Abstract
Most tumors have an aberrantly activated lipid metabolism1,2, which enables them to synthesize, elongate and desaturate fatty acids to support proliferation. However, only particular subsets of cancer cells are sensitive toward approaches targeting fatty acid metabolism, and in particular fatty acid desaturation3. This suggests that many cancer cells harbor an unexplored plasticity in their fatty acid metabolism. Here, we discover that some cancer cells can exploit an alternative fatty acid desaturation pathway. We identify various cancer cell lines, murine hepatocellular carcinomas (HCC), and primary human liver and lung carcinomas that desaturate palmitate to the unusual fatty acid sapienate to support membrane biosynthesis during proliferation. Accordingly, we found that sapienate biosynthesis enables cancer cells to bypass the known stearoyl-CoA desaturase (SCD)-dependent fatty acid desaturation. Thus, only by targeting both desaturation pathways the in vitro and in vivo proliferation of sapienate synthesizing cancer cells is impaired. Our discovery explains metabolic plasticity in fatty acid desaturation and constitutes an unexplored metabolic rewiring in cancers.
Keywords: sapienate, SCD, FADS2, cancer, fatty acid metabolism, fatty acid desaturation, liver cancer, lung cancer, lipid metabolism
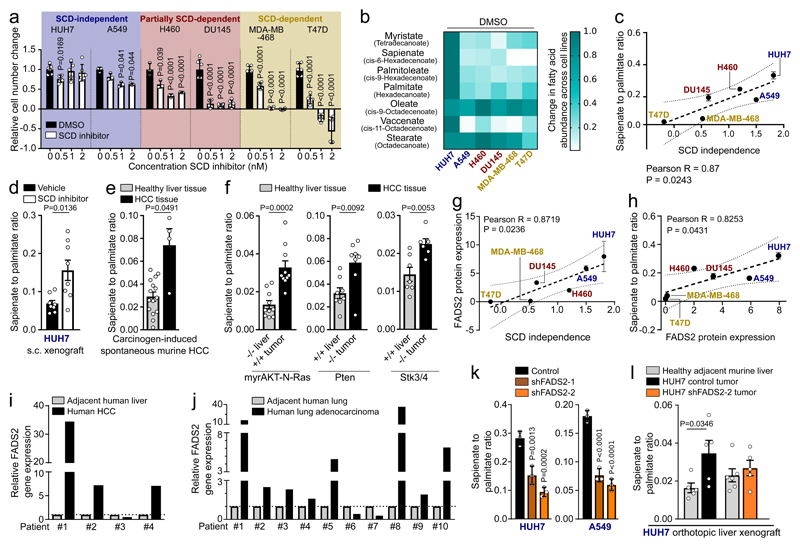
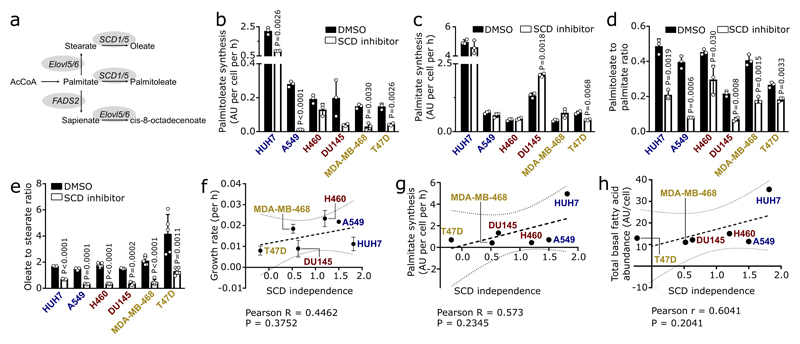
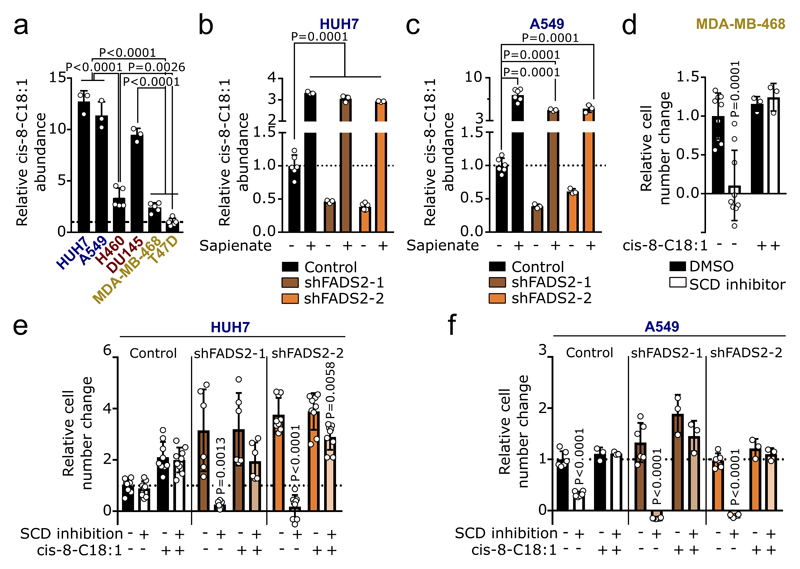
Cancer cells display differential usage of SCD-dependent fatty acid desaturation (Extended Data Figure 1a)3. To illuminate this plasticity, we treated different cancer cells (liver: HUH7; lung: A549 and H460; prostate: DU145; breast: MDA-MB-468 and T47D) with the SCD inhibitor Merck Frosst Cpd 3j4, in conditions of low extracellular fatty acid availability. We observed that they exhibited a broad sensitivity profile towards SCD inhibition (Figure 1a). Based on this proliferation response, we classified these cancer cells into SCD-dependent (proliferation inhibition or cell death), partially SCD-dependent (less than 50% proliferation), or SCD-independent (more than 50% proliferation). Next, we excluded that the differential dependency of cancer cells on SCD activity was a result of their individual growth rate, fatty acid synthesis rate, or the degree with which Merck Frosst Cpd 3j inhibited SCD (Extended Data Figure 1b-g). Based on these results, we concluded that some cancer cells are SCD-independent, which cannot be explained by their known fatty acid metabolism. Consequently, we hypothesized that SCD-independent and partially SCD-dependent cancer cells exploit an alternative desaturation pathway. We reasoned that the presence of such an alternative pathway must result in the synthesis of unusual monounsaturated fatty acids and therefore measured saturated and monounsaturated C12 to C18 fatty acids. In general, the different cancer cells presented a wide range in total fatty acid abundance, which did not correlate with SCD independence (Extended Data Figure 1h; Supplementary Table 1a, b). Surprisingly, we discovered an elevated abundance of the unusual fatty acid sapienate (cis-6-C16:1) in SCD-independent and partially SCD-dependent cancer cells that increased upon SCD inhibition (Figure 1b; Extended Data Figure 2a; Supplementary Table 1a, b).
Figure 1. Some cancer cells desaturate palmitate to sapienate via FADS2.
(a) Sensitivity profile of cancer cells treated Merck Frosst Cpd 3j normalized to control (HUH7 n=6; A459 n=3, H460 n=3, DU145 n=6, MDA-MB-468 n=6, T47D n=6). Two-way ANOVA with Dunnett’s multiple comparisons.
(b) Heat map representing fatty acid abundances normalized to highest abundance of each fatty acid across all cell lines. Over 90% reduction: white, no reduction: dark green.
(c) Correlation between SCD independence defined as area under the cell number curve of (a) and desaturation activity to sapienate (Extended Data Figure 2b). Trend line (dashed line); 95% confidence intervals (dotted lines). n=3.
(d-f) Sapienate to palmitate ratio of HUH7 subcutaneous xenografts treated with vehicle (n=8) or Merck Frosst Cpd 3j (n=8; 1.5 mg per kg twice daily with oral (p.o.) gavage) and in diethylnitrosamine (DEN) or genetically-induced HCC (normal n=15; DEN n=4); myrAKT-N-Ras (n=10), Pten (n=8) and Stk (normal n=7; HCC n=6). Unpaired two-sided Student’s T-test with Welch’s correction.
(g,h) Correlation between FADS2 protein expression and SCD independence or desaturation activity to sapienate (Extended Data Figure 2b). Trend line (dashed line); 95% confidence intervals (dotted lines). n=3.
(i,j) FADS2 gene expression in paired samples of human HCC (n=4) and non-small cell lung adenocarcinoma (n=10) versus normal adjacent tissue.
(k) Desaturation activity to sapienate in HUH7 and A549 cells with a non-targeting shRNA or shRNAs targeting FADS2 (n=3). One-way ANOVA with Dunnett’s multiple comparisons.
(l) Sapienate to palmitate ratio in normal adjacent liver and HUH7 orthotopic liver tumors with non-targeting shRNA or shRNA targeting FADS2 (n=5). Two-way ANOVA with Sidak’s multiple comparisons.
Experiments were performed in low FBS (1%: HUH7; 0.5%: other) with treatment of 72 h. Error bars represent SD (in vitro) or SEM (in vivo) from mean of biological independent samples (in vitro) or animals (in vivo).
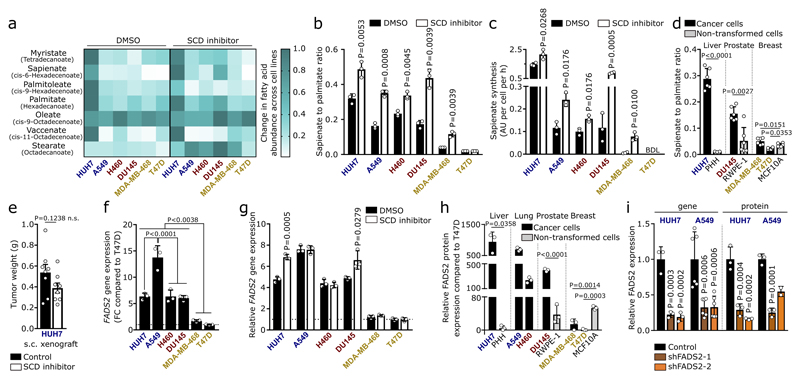
Sapienate is a major component of human sebum and is to date considered a specific marker of sebocyte metabolism in the sebaceous glands5,6. Since sebocytes produce sapienate from palmitate, we determined the desaturation activity from palmitate to sapienate by assessing the sapienate to palmitate ratio and sapienate biosynthesis. Both measures increased upon SCD inhibition (with the exception of SCD-dependent T47D cells; Extended Data Figure 2b, c). Moreover, the sapienate to palmitate ratio correlated with SCD independence and was higher in SCD-independent (HUH7) and partially SCD-dependent (DU145) cancer cells compared to non-transformed cells of the same tissue origin (Figure 1c, Extended Data Figure 2d). Subsequently, we determined the desaturation activity to sapienate in the HUH7 tumor xenografts upon SCD inhibition. In agreement with our in vitro data, we found that SCD inhibition did not significantly alter final tumor weight, but increased the desaturation activity to sapienate (Figure 1d, Extended Data Figure 2e). Accordingly, we observed that (diethylnitrosamine)- and genetically-induced murine HCC exhibited a significantly elevated desaturation activity to sapienate compared to normal liver (Figure 1e, f). These data collectively show that cancer cells, and in particular HCC, can produce sapienate both in vitro and in vivo.
