Abstract
The early 1980s revelation of cis-acting genomic elements, known as transcriptional enhancers, is still regarded as one of the fundamental discoveries in the genomic field. However, only with the emergence of genome-wide techniques has the genuine biological scope of enhancers begun to be fully uncovered. Massive scientific efforts of multiple laboratories rapidly advanced the overall perception that enhancers are typified by common epigenetic characteristics that distinguish their activating potential. Broadly, chromatin modifiers and transcriptional regulators lay down the essential foundations necessary for constituting enhancers in their activated form. Basing on genome-wide ChIP-sequencing of enhancer-related marks we identified myogenic enhancers before and after muscle differentiation and discovered that MyoD was bound to nearly a third of condition-specific enhancers. Experimental studies that tested the deposition patterns of enhancer-related epigenetic marks in MyoD-null myoblasts revealed the high dependency that a specific set of muscle enhancers have towards this transcriptional regulator. Re-expression of MyoD restored the deposition of enhancer-related marks at myotube-specific enhancers and partially at myoblasts-specific enhancers. Our proposed mechanistic model suggests that MyoD is involved in recruitment of methyltransferase Set7, acetyltransferase p300 and deposition of H3K4me1 and H3K27ac at myogenic enhancers. In addition, MyoD binding at enhancers is associated with PolII occupancy and with local noncoding transcription. Modulation of muscle enhancers is suggested to be coordinated via transcription factors docking, including c-Jun and Jdp2 that bind to muscle enhancers in a MyoD-dependent manner. We hypothesize that distinct transcription factors may act as placeholders and mediate the assembly of newly formed myogenic enhancers.
Keywords: muscle enhancers, skeletal muscle, myogenesis, transcription, MyoD, placeholders, ChIP-seq
TRANSCRIPTIONAL ENHANCERS AS GENERATORS OF DIVERSIFICATION
One of the major milestones in the field of genomics over the last half century is the discovery of transcriptional enhancers, made by two independent laboratories in the early 1980s [Banerji et al., 1981; Moreau et al., 1981]. Originally identified in the DNA of the mammalian simian virus (SV40), enhancers were established as cis-acting DNA segments that could stimulate Pol-II-mediated transcription over long-ranges, independently of their orientation relative to their corresponding promoter site. The discovery was demonstrated to be a widespread genomic phenomenon when Schaffner et al. showed for the first time that the 72 base pairs (bp) repeated viral sequence element, which they identified in the SV40 genome, could amplify the transcription of the rabbit β-globin gene in human Hela cells by nearly 200-times [Banerji et al., 1981]. Due to their capability to enhance the transcription of their respective distally located target gene these genomic elements were labeled by Schaffner and his colleagues as “enhancers” [Banerji et al., 1981]. Shortly afterward, the same group of scientists identified a novel enhancer within a murine immunoglobulin heavy chain gene, and demonstrated that transcriptional enhancers exist in the genome of metazoan organisms as well [Banerji et al., 1983]. Soon after this, the Tjian laboratory provided new mechanistic insights regarding the function of transcriptional enhancers by demonstrating that enhancer activation is governed by specific transcription factors that selectively bind distinct recognition sequences within enhancers and control their activity [Lee et al., 1987].
In the following decade, a new question was posed regarding the genomic mechanism that restrains enhancers from activating improper genes. This conundrum was elegantly addressed by Kellum and Schedl, using their early identification of novel “specialized chromatin structures” (also known as “scs domains”) flanking the Drosophila melanogaster heat shock cytogenetic locus 87A7. They then developed an enhancer-blocking assay, which demonstrated that the stimulatory activity of the yolk protein-1 enhancer could be blocked by several scs-like elements that were cloned between the enhancer region and its corresponding promoter [Kellum and Schedl, 1992]. The functional concept of ‘genomic insulators’ was then coined and delineated the strong linkage between long-range chromatin domains and transcriptional control. Nevertheless, it was only with the advent of state-of-the-art genome-wide approaches such as chip-on-ChIP and ChIP-seq (by the years 1999 and 2009, respectively) that the scope of transcriptional enhancers could be vastly explored to reflect their unequivocal importance in phenotypic diversification and lineage specification, manifested by cell-type-specific gene expression, as discussed further below.
