 A new series of quinolone-isoniazid hybrid compounds were designed, synthesised and studied for their potential anti-mycobacterial tuberculosis activity in vitro.
A new series of quinolone-isoniazid hybrid compounds were designed, synthesised and studied for their potential anti-mycobacterial tuberculosis activity in vitro.
Abstract
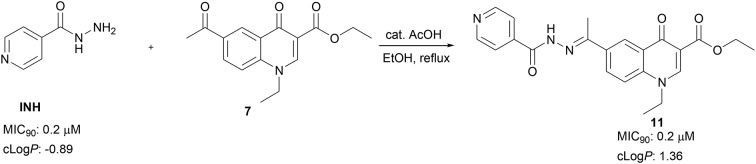
Herein, we propose novel quinolones incorporating an INH moiety as potential drug templates against TB. The quinolone-based compounds bearing an INH moiety attached via a hydrazide–hydrazone bond were synthesised and evaluated against Mycobacterium tuberculosis H37Rv (MTB). The compounds were also evaluated for cytotoxicity against HeLa cell lines. These compounds showed significant activity (MIC90) against MTB in the range of 0.2–8 μM without any cytotoxic effects. Compounds 10 (MIC90; 0.9 μM), 11 (MIC90; 0.2 μM), 12 (MIC90; 0.8 μM) and compound 15 (MIC90; 0.8 μM), the most active compounds in this series, demonstrate activities on par with INH and superior to those reported for the fluoroquinolones. The SAR analysis suggests that the nature of substituents at positions –1 and –3 of the quinolone nucleus influences anti-MTB activity. Aqueous solubility evaluation and in vitro metabolic stability of compound 12 highlights favourable drug-like properties for this compound class.
Introduction
Tuberculosis (TB), an air borne infectious disease,1 has been a scourge to the human race for thousands of years.2 The disease is caused by the bacterium, Mycobacterium tuberculosis (MTB),3 with infections resulting in both latent TB and active pulmonary TB.4 Approximately 2–3 billion people worldwide carry the latent form of the disease,5 and about 10% of this group will eventually develop active pulmonary TB in their life time.6 Risk factors associated with conversion of latent to the active form of the disease include HIV infection, diabetes, smoking, malnutrition,7 and rheumatoid arthritis.8 It is important to note that the latent form of TB is void of signs and symptoms of the disease.
An estimated 10.0 million people worldwide suffered from active pulmonary TB, and about 1.3 million deaths were recorded in 2017. In the same year, an additional 300 000 people died from TB and HIV co-infections.9 The recommended treatment regimen for TB consists of a cocktail of four drugs; isoniazid (INH), ethambutol, pyrazinamide, and rifampicin,10 which must be administered daily at a total dose of approximately 5 grams for two months followed by isoniazid and rifampicin daily for another four months.11 These drugs act on different targets within MTB.12 Despite this combination therapy approach, the present scenario of TB treatment has been aggravated by the emergence and spread of several forms of drug-resistant MTB.13 Three resistance types, classified based on drug resistance profiles, includes; multi-drug resistant (MDR) strain, which is resistant to isoniazid and rifampicin but susceptible to fluoroquinolones,14 extensively drug resistant (XDR) strain, which is resistant to isoniazid, rifampicin, fluoro-quinolones and at least one of the three second line injectable anti-TB drugs (amikacin, kanamycin and capreomycin),15 and totally drug resistant (XXDR or TDR) strain, which is resistant to all anti-TB agents except newly approved agents such as bedaquiline.16 In 2017, there were 457 560 cases of MDR-TB and 6–11% of these cases were XDR-TB. There is low treatment success rate for drug(s) resistant TB, reported to be 55% globally.9 Poor patient adherence, partly due to the cocktail of drugs and lengthy period required to take them,17 has been attributed to the drug resistance crisis.18 Also suggested to be implicated in the emergence of resistant strains is the issue of mis-matched pharmacokinetics (PK) and pharmacodynamic (PD) properties of the various drugs employed in the first line regimen, creating a scenario wherein bacilli in different compartments and/or stages are often not exposed to lethal concentrations of some drugs.19
