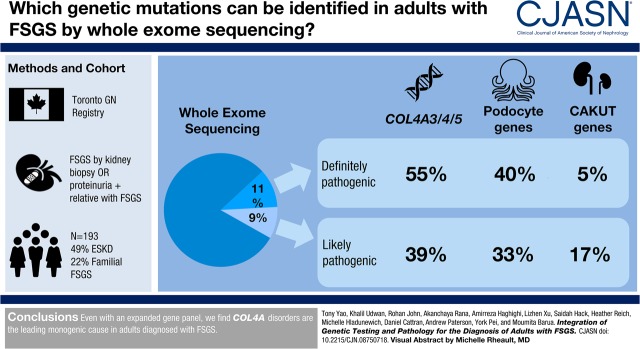
Visual Abstract
Keywords: FSGS; idiopathic nephrotic syndrome; human genetics; type 4A collagen; renal development; Glomerular Basement Membrane; Podocytes; nephrotic syndrome; Whole Exome Sequencing; Urogenital Abnormalities; vesico-ureteral reflux; kidney; glomerulonephritis; Kidney Failure, Chronic; Renal Insufficiency; Genetic Testing; Registries; Cohort Studies
Abstract
Background and objectives
FSGS and nephrotic syndrome studies have shown that single gene causes are more likely to be found in pediatric cases than adults. Consequently, many studies have examined limited gene panels in largely pediatric cohorts.
Design, setting, participants, & measurements
Whole-exome sequencing was performed in adults with FSGS diagnosed between 1976 and 2017 in the Toronto GN Registry. An expanded panel of 109 genes linked to FSGS, glomerular basement membrane abnormalities, as well as causes of pediatric ESKD including congenital abnormalities of the kidney and urinary tract (CAKUT) and nephronophthisis, were examined.
Results
The cohort was composed of 193 individuals from 179 families. Nearly half (49%) developed ESKD at a mean age of 47±17 years. The genetic diagnostic rate was 11%. Of definitely pathogenic variants, 55% were in COL4A (A3/A4/A5), 40% were in podocyte genes, and 5% were in CAKUT genes. Many, but not all individuals with COL4A definitely pathogenic variants had some evidence of glomerular basement membrane abnormalities. The estimated mean survival/age of kidney failure for individuals with COL4A definitely pathogenic variants was 58 years (95% confidence interval, 49 to 69), far later than what has been reported in the literature. Likely pathogenic variants were identified in an additional 9% of the cohort, with most in COL4A. Correlation with glomerular basement membrane morphology suggested a causal role for at least some of these likely pathogenic variants.
Conclusions
Even with an expanded gene panel, we find that COL4A disorders are the leading monogenic cause in adults diagnosed with FSGS.
Podcast
This article contains a podcast at https://www.asn-online.org/media/podcast/CJASN/2019_01_15_CJASNPodcast_19_02_.mp3
Introduction
FSGS is a clinicopathologic entity characterized by proteinuria, with or without features of nephrotic syndrome, glomerulosclerosis and podocyte foot process effacement. It is divided into primary and secondary forms, but overlapping clinical and histopathologic features make precise etiologic diagnosis challenging. As a result, many cases of FSGS treated with immunosuppression are unresponsive, progressing to ESKD necessitating dialysis and/or transplantation (1).
The study of hereditary forms of FSGS has helped inform our understanding of its molecular pathogenesis (2–4). Most genes in which specific variants are reported to cause FSGS have protein products that are important regulators of the podocyte actin cytoskeleton or slit diaphragm components, in keeping with the current paradigm that FSGS represents a primary podocyte disorder (2–6). Much of the literature where this viewpoint arises is focused on pediatric patients and limited to sequencing gene panels composed mostly of genes associated with disease that are also expressed in the podocyte. However, such a list can quickly become incomplete in the next-generation era of rapid disease gene discovery.
Several reports have implicated pathogenic variants in COL4A3/A4/A5 in FSGS, with at least one indicating that it is the leading single gene (i.e., monogenic) cause in adults with disease (7–13).These reports consist of smaller sample sizes, cohorts comprising many related individuals, and examined only select genes. Previous work has also demonstrated that pathogenic variants in genes associated with other kidney diseases, such as congenital abnormalities of the kidney and urinary tract (CAKUT) and not expressed in the podocyte, are also found in individuals clinically diagnosed with FSGS (14,15).
We performed whole-exome sequencing in a large cohort of mostly unrelated cases comprising 193 adults from 179 families with FSGS. We examined an expanded subset of 109 genes associated with FSGS, basement membrane abnormalities, as well as causes of pediatric ESKD including CAKUT and nephronophthisis, to determine the distribution of pathogenic variants in a genetically determined, predominantly European, adult population.
Materials and Methods
Patient Ascertainment
Patients were recruited at University Health Network, Toronto, Ontario, Canada, with the majority from the Toronto GN Registry from 1976 to 2017, after receiving informed consent in accordance with the hospital Research Ethics Board. We obtained longitudinal clinical data and eventually blood, saliva, or isolated DNA. Clinical information was obtained from telephone interviews, questionnaires, and physician reports. Genomic DNA was extracted from blood or saliva samples using standard procedures.
Clinical Definitions
Study participants were probands with a pathologic diagnosis of FSGS as a result of (1) segmental and/or global glomerular sclerosis, (2) podocyte effacement, or (3) nonspecific immunofluorescence, and without any other diagnoses such as SLE nephritis, Henoch–Schonlein purpura/IgA, membranoproliferative GN/C3, membranous nephropathy, and HIV-associated nephropathy. Relatives of probands with >500 mg of protein excretion per day were considered as affected and also included. Familial cases were defined as two or more affected individuals that were not separated by more than two meiotic events. Complete remission was defined as reduction of proteinuria to <0.3 g/d, associated with stable kidney function (change in serum creatinine <25% from baseline) after 3–6 months of immunosuppressive treatment including prednisone, cyclophosphamide, cyclosporine, tacrolimus, mycophenolate mofetil, azathioprine, and rituximab, either alone or in combination. Partial remission was defined as a reduction of proteinuria to 0.3–3 g/d and stable serum creatinine. Resistance was defined as no complete or partial remission. Clinical data are reported as percentage of all individuals with available information.