In sebocytes, sapienate is produced by fatty acid desaturase (FADS) 25 (Extended Data Figure 1a). Therefore, we investigated whether cancer cells exploit FADS2 to synthesize sapienate. We found that FADS2 gene expression was increased in SCD-independent and partially SCD-dependent cancer cells compared to SCD-dependent cells, and in liver and prostate cancer cells upon SCD inhibition (Extended Data Figure 2f, g). Consistently, FADS2 protein expression correlated with SCD independence and desaturation activity to sapienate in cancer cells (Figure 1g, h). Moreover, FADS2 protein and gene expression was elevated in HUH7 and DU145 cancer cells compared to corresponding non-transformed cells (Extended Data Fig. 2h). Similarly, FADS2 gene expression was increased in matched pairs of cancer versus adjacent non-cancerous tissue of HCC (3 out of 4) and non-small cell lung cancer (8 out of 10) from human patients (Fig. 1i, j). These data suggest an involvement of FADS2 in sapienate biosynthesis. Accordingly, FADS2 silencing resulted in a decreased desaturation activity to sapienate in vitro and in vivo (Figure 1k, l; Extended Data Figure 2i). These findings demonstrate that some cancer cells exploit FADS2 to produce sapienate.
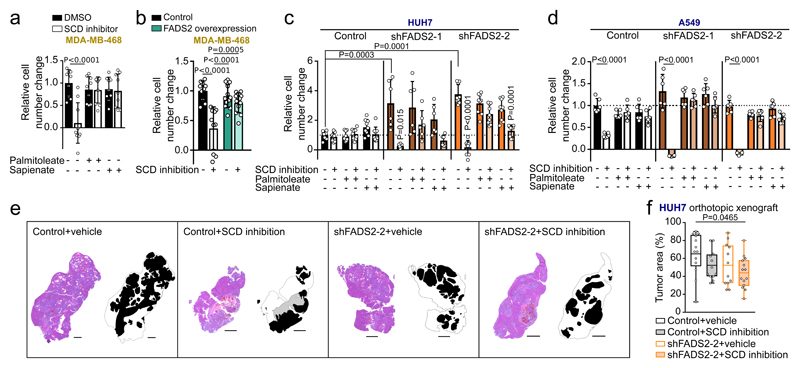
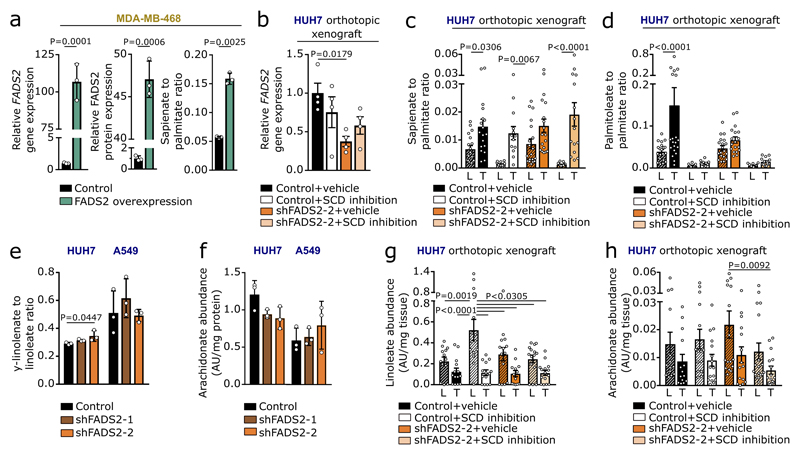
Next, we investigated whether sapienate biosynthesis causes SCD-independence. Indeed, sapienate supplementation or FADS2 overexpression in SCD-dependent MDA-MB-468 cells restored proliferation upon SCD inhibition, i.e. resulted in SCD-independence (Figure 2a, b; Extended Data Figure 3a). Moreover, FADS2 silencing combined with SCD inhibition caused proliferation inhibition or cell death in HUH7 and A549 cells, respectively (Figure 2c, d), whereas sole FADS2 knockdown seem to increase proliferation in HUH7 cells. These findings indicate that some cancer cells might rely on the metabolic plasticity provided through simultaneous SCD and FADS2 desaturation activity at the expense of maximized proliferation - a phenomenon that has been described before7. Subsequently, we assessed dual inhibition of SCD- and FADS2-dependent desaturation in HUH7 orthotopic liver xenografts. We found that only dual inhibition of SCD and FADS2 resulted in a significantly smaller tumor area compared to control tumors (Figure 2e, f). Differently to the in vitro results, no full inhibition of tumor growth was achieved in vivo, likely due to the lower in vivo knockdown efficiency and a partial compensation through extracellular sapienate uptake (Extended Data Figure 3b-d). An involvement of linoleate (known substrate of FADS2 in polydesaturation) metabolization in the observed SCD-independence was excluded (Extended Data Figure 3e-h). Taken together, these data demonstrate that dual activity of SCD- and FADS2-dependent desaturation can provide metabolic plasticity supporting proliferation, which can be impaired in vitro and in vivo by combined inhibition of both pathways.
Figure 2. Sapienate synthesis via FADS2 causes independence from the known SCD-catalyzed fatty acid desaturation.
(a,b) Relative proliferation of MDA-MB-468 control (with or without sapienate) and FADS2 overexpression cells upon treatment 0.5 nM Merck Frosst Cpd 3j normalized to control (a: n=9; b: control n=10, overexpression n=12). Two-way ANOVA with Tukey’s multiple comparisons.
(c,d) Relative proliferation of HUH7 and A549 cells (with or without sapienate) upon FADS2 knockdown with(out) 2 nM Merck Frosst Cpd 3j normalized to control (c: control n=9; shFADS2-1 n=6; shFADS2-2 n=6; d: n=6). Two-way ANOVA with Tukey multiple comparisons (within different cell lines); one-way ANOVA with Dunnett’s multiple comparisons (across different cell lines). Only pair-wise comparisons are depicted.
(e,f) Representative images of hematoxylin and eosin stain and relative area of resected tumor nodules derived from HUH7 control (non-targeting shRNA) or FADS2 knockdown (shFADS2-2) orthotopic liver xenografts in mice treated with vehicle or Merck Frosst Cpd 3j (1.5 mg per kg twice daily per oral; p.o.; control+vehicle n=13; control+SCD inhibition n=12; shFADS2-2+vehicle n=12, shFADS2-2+SCD inhibition n=14 of one experiment). Masks in (e) show the tissue contour and indicating tumor (black), necrotic (grey) and liver (white) area. Scale bar represents 1,000 µm in all cases. Box blots in (f) show box extending from the 25th to 75th percentiles, whiskers indicating the minimum and maximum, and a line indicating the mean. One-way ANOVA with Tukey’s multiple comparisons.
Experiments were performed in low FBS (1%: HUH7; 0.5%: others) with treatment of 72 h. Error bars represent SD from mean of biological independent samples (in vitro) or animals (in vivo), unless stated otherwise.
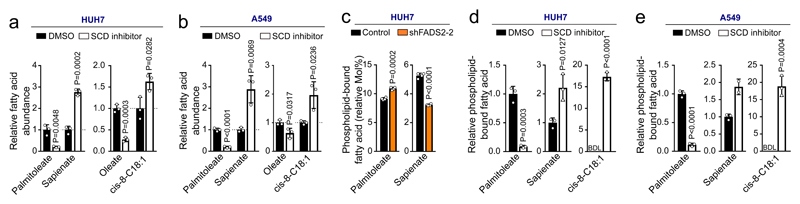
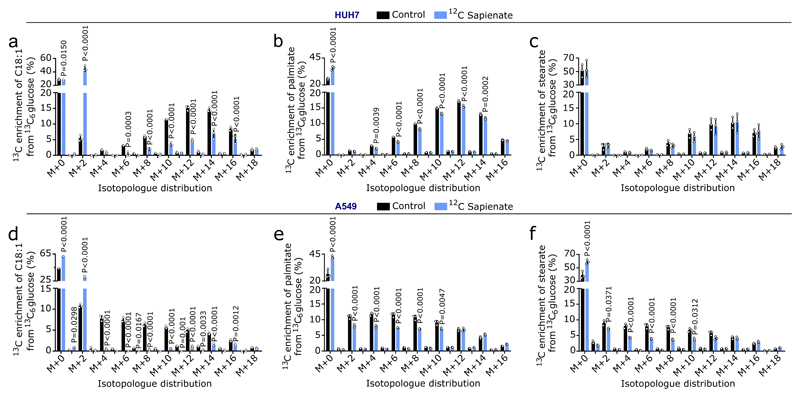
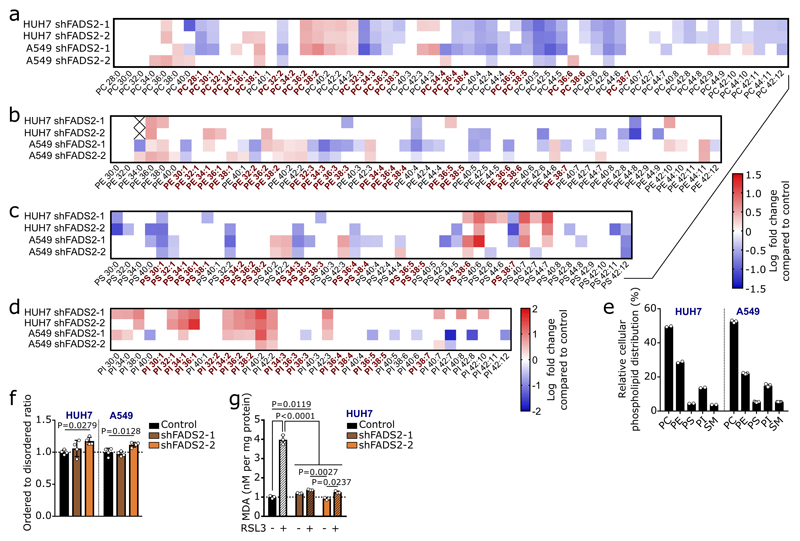
An important fate of fatty acids is membrane synthesis, for which fatty acids are often elongated. Accordingly, we observed carbons of sapienate in its elongation product cis-8-octadecenoate (cis-8-C18:1; Extended Data Figure 4). In line with sapienate elongation, cis-8-octadecenoate abundance was higher in SCD-independent cells than in SCD-dependent cells, increased in HUH7 and A549 cells upon sapienate supplementation or SCD inhibition, and decreased upon FADS2 silencing (Figures 3a, b; Extended Data Figure 5a-c; Supplementary Table 1c, d). Consistently, cis-8-octadecenoate supplementation rescued the proliferation of SCD-dependent MDA-MB-468 cells and FADS2 knockdown cells (HUH7, A549) upon SCD inhibition (Extended Data Figure 5d-f). We then determined whether sapienate is used for membrane synthesis. FADS2 silencing altered the overall composition of membrane-bound phospholipids, decreased the fraction of phospholipids built from sapienate and increased phospholipids built from the SCD product palmitoleate in HUH7 and A549 cells (Figure 3c; Extended Data Figure 6a-e; Supplementary Table 1e). The opposite change occurred upon SCD inhibition (Figure 3d, e). Functionally, these changes in membrane composition resulted in a trend toward decreased membrane fluidity and significantly increased resistance to lipid peroxidation in FADS2 knockdown cells (Extended Data Figure 6f, g). Thus, these data show that some cancer cell lines elongate sapienate and use it for membrane biosynthesis.
Figure 3. Sapienate and its elongation product cis-8-octadecenoate are used in membrane synthesis.
(a,b) Relative fatty acid abundances in HUH7 and A549 cells treated 2 nM Merck Frosst Cpd 3j (n=3). Data were normalized to the respective controls.
(c) Palmitoleate and sapienate abundances in membrane phospholipids in HUH7 cells with non-targeting shRNA and an shRNA targeting FADS2 (n=4).
