Abstract
Coccidioidomycosis is an emerging fungal infection in Washington, USA, and the epidemiology of the disease in this state is poorly understood. We used whole-genome sequencing to differentiate locally acquired cases in Washington on the basis of the previously identified phylogeographic population structure of Coccidioides spp. Clinical isolates from coccidioidomycosis cases involving possible Washington soil exposure were included. Of 17 human infections with epidemiologic evidence of possible local acquisition, 4 were likely locally acquired infections and 13 were likely acquired outside Washington. Isolates from locally acquired cases clustered within the previously established Washington clade of C. immitis. Genetic differences among these strains suggest multiple environmental reservoirs of C. immitis in the state.
Keywords: Coccidioides, coccidioidomycosis, valley fever, fungi, fungal infections, whole-genome sequencing, surveillance, public health, Washington, United States
Coccidioidomycosis, also known as Valley fever, is a disease of growing public health concern and is caused by 2 closely related fungal species, Coccidioides immitis and C. posadasii. Coccidioides spp. are dimorphic, forming mycelia in soil and arthroconidia capable of infecting humans and certain other mammals, and spherules in mammalian tissue (1). This mycosis causes a wide spectrum of conditions, ranging from asymptomatic infection and mild pulmonary disease to severe pulmonary and disseminated disease, which can be life-threatening (1,2). Infection is generally caused by inhalation of pathogenic arthroconidia from disturbed soil or dust, such as through occupational or recreational activities or weather events that raise dust (2–7).
Coccidioides spp. are endemic to warm, arid regions of the Western Hemisphere that have low rainfall (1,8). The 2 species have colonized different geographic locations: C. posadasii is largely found in Arizona, Texas, Mexico, and Central and South America; and C. immitis is primarily found in central and southern California. Coccidioidomycosis causes a major burden of disease in many of these areas; the annual incidence in Arizona is >75 cases/100,000 population (9). Case counts have recently increased in California; >5,300 cases were reported in 2016 (13.7 cases/100,000 population (10). Climate suitability projections predict expansion of the suitable environment for coccidioidomycosis and increasing incidence in areas that already sustain Coccidioides growth (11).
During 2010–2011, three cases of coccidioidomycosis were identified and reported in southeastern Washington (7). Subsequently, Coccidioides DNA and viable propagules were isolated from soil at a location of suspected exposure for 1 of these cases (12,13). Whole-genome sequencing (WGS) analysis of the recovered isolates demonstrated near identical genetic identity between soil and clinical isolates, further confirming endemic presence of infectious C. immitis in Washington (13).
Public health surveillance for coccidioidomycosis was implemented statewide in Washington in April 2014. Before 2014, cases of coccidioidomycosis were reported sporadically, but no standard reporting procedure existed. Since 2014, the number of reported cases of coccidioidomycosis has increased each year; reported cases in Washington suspected to be locally acquired have higher rates of hospitalization and death compared with cases from other disease-endemic regions (14).
Because most persons infected with Coccidioides spp. are asymptomatic or have only mild illness, and most illness self-resolves, we believe that only the most severe cases of coccidioidomycosis with exposure in Washington are being identified and reported to public health authorities. In addition, Coccidioides spp. infections might be asymptomatic or only manifest as subclinical disease until a reactivation or complication develops later. Also, most residents of Washington have some travel history to historically disease-endemic areas, such as Arizona, California, or Mexico. These findings create a challenge in differentiating between autochthonous and imported cases of coccidioidomycosis, which is a useful distinction for public health surveillance in Washington.
To better determine the nature of exposure for reported cases of coccidioidomycosis in Washington, the Washington Department of Health, in collaboration with the Centers for Disease Control and Prevention (CDC; Atlanta, GA, USA) and the Translational Genomics Research Institute (Flagstaff, AZ, USA), initiated enhanced surveillance by incorporating WGS of clinical isolates aimed at determining the geographic origins of Coccidioides spp. strains. This approach is based on previous population structure studies providing evidence that C. immitis and C. posadasii have well-defined geographic structures (15–19). Subsequently, Engelthaler et al. demonstrated that isolates can generally be assigned to the specific geographic populations on the basis of WGS analysis (20). This approach is consistent with a general trend in molecular epidemiology that uses WGS and a One Health approach to ascertain clusters or outbreaks of bacterial, fungal, viral, and parasitic diseases (21–23).