In addition, the first line treatment regimen is not very safe: INH and rifampicin – the main drugs in the regimen – have been reported to cause hepatotoxicity and other serious side effects.20
The cumbersome first line treatment and the growing resistance necessitate the adoption and implementation of new measures to treat TB effectively and to limit the rising prevalence of drug-resistant disease. To this effect, several strategies, including drug repurposing,21 derivatization of existing drugs,22 mechanism and structure based designed, high-throughput screening,23 all aimed particularly at the development of new molecules exhibiting unique and novel modes of action, have been exploited.24 This has culminated in new drug candidates (bedaquiline, pretomanid, sutezolid, Q203, SQ109, PBTZ169) entering preclinical and clinical studies.25 Bedaquiline acts on ATP synthase and interferes with energy production leading to death of both dormant and replicating bacilli.26 SQ109 target MmpL3 and prevent the export of trehalose monomycolate which is required for mycolic acid synthesis. This ultimately leads to disruption of cell wall synthesis.27 Although novel in their mode of actions, these new compounds will eventually enter into combinations wherein mis-matched PK/PD is likely to be an issue. Another strategy worth exploiting is hybridizing chemical motifs with experimentally proven anti-TB molecules to generate new molecular entities with the hope of interfering with multiple biochemical pathways within MTB;28 this might also slow the emergence of drug-resistance. Compounds containing a hydrazide–hydrazone structural motif (1, Fig. 1) have been reported to exhibit anti-TB activity, as well as activities against other infectious diseases like malaria and trypanosomiasis,29 thereby demonstrating an inherent ability to interfere with essential biochemical pathways within infectious agents.
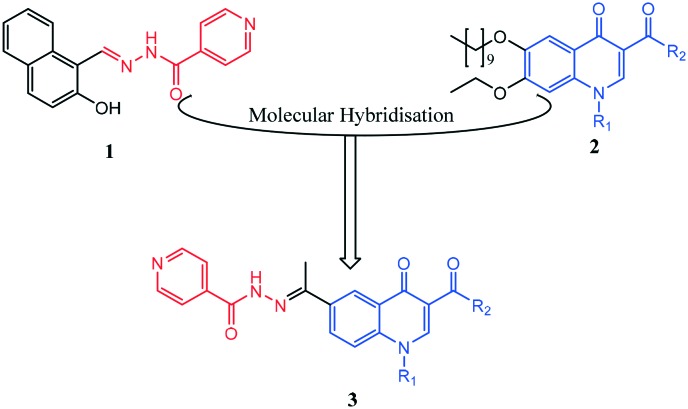
Fig. 1. Structural design strategy. (1) Iron chelating hydrazide–hydrazone containing compound,36 (2) quinolones exhibiting anti-TB activity,35 and (3) target hybrid.
Compound 1 has been reported to exhibit anti-TB activity by inhibiting a novel target, methionine aminopeptidase.30
On the other hand, the quinolone nucleus (2, Fig. 1) is a versatile and efficacious anti-TB agent.31 Previous literature highlights this compound class as a prominent source of anti-TB agents that can be tailored via structural modification to generate novel compounds capable of targeting different biochemical pathways within the bacillus. The fluoroquinolones, exemplified by ciprofloxacin, are an important second-line treatment option for treating multidrug-resistant MTB32 and work by inhibiting DNA gyrase and topoisomerase IV.33 Hong et al. demonstrated that incorporating lipophilic units at position –2 of the quinolone ring leads to compounds that target the QcrB subunit of the cytochrome bc1 complex in the mycobacterial electron transport chain.34 Moreover, another set of quinolone molecules (2), with potent anti-TB activity has been reported.35 This set differs structurally from fluoroquinolones and quinolones reported by Hong et al. and, as such, may present us with a novel mode of action not yet reported with other quinolones. To exploit the hybridisation strategy in TB chemotherapy, our approach is to imprint the hydrazide–hydrazone structural motif of compound 1 (Fig. 1) onto a reputable anti-biotic class of compound, the quinolone, with intent to generate quinolone-isoniazid hybrid molecules 3 with potential antimycobacterial activity against MTB H37Rv strain.