(d,e) Relative phospholipid-bound palmitoleate, sapienate and cis-8-octadecenoate abundances in HUH7 and A549 cells treated with control or 2 nM Merck Frosst Cpd 3j normalized to control (n=2 in e sapienate+SCD inhibitor; n=3 in all other cases,). BDL denotes ‘below detection limit’. When the respective control was BDL, the data were normalized to the total abundance of all fatty acids measured and are represented in arbitrary units.
Experiments were performed in low FBS (1% : HUH7; 0.5% : others) with treatment of 72 h. Error bars represent SD from mean of biological independent samples. Unpaired two-sided Student’s T-tests.
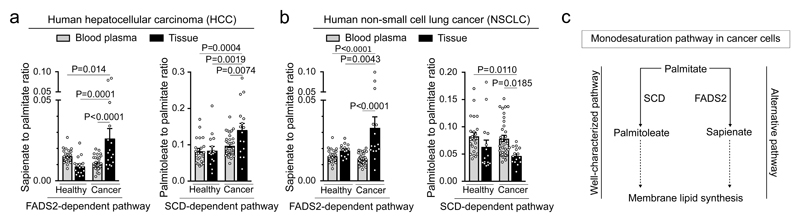
Finally, we asked whether sapienate metabolism also occurs in primary human cancers. We measured the ratio of sapienate to palmitate in cancer and normal lung and liver tissue as well as blood plasma from human subjects. Blood plasma was from healthy volunteers or cancer patients, while normal lung and liver was obtained from adjacent non-cancerous tissue from cancer patients and non-transplanted donor organs, respectively. In addition, we determined the palmitoleate to palmitate ratio in the same tissues and blood plasma samples as a readout of the SCD-dependent fatty acid desaturation pathway. Strikingly, we found that only in cancer tissue, but not in normal tissue, the sapienate to palmitate ratio was significantly increased compared to the blood plasma ratio (Figure 4a, b). Accordingly, the sapienate to palmitate ratio was higher in cancer tissue compared to normal tissue (Figure 4a, b). The increase in the sapienate to palmitate ratio was more pronounced than the corresponding change in the palmitoleate to palmitate ratio when comparing cancer and normal tissue (Figure 4a, b). This suggests a specific increase in sapienate biosynthesis in these cancers, rather than a general increase in the synthesis of monounsaturated fatty acids. Taken together, these data provide evidence that sapienate metabolism occurs in vivo in primary lung and liver carcinomas from human patients.
Figure 4. Evidence for sapienate synthesis in primary human cancers.
(a) Sapienate to palmitate and palmitoleate to palmitate ratios in HCC and normal liver tissue as well as in blood plasma from humans. Blood plasma was from healthy volunteers or liver cancer patients, while normal liver was adjacent non-cancerous tissue from liver cancer patients and non-transplanted donor livers (normal: blood plasma n=23, tissue n=16 and cancer: blood plasma n=33, tissue n=16). Grey indicates blood plasma and black indicates tissue. Notably, blood plasma ratios from healthy volunteers are the same as in Figure 4b.
(b) Sapienate to palmitate and palmitoleate to palmitate ratios in lung cancers and normal lung tissue as well as in blood plasma from humans. Blood plasma was from healthy volunteers or cancer patients, while normal lung was adjacent non-cancerous tissue from cancer patients (normal: blood plasma n=23, tissue n=15 and cancer: blood plasma: n=34, tissue n=15). Grey indicates blood plasma and black indicates tissue. Notably, blood plasma ratios from healthy volunteers are the same as in Figure 4a.
(c) Sapienate metabolism is an alternative monodesaturation pathway.
Two-Way ANOVA with Tukey multiple comparisons. Error bars represent SEM of mean from different individuals.
So far, the well-characterized SCD-dependent fatty acid desaturation pathway was considered to be the only source of de novo generated monounsaturated fatty acids in cancer cells8. Here, we discovered that cancer cells can rewire their fatty acid metabolism and desaturate palmitate to the unusual fatty acid sapienate (Figure 4c). Particularly, we find evidence for sapienate metabolism in human lung and liver carcinoma. This finding can explain metabolic plasticity and demonstrates heterogeneity in the fatty acid desaturation metabolism of cancer cells. While we find that sapienate and cis-8-octadecenoate can support membrane synthesis, it is tempting to speculate that sapienate and its elongation products impact the known fatty acid and lipid signaling networks of cancer cells9–11. Consequently, this could provide cancer cells with a hitherto unexplored possibility to deregulate signaling networks, opening new opportunities to understand and target them. In conclusion, our discovery increases the current understanding of fatty acid metabolism in cancers and suggests sapienate biosynthesis as an alternative source of monounsaturated fatty acids.
Experimental Procedures
Cell lines, cell culture and chemicals
All cell lines were confirmed to be mycoplasma free based on the MycoAlert™ Mycoplasma Detection Kit (Lonza, Basel, Switzerland). Human HEK293T epithelial cells, RWPE-1 prostate cells, MCFl0A breast cells, A549 and H460 lung carcinoma, MDA-MB-468 and T47D breast adenocarcinoma, and DU145 prostate carcinoma cell lines were obtained from ATCC (Manassas, VA, USA). HUH7 liver carcinoma cell line was obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank (Osaka, Japan). Cell lines have not been authenticated with the exception of MDA-MB-468 cells which were authenticated by fingerprinting. RWPE-1 cells were cultured in keratinocyte serum-free medium (K-SFM), supplemented with 0.05 mg per mL bovine pituitary extract, 5 ng per mL epidermal growth factor, 1% penicillin (50 U per mL) and 1% streptomycin (50 µg per mL) (all Life Technologies, CA, USA). MCF10A cells were cultured in Dulbecco's modified Eagle's medium-F12 (DMEM-F12) (Life Technologies, CA, USA), supplemented with 5% horse serum (Life Technologies, CA, USA), 1% penicillin (50 U per mL) (Life Technologies, CA, USA), 1% streptomycin (50 µg per mL) (Life Technologies, CA, USA), 0.5 μg per mL hydrocortisone (Sigma-Aldrich, MO, USA), 100 ng per mL cholera toxin (Sigma-Aldrich, MO, USA), 10 μg per mL insulin (Sigma-Aldrich, MO, USA), and 20 ng per mL recombinant human epidermal growth factor (PeproTech EC, London, UK). Other cells were cultured in high glucose (4.5 g per L) Dulbecco's modified Eagle's medium (DMEM) (Life Technologies, CA, USA) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, MA, USA), 1% penicillin (50 U per mL) (Life Technologies, CA, USA) and 1% streptomycin (50 µg per mL) (Life Technologies, CA, USA). For growth and labeling experiments, low serum conditions (0.5-1% FBS) were applied. 13C6-glucose (CLM-1396 Cambridge Isotope Laboratories, MA, USA) was used for labeling experiments. Hygromycin B and puromycin dihydrochloride (Life Technologies, CA, USA) were added to the growth medium for selection of overexpression and knockdown cell lines, respectively. Merck Frosst Cpd 3j4 was used as an SCD inhibitor, as described in the patent application WO2006/130986. The fatty acids palmitoleate (16:1) and oleate (18:1) were purchased from Sigma-Aldrich (MO, USA). Cis-8-octadecenoate (18:1) was purchased from Larodan (Solna, Sweden) and sapienate (16:1, hexadecenoic acid cis-6) from Matreya LLC. Solvents for metabolite extraction and mass spectrometry were HPLC grade from Sigma-Aldrich (MO, USA).
Knockdown and overexpression strategies
FADS2 knockdown cell lines were generated using the shRNA-expressing lentiviral pLKO1-puro vector with a puromycin selection cassette (Plasmid #8453; Addgene, MA, USA). Clone IDs for shRNAs were as follows: shFADS2-1 (TRCN0000064755; sequence: CCGGCCACGGCAAG-AACTCAAAGATCTCGAGATCTTTGAGTTCTTGCCGTGGTTTTTG) and shFADS2-2 (TRCN0000064757; sequence: CCGGCCACCTGTCTGTCTACAGAAACTCGAGTTTCTGTAGA-CAGACAGGTGGTTTTTG) (Sigma-Aldrich, MO, USA). A scrambled shRNA, i.e. TRC1 sequence: AACAAGATGAAGAGCACCAA was used as a negative control for FADS2 knockdown cells. FADS2 overexpression cell lines were generated using the pLVX-IRES-Hyg vector with a hygromycin selection cassette (Clontech Laboratories Inc., CA, USA). An empty pLVX-IRES-Hyg vector served as a negative control for FADS2 overexpression. Lentiviruses were produced by transfection of HEK293T cells. Transduction of cells was performed overnight and the medium was replaced the next day. Poly-clonal cells were selected for 1-2 weeks with puromycin in the case of knockdown cells or with hygromycin in the case of overexpression before experiments were performed. All knockdown and overexpression cell lines were validated by quantitative real-time PCR (qRT-PCR) and proteomic analysis (Extended Data Figure 2i, 3a). Primers for FADS2 were designed to amplify a cDNA segment in the sequence as follows: forward primer 5’-gaccacggcaagaactcaaag-3’ and reverse primer 5’-gagggtaggaatccagccatt-3’. For SCD1, the forward and reverse primer used were 5’-tctctgctacacttgggagc-3’ and 5’-gagctttgtaagagcggtgg-3’, respectively. Relative gene transcript levels were compared to the control gene RPL-19, with 5’-attggtctcattggggtctaac-3’ and 5’-agtatgctcaggcttcagaaga-3’ as forward and reverse primer, respectively. Real-time PCR reactions were performed on a 7,500 Fast Real-Time PCR System (Applied Biosystems, Life Technologies). Amplification was performed at 95 °C for 10 min, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Samples were assayed in triplicates.
Quantification of FADS2 gene expression in primary tumors
Human HCC and adjacent normal liver tissue samples were retrieved from the archive of the Institute of Pathology of the LMU Munich with approval of the LMU Munich’s ethics committee (approval no. 307-16 UE). Representative tumor and normal tissue areas were reviewed by a resident pathologist in representative tissue slides stained with hematoxylin and eosin (H&E). Corresponding areas were micro-dissected from paraffin blocks using biopsy punches (2 mm diameter). Deparaffinization of the tissue and RNA isolation was carried out using the Formapure Total Kit (Beckman Coulter). cDNA synthesis was performed using 1 µg of RNA with the MultiScribe Reverse Transcription kit (Applied Biosystems). Analysis of the gene expression of FADS2 was performed by qRT-PCR using SYBR-Green Master Mix (Applied Biosystems) in a CFX Connect Real-Time System (Bio-Rad). Primers were used at a concentration of 0.5 µM in a final reaction volume of 15 µl. Three technical replicates were analyzed per sample. RPLP0 expression was used as a control for normalization. Primer sequences were as follows: FADS2 forward primer 5′-gaccacggcaagaactcaaag-3′ and reverse primer 5′-gagggtaggaatccagccatt-3′; RPLP0 forward primer 5′- gaaactctgcattctcgcttc-3′ and reverse primer 5′- ggtgtaatccgtctccacag -3.