We report WGS analyses of isolates from 17 recent coccidioidomycosis cases in Washington, and demonstrate that 13 (76%) cases involved coccidioidomycosis most likely acquired outside Washington; 4 cases (24%) likely involved local acquisition. We discuss the utility of the WGS method to enhance epidemiologic surveillance.
Materials and Methods
Cases and Isolates
We identified coccidioidomycosis cases through passive reporting to local health jurisdictions from healthcare providers in Washington and laboratories performing testing for residents of Washington. Confirmed cases, as classified according to the Council of State and Territorial Epidemiologists case definition (24), reported during 2014–2017 were included in the analysis. Reported case-patients were interviewed by local health jurisdictions to determine clinical course, travel history, and any potential soil or dust exposures. Medical records were requested when possible for complete data abstraction to case reporting forms. When the exposure was suspected to have occurred in the disease-endemic area of Washington or exposure history was unknown, available clinical isolates were sent to CDC for confirmation.
At CDC, all isolates were grown on brain heart infusion (BHI) agar at 25°C for 10 days. Genomic DNA was extracted by using the DNeasy Blood and Tissue kit (QIAGEN, https://www.qiagen.com/us) according to the manufacturer’s recommendations. Genomic DNA was stored at −20°C until further use. The Washington State and CDC Institutional Review Boards determined this project to be enhancing surveillance for a notifiable condition and did not require human subjects review.
Genome Sequencing, Assembly, and Analyses
We sequenced genomes of 18 isolates from 17 patients using Illumina (https://www.illumina.com) HiSeq and MiSeq sequencing platforms, as described (20,25). We prepared DNA samples for paired-end sequencing by using the Kapa Biosystems (https://www.kapabiosystems.com) Library Preparation Kit protocol with an 8-bp index modification and sequenced to a read length of 250 bp on the Illumina HiSeq or 250 bp on the Illumina MiSeq. All WGS data files have been deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProject PRJNA472461.
We performed genome assembly and analyses as described (20). In brief, the Washington-1(B10637) strain was used as the reference for C. immitis single-nucleotide polymorphism (SNP) matrices, and the B10813_Tx strain was used as a reference in the C. posadasii SNP matrix. We aligned Illumina read data against the respective reference assemblies by using Burrow-Wheeler Aligner version 0.7.7 (http://bio-bwa.sourceforge.net) and identified SNP variants by using SamTools version 0.1.19 (http://samtools.sourceforge.net). We filtered SNP calls by using a publicly available SNP analysis pipeline (26) (http://tgennorth.github.io/NASP), as described, to remove positions with <10 times coverage with <90% variant allele calls, or identified by using Nucmer (http://nebc.nox.ac.uk/bioinformatics/docs/nucmer.html) as being within duplicated regions in the reference (20,25). We constructed a maximum-parsimony phylogeny on the basis of SNP matrices by using MEGA7 (http://www.megasoftware.net), including 17 described strains from various geographic areas for comparison; we determined bootstrap support for the tree by using 1,000 reiterations (20).
We extracted candidate SNPs that distinguish Washington strains from non-Washington strains (Appendix Table) from whole-genome assemblies by using Samtools version 1.8, EMBOSS version 6.5.7, and NCBI-toolkit version 7.0.0 (27–29). We chose SNPs if their positions were consecutive or no more than 1 nt apart and had >10 times coverage. In brief, we extracted an ≈1-kb region surrounding candidate SNP positions from all draft assemblies, aligned it to Washington-1(B10637) reference, and designed degenerate primers by using Primer3 version 3.0 (30). We compared regions with those of C. immitis strain H538–4 to determine genomic loci annotation.