Comparative genomic studies in metazoans pointed to a paradoxical anti-correlation between the number of genes per organism (genome size) and the relative level of phenotypic complexity, characterized by the diversity of the organism’s behavior and the number of different cell types present. Thus for example, both the Caenorhabditis elegans nematode and the Homo sapiens genomes harbor approximately 20,000–25,000 genes each, and yet the Human genome is roughly 30 times larger than that of the nematode, implying that the developmental complexity of the Human and other complex mammalian genomes must be achieved through alternative mechanisms related to the noncoding portion of the genome rather than by a simple increment in the number of genes [Bulger and Groudine, 2011]. In addition, only a small fraction (less than 2%) of the mammalian genome encodes for protein-coding genes, suggesting that the surrounding noncoding regions acquired through evolution in higher organisms, must carry vital attributes that can expand the potential of information concealed within the coding dimension. Essentially, it is now being recognized that the utilization of protein-coding genes is completely impacted by the non-coding dimension, which confers phenotypic subtleties that ultimately yield the profound differences that distinguish higher organisms from lower ones [Salmena et al., 2011]. At the same event, it is important to note that these noncoding genomic portions are generally not conserved among species (as the total genome size among species may differ significantly), and could reflect an important functional layer that contradicts the central dogma linking evolutionary conservation with function. Therefore, given that the noncoding dimension may play a vital role in dictating the uniqueness of complex genomes, and that unlike the coding genes this portion is not conserved among all species, it is important to be cautious when attempting to extrapolate between results obtained by mouse models to the phenomena manifested by Human, especially when the observations are related to the noncoding dimension. Thus, from a translational research perspective, it would become more coherent and informative, in general, to utilize mice model for studying the coding portion of the genome rather than the noncoding portion.
Indeed, genomic mechanisms that can boost diversification of transcriptome and proteome by generating numerous mRNA products from a single gene and expanding the usage of the template code have been characterized, and include alternative splicing [Kim et al., 2007], epigenetic signals [Bonasio, 2014], RNA editing [Farajollahi and Maas, 2010], microRNAs [Heimberg et al., 2008], and ceRNAs [Salmena et al., 2011]. In this respect it is important to note to a comparative multi-species study that revealed a higher frequency of alternative splicing in vertebrates as compared to invertebrates, suggesting that introns accumulation confers an evolutionary advantage [Kim et al., 2007]. Nevertheless, alongside these genomic mechanisms, temporal and spatial diversification of the transcriptional program can contribute enormously to the uprising of phenotypic complexity within a specific organism. In particular, in metazoans cellular differentiation of multiple lineage-specific embryonic stem cells (ES cells) demands a precise and firmly controlled gene expression patterning of developmental genes. For facilitating the complex demand of cellular differentiation, including the temporal and spatial arrangement of transcription factors, combinatorial transcriptional regulatory dynamics that involve distal cis-regulatory transcriptional enhancers that decoupled from the confines of the promoter-proximal region, have been evolved [Bulger and Groudine, 2011]. By distributing the regulatory sites further away, up-stream or down-stream from the promoter regions complex organisms were able to circumvent the intrinsic bottleneck existing in more primitive organisms in which transcription is exclusively regulated by the promoter region alone.