Human lung adenocarcinoma (NSCLC) and adjacent normal lung samples were retrieved from patients enrolled in an IRB-approved protocol, after obtaining informed consent (ClinicalTrials.gov Identifier: NCT02095808). Based on pre-operative imaging and gross inspection at resection, viable fragments of tumor and lung were sampled. RNA isolation was carried out using the Trizol reagent (Thermo Fisher Scientific, MA, USA) and FastPrep®-24 (MPbio, CA, USA). cDNA synthesis was performed using 1 µg of RNA with the qScript cDNA Synthesis Kit (QuantaBio, MA, USA). Analysis of FADS2 gene expression was performed by qRT-PCR using Platinum® SYBR® Green qPCR SuperMix-UDG (Thermo Fisher Scientific, MA, USA) on a Viia7 Real Time PCR system (Applied Biosystems, Life Technologies). Amplification was performed at 95°C for 10 min, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Primers for FADS2 were designed to amplify a cDNA segment in the sequence as follows: forward primer 5’-gaccacggcaagaactcaaag-3’ and reverse primer 5’-gagggtaggaatccagccatt-3’. Relative gene transcript levels were compared to the control gene RPL-19, with 5’-attggtctcattggggtctaac-3’ and 5’-agtatgctcaggcttcagaaga-3’ as forward and reverse primer, respectively. Samples were assayed in triplicates.
Growth and labeling experiments
Cancer cells were seeded in the wells of 12-well plates (Corning, NY, USA) at either 7×104 cells per well (HUH7, A549, H460 and DU145), 1.5×105 cells per well (MDA-MB-468) or 2×105 cells per well (T47D) in low FBS DMEM (1.5 mL per well) and grown in a humidified environment at 37 °C with 5% CO2. For HUH7, 1% FBS was considered low FBS DMEM; for all other cell lines, 0.5% FBS was used. Low FBS (0.5-1%) DMEM contains a total of 4.31-8.62 µM fatty acids, with palmitate, oleate and stearate being the most abundant fatty acids (Extended Data Figure 7a). After 24 h, the medium was aspirated, cells were washed with DPBS, and low FBS DMEM (4.5 g per L glucose for growth experiments or 4.5 g per L 13C6-glucose for labeling experiments, including fatty acid synthesis assessment) supplemented with 0.1% DMSO (control) or Merck Frosst Cpd 3j (either at 0.5 nM, 1 nM or 2 nM concentration dissolved in DMSO) was added to the wells (1.5 mL per well). Treatment was carried out for 72 h, during which cells were grown in a humidified environment at 37 °C with 5% CO2. For growth experiments, cells were counted prior to treatment (initial count) and 72 h after treatment (final count) using a MoxiTM Z Mini Automated Cell Counter (Orflo Technologies, ID, USA) or a Countess™ II Automated Cell Counter (Thermo Fisher Scientific, MA, USA) following trypsinization. Cell number change was calculated by subtracting the initial count from the final count, and was subsequently normalized to control condition, unless stated otherwise. For labeling experiments, cells were washed with saline solution after treatment and metabolism was quenched by flash-freezing the plates in liquid nitrogen. Plates were stored at –80 °C until metabolite extraction. All experiments were performed in triplicates. Key experiments were confirmed in a more physiological blood-like medium (BLM)12,13 with low FBS (Extended Data Figure 7b-h).
For rescue experiments, cells were seeded and grown as described above. After 24 h, the medium was replaced by medium containing 0.1% DMSO (control) or Merck Frosst Cpd 3j (0.5 nM, 1 nM or 2 nM), supplemented with either 1% ethanol (control) or 20 µM palmitoleate (cis-9-16:1), sapienate (cis-6-16:1), oleate (cis-9-18:1) or cis-8-octadecenoate (cis-8-C18:1) dissolved in ethanol. Cells were counted prior to treatment and 72 h after treatment using a MoxiTM Z Mini Automated Cell Counter (Orflo Technologies, ID, USA) or a Countess™ II Automated Cell Counter (Thermo Fisher Scientific, MA, USA) following trypsinization, and relative cell number changes were calculated as described above. All experiments were performed in triplicates. Changes in intracellular fatty acid abundance upon supplementation were confirmed by mass spectrometry (Supplementary Table 1c, d).
Primary hepatocytes of one donor were obtained from the Hepatocytes and Liver Stem Cell Bank, Cliniques Universitaires St Luc, Brussels, Belgium. Cells were thawed using the Corning® Gentest™ High Viability CryoHepatocyte Recovery Kit (Corning, NY, USA). Cells were then plated on collagen I-coated 6-well plates at a density of 1.4×106 cells per well, and left to attach for 8 h in Corning Plating Medium (Corning, NY, USA).
Reverse labeling experiments
HUH7 and A549 cells were seeded at 1×106 cells per flask in T75 flasks in low FBS (1% FBS, HUH7) or 10% FBS (A549) DMEM and grown as described above. After 24 h, the medium was aspirated, cells were washed with DPBS and 1% (HUH7) or 10% (A549) dialyzed FBS DMEM (4.5 g per L 13C6-glucose) was added to the cells. Cells were cultured for 1 week (medium was replaced every 3 d) to fully label all cellular metabolites. Next, cells were trypsinized, washed and seeded in the wells of a 12-well plate at 7×104 cells per well in low FBS DMEM containing 4.5 g per L 13C6-glucose (1% FBS for HUH7 and 0.5% FBS for A549). After 24 h, the medium was aspirated, cells were washed with DPBS and low FBS DMEM containing 4.5 g per L 13C6-glucose (1% FBS for HUH7 and 0.5% FBS for A549) supplemented with either 1% ethanol (control) or 20 µM 12C sapienate (cis-6-16:1) dissolved in ethanol was added to the cells (1.5 mL per well). Treatment was carried out for 72 h. After treatment, cells were washed with saline solution and metabolism was quenched by flash-freezing the plates in liquid nitrogen. Plates were stored at -80 °C until metabolite extraction. All experiments were performed in triplicates.
If fully labelled cells use 12C sapienate, a decrease in the 13C enrichment will be observed in the metabolites into which sapienate is incorporated, such as cis-8-octadecenoate. Since our GC-MS setup does not allow for separation of cis-8 and cis-9-octadecenoate, and our GC-FID setup does not allow for assessment of 13C enrichment, we opted to assess the 13C enrichment in the pool of octadecenoate (consisting of cis-8 and cis-9-octadecenoate) via GC-MS. Note that only cis-8-octadecenoate, and not cis-9-octadecenoate, can result from sapienate metabolism. Hence, a decrease in 13C enrichment in the octadecenoate pool indicates the incorporation of 12C sapienate into cis-8-octadecenoate.
Metabolite extraction and metabolite measurement
Metabolite extractions were performed using the methods described in Christen et al. and Lorendeau et al. 14,15. Briefly, for 12-well cell culture plates containing 1-5×105 cells per well, the medium was aspirated, cells were washed with blood bank saline, and cell metabolism was quenched by flash-freezing the plates in liquid nitrogen. Next, 400 µL –20 °C cold 65% methanol was added to the wells, cells were scraped with a pipet tip and suspensions were transferred to Eppendorf tubes. Next, 250 µL –20°C cold chloroform was added and samples were vortexed at 4 °C for 10 min to extract metabolites. Phase separation was achieved by centrifugation at 4 °C for 10 min, after which the chloroform phase (containing the total fatty acid content) was separated and dried by vacuum centrifugation. For tissue samples, tissues were weighed (5-10 mg) and pulverized (Cryomill, Retsch) under liquid nitrogen conditions. For plasma samples, 10 µL (murine) or 50 µL (human) of cold plasma was transferred to an Eppendorf tube. Next, 800 µL –20 °C cold 65% methanol was added to the samples, followed by 500 µL –20 °C cold chloroform. Samples were then handled as described above. Dried fatty acid samples were immediately processed to fatty acid methyl esters as described below, thereby avoiding degradation.
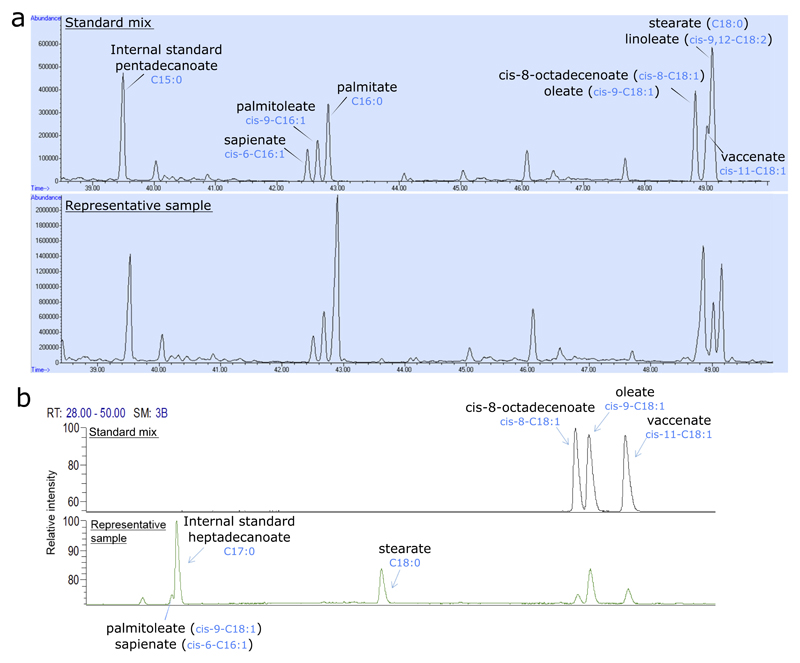
Total fatty acid samples were esterified with 500 µL 2% sulphuric acid in methanol for 180 min at 60 °C or overnight at 50°C and extracted by addition of 600 µL hexane and 100 µL saturated aqueous NaCl. Samples were centrifuged for 5 min and the hexane phase was separated and dried by vacuum centrifugation. Samples were resuspended in hexane, after which isotopologue distributions of fatty acids were measured with a 7890A GC system (Agilent Technologies, CA, USA) combined with a 5975C or a 7000 inert MSD system (Agilent Technologies, CA, USA). One microliter of each sample was injected in splitless (5975C) or 5:1 split (7000) mode with an inlet temperature of 270 °C onto a DB35MS column (Agilent Technologies, CA, USA). Helium was used as a carrier gas with a flowrate of 1 mL per min. The oven was held at 80 °C for 1 min and ramped with 5 °C per min to 300 °C. The MS system was operated under electron impact ionization at 70 eV and a mass range of 100-650 amu was scanned. Evidence for separation of cis-6-C16:1 and cis-9-C16:1 is presented in Extended Data Figure 8a. Isotopologue distributions were extracted from the raw chromatograms using an in-house Matlab script. Correction for naturally occurring isotopologues was achieved using Isocor software. Palmitate, palmitoleate and sapienate synthesis were calculated based on isotopologue distributions of the according fatty acid, using Isotopomer Spectral Analysis (ISA)16 and an in-house Matlab script. For determination of relative metabolite abundances, the total ion counts were normalized to an internal standard (pentadecanoate or heptadecanoate) and the cell number or protein content for cell extracts. For tissue extracts, the total ion counts were normalized to the internal standard (pentadecanoate or heptadecanoate) and tissue weight. The desaturation activity from palmitate to sapienate or palmitoleate was calculated by dividing the normalized total ion count of sapienate or palmitoleate by the normalized total ion count of palmitate.
Cis-8-octadecenoate abundances were measured with a gas chromatography-flame ionization detector (GC-FID) system (TRACE GC ULTRA, Thermo Fisher Scientific Inc., Waltham, MA). One microliter of each sample was injected onto a SLB-IL111 capillary column (100 m × 0.25 mm ID, 0.20µm thickness, Supelco, Bellefonte, PA) with an inlet temperature of 250 °C. Helium was used as a carrier gas at 1.2 mL per min and the oven temperature was maintained isothermally at 140 °C. cis-C18:1 isomers (∆8,9,11) were synthesized as described before17, processed to fatty acid methyl esters, and used for validation of the separation of cis-C18:1 isomers and confirmation of the retention times of respective cis-C18:1 fatty acid methyl esters. Evidence for separation of cis-8-C18:1 and cis-9-C18:1 is presented in Extended Data Figure 8b. For determination of relative cis-8-octadecenoate abundances, the total ion counts were normalized to the internal standard (pentadecanoate or heptadecanoate). For cell extracts, total ion counts were further normalized to the protein content or to the total fatty acid abundance. Total ion counts in tissue samples were further normalized to tissue weight.