Clinicopathologic parameters were compared in pairwise combinations of patients with pathogenic type 4 collagen variants, patients with other pathogenic variants, and patients with no proven genetic cause. For all percentages shown, the denominator was adjusted to reflect missing data. Statistical significance was determined by the two-sample t test (two-tailed) and two-proportion Z test (two-tailed). The Kaplan–Meier method was used to analyze censored events over time to estimate mean age at disease onset and ESKD. Kaplan–Meier survival plots were generated with IBM SPSS Statistics version 25.0 (IBM, Armonk, NY). Statistical significances were determined by the log rank (Mantel–Cox) test.
Exome Capture and Next-Generation Sequencing
Whole-exome sequencing was performed by The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Ontario, Canada. A shotgun library was made from each sample and captured using the Agilent SureSelect Human All Exon V5 (Santa Clara, CA) according to protocol. The manufacturer’s specifications state that the capture regions total approximately 180,000 exons from approximately 18,700 genes or 54 Mb. Enriched libraries were then sequenced by 150 bp paired-end read sequencing on Illumina HiSeq 2500 (Illumina Inc., San Diego, CA).
In Silico Data Processing
Reads were mapped to the hg19 reference sequence using the BWA-backtrack algorithm from BWA v0.5.9 (16). Duplicate reads were removed using MarkDuplicates from Picard v1.79. Local read realignment around insertions and deletions (indels), base quality score recalibration, and variant calling with UnifiedGenotyper, were accomplished using GATK v1.1–28 (17,18). Single nucleotide polymorphism calls were subjected to variant quality score recalibration. Indels were discarded if they overlapped repeat masked regions, and hard-filtered by variant call annotations QualByDepth (QD <10.0), ReadPosRankSumTest (ReadPosRankSum <−20.0), and Strand Bias (SB >−0.01). Base calling was performed using CASAVA v1.8.2. Copy number variants were identified using XHMM after filtering out regions with extreme GC-content and repeat-masked regions (19,20).
Genetic ancestry of the study cohort was analyzed using iAdmix software (https://bansal-lab.github.io/software/iadmix.html) with population allele frequencies from HapMap Phase III data (http://hapmap.ncbi.nlm.nih.gov/) (21,22).For each individual, admixture proportions were calculated then summed into three continental groups: European, African, and East Asian. Individuals with estimated >90% ancestry from one continental population were classified as belonging to the respective continental group, whereas the remaining individuals were considered admixed.
Variant calls were compared against ethnically matched controls. The following minor allele frequency cut-offs, as determined in gnomAD (http://gnomad.broadinstitute.org/), were used for dominant and recessive disease genes respectively: 0.00005, and 0.005 (accessed February 22, 2018) (23). In addition, variants meeting these criteria and that were shared by affected relatives where applicable were also kept. Copy number variants were kept if absent from the Database of Genomic Variants (http://dgv.tcag.ca/; accessed June 25, 2018) (24).
Assignment of Pathogenicity Categories
Recommendations by the American College of Medical Genetics (ACMG) were followed to categorize variants (25). Variants in 109 genes associated with FSGS, nephrotic syndrome, CAKUT, or nephronophthisis were examined. Zygosity of each variant in the patient had to match the corresponding gene’s known pattern of inheritance. Missense variants predicted to be deleterious by more than half of all in silico algorithms in dbNSFP v3.0 were considered “consistently predicted to be deleterious” (26,27). Human Splicing Finder 3.1 (http://www.umd.be/HSF3/HSF.shtml) was used to analyze the effect of splice site variants (28).
The following types of variants were considered “definitely pathogenic”: variants previously reported to cause FSGS or related phenotypes, and de novo variants. The following types of variants were considered “likely pathogenic”: nonsense, frameshift, and canonical splice site variants in genes in which loss of function is a known pathogenic mechanism; or variants affecting the same codon as a previously reported pathogenic mutation and consistently predicted to be deleterious by in silico algorithms. The following types of variants were considered “possibly pathogenic”: variants residing in well established functional domains that contain previously reported pathogenic mutations and consistently predicted to be deleterious by in silico algorithms, or variants in trans with a previously reported pathogenic mutation and consistently predicted to be deleterious by in silico algorithms.
The remaining rare variants were considered variants of unknown significance.
Our definitions of possibly pathogenic falls within likely pathogenic variant ACMG criteria. However, we have added the category of possibly pathogenic to reflect the different levels of evidence that is being satisfied.
De Novo Variation and Confirming Biologic Identity of Parents
The PCR AmpFLSTR Identifiler kit was used to test 15 short tandem repeats. The number of repeats for each specified locus was evaluated for concordance with parental genotypes.
Results
Demographics
The cohort was composed of 193 individuals from 179 families. Forty three individuals were familial cases belonging to 29 pedigrees, with the remaining 150 cases being sporadic. Sex distribution within the cohort was 56% male (Supplemental Table 1, Table 1). In familial cases, 51% were male, whereas in sporadic cases, 57% were male (P=0.49). Population substructure revealed that 65% were European, 4% were African, 12% were East Asian, and 19% were of admixed descent (Table 1). The mean age of onset was 34±16 (SD) years, and most (78%) individuals presented with normal kidney function (Table 1). A total of 49% of patients developed ESKD at mean age of 47±17 years at the time of last follow-up (Table 1). Of these, 59% underwent kidney transplantation, with 10% showing disease recurrence (Table 1).
Table 1.