Considering the above observations and our efforts to identify novel anti-TB agents that interfere with multiple biochemical pathways,35 a series of quinolone aroylhydrazone conjugates was synthesised based on quinolone pharmacophore, and the resulting compounds screened in vitro against MTB to determine their anti-TB potential. In addition, these compounds were also screened against a HeLa cell lines to investigate potential cytotoxicity.
Aroylhydrazones have been established to exhibit a wide range of biological properties, which are usually attributed to their ability to chelate copper or iron.36 Iron chelators have been shown to retard or completely halt MTB growth.37 The quinolone scaffold is a notable compound class that can be chemically tailored to probe different cellular targets within MTB. By imprinting the aroylhydrazones fragment of compound 1 on to new quinolone structures, we hoped to generate novel compounds capable of inhibiting the growth of MTB. The aroyl hydrazine utilised for synthesis of target aroylhydrazones is INH, a known anti-TB agent.4 Besides forming a structural motif capable of eliciting an additional pharmacological response, functionalisation of the hydrazine unit in INH into aroylhydrazone was predicted to avoid the fast acetylation of INH which has been implicated in its toxicity38 and, in addition, incorporates INH in to a molecular framework of higher lipophilicity. Higher lipophilicity is very much desirable to effectively target MBT within lung granulomas.38,39
Results and discussion
Chemistry
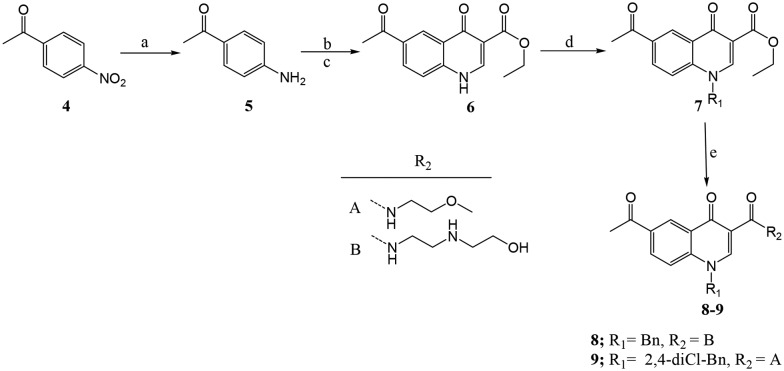
Target compounds were achieved following the synthetic routes presented in Schemes 1 and 2. The p-nitroacetophenone (4) was reduced to p-aminoacetophenone (5) using reduced iron powder and acetic acid under ultra sound conditions. Treatment of 5 in acetonitrile with diethyl ethoxymethylenemalonate under reflux generated condensed methylenemalonate ester, which was isolated upon in vacuo evaporation of solvents. Cyclisation of methylenemalonate esters was carried out in boiling diphenyl ether at 245–250 °C for five minutes to afford quinolone (6) in 55% yield (Scheme 1).
Scheme 1. aReagents and conditions: (a) reduced Fe powder, AcOH; (b) acetonitrile, diethyl ethoxylmethylenemalonate, reflux 12 h; (c) diphenyl ether, 245–250 °C, 5 min; (d) K2CO3, CHCl3/THF (2 : 1), alkylhalide (1.2 equiv.), 12 h; (e) amine (5 equiv.), DBU (1.2 equiv.), CHCl3, reflux 12 h.