The advent of massively parallel sequencing and the fact that enhancer regions could be identified by their well-characterized epigenetic signature, as will be further discussed below, has enabled the elucidation and mapping of enhancers in numerous cell-types and uncovered their complex but largely invariant chromatin arrangement. Seminal next-generation sequencing (NGS) studies by the laboratories of Ren and Pennacchio have brought about the revolutionary finding that chromatin states at promoters are highly correlated even among distinct cell types, whereas at enhancers histone modifications are largely cell type-specific and highly associated with transcriptional patterns [Heintzman et al., 2009; Visel et al., 2009]. These novel observations, certainly marking a crucial milestone in the genomic field, prompted scientists to consider transcriptional enhancers as a fundamental determinant that markedly influences cell type-specific transcription patterning in the genomes of complex creatures. Indeed, a body of scientific evidence suggests that due to the central role that enhancers play in orchestrating transcription they also emerge as prominent players in a range of molecular networks influencing major cellular characteristics such as nuclear and chromosomal architecture [Chepelev et al., 2012] and stem cell pluripotency [Creyghton et al., 2010]. Accordingly, transcriptional enhancers have been distinguished as an essential layer of cellular regulation, which contributes immensely to acceleration of behavioral and morphological diversification in multicellular organisms.
UNIQUE EPIGENETIC CHARACTERISTICS OF ENHANCERS ENABLE THEIR GENOME-WIDE MAPPING
Due to the imperative role of transcriptional enhancers in regulation of tissue-specific genes, it is essential to obtain a full assembly of their genomic locations in each cell type. For doing so it is important to recognize certain enhancer characteristics that provide the basis for enhancer identification in recent genome-wide studies. Similarly to promoters, enhancers are typified by sets of DNA sequences (also referred to as “binding sites”) that attract multiple transcription factors that can activate or suppress transcription. Concomitantly with the recruitment of transcription factors to their cognate binding sites, the recruitment of chromatin-modifying coactivators or corepressors take place, facilitating the way for decoration of the enhancer region with unique histone modifications, as further described below (reviewed in [Bulger and Groudine, 2011; Calo and Wysocka, 2013; Ong and Corces, 2011; Spitz and Furlong, 2012]). Historically, the main characteristic hallmark of transcriptional enhancers that has been repeatedly validated and determined by changes in sensitivity of chromatin to Dnase I, restriction enzymes, or enzyme micrococcal nuclease (MNase), is that enhancer regions are typically devoid of nucleosome − the basic structural unit of eukaryotic chromatin [Crawford et al., 2006]. This unique structural arrangement of nucleosome-free regions allows the DNA fiber to become accessible to the binding of various transcription factors. Lately, genome-wide studies paved the way for uncovering specific enhancer blue prints that are currently being applied in defining active cis-regulatory elements in a tissue-selective manner. Nucleosomes that flank the genomic region of active enhancers often carry post-translational modifications. It is now clear that monomethylated H3K4 (H3K4me1) deposition, coupled with high levels of acetylation of histone H3K27 (H3K27ac), form a predominant epigenetic signature for enhancers associated with actively transcribed genes [Heintzman et al., 2007; Shlyueva et al., 2014]. In addition, several studies have identified a disparity in typical chromatin patterns that appear on active enhancers and promoters. For example, trimethylation of histone H3 on lysine 4 (H3K4me3) is highly enriched across the promoters of active genes, while at activated enhancers this mark is typically absent. By contrast, unlike active enhancers that are robustly marked by H3K4me1, active promoters often lack this mark [Koch et al., 2007; Rada-Iglesias et al., 2011]. Indeed, an important study recently revealed an unexpected roles for H3K4me1 in gene repression and also restricting the recruitment of chromatin-modifying complexes to specific locations within promoters [Cheng et al., 2014]. It is interesting however to note that both enhancers and promoters at their repressed or silent states are both generally occupied by H3K27me3-modified nucleosomes, a process mediated by the Polycomb repressive complex [Simon and Kingston, 2009]. By utilizing the virtue of predictive histone mark signature of active enhancers many genome-wide studies were able to re-confirmed the firm concordance between epigenetic patterns at enhancers and their transcriptionally active state that was confirmed by activity assays [Arnold et al., 2013; Bonn et al., 2012].