Baseline characteristics including pathologic features of the sequenced cohort
| Characteristic | Percentage (%) of Available Data | No. of Patients |
|---|---|---|
| Sex | 193 | |
| Male | 56 | 109 |
| Female | 44 | 84 |
| Ethnicity | 193 | |
| European | 65 | 125 |
| African | 4 | 7 |
| East Asian | 12 | 24 |
| Admixed | 19 | 37 |
| Pathology where electron microscopy is available | 126 | |
| Glomerular basement membrane abnormalities | 30 | 37 |
| Podocyte effacement >50% | 73 | 91 |
| Patients on therapy | 154 | |
| Prednisolone alone | 23 | 35 |
| Other immunosuppression alone | 4 | 6 |
| Prednisolone and other immunosuppression | 43 | 66 |
| ACEi+ARB alone | 31 | 47 |
| Remission rate on steroid or immunosuppression throughout the course of the disease | 82 | |
| Partial remission | 45 | 37 |
| Complete remission | 29 | 24 |
| No remission | 26 | 21 |
| Patients with known status for ESKD | 147 | |
| Progression to ESKD | 49 | 72 |
| Kidney transplant (% of transplant in patients with ESKD) | 59 | 42 |
| Recurrence of disease after kidney transplant (within the first yr) | 10 | 4 |
| Mean age±SD (years) | No. of Patients | |
| Onset of kidney disease | 34±16 | 161 |
| ESKD | 47±17 | 70 |
A total of 193 individuals from 179 families had exome sequencing performed but data were not available for all participants. Number of individuals in which data are available or satisfying the criteria is indicated in the last column. ACEi, angiotensin converting enzyme inhibitors; ARB, angiotensin receptor blockers.
One hundred forty-eight participants had at least one kidney biopsy, with a pathologic diagnosis that was consistent with FSGS. Nineteen individuals were affected relatives to biopsied probands, with no kidney biopsy. Of biopsied individuals, 145 reports were available for review, 126 of which had electron micrograph descriptions/interpretations. Thirty percent had some comment on glomerular basement membrane abnormalities, typically focal thinning, and 73% had >50% podocyte foot process effacement (Table 1). Where data were available, we found that 69% (107 out of 154) of patients were treated with some form of immunosuppressive medication (Table 1). Of the 107 treated patients, the remission status was known for 82 individuals. A partial and complete remission was reported in 45% (37 out of 82) and 29% (24 out of 82), respectively, and no remission was reported in 26% (21/82).
Pathogenic Variants
Massively parallel sequencing resulted in an average of 52,378,976 uniquely aligned 150 bp paired-end reads per individual, with a mean target coverage of 88× and 89% exomic coverage. An examination of 109 genes associated with FSGS, basement membrane abnormalities, CAKUT, and nephronophthisis was done with gene-specific mean depth of coverage across all samples provided in Supplemental Table 2. Most pathogenic variants were categorized as such because they were previously reported in the literature and had a higher prevalence in cases compared with controls, as per ACMG guidelines (25). In one case, we were able to identify a de novo variant in INF2, after confirming parental status with short tandem repeat profiling.
The overall genetic diagnostic rate was 11% (20 out of 179). Genetic testing yielded a diagnosis in 28% (eight out of 29) of families with kidney disease and in 8% (12 out of 150) of sporadic cases (Supplemental Figure 1). Pathogenic variants in COL4A3/A4/A5 accounted for 55% (11 out of 20) of cases attributed to a single gene cause, with most found in COL4A5 (Figure 1, Table 2) . Interestingly, we found COL4A5 pathogenic variants in a nearly equal distribution of males and females. Pathogenic variants in podocyte genes accounted for 40% (eight out of 20) of cases with a genetic cause, including in those frequently reported in pediatric patients such as NPHS1, NPHS2, and LMX1B (Figure 1, Table 3). One case was found to be due to a pathogenic variant in BMP4, which has been associated with CAKUT (Figure 1, Table 3).
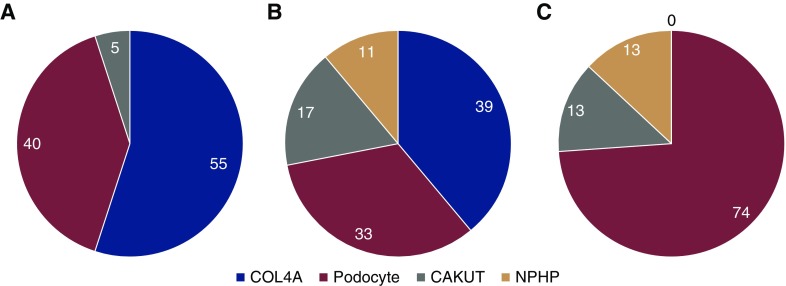
Figure 1.
Distribution of gene groups in definitely and likely pathogenic variants reveal that COL4A is leading single gene cause. The overall genetic diagnostic rate was 11% (20 out of 179) in the case series when only considering definitely pathogenic variants. There were (A) 20 definitely pathogenic, (B) 18 likely pathogenic, and (C) 15 possibly pathogenic variants identified. Of the (A) definitely pathogenic and (B) likely pathogenic variants, the highest percentage was in COL4A3/A4/A5, followed by podocyte then CAKUT genes. A minority of likely pathogenic and (C) possibly pathogenic variants were also found in the NPHP genes. Numbers in graph represent percentage.
Table 2.
