Abstract
Background
To understand the roles of serum exosomes in rheumatoid arthritis (RA), we comprehensively investigated the protein profiles of serum exosomes in patients with RA.
Methods
Exosomes were isolated from serum samples obtained from 33 patients (12 with active RA [aRA], 11 with inactive RA [iRA], 10 with osteoarthritis [OA]) and 10 healthy donors (HLs). Proteins extracted from the exosomes were separated by two-dimensional differential gel electrophoresis (2D-DIGE) and identified by mass spectrometry.
Results
In total, 204 protein spots were detected by 2D-DIGE. In the aRA, iRA, and OA groups, 24, 5, and 7 spots showed approximately ≥ ±1.3-fold intensity differences compared with the HL group, respectively. We were able to identify proteins in six protein spots. Among them, the protein spot identified as Toll-like receptor 3 (TLR3) showed approximately 6-fold higher intensity in the aRA group than in the other groups.
Conclusions
Patients with active RA possessed considerably different protein profiles of serum exosomes from patients with iRA, patients with OA, and healthy donors. The unique protein profile of serum exosomes, such as the possession of abundant TLR3 fragments, may reflect the pathophysiology of active RA.
Electronic supplementary material
The online version of this article (10.1186/s41927-018-0041-8) contains supplementary material, which is available to authorized users.
Keywords: Exosome, Proteomics, Rheumatoid arthritis, Osteoarthritis, Toll like receptor 3
Background
Exosomes are nanometer-sized (50–100 nm) membranous extracellular vesicles secreted from various types of cells [1]. Exosomes are initially generated by inward budding of late endosomes, called “multivesicular bodies (MVBs)”, in cells and then released from the cells by the fusion of MVBs with plasma membranes [1]. Exosomes are involved in intercellular communications via the delivery of proteins, lipids, and RNAs (mRNA and microRNA) through exosome-cell fusion and via signal delivery through exosome-cell interactions [2, 3].
Immunologically, exosomes play roles in the presentation of antigens to T cells [4]. Exosomes derived from antigen-presenting cells (APCs) were reported to possess major histocompatibility complex (MHC) class I and II molecules, co-stimulatory molecules of CD80 and CD86, and adhesion molecule of intercellular adhesion molecule (ICAM)-1, which are essential to antigen presentation [5, 6]. In fact, exosomes derived from dendritic cells (DCs) have been shown to induce CD8+ T cells to produce interferon (IFN)-γ and tumor necrosis factor (TNF)-α in an exosomal MHC class I-dependent manner [7]. Furthermore, mature DC-derived exosomes have been reported to induce the proliferation of CD4+ T cells in an antigen-specific manner [8].
In the field of cancer, the immunosuppressive and immunostimulatory properties of exosomes have been demonstrated [4]. For example, tumor-derived exosomes were reported to induce T cell apoptosis via Fas ligand [9] and to inhibit interleukin (IL)-2-induced proliferation of T cells [10]. Quite recently, exosomal programmed death-ligand 1 (PD-L1) was shown to inhibit the function of CD8+ T cells and facilitate the progression of melanoma [11].
Collectively, exosomes are thought to play immuno-pathological roles in various diseases. Rheumatoid arthritis (RA) is a systemic autoimmune disorder characterized by inflammation and proliferation of synovial tissue, destruction of cartilage and bone, and production of autoantibodies, such as anti-citrullinated peptide antibodies [12]. Although the pathogenesis of RA remains unclear, the pathophysiology is thought to be as follows: First, innate immunity activates DCs [13]. Subsequently, DCs migrate into central lymphoid organs and present autoantigens, such as type II collagen and citrullinated proteins, to T cells [13]. The activated T cells then induce B cells to produce autoantibodies [14]. These lymphocytes migrate into synovial tissue and enhance adaptive immune responses, leading to synovial angiogenesis and hyperplasia of synovial tissue [14]. Inflammatory cytokines such as IL-1, IL-6, IL-17, and TNF-α produced in this process amplify osteoclast differentiation and activation, which results in the destruction of cartilage and bone [12].
As mentioned above, exosomes have various immunological functions and are thus expected to play roles in the pathophysiology of RA. For instance, fibroblast-like synoviocyte (FLS)-derived exosomes were reported to contain citrullinated proteins [15] and a membrane form of TNF-α [16]. IL-1β-stimulated FLS-derived exosomes were reported to up-regulate the matrix metalloproteinase (MMP)-13 expression in chondrocytes [17]. We showed that IL-1β and anti-rheumatic drugs of salazosulfapyridine (SASP) and methotrexate (MTX) considerably altered the protein profiles of exosomes derived from SW982 of synovial sarcoma cell line [18]. Recently, it was reported that some serum exosomal proteins in patients with RA might be useful parameters of disease activity [19]. In terms of bone formation and resorption, osteoclast-derived exosomes containing microRNA (miR-214) were reported to play an inhibitory role in osteoblast activity [20, 21]. However, at present, the roles of exosomes in RA are largely unclear.
These previous findings suggest that characterizing the exosomes generated in patients with RA would be quite useful. We therefore characterized the protein profile of serum exosomes in patients with RA by two-dimensional differential gel electrophoresis (2D-DIGE) and a subsequent mass spectrometry (MS) analysis. We found that the serum exosomes in patients with active RA (aRA) possessed considerably different protein profiles from those in patients with inactive RA (iRA), patients with osteoarthritis (OA), and healthy donors (HLs). In particular, the serum exosomes in patients with aRA contained abundant Toll-like receptor (TLR) 3 fragments. Our data will help promote the understanding of the roles of exosomes in the pathophysiology of RA.
Materials and methods
Preparation of blood samples
Blood samples were obtained from 33 patients (12 patients with aRA, 11 with iRA, 10 with OA) and 10 HLs. Clinical information on the patients with RA is shown in Table 1. The diagnoses of RA and OA were made according to the respective criteria by American College of Rheumatology [22, 23]. We defined aRA as “Disease Activity Score 28 using C-reactive protein (DAS28-CRP) > 2.7” and iRA as “DAS28-CRP < 2.3” [24]. Blood samples were drawn into vacuum tubes (Venoject II, VPAS109K50; Terumo Corporation, Tokyo, Japan) and centrifuged at 1500 g for 10 min to separate the sera at room temperature. The separated serum samples were then stored at − 80 °C until use.
Table 1.
