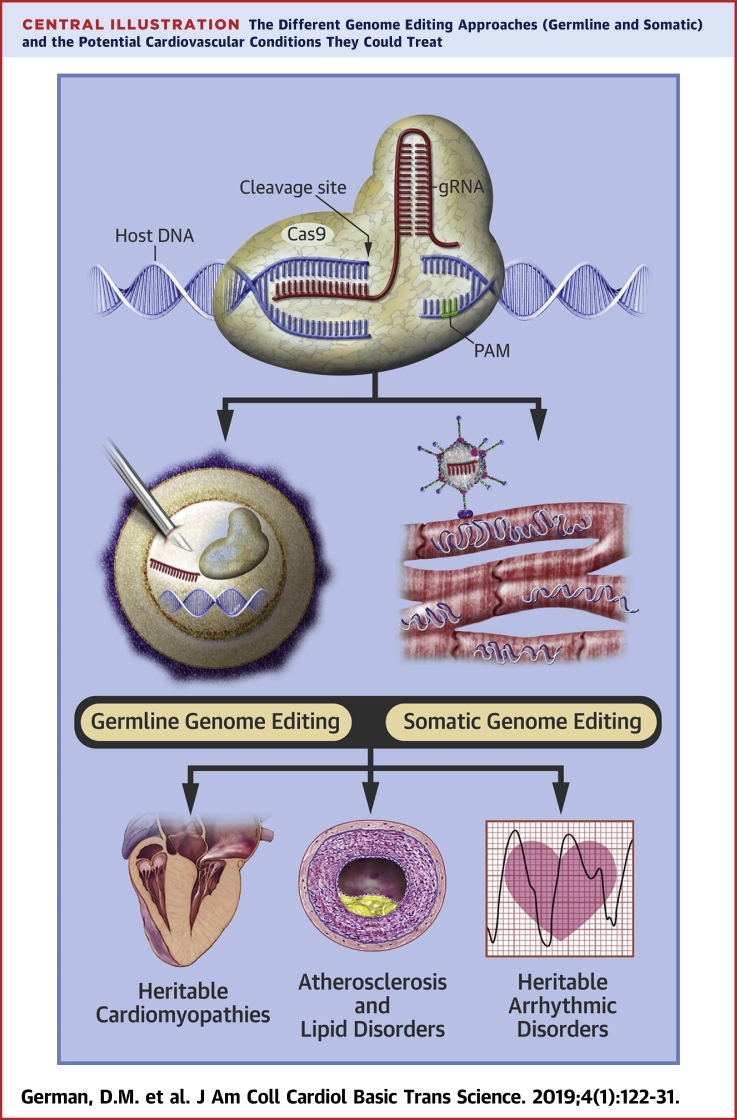
Using the abilities of the CRISPR-Cas9 system requires introduction of a Cas9 nuclease and a segment of guide RNA (gRNA) into the cell(s) of interest, either through direct injection or via viral or nonviral vectors. In practice, the Cas9 nuclease most often comes from Streptococcus pyogenes (SpCas9), but Staphylococcus aureus Cas9 (SaCas9) is also frequently used because of its smaller size (thus, ease of entry into smaller vectors). The gRNA is approximately 100-nucleotides long and contains a 20-nucleotide protospacer sequence that hybridizes to complementary DNA, as well as a shorter 2 to 6 nucleotide protospacer-adjacent motif (PAM) that facilitates binding of host DNA to the Cas9 nuclease (Central Illustration). It is the customization of the protospacer that allows for Cas9 nuclease to create a specific double-stranded break (DSB) in host DNA at user-specified sites (1). However, the PAM sequence is necessary for host DNA recognition and binding by Cas9, as well as subsequent hybridization with the protospacer region of the gRNA and eventual DNA cleavage (2). Thus, it represents another key element required for target sequence specificity. For example, Cas9 from S. pyogenes recognizes a 5′-NGG-3′ PAM sequence, the requirement of which significantly limits target specificity while also increasing the probability of off-target activity. Theoretically, such a specific PAM requirement could limit the therapeutic potential of the technology. However, Cas9 enzymes from other bacterial species (e.g., Streptococcus thermophilus) 3, 4, as well as those obtained via protein engineering (5), bind to a variety of PAMs with enhanced nucleotide specificity and fewer nonspecific bindings or cleavage. These new Cas9 versions are likely to render concerns over PAM specificity obsolete in the near future.
Central Illustration.