Results
During 2014–2017, a total of 167 confirmed cases of coccidioidomycosis were reported in Washington. For these cases, 18 isolates (Table) from 17 cases were available. Travel and exposure histories suggested local acquisition in Washington (9 case-patients) or were unknown at the time of specimen collection (8 case-patients).
Table. Characteristics of 18 clinical isolates subjected to whole-genome sequencing analysis from patients with coccidioidomycosis, Washington, USA, 2014–2017*.
| CDC-ID | Travel history of patient | Coccidioides species | Collection date | Specimen source site | Phylogenetic population |
|---|---|---|---|---|---|
| B10917 | Unknown | C. posadasii | 2014 | Sputum | Non-Washington |
| B10918 | AZ, MT, SD, OR | C. posadasii | 2014 | Sputum | Non-Washington |
| B11036 | AZ | C. posadasii | 2015 | Lung | Non-Washington |
| B11034 | AZ | C. immitis | 2014 | Lung | Washington |
| B11019 | None | C. immitis | 2014 | BAL | Washington† |
| B11035 | CA | C. immitis | 2015 | Cerebrospinal fluid | Non-Washington |
| B11080 | Mexico, CA | C. immitis | 2015 | Neck | Non-Washington |
| B11198 | Mexico, CA | C. immitis | 2015 | Tissue | Non-Washington |
| B11299 | AZ | C. posadasii | 2015 | Bronchial washings | Non-Washington |
| B11517 | CA | C. immitis | 2016 | Throat | Non-Washington |
| B11518 | None | C. immitis | 2016 | Testicle | Washington† |
| B11587 | CA | C. immitis | 2016 | Tissue | Non-Washington |
| B11863 | HI, Costa Rica | C. immitis | 2016 | Sputum | Non-Washington |
| B11873 | CA | C. immitis | 2016 | Bronchial washings | Non-Washington |
| B12398 | None | C. immitis | 2016 | Blood | Washington |
| B12495 | CA | C. immitis | 2016 | Cerebrospinal fluid | Non-Washington |
| B12496 | Mexico, CA | C. immitis | 2016 | Knee wound | Non-Washington |
| B13956 | None | C. immitis | 2017 | BAL | Washington |
*BAL, bronchoalveolar lavage; CDC, Centers for Disease Control and Prevention; ID, identification. †These isolates were from the same patient.
We speciated 14 isolates as C. immitis and 4 as C. posadasii and subjected these isolates to WGS with 25–160 times sequencing coverage. A total of 68,717 SNPs were identified by comparison with the reference isolate. Phylogenetic analysis and comparison with sequenced isolates of C. immitis demonstrated that 5 isolates from 4 patients in Washington were highly genetically related to each other and to isolates from the identified Washington clade that contained human and soil isolates (13). No more than 354 SNPs differentiated any 2 isolates within the Washington cluster, which was also supported by 100% bootstrap values (Figure). In contrast, >10,000 SNPs differentiated the Washington cluster from its nearest neighbor, an isolate from another case-patient in Washington. This case-patient (B12496) reported travel history to disease-endemic regions in California and Mexico. Tens of thousands of SNPs differentiated Washington cluster isolates from those from other states. No more than 3 SNPs were identified among multiple soil isolates collected 4 years apart from a single sampling site, and 4 SNPs were identified between isolates collected 2 years apart from 1 patient in Washington who had a chronic infection. However, 234–324 SNPs were identified in pairwise comparisons between isolates from different patients in the Washington clade (Figure, panel B).
Figure.