Clinical information of patients with aRA, iRA, and OA and of HL
| Active RA (n = 12) | Inactive RA (n = 11) | p Value (aRA vs. iRA) | OA (n = 10) | HL (n = 10) | |
|---|---|---|---|---|---|
| Age (years), mean (range) | 62.2 (31 to 79) | 63.4 (41 to 80) | 0.85 | 62.5 (41 to 74) | 60.5 (41 to 82) |
| Sex, male/female | 2/10 | 3/8 | 0.93 | 1/9 | 2/8 |
| Disease duration (years), mean (range) | 5.6 (0.25 to 20) | 10.9 (0.25 to 38) | 0.22 | (−) | (−) |
| ESR (mm/h), mean (range) | 56.3 (16 to 100) | 21.0 (2 to 41) | < 0.01 | (−) | (−) |
| CRP (mg/L), mean (range) | 3.19 (0.07 to 7.16) | 0.14 (0.02 to 0.33) | < 0.01 | (−) | (−) |
| MMP-3 (ng/mL), mean (range) | 252.0 (18.8 to 602.4) | 52.7 (10 to 121.5) | < 0.01 | (−) | (−) |
| SJC, mean (range) | 3.7 (1 to 8) | 0 | < 0.01 | (−) | (−) |
| TJC, mean (range) | 6.3 (0 to 17) | 0.8 (0 to 3) | < 0.01 | (−) | (−) |
| RF positive (%) | 83.3 | 72.7 | 0.56 | (−) | (−) |
| ACPA positive (%) | 90.0 | 77.8 | 0.49 | (−) | (−) |
| DAS28-ESR, mean (range) | 4.80 (4.21 to 5.72) | 2.33 (0.51 to 3.02) | < 0.01 | (−) | (−) |
| DAS28-CRP, mean (range) | 3.84 (3.10 to 4.64) | 1.49 (0.98 to 2.2) | < 0.01 | (−) | (−) |
| Medication | |||||
| DMARDs (%) | 75.0 | 90.9 | 0.34 | (−) | (−) |
| Methotrexate (%) | 50.0 | 81.8 | 0.11 | (−) | (−) |
| Biologics (%) | 8.0 | 18.1 | 0.51 | (−) | (−) |
| Prednisolone (%) | 41.7 | 0 | 0.01 | (−) | (−) |
ESR erythrocyte sedimentation rate, CRP C-reactive protein, MMP-3 matrix metalloproteinase-3, SJC swollen joint count, TJC tender joint count, RF rheumatoid factor, ACPA anti-cyclic citrullinated peptide antibody, DAS28-ESR disease activity score 28 joints using erythrocyte sedimentation rate, DAS28-CRP disease activity score 28 joints using C-reactive protein, DMARDs disease-modifying anti-rheumatic drugs
This study was approved by the ethics committee of Sagamihara National Hospital and that of St. Marianna University School of Medicine. All of the blood samples were obtained with written informed consent.
Preparation of serum exosomal proteins
Exosomes were isolated from the serum samples using ExoQuick® (System Bioscience, Mountain View, CA, USA) according to the manufacturer’s instructions. The collected exosomes were dissolved in a cell lysis buffer (30 mM Tris-HCl pH 8.0, 7 M Urea, 2 M Thiourea, 4% 3-(3-cholamidepropyl) dimethylammonio-1-propanesulfonate [CHAPS]). The protein concentrations of the resulting exosome lysates were then determined by the Bradford assay. A portion of the collected exosomes was suspended in phosphate-buffered saline (PBS) for a transmission electron microscopic analysis.
Transmission electron microscopic analyses
A 5-μl aliquot of PBS-suspended exosomes was placed onto a support grid (Cu, 200 mesh). After 60 s, the grid was washed twice in double distilled water and incubated in 5% aqueous solution of phosphotungstic acid for 10 s. The size and morphology of the collected exosomes were examined using a transmission electron microscope (JEOL Ltd., Tokyo, Japan).
The 2D-DIGE analysis and protein identification
The exosome lysates were separated by 2D-DIGE, as described previously [25]. In brief, each protein sample was labeled with Cy5 saturation dye. Similarly, 2.5 μg of an internal standard sample (a mixture of equal amounts of all 43 samples) was labeled with Cy3 saturation dye. Then, 2.5 μg of the Cy3-labeled internal standard sample and 2.5 μg of the individual Cy5-labeled protein samples were mixed and subjected to 2D-DIGE. Separated proteins were detected using an image analyzer (Typhoon 9400 Imager; GE Healthcare UK, Buckinghamshire, UK). Acquired gel images were analyzed using the Progenesis software program (PerkinElmer, Waltham, MA, USA), in which the intensity of the protein spots in the gel image of each sample was corrected using that of the internal standard sample.
Proteins were identified by a mass spectrometric analysis as described previously [25]. In brief, the gel fragments that corresponded to protein spots of interest were recovered from 2D gels. Peptides generated by in-gel digestion with trypsin were extracted from the gel fragments. The extracted peptides were then subjected to matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry (Ultraflex; Bruker Daltonics, Bremen, Germany). The determined peptide masses were compiled to allow a search of the National Center for Biotechnology Information protein database using the Mascot software program (Matrix Science, London, UK).
Detection of TLR3 by Western blotting (WB)
Proteins were extracted from the purified exosomes into lysis buffer (7 M urea, 2 M thiourea, and 4% CHAPS) for WB. The extracted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or by 2D electrophoresis (2DE). In 2DE, the protein samples were first subjected to isoelectric focusing (IEF) using gels with the range of pI3.0–11.0. The IEF-separated proteins were then further separated by 12.5% SDS-PAGE. The proteins separated by SDS-PAGE or 2DE were transferred onto polyvinylidene difluoride membranes. In WB, the membranes were incubated with rabbit anti-human TLR3 polyclonal antibodies (Abcam, Cambridge, UK) followed by horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G antibodies (Dako, Glostrup, Denmark). The bound antibodies were visualized using ImmunoStar LD (Wako Pure Chemical Industries, Ltd., Tokyo, Japan). In each membrane, the intensity of a blank area in each membrane was defined as 1.0.
Statistical analyses
The statistical significance of differences in parameters clinical features between the aRA and iRA groups and that of differences in protein spot intensity in 2D-DIGE among the 4 groups was calculated by Student’s t-test and a one-way analysis of variance, respectively. The expression of TLR3 fragments (17–18 kDa) in the 4 groups was statistically evaluated by Fisher’s exact test. P-values of less than 0.05 were considered to indicate statistical significance.
Results
Clinical information of the participants
Clinical information of the 43 participants is shown in Table 1. No significant differences were found between the aRA and iRA groups with regard to age, sex, disease duration, and test items other than markers for inflammatory condition (Table 1). Regarding the medication, the percentage of patients receiving oral corticosteroids in the aRA group (41.7%) was significantly higher than that in the iRA group (0%) (Table 1).
Isolation of exosomes from serum samples
We isolated exosomes from serum samples and checked for successful isolation of exosomes using a transmission electron microscopic analysis. Almost all of the isolated extracellular vesicles were found to have a diameter of 50–100 nm, as shown in Fig. 1. The observed sizes were compatible with the definition of exosomes [1, 2].
Fig. 1.