The Different Genome Editing Approaches (Germline and Somatic) and the Potential Cardiovascular Conditions They Could Treat
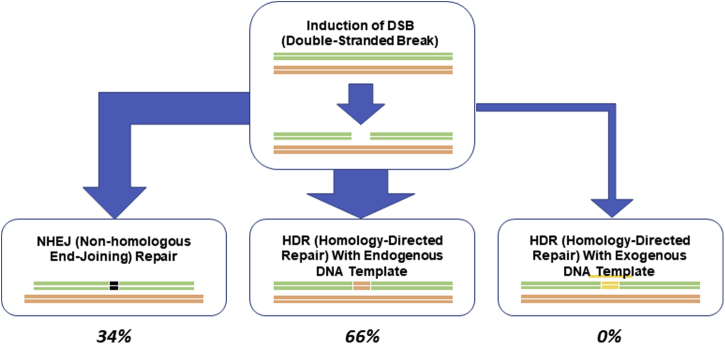
After cleavage by CRISPR-Cas9, DSBs can be repaired by 1 of 2 endogenous mechanisms. Nonhomologous end-joining (NHEJ) is the dominant mechanism in nonproliferating cells, including human myocardial cells, but it is error prone because the affected DNA segment is reconstructed without use of a DNA template (Figure 1). This method is highly vulnerable to production of insertion-deletion mutations (indels), and, in general, would not be an acceptable component of genome editing when the desired outcome is replacement of a dysfunctional gene with a functional one. However, if knockout is all that is desired, production of indel mutations via NHEJ may be a reasonable therapeutic approach, even in somatic cells.
Figure 1.
Types and Frequencies of DNA Repair in Somatic Cells After Gene Deletion by CRISPR
Homolog template is almost never used, and externally provided template is used infrequently. Almost all the repair is performed using NHEJ. CRISPR = clustered regularly interspersed short palindromic repeats; DSB = double-stranded break; HDR = homology-directed repair; NHEJ = nonhomologous end-joining.
Homology-directed repair (HDR) is the second but much less frequent means by which DSBs can be fixed in somatic cells. HDR machinery rebuilds the DSB site using either homologous chromosomal DNA or an exogenous template DNA strand (a single-stranded oligodeoxynucleotide [ssODN]) (Figure 1). The clinical applicability of CRISR-Cas9 in humans is hampered, in part, by the low rates of HDR compared with NHEJ in somatic gene editing.
In theory, genome editing techniques can be applied to both developing embryos and intact mature organisms to either produce loss-of-function in a gene with deleterious downstream effects (knockout) or to restore function to a mutated gene (knock in). In somatic genome editing, genetic modifications are created in differentiated cells in a developing or adult organism, which allows for treatment of established disease or for disease prophylaxis in those genetically at-risk. Cas9 and gRNA are typically delivered in vivo by a viral vector; adenovirus, adeno-associated virus (AAV), and lentivirus have been used most commonly in animals, and each has relative advantages and disadvantages.
Germline genome editing is the process of transforming embryonic or germ cell DNA and can be successfully accomplished ex vivo via direct co-injection of Cas9 and gRNA with sperm into human oocytes (6). It allows for complete and permanent transformation of the human genome, affecting all subsequent generations.
Therapeutic Applications
One of the promises of genome editing is that it will allow for treatment of monogenic diseases that currently have either ineffective or minimally effective therapies. In cardiovascular medicine, heritable cardiomyopathies, such as hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy, and Duchenne muscular dystrophy (DMD), as well as heritable arrhythmic disorders, vasculopathies such as Marfan’s syndrome, and infiltrative diseases such as transthyretin amyloidosis, are potential candidates for clinical applications of germline genome editing techniques (Central Illustration). Editing of the germline in these types of disorders, in which a single gene mutation is responsible for disease manifestation, is capable of permanent correction of the disorder in descendants of affected individuals or those carrying a deleterious mutation.
For many disorders, germline genome editing is unlikely to find a major role in treatment and prevention because of the interplay of genetic and environmental factors in contributing to disease manifestation. For complex disorders such as coronary artery disease and atherosclerosis, somatic genome editing may eventually play a role in treatment. In addition, somatic genome editing may also be useful for post-natal treatment of monogenic disorders, especially because clinical application of germline genome editing is likely to remain ethically more controversial than somatic genome editing for some time. Nonetheless, with close to 7,500 monogenic disorders affecting approximately 780 million people, germline editing has the theoretical potential for permanently eliminating these diseases.