CHROMATIN MODIFYING ENZYMES AND TRANSCRIPTION FACTORS DICTATE THE ACTIVATION STATUS OF ENHANCERS
Although there are remarkable differences between promoters and enhancers, both recruit an extremely diverse set of chromatin-modifying enzymes to promote the formation of distinctive patterns of histone modifications. Deposition of H3K4me1 at enhancer regions is largely catalyzed by the Trr/MLL3/MLL4 COMPASS-like complexes [Herz et al., 2012; Hu et al., 2013; Lee et al., 2013], although it has also been shown that additional methyltransferase enzymes, including MLL1/2 [Jeong et al., 2011; Kawabe et al., 2012] and Set7/9 [Blum et al., 2012; Tao et al., 2011] can mediate H3K4 monomethylation within enhancer regions in specific cellular contexts as well. It is known that acetylation of H3K27ac is mainly mediated by the two homologous acetyltransferases - CREB binding protein (CBP) and p300 [Jin et al., 2011; Pasini et al., 2010; Tie et al., 2009]. Importantly, deposition of H3K27ac at distal enhancers demarcates enhancers in their active state and distinguishes them from poised enhancers. Thus, through the recent years numerous laboratories, studying various cell types, have performed genome-wide mapping of enhancer regions based on the characteristic localization patterns of these two enzymes [Ghisletti et al., 2010; Heintzman et al., 2009; Kim et al., 2010; Rada-Iglesias et al., 2011]. Altogether, these studies captured unique set of enhancers that uncover tissue-specific transcriptional patterns, suggesting that tissue-specific occupancy of enhancers by p300 or CBP governs unique developmental circuits in numerous tissues [Heintzman et al., 2009]. It is important to note that although H3K27ac is one of the main substrates of CBP/p300, other lysine residues such as H3K18 could be subjected to acetylation mediated by these enzymes [Jin et al., 2011] and are indeed detectable at enhancer regions as well [Wang et al., 2008b]. Along these lines, recent studies revealed that in metazoans the acetyltransferase GCN5/PCAF complex, ATAC is targeted to p300-unbound enhancers, defining a separate class of enhancers modified independently of p300 [Krebs et al., 2011]. In accordance to these findings, H3K9ac, which is mainly deposited by GCN5/PCAF, was detected together with H3K14ac on a specific subset of active enhancers in embryonic stem cells [Karmodiya et al., 2012].
The systematic profiling of histone modifications at enhancers revealed the presence of a histone modification, H3K79me3, which was previously characterized as localizing to gene bodies, [Barski et al., 2007] to be at the core of developmental enhancers [Bonn et al., 2012]. This finding suggests that active enhancers may become transcribe by themselves as indeed, distal enhancers have been shown to recruit RNA polymerase II, and to be associated with transcription of large noncoding RNAs (ncRNAs) and enhancer-derived RNAs (eRNAs) [De Santa et al., 2010; Kim et al., 2010]. The mechanistic role of eRNAs is still ambiguous, as studies suggest their involvement in long-range transcriptional activation through association with the Mediator complex [Lai et al., 2013], or a functional role in the maintenance of accessible chromatin [Natoli and Andrau, 2012], while other studies claim that the generation of eRNAs is part of non-specific events that could be best described as the “ripple effect” [Ebisuya et al., 2008].
These canonical and non-canonical observations, triggered by the applicability of different forms of massively parallel sequencing methods for identifying enhancers, such as DNase-seq [Boyle et al., 2008], MNase-seq [Yuan et al., 2005], FAIRE-seq [Giresi et al., 2007], and FIREWACh [Murtha et al., 2014], have expedited enhancer profiling and investigation. As a result, genomic scientists in many laboratories around the world have acquired the precise mappings of enhancers in numerous cell types, including heart [May et al., 2012; Narlikar et al., 2010], forebrain [Visel et al., 2013], cortical neurons [Kim et al., 2010], skeletal muscle [Blum et al., 2012], adipocytes [Mikkelsen et al., 2010], and macrophages of the bone marrow [Ghisletti et al., 2010].