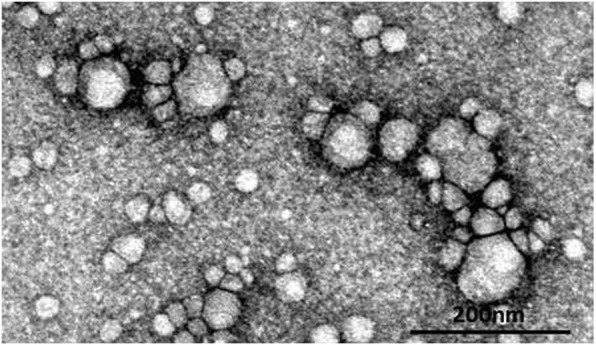
A transmission electron micrograph of exosomes isolated from serum samples. Serum exosomes isolated using Exoquick® were analyzed by transmission electron microscopy. A representative result from the serum exosome samples is shown
The comparison of the protein profiles of serum-derived exosomes among patients with RA and OA and HLs
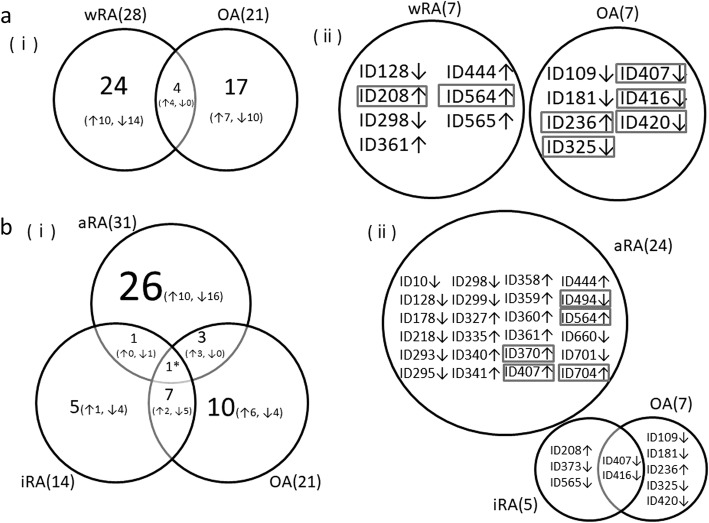
We investigated the protein profiles of exosomes in each of the 43 serum samples by 2D-DIGE. As a result, a total of 204 protein spots were detected on the gels, as shown in Fig. 2. The spot intensities in each group are shown in Additional file 1: Table S1. We first compared the intensity of the separated protein spots between the whole RA (wRA; aRA + iRA) and HL groups and between the OA and HL groups. In the comparison between the wRA and HL groups, 28 of the 204 protein spots showed significantly different intensities (p < 0.05) (Table 2A). Of these 28 spots, 7 showed a difference in intensity exceeding ±1.3-fold (Table 2A). In the comparison between the OA and HL groups, 21 protein spots showed different intensities (p < 0.05) (Table 2D). Of these 21 spots, 7 showed a difference in intensity exceeding ±1.3-fold (Table 2D). As shown in Fig. 3a-(i), only 4 spots overlapped between the 28 spots that differed in the wRA group and the 21 spots that differed in the OA group (14% and 19%, respectively). Furthermore, no spots overlapped between these 2 groups when we focused on the spots with a difference in intensity exceeding ±1.3-fold (Fig. 3a-[ii]). These results indicate that the protein profiles of exosomes are quite different between the wRA and OA groups.
Fig. 2.
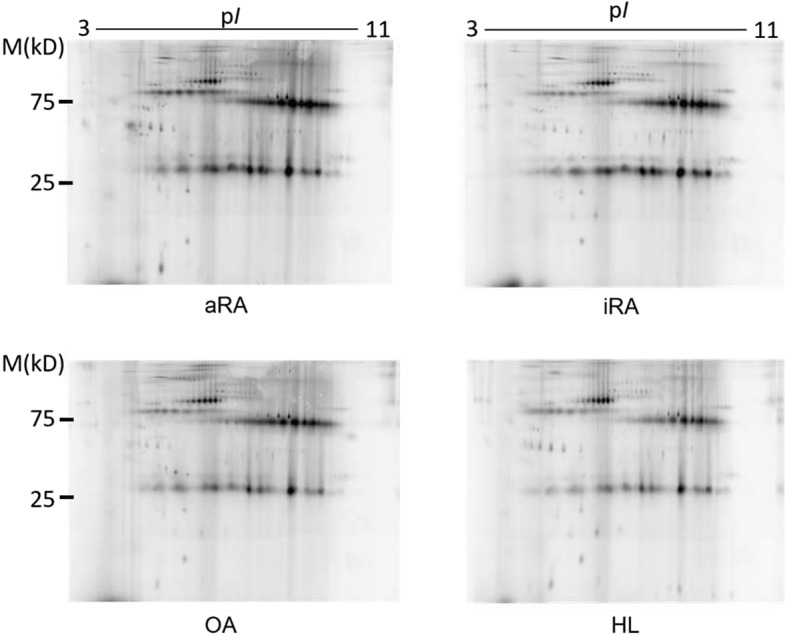
Representative 2D-DIGE images showing protein profiles of exosomes isolated from the serum samples of the aRA, iRA, OA, and HL groups. A representative image from each group is shown
Table 2.
Differences in the exosomal protein profiles among the disease categories detected by 2D-DIGE
| A. wRA vs. HL | |||
| Total number of detected protein spots | Number of spots with different intensities (p < 0.05) | fold difference (wRA/HL) | number of spots |
| 204 | 28 | X ≥ 1.3 | 5 |
| 1.3 > X > −1.3 | 21 | ||
| −1.3 ≥ X | 2 | ||
| B. aRA vs. HL | |||
| Total number of detected protein spots | Number of spots with different intensities (p < 0.05) | fold difference (aRA/HL) | number of spots |
| 204 | 31 | X ≥ 1.3 | 13 |
| 1.3 > X > −1.3 | 7 | ||
| − 1.3 ≥ X | 11 | ||
| C. iRA vs. HL | |||
| Total number of detected protein spots | Number of spots with different intensities (p < 0.05) | fold difference (iRA/HL) | number of spots |
| 204 | 14 | X ≥ 1.3 | 1 |
| 1.3 > X > −1.3 | 9 | ||
| −1.3 ≥ X | 4 | ||
| D. OA vs. HL | |||
| Total number of detected protein spots | Number of spots with different intensities (p < 0.05) | fold difference (OA/HL) | number of spots |
| 204 | 21 | X ≥ 1.3 | 1 |
| 1.3 > X > −1.3 | 14 | ||
| −1.3 ≥ X | 6 | ||
wRA whole RA, aRA active RA, iRA inactive RA, OA osteoarthritis, HL healthy donor
Fig. 3.