Pathogenic variants in COL4A genes
| Patient ID (Family ID) | Sex | Ethnicity | Age at Disease Onset | Age at ESKD | Exon Number | Nucleotide Change | Protein Effect | Allele Frequency | Zygosity | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Pathogenic COL4A3 variants | ||||||||||
| 7215 (F23) | F | EUR | Early teens | n/a (34) | 31 | c.2452G>A | Gly818Arg | 1.17E-05 | Het | S17, S18 |
| 2555 (s) | F | EUR | 36 | 48 | 42 | c.3655G>T | Gly1219Cys | 0 | Het | S19 |
| 6062 (s) | F | EUR | 30 | unknown | 21 | c.1219G>C | Gly407Arg | 0 | Het | S20 |
| Pathogenic COL4A4 variants | ||||||||||
| 6329 (F14) | M | EUR | 42 | n/a (48) | 32 | c.2906C>G | Ser969stop | 6.49E-05 | Het | S18, S21–S24 |
| Pathogenic COL4A5 variants | ||||||||||
| 5515 (F8) | M | EUR | 32 | n/a (42) | 50 | c.4946T>G | Leu1649Arg | 0 | Hemi | S25 |
| 5519 (F8) | M | EUR | 25 | n/a (44) | 50 | c.4946T>G | Leu1649Arg | 0 | Hemi | S25 |
| 2594 (F16) | F | EUR | 28 | 40 | 20 | c.1276G>A | Gly426Arg | 0 | Het | S26, S27 |
| 1590 (s) | F | EUR | 28 | n/a (56) | 35 | c.3017G>T | Gly1006Val | 0 | Het | S28 |
| 2480 (s) | F | Admixed | Unk | 30 | 33 | c.2804G>A | Gly935Asp | 0 | Het | S27 |
| 4976 (s) | M | EUR | 41 | n/a (56) | 25 | c.1781G>A | Gly594Asp | 0 | Hemi | S29 |
| 6223 (s) | F | EUR | Unk | unknown | 31 | c.2605G>A | Gly869Arg | 0 | Het | S26, S30–S35 |
| 5269 (s) | M | EUR | 57 | 66 | 39 | c.3508G>A | Gly1170Ser | 0 | Hemi | S17, S36–S39 |
The following minor allele frequency (MAF) cut-offs as determined in gnomAD (http://gnomad.broadinstitute.org/) were used for dominant and recessive disease genes respectively: 0.00005 and 0.005 (accessed February 22, 2018). The MAF of the only COL4A4 variant exceeds the cut-off, but it is a well established founder mutation. Patients 5515 and 5519 are brothers. (F) designates family pedigree number whereas (s) indicates a sporadic case. Het indicates heterozygous and hemi indicates hemizygous. All references refer to the supplemental reference list, which can be found in the Supplemental Material, and indicate previous reports of the variant. Age at ESKD was indicated as n/a for patients without ESKD, followed by their age at time of analysis in parentheses.. F, female; EUR, European; M, male.
Table 3.
Pathogenic variants in non-COL4A genes
| Patient ID (Family ID) | Sex | Ethnicity | Age at Disease Onset | Age at ESKD | Gene Symbol | Inheritance | Exon Number | Nucleotide Change | Protein Effect | Allele Frequency | Zygosity | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathogenic podocyte gene variants | ||||||||||||
| 2517 (F4) | M | Admixed | unk | 14 | LMX1B | AD | 4 | c.668G>A | Arg223Gln | 0 | Het | S40–S44 |
| 2518 (F4) | M | Admixed | unk | 18 | LMX1B | AD | 4 | c.668G>A | Arg223Gln | 0 | Het | S40–S44 |
| 5496 (F7) | M | EAS | 16 | 22 | COQ8B | AR | 15 | c.1356_1362del | Gln452Hisfs | 0 | Homo | S45 |
| 5497 (F7) | F | EAS | 21 | n/a (30) | COQ8B | AR | 15 | c.1356_1362del | Gln452Hisfs | 0 | Homo | S45 |
| 5935 (F9) | F | EUR | unk | unk | INF2 | AD | 2 | c.312C>G | Cys104Trp | 0 | Het | S46 |
| 5936 (F9) | F | EUR | unk | unk | INF2 | AD | 2 | c.312C>G | Cys104Trp | 0 | Het | S46 |
| 6996 (F21) | M | Admixed | 25 | n/a (51) | NPHS1 | AR | 22 | c.2928G>T | Arg976Ser | 4.87E-05 | Homo | S47–S51 |
| 2378 (s) | M | EUR | 22 | 23 | INF2 | AD | 2 | c.317G>C | Arg106Pro | 0 | Het | S46 |
| 2745 (s) | F | EAS | 11 | n/a (20) | LMX1B | AD | 4 | c.737G>A | Arg246Gln | 0 | Het | S52–S54 |
| 5601 (s) | F | EUR | 24 | unk | LMX1B | AD | 4 | c.737G>A | Arg246Gln | 0 | Het | S52–S54 |
| 6251 (s) | F | EUR | 40 | n/a (49) | NPHS2 | AR | 7 | c.868G>A | Val290Met | 1.20E-04 | Homo | S55–S58 |
| Pathogenic CAKUT gene variants | ||||||||||||
| 7942 (s) | M | EUR | 55 | n/a (59) | BMP4 | AD | 2 | c.272C>G | Ser91Cys | 1.90E-04 | Het | S59, S60 |
The following minor allele frequency (MAF) cut-offs as determined in gnomAD (http://gnomad.broadinstitute.org/) were used for dominant and recessive disease genes respectively: 0.00005 and 0.005 (accessed February 22, 2018). The INF2 variant in 2378 was shown to be de novo by examining parental sequence data. The MAF of the BMP4 variant exceeds the cut-off, but it has been shown to be functionally hypomorphic. Patients 2517 and 2518 are son and father. Patients 5496 and 5497 are siblings. The relationship of patients 5935 and 5936 is unknown. (F) designates family pedigree number whereas (s) indicates a sporadic case. Het indicates heterozygous and homo indicates homozygous. All references refer to the supplemental reference list, which can be found in the Supplemental Material, and indicate previous reports of the variant and indicate previous reports of the variant. Age at kidney disease was indicated as n/a for patients without kidney disease, followed by their age at time of analysis in parentheses. Unk indicates that data were unavailable. M, male; EAS, East Asian; F, female; EUR, European.
Likely Pathogenic, Possibly Pathogenic, and Digenic Variants
Eighteen likely pathogenic variants were identified in 17 patients, which is an additional 9% (17 out of 179) of the cohort. Of these, 14 were sporadic and three were familial cases (Supplemental Tables 3 and 4). Thirty nine percent (seven out of 18) of variants were in COL4A3/A4/A5 (Figure 1). Possibly pathogenic variants were found in further 6% (11 out of 179) of the cohort (Supplemental Table 5). Ten were in sporadic cases and one was a familial case.
Three cases with two definitely or likely pathogenic variants (called digenic) were discovered. Patient 2378 had a definitely pathogenic variant in INF2 (c.317G>C, p.R106P) and a likely pathogenic variant in COL4A3 (c.2172delA, p.G724fs). Patient 7939 had two likely pathogenic variants in LMX1B (c.879delG, p.L293fs) and COL4A3 (c.84delC, p.S28fs). Patient 7276 had a likely pathogenic variant in LAMA5 (c.6065–1G>T, abolished exon 46 splice acceptor) and a possibly pathogenic variant in INF2 (c.500A>C, p.H167P).