Another crucial layer of active enhancers is portrayed by enhancer-binding tissue-specific transcription factors, which facilitating in the recruitment of histone-modifying enzymes [Creyghton et al., 2010]. ChIP-seq coupled with the computational survey of putative binding motifs across enhancer regions uncovered the localization of multiple cell type-specific transcription factors to enhancers. These factors, formerly discovered and characterized as crucial regulators of lineage-specific differentiation were recently bestowed a renewed attention due to their rediscovery in the context of enhancer regulation. These factors, including PU.1 in pro-B lymphocytes [Ghisletti et al., 2010], Foxa1 and Foxa2 in hepatocytes (liver cells) [Creyghton et al., 2010; Xu et al., 2009], STAT1, 4 and 6 proteins in T lymphocytes [Vahedi et al., 2012], and Rfx1 in neural progenitors [Creyghton et al., 2010], among others. Therefore, these factors are now referred to as master regulators of enhancer activity. Future basic studies as well as translational studies exploring therapeutic strategies are on the horizon to uncover their function and implication in genomic regulation.
Epigenetic landscape studies of enhancer elements during differentiation revealed that depending on the combinatorial pattern of histones at enhancers in a given state, transcriptional enhancers could be segregated into a few functional groups: enhancers in their active state (“activated” enhancers), deposited with H3K4me1 and H3K27ac; enhancers in their inactive state (“poised” enhancers), deposited with H3K4me1 but lack H3K27ac; and “latent” enhancers that in terminally differentiated cells, which are devoid of any of the hallmarks of enhancers but are still competent of repossessing these histone features and transcription factor binding upon selective cellular stimulation [Bogdanovic et al., 2012; Creyghton et al., 2010; Ostuni et al., 2013; Rada-Iglesias et al., 2011]. Accordingly, prior to terminal differentiation, in embryonic stem cells (ESC) the enhancers regulating lineage specific genes are found in a poised state. These, upon receiving of proper differentiation stimuli turn active and allow the transcriptional stimulation of their cognate developmental genes [Creyghton et al., 2010]. Complementary insights obtained by recent developmental studies disclosed intriguing features related to poised enhancers. Similar to the bivalent domains marked by a combination of H3K4me3 and H3K27me3 at promoters of developmental genes in embryonic stem cells [Bernstein et al., 2006], poised enhancers in embryonic stem cells or undifferentiated cells were established as possessing both, H3K27me3 and H3K4me1 modifications, which associated with active and repressed states, respectively [Asp et al., 2011; Rada-Iglesias et al., 2011].
Finally, developmental studies that focused on epigenetic regulation in ES cells have demonstrated that through the earliest phase of development, poised enhancers are accessed by pioneering factors (also called “placeholders”), which are capable of directly binding to nucleosomal DNA and remodeling of the enhancer region, leading to recruitment of activating transcription factors. Triggering transcriptional competency at conjugated promoters by enhancers is a sequential process that typically includes nucleosomes repositioning, DNA demethylation, and recruitment of chromatin modifiers that subsequently induce histone methylation and acetylation [Ghisletti et al., 2010; Serandour et al., 2011; Zaret and Carroll, 2011]. Following differentiation and dependent on the lineage trajectory that is selected, the occupancy of pioneer factors at enhancer regions declines, and coordinately with their replacement by tissue-specific master regulators, the enhancer is prompt to its active state. Thus for an instance, upon adaptation of the endodermal lineage, the levels of pioneering factor FoxD3 in differentiating ES cells are greatly reduced, as this factor is superseded by the activating transcription factor FoxA1 [Xu et al., 2009]. Similarly, in differentiating neurons Sox2 is replaced at enhancers by Sox3 and Sox11 [Bergsland et al., 2011], and in macrophages binds primarily to de-novo enhancers bound by the pioneering factors PU.1 and C/EBPa [Kaikkonen et al., 2013]. As would be further discussed below, an important facet of the biology of placeholders is the replacement of these transcription factors in a timely dependent manner by enhancer activating master regulators.
In this article, we report on the theoretical basis and the scientific process that led us to the genome-wide identification of active enhancers in skeletal muscle cells, before and after terminal differentiation. We uncover general aspects related to enhancer biology and specific details related to the myogenic system in particular that enable us to place our discoveries in a relevant functional context. We further propose that studies of placeholders that regulate myogenic enhancer activation could contribute greatly to understanding of enhancer-regulated myogenesis and advance our understanding of muscle-related malignancies such as rhabdomyosarcoma.
