Abstract
Cyrtosia septentrionalis is an achlorophyllous mycoheterotrophic orchid in the subfamily Vanilloideae (Orchidaceae). This article reports C. septentrionalis’s complete plastome sequence and compare it with other orchid plastomes with a same mycoheterotrophic nutritional mode. The C. septentrionalis plastome has decreased to 96,859 bp in length, but it still maintains a quadripartite structure. The C. septentrionalis plastome contains 38 protein-coding genes, 25 tRNA genes, and four ribosomal RNA genes. Most genes related to photosynthesis have been lost, whereas the majority of housekeeping genes remain; this pattern corresponds to the end of stage 3 gene degradation. The inverted repeat regions of the C. septentrionalis plastome have decreased to 10,414 bp and mainly contain the gene ycf2. A block consisting of four rrn genes and rps7 and rps12 has shifted to a small single-copy region. As a result, the small single-copy region was found to be expanded, despite the loss of all ndh genes in the region. Three inversion mutations are required to explain the C. septentrionalis plastome’s current gene order. The species is endangered, and these results have implications for its conservation.
Keywords: plastome, gene loss, inverted repeat contraction, gene relocation, Cyrtosia septentrionalis, mycoheterotrophy
Introduction
The family Orchidaceae consists of 736 genera, comprising 28,000 species (Christenhusz and Byng 2016), of which the plastome sequences of 116 species from 38 genera have been completely decoded (NCBI database, July 7, 2018). Although most Orchidaceae species have a photosynthetic nutritional mode that is generally similar to other plants, 232 species belonging to 43 genera do not photosynthesize—or they photosynthesize at insignificant levels—and instead rely on mycoheterotrophy for nutrition (Merckx et al. 2013). Among the five subfamilies of Orchidaceae, complete mycoheterotrophism is observed in three subfamilies—Vanilloideae, Orchidoideae, and Epidendroideae—but not in Cypripedioideae or Apostasioideae.
To date, plastome studies on mycoheterotrophic orchids have been performed for 20 species from 10 genera. All of these genera belong to the Epidendroideae subfamily, except for one Rhizanthella species (Delannoy et al. 2011), which belongs to Orchidoideae. These include one Aphyllorchis species (Feng et al. 2016), one Cephalanthera species (Feng et al. 2016), seven Corallorhiza species (Barrett and Davis 2012; Barrett et al. 2018), one Cymbidium species (Kim et al. 2017; Kim et al. 2018), two Epipogium species (Schelkunov et al. 2015), one Eulophia species (Huo et al. 2017), one Gastrodia species (Yuan et al. 2018), one Hexalectris species (Barrett and Kennedy 2018), and four Neottia species (Logacheva et al. 2011; Feng et al. 2016). The plastomes of Platanthera japonica and Cremastra appendiculata have also been decoded (Dong et al. 2018), revealing typical photosynthetic plastomes, even some other congeneric species are mycoheterotrophy.
Plastomes of mycoheterotrophic plants that have lost their photosynthetic ability have been reported to degrade rapidly (Braukmann and Stefanović 2012; Wicke et al. 2013; Wu et al. 2017). Mycoheterotrophic orchids also lost their photosynthetic ability and various levels of gene loss have been observed in the orchid plastomes of 22 species in 10 genera (Logacheva et al. 2011; Barrett and Davis 2012; Schelkunov et al. 2015; Feng et al. 2016; Barrett et al. 2018). With regard to gene loss patterns in these plastomes, ndh genes are usually lost first, followed by photosynthetic genes (atp, psa, psb, and pet gene classes) and housekeeping genes (rps, rpl, trn, and rrn gene classes), in order of precedence (Barrett and Davis 2012; Wicke et al. 2016; Graham et al. 2017).