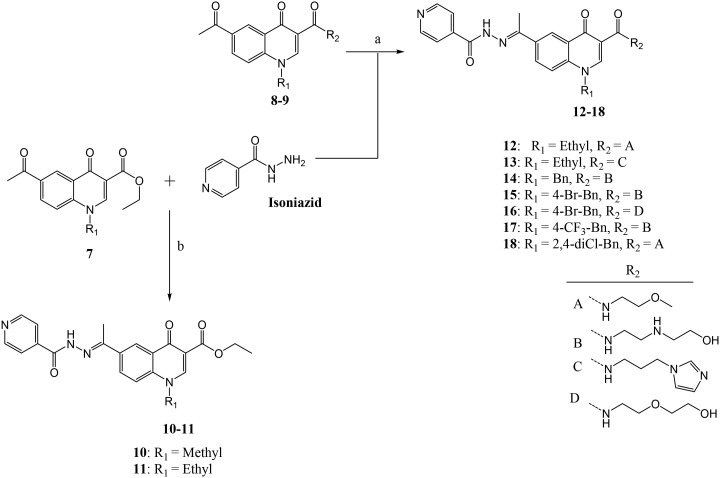
Scheme 2. aReagents and conditions: (a) and (b) isoniazid, AcOH (cat), EtOH, reflux overnight.
The temperatures and reaction times needed to be carefully controlled during the cyclisation process. We observed that cyclisation failed to occur at temperatures below 240 °C, and temperatures above 255 °C led to decarboxylation of the ester group. Also heating at 245–250 °C for more than five minutes produced a mixture of two compounds; one having the ester group intact and a decarboxylated compound. Treatment of compound 6 with K2CO3 and alkyl/arylhalides under reflux overnight resulted in N-alkylation of the secondary amine. After reaction completion as indicated by TLC, the reaction mixture was filtered and the filtrate evaporated to dryness in vacuo and later washed with petroleum ether to afford intermediate 7 (in 50% yield), the quinolone fragment containing a ketone required for further functionalisation with aroylhydrazine (isoniazid). The ethylester functionality in 7 underwent aminolysis in the presence of respective amines and DBU as a base under refluxing conditions to generate amide containing compounds exemplified by 8 and 9 in 40–60% yields.
Treatment of compounds 7 and 8 in ethanol with INH and catalytic amount of acetic acid under reflux overnight resulted in compounds 10–18 in 30–70% yields, the compounds precipitated out of the reaction mixture and were isolated by filtration (Scheme 2). All compounds were confirmed using 1H and 13C NMR, and HRMS. The 1H NMR spectra of all compounds show a singlet at ca. 2.5 ppm, which is assigned to the –CH3– of methylketone or methylimine unit. Apart from compounds 10 and 11, having an ethylester at position –3, the 1H NMR of all compounds 10–18 show a singlet at ca. 11 ppm on their respective 1H NMR spectra, and this signal suggests the successful formation of hydrazone–hydrazide bond (–C( O)NH–N–).
Comparing the 13C NMR spectra of compounds 8 and 9 against compounds 10–18 further confirmed this observation. The 13C NMR spectra of compounds 8 and 9 showed a peak at ca. 197 ppm which was assigned to the –C O unit of the methylketone, and its disappearance and the appearance of a new peak at ca. 111 ppm in compounds 10–18 further confirmed the transformation of a carbonyl unit into an imine. The purity of all compounds was determined to be greater than 95% using preparative HPLC.
Evaluation of cytotoxicity
Target compounds were screened in vitro against human cervix adenocarcinoma (HeLa) cell lines to investigate potential cytotoxicity effects at 20 μM. Emetine (which induces cell apoptosis) was used as positive control drug and yielded an IC50 value of 0.015 μM. Compounds exhibiting <50% cell viability were considered to pose cytotoxicity risk, while compounds exhibiting >50% cell viability were assumed to have IC50 values ≥20 μM and as such pose little cytotoxicity risk. None of the compounds in this study reduced HeLa cell viability to below 75% during a 24 hour incubation (Fig. 2), which suggested they possessed little overt cytotoxicity risk at the concentrations used for anti-MTB evaluation.
Fig. 2. Percentage HeLa cell viability after incubation with test compounds at 20 μM for 24 hours. The values are the mean ±SD of experiments performed in duplicate, and none of the compounds showed any cytotoxicity effects. The SD values were calculated using the expression.40.