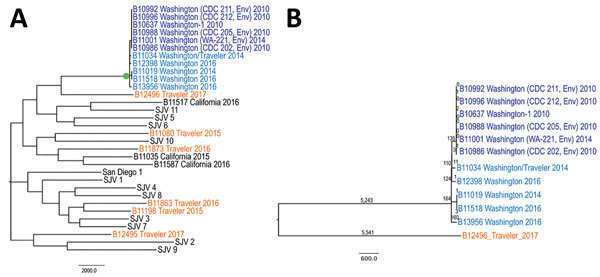
Genetic relationships among Coccidioides immitis isolates. A) Isolates from patients in Washington, USA, compared with isolates from other locations. B) Genetic relationships among C. immitis isolates from the Washington clade; single-nucleotide polymorphism numbers are shown above the branches. Dark blue indicates previously described environmental and human isolates (13) from Washington, light blue indicates isolates from new cases that were likely acquired in Washington, and orange indicates isolates from cases that were likely acquired outside Washington. Isolates from patients with the documented travel histories are indicated as Traveler. Travel history is detailed in the Table. Isolates with SJV names are from the San Joaquin Valley, California, as described (20). Bootstrap values shown in green circle indicate 100% support. Scale bars indicate nucleotide substitutions per site. CDC, Centers for Disease Control and Prevention; Env, environment.
Three of the 4 case-patients infected with Washington clade strains reported no travel history outside Washington. Conversely, 8 of 9 C. immitis isolates that did not cluster with the Washington clade were isolated from patients who reported travel to California.
Washington strains can be distinguished from non-Washington strains in 3 genomic regions on the basis of annotations from C. immitis strain H538-4; locus DS016982: adipocyte-derived leucine aminopeptidase; locus DS01702: hypothetical protein; and locus DS016985: glyoxal oxidase (Appendix). The DNA fragment from locus DS016982 contains SNP GG at positions 94460–94461 for Washington strains and SNP CA for non-Washington strains and can be amplified by using forward primer 5′-GGTACGTCACAAGTCCCCAG-3′ and reverse primer 3′-AAGAGTACTCGCGAAGGAAGC-5′. For locus DS017021, the DNA fragment amplified with forward primer 5′-CTTGACTGTGCAGGGCCTTA-3′ and reverse primer 3′-ACCGGCCTAACTCCATGGTA-5′ encompasses SNPs GGT for Washington strains and TGC for non-Washington strains at positions 105566–105568) The fragment of locus DS016985 can be amplified by using primer pairs 5′-TTCCGCTTGATGGCTGAAGT-3′/3′-TGTGGCCCTCCTATTGCTTG-5′ and contains consecutive SNP CC for Washington strains and SNP GA for non-Washington strains (positions 232442–232443).
We identified 4 case-patients who had C. posadasii infections by using internal transcribed spacer sequencing and confirmed by WGS. Isolates from the 4 case-patients clustered with described C. posadasii populations from Arizona (data not shown). Three of those case-patients had traveled to Arizona, and 1 case-patient had an unknown travel history.
Discussion
Determining the potential sources of exposure and geographic distribution of Coccidioides in areas where coccidioidomycosis is uncommon is a surveillance challenge because patients often report travel histories to other disease-endemic areas. To address this problem, we implemented WGS to better determine the likely geographic origin of isolates obtained from patients in Washington who had suspected local acquisition or unknown exposure histories. This method is based on the observation that Coccidioides populations have well defined geographic structure, and most isolates can be assigned to specific geographic areas on the basis of their WGS genotypes, such as Arizona, Texas, Mexico, central California, or Washington (20). We demonstrated the utility of this tool for investigating the epidemiology of coccidioidomycosis in Washington.
Of 18 isolates from 17 patients included in this study, 5 (28%) clustered with strains isolated from soil in Washington, which was consistent with local exposure. The remaining 13 (72%) isolates clustered with populations from other areas, such as Arizona, California, or Mexico, which was generally consistent with reported travel histories of patients. The only discordance between WGS and epidemiologic data was observed for isolate B11863, which clustered with C. immitis isolates from California. The patient from whom this isolate originated reported travel out of state but also reported extensive soil exposure in a county in Washington that had previously reported autochthonous cases. Specifically, the only reported travel was to Hawaii 2 months before disease onset and to Costa Rica 1 year before disease onset, with a layover in California during which the patient did not leave the airport. In addition, the patient reported 1 trip to southern California 6 years before disease onset. This discordance was an unexpected finding and might represent more diversity among Coccidioides spp. in Washington than previously understood, reactivated infection because of previous travel, or infection from a fomite or other carrier source of C. immitis. Further investigation and surveillance data are needed to determine which hypothesis is most plausible.