Of the 14 genera (245 species) in the Vanilloideae subfamily (Chase et al. 2015), five genera (41 species) are classified as mycoheterotrophic orchids (Merckx et al. 2013). However, only plastomes of photosynthetic Vanilla species have been reported (Lin et al. 2015; Amiryousefi et al. 2017; Niu et al. 2017), and there is no complete plastome sequence of a mycoheterotrophic vanilloid orchid. Therefore, this study aimed to decode the plastome of Cyrtosia septentrionalis (Rchb.f.) Garay, a mycoheterotrophic Vanilloideae.
The tribe Vanilleae of subfamily Vanilloideae consists of 9 genera, 169 species (Chase et al. 2015). The phylogenetic position of Cyrtosia in Vanilleae has been well established using various molecular markers such as plastid psaB, rbcL, and psbC (Cameron 2004; Cameron and Carmen Molina 2006), mitochondrial atpA and nad1B-C (Cameron 2009), and nuclear ribosomal RNA and xdh sequences (Cameron 2009; Górniak et al. 2010). These studies suggest that Cyrtosia, Erythrorchis, and Pseudovanilla form a clade that is sister to Vanilla. Complete plastome sequence data are available for Vanilla but not for the other three genera. Therefore, we selected the Vanilla plastome as a reference sequence to compare the structure and gene contents of the Cyrtosia plastome for this study.
Five species of Cyrtosia are distributed throughout the tropical and subtropical regions of Southeast Asia (Merckx et al. 2013); among them, C. septentrionalis is found in warm areas of Japan, China, and South Korea (Lee 2011). Cyrtosia septentrionalis is a perennial plant that grows to around 50 cm tall. It is a nonphotosynthetic orchid that has white subterranean rhizomes, red aerial stems, and no leaves. Its flowers bloom in early summer, and its red fruits ripen from summer through autumn (Lee 2011). Its pharmacological value makes the species a target for overcollecting, and as a result it has been designated a legally protected species and is protected by the Korean government under the Biodiversity Conservation Act.
This study completely decoded the plastome of C. septentrionalis using next-generation sequencing. These data revealed outstanding cases of gene loss patterns, inverted repeat (IR) contraction and expansion, and gene relocation, and these are discussed in terms of mycoheterotrophy orchid plastomes in general. Furthermore, data on simple sequence repeats (SSRs) with large variability are also presented. These findings offer strategies not only for future plastome studies but also for population genetic studies and efforts to conserve this endangered species.
Materials and Methods
A living C. septentrionalis individual was collected from Jinan-gun, Jeollabuk-do, South Korea, with a collection permit. Genomic DNA was extracted using a G-spinII Plant Genomic DNA extraction kit (iNtRON, Seongnam, Korea). The extracted DNA was deposited in the Plant DNA Bank in Korea under accession number PDBK2016-1045.
Approximately 100 ng of extracted DNA (270.30 ng/µl) was used for library construction and raw sequence reads were generated using Illumina MiSeq (San Diego, CA). For trimming and normalization of raw reads, BBDuk version 37.64 and BBNorm version 37.64—both implemented in Geneious 11.1.2 (Kearse et al. 2012)—were used with kmer length of 27. The trimmed reads were assembled de novo in the Geneious assembler using two different methods. For the first method, plastid reads were filtered and collected from trimmed reads using Vanilla planifolia as a reference (GenBank accession number NC036809); the collected plastid reads were then subjected to de novo assembly. The reference-guided assembly method was used to assemble de novo results into complete plastome sequences. For the second method, all redundant reads were removed from trimmed reads by the normalization process. The normalized reads were subjected to de novo assembly and eight plastome contigs were recovered. All redundant reads were mapped to the plastome contigs and finally a single plastome contig was recovered. Possible sequence errors were corrected using redundant reads.
The two de novo assembly methods generated a single, identical contig. The plastome was annotated using the National Center for Biotechnology Information’s (NCBI) BLAST and Geneious 11.1.2 and tRNAscan-SE (Lowe and Chan 2016).