Venn diagrams showing the numbers of protein spots with significantly different intensities between the HL group and each disease group. (a) (i) Numbers of protein spots with significantly different intensities between the wRA and HL groups and between the OA and HL groups (p < 0.05). Up and down arrows indicate the spots whose intensity was higher and lower, respectively, in the disease groups than that in HL group. (ii) IDs of protein spots with an intensity ≥ ±1.3-fold different in the diagram (i). IDs of protein spots with a significantly different intensity between the wRA and OA groups are boxed. Up and down arrows indicate the spots whose intensity was higher and lower, respectively, compared to the intensity of the identical spots in the HL group. (b) (i) Numbers of protein spots with significantly different intensities between the aRA and HL groups, between the iRA and HL groups, and between the OA and HL groups (p < 0.05). Up and down arrows indicate the spots whose intensity was higher and lower, respectively, in the disease groups than that in HL group. The spots with “*” showed a higher intensity in the aRA group and a lower intensity in the iRA and OA groups than in the HL group. (ii) IDs of protein spots with an intensity ≥ ±1.3-fold different in the diagram (i). IDs of protein spots with a significantly different intensity between the aRA and OA groups and between the aRA and iRA groups are boxed. Up and down arrows indicate the spots whose intensity was higher and lower, respectively, compared to the intensity of the identical spots in the HL group
We next compared the protein profiles between the aRA and HL groups and between the iRA and HL groups. On comparing the aRA and HL groups, 31 spots showed different intensities (p < 0.05) (Table 2B). Of these 31 spots, 24 showed a difference in intensity exceeding ±1.3-fold (Table 2B). On comparing the iRA and HL groups, 14 protein spots showed different intensities (p < 0.05) (Table 2C). Of these 14 spots, 5 showed a difference in intensity exceeding ±1.3-fold (Table 2C). Of note, as shown in Fig. 3b-(i), only 2 spots overlapped between the 31 spots that differed in the aRA group and the 14 spots that differed in the iRA group (6.5% and 14%, respectively). Furthermore, no spots overlapped between the 2 groups when we focused on the spots with a difference in intensity exceeding ±1.3-fold (Fig. 3b-[ii]). These results indicate that the protein profiles of exosomes differ markedly between the aRA and iRA groups. In addition, only 4 spots overlapped between the 31 spots that differed in the aRA group and the 21 spots that differed in the OA group (13% and 19%, respectively) (Fig. 3b-[i]). No spots overlapped between the 2 groups when we focused on the spots with a difference in intensity exceeding ±1.3-fold (Fig. 3b-[ii]). These results indicate that the protein profiles of exosomes differ markedly between the aRA and OA groups. In contrast, 6 spots overlapped between the 14 spots that differed in the iRA group and the 21 spots that differed in the OA group (43% and 29%, respectively) (Fig. 3b-[i]). The location of spots that showed an intensity with more than ±1.3-fold difference (p < 0.05) compared to that in the HL group were marked on the 2D-DIGE images of the aRA, iRA, and OA groups (Additional file 1: Figure S1).
Identification of proteins in the spots with different intensity
We then compared the spot intensity of the 14 spots shown in Fig. 3a-(ii) between the wRA and OA groups. As a result, 7 spots (ID208, ID564, ID407, ID416, ID420, ID236, and ID325, rectangle in Fig. 3a-[ii]) showed different intensities (p < 0.05) between the wRA and OA groups. Similarly, we compared the spot intensity of the 34 spots (including 2 overlapping spots) shown in Fig. 3b-(ii) between the aRA and OA groups and between the aRA and iRA groups. As a result, 5 spots (ID370, ID407, ID494, ID564, and ID704, rectangle in Fig. 3b[ii]) showed different intensity (p < 0.05) between the aRA and OA groups and between the aRA and iRA groups.
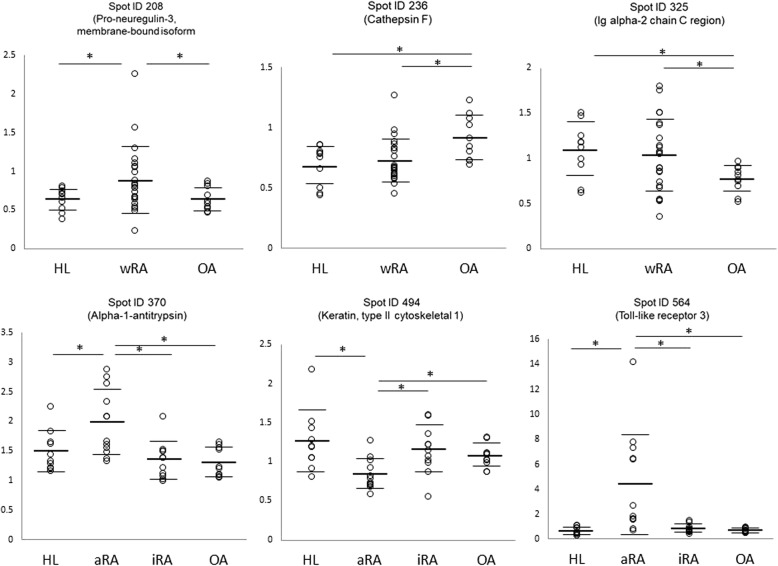
We tried to identify the protein names of the former 7 spots and the latter 5 spots (actually 10 spots because of 2 overlapped spots [ID407 and ID564]) by a mass spectrometric analysis (Fig. 4). As a result, we were able to name 6 out of the 10 protein spots, as listed in Table 3. ID208 (increased only in the wRA group) was found to be a membrane-bound isoform of Pro-neuregulin-3. ID564 (increased in the wRA and aRA groups) was found to be TLR3. ID370 (increased only in the aRA group) was found to be alpha-1-antitrypsin. ID494 (decreased only in the aRA group) was found to be keratin of type II cytoskeletal 1. ID236 (increased only in the OA group) was found to be cathepsin F. ID 325 (decreased only in the OA group) was found to be Ig alpha-2 chain C region. We confirmed that these 6 identified proteins were registered in Vesiclepedia, a database of extracellular vesicle components [26].
Fig. 4.
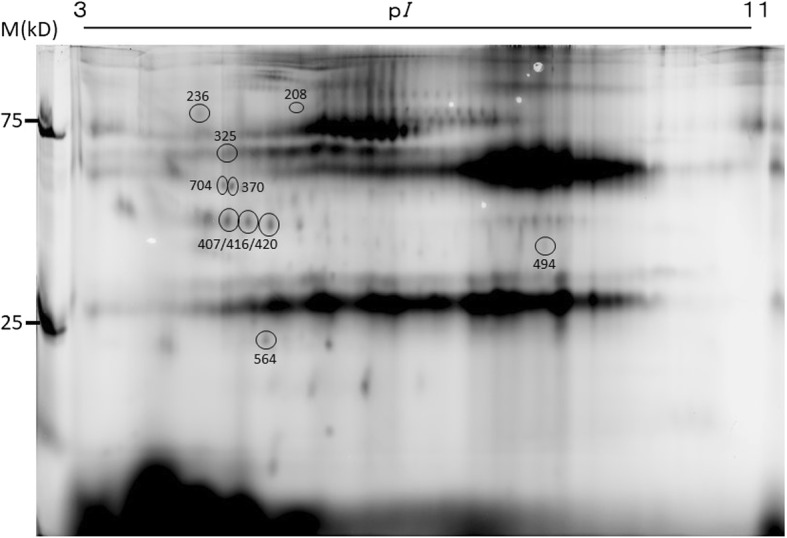
Location and IDs of exosomal protein spots subjected to identification
Table 3.