MYOD – A MASTER REGULATOR OF MUSCLE ENHANCER ACTIVATION
Development and formation of skeletal muscle is primarily regulated by the MyoD family of myogenic regulators, also known as MRFs (myogenic regulatory factors). The four members of this basic-helix-loop-helix (bHLH) family of transcription factors include MyoD, Myf5, Mrf4 and Myogenin, are all capable of activating the myogenic program when forcedly expressed in non-muscle cells [Moncaut et al., 2013]. The discovery that MyoD is capable of imposing myogenic trans-differentiation was historically the first demonstration of cellular reprogramming. The first experimental exemplification was performed in fibroblasts [Davis et al., 1987] and subsequently was followed by additional demonstrations performed in various cell types, including retinal epithelial cells, adipocytes, hepatoma, neuroblastoma, and melanoma [Weintraub et al., 1989]. Nevertheless, in the absence of the first three master regulators skeletal muscle is not formed, whereas in the absence of myogenin myoblasts form but fully functional muscle does not [Buckingham and Rigby, 2014], suggesting that while MyoD, Myf5 and Mrf4 are responsible of determination of myogenic lineage, myogenin is in charge of dictating more advanced phases of myogenesis [Nabeshima et al., 1993]. The greater efficiency of the three master regulators to impose differentiation through the myogenic lineage, as compared to myogenin, is explained by their possession of a C-terminal domain [Gerber et al., 1997], which enables to recruit the ATP-dependent chromatin remodeling enzymes SWI/SNF for induction of myogenic target genes [de la Serna et al., 2005].
To become transcriptionally active at the onset of muscle differentiation, MyoD forms heterodimers with E-proteins (including E12, E47, E2–2, and HEB), which allow its docking to a minimal DNA binding site that composed of two E-boxes motifs. The E-box consensus motif, comprised of the VCASCTGT sequence (V=A/C/G, S=C/G) is enriched within DNA sequences of both, promoter and enhancer regions of muscle-lineage genes [Blais et al., 2005; Cao et al., 2010]. The cooperative binding of MyoD and E-proteins, which confers synergistic affinity, allows MyoD to become stably bound to DNA and recognizable by the transcription machinery [Weintraub et al., 1990]. Concomitantly with its DNA-docking, MyoD recruits histone acetyltransferase enzymes and chromatin remodeling enzymatic complexes, which increase histone acetylation and chromatin accessibility, respectively [de la Serna et al., 2005; Forcales et al., 2012]. In addition, functional studies have shown that MyoD, through its helix 3 domain interacts with the homeodomain proteins Pbx and Meis, which tether it to inaccessible E-box domains [Berkes et al., 2004]. Thus, prior to myogenic differentiation, at the myoblastic stage, Pbx occupies specific docking sites within the myogenin promoter and recruits MyoD during the entry into the differentiation stage. Interestingly, suppression of Pbx function by morpholino injection to zebrafish fast muscle resulted in repression of myogenin and fast-muscle genes expression [Maves et al., 2007].
Despite the fact that numerous studies have been focused primarily on the mechanistic aspects related to the function of MyoD at promoter regions, traditional enhancer assays, utilizing enhancer-driven expression of a reporter gene, have demonstrated that MyoD plays a crucial role in the activation of a specific set of enhancers linked to myogenic genes. Such myogenic enhancers, enriched by E-box binding motifs that are necessary for the recruitment of MyoD were found in the distal regions of several well recognized myogenic genes including MyoD itself [Goldhamer et al., 1995], myogenin [Yee and Rigby, 1993], Myf5 and Mrf4 [Carvajal et al., 2008], Ckm [Johnson et al., 1989], γ-sarcoglycan [Wakabayashi-Takai et al., 2001], and myosin chain 1 [Wentworth et al., 1991].