Twenty-nine orchid plastome sequences were downloaded from the NCBI to compare genes (supplementary table S1, Supplementary Material online). Genomic tandem repeats were identified using Phobos v3.3.12 (Mayer 2010). Only perfect repeats with a minimum total length of 10 bp were located. The plastome of C. septentrionalis was aligned with two sequences, V. planifolia and Habenariaradiata (NC035834), using the progressiveMAUVE (Darling et al. 2010) method to detect genomic rearrangement. A circular plastome map was visualized in OGDraw (Lohse et al. 2007).
For phylogenetic tree construction, 79 protein-coding gene and four rRNA gene sequences were aligned using the MUSCLE v.3.8.425 program (Edgar 2004), which was implemented in Geneious 11.1.2. The aligned sequences were 77,315 bp in length. A maximum likelihood analysis was conducted using RAxML v 7.7.1 (Stamatakis et al. 2008) with GTR base substitution model, which was suggested by PAUP modeltest (Posada and Crandall 1998).
Results and Discussion
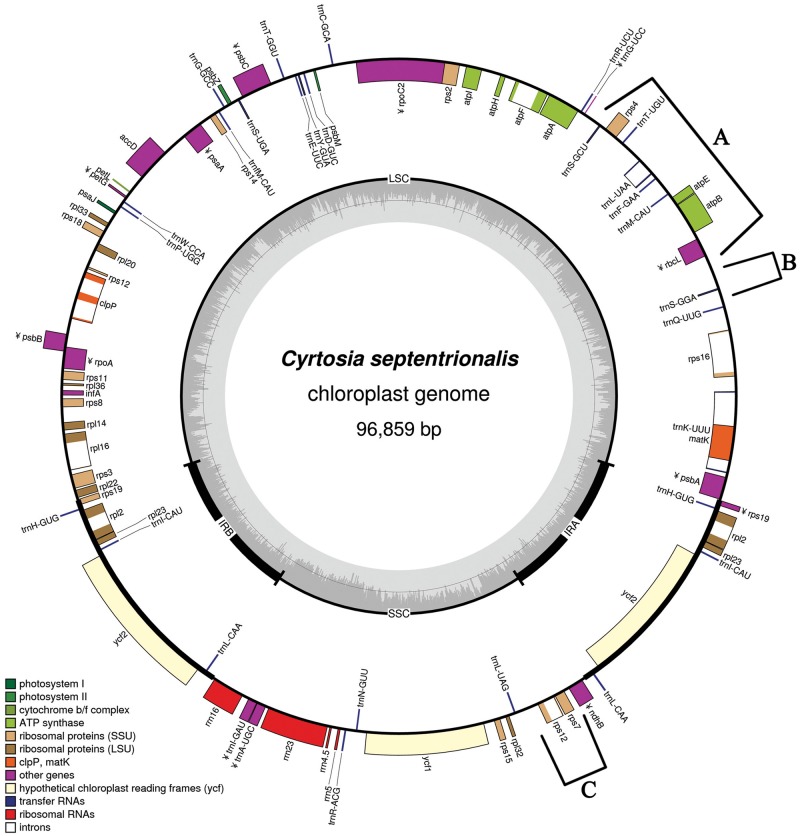
A total of 12,363,464 trimmed and 1,851,162 normalized reads were recovered from 12,551,898 raw reads, which were an average of 301 bp in length. Of these, 316,705 reads (2.76%) were plastid reads. Average coverage depth was 709 times for each site. The complete plastome of C. septentrionalis was found to be 96,859 bp in length (fig. 1). The plastome shows a typical quadripartite structure, with 58,085 bp in large single copy (LSC), 17,946 bp in small single copy (SSC), and 10,414 bp in IR regions (fig. 2 and supplementary table S1, Supplementary Material online). The plastome size of nonphotosynthetic C. septentrionalis was 65% that of the photosynthetic Vanilla planitifolia, and both belong to tribe Vanilleae.
Fig. 1.
—Plastid circle map of Cyrtosia septentrionalis. Pseudogenes are marked with the letter ¥. Three inversion regions along the C. septentrionalis plastome are marked on the circle map (A, 7.7-kb inversion; B, 1.4-kb inversion; C, 1.8-kb inversion).