Hypertrophic cardiomyopathy
HCM is a disease of cardiac muscle that results in ventricular hypertrophy and has a propensity for arrhythmias, syncope, and heart failure. Ventricular outflow tract obstruction and associated systolic anterior motion of the mitral leaflet and resulting mitral regurgitation, as well enhanced myocardial stiffness from fibrosis, are accompanying findings. A variety of sarcomeric gene mutations have been implicated in the disorder. Mutations in MYBPC3 account for approximately one-third of all HCM in humans, as well as a significant number of cases of inherited dilated and noncompaction cardiomyopathy (7). Ma et al. (6) recently demonstrated the successful correction of a MYBPC3 mutation in human germ cells using CRISPR-Cas9. They microinjected recombinant Cas9 protein with gRNA and ssODN DNA into human zygotes produced by fertilization of healthy donor oocytes with sperm from a male donor who was heterozygous for MYBPC3 mutation. Most (66.7%) of the embryos injected in this way exhibited a homozygous wild-type genotype, as opposed to 47.4% of control embryos. However, 24% of the embryos exhibited mosaicism, and 9.3% had a persistent heterozygous mutant genotype. The mosaicism was attributed to the inability of CRISPR to correct all mutant genes after cell division occurred.
When the investigators co-injected Cas9 with sperm into M-phase oocytes, 72.4% of the 68 resulting embryos exhibited a homozygous wild-type genotype, and there were no mosaic or mutant embryos. The absence of mosaicism was attributed to gene correction before the fertilized egg started dividing. The remaining 27.6% of embryos were uniformly heterozygous for the wild-type allele and a NHEJ-mediated repair. In addition, genome sequencing of CRISPR-Cas9−targeted blastomeres failed to reveal significant off-target effects. The study demonstrated, for the first time, that CRISPR-Cas9 could be used to abolish disease-causing mutations in human embryos and that modifications to the timing of Cas9 injection during embryogenesis resulted in significant increases in HDR efficiency (Figure 2) (6). Furthermore, in mouse embryos, it was shown that adding RAD51 significantly increased the chances of HDR repair. This high incidence of HDR seems to be inherent in embryos and is probably meant to prevent spontaneous mutations from germline transmission. This method, in combination with pre-implantation genetic diagnosis, may be useful clinically in the prevention of transmission of HCM and other monogenic disorders.
Figure 2.
Types and Frequencies of DNA Repair in Germline Cells After Gene Deletion by CRISPR
Unlike somatic cells, the externally provided template is not used, and NHEJ is used only one-third of the time. The homolog template (from the normal parent) is used for repair in two-thirds of cases. This form of repair is exclusive to the germline. Methods are being developed to increase homolog repair to 100% to obviate the need for pre-natal genetic diagnosis before in vitro implantation of the corrected embryo. Abbreviations as in Figure 1.
Somatic genome editing might be feasible in HCM as well, because the development of cardiac hypertrophy, myocardial fibrosis, and symptomatic disease is generally a gradual process. Mearini et al. (8) administered nonmutant Mybpc3 cDNA to Mybpc3 knockout mice (without use of CRISPR-Cas9) via an AAV vector (specifically AAV9, the most cardiotropic AAV serotype) and found that this therapy successfully increased expression of functional cMyBP-C (to ∼60% of wild-type levels). This prevented cardiac hypertrophy and cardiac functional impairment in young mice (8). Although AAV vectors are generally too small to package SpCas9, use of other Cas9 enzymes may allow for testing of somatic genome editing of this disease. Several other naturally occurring and engineered Cas9 enzymes are smaller in size, which may facilitate such viral delivery (5). However, it must be realized that HDR is uncommon in somatic cells (Figure 2, top panel); thus, gene correction in the human myocardium may not be possible.
Duchenne muscular dystrophy
DMD is a relatively common X-linked disease that leads to progressive skeletal muscle weakness and fatal cardiomyopathy. It is caused by mutations in the DMD gene that codes for dystrophin. The gene is long, with 79 exons, and mutations anywhere along the length of the gene can cause the entire protein to be dysfunctional. Because of the heritability of the disorder and the lack of effective therapies, DMD is an appealing candidate for germline genome editing.
Investigators injected Cas9, gRNA targeting exon 23, and template ssODN DNA into zygotes of mice with a nonsense Dmd mutation and implanted the modified zygotes into female mice. Sequencing of Dmd exon 23 in these corrected mice revealed mosaicism in most animals, although a majority of them showed improvement in muscle function even when only a subset of cells had functional dystrophin that was restored through either NHEJ or HDR (9). Modifications of methods to include nuclease and gRNA injection into M-phase oocytes with sperm, as was done by Ma et al. (6), might further improve these results.