Of the 4 patients infected with the local strains from Washington, 3 patients reported no travel history outside Washington. The fourth patient reported travel to Arizona 1 year before the onset of coccidioidomycosis but also reported extensive exposure to local soils in Washington and dust during hiking and gardening. This result indicates that compatible travel history alone does not exclude the possibility of local exposure and demonstrates the added value of genomic approaches to determining exposure location. Although isolates in the Washington clade were genetically related (Figure, panel A), considerably higher genetic diversity was observed for new strains compared with that detected for the original isolates from Washington identified in 2013 (13). Specifically, 245–299 pairwise SNPs were identified among the newly identified strains from Washington, compared with <5 SNPs detected among previously described isolates from Washington from the same exposure site (Figure, panel B). This small but apparent genetic divergence of the new isolates indicates that new infections were acquired from different local C. immitis populations, likely at different sites, and suggests the presence of multiple environmental loci for Coccidioides spp. in Washington. The divergent isolates were collected from soil in a location ≈70 miles from the suspected exposure location of 1 of the patients. However, in sharp contrast to the populations of Coccidioides spp. in Arizona or California, where isolates are differentiated by thousands of SNPs, the overall low genetic variability in the Washington population is consistent with a relatively small population size, strongly suggesting a recent common ancestry of Washington strains and a relatively recent expansion of C. immitis to Washington, (Figure).
Our study has 2 main limitations. First, there might be additional genetically diverse populations of Coccidioides in Washington that have not yet been discovered or proven; therefore, some genotypes that have been deemed as acquired outside Washington might have been acquired locally. For example, 1 isolate from a patient who was highly suspected of having local acquisition clustered with isolates from California. Likewise, there might be genetically similar populations of Coccidioides spp. in Oregon, California, or other nearby states that have not yet been documented. These genetically similar populations might add complexity to determination of location of exposure. However, to date, no clinical or environmental isolates with strong epidemiologic links to locations outside Washington that cluster with isolates from Washington have been identified. This finding is consistent with those of previous studies that demonstrated that C. immitis and C. posadasii have well-defined geographic structures (15–19).
Second, all patients in our study were given diagnoses of rather atypical cases of coccidioidomycosis that were either asymptomatic (diagnosed during biopsy), cutaneous, chronic, or unusually severe. In general, the acute primary pulmonary form of coccidioidomycosis constitutes the most common manifestation in highly endemic areas and often represents the earliest manifestation of the disease. Conversely, reactivation of latent infections can occur months to years after initial exposure, typically in immunosuppressed persons. Many complicated infections might follow a subacute or chronic disease progression. This lack of acute respiratory cases and preponderance of atypical disease in Washington indicates the likely gap between early identification and delayed diagnosis because of lack of awareness and might also explain the high prevalence of the out-of-state strains of C. immitis in our study. However, even in highly endemic regions, acute pulmonary disease is often missed or delayed in diagnosis for months (31,32), supporting the possibility that an underlying background of typical acute disease might remain unidentified in Washington (33).
Our results indicate that WGS is a useful tool to assist in determining exposure location in surveillance situations in which exposure histories are unclear or unknown. For patients in this study, coccidioidomycosis was more commonly associated with travel to other disease-endemic areas compared with local exposure; however, travel to endemic regions does not preclude local acquisition of the disease. Our results also indicate that coccidioidomycosis is likely to be underdiagnosed and underreported in Washington on the basis of atypical disease manifestations for locally acquired cases. More research is needed to determine the true prevalence of locally acquired coccidioidomycosis in Washington.
Additional information on utility of whole-genome sequencing to ascertain locally acquired cases of coccidioidomycosis, Washington, USA.
Acknowledgments
We thank Denny Flodin-Hursh, Jill Stiffler, Cindy Ralston, Heather Hill, and all other local health jurisdiction and healthcare facility staff who supported this study by obtaining isolates and interviewing patients, and Joyce Peterson and Ngoc Le for assistance with processing and identification of isolates.