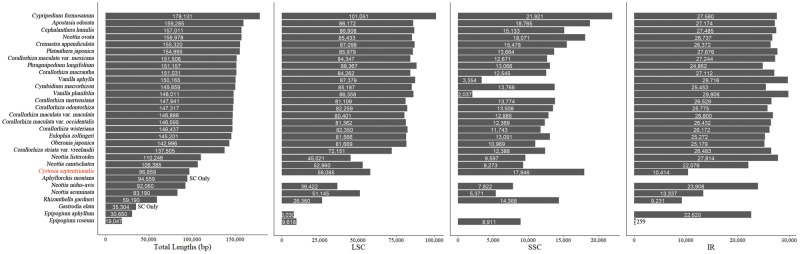
Fig. 2.
—General features of Cyrtosia septentrionalis and 29 other orchid plastomes. All 21 of the decoded mycoheterotrophic orchid plastomes are included except Hexalectris warnockii, which was not available in the NCBI database. Species are organized by plastid size.
The C. septentrionalis plastome size shows a medium range of variation compared with other nonphotosynthetic orchids (fig. 2). Initially, it seems similar to the plastomes of Aphyllorchis montana and Neottia nidus-avis (fig. 2), but a detailed comparison revealed quite different characteristics between the three species. Although C. septentrionalis and N. nidus-avis show quadripartite structures, A. montana only contains the SC region, as the IR region has been lost. In addition, the sizes of the LSC, IR, and SSC regions are remarkably different between the plastomes of C. septentrionalis and N. nidus-avis. When only considering the size of each region, the plastome of C. septentrionalis shows features that are not found in any other orchid. This can be explained by the following three evolutionary phenomena, which we discuss below: gene loss, IR boundary shift, and gene relocation.
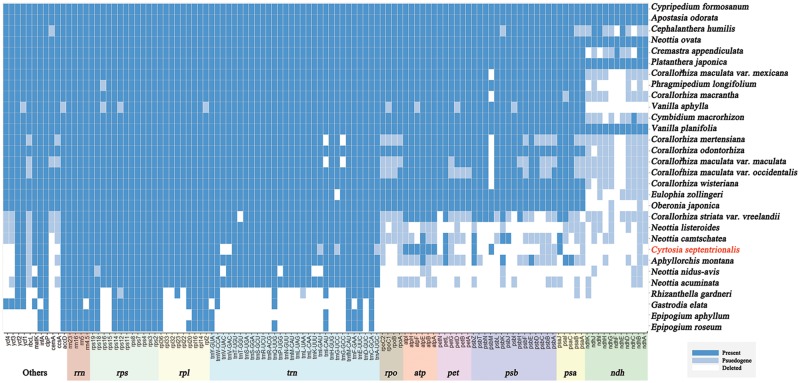
The plastomes of photosynthetic land plants usually contain about 113 genes. These include 79 protein-coding genes, 30 tRNA genes, and four ribosomal RNA genes (Shinozaki et al. 1986; Kim and Lee 2004). However, the plastome of C. septentrionalis only has 38 protein-coding genes, 25 tRNA genes, and four ribosomal RNA genes (fig. 1 and supplementary table S2, Supplementary Material online). This means that around 52% of the protein-coding genes and 17% of the tRNA genes have been lost or pseudogenized. Most of the lost genes are those involved in photosynthesis. Initially, all 11 ndh genes were lost (fig. 3). Of the 26 genes involved in photoelectron transfer (psa, psb, and pet), only four—psaJ, psbM, psbZ, and petL—remain. However, all six genes that form ATP synthase are present in a functional form. Furthermore, ccsA, cemA, rbcL, ycf3, and ycf4 genes, which are directly or indirectly involved in photosynthesis, were also lost. However, accD, clpP, matK, infA, ycf1, and ycf2, involved in plastid metabolism and housekeeping, are still present. In addition, all 25 genes (rps, rpl, and rrn) that make ribosomal proteins and ribosomal RNA are present, and 25 out of 30 tRNA genes (trn) are present. On the other hand, all four RNA polymerase genes were lost.