Achieving effective in vivo genome editing of cardiac muscle seems a more formidable task because of the difficulties of delivering Cas9 and gRNA to cardiac tissue. However, in a mouse model of DMD, El Refaey et al. (10) showed that systemic administration of S. aureus Cas9 and gRNA in an AAV vector led to restoration of the DMD reading frame, and thus, expression (in ∼40% of cardiac muscle fibers), and ultimately to improvements in cardiac myofiber architecture, cardiac fibrosis, and papillary muscle contractility. Recently, Amoasii et al. (11) showed an increased expression of cardiac and skeletal muscle dystrophin in a dog model of DMD after treatment with intravenous AAV9 that contained Cas9 and gRNA. These findings demonstrated that, in cases such as DMD, partial effectiveness also has a significant potential to improve symptoms and clinical outcome. Additional studies will be required to evaluate long-term safety and efficacy in larger animals, and ultimately, humans.
Other nonischemic cardiomyopathies
A wide variety of additional causes of nonischemic cardiomyopathy will likely one day be treatable via either germline or somatic genome editing. For example, phospholamban regulates intracellular calcium concentrations through its inhibitory actions on sarcoplasmic reticulum calcium−adenosine triphosphatase (SERCA2), and mutations in the gene for phospolamban (PLN) have been identified as a cause of dilated nonischemic cardiomyopathy (12). Kaneko et al. (13) performed germline genome editing via CRISPR-Cas9 to knockout the PLN gene in a mouse model of severe heart failure (calsequestin [CSQ] overexpressing mice). Compared with control heart failure mice, PLN knockout mice survived longer and had improved cardiac size and function (13).
Dyslipidemia and atherosclerotic cardiovascular disease
Lipid metabolism is an attractive target for somatic genome editing for several reasons. Editing of genes involved in the development of dyslipidemia has the potential to affect not only individuals with monogenic disorders (e.g., familial hypercholesterolemia) but also the large population of individuals without monogenic lipid disorders who have established atherosclerotic disease or elevated risk for cardiovascular events. In addition, although progress in somatic gene editing has thus far been hampered by challenges in gene delivery to tissues of interest, the liver is one particular tissue where success has already been achieved. For instance, AAV vectors have been used successfully in human trials for gene transfer and treatment of hemophilia A and B 14, 15, and additional pre-clinical studies have used them for somatic genome editing for lipid disorders.
Proprotein convertase subtilisin/kexin type 9 (PCSK9) became a novel therapeutic target for prevention of atherosclerotic cardiovascular disease when it was observed that loss-of-function mutations in PCSK9 were associated with reduced low-density lipoprotein cholesterol and reduced risk for coronary heart disease, with no clear adverse clinical consequences (16). Pharmacological inhibition of PCSK9 in vivo also dramatically lowers low-density lipoprotein cholesterol and reduces cardiovascular events in subjects with an elevated baseline risk (17). Ding et al. (18) showed that when Cas9 and gRNA targeting Pcsk9 was delivered in vivo to mice via an adenovirus vector, approximately 50% of the Pcsk9 alleles in the liver tissue were successfully edited, with a wide variety of loss-of-function indels generated via NHEJ (Figure 1). No significant off-target effects were seen. Furthermore, this experiment resulted in substantially (∼90%) lower levels of plasma PCSK9 levels and total plasma cholesterol (35% to 40% reduction) in the edited mice (18). Similar results were achieved using mice with transplanted human hepatocytes using an adenovirus vector (19). Although these studies used adenovirus vectors to deliver Cas9 and gRNA to the liver, adenovirus is not a suitable vector for use in human therapies because of its substantial immunogenicity. Because of its size, S. pyogenes Cas9 cannot be packaged in smaller, less immunogenic vectors such as AAV. However, Ran et al. (20) produced knockout of Pcsk9 in a mouse liver using an AAV vector packaged with S. aureus Cas9, which resulted in ∼95% decrease in blood PCSK9 and ∼40% decrease in blood cholesterol levels with no major off-target effects. These studies provided proof-of-concept that it might soon be possible to immunize patients against atherosclerotic cardiovascular disease by using somatic genome editing to permanently lower plasma lipid levels. Additional work will need to ensure these methods are safe and effective in humans.
Although some of the results of these early investigations in somatic genome editing in cardiovascular disease have been encouraging, most of these studies have depended on NHEJ-mediated DSB repair (Figure 1). Because HDR tends to be inefficient and to only operate in proliferating cells, these techniques would not be useful for producing gain-of-function genome edits in mature cardiac or vascular tissues. In addition, off-target effects have the potential to be more damaging if NHEJ is used (potentially producing activating or inactivating indel mutations in proto-oncogenes or tumor suppressor genes, or producing loss of function mutations in other essential genes).