FADS2 protein analysis
Human cell pellets (1-4×107 cells per pellet) in triplicate were lysed in a urea lysis buffer containing 8 M urea and 20 mM HEPES pH 8.0. Cell pellets were homogenized by sonication on a Diagenode Bioruptor Plus instrument using the following settings: high intensity power output, 10 cycles of 30 sec ON/30 sec OFF pulses, 4 °C water bath. The protein concentration in each lysate was measured by Bradford (Bio-Rad Laboratories, CA, USA) and aliquots containing 100 µg of protein were used for further analysis. Proteins were reduced by addition of DTT to a concentration of 5 mM and incubation for 30 min at 55 °C. Next, proteins were alkylated by addition of iodoacetamide to a concentration of 10 mM for 15 min at room temperature in the dark. Samples were diluted with 20 mM HEPES pH 8.0 to a urea concentration of 4 M and proteins were digested with 1 µg lysyl endopeptidase (Wako Chemicals, DA, USA) (1/100, w/w) for 4 h at 37 °C. Samples were further diluted with 20 mM HEPES pH 8.0 to a final urea concentration of 2 M and proteins were digested with 1 µg trypsin (Promega, WI, USA) (1/100, w/w) overnight at 37 °C. The resulting peptide mixture was purified using OMIX C18 pipette tips (Agilent Technologies, CA, USA) or SampliQ SPE C18 cartridges (Agilent Technologies, CA, USA) and 100 µg of peptides of each sample were dried completely by vacuum drying and stored at –20 °C until further use.
Peptides were re-suspended in loading solvent (0.1% TFA in water/acetonitrile, 98/2 (v/v)) and 3 µg was injected for LC−MS/MS analysis on an Ultimate 3000 RSLC nano LC (Thermo Fisher Scientific, MA, USA) in-line coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, MA, USA). The peptides were first loaded on a trapping column (made in-house, 100 μm internal diameter (I.D.) × 20 mm, 5 μm beads C18 Reprosil-HD, Dr. Maisch, Germany) and after flushing from the trapping column, peptides were separated on an analytical column in the needle (made in-house, 75 μm I.D. × 400 mm, 1.9 μm beads C18 Reprosil-HD, Dr. Maisch) using a non-linear 150 min gradient of 2 to 56% solvent B (0.1% formic acid in water/acetonitrile, 20/80 (v/v)) at a constant flow rate of 250 nL per min and at a constant temperature of 50 °C (CoControl 3.3.05, Sonation). Following a 10 min wash reaching 99% solvent B, the column was re-equilibrated with solvent A (0.1% formic acid in water).
The mass spectrometer was operated in data-dependent, positive ionization mode, automatically switching between MS and MS/MS acquisition for the 16 most abundant peaks in a given MS spectrum. The source voltage was set to 2.5 kV and the capillary temperature was 250 °C. One MS1 scan (m/z 375-1500, AGC target 3E6 ions, maximum ion injection time of 60 ms), acquired at a resolution of 60,000 (at 200 m/z) was followed by up to 16 tandem MS scans, acquired at a resolution of 15,000 (at 200 m/z) of the most intense ions fulfilling predefined selection criteria: AGC target 1E5 ions, maximum ion injection time of 80 ms, isolation window of 1.5 m/z, fixed first mass of 145 m/z, spectrum data type: centroid, under fill ratio 2%, intensity threshold 1.3×104, exclusion of unassigned and singly charged precursors, peptide match preferred, exclude isotopes on, dynamic exclusion time of 12 s. The normalized collision energy was set to 28% and the polydimethylcyclosiloxane background ion at 445.12003 Da was used for internal calibration (lock mass).
Data analysis was performed by MaxQuant (version 1.6.1.0) using the Andromeda search engine with default search settings including a false discovery rate set at 1% on both the peptide and protein level. Spectra were searched against the human proteins in the Swiss-Prot database (database release version of December 2017 containing 20,243 human protein sequences, downloaded from www.uniprot.org). The mass tolerance for precursor and fragment ions was set to 4.5 and 20 ppm, respectively, during the main search. Enzyme specificity was set as C-terminal to arginine and lysine (trypsin), also allowing cleavage at arginine/lysine-proline bonds with a maximum of two missed cleavages. Carbamidomethylation of cysteine residues was set as a fixed modification and variable modifications were set to oxidation of methionine residues (to sulfoxides) and acetylation of protein N-termini. Proteins were quantified by the MaxLFQ algorithm integrated in the MaxQuant software. Only proteins with at least one unique or razor peptide were retained for identification, while a minimum ratio count of two unique peptides was required for quantification. The obtained LFQ intensity values of FADS2 were used to quantify the protein and compare FADS2 levels between different samples.
Analysis of phospholipid-bound sapienate and palmitoleate
HUH7 cells carrying a non-targeting shRNA or a shRNA targeting FADS2 (shFADS2-2) were seeded in T75 flasks, allowed to attach for 24 h and subsequently grown for 72 h in 1% FBS DMEM. Cells were trypsinized, washed with blood bank saline and re-suspended in TAG lysis buffer (1% IGEPAL® CA-630 (Nonidet P-40), 50 mM Trizma® hydrochloride and 150 mM sodium chloride). Lipids were extracted from cell lysate according to the Folch method18. An internal standard containing a known concentration of 1,2-Diheptanoyl-sn-glycero-3-phosphocholine (17:0) was added to samples prior to extraction to allow the quantification of total phospholipids. Lipid fractions were separated by thin-layer chromatography and fatty acid methyl esters (FAMEs) were prepared as previously described19,20. Separation and detection of total phospholipid FAMEs was achieved using a 6890N Network GC System (Agilent Technologies; CA, USA) with flame ionization detection. FAMEs were identified by their retention times compared to a standard containing 31 known fatty acids and quantified in micromoles from the peak area based on their molecular weight. The micromole quantities were then totaled and each fatty acid was expressed as a percentage of this value (molar percentage; mol%) or µg fatty acids normalized to cellular protein concentration.
Analysis of phospholipid species
HUH7 and A549 cells carrying a non-targeting shRNA or a shRNA targeting FADS2 (shFADS2-1 and shFADS2-2) were seeded in T75 flasks, allowed to attach for 24 h and subsequently grown for 72 h in low FBS DMEM (1% FBS for HUH7; 0.5% FBS for A549). Cells were trypsinized, washed three times with cold DPBS, and cell pellets were re-suspended in 0.8 mL DPBS. Lipid extraction and MRM-based phospholipid (semi)quantification analysis was performed as described previously21. Briefly, 0.7 mL of homogenized cells were mixed with 0.9 mL MeOH:HCl (1N) (8:1), 0.8 mL CHCl3 and 200 μg per mL of the antioxidant 2,6-di-tert-butyl-4-methylphenol (Sigma). The organic fractions were evaporated under vacuum using a Savant Speedvac spd111v (Thermo Fisher Scientific) at room temperature and the remaining lipid pellet was stored at -20 °C under argon. Prior to mass spectrometry analysis, lipid pellets were reconstituted in running solution (CH3OH:CHCl3:NH4OH; 90:10:1.25; v/v/v). Lipid standards PC25:0, PC43:6, SM30:1, PE25:0, PE43:6, PI25:0, PI31:1, PI43:6, PS25:0, PS31:1 and PS37:4 (Avanti Polar Lipids) were added based on the amount of DNA of the original sample. Phospholipids were analyzed by electrospray ionization tandem mass spectrometry (ESI-MS/MS) on a hybrid quadrupole linear ion trap mass spectrometer (4000 QTRAP system, AB SCIEX) equipped with a TriVersa NanoMate robotic nanosource (Advion Biosciences) for automated sample injection and spraying as described22. Phospholipid profiling was executed by (positive or negative) precursor ion or neutral loss scanning at a collision energy of 50 eV/45 eV, 35 eV, −35 eV and −60 eV for precursor 184 [sphingomyelin (SM)/phosphatidylcholine (PC)], neutral loss 141 [phosphatidylethanolamine (PE)], neutral loss 87 [phosphatidylserine (PS)] and precursor 241 [phosphatidylinositol (PI)], respectively. Phospholipid quantification was performed by multiple reaction monitoring (MRM), the transitions being based on the neutral losses or the typical product ions as described above. Typically, a 3 min period of signal averaging was used for each spectrum. The data were corrected for carbon isotope effects and chain length and analyzed using in house-developed software (RALP). As a background, the intensities of species detected in the ‘internal standards only’ spectra were considered after being divided by the ion suppression factor of each sample. The ion suppression factor was calculated for each sample separately by dividing the intensity of the standards in the ‘internal standards only’ spectrum by the intensity of the standards in the sample spectrum. Only the phospholipid species displaying an intensity of at least 5 times the blank value were taken into account. In order to quantify the total amount of phospholipids in a phospholipid class, the abundances of individually measured species within the phospholipid class were totalled. Data were normalized based on DNA amount. MRM-based analysis of the C16:1 containing phospholipids was based on Ekroos et al23.
Analysis of lipid peroxidation sensitivity
3×107 HUH7 control and FADS2 knockdown cells were seeded in 15 cm Petri dishes in 1% FBS DMEM. After 24 h, cells were treated with control or 5 µM RSL3, the latter inhibiting glutathione peroxidase 4 and inducing lipid peroxidation. Lipid peroxidation was quantified using the MDA assay kit (Sigma) according to manufacturer’s instructions with some exceptions. Briefly, 3×107 cells were collected in BHT supplemented PBS. TBA-acetic acid solution was buffered to pH 3.5. Plates were read using an EnSpire Multimode Plate Reader (PerkinElmer). Signal was normalized to total amount of sample DNA.
Analysis of membrane fluidity
HUH7 and A549 control and FADS2 knockdown cells were grown on glass coverslips (n=4) in low FBS DMEM (1% FBS for HUH7; 0.5% FBS for A549) for 3 d and subsequently fixed for 15-30 min in 4% PFA at room temperature. For lipid phase analysis, cells were stained with di-4-ANEPPDHQ (Thermo Fisher Scientific, MA, USA) according to the manufacturer's specifications. For imaging, a Nikon A1R confocal microscope attached to Ti eclipse outfitted with a Plan Apo VC 60x lens oil immersion lens with an NA of 1.4 was used (Nikon Instruments, Tokyo, Japan). The spectral detector was set to 530-590 and 590-650 nm to image the spectral shift of the dye from lipid ordered to disordered phase. Resulting images were analyzed with NIS software (Nikon Instruments, NY, USA), thereby segmenting the cells in the images and calculating the ordered to disordered ratio24. The higher the ordered to disordered ratio, the more saturated lipids are present in the membrane. Per coverslip, 10 fields of views were imaged.
Mouse models
All animal experiments were approved by the local authorities in compliance with all relevant ethical regulations (including but not limited to tumor size). For injection models, mice were randomized before injection of cancer cells. All samples were analyzed blinded. Sample size was determined using power calculations with B=0.8 and P<0.05 based on preliminary data and in compliance with the 3R system: Replacement, Reduction, Refinement.