Antitubercular activity against Mtb H37Rv
Target compounds 10–18 and quinolone fragments 8 and 9 were evaluated for antimycobacterial activity in vitro. The compounds were screened in a broth microdilution assay against Mycobacterium tuberculosis H37Rv, drug-susceptible laboratory strain, with rifampicin and INH included as standards. The antimycobacterial activities are reported as minimum inhibitory concentration (MIC90) required to inhibit 90% of mycobacterial growth. Antimycobacterial activities of target compounds are summarised in Table 1. All tested compounds exhibited antimycobacterial activity, with most of the compounds exhibiting potent activity, having MIC90 values less 3 μM. This activity profile is comparable to those of INH and fluoroquinolones currently deployed in the treatment of TB.
Table 1. In vitro antimycobacterial activity, cytotoxicity and calculated clog P of target compounds in comparison with standard drugs.
| Compound | MIC90 (μM) | clog P a |
| H37Rv | ||
| 8 | 43.4 | 0.5 |
| 9 | 125 | 2.6 |
| 10 | 0.9 | 0.8 |
| 11 | 0.2 | 1.3 |
| 12 | 0.8 | 0.1 |
| 13 | 7.8 | 0.2 |
| 14 | 2.0 | 0.5 |
| 15 | 0.8 | 1.3 |
| 16 | 2.7 | 1.5 |
| 17 | 1.8 | 1.1 |
| 18 | 4.8 | 2.6 |
| INH | 0.2 | –0.8 |
| Rif | 0.06 | ND |
aclog P values are calculated from ACD Chemsketch freeware version 12.0. INH = isoniazid, Rif = rifampicin, ND = not determined,
As has been noted earlier that the nature of substituents around the quinolone ring determines activity as well as target within the bacterium, to this effect, our library was designed to incorporate target compounds bearing different substituents at positions –1 and –3 of the quinolone ring. SAR analysis of this series suggested that an ethylester at position –3 of the quinolone nucleus promoted activity better than amides, this was evident when comparing compound 11 (MIC90; 0.2 μM) bearing an ethylester with compound 12 (MIC90; 0.8 μM) and compound 13 (MIC90; 7.8 μM) – both having an amide at position –3 of the quinolone ring. Furthermore, comparing compounds 12 (MIC90; 0.8 μM) and 18 (MIC90; 4.8 μM), which differ only on the nature of substituent at position –1 of the quinolone ring, suggested that the ethyl unit at this position was favoured for anti-TB activity compared to a phenyl ring. It is, however, important to note that compound 15 which bears a substituted phenyl ring and an amide at position –1 and –3, respectively of the quinolone ring demonstrated potent activity (MIC90; 0.8 μM) comparable to that of INH and superior to that reported for most fluoroquinolones. The substitution pattern on the phenyl ring also seemed to greatly influence activity, with para bromo substituents giving the best activity. This was evident when comparing the structure and activity of 15 (MIC90; 0.8 μM) against compounds 14 (MIC90; 2.0 μM) and 17 (MIC90; 1.8 μM).
Comparison of the structure and activity of compound 8 (MIC90; 43.4 μM) against the structure and activity of compound 14 (MIC90; 2.0 μM) suggested that anti-TB activity increased significantly with the incorporation of the hydrazone–hydrazide unit. This was also evident when comparing compound 9 (MIC90; 125 μM) against compound 18 (MIC90; 4.8 μM). Comparing compounds 12 (MIC90; 0.8 μM) and 13 (MIC90; 7.8 μM) suggested that the nature of the amide moiety at position –3 greatly affected anti-TB activity, an observation also evident when comparing compounds 15 (MIC90; 0.8 μM) and 16 (MIC90; 2.7 μM), which differ only in their amide moiety at position –3.