One promising technique that may be able to overcome this limitation is in vivo base editing 21, 22. Base editors are CRISPR-Cas9 systems that have been modified to alter single base pairs rather than induce DSBs. For example, base editor 3 (BE3) is a S. pyogenes Cas9 mutated to induce only a single-strand break, and it is attached to a cytosine deaminase domain that exchanges a cytosine base at the nick site for a uracil base. This modified Cas9 enzyme then removes the corresponding guanine base, and the result is a C-G pair replaced with a U-A pair; the uracil is ultimately permanently replaced with thymine. Chadwick et al. (23) used an adenovirus vector and BE3 to show that this method could also be used to disrupt mouse Pcsk9, and again found significant reductions in plasma PCSK9 and cholesterol levels in treated mice.
These techniques were extended to modification of ANGPTL3 (angiopoietin-like 3), which codes for a protein that regulates blood lipid levels. Loss-of-function mutations in ANGPTL3 are associated with reduced low-density lipoprotein cholesterol and triglycerides, as well as a reduced risk for coronary heart disease 24, 25. Chadwick et al. (26) again used BE3, introduced via an adenovirus vector, to produce cytosine-to-thymine nonsense mutations in Angptl3 in mice. Treated mice had a 31% decline in plasma triglycerides and a 19% decline in plasma cholesterol. There were no significant signs of off-target mutagenesis (26). Although the current usefulness of base-editing techniques is limited to generating specific point mutations largely confined to the generation of knockouts, and further limited by challenges in packaging larger components into small viral vectors, future advances may ameliorate some of these problems. In addition to PCSK9 and ANGPTL3, somatic genome editing may be useful in addressing cardiovascular risk carried by LPA, APOB, or other genetic mutations. Germline genome editing may also be useful in treating heritable monogenic dyslipidemias.
Age-related clonal hematopoiesis
Somatic genome editing has the additional potential of providing treatment for diseases that are only now beginning to be understood. Clonal hematopoiesis (of indeterminate potential, CHIP), which is also known as age-related clonal hematopoiesis (ARCH), has recently become widely appreciated as a possible contributor to cardiovascular disease. It refers to the clonal amplification of hematopoietic cells due to the accumulation of somatic mutations that confer competitive advantages to these cells at the expense of other cell types (27). Its prevalence increases with age, and it is associated with increased risk for atherosclerotic cardiovascular disease, stroke, and death (28). Although mutations in a large variety of driver genes have been implicated in the development of ARCH, TET2 and DNMT3A are believed to play a major role.
Sano et al. (29) used lentivirus vectors carrying Cas9 and gRNA to inactivate Tet2 and Dnmt3a (via indel mutations) in mouse bone marrow cells and then implanted those cells into irradiated mice. Mice with Tet2 or Dnmt3a inactivation displayed a greater decline in cardiac function as well as increased cardiac size and fibrosis when challenged with angiotensin II infusion compared with those without inactivation of those genes. In addition, inactivation of both genes led to increased expression of pro-inflammatory cytokines (29). These results further implicated these genes in ARCH−mediated cardiovascular disease, and potentially suggest a future therapy. If methods can be developed to reliably knock in somatic gene mutations, it may be possible to reverse driver gene mutations using a vector that targets hematopoietic stem and/or progenitor cells. Consequently, somatic genome editing may be a useful approach to treating ARCH in humans, thus reducing cardiovascular risk. These approaches may be preferable to bone marrow transplantation.
Transthyretin cardiac amyloidosis
One of the challenges of somatic genome editing has been defining the optimal method for delivery of the CRISPR-Cas9 components to the cells of interest. Viral vectors are not optimal for this purpose. Finn et al. (30) demonstrated that, under the right conditions, nonviral vectors may be able to successfully deliver somatic genome editing tools to desired tissues. Transthyretin (TTR) cardiac amyloidosis is an infiltrative disease of cardiac muscle that results from deposition of abnormal pre-albumin (transthyretin) protein and is caused by either heritable TTR gene mutations or accumulation of wild-type transthyretin. It may be an attractive candidate for somatic genome editing because most transthyretin is produced in the liver.