Seven cases were found to be carriers of previously reported pathogenic variants in genes associated with autosomal recessive inheritance patterns (Supplemental Table 6). Two of these cases had an additional definitely, likely, or possibly pathogenic variant (Supplemental Table 6). Patient 2517, a familial case with a segregating definitely pathogenic variant in LMX1B (c.668G>A, p.R223Q), was also found to be heterozygous for a reported definitely pathogenic variant in ZMPSTE24 (c.794A>G, p.N265S) that was not found in his affected father. Patient 7596 had possibly pathogenic variants in CUBN (c.6928_6934delTAACCTC, p.E2310Cfs; c.7968_7969delinsGTTATATAAGGTATAACA, p.L2656_P2657delinsFVIPYIT) and a heterozygous definitely pathogenic variant in NPHS1 (c.1868G>T, p.C623F).
Copy number analysis from exome data revealed a heterozygous deletion not in other cases or controls spanning intron 16 to intron 30 of COL4A4 in patient 6084, who additionally has a variant of uncertain significance in COL4A4 (c.4708G>A, p.E1570K). The breakpoints of the deletion could not be determined, limited by having only coding sequence.
Clinical and Pathologic Characteristics associated with Definitely and Likely Pathogenic Variants
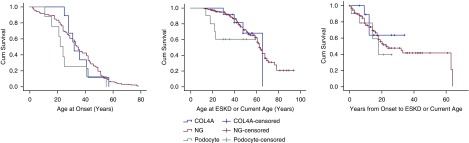
Clinical features were analyzed in patients with definitely pathogenic variants in COL4A, podocyte, CAKUT, and nephronophthisis genes (Supplemental Table 7, Table 4). Patients with definitely pathogenic variants in genes expressed in podocytes or associated with developmental defects had a trend toward earlier age at disease onset (26 years; 95% confidence interval [95% CI], 17 to 37) than both those with COL4A (36 years; 95% CI, 29 to 42) and no proven genetic basis (34 years; 95% CI, 32 to 37) subgroups (P=0.15 and 0.11, respectively; Figure 2, Table 4); and similarly, an earlier estimated mean survival/age of ESKD (mean age, 43 years; 95% CI, 31 to 55) than COL4A (58 years; 95% CI, 49 to 69) and no proven genetic basis (62 years; 95% CI, 58 to 66) subgroups (P=0.26 and P=0.06, respectively; Figure 2, Table 4). An earlier age of estimated mean survival/age of ESKD was also observed for COL4A (58 years; 95% CI, 49 to 69) compared with the no proven genetic basis (62 years; 95% CI, 58 to 66) subgroup (P=0.06). None of the comparisons were significant but the lowest P values achieved were for ESKD with podocyte/developmental defects compared with no proven genetic basis subgroup (P=0.06), and COL4A compared with no proven genetic basis subgroup (P=0.06). The analysis was also performed evaluating males and females separately (Supplemental Table 8).
Table 4.
Clinical characteristics of COL4A, other genetic cause, and no proven genetic cause subgroups
| Clinical characteristic | COL4A (n=12) | Other Genetic (n=12) | No Proven Genetic (n=169) |
|---|---|---|---|
| % Male | 42 | 67 | 58 |
| Mean age of onset of kidney disease (95% CI) | 36 (29 to 42) | 26 (17 to 37) | 34 (32 to 37) |
| % with hematuria | 60 | 25 | 29 |
| % of patients with family history of kidney disease | 46 | 44 | 12.2 |
| % Partial remission | 29 | 30 | 26 |
| % Complete remission | 0 | 0 | 20 |
| % No remission | 14 | 20 | 15 |
| % Unknown status of remission | 57 | 50 | 39 |
| % ESKD | 46 | 44 | 50 |
| % with only global glomerulosclerosis on light microscopy | 11 | 50 | 9 |
| % Glomerular basement membrane abnormalities on electron microscopy | 56 | 25 | 27 |
| % with >50% podocyte foot process effacement on electron microscopy | 100 | 75 | 72 |
| No. of patients where pathology report with electron microscopy description is available | 9 | 4 | 113 |
| Mean age at ESKD (95% CI) | 58 (49 to 69) | 43 (31 to 55) | 62 (58 to 66) |
| % Kidney transplant in patients with ESKD | 100 | 75 | 54 |
| % of recurrence of disease after kidney transplant (within the first year) | 0 | 0 | 13 |
Patients in the COL4A, podocyte/kidney development (called other), and no proven genetic basis subgroups had disease onset at 36 years (95% CI, 29 to 42), 26 years (95% CI, 17 to 37), and 34 years (95% CI, 32 to 37). The estimated mean survival/age at ESKD was 58 years (95% CI, 49 to 69), 43 years (95% CI, 31 to 55), and 62 years (95% CI, 58 to 66). The only statistically significant P values were in comparing COL4A with the no proven genetic basis subgroup for hematuria (P=0.02), glomerular basement membrane abnormalities (P=0.03), and >50% effacement (P=0.03). Number of patients where the pathology report was available is indicated. Percentages represent of available data. Absolute numbers are presented in Supplemental Table 9. 95% CI, 95% confidence interval.
Figure 2.
Kaplan-Meier plots demonstrating that patients with pathogenic variants in genes expressed in the podocyte or associated with development defects had a trend toward an earlier age at disease onset and ESKD compared to other subgroups, although none of the pairwise comparisons were significant. The lowest P values achieved were for age at ESKD for podocyte/developmental defects compared with no proven genetic basis subgroup (P=0.06), and COL4A compared with no proven genetic basis subgroup (P=0.06). COL4A, COL4A subgroup; NG, no proven genetic basis subgroup; Podocyte, podocyte and kidney development defect subgroup.
For other clinicopathologic characteristics, the statistically significant comparisons were for COL4A compared with the no proven genetic basis subgroup for hematuria (P=0.02), glomerular basement membrane abnormalities (P=0.03), and >50% podocyte effacement (P=0.03) (Figure 3, Table 4). No individuals with an identified genetic cause had a complete remission when treated with immunosuppression (Table 4). Those with likely pathogenic variants in COL4A revealed GBM abnormalities in one of four cases where pathology was available, supportive of a deleterious role of at least some associated likely pathogenic COL4A variants (Figure 4).