Aqueous solubility and in vitro microsomal stability
Compound 12 having an MIC90 value of 0.8 μM and clog P of 0.1 was selected for aqueous solubility evaluation in phosphate buffered saline (PBS) at pH 6.5. The compound showed moderate aqueous solubility and was then subjected to metabolic turnover stability studies in human, rat, and mouse liver microsomes (Table 2). The data suggested that compound 12 was stable across the three species, with more than 70% of the compounds remaining after 80–150 minutes (t1/2 >150 min). This good stability profile was further confirmed by a low hepatic extraction (EH) ratio of <0.5 across the three species.
Table 2. Aqueous solubility (Sw) and in vitro metabolic stability evaluation of compound 12 in human, rat and mouse liver microsomes.
| Compound | MIC90 (μM) | b S w (μM) pH 6.5 |
a
HLM/RLM/MLM |
|||
| % remaining | CLint μg min–1 mg–1 protein | t 1/2 (min) | c E H | |||

|
0.8 | 95 | 90.6/87/77 | 13.3/18.7/35.3 | >150/>150/79.7 | <0.42/<0.3/<0.49 |
aHLM/RLM/MLM are human, rat and mouse liver microsomes.
bAqueous solubility estimated by nephelometry.
cPredicted hepatic extraction (EH) ratio based on in vitro intrinsic clearance (CLint).
Conclusions
This work presents novel quinolones incorporating an INH moiety through a hydrazide–hydrazone functional unit. Target compounds were realised through a simple hybridisation approach and were tested against the HeLa cell line and H37Rv strain of MTB to evaluate for cytotoxicity and anti-TB potentials, respectively. While none of the compounds showed a potential cytotoxicity risk, they all exhibited potent anti-TB activity (MIC90 in the range of 0.2–8 μM). An ethylester at position –3 seems to promote activity better than amides, while an aliphatic unit at position –1 seems to favour anti-TB activity compared to having a benzyl unit at this position. Besides having an in vitro activity profile comparable to INH, our hit compounds present an advantage over INH in that they have higher lipophilicity and lack the free hydrazine unit – the feature to which INH toxicity has been attributed. In summary, these observations support the design strategy adopted and suggest that the approach is worth further exploitation in search of more potent and safe anti-TB agents.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors acknowledge the financial support of the National Research Foundation of South Africa (SDK, 107270), Rhodes University for a Postdoctoral Fellowship and RC grant (RMB) as well as Rhodes University Sandisa Imbewu (SDK, HCH) towards this research. The cytotoxicity component of the project was funded by the South African Medical Research Council (SAMRC) with funds from National Treasury under its Economic Competitiveness and Support Package awarded to HCH. All TB screening work was conducted in the MMRU (UCT) with the support of the SAMRC through the Strategic Health Innovation Partnerships (SHIP) initiative (to DFW). Aqueous solubility studies and in vitro metabolic stability studies were performed at H3D, University of Cape Town.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8md00480c
References
- Tomás B. A., Pell C., Cavanillas A. B., Solvas J. G., Pool R., Roura M. PLoS One. 2013;12:e82440. doi: 10.1371/journal.pone.0082440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. K., Mohan A. Indian. J. Med. Res. 2013;137:455–493. [PMC free article] [PubMed] [Google Scholar]
- Bailo R., Bhatt A., Aínsa J. A. Biochem. Pharmacol. 2015;96:159–167. doi: 10.1016/j.bcp.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Ai J. W., Ruan Q. L., Liu Q. H., Zhang W. H. Emerging Microbes Infect. 2016;5:e10. doi: 10.1038/emi.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I., Gagneux S. PLoS Pathog. 2009;5:e1000600. doi: 10.1371/journal.ppat.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza L. D., Ogusku M. M., Sadahiro A., Pontillo A. Infect., Genet. Evol. 2016;41:240–244. doi: 10.1016/j.meegid.2016.04.015. [DOI] [PubMed] [Google Scholar]
- Fogel N. Tuberculosis. 2015;95:527e531. doi: 10.1016/j.tube.2015.05.017. [DOI] [PubMed] [Google Scholar]
- Ai J. W., Zhang S., Ruan Q. L., Yu Y. Q., Zhang B. Y., Liu Q. H., Zhang W. H. J. Rheumatol. 2015;42:2229–2237. doi: 10.3899/jrheum.150057. [DOI] [PubMed] [Google Scholar]
- WHO, Global Tuberculosis report 2018, http://www.who.int/tb/publications/global_report/en/accessed on 10.11.18.