Proteins identified by mass spectrometry
| Spot ID | Observed | Fold difference | Protein | Theoretical | Masscot Score | Coverage (%) | Confirmed Sequences (Masscot ion score) | Search log | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MW | pI | wRA | OA | HL | MW | pI | |||||||
| aRA | iRA | ||||||||||||
| 208 | 90 | 5.3 | 1.4* | 1.0 | 1.0 | Pro-neuregulin-3, membrane-bound isoform | 78 | 7.8 | 61 | 12 | 388QAKQIQEQLK397(19) | 38,220 | |
| NRG3_HUMAN | |||||||||||||
| 236 | 87 | 5.1 | 1.1 | 1.3* | 1.0 | Cathepsin F | 53 | 8.5 | 26 | 3 | 103TLLCSFQVLDELGRHVLLR121(26) | 38,210 | |
| CATF_HUMAN | peptide summary | ||||||||||||
| 325 | 65 | 5.1 | 0.9 | 0.7* | 1.0 | Ig alpha-2 chain C region | 37 | 5.7 | 132 | 10 | 251WLQGSQELPR260(53) | 38,207 | |
| IGHA2_HUMAN | 270QEPSQGTTTFAVTSILR286(70) | ||||||||||||
| 370 | 53 | 5.1 | 1.3* | 0.9 | 0.9 | 1.0 | Alpha-1-antitrypsin | 47 | 5.4 | 170 | 33 | 50ITPNLAEFAFSLYR63(79) | 38,190 |
| A1AT_HUMAN | 216GKWERPFEVK225(10) | ||||||||||||
| 494 | 40 | 7.0 | 0.7* | 0.9 | 0.9 | 1.0 | Keratin, type II cytoskeletal 1 | 66 | 8.2 | 62 | 9 | 212WELLQQVDTSTR223(14) | 38,206 |
| K2C1_HUMAN | 224THNLEPYFESFINNLR239(41) | ||||||||||||
| 564 | 24 | 5.2 | 6.3* | 1.2 | 1.1 | 1.0 | Toll-like receptor 3 | 104 | 6.7 | 68 | 19 | 532LEILDLQHNNLAR544(7) | 38,215 |
| TLR3_HUMAN | |||||||||||||
MW molecular weight, wRA whole RA, aRA active RA, iRA inactive RA, OA osteoarthritis, HL healthy donor
Fold differences are shown with the intensity of the protein spots in HLs defined as 1.0. Asterisks indicate that the spot intensity is significantly different (p < 0.05) from that in HLs
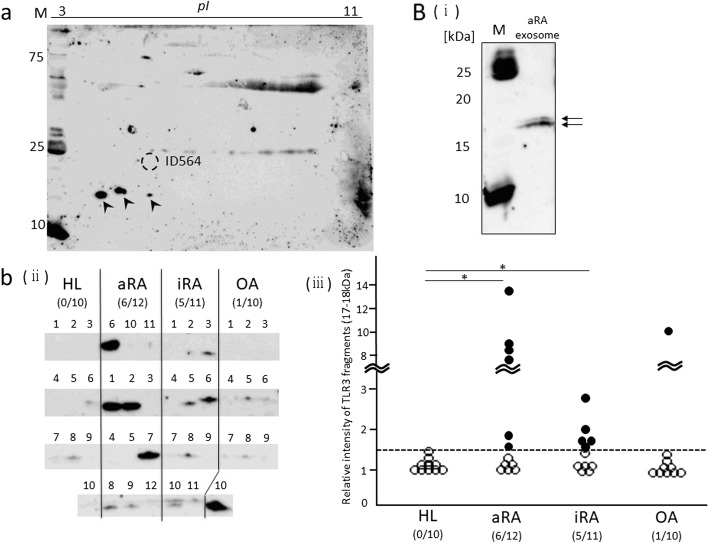
Among these six protein spots, the difference in ID564 (assigned as TLR3) was most drastic, as shown in Fig. 5. Since the observed molecular weight (MW) (24 kD) of ID564 was rather small compared to the theoretical MW (104 kD) of full-length TLR3, ID564 was thought to be a part of TLR3. Mass spectrometry detected a peptide corresponding to the 532–544 amino acid residues of TLR3 (Table 3), indicating that the spot ID564 was a middle part of TLR3.
Fig. 5.
Differences in the relative spot intensities of the identified proteins. Open circles show the relative intensity of individual samples. Bars show the mean ± SD. *, p < 0.05
Detection of the TLR3 fragment by 1DE- and 2DE-WB
We tried to detect the TLR3 fragment (24 kDa) corresponding to the ID564 spot using 2DE-WB and a mixture of 4 aRA exosomal protein samples with a relatively high intensity of ID564 using antibodies raised against the human TLR3 fragment of amino acid 464–647. However, the ID564 spot was not detected by the antibody; instead, 3 protein spots of 17–18 kDa were detected (Fig. 6a). We therefore focused on the TLR3 fragments (17–18 kDa) and compared the expression of these fragments among the HL (n = 10), aRA (n = 12), iRA (n = 11), and OA (n = 10) groups by 1DE-WB. The 17- to 18-kDa TLR3 spots were ultimately detected as doublet bands on 1DE-WB (Fig. 6b [i]). In 6 of the 12 aRA samples, 5 of the 11 iRA samples and 1 of the 10 OA samples, the band intensity of the TLR3 fragments was higher than the mean of that in the HL group + 2.5 standard deviations (Fig. 6b [ii] and [iii]). In the aRA and iRA groups, the ratios of samples with a high band intensity of TLR3 fragments (17–18 kDa) were significantly higher than in the HL group (Fig. 6b [ii] and [iii]). These results suggest that patients with RA more frequently have large amounts of TLR3 fragments (17–18 kDa) in serum-derived exosomes than HLs.
Fig. 6.
Detection of TLR3 in the serum-derived exosome protein. (a) TLR3 was detected by 2DE-WB from the exosomal protein sample mixture from 4 aRA patients (aRA 1, 6, 10, and 11) with a relatively high ID564 intensity. Arrowheads, TLR3 fragments (17–18 kDa). (b) (i) The TLR3 fragments (17–18 kDa) in the exosomal protein samples described in (a) were detected by 1DE-WB. (ii) The TLR3 fragments (17–18 kDa) in the exosomal protein samples from the HLs (n = 10) and patients with aRA (n = 12), iRA (n = 11), and OA (n = 10) were detected by 1DE-WB. (iii) The intensity of the bands was measured using a densitometer. The intensity of the blank area in each membrane was defined as 1.0. The obtained relative expression of the TLR3 fragments (17–18 kDa) was compared among the 4 groups. The dotted line indicates the mean + 2.5SD of the band intensity in the HL group. The intensity of the TLR3 fragments (17–18 kDa) was higher in 6 of the 12 aRA samples, 5 of the 11 iRA samples, and 1 of the 10 OA samples than the mean of that in HL groups + 2.5SD (filled circles). The ratios of samples with a high intensity of TLR3 fragments (17–18 kDa) were significantly higher in the aRA and iRA groups than in the HL group. * p < 0.05
Discussion
To our knowledge, this is the first report to compare the protein profiles of serum exosomes among patients with RA, those with OA, and HLs.
In the aRA group, ID564, identified as TLR3, showed about 6-fold higher intensity than the other groups. Given the observed small MW (24 kDa) of ID564 and the mass spectrometric data, ID564 was thought to represent a middle part fragment of TLR3. Furthermore, patients with RA more frequently had large amounts of the TLR3 fragments (17–18 kDa) in serum-derived exosomes than HLs.