Figure 3.
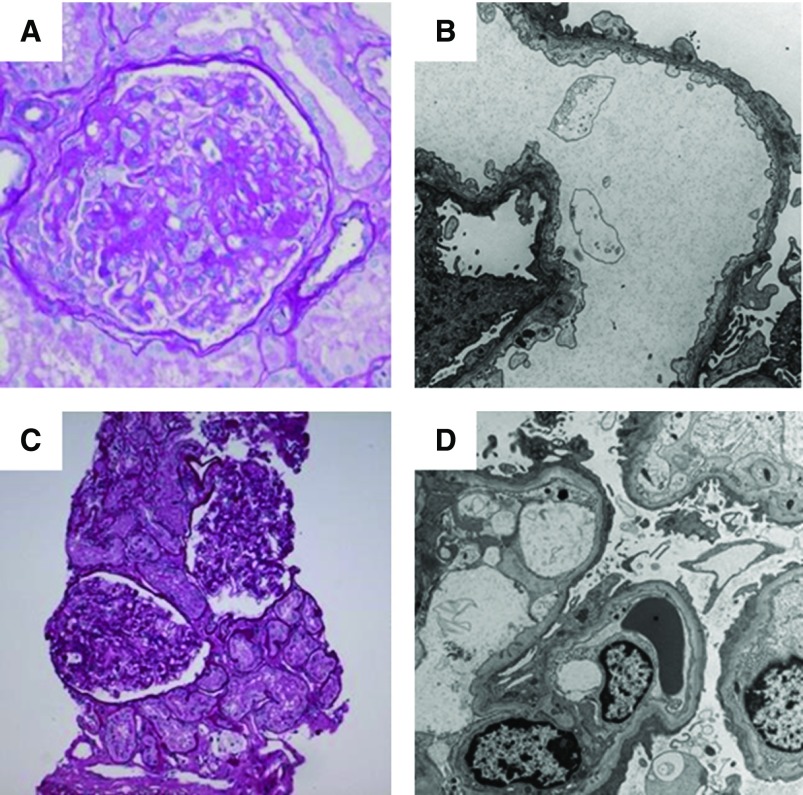
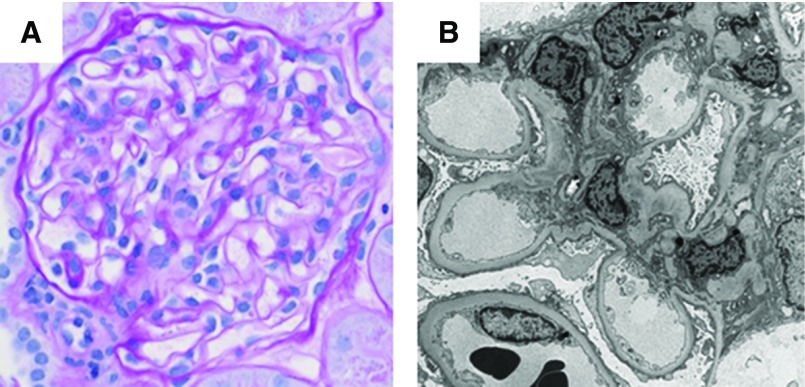
Kidney pathology in COL4A-associated FSGS often demonstrate glomerular basement membrane abnormalities. Female patient (6062) with COL4A3-associated disease showing (A) segmental scarring (periodic acid–Schiff staining, ×20 magnification), and on (B) ultrastructural examination, diffusely thin glomerular basement membranes, and extensive podocyte foot process effacement, at approximately 80% (×10,000 magnification). Female patient (6251) with NPHS2-associated disease showing (C) enlarged glomeruli (periodic acid–Schiff staining, ×10 magnification) and one glomerulus with a segmental scar (not shown). (D) Ultrastructural examination shows normal thickness glomerular basement membranes and diffuse podocyte foot process effacement, at approximately 90% (×6000 magnification).
Figure 4.
Genetic testing and pathology can be complementary tests to improve interpretation. Kidney pathology in a female (7939) with a likely pathogenic variant in COL4A3. (A) The glomeruli show mildly thickened capillary walls and no segmental scarring (in 53 glomeruli available for light microscopy; periodic acid-Schiff staining, x40 magnification). (B) Ultrastructural examination confirms thickened glomerular basement membranes, at approximately two-fold, and demonstrates extensive podocyte foot process effacement, at approximately 80% (×4000 magnification).
Discussion
We examine a large panel of genes associated with FSGS, glomerular basement membrane disease, CAKUT, and nephronophthisis available from exome data in 193 mostly sporadic adult FSGS cases, achieving an overall genetic diagnostic rate of 11%. However, we are likely underestimating genetic contribution by the stringency of the criteria we use. Our findings demonstrate FSGS is commonly the end point of COL4A disorders as well as podocyte and kidney developmental defects.
The majority of kidney pathologies associated with definitely pathogenic COL4A3/A4/A5 variants had evidence of glomerular basement membrane abnormalities including thinning, thickening, irregularities, and lamellation, usually focal and sometimes subjectively described as minor and presumed not to be clinically significant. However, we also identified a minority of definitely pathogenic COL4A3/A4/A5 variants that do not have any obvious glomerular basement membrane abnormalities. Furthermore, we found cases with no proven genetic cause but that are associated with extensive glomerular basement membrane abnormalities. This suggests that our method for genetic testing has missed some definitely pathogenic variants and/or that morphologic glomerular basement membrane abnormalities can be nonspecific. Subgenomic capture strategy, as is used for exome sequencing, does not capture the entirety of the coding sequence; in our case, 95% (with at least 10× coverage) of COL4A. Variants and copy number changes in intronic regions will also not be detected with this methodology. Additionally, in cases with likely pathogenic variants in COL4A not considered in the diagnostic rate, we found one case with glomerular basement abnormalities, supporting a possible deleterious role for at least some of these variants.