Fig. 3.
—Summary of gene content for Cyrtosia septentrionalis and 29 other orchid plastomes. Blue-colored boxes represent active genes, light blue-colored boxes indicate pseudogenes, and blank boxes denote loss of the gene. Species are organized by plastome size.
In summary, most of C. septentrionalis’s plastome genes involved in photosynthesis have been lost, whereas the majority of its housekeeping genes are still present (fig. 3). Gene losses occurred at levels similar to those reported for Corallorhiza striata and Neottia camtschatea (Barrett and Davis 2012; Feng et al. 2016). Compared with the reported plastome degradation progression stages of nonphotosynthetic plants (Barrett and Davis 2012; Wicke et al. 2016; Graham et al. 2017), the plastome of C. septentrionalis is considered to correspond to the end of plastome degradation stage 3. Eight genes have intron(s) in the C. septentrionalis plastome: atpF, clpP, rps12, rps16, rpl16, rpl2, trnK(UUU), and trnL(UAA). Among them, the type IIA intron trnK(UUU) is often absent in other nonphotosynthetic plastomes such as Cuscuta and Pilostyles (McNeal et al. 2009; Graham et al. 2017), but it is present in the C. septentrionalis plastome. The same intron is also found in the various orchid plastomes, such as A.montana, Corallorhiza striata var. vreelandii, Neottia acuminata, N. camtschatea, and Neottialisteroides (Barrett and Davis 2012; Feng et al. 2016).
The plastome of C. septentrionalis has a general quadripartite structure because it contains an IR region. However, the IR region has been drastically reduced to 10,414 bp, most of which is occupied by ycf2, rpl23, rpl2, and three trn genes (figs. 1 and 2). This is markedly different from most common land plant plastomes and is even significantly different from plastomes of plants from the same family (Orchidaceae). The IR region usually contains four rrn genes, rps7, rps12, three to five trn genes, and ycf2. However, in the C. septentrionalis plastome, all of those genes are located in the SSC region. When all ndh genes located in the SSC region have been lost, the SSC region is usually shortened (Lin et al. 2015; Feng et al. 2016). However, C. septentrionalis lost all of its ndh genes, but the SSC region had actually extended to 17,946 bp whereas the IR region was greatly shortened. This phenomenon is related to the fact that IR contraction occurred at the SSC boundary. This can be explained as the result of gene loss and IR expansion/contraction. The IR region is usually maintained, even in the case of mycoheterotrophic orchids in which various plastid gene losses have occurred. This can be attributed to the fact that, although photosynthetic function has been lost, the plastid’s basic function remains (Schelkunov et al. 2015; Feng et al. 2016; Barrett et al. 2018). The presence of the IR is believed to contribute to the plastome’s stability (Palmer and Thompson 1982). In nonphotosynthetic orchids, the IR has only been reported to be lost in the plastomes of Gastrodia elata and Aphylloorchis montana (Feng et al. 2016; Yuan et al. 2018).