- Arbex M. A., Marília C. L., Siqueira H. R., Mello F. A. J. Bras. Pneumol. 2010;36:626–640. doi: 10.1590/s1806-37132010000500016. [DOI] [PubMed] [Google Scholar]
- Onajole O., Pieroni M., Tipparaju S., Lun S., Stec J., Chen G., Gunosewoyo H., Guo H., Ammerman N., Bishai W., Kozikowski A. J. Med. Chem. 2013;56:4093–4103. doi: 10.1021/jm4003878. [DOI] [PubMed] [Google Scholar]
- Marriner G. A., Nayyar A., Uh E., Wong S. Y., Mukherjee T., Via L. E., Carroll M., Edwards R. L., Gruber T. D., Choi I., Lee J., Arora K., England K. D., Boshoff H. M., Barry III C. L. Top. Med. Chem. 2011;7:47–124. [Google Scholar]
- Tiwari R., Möllmann U., Cho S., Franzblau S. G., Miller P. A., Miller M. J. ACS Med. Chem. Lett. 2014;5:587–591. doi: 10.1021/ml500039g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T., Jenkins H. E., Lu C., McLaughlin M., Floyd K., Zignol M. Drug Resist. Updates. 2014;17:105–123. doi: 10.1016/j.drup.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor R., Akhter S., Rahman F., Munshi S. K., Kamal S. M., Feroz F. J. Infect. Chemother. 2013;19:243–248. doi: 10.1007/s10156-012-0490-8. [DOI] [PubMed] [Google Scholar]
- Velayati A., Farnia P., Masjedi M. Int. J. Mycobact. 2013;2:71–72. doi: 10.1016/j.ijmyco.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Boru C. G., Shimels T., Bilal A. I. J. Infect. Public Health. 2017;10:527–533. doi: 10.1016/j.jiph.2016.11.018. [DOI] [PubMed] [Google Scholar]
- Shukla M., Sharma A., Jaiswal S., Lal J. Expert Opin. Drug Metab. Toxicol. 2016;12:765–778. doi: 10.1080/17425255.2016.1183643. [DOI] [PubMed] [Google Scholar]
- Srivastava S., Pasipanodya J., Meek C., Leff R., Gumbo T. J. Infect. Dis. 2011;204:1951–1959. doi: 10.1093/infdis/jir658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Sasi P., Rai G., Gupta V. H., Amarapurkar D., Wangikar P. P. Med. Chem. Res. 2011;20:1611–1615. [Google Scholar]
- Sharma D., Dhuriya Y., Deo N., Bisht D. Front. Microbiol. 2017;8:2452. doi: 10.3389/fmicb.2017.02452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles D. J. Trends Microbiol. 1997;5:379. doi: 10.1016/S0966-842X(97)01128-1. [DOI] [PubMed] [Google Scholar]
- Mdluli K., Kaneko T., Upton A. Cold Spring Harbor Perspect. Med. 2015;5:a021154. doi: 10.1101/cshperspect.a021154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coxon G. D., Cooper C. B., Gillespie S. H., McHugh T. D. J. Infect. Dis. 2012;15:S258–S264. doi: 10.1093/infdis/jis191. [DOI] [PubMed] [Google Scholar]
- Lohrasbi V., Talebi M., Bialvaei A. Z., Fattorini L., Drancourt M., Heidary M., Darban-Sarokhalil D. Tuberculosis. 2018;109:17–27. doi: 10.1016/j.tube.2017.12.002. [DOI] [PubMed] [Google Scholar]