Finn et al. (30) administered a lipid nanoparticle packaged with Cas9 mRNA and gRNA targeted to the Ttr gene in mice and found a significant (>97%) drop in serum TTR levels. It is unclear whether this approach will translate into disease phenotype rescue or prevention. Additional studies will be needed to investigate the effects on cardiac tissue and to determine whether the lipid nanoparticle vector can be used successfully for other conditions. Other nonviral vectors (e.g., microbubbles) that can be selectively destroyed in the tissue of interest by ultrasound to introduce the vector locally also hold promise (31).
Inherited arrhythmic disorders
Channelopathies and other inherited arrhythmic disorders are another area of potential therapeutic application of genome editing. Although CRISPR-Cas9 is already proving useful in the characterization of ion channel protein function and drug−gene interactions in induced pluripotent stem cell−based cell cultures, genome editing will also likely provide the opportunity to treat these rare disorders.
PRKAG2 syndrome is a rare familial disorder characterized by abnormal glycogen storage, ventricular pre-excitation, recurrent arrhythmias, and cardiac hypertrophy. It is caused by mutations in the PRKAG2 gene coding for a regulatory subunit of the adenosine monophosphate-activated protein kinase (AMPK) (32). Xie et al. (33) showed that post-natal correction of the disorder was achieved in mice via a single administration of Cas9 and gRNA in an AAV9 vector, with significant reduction in left ventricular wall thickness, decreased myocardial glycogen content, normalization of the QRS width and PR intervals, and improvements in myofibril organization and ventricular function. Further work is required to translate these results to different types of PRKAG2 mutations and to larger animals to better define the corrected phenotype.
Long QT syndrome (LQTS), which predisposes individuals to life-threatening cardiac arrhythmias, would potentially be an attractive candidate for germline and somatic genome editing. Rare cases of LQTS are caused by mutations in genes that code for calmodulin (CALM1, CALM2, and CALM3), a protein that binds calcium and interacts closely with L-type calcium channels in cardiomyocytes. When this protein is over-expressed, it causes action potential prolongation. CRISPR interference is a novel method of modulating gene expression without permanently modifying the genome; gene expression is modified by using dCas9 (“dead” Cas9, which lacks endonuclease activity) along with a transcriptional activator or suppressor. Limpitikul et al. (34) cultured cardiomyocytes from induced pluripotent stem cells derived from a patient with a disease-causing CALM2 mutation, and demonstrated that these cells recapitulated the cellular phenotype of LQTS. Treatment with CRISPR interference significantly lowered levels of CALM2 mRNA, and calmodulin protein, and also reduced action potential duration. In addition, these investigators showed that expression of CALM1 or CALM3 could also be selectively reduced in this way (34). It remains to be seen whether this approach might be effective in vivo or applicable to other variants of LQTS or other cardiac conditions.
Marfan syndrome
Base editing techniques may be one way of more precisely correcting pathogenic single nucleotide mutations with perhaps fewer concerns about the inefficiencies of HDR and about off-target effects. Marfan syndrome is a connective tissue disorder associated with abnormalities in multiple organ systems, but for which morbidity and mortality are most closely tied to the risk of thoracic aortic aneurysm and aortic dissection. It is most often caused by autosomal dominant mutations in the gene FBN1, which codes for the extracellular matrix protein fibrillin-1 (35). Zeng et al. (36) recently identified an individual with Marfan syndrome who was heterozygous for a pathogenic T7498C mutation in the FBN1 gene and cultured human cells with an identical mutation in FBN1. After those cells were transfected with gRNA and BE3, sequencing revealed that 10 of 20 clones had been edited, 8 with a desirable C-to-T correction and 2 others with unwanted base pair conversions. The investigators then tested the technique in human embryos; zygotes produced via in vitro fertilization of donor oocytes with sperm from the same Marfan syndrome patient were subsequently microinjected with BE3 and gRNA. In 100% of the 7 treated embryos, the FBN1 T7498C mutation was corrected, compared with 50% of control embryos, although there was 1 unwanted base conversion in 1 of the treated embryos. No unintended corrections were discovered via screening of potential off-target sites in corrected embryos (36). The results provided proof-of-concept that base editing techniques might be applicable to the correction of pathogenic gene mutations in the human germline, especially if problems of off-target conversions could be more thoroughly resolved.
Ethical Considerations
Several ethical concerns must be considered with germline gene editing because any intended and unintended changes would be transmitted to subsequent generations. Mosaicism is also a cause of major concern. However, as shown by Ma et al. (6), mosaicism can be largely prevented if CRISPR is introduced before cell division starts. Furthermore, detailed analysis demonstrated no off-target effects. However, the gene edited in this case was only 4 base pairs long, and more off-target effects may occur when editing larger genes (37). Whether these off-target effects are automatically recognized and corrected in the human embryo is not known. Consequently, more studies need to confirm the absence of off-target effects in human genome editing, and to further develop novel techniques such as base editing that might allow for more precise mutation correction in select cases.