Our genetic–pathologic correlation supports the notion that diffuse podocyte effacement can occur as a result of GBM abnormalities. In the absence of a complementary test (i.e., genetic testing) at the time of presentation, as well as other clinical characteristics that decreased suspicion for a collagen 4A defect such as a lack of family history, being female, and diffuse podocyte foot effacement on pathology, many of these individuals were tried on immunosuppressive therapies and were unresponsive. For other genetic causes such as CAKUT, biopsy features including glomerulomegaly could similarly raise suspicion for likely pathogenic variant disease causality. Thus, our experience highlights the complementary nature of pathology and genetic testing to improve interpretation of each test.
A previous report suggests that the risk of developing ESKD in heterozygous COL4A females is low before 40 years of age, but increases after 60 years of age (29). Twice as many women are affected by X-linked diseases, but are commonly undiagnosed (30). Patients with pathogenic variants in genes expressed in podocytes or associated with developmental defects had an earlier age at disease onset and ESKD than both the COL4A and no proven genetic basis subgroups. Our estimated mean survival/age for ESKD in COL4A5 was 58 years (95% CI, 46 to 70), which is later than the 24 years reported in the literature, which is likely driven by the preponderance of females with deleterious COL4A5 variants, influencing a milder clinical course than what has been previously reported for collagen 4A disorders (31).
COL4A5 disease has been reported to have X-linked recessive inheritance. We report COL4A5 variants that have been found in other cases, regardless of sex, but cannot definitively conclude causality as we did not perform X-chromosome inactivation studies. Although X-inactivation ratios of 50:50 are expected in a normal population of cells, ratios can deviate from modifier genes or variant selection advantages (32). The Genotype-Tissue Expression (GTEx project) includes high-coverage RNA-sequencing data from diverse human tissues to investigate male–female differences in expression and shows that COL4A5 is indeed subject to inactivation (33).
The cases included in this study are patients recruited from specialty clinics at a quaternary-level hospital in Toronto, Ontario, Canada, and may not be representative of a population-based study. Furthermore, we have acquired a predominantly case cohort, recruiting only affected individuals. As a result, we are unable to determine parental phase for two variants in the same individual, identify de novo variants, and define the penetrance of rare variants that are found. Additionally, for COL4A cases, we do not have formalized ocular and auditory assessments to report.
Our experience suggests that genetic–pathologic correlation improves diagnostic accuracy in FSGS. Nearly all variants we designated as definitely pathogenic are the exact same variants that have been previously reported. Inherently, designation of pathogenicity is biased toward the most studied genes. We suspect that we are still underestimating genetic contribution because of our inability to resolve the significance of many rare variants, highlighting the need for genetic, clinical, and pathologic data sharing as well as large-scale functional libraries.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors thank the patients and their families for their participation; Sergio Pereira, Daniel Merico, Wilson Sung, Liz Weili, and Bhooma Thiruvahindrapuram of The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Ontario, Canada for assistance with sequencing and variant calling; and Winnie Chan, Xuewen Song, and Ning He for assistance with patient recruitment and technical advice.
M.B. received a University of Toronto McLaughlin Centre Accelerator grant in Genomic Medicine in 2017, a NephCure Kidney International-Neptune Ancillary Studies grant in 2016, and a health research grant (14-04) from Physician’s Services Incorporated in 2015, which funded this work. M.B. is supported by a new investigator award from the Kidney Research Scientist Core Education and National Training Program.
Footnotes
T.Y., K.U., Y.P., and M.B. contributed equally to this work
Published online ahead of print. Publication date available at www.cjasn.org.
Supplemental Material
This article contains the following supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.08750718/-/DCSupplemental.
Supplemental Table 1. Additional baseline clinical characteristics of the sequenced cohort.
Supplemental Table 2. List of genes associated with FSGS and related phenotypes.
Supplemental Table 3. Likely pathogenic variants in COL4A genes.
Supplemental Table 4. Likely pathogenic variants in non-COL4A genes.
Supplemental Table 5. Possibly pathogenic variants.
Supplemental Table 6. Previously reported variants with uncertain pathogenic significance.
Supplemental Table 7. Additional clinical characteristics for patient subgroups.
Supplemental Table 8. Tables demonstrating ages at disease onset and estimated mean survival/age of kidney failure by subgroup, with sexes analyzed separately.
Supplemental Table 9. Clinical characteristics of COL4A, other genetic cause, and no proven genetic cause subgroups.
Supplemental Figure 1. Pedigrees of families with definitely pathogenic variants.