- Andries K., Verhasselt P., Guillemont J., Gohlmann H., Neefs J. M., Winkler H., Van Gestel J., Timmerman P., Zhu M., Lee E., Williams P., de Chaffoy D., Huitric E., Hoffner S., Cambau E., Truffot-Pernot C., Lounis N., Jarlier V. Science. 2005;307:223. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- Tahlan K., Wilson R., Kastrinsky D., Arora K., Nair V., Fischer E., Barnes S., Walker J., Alland D., Barry 3rd C., Boshoff H. Antimicrob. Agents Chemother. 2012;56:1797–1809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Majewski M. W., Tiwari R., Miller P. A., Cho S., Franzblau S. G., Miller M. J. Bioorg. Med. Chem. Lett. 2016;26:2068–2071. doi: 10.1016/j.bmcl.2016.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Velezheva V., Brennan P., Ivanov P., Kornienko A., Lyubimov S., Kazarian K., Nikonenko B., Majorov K., Apt A. Bioorg. Med. Chem. Lett. 2016;26:978–985. doi: 10.1016/j.bmcl.2015.12.049. [DOI] [PubMed] [Google Scholar]
- Popiołek L. Med. Chem. Res. 2017;26:287–301. doi: 10.1007/s00044-016-1756-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S., Aniemeke E., Ha N., Chong C., Gu P., Zhou J., Zhang Y., Graviss E., Liu J., Olaleye O. Tuberculosis. 2016;101:S73–S77. doi: 10.1016/j.tube.2016.09.025. [DOI] [PubMed] [Google Scholar]
- Asif M. Sci. Int. 2013;1:336–342. [Google Scholar]
- Tiberi S., Scardigli A., Centis R., D'Ambrosio L., Muñoz-Torrico M., Salazar-Lezama M. A., Spanevello A., Visca D., Zumla A., Migliori G. B., Luna J. A. Int. J. Infect. Dis. 2017;56:181–184. doi: 10.1016/j.ijid.2016.10.026. [DOI] [PubMed] [Google Scholar]
- Aldred K. J., Kerns R. J., Osheroff N. Biochemistry. 2014;53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W. D., Gibbons P. D., Leung S. C., Amewu R., Stocks P. A., Stachulski A., Horta P., Cristiano M. S., Shone A. E., Moss D., Ardrey A., Sharma R., Warman A. J., Bedingfield P. P., Fisher N. E., Aljayyoussi G., Mead S., Caws M., Berry N. G., Ward S. A., Biagini G. A., O'Neill P. M., Nixon G. L. J. Med. Chem. 2017;60:3703–3726. doi: 10.1021/acs.jmedchem.6b01718. [DOI] [PubMed] [Google Scholar]
- Beteck R. M., Seldon R., Coertzen D., van der Watt M. E., Reader J., Mackenzie J. S., Lamprecht D. A., Abraham M., Eribez K., Müller J., Rui F., Zhu G., de Grano R. V., Williams I. D., Smit F. J., Steyn A. J., Winzeler E. A., Hemphill A., Birkholtz L. M., Warner D. F., N'Da D. D., Haynes R. K. Commun. Chem. 2018;1:62. [Google Scholar]
- Hruskov K., Potuckov E., Hergeselov T., Liptakov L., Haskov P., Mingas P., Kovaríkov P., Simunek T., Vavrov K. Eur. J. Med. Chem. 2016;120:97–110. doi: 10.1016/j.ejmech.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Dragset M. S., Poce G., Alfonso S., Padilla-Benavides T., Ioerger T. R., Kaneko T., Sacchettini J. C., Biava M., Parish T., Argüello J. M., Steigedal M., Rubina E. J. Antimicrob. Agents Chemother. 2015;59:2256–2264. doi: 10.1128/AAC.05114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumara K., Khare G., Beena, Kidwai S., Tyagi A. K., Singh R., Rawat D. S. Med. Chem. Commun. 2015;6:131–137. [Google Scholar]
- Machado D., Girardini M., Viveiros M., Pieroni M. Front. Microbiol. 2018;9:1367. doi: 10.3389/fmicb.2018.01367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synek V. Accredit. Qual. Assur. 2008;13:335–337. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.