One can argue that as long as we have pre-implantation genetic diagnosis we do not need germline gene editing because we can select the unaffected embryos for implantation. However, this argument does not hold for polygenic diseases, or when both parents are homozygous for a gene variant. It has also been argued that applications of germline gene editing could widen inappropriately and be used for purposes other than therapy. However, we must address these concerns with effective policies rather than prevention of the development of potential therapies. International consensus and tight regulation will be critical to ensure that this technology is only used for necessary treatment purposes rather than creating “designer” babies or providing nonessential treatment. Indeed, recent reports suggest that CRISPR-Cas9 has already been used to create babies in China, sparking an international debate and stressing the immediate importance of setting up such regulatory systems (38). The earlier these deliberations begin, the better prepared society will be for such treatments when they become available.
Another concern is that the expense associated with germline gene editing and in vitro fertilization will result in the treatment only benefiting wealthy patients. Coverage by insurance could mitigate this concern, and governmental health care services should take this possibility into account when instituting future policies. In many instances, the cost of germline gene editing and in vitro fertilization would be significantly less than the lifelong pharmacological treatment of the condition, therefore making these treatments economically more attractive.
Finally, objections to germline gene editing on religious grounds will continue to exist and people with such objections can decline this therapy. Accordingly, the National Academy of Medicine has recommended that attempts to make gene therapy safe should continue (39).
Conclusions
It is rapidly becoming apparent that a wide variety of cardiovascular diseases may one day be curable using CRISPR-Cas9 or similar technology, including many that heretofore have been entirely untreatable. Germline genome editing promises to permanently resolve monogenic cardiovascular disorders for the offspring and subsequent generations of affected individuals. Although technically easier and likely to be ready for implementation in humans in the near future, this approach remains ethically controversial. Public debate and public policy determinations will need to proceed rapidly to allow decisions to be made regarding how and when these therapies may be used clinically. In addition, further technical matters will need to be more fully resolved, including those of long-term risks, off-target effects, mosaicism, and applicability to a wider variety of mutations and cardiovascular conditions.
Although currently beset by several technical challenges, and not yet past small animal models, somatic genome editing may also be useful for a variety of cardiovascular disorders. It potentially avoids ethical concerns about permanent editing of the germline and allows treatment of already diseased individuals. If technical challenges of Cas9-gRNA delivery (viral vector immune response, nonviral vector delivery, size of the vector) can be worked out in large animals and humans, then CRISPR-Cas9 may have a significant place in the treatment of a wide variety of disorders in which partial or complete gene knockout is desired (e.g., for PCSK9 and DMD). More challenging (and perhaps unachievable) will be knock in via HDR or base editing in nonproliferating cells. Off-target effects remain a concern in somatic genome editing as well, although small animal studies thus far have been reassuring. Some of the therapies mentioned in this review will be ready for small clinical trials in the near future (perhaps soonest in high-risk patients with hereditary lipid disorders).
Footnotes
The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
All authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and US Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC: Basic to Translational Scienceauthor instructions page.