TLR3 is a member of the TLR family of pattern recognition receptors of the innate immune system [27]. TLR3 is an endosomal TLR that recognizes double-stranded RNA (dsRNA), a molecular pattern associated with viral infection [27, 28]. TLR3 signal activates transcription factors of NF-κB and interferon regulatory factor (IRF) 3, which leads to the secretion of type I interferons and proinflammatory cytokines, such as IL-6 and IL-8 [27, 28]. In the intracellular trafficking and maturation of TLR3, a precursor form of TLR3 is transported to endosomes through the endoplasmic reticulum (ER) and Golgi apparatus [29]. In the endosomes, the precursor form of TLR3 is cleaved to produce a C-terminal 70-kDa fragment of TLR3 that can deliver signals [29]. The amino acid sequences of ID564 identified by the mass spectrometric analysis were located in the range of the C-terminal 70-kDa fragment (Table 3). We therefore considered that the 24-kDa fragments of TLR3 had been generated by additional proteolytic cleavage of the C-terminal 70-kDa fragment in endosomes and then taken up into exosomes after the cleavage.
Several reports have suggested that the expression of TLR3 and the activation of its pathways are associated with the pathogenesis of RA. For example, TLR3 was found to be more highly expressed in RA synovial tissue than in OA synovial tissue [30]. The stimulation of cultured fibroblast-like synoviocytes (FLSs) with poly (I-C), a synthetic TLR3 ligand, up-regulated the TLR3 expression in FLSs [30]. RNA released from necrotic synovial fluid cells of RA patients activated the TLR3 pathway in FLSs and up-regulated the expression of IFN-β mRNA and the production of IL-6 [30]. It was recently reported that poly (I:C) of a TLR3 ligand associated with extracellular vesicles (EVs) was able to selectively activate antiviral and proinflammatory responses in synovial fibroblasts [31]. In addition, it has been reported that the expression of TLR3 is increased in the spleen of rats with collagen-induced arthritis as well as in those with pritane-induced arthritis, and that the increased expression of TLR3 was decreased by treatment with MTX [32]. Thus, TLR3, which is highly expressed in synoviocytes and possibly in the spleen of patients with active RA as well, is thought to play a role in the production of proinflammatory cytokines. The markedly high levels of the TLR3 fragment in serum exosomes may be caused by the high expression of TLR3 in FLSs in patients with active RA. From this viewpoint, the 24- and the 17–18 kDa TLR3 fragments in serum exosomes may be useful markers of the activation of FLSs in patients with RA.
While the TLR3 fragments (17–18 kDa and 24 kDa) are likely non-functional, they may still have some pathological significance. We previously reported that a degradation product of Apo-AI has a biological function of promoting IL-8 secretion from cells [33]. The TLR3 fragments detected in the present study might therefore perform some biological functions that differ from those of full-length TLR3. Further studies will be needed to investigate this possibility.
A membrane-bound isoform of pro-neuregulin-3 showed 1.4-fold higher expression in the wRA group than in the OA and HL groups (Table 3, Fig. 5). Pro-neuregulin-3 is a precursor of neureglin-3, which is a ligand for ErbB4, a member of the epidermal growth factor receptor (EGFR) family [34]. It has been reported that the binding of neureglin-3 to ErbB4 promotes the proliferation, migration, and differentiation of neuroblasts [34, 35]. Although little is known about the involvement of neuregulin in the pathogenesis of rheumatic diseases, it has been reported that EGFR is expressed in fibroblasts and vascular endothelial cells and that EGFR signaling induces FLS proliferation and cytokine production in patients with RA [36]. Given the high expression of pro-neuregulin-3 in serum-derived exosomes of patients with RA detected in our study, the neuregulin-3-ErbB4 pathway may be activated to promote proliferation in FLSs. Furthermore, pro-neuregulin-3 in serum exosomes may be converted to neuregulin-3 and act as a ligand for ErbB4 in FLSs. Further studies are needed to elucidate these points.
The intensity of the spot ID236, identified as cathepsin F, was higher in the OA group than in the other three groups. Cathepsin F is a member of the papain family cysteine proteinases [37]. Given that another member of cathepsin K was reported to be involved in the pathogenesis of OA [38, 39], cathepsin F might also be involved in the pathogenesis of OA. This point needs to be investigated in the future.
The observed MWs of the proteins identified in this study were slightly different from their theoretical MWs (Table 3). Post-translational modifications, such as glycosylation, have been reported for the spots ID208 (pro-neuregulin-3, membrane-bound isoform), ID236 (cathepsin F), and ID325 (Ig alpha-2 chain C region), which may have contributed to this discrepancy in MW findings [40–42]. For spot ID494 (Keratin, type II cytoskeletal), the observed MW was smaller than the theoretical MW, suggesting that ID494 might be a keratin degradate, similar to the case of TLR3.
Exosomal lymphatic vessel endothelial hyaluronic acid receptor-1 (LYVE-1) was very recently reported to be a candidate marker of RA activity by a 2D-LC-MS/MS (two-dimensional liquid chromatography-tandem mass spectrometry) analysis of serum exosomal proteins from RA patients with complete remission and those with non-complete remission [19]. However, in that study, none of the six proteins we identified in our own were found to be candidate markers of RA activity [19]. Similarly, LYVE-1 was not identified as a differently expressed protein in our study [19]. This difference may be due to differences in the serum donors (we analyzed exosomal samples from OA patients and HLs in addition to patients with RA) and different methods used in these two studies (2D-LC-MS/MS vs. 2D-DIGE followed by MS/MS).
In addition, we isolated serum exosomes using Exoquick®, which was useful to isolate exosomes from a large number of blood samples. However, similarly as other methods, contamination of serum proteins cannot be completely avoided. It would be needed to investigate characters and functions of the individual identified proteins as exosomal proteins.
Conclusions
In the present study, we demonstrated that the serum exosomes of patients with aRA have different protein profiles from those of patients with iRA, patients with OA, and HLs. The 24- and the 17- to 18-kDa TLR3 fragments, which were found to be markedly increased in patients with aRA, may reflect the inflammatory condition of FLSs. Our findings may aid in the clarification of the roles of exosomes in the pathophysiology of RA.
Additional file
Figure S1. Location of differently enriched protein spots. Table S1. Averages of spot intensities of each group. (DOCX 101 kb)
Acknowledgements
We would like to thank Professor M. Takagi and Mr. Y. Natsuki for their help in the transmission electron microscopic analysis, Professor N. Fukui for the collection of the sera of the patients with OA, and Ms. M. Katano for her excellent technical assistance.