This study was supported by an Advanced Molecular Detection Initiative and Epidemiology and Laboratory Capacity grant from CDC.
Biography
Ms. Oltean is a zoonotic and vectorborne disease epidemiologist in the Office of Communicable Disease Epidemiology, Washington State Department of Health, Shoreline, WA. Her primary research interests are zoonotic disease surveillance system design, implementation, and evaluation.
Footnotes
Suggested citation for this article: Oltean HN, Etienne KA, Roe CC, Gade L, McCotter OZ, Engelthaler DM, et al. Utility of whole-genome sequencing to ascertain locally acquired cases of coccidioidomycosis, Washington, USA. Emerg Infect Dis. 2019 Mar [date cited]. https://doi.org/10.3201/eid2503.181155
References
- 1.Brown J, Benedict K, Park BJ, Thompson GR III. Coccidioidomycosis: epidemiology. Clin Epidemiol. 2013;5:185–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams D, Fullerton K, Jajosky R, Sharp P, Onweh D, Schley A, et al. Summary of notifiable infectious diseases and conditions—United States, 2013. MMWR Morb Mortal Wkly Rep. 2015;62:1–122. [DOI] [PubMed] [Google Scholar]
- 3.Cordeiro RA, Brilhante RS, Rocha MF, Bandeira SP, Fechine MA, de Camargo ZP, et al. Twelve years of coccidioidomycosis in Ceará State, Northeast Brazil: epidemiologic and diagnostic aspects. Diagn Microbiol Infect Dis. 2010;66:65–72. [DOI] [PubMed] [Google Scholar]
- 4.Wilken JA, Sondermeyer G, Shusterman D, McNary J, Vugia DJ, McDowell A, et al. Coccidioidomycosis among workers constructing solar power farms, California, USA, 2011–2014. Emerg Infect Dis. 2015;21:1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pappagianis D; Coccidioidomycosis Serology Laboratory. Coccidioidomycosis in California state correctional institutions. Ann N Y Acad Sci. 2007;1111:103–11. [DOI] [PubMed] [Google Scholar]
- 6.Colson AJ, Vredenburgh L, Guevara RE, Rangel NP, Kloock CT, Lauer A. Large-scale land development, fugitive dust, and increased coccidioidomycosis incidence in the antelope valley of California, 1999–2014. Mycopathologia. 2017;182:439–58. [DOI] [PubMed] [Google Scholar]
- 7.Marsden-Haug N, Goldoft M, Ralston C, Limaye AP, Chua J, Hill H, et al. Coccidioidomycosis acquired in Washington State. Clin Infect Dis. 2013;56:847–50. [DOI] [PubMed] [Google Scholar]
- 8.Galgiani JN, Thompson G III. Valley fever (coccidioidomycosis), tutorial for primary care professionals, 2016. Valley Fever Center for Excellence, The University of Arizona [cited 2018 Nov 30]. https://vfce.arizona.edu/sites/default/files/9-valley_fever_tutorial.pdf
- 9.Arizona Department of Health Services (AZDoH). Valley fever 2016 annual report, 2017. [cited 2018 Nov 30]. https://www.azdhs.gov/documents/preparedness/epidemiology-disease-control/valley-fever/reports/valley-fever-2016.pdf
- 10.Cooksey GS, Nguyen A, Knutson K, Tabnak F, Benedict K, McCotter O, et al. Notes from the field: increase in coccidioidomycosis—California, 2016. MMWR Morb Mortal Wkly Rep. 2017;66:833–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorris M, Cat L, Zender C, Treseder K, Randerson J. Coccidioidomycosis dynamics in relation to climate in the southwestern United States. GeoHealth. 2018;2:6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsden-Haug N, Hill H, Litvintseva AP, Engelthaler DM, Driebe EM, Roe CC, et al. ; Centers for Disease Control and Prevention (CDC). Coccidioides immitis identified in soil outside of its known range - Washington, 2013. MMWR Morb Mortal Wkly Rep. 2014;63:450. [PMC free article] [PubMed] [Google Scholar]
- 13.Litvintseva AP, Marsden-Haug N, Hurst S, Hill H, Gade L, Driebe EM, et al. Valley fever: finding new places for an old disease: Coccidioides immitis found in Washington State soil associated with recent human infection. Clin Infect Dis. 2015;60:e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Washington State Department of Health (WADoH). Washington State annual report on fungal disease, 2014. [cited 2018 Nov 30]. https://www.doh.wa.gov/Portals/1/Documents/5100/420-146-FungalDiseaseAnnualReport.pdf