Subcutaneous HUH7 xenograft model
Mice were fed ad libitum a CRM (E) expanded low-fat diet (Special Diets Services 801730) one week prior to the start of the experiments until sacrificing. 1×106 HUH7 cells were subcutaneously injected in 50% Matrigel™, (BD Biosciences), 50% cell culture medium without FCS into 8-9-week old female immunocompromised NMRInu/nu mice (Taconic M&B AS, Ejby, Denmark). Tumors were allowed to grow up to a mean tumor size of 30 mm2 (length × width), before mice were allocated to treatment and control group by stratified randomization based on their primary tumor size (8 animals per group). Tumor-bearing mice were either treated with vehicle (10% v/v ethanol, 40% v/v solutol) alone or with 1.5 mg per kg SCD inhibitor Merck Frosst Cpd 3j for six consecutive days twice daily per oral (p.o.). After the last treatment, mice were euthanized and tumors were harvested and snap-frozen for further analyses. Humane endpoints were determined as a tumour size of 225 mm2. Following symptoms were monitored and upon detection of one of the symptoms the animal was euthanized: Ulcerated tumours, loss of 20 % body weight, self-mutilation, constant circling, constant curved posture, blood attachment at body openings, bloody diarrhea, abnormal breathing, constant eye and nasal discharge, apathic, constantly cramps, constant tremble, crouching posture, self-isolation. All animal experiments were conducted in accordance with the European Union directives and the German animal welfare act and approved by the Landesamt für Gesundheit und Soziales (LAGeSo, Berlin, Germany).
DEN-induced hepatocellular carcinoma model
Male C57Bl/6N mice were obtained from the KU Leuven animal facility and injected with diethylnitrosamine (DEN) (25 mg per kg) intraperitoneally at the age of 14 d. After 4 weeks, mice were fed with a 13 kJ% fat diet (Ssniff S8655-E220) until sacrificed and analyzed at 20, 30, 31 or 32 weeks after DEN administration. Tumor and non-tumor tissues were collected and rapidly frozen for metabolomic analysis using a liquid nitrogen cooled Biosqueezer (Biospec Products). Tissues were weighed (5-10 mg) and pulverized (Cryomill, Retsch) under liquid nitrogen conditions. The pulverized tissues were extracted for GC-MS analysis as described above. Humane endpoints were determined as follows: Liver tumours can be detected by palpation as hard masses in otherwise soft abdominal area or by an enlarged abdomen at later stages. A 20% increase in the normal abdomen diameter was considered as a humane endpoint. Following symptoms were monitored and upon detection of one of the symptoms the animal was euthanized: loss of ability to ambulate, loss of skin elasticity, laboured respiration, or weight loss over 20% of initial body weight. Housing and experimental animal procedures were approved by the Institutional Animal Care and Research Advisory Committee of the KU Leuven, Belgium.
Hepatocyte-specific Pten and Stk knockout hepatocellular carcinoma model
mice were fed standard chow ad libitum throughout the procedure. C57BL/6 mice carrying Pten conditional knockout alleles were crossed with Albumin (Alb)-Cre-transgenic mice. Control animals were PtenloxP/loxP; Alb-Cre—, whereas the experimental mice were PtenloxP/loxP; Alb-Cre+ (HepPten—). For hepatocyte-specific Stk3 and Stk4 deletion, mice carrying Stk3 and Stk4 conditional knockout alleles (in a mixed background of CD1, C57BL/6 and 129) were crossed with Albumin (Alb)-Cre-transgenic mice. Control animals were Stk3loxP/loxP; Stk4loxP/loxP; Alb-Cre—, whereas the experimental mice were Stk3loxP/loxP; Stk4loxP/loxP; Alb-Cre+. At necropsy (approximately 12 months for Pten and 4-6 months for Stk), tumors and liver tissues were harvested and snap-frozen for further analysis from male mice. Humane endpoints were determined as follows: Maximum tumour size was based on linear or volumetric measurements. When linear measurements are used, the longest dimension of the tumour should not exceed 2.0 cm in mice. If multiple tumours are present, the combination of the two longest diameters may not exceed 2.0 cm for mice. Tumor volume (TV) can also be used to evaluate tumour size using the following formula: TV = (width)2 x length/2. Maximum single tumour size or tumour burden for mice is 2000 mm3. All animal procedures were approved by and carried out in accordance with the policies and regulations set forth by the Institutional Animal Care and Use Committee (IACUC) at MD Anderson Cancer Center.
Hepatocyte-specific myrAKT-N-Ras overexpression hepatocellular carcinoma model
wild-type FVB/N mice were obtained from Charles River (Wilmington, MA) and subjected to hydrodynamic injection. Briefly, 10 μg of the plasmids encoding myr-AKT1 and/or N-RasV12 along with sleeping beauty transposase in a ratio of 25:1 were diluted in 2 mL saline (0.9% NaCl), filtered through a 0.22 μm filter, and injected into the lateral tail vein of 6 to 8-week-old FVB/N male mice in 5 to 7 sec. After 6-9 weeks, tumors were harvested and snap-frozen for further analysis. Humane endpoints were determined as follows: Liver tumours can be detected by palpation as hard masses in otherwise soft abdominal area or by an enlarged abdomen at later stages. When a tumour was first noticed it was recorded. A 20% increase in the normal abdomen diameter was considered as a humane endpoint. Mice were housed, fed, and monitored in accordance with protocols approved by the Committee for Animal Research at the University of California, San Francisco.
Orthotopic HUH7 liver cancer model
Mice were fed ad libitum a CRM (E) expanded low-fat diet (Special Diets Services 801730) one week prior to the start of the experiments until sacrificing. Next, 0.5x106 HUH7 control or shFADS2-2 cells were orthotopically injected in 100% Matrigel™, (BD Biosciences) into the left liver lobe of anesthetized (3% isoflurane, 2% oxygen) 6-week-old male immunocompromised NMRInu/nu mice (Taconic M&B AS, Ejby, Denmark). For analgesia, mice were given 5 mg per kg carprofen subcutaneous before and after surgery and for the following 3 d. After 8 d, mice were either treated with vehicle (10% v/v ethanol, 40% v/v solutol) alone or with 1.5 mg per kg SCD inhibitor Merck Frosst Cpd 3j for six consecutive days twice daily per oral (p.o.). Fourteen hours after the last treatment, mice were euthanized, the tumor nodule was resected, and blood and normal liver tissue were sampled. For metabolomic analysis, half of the tumor nodule and non-tumor tissues were rapidly frozen using a liquid nitrogen cooled Biosqueezer (Biospec Products). Tissues were then weighed (5-10 mg) and pulverized (Cryomill, Retsch) under liquid nitrogen conditions. The pulverized tissues were extracted for GC-MS analysis as described above. For histological quantification of the tumor area, half of the nodule was formalin-fixed and paraffin embedded (FFPE). From these FFPE tissue blocks, 4 µm sections were cut and stained with hematoxylin and eosin. The mean tumor area in percent of the total tissue area was determined in low-power magnifications by two independent physicians trained in histopathology. Humane endpoints were determined as a tumour size of 2 cm3. Following symptoms were monitored and upon detection of one of the symptoms the animal was euthanized: Loss of ability to ambulate, laboured respiration, surgical infection or weight loss over 10 % of initial body weight. Housing and experimental animal procedures were approved by the Institutional Animal Care and Research Advisory Committee of the KU Leuven, Belgium.
Collection of clinical samples
Human samples were collected upon ethical approval of local authorities. Analysis was performed blinded. Patient information is provided in Supplementary Table 2.
Liver
Liver and/or liver cancer samples were obtained from Indivumed GmbH (Hamburg, Germany), from the Laboratory of Hepatology (Commissie Medische Ethiek UZ Leuven – KU Leuven, Belgium), and the archive of the Institute of Pathology of the LMU Munich with approval of the LMU Munich’s ethics committee (approval no. 307-16 UE), respecting patients’ rights. Blood samples from healthy volunteers and HCC patients were collected in collaboration with the Laboratory of Hepatology (UZ Leuven – KU Leuven, Belgium) after obtaining informed consent. Freshly isolated primary hepatocytes (donor F125) were obtained from the Hepatocytes and Hepatic Stem Cells Bank from the Cliniques Universitaires St Luc, Brussels, Belgium. An agreement from the Belgian Ministry of Health was obtained for the Hepatocytes and Hepatic Stem Cells Bank. A written and signed informed consent has been obtained for collection of the cells.
Lung
Patients with non-small cell lung cancer were enrolled in an IRB-approved protocol after obtaining informed consent (ClinicalTrials.gov Identifier: NCT02095808). Study eligibility included pulmonary masses measuring 1 cm or more in diameter. Standard surgical procedures were followed, with the majority of cases being robotic lobectomies. Based on pre-operative imaging and gross inspection at resection, viable fragments of tumor and lung were sampled. Plasma samples from lung cancer patients were drawn primarily from an arterial line throughout the procedure.
Statistical analysis and software
Statistical data analysis was performed using GraphPad Prism 7 (GraphPad Software Inc., CA, USA) on n≥3 biological replicates. Details on statistical tests and post-tests are presented in the figure legends. Detection of mathematical outliers was performed using Grubb's test. Sample size for all in vitro experiments was chosen empirically. For in vivo experiments, sample size was determined using power calculations with β=0.8 and P<0.05, based on preliminary data. Data are presented as mean ± SD, or as mean ± SEM, as indicated in the figure legends. Where applicable custom codes are available from the corresponding author upon reasonable request.
Extended Data
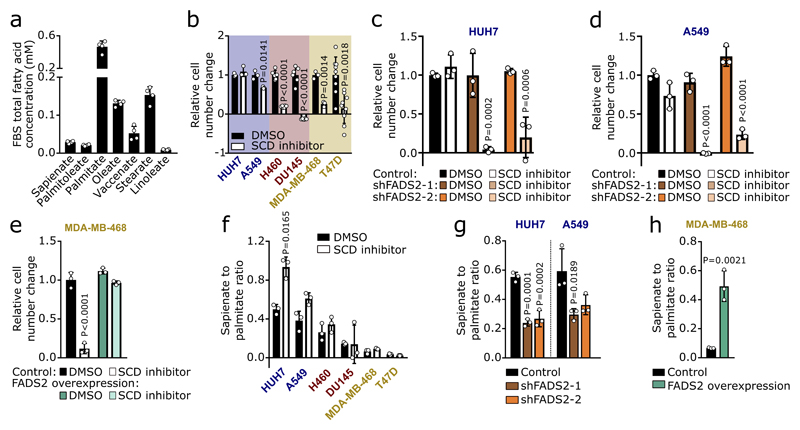
Extended Data Figure 1. SCD-independent cancer cells produce sapienate.
(a) Schematic overview of fatty acid metabolism. AcCoA: Acetyl-coenzyme A; SCD1/5: Stearoyl-CoA desaturase 1 and 5; Elovl5/6: elongation of very long chain fatty acids protein 5 and 6.
(b-e) SCD desaturation activity based on the palmitoleate to palmitate ratio, oleate to stearate ratio, palmitoleate and palmitate synthesis upon Merck Frosst Cpd 3j treatment (HUH7, A549: 2 nM; H460, DU145: 1 nM; MDA-MB-468, T47D: 0.5 nM; panel b-d: n=3; panel e: HUH7 n=3, A459 n=3, H460 n=6 (control) n=4 (SCD inhibitor), DU145 n=3, MDA-MB-468 n=5, T47D n=5 (control) n=6 (SCD inhibitor)). Unpaired two-sided Student’s T-tests with Holm-Sidak multiple comparisons.
(f-h) Correlation between SCD independence and palmitate synthesis, growth rate or total fatty acid abundance (n=3). SCD independence was defined as area under the cell number curve of Figure 1a. Palmitate synthesis was derived from (e). Total fatty acid abundance was derived from Extended Data Figure 2a. Trend line (dashed line) and 95% confidence intervals (dotted lines) are depicted.