Funding
No funding was obtained for this study.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- 2D-DIGE
Two-dimensional differential image gel electrophoresis
- 2D-LC-MS/MS
Two-dimensional liquid chromatography-tandem mass spectrometry
- ACPA
Anti-cyclic citrullinated peptide antibody
- APC
Antigen-presenting cell
- aRA
Active RA
- CHAPS
3-(3-cholamidepropyl) dimethylammonio-1-propanesulfonate
- CRP
C-reactive protein
- DAS28-CRP
Disease activity score 28 joints using C-reactive protein
- DAS28-ESR
Disease activity score 28 joints using erythrocyte sedimentation rate
- DC
Dendritic cell
- DMARDs
Disease-modifying anti-rheumatic drugs
- dsRNA
Double-stranded RNA
- EGFR
Epidermal growth factor receptor
- ER
Endoplasmic reticulum
- ESR
Erythrocyte sedimentation rate
- FLS
Fibroblast-like synoviocyte
- HL
Healthy control
- ICAM
Intercellular adhesion molecule
- IFN
Interferon
- IL
Interleukin
- iRA
Inactive RA
- IRF
Interferon regulatory factor
- LYVE-1
Lymphatic vessel endothelial hyaluronic acid receptor-1
- MHC
Major histocompatibility complex
- MMP
Matrix metalloproteinase
- MS
Mass spectrometry
- MTX
Methotrexate
- MVBs
Multivesicular bodies
- MW
Molecular weight
- NF-κB
Nuclear factor-kappa B
- OA
Osteoarthritis
- PBS
Phosphate-buffered saline
- PD-L1
Programmed death-ligand 1
- RA
Rheumatoid arthritis
- RF
Rheumatoid factor
- SASP
Salazosulfapyridine
- SJC
Swollen joint count
- TJC
Tender joint count
- TLR
Toll-like receptor
- TNF
Tumor necrosis factor
- wRA
Whole RA
Authors’ contributions
HT participated in the study design, performed the experiments and drafted the manuscript. MA participated in its design, contributed to the interpretation of the results, and helped draft the manuscript. NS, TS, and MS participated in the acquisition and analysis of the data and contributed to editing the manuscript. KO (Kazuki Omoteyama) participated in the acquisition and analysis of the data and contributed to editing the manuscript. KO (Kazuki Okamoto) participated in the acquisition and analysis of the data and contributed to editing the manuscript. AH, TM, and ST participated in the acquisition and analysis of the data and helped edit the manuscript. MSK participated in the study design and contributed to the interpretation of the results and editing the manuscript. TK conceived the study, participated in its design and coordination, and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the ethics committee of Sagamihara National Hospital and that of St. Marianna University School of Medicine. All of the blood samples were obtained with written informed consent.
Consent for publication
Not applicable.
Competing interests
AH has received research funding from Chugai Pharmaceutical Co., Ltd. TM was a member of the Department of Lifetime Clinical Immunology (LCI) at Tokyo Medical and Dental University (TMDU). TMDU received unrestricted research grants for the Department of LCI from AbbVie GK, Ayumi Pharmaceutical Corporation, Chugai Pharmaceutical Co., Ltd., CSL Behring K.K., Japan Blood Products Organization, Mitsubishi Tanabe Pharma Corporation, Nippon Kayaku Co., Ltd., Ono Pharmaceutical Co., Ltd., Towa Pharmaceutical Co., Ltd., and UCB Japan Co. Ltd., with which TMDU paid the salary of TM. ST was supported by research grants from nine pharmaceutical companies: Abbott Japan Co., Ltd., Astellas Pharma Inc., Chugai Pharmaceutical Co., Ltd., Eisai Co., Ltd., Mitsubishi Tanabe Pharma Corporation, Pfizer Japan Inc., Takeda Pharmaceutical Company Limited, and Teijin Pharma Limited. ST received honoraria from Asahi Kasei Pharma Corporation, Astellas Pharma Inc., AbbVie GK., Chugai Pharmaceutical Co., Ltd., Ono Pharmaceutical Co., Ltd., Mitsubishi Tanabe Pharma Corporation, and Pfizer Japan, Inc. The other authors have no financial or commercial conflicts of interest to declare.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hirotaka Tsuno, Phone: +81-42-742-8311, Email: h-tsuno@sagamihara-hosp.gr.jp.
Mitsumi Arito, Email: m-ari@marianna-u.ac.jp.
Naoya Suematsu, Email: n2sue@marianna-u.ac.jp.
Toshiyuki Sato, Email: stoshi@marianna-u.ac.jp.
Atsushi Hashimoto, Email: a-hashimioto@sagamihara-hosp.gr.jp.
Toshihiro Matsui, Email: t-matsui@sagamihara-hosp.gr.jp.
Kazuki Omoteyama, Email: omoteyama@marianna-u.ac.jp.
Masaaki Sato, Email: msato@marianna-u.ac.jp.
Kazuki Okamoto, Email: k2oka@sanken.osaka-u.ac.jp.
Shigeto Tohma, Email: touma-shigeto@tokyo-hosp.jp.
Manae S. Kurokawa, Email: manae@marianna-u.ac.jp
Tomohiro Kato, Email: t3kato@marianna-u.ac.jp.
References
- 1.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 2.Record M, Subra C, Silvente-Poirot S, Poirot M. Exosomes as intercellular signalosomes and pharmacological effectors. Biochem Pharmacol. 2011;81:1171–1182. doi: 10.1016/j.bcp.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 4.Chaput N, Théry C. Exosomes: immune properties and potential clinical implementations. Semin Immunopathol. 2011;33:419–440. doi: 10.1007/s00281-010-0233-9. [DOI] [PubMed] [Google Scholar]
- 5.Clayton A, Court J, Navabi H, Adams M, Mason MD, Hobot JA, et al. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods. 2001;247:163–174. doi: 10.1016/S0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- 6.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Admyre C, Johansson SM, Paulie S, Gabrielsson S. Direct exosome stimulation of peripheral human T cells detected by ELISPOT. Eur J Immunol. 2006;36:1772–1781. doi: 10.1002/eji.200535615. [DOI] [PubMed] [Google Scholar]
- 8.Segura E, Nicco C, Lombard B, Véron P, Raposo G, Batteux F, et al. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood. 2005;106:216–223. doi: 10.1182/blood-2005-01-0220. [DOI] [PubMed] [Google Scholar]
- 9.Andreola G, Rivoltini L, Castelli C, Huber V, Perego P, Deho P, et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med. 2002;195:1303–1316. doi: 10.1084/jem.20011624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67:7458–7466. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 11.Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 13.Gary SF. Etiology and pathogenesis of rheumatoid arthritis. In: Gary SE, Ralph CB, Ted H, Ian BM, Shaun R, John SS, editors. Kelley’s textbook of rheumatology. 8. Philadelphia: Saunders; 2009. pp. 1035–1087. [Google Scholar]
- 14.Gary SF. Pathogenesis of rheumatoid arthritis. In: Paul LR, editor. UpToDate. Waltham, MA: UpToDate; 2016. [Google Scholar]
- 15.Skriner K, Adolph K, Jungblut PR, Burmester GR. Association of citrullinated proteins with synovial exosomes. Arthritis Rheum. 2006;54:3809–3814. doi: 10.1002/art.22276. [DOI] [PubMed] [Google Scholar]