- 15.Barker BM, Jewell KA, Kroken S, Orbach MJ. The population biology of coccidioides: epidemiologic implications for disease outbreaks. Ann N Y Acad Sci. 2007;1111:147–63. [DOI] [PubMed] [Google Scholar]
- 16.Neafsey DE, Barker BM, Sharpton TJ, Stajich JE, Park DJ, Whiston E, et al. Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 2010;20:938–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher MC, Rannala B, Chaturvedi V, Taylor JW. Disease surveillance in recombining pathogens: multilocus genotypes identify sources of human Coccidioides infections. Proc Natl Acad Sci U S A. 2002;99:9067–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher MC, Koenig GL, White TJ, Taylor JW. Pathogenic clones versus environmentally driven population increase: analysis of an epidemic of the human fungal pathogen Coccidioides immitis. J Clin Microbiol. 2000;38:807–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teixeira MM, Barker BM. Use of population genetics to assess the ecology, evolution, and population structure of Coccidioides. Emerg Infect Dis. 2016;22:1022–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelthaler DM, Roe CC, Hepp CM, Teixeira M, Driebe EM, Schupp JM, et al. Local population structure and patterns of Western Hemisphere dispersal for Coccidioides spp., the fungal cause of Valley fever. MBio. 2016;7:e00550–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salipante SJ, SenGupta DJ, Cummings LA, Land TA, Hoogestraat DR, Cookson BT. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol. 2015;53:1072–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allard MW, Bell R, Ferreira CM, Gonzalez-Escalona N, Hoffmann M, Muruvanda T, et al. Genomics of foodborne pathogens for microbial food safety. Curr Opin Biotechnol. 2018;49:224–9. [DOI] [PubMed] [Google Scholar]
- 23.Gardy JL, Loman NJ. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat Rev Genet. 2018;19:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Centers for Disease and Prevention. Coccidioidomycosis/Valley fever (Coccidioides spp.), 2011. Case definition. CSTE position statement. 10-ID-04 [cited 2018 Nov 30] https://wwwn.cdc.gov/nndss/conditions/coccidioidomycosi s/case-definition/2011
- 25.Etienne KA, Roe CC, Smith RM, Vallabhaneni S, Duarte C, Escadon P, et al. Whole-genome sequencing to determine origin of multinational outbreak of Sarocladium kiliense bloodstream infections. Emerg Infect Dis. 2016;22:476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, et al. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genom. 2016;2:e000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vakatov D. The NCBI C++ toolkit book, 2004. National Center for Biotechnology Information [cited 2018 Nov 30]. https://ncbi.github.io/cxx-toolkit
- 29.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–7. [DOI] [PubMed] [Google Scholar]
- 30.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valdivia L, Nix D, Wright M, Lindberg E, Fagan T, Lieberman D, et al. Coccidioidomycosis as a common cause of community-acquired pneumonia. Emerg Infect Dis. 2006;12:958–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang DC, Anderson S, Wannemuehler K, Engelthaler DM, Erhart L, Sunenshine RH, et al. Testing for coccidioidomycosis among patients with community-acquired pneumonia. Emerg Infect Dis. 2008;14:1053–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tartof SY, Benedict K, Xie F, Rieg GK, Yu KC, Contreras R, et al. Testing for coccidioidomycosis among community-acquired pneumonia patients, southern California, USA. Emerg Infect Dis. 2018;24:779–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information on utility of whole-genome sequencing to ascertain locally acquired cases of coccidioidomycosis, Washington, USA.