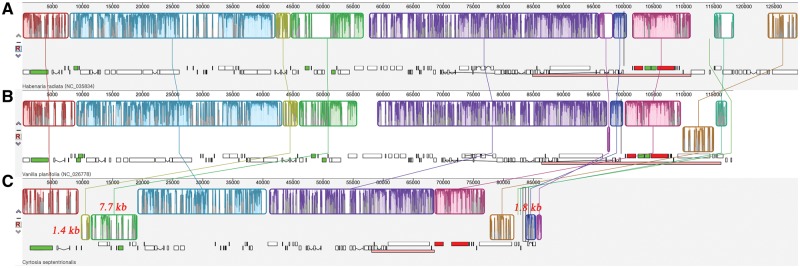
To explain why the gene order of the C. septentrionalis plastome differs from that of V.planifolia, even though they belong to the same tribe (supplementary fig. S1, Supplementary Material online), three inversions must be assumed (fig. 4). Based on the results of comparative studies, it can be inferred that two inversions occurred in the LSC region and one in the SSC region. The 7.7-kb inversion in the LSC region includes the pseudo rbcL–atpB–atpE–rps4 gene block, which is located alongside another 1.4-kb inversion in the ycf3 and trnS-GGA gene region. The third inversion is 1.8 kb long, is found in the SSC region, and includes the rps7 and rps12 genes (fig. 4). The three inversions found in the C. septentrionalis plastome are unique to this species; however, they are not special in terms of nonphotosynthetic plants, as gene losses are usually accompanied by gene relocations, for example, the atpF–atpH region inversion in A.montana (Feng et al. 2016), 11-kb inversion between psbA–rps2 in N.acuminata (Feng et al. 2016), 16-kb inversion between petB–cemA in Corallorhiza maculata (Barrett et al. 2014), and 29-kb inversion between ycf3–trnS-GCU in Hexalectris warnockii (Barrett and Kennedy 2018).
Fig. 4.
—Plastome alignment using progressive MAUVE including (A) Habenaria radiata (NC035834), (B) Vanilla planifolia (NC026778), and (C) Cyrtosia septentrionalis sequences. Inversions between C. septentrionalis and V. planifolia are indicated as a red letter with its size. Vanilla and Cyrtosia are phylogenetically closely related genera in the tribe Vanilleae, but are not sister group (Cameron 2004, 2009; Cameron and Carmen Molina 2006; Górniak et al. 2010).
A large number of SSRs are present in the C. septentrionalis plastome. Of the 135 SSRs, the majority (85) are pentanucleotide repeats, followed by 26 mononucleotide, 15 dinucleotide, seven tetranucleotide, and two trinucleotide repeats (supplementary table S3, Supplementary Material online). In nonphotosynthetic orchid plastomes, phenomena such as gene loss, IR contraction, and gene relocation seem to occur in a complex manner during evolutionary processes. These processes are considered to be relaxation processes necessary to facilitate gene exclusion while maintaining the function of the remaining genes.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the Saemangeum Regional Environmental Office of Korea for the Cyrtosia septentrionalis collection permit. This work was supported by the National Research Foundation of Korea (NRF) under Grant No. NRF-2015M3A9B8030588 and by the National Institute of Biological Resources (NIBR) under the genetic diversity research program (2016) for endangered species to K.J.K. We thank the two anonymous reviewers for their helpful suggestions, which improved the manuscript.
Footnotes
Data deposition: Data have been deposited at NCBI GenBank under the accession MH615835.
Literature Cited
- Amiryousefi A, Hyvönen J, Poczai P.. 2017. The plastid genome of Vanillon (Vanilla pompona, Orchidaceae). Mitochondrial DNA B Resour. 2(2):689–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CF, et al. 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 31(12):3095–3112. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Davis JI.. 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 99(9):1513–1523. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Kennedy AH.. 2018. Plastid genome degradation in the endangered, mycoheterotrophic, North American orchid Hexalectris warnockii. Genome Biol Evol. 10(7):1657–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CF, Wicke S, Sass C.. 2018. Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex. New Phytol. 218(3):1192–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braukmann T, Stefanović S.. 2012. Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol Biol. 79(1–2):5–20. [DOI] [PubMed] [Google Scholar]
- Cameron KM. 2004. Utility of plastid psaB gene sequences for investigating intrafamilial relationships within Orchidaceae. Mol Phylogenet Evol. 31(3):1157–1180. [DOI] [PubMed] [Google Scholar]
- Cameron KM. 2009. On the value of nuclear and mitochondrial gene sequences for reconstructing the phylogeny of vanilloid orchids (Vanilloideae, Orchidaceae). Ann Bot. 104(3):377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron KM, Carmen Molina M.. 2006. Photosystem II gene sequences of psbB and psbC clarify the phylogenetic position of Vanilla (Vanilloideae, Orchidaceae). Cladistics 22(3):239–248. [Google Scholar]
- Chase MW, et al. 2015. An updated classification of Orchidaceae. Bot J Linn Soc. 177(2):151–174. [Google Scholar]