- 16.Zhang HG, Liu C, Su K, Yu S, Zhang L, Zhang S, et al. A membrane form of TNF-alpha presented by exosomes delays T cell activation-induced cell death. J Immunol. 2006;176:7385–7393. doi: 10.4049/jimmunol.176.12.7385. [DOI] [PubMed] [Google Scholar]
- 17.Kato T, Miyaki S, Ishitobi H, Nakamura Y, Nakasa T, Lotz MK, et al. Exosomes from IL-1β stimulated synovial fibroblasts induce osteoarthritic changes in articular chondrocytes. Arthritis Res Ther. 2014;16:R163. doi: 10.1186/ar4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuno H, Suematsu N, Sato T, Arito M, Matsui T, Iizuka N, et al. Effects of methotrexate and salazosulfapyridine on protein profiles of exosomes derived from a human synovial sarcoma cell line of SW982. Proteomics Clin Appl. 2016;10:164–171. doi: 10.1002/prca.201500064. [DOI] [PubMed] [Google Scholar]
- 19.Yoo J, Lee SK, Lim M, Sheen D, Choi EH, Kim SA. Exosomal amyloid a and lymphatic vessel endothelial hyaluronic acid receptor-1 proteins are associated with disease activity in rheumatoid arthritis. Arthritis Res Ther. 2017;19:119. doi: 10.1186/s13075-017-1334-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun W, Zhao C, Li Y, Wang L, Nie G, Peng J, et al. Osteoclast-derived microRNA-containing exosomes selectively inhibit osteoblast activity. Cell discov. 2016;2:16015. doi: 10.1038/celldisc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, Liu J, Guo B, Liang C, Dang L, Lu C, et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat Commun. 2016;7:10872. doi: 10.1038/ncomms10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 23.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, et al. The American College of Rheumatology criteria for the classification and reporting of osteoarthritis of the knee. Arthritis Rheum. 1986;29:1039–1049. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 24.Inoue E, Yamanaka H, Hara M, Tomatsu T, Kamatani N. Comparison of disease activity score (DAS)28- erythrocyte sedimentation rate and DAS28- C-reactive protein threshold values. Ann Rheum Dis. 2007;66:407–409. doi: 10.1136/ard.2006.054205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimada S, Nakamura M, Tanaka Y, Tsutsumi K, Katano M, Masuko K, et al. Crosslinking of the CD69 molecule enhances S100A9 production in activated neutrophils. Microbiol Immunol. 2007;51:87–98. doi: 10.1111/j.1348-0421.2007.tb03878.x. [DOI] [PubMed] [Google Scholar]
- 26.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 28.Botos I, Liu L, Wang Y, Segal DM, Davies DR. The toll-like receptor 3: dsRNA signaling complex. Biochim Biophys Acta. 2009;1789:667–674. doi: 10.1016/j.bbagrm.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Cattaneo A, Gobert FX, Müller M, Toscano F, Flores M, Lescure A, et al. Cleavage of toll-like receptor 3 by cathepsins B and H is essential for signaling. Proc Natl Acad Sci U S A. 2012;109:9053–9058. doi: 10.1073/pnas.1115091109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brentano F, Schorr O, Gay RE, Gay S, Kyburz D. RNA released from necrotic synovial fluid cells activates rheumatoid arthritis synovial fibroblasts via toll-like receptor 3. Arthritis Rheum. 2005;52:2656–2665. doi: 10.1002/art.21273. [DOI] [PubMed] [Google Scholar]
- 31.Frank-Bertoncelj M, Pisetsky DS, Kolling C, Michel BA, Gay RE, Jüngel A, et al. TLR3 ligand poly (I:C) exerts distinct actions in synovial fibroblasts when delivered by extracellular vesicles. Front Immunol. 2018;9:28. doi: 10.3389/fimmu.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu W, Meng L, Jiang C, Xu J, Wang B, Han Y, et al. Overexpression of toll-like receptor 3 in spleen is associated with experimental arthritis in rats. Scand J Immunol. 2012;76:263–270. doi: 10.1111/j.1365-3083.2012.02724.x. [DOI] [PubMed] [Google Scholar]
- 33.Takakuwa Y, Kurokawa MS, Ooka S, Sato T, Nagai K, Arito M, et al. AC13, a C-terminal fragment of apolipoprotein A-I, is a candidate biomarker for microscopic polyangiitis. Arthritis Rheum. 2011;63:3613–3624. doi: 10.1002/art.30560. [DOI] [PubMed] [Google Scholar]
- 34.Zhang D, Sliwkowski MX, Mark M, Frantz G, Akita R, Sun Y, et al. Neuregulin-3 (NRG3): a novel neural tissue-enriched protein that binds and activates ErbB4. Proc Natl Acad Sci U S A. 1997;94:9562–9567. doi: 10.1073/pnas.94.18.9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kao WT, Wang Y, Kleinman JE, Lipska BK, Hyde TM, Weinberger DR, et al. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc Natl Acad Sci U S A. 2010;107:15619–15624. doi: 10.1073/pnas.1005410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan FL, Li X, Lu WG, Sun JM, Jiang DL, Xu RS. Epidermal growth factor receptor (EGFR) as a therapeutic target in rheumatoid arthritis. Clin Rheumatol. 2013;32:289–292. doi: 10.1007/s10067-012-2119-9. [DOI] [PubMed] [Google Scholar]
- 37.Wang B, Shi GP, Yao PM, Li Z, Chapman HA, Bromme D. Human cathepsin F Molecular cloning, functional expression, tissue localization, and enzymatic characterization. J Biol Chem. 1998;273:32000–32008. doi: 10.1074/jbc.273.48.32000. [DOI] [PubMed] [Google Scholar]
- 38.Kozawa E, Nishida Y, Cheng XW, Urakawa H, Arai E, Futamura N, Shi GP, Kuzuya M, Hu L, Sasaki T, Ishiguro N. Osteoarthritic change is delayed in a Ctsk-knockout mouse model of osteoarthritis. Arthritis Rheum. 2012;64:454–464. doi: 10.1002/art.33398. [DOI] [PubMed] [Google Scholar]
- 39.Kozawa E, Cheng XW, Urakawa H, Arai E, Yamada Y, Kitamura S, Sato K, Kuzuya M, Ishiguro N, Nishida Y. Increased expression and activation of cathepsin K in human osteoarthritic cartilage and synovial tissues. J Orthop Res. 2016;34:127–134. doi: 10.1002/jor.23005. [DOI] [PubMed] [Google Scholar]
- 40.Carteron C, Ferrer-Montiel A, Cabedo H. Characterization of a neural-specific splicing form of the human neuregulin 3 gene involved in oligodendrocyte survival. J Cell Sci. 2006;119:898–909. doi: 10.1242/jcs.02799. [DOI] [PubMed] [Google Scholar]
- 41.Somoza JR, Palmer JT, Ho JD. The crystal structure of human cathepsin F and its implications for the development of novel immunomodulators. J Mol Biol. 2002;322:559–568. doi: 10.1016/S0022-2836(02)00780-5. [DOI] [PubMed] [Google Scholar]
- 42.Picariello G, Ferranti P, Mamone G, Roepstorff P, Addeo F. Identification of N-linked glycoproteins in human milk by hydrophilic interaction liquid chromatography and mass spectrometry. Proteomics. 2008;8:3833–3847. doi: 10.1002/pmic.200701057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Location of differently enriched protein spots. Table S1. Averages of spot intensities of each group. (DOCX 101 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.