Cancer cell experiments were performed in low FBS DMEM (1%: HUH7; 0.5%: others) with treatment of 72 h. Error bars represent mean ± SD from biological independent samples.
Extended Data Figure 2. Sapienate is produced via FADS2 in cancer cells.
(a) Heat map representing fatty acid abundances with(out) Merck Frosst Cpd 3j treatment (HUH7, A549: 2 nM; H460, DU145: 1 nM; MDA-MB-468, T47D: 0.5 nM) normalized to highest abundance of each fatty acid across all cell lines/conditions (Figure 1b, Supplementary Table 1a). Over 90% reduction: white, no reduction: dark green.
(b,c) Desaturation activity to sapienate upon Merck Frosst Cpd 3j treatment (HUH7, A549: 2 nM; H460, DU145: 1 nM; MDA-MB-468, T47D: 0.5 nM; n=3). Unpaired two-sided Student’s T-tests with Holm-Sidak multiple comparisons.
(d) Sapienate to palmitate ratio in HUH7 (n=6) versus freshly isolated primary human hepatocytes (PHH; n=3), DU145 (n=6) versus RWPE-1 (n=6) prostate cells, and MDA-MB-468 (n=6) and T47D (n=6) versus MCF10A (n=6) breast cells. Unpaired Student’s T-tests and Welch’s correction (HUH7 versus PHH; DU145 versus RWEP-1); one-way ANOVA with Dunnett’s multiple comparisons (MDA-MB-468, T47D versus MCF10A).
(e) Tumor weight of HUH7 subcutaneous xenografts treated with(out) Merck Frosst Cpd 3j (n=8 one experiment; 1.5 mg per kg twice daily p.o.). Unpaired Student’s T-test with Welch’s correction.
(f,g) FADS2 gene expression in cells with(out) Merck Frosst Cpd 3j as described in (b,c) normalized to T47D cells (n=3). One-way ANOVA with Tukey’s multiple comparisons (f); unpaired Student’s T-tests with Holm-Sidak multiple comparisons (g).
(h) FADS2 protein expression in the same conditions as in (d). Statistics as described in (d). n=3.
(i) FADS2 gene/protein expression in HUH7 and A549 cells upon FADS2 silencing normalized to control (Gene: HUH7 n=3, A549 n=6; protein n=3 except for A549 shFADS2-2 n=2). One-way ANOVA with Dunnett’s multiple comparisons.
Cancer cell experiments were performed in low FBS DMEM (1%: HUH7; 0.5%: others) with treatment of 72 h. Error bars represent SD (in vitro) or SEM (in vivo) from mean of biological independent samples (in vitro) or animals (in vivo).
Extended Data Figure 3. Sapienate rather than arachidonate metabolism causes SCD-independence.
(a) Relative FADS2 gene/protein expression and desaturation activity to sapienate in MDA-MB-468 control and FADS2 overexpression cells with DMSO or 0.5 nM Merck Frosst Cpd 3j normalized to control (n=3). Unpaired two-sided Student’s T-test.
(b) Relative FADS2 gene expression in tumor nodules from HUH7 control or FADS2 knockdown orthotopic xenografts with vehicle or Merck Frosst Cpd 3j (1.5 mg per kg twice daily per oral; p.o.; n=4; one experiment) normalized to control. One-way ANOVA with Tukey’s multiple comparisons.
(c,d) Relative desaturation activity from palmitate to sapienate or palmitoleate in normal adjacent liver (L) and tumor nodules (T) in the same model as described in (f) normalized to normal control livers. Control+vehicle-L n=18 (c) n=20 (d); control+vehicle-T n=18 (c) n=20 (d); control+SCD inhibition-L n=14 (c, d); control+SCD inhibition-T n=13 (c) n=14 (d); shFADS2-2+vehicle-L n=19 (c, d); shFADS2-2+vehicle-T n=18 (c, d); shFADS2-2+SCD inhibition-L n=15 (c) n=16 (d); shFADS2-2+SCD inhibition-T n=15 (c, d); two experiments. Two-way ANOVA with Sidak’s multiple comparisons.
(e,f) Desaturation activity from linoleate to γ-linolenate based on the γ-linolenate to linoleate ratio and arachidonate abundance in HUH7 and A549 control (non-targeting shRNA) and FADS2 knockdown (shFADS2) cells (n=3). One-way ANOVA with Dunnett’s multiple comparisons.
(g,h) Linoleate and arachidonate abundance in normal adjacent murine liver and tumor nodules from HUH7 control (non-targeting shRNA) or FADS2 knockdown (shFADS2) orthotopic xenografts treated with vehicle or Merck Frosst Cpd 3j (1.5 mg per kg twice daily per oral; p.o.). Control+vehicle-L n=12 (g) n=14 (h); control+vehicle-T n=13 (g) n=14 (h); control+SCD inhibition-L n=14 (g) n=15 (h); control+SCD inhibition-T n=14 (g) n=16 (h); shFADS2-2+vehicle-L n=14 (g) n=16 (h); shFADS2-2+vehicle-T n=13 (g) n=15 (h); shFADS2-2+SCD inhibition-L n=15 (g) n=18 (h); shFADS2-2+SCD inhibition-T n=15 (g) n=16 (h); two experiments. Two-way ANOVA with Tukey’s multiple comparisons.
Cancer cell experiments were performed in low FBS DMEM (1% : HUH7; 0.5% : others) with treatment of 72 h. Error bars represent SD (in vitro) or SEM (in vivo) from mean of biolocial independent samples (in vitro) or animals (in vivo).
Extended Data Figure 4. Carbons from sapienate are detected in octadecenoate.
(a-f) 13C enrichment of palmitate or stearate from 13C6 glucose in HUH7 or A549 cells in control condition (ethanol, black) or upon 12C sapienate supplementation (blue). Cells were grown in 10% dialyzed FBS DMEM containing 4.5 g per L 13C6 glucose for 1 week, after which cells were grown for 72 h in 0.5% FBS DMEM containing 4.5 g per L 13C6 glucose supplemented with ethanol or 20 µM 12C sapienate.
The purpose of this experiment was to trace the incorporation of carbons from sapienate into cis-8-octadecenoate. Palmitate and stearate were measured as controls. Since 13C-labeled sapienate is not commercially available, we performed a reverse labeling in which we pre-labeled HUH7 and A549 cells with 13C6-glucose to enrich cis-8-octadecenoate with 13C. Then, we supplemented these cells with unlabeled sapienate in the presence of 13C6-glucose and determined the 13C enrichment of octadecenoate. If sapienate is elongated to cis-8-octadecenoate, we expect a shift in the 13C enrichment from higher to lower octadecenoate isotopologues. Indeed, we found that supplementation of unlabeled sapienate shifted the 13C enrichment accordingly (a, d). Moreover, the largest 13C enrichment increase was found in the M+2 isotopologue, indicating the elongation of unlabeled sapienate to octadecenoate with 13C labeled acetyl-CoA. As expected, sapienate supplementation did not or only marginally change the 13C enrichment of palmitate and stearate (b,c,e,f).
Unpaired two-sided Student’s T-tests; n=3. Error bars represent mean ± SD from biological independent samples.
Extended Data Figure 5. Sapienate is elongated to cis-8-octadecenoate.
(a) Relative cis-8-octadecenoate abundances in cancer cells normalized to T47D cells. HUH7 n=3; A549 n=3, H460 n=5, DU145 n=3, MDA-MB-468 n=5, T47D n=5. One-way ANOVA with Tukey’s multiple comparisons.
(b,c) Relative cis-8-octadecenoate abundances in HUH7 and A549 control (non-targeting shRNA) and FADS2 knockdown (shFADS2) cells in control condition (ethanol) or upon 20 µM sapienate supplementation normalized to control. HUH7: control n=6 (ethanol) n=3 (sapienate); shFADS2-1 n=3; shFADS2-2 n=6 (ethanol) n=3 (sapienate); A549: control n=6; shFADS2-1 n=3; shFADS2-2 n=3. Data values are shown in Supplementary Table 1c, d. Two-way ANOVA with Tukey’s multiple comparisons.
(d) Relative proliferation of MDA-MB-468 cells with ethanol (n=9) or 20 µM cis-8-octadecenoate (n=3) upon treatment with DMSO or 0.5 nM Merck Frosst Cpd 3j. Data were normalized to control with error bars representing SEM. Two-way ANOVA with Tukey multiple comparisons.
(e,f) Relative proliferation of HUH7 and A549 control (non-targeting shRNA) and knockdown (shFADS2) cells with ethanol or 20 µM cis-8-octadecenoate upon treatment with DMSO or 2 nM Merck Frosst Cpd 3j. HUH7: control n=9; shFADS2-1 n=6; shFADS2-2 n=9; A549: EtOH n=6; cis-8-C18:1 n=3. Data were normalized to control. Two-way ANOVA with Tukey multiple comparisons. Only statistics for pair-wise comparisons are depicted.
Cancer cell experiments were performed in low FBS DMEM (1%: HUH7; 0.5%: all other cancer cells) with treatment of 72 h. Error bars represent mean ± SD from biological independent samples, unless otherwise noted.
Extended Data Figure 6. Sapienate and cis-8-octadecenoate are used in membranes.
(a-d) Heat map representing abundance changes of phosphatidylcholine (a), phosphatidylethanolamine (b), phosphatidylserine (c) and phosphatidylinositol (d) species in control and FADS2 knockdown HUH7 and A549 cells relative to control. HUH7: control n=3; shFADS2-1 n=4; shFADS2-2 n=5; A549: n=5. Only significant differences are depicted as log2 fold change compared to control. X denotes blank or excluded values. Phospholipid species carrying sapienate or palmitoleate are depicted in bold red and listed in Supplementary Table 1e. Two-way ANOVA with Dunnett’s multiple comparisons.
(e) Relative distribution of phospholipid species in HUH7 (n=2) and A549 (n=5) cell with non-targeting shRNA (control). PC: phosphatidylcholine; PE: phosphatidylethanolamine; PS: phosphatidylserine; PI: phosphatidylinositol; SM: sphingomyelin.
(f) Membrane fluidity based on the ordered to disordered ratio in HUH7 and A549 with a non-targeting shRNA (control; black) or two different shRNA targeting FADS2 (brown and orange) normalized to control (n=4). The higher the ordered to disordered ratio, the more saturated lipids are present in the membrane. One-way ANOVA with Dunnett’s multiple comparisons.
(g) Lipid peroxidation sensitivity via MDA assay in HUH7 with a non-targeting shRNA (control; black) or two different shRNA targeting FADS2 (brown and orange) normalized to control (n=3). Cells were treated with vehicle or 5 µM RSL3, the latter inhibiting glutathione peroxidase 4 and inducing lipid peroxidation. Two-way ANOVA with Sidak’s multiple comparisons.
Cancer cell experiments were performed in low FBS DMEM (1%: HUH7; 0.5%: all other cancer cells) with treatment of 72 h. Data are presented as mean ± SD from biological independent samples.
Extended Data Figure 7. SCD independence and sapienate metabolism occur in medium with glucose and amino acid concentrations that are similar to physiological conditions.
(a) Fatty acid concentrations of FBS (fetal bovine serum; n=4). Low FBS condition (0.5-1% FBS) corresponds to a total fatty acid concentration of 4.31-8.62 µM.