- Christenhusz MJM, Byng JW.. 2016. The number of known plants species in the world and its annual increase. Phytotaxa 261(3):201–217. [Google Scholar]
- Darling AE, Mau B, Perna NT.. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5(6):e11147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E, Fujii S, Colas Des Francs-Small C, Brundrett M, Small I.. 2011. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 28(7):2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong WL, et al. 2018. Molecular evolution of chloroplast genomes of orchid species: insights into phylogenetic relationship and adaptive evolution. Int J Mol Sci. 19(3):716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YL, et al. 2016. Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol Evol. 8(7):2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górniak M, Paun O, Chase MW.. 2010. Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, xdh: congruence with organellar and nuclear ribosomal DNA results. Mol Phylogenet Evol. 56(2):784–795. [DOI] [PubMed] [Google Scholar]
- Graham SW, Lam VKY, Merckx VSFT.. 2017. Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes. New Phytol. 214(1):48–55. [DOI] [PubMed] [Google Scholar]
- Huo X, Zhao Y, Qian Z, Liu M.. 2018. Characterization of the complete chloroplast genome of Eulophia zollingeri, an endangered orchid in China. Conserv Genet Resour. 10(4):817–819. [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Shin CH, Sun H, Kim JH.. 2018. Sequencing of the plastome in the leafless green mycoheterotroph Cymbidium macrorhizon helps us to understand an early stage of fully mycoheterotrophic plastome structure. Plant Syst Evol. 304(2):245–258. [Google Scholar]
- Kim K-J, Lee H.. 2004. Complete chloroplast genome sequences from Korean ginseng (Panax ginseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11(4):247–261. [DOI] [PubMed] [Google Scholar]
- Kim YK, et al. 2017. The complete plastome sequence of the endangered orchid Cymbidium macrorhizon (Orchidaceae). Mitochondrial DNA B Resour. 2(2):725–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NS. 2011. Illustrated flora of Korean orchids. Seoul (Korea: ): Ewha Womans University Press (in Korean; ). [Google Scholar]
- Lin CS, et al. 2015. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci Rep. 5:9040.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Penin AA.. 2011. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol Evol. 3(1):1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R.. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Chan PP.. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C. 2010. Phobos 3.3.11. Available from: http://www.ruhr-uni-bochum.de/ecoevo/cm/cm_phobos.htm, last accessed July 25, 2018.
- McNeal JR, Kuehl JV, Boore JL, Leebens-Mack J, dePamphilis CW.. 2009. Parallel loss of plastid introns and their maturase in the genus Cuscuta. PLoS One. 4(6):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckx VSFT, et al. 2013. Taxonomy and classification In: Merckx VSFT, editor. Mycoheterotrophy: the biology of plants living on fungi. New York: Springer; p. 19–101. [Google Scholar]
- Niu Z, et al. 2017. The complete plastome sequences of four orchid species: insights into the evolution of the Orchidaceae and the utility of plastomic mutational hotspots. Front Plant Sci. 8:715.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Thompson WF.. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29(2):537–550. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA.. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14(9):817–818. [DOI] [PubMed] [Google Scholar]
- Schelkunov MI, et al. 2015. Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol Evol. 7(4):1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 5(9):2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J.. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57(5):758–771. [DOI] [PubMed] [Google Scholar]
- Wicke S, et al. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25(10):3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, et al. 2016. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc Natl Acad Sci U S A. 113(32):9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-S, Wang T-J, Wu C-W, Wang Y-N, Chaw S-M.. 2017. Plastome evolution in the sole hemiparasitic genus laurel dodder (Cassytha) and insights into the plastid phylogenomics of Lauraceae. Genome Biol Evol. 9(10):2604–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, et al. 2018. The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat Commun. 9(1):1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.