References
- 1.Falk R: JC, Nachman P: Primary glomerular disease. In: The Kidney, edited by Brenner BM, Philadelphia, W. B. Saunders Company, 2000, pp 1263–1332 [Google Scholar]
- 2.Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, Mathis BJ, Rodríguez-Pérez JC, Allen PG, Beggs AH, Pollak MR: Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet 24: 251–256, 2000 [DOI] [PubMed] [Google Scholar]
- 3.Reiser J, Polu KR, Möller CC, Kenlan P, Altintas MM, Wei C, Faul C, Herbert S, Villegas I, Avila-Casado C, McGee M, Sugimoto H, Brown D, Kalluri R, Mundel P, Smith PL, Clapham DE, Pollak MR: TRPC6 is a glomerular slit diaphragm-associated channel required for normal renal function. Nat Genet 37: 739–744, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown EJ, Schlöndorff JS, Becker DJ, Tsukaguchi H, Tonna SJ, Uscinski AL, Higgs HN, Henderson JM, Pollak MR: Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet 42: 72–76, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winn MP, Conlon PJ, Lynn KL, Farrington MK, Creazzo T, Hawkins AF, Daskalakis N, Kwan SY, Ebersviller S, Burchette JL, Pericak-Vance MA, Howell DN, Vance JM, Rosenberg PB: A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science 308: 1801–1804, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Kestilä M, Lenkkeri U, Männikkö M, Lamerdin J, McCready P, Putaala H, Ruotsalainen V, Morita T, Nissinen M, Herva R, Kashtan CE, Peltonen L, Holmberg C, Olsen A, Tryggvason K: Positionally cloned gene for a novel glomerular protein–nephrin–is mutated in congenital nephrotic syndrome. Mol Cell 4: 575–582, 1998 [DOI] [PubMed] [Google Scholar]
- 7.Wu Y, Hu P, Xu H, Yuan J, Yuan L, Xiong W, Deng X, Deng H: A novel heterozygous COL4A4 missense mutation in a Chinese family with focal segmental glomerulosclerosis. J Cell Mol Med 20: 2328–2332, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malone AF, Phelan PJ, Hall G, Cetincelik U, Homstad A, Alonso AS, Jiang R, Lindsey TB, Wu G, Sparks MA, Smith SR, Webb NJA, Kalra PA, Adeyemo AA, Shaw AS, Conlon PJ, Jennette JC, Howell DN, Winn MP, Gbadegesin RA: Rare hereditary COL4A3/COL4A4 variants may be mistaken for familial focal segmental glomerulosclerosis. Kidney Int 86: 1253–1259, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braunisch MC, Büttner-Herold M, Günthner R, Satanovskij R, Riedhammer KM, Herr PM, Klein HG, Wahl D, Küchle C, Renders L, Heemann U, Schmaderer C, Hoefele J: Heterozygous COL4A3 variants in histologically diagnosed focal segmental glomerulosclerosis. Front Pediatr 6: 171, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gast C, Pengelly RJ, Lyon M, Bunyan DJ, Seaby EG, Graham N, Venkat-Raman G, Ennis S: Collagen (COL4A) mutations are the most frequent mutations underlying adult focal segmental glomerulosclerosis. Nephrol Dial Transplant 31: 961–970, 2016 [DOI] [PubMed] [Google Scholar]
- 11.Xie J, Wu X, Ren H, Wang W, Wang Z, Pan X, Hao X, Tong J, Ma J, Ye Z, Meng G, Zhu Y, Kiryluk K, Kong X, Hu L, Chen N: COL4A3 mutations cause focal segmental glomerulosclerosis. J Mol Cell Biol 7: 184, 2015 [DOI] [PubMed] [Google Scholar]
- 12.Pierides A, Voskarides K, Athanasiou Y, Ioannou K, Damianou L, Arsali M, Zavros M, Pierides M, Vargemezis V, Patsias C, Zouvani I, Elia A, Kyriacou K, Deltas C: Clinico-pathological correlations in 127 patients in 11 large pedigrees, segregating one of three heterozygous mutations in the COL4A3/COL4A4 genes associated with familial haematuria and significant late progression to proteinuria and chronic kidney disease from focal segmental glomerulosclerosis. Nephrol Dial Transplant 24: 2721–2729, 2009 [DOI] [PubMed] [Google Scholar]
- 13.Voskarides K, Damianou L, Neocleous V, Zouvani I, Christodoulidou S, Hadjiconstantinou V, Ioannou K, Athanasiou Y, Patsias C, Alexopoulos E, Pierides A, Kyriacou K, Deltas C: COL4A3/COL4A4 mutations producing focal segmental glomerulosclerosis and renal failure in thin basement membrane nephropathy. J Am Soc Nephrol 18: 3004–3016, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Barua M, Stellacci E, Stella L, Weins A, Genovese G, Muto V, Caputo V, Toka HR, Charoonratana VT, Tartaglia M, Pollak MR: Mutations in PAX2 associate with adult-onset FSGS. J Am Soc Nephrol 25: 1942–1953, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ovunc B, Otto EA, Vega-Warner V, Saisawat P, Ashraf S, Ramaswami G, Fathy HM, Schoeb D, Chernin G, Lyons RH, Yilmaz E, Hildebrandt F: Exome sequencing reveals cubilin mutation as a single-gene cause of proteinuria. J Am Soc Nephrol 22: 1815–1820, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA: The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ: A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, Handsaker RE, McCarroll SA, O’Donovan MC, Owen MJ, Kirov G, Sullivan PF, Hultman CM, Sklar P, Purcell SM: Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet 91: 597–607, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fromer M, Purcell SM: Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet 81: 7.23.1–7.23.21, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bansal V, Libiger O: Fast individual ancestry inference from DNA sequence data leveraging allele frequencies for multiple populations. BMC Bioinformatics 16: 4, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE; International HapMap 3 Consortium : Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG; Exome Aggregation Consortium : Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW: The Database of Genomic Variants: A curated collection of structural variation in the human genome. Nucleic Acids Res 42: D986–D992, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL: Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Jian X, Boerwinkle E: dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat 32: 894–899, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Wu C, Li C, Boerwinkle E: dbNSFP v3.0: A one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat 37: 235–241, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desmet F-O, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C: Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37: e67, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jais JP, Knebelmann B, Giatras I, De Marchi M, Rizzoni G, Renieri A, Weber M, Gross O, Netzer KO, Flinter F, Pirson Y, Dahan K, Wieslander J, Persson U, Tryggvason K, Martin P, Hertz JM, Schröder C, Sanak M, Carvalho MF, Saus J, Antignac C, Smeets H, Gubler MC: X-linked Alport syndrome: Natural history and genotype-phenotype correlations in girls and women belonging to 195 families: A “European Community Alport Syndrome Concerted Action” study. J Am Soc Nephrol 14: 2603–2610, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Savige J, Colville D, Rheault M, Gear S, Lennon R, Lagas S, Finlay M, Flinter F: Alport syndrome in women and girls. Clin J Am Soc Nephrol 11: 1713–1720, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savige J, Storey H, Il Cheong H, Gyung Kang H, Park E, Hilbert P, Persikov A, Torres-Fernandez C, Ars E, Torra R, Hertz JM, Thomassen M, Shagam L, Wang D, Wang Y, Flinter F, Nagel M: X-linked and autosomal recessive alport syndrome: Pathogenic variant features and further genotype-phenotype correlations. PLoS One 11: e0161802, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Migeon BR: Non-random X chromosome inactivation in mammalian cells. Cytogenet Cell Genet 80: 142–148, 1998 [DOI] [PubMed] [Google Scholar]
- 33.Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, Cummings BB, Castel SE, Karczewski KJ, Aguet F, Byrnes A, Lappalainen T, Regev A, Ardlie KG, Hacohen N, MacArthur DG; GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz : Landscape of X chromosome inactivation across human tissues. Nature 550: 244–248, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.