(b) Sensitivity profile of cancer cells to Merck Frosst Cpd 3j (white; HUH7, A549: 2 nM; H460, DU145: 1 nM; MDA-MB-468, T47D: 0.5 nM) in blood-like medium (BLM)12,13 normalized to control. HUH7 n=3; A549 n=3; H460 n=6; DU145 n=6, MDA-MB-468 n=3; T47D n=9. Two-way ANOVA with Dunnett’s multiple comparisons.
(c, d) Sensitivity profile of HUH7 and A549 control (non-targeting shRNA; black) and knockdown (shFADS2; brown and orange) cells treated with DMSO (dark bars) or 2 nM Merck Frosst Cpd 3j (light bars) in blood-like medium (BLM)12,13 normalized to control (n=3). Two-way ANOVA with Holm-Sidak multiple comparisons.
(e) Sensitivity profile of MDA-MB-468 control (black) and FADS2 overexpression (green) cells DMSO (dark bars) or 0.5 nM Merck Frosst Cpd 3j (light bars) in blood-like medium (BLM)12,13 normalized to control (n=3). Two-way ANOVA with Holm-Sidak multiple comparisons.
(f) Desaturation activity to sapienate based on the sapienate to palmitate ratio in cancer cells in conditions as described in (b). n=3. Unpaired two-sided Student’s T-tests with Holm-Sidak multiple comparisons.
(g,h) Desaturation activity from palmitate to sapienate based on the sapienate to palmitate ratio in the same conditions as described in (c-f). n=3. One-way ANOVA with Dunnett’s multiple comparisons (g); unpaired Student’s T-test (h).
Cancer cell experiments were performed in low FBS BLM (1%: HUH7; 0.5%: all other cancer cells) with treatment of 72 h. Data are presented as mean ± SD from biological independent samples.
Extended Data Figure 8. Separation and detection of sapienate and cis-8-octadecenoate.
(a) Separation and detection of sapienate (cis-6-hexadecenoate) and palmitoleate (cis-9-hexadecenoate) via gas chromatography mass spectrometry. Separation was optimized using a standard mix containing pentadecanoate, sapienate, palmitoleate, palmitate, cis-8-octadecenoate, oleate, vaccinate, linoleate and stearate (upper panel), and the method was subsequently validated by measurement of these fatty acids in biological samples. A representative biological sample is presented in the lower panel.
(b) Separation and detection of cis-8-octadecenoate, oleate (cis-9-octadecenoate) and vaccinate (cis-11-octadecenoate) via gas chromatography flame ionization detector. Separation was optimized using a standard mix containing cis-8-, cis-9- and cis-11-octadecenoate (upper panel), and the method was subsequently validated by measurement of these fatty acids in biological samples. A representative biological sample is presented in the lower panel.
Supplementary Material
Acknowledgements
We thank all patients and volunteers as well as Prof. Van Pelt, Ingrid Vander Elst and Petra Windmolders for advice on the orthotopic injections and patient sample collection; Dr. Impens and Delphi Van Haver (VIB Proteomics Core); Dr. van Hoef (VIB-CCB Bioinformatics Expertise Center); and Dr. Nittner (VIB-CCB Histology Expertise Center). SP is supported by a VIB International PhD student fellowship. GR is supported by Kom op tegen Kanker and FWO fellowships. RS, MR, JFG and JAGD are supported by FWO fellowships. RJD, a Howard Hughes Medical Institute Investigator, Joel B. Steinberg, M.D. Chair in Pediatrics and Robert L. Moody, Sr. Faculty Scholar at UT Southwestern, is funded by CPRIT (RP160089) and the National Cancer Institute (R35CA22044901). TGPG is funded by the German Cancer Aid (DKH-111886, DKH-70112257), LMUexcellent, Bettina-Bräu-Stiftung, Dr. Leopold und Carmen Ellinger, Matthias-Lackas, Walter Schulz, Wilhelm Sander (2016.167.1), Gert & Susanna Mayer Foundations, and the Deutsche Forschungsgemeinschaft (DFG 391665916). SMF is funded by the European Research Council under the ERC Consolidator Grant Agreement n.771486–MetaRegulation and Marie Curie CIG n.617727–MetabolismConnect, FWO Odysseus II, KU Leuven Methusalem Co-funding, and Bayer AG.
Footnotes
Author Contributions
Investigation and Validation: KV, StC, SP generated knockdown and overexpression cell lines; KV, StC, SP, DB, CEN and LRP quantified FADS2 gene expression; KV, StC and SP performed growth and labeling experiments – growth experiments were reproduced independently; KV and StC performed reverse labeling experiments; KV, StC, SP, DB and RS performed metabolite extractions; KV, StC, RS and KY performed metabolite measurement; KV, SP, GR and MR analyzed FADS2 protein data; TC, CC and LH analyzed phospholipid-bound sapienate and palmitoleate; AT and JD performed phospholipidomic analysis and assessed lipid peroxidation sensitivity; AK analyzed membrane fluidity; KV, DB, RS, CCK performed mouse experiments; KV, MFO, BF, RJD, DC, ChV and TGPG collected and assessed human clinical samples. TGPG and MFO performed H&E staining. Formal Analysis: KV, StC, and SMF interpreted all data except of H&E staining that were interpreted by TGPG and MFO. Resource: MN and ES provided primary hepatocytes. RB and CaV advised on genetic engineering and primary hepatocytes. BF and RJD provided human lung tissue and plasma samples. DC, ChV, SvC, SG, AS, TGPG provided human liver tissue and plasma samples. NG, JVS and LH advised on lipid and fatty acid analysis. SM advised on membrane fluidity. RD, JFG and JAGD support mass spectrometry method development. AM, MY, SYK and LB provided liver tissue and plasma from genetic mouse models. AS, PS, AH, JAVG, SvC, SG, AS, TC, KKW provided reagents. SMF advised on experiments and analysis. Conceptualization: KV, StC, SMF. Visualization: KV. Writing – Original Draft: SMF. Writing – Review & Editing: KV, StC, SMF. Supervision: SMF. Funding – SMF.
Author Information
AH, CCK, AS, PS, SvC and SG have competing interests as employees of Bayer AG. KKW is a founder and equity holder of G1 Therapeutics and he has Consulting/Sponsored Research Agreements with AstraZeneca, Janssen, Pfizer, Array, Novartis, Merck, Takeda, Ono, Targimmune and BMS. SMF has received funding from Bayer AG and Merck.
Data availability
The authors declare that all data supporting the findings of this study are available within the article, its extended data files, source data or from the corresponding author upon reasonable request.
References
- 1.Elia I, Schmieder R, Christen S, Fendt S-M. Organ-Specific Cancer Metabolism and Its Potential for Therapy. Handbook of Experimental Pharmacology. 2016;233:321–353. doi: 10.1007/164_2015_10. [DOI] [PubMed] [Google Scholar]
- 2.Rohrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 3.Peck B, Schulze A. Lipid desaturation – the next step in targeting lipogenesis in cancer? The FEBS Journal. 2016;283:2767–2778. doi: 10.1111/febs.13681. [DOI] [PubMed] [Google Scholar]
- 4.Ramtohul YK, et al. SAR and optimization of thiazole analogs as potent stearoyl-CoA desaturase inhibitors. Bioorganic & medicinal chemistry letters. 2010;20:1593–1597. doi: 10.1016/j.bmcl.2010.01.083. [DOI] [PubMed] [Google Scholar]
- 5.Ge L, Gordon JS, Hsuan C, Stenn K, Prouty SM. Identification of the Δ-6 Desaturase of Human Sebaceous Glands: Expression and Enzyme Activity. Journal of Investigative Dermatology. 2003;120:707–714. doi: 10.1046/j.1523-1747.2003.12123.x. [DOI] [PubMed] [Google Scholar]
- 6.Prouty SM, Pappas A. In: Lipids and Skin Health. Pappas Apostolos., editor. Springer International Publishing; 2015. pp. 139–157. [Google Scholar]
- 7.Fischer E, Sauer U. Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nature genetics. 2005;37:636–640. doi: 10.1038/ng1555. [DOI] [PubMed] [Google Scholar]
- 8.Zoeller RA, Wood R. The importance of the stearoyl-CoA desaturase system in octadecenoate metabolism in the Morris hepatoma 7288C. Biochim Biophys Acta. 1985;845:380–388. doi: 10.1016/0167-4889(85)90202-2. [DOI] [PubMed] [Google Scholar]
- 9.Currie E, Schulze A, Zechner R, Walther TC, Farese RV. Cellular fatty acid metabolism and cancer. Cell metabolism. 2013;18 doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ackerman D, Simon MC. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends in Cell Biology. 2014;24:472–478. doi: 10.1016/j.tcb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dall'Armi C, Devereaux Kelly A, Di Paolo G. The Role of Lipids in the Control of Autophagy. Current Biology. 2013;23:R33–R45. doi: 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Garcia J, Fendt SM. Assessing the Impact of the Nutrient Microenvironment on the Metabolism of Effector CD8(+) T Cells. Methods in molecular biology (Clifton, N.J.) 2019;1862:187–216. doi: 10.1007/978-1-4939-8769-6_14. [DOI] [PubMed] [Google Scholar]
- 13.Tardito S, et al. Glutamine Synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nature cell biology. 2015;17:1556–1568. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christen S, et al. Breast Cancer-Derived Lung Metastases Show Increased Pyruvate Carboxylase-Dependent Anaplerosis. Cell Rep. 2016;17:837–848. doi: 10.1016/j.celrep.2016.09.042. [DOI] [PubMed] [Google Scholar]
- 15.Lorendeau D, et al. Dual loss of succinate dehydrogenase (SDH) and complex I activity is necessary to recapitulate the metabolic phenotype of SDH mutant tumors. Metabolic engineering. 2016 doi: 10.1016/j.ymben.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Kharroubi AT, Masterson TM, Aldaghlas TA, Kennedy KA, Kelleher JK. Isotopomer spectral analysis of triglyceride fatty acid synthesis in 3T3-L1 cells. The American journal of physiology. 1992;263:E667–675. doi: 10.1152/ajpendo.1992.263.4.E667. [DOI] [PubMed] [Google Scholar]
- 17.Gunstone FD. Lipid Synthesis and Manufacture. Sheffield Academic Press; 1999. [Google Scholar]
- 18.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. The Journal of biological chemistry. 1957;226:497–509. [PubMed] [Google Scholar]
- 19.Holub BJ, Skeaff CM. Nutritional regulation of cellular phosphatidylinositol. Methods in enzymology. 1987;141:234–244. doi: 10.1016/0076-6879(87)41071-9. [DOI] [PubMed] [Google Scholar]
- 20.Hodson L, Skeaff CM, Wallace AJ, Arribas GL. Stability of plasma and erythrocyte fatty acid composition during cold storage. Clinica chimica acta; international journal of clinical chemistry. 2002;321:63–67. doi: 10.1016/s0009-8981(02)00100-6. [DOI] [PubMed] [Google Scholar]
- 21.Marien E, et al. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. International Journal of Cancer. Journal International du Cancer. 2015;137:1539–1548. doi: 10.1002/ijc.29517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rysman E, et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer research. 2010;70:8117–8126. doi: 10.1158/0008-5472.can-09-3871. [DOI] [PubMed] [Google Scholar]
- 23.Ekroos K, et al. Charting molecular composition of phosphatidylcholines by fatty acid scanning and ion trap MS3 fragmentation. Journal of lipid research. 2003;44:2181–2192. doi: 10.1194/jlr.D300020-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Owen DM, Rentero C, Magenau A, Abu-Siniyeh A, Gaus K. Quantitative imaging of membrane lipid order in cells and organisms. Nature protocols. 2011;7:24–35. doi: 10.1038/nprot.2011.419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.