References
- 1.Mali P., Yang L., Esvelt K.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders C., Niewoehner O., Duerst A., Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleinstiver B.P., Prew M.S., Tsai S.Q. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedland A.E., Baral R., Singhal P. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ribeiro L.F., Ribeiro L.F.C., Barreto M.Q., Ward R.J. Protein engineering strategies to expand CRISPR-Cas9 applications. Int J Genomics. 2018;2018:1652567. doi: 10.1155/2018/1652567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma H., Marti-Gutierrez N., Park S.W. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413–419. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 7.Schlossarek S., Mearini G., Carrier L. Cardiac myosin-binding protein C in hypertrophic cardiomyopathy: mechanisms and therapeutic opportunities. J Mol Cell Cardiol. 2011;50:613–620. doi: 10.1016/j.yjmcc.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Mearini G., Stimple D., Geertz B. Mybpc3 gene therapy for neonatal cardiomyopathy enables long-term disease prevention in mice. Nat Commun. 2014;5:5515. doi: 10.1038/ncomms6515. [DOI] [PubMed] [Google Scholar]
- 9.Long C., McAnally J.R., Shelton J.M. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Refaey M., Xu L., Gao Y. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ Res. 2017;121:923–929. doi: 10.1161/CIRCRESAHA.117.310996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amoasii L., Hildyard J.C.W., Li H. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitt J.P., Kamisago M., Asahi M. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 13.Kaneko M., Hashikami K., Yamamoto S., Matsumoto H., Nishimoto T. Phospholamban ablation using CRISPR/Cas9 system improves mortality in a murine heart failure model. PLoS One. 2016;11:e0168486. doi: 10.1371/journal.pone.0168486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George L.A., Sullivan S.K., Giermasz A. Hemophilia B gene therapy with a high-specific-activity factor IX variant. N Engl J Med. 2017;377:2215–2227. doi: 10.1056/NEJMoa1708538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rangarajan S., Walsh L., Lester W. AAV5-factor VIII gene transfer in severe hemophilia A. N Engl J Med. 2017;377:2519–2530. doi: 10.1056/NEJMoa1708483. [DOI] [PubMed] [Google Scholar]
- 16.Cohen J.C., Boerwinkle E., Mosley T.H., Jr., Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 17.Sabatine M.S., Giugliano R.P., Keech A.C. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 18.Ding Q., Strong A., Patel K.M. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Raghavan A., Chen T. CRISPR-Cas9 targeting of PCSK9 in human hepatocytes in vivo-brief report. Arterioscler Thromb Vasc Biol. 2016;36:783–786. doi: 10.1161/ATVBAHA.116.307227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ran F.A., Cong L., Yan W.X. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaudelli N.M., Komor A.C., Rees H.A. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;55:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chadwick A.C., Wang X., Musunuru K. In vivo base editing of PCSK9 (proprotein convertase subtilisin/kexin type 9) as a therapeutic alternative to genome editing. Arterioscler Thromb Vasc Biol. 2017;37:174–177. doi: 10.1161/ATVBAHA.117.309881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musunuru K., Pirruccello J.P., Do R. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dewey F.E., Gusarova V., Dunbar R.L. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N Engl J Med. 2017;377:211–221. doi: 10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chadwick A.C., Evitt N.H., Lv W., Musunuru K. Reduced blood lipid levels with in vivo CRISPR-Cas9 base editing of ANGPTL3. Circulation. 2018;137:975–977. doi: 10.1161/CIRCULATIONAHA.117.031335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuster J.J., Walsh K. Somatic mutations and clonal hematopoiesis: unexpected potential new drivers of age-related cardiovascular disease. Circ Res. 2018;122:523–532. doi: 10.1161/CIRCRESAHA.117.312115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaiswal S., Natarajan P., Silver A.J. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;37:111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sano S., Oshima K., Wang Y., Katanasaka Y., Sano M., Walsh K. CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. Circ Res. 2018;123:335–341. doi: 10.1161/CIRCRESAHA.118.313225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn J.D., Smith A.R., Patel M.C. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Christiansen J.P., French B.A., Klibanov A.L., Kaul S., Lindner J.R. Targeted tissue transfection with ultrasound destruction of plasmid-bearing cationic microbubbles. Ultrasound Med Biol. 2003;29:1759–1767. doi: 10.1016/s0301-5629(03)00976-1. [DOI] [PubMed] [Google Scholar]
- 32.Porto A.G., Brun F., Severini G.M. Clinical spectrum of PRKAG2 syndrome. Circ Arrhythm Electrophysiol. 2016;9:e003121. doi: 10.1161/CIRCEP.115.003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie C., Zhang Y.P., Song L. Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res. 2016;26:1099–1111. doi: 10.1038/cr.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Limpitikul W.B., Dick I.E., Tester D.J. A precision medicine approach to the rescue of function on malignant calmodulinopathic long-QT syndrome. Circ Res. 2017;120:39–48. doi: 10.1161/CIRCRESAHA.116.309283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landis B.J., Veldtman G.R., Ware S.M. Genotype-phenotype correlations in Marfan syndrome. Heart. 2017;103:1750–1752. doi: 10.1136/heartjnl-2017-311513. [DOI] [PubMed] [Google Scholar]
- 36.Zeng Y., Li J., Li G. Correction of the Marfan syndrome pathogenic FBN1 mutation by base editing in human cells and heterozygous embryos. Mol Ther. 2018;26:2631–2637. doi: 10.1016/j.ymthe.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Y., Foden J.A., Khayter C. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.How to respond to CRISPR babies? Available at: https://www.nature.com/magazine-assets/d41586-018-07634-0/d41586-018-07634-0.pdf. Accessed January 9, 2019.
- 39.Human Genome Editing. Available at: https://www.nap.edu/catalog/24623/human-genome-editing-science-ethics-and-governance. Accessed January 2019.