To obtain a comprehensive assembly of active enhancers that drive the myogenic transcriptional program prior to and post differentiation, and to uncover their prominent molecular mechanisms, we recently conducted genome-wide ChIP-seq and gene expression studies. To enhance the rigor of our scientific observations, we identified active enhancers based on the net assembly of five well-defined enhancer markers, which include three histone marks (H3K4me1, H3K18ac and H3K27ac) and two transcriptional enzymes (p300 and PolII) [Blum et al., 2012]. To focus on genomic regions that are likely to represent active enhancers we generated ten subsets, each composed of genomic regions that associate with a different combination of at least three of the five enhancer marks that were originally mapped. Because genomic regions that fulfill regulatory roles, such as enhancers, are subjected to evolutionary selection [Siepel et al., 2005] we determined the conservation level of each of the ten subsets and pooled the four subsets that were the most highly conserved. Our analysis indicated that the strongest conservation sequences were arrayed in the central portion of enhancer regions that were typically marked by a combination of H3K4me1, H3K27ac, p300 or PolII. In line with other tissue-specific genomic regions whose evolution occurs in a rapid manner [Prabhakar et al., 2006], our analysis pointed to a strong, but not ultra-strong conservation among the genomic sequences of the newly identified muscle-specific enhancers [Blum et al., 2012]. The deposition of the four enhancer marks (H3K27ac, H3K4me1, PolII and p300), which were previously shown to be associated with active enhancers in ES cells [Creyghton et al., 2010], neuronal cells [Kim et al., 2010] and macrophages [De Santa et al., 2010], was validated by conventional ChIP assays, and the activating potential of a representative subset of myogenic enhancers deposited with combinations of these active marks was validated by a set of traditional enhancer-reporter assays.
Several studies have demonstrated that the number of active enhancers is increases throughout differentiation. In zebra fish for example, the number of detectable active enhancers was increased by more than 10-fold between the blastula stage and the gastrula stage [Bogdanovic et al., 2012], and a similar increase was observed within differentiation of embryonic stem cells (ESC) to neuronal progenitor cells (NPC) [Creyghton et al., 2010]. In accordance, an overall expansion in the number of active muscle enhancers was found to characterize the myogenic differentiation program − as the number of mappable enhancers was increased by nearly 150% between the myoblastic stage (4315 enhancer) to the myotube stage (6313 enhancers). Of these, approximately 65% of myoblast and 75% of myotube enhancers were active in a condition exclusive manner, while the remaining was marked as constitutively active. These findings imply that the complex transcriptional program associated with the transition from pre-differentiated cells to fully differentiated cells is propelled by a potent increase in distal enhancer activity. Notably, our genome-wide inventory of muscle enhancers recovered multiple genomic segments previously established as enhancer regions, and broadly there was a significant correlation between the transcription levels of genes and the activity of their adjacent condition-specific enhancers, indicating that myogenic enhancers contribute to transcriptional amplification of thousands of target genes. Interestingly, the promoter-enhancer distances of myotube-specific enhancers (~40 kb) were significantly shorter compared to the distances of myoblast-specific enhancers (~53 kb), suggesting that shortening in the enhancer-promoter ranges may reflect a structural attribute of chromatin associated with muscle differentiation.
Similarly to other studies who showed the concomitant expression of noncoding RNAs emanating from a subset of enhancer regions [De Santa et al., 2010; Kim et al., 2010], roughly 10% of condition-specific myogenic enhancers paired with noncoding transcripts [Trapnell et al., 2010] and nearly 60% of these enhancers were loaded with distinct levels of PolII. Computational analysis revealed that approximately half of the myoblast-specific and roughly 80% of the myotube-specific enhancers harbor putative MyoD binding sites [Blum et al., 2012]. Nevertheless, superimposition of our myogenic enhancer catalog against previously mapped MyoD-binding sites [Cao et al., 2010] indicated that only third of the condition-specific enhancers were bound by MyoD. This finding strengthens the notion that MyoD recruitment to myogenic enhancers is dependent and limited by a combinatorial contribution of sequence-specific regulators, histone modifiers, and chromatin formation.
The ability of MyoD to activate muscle enhancers is likely modulated through synergistic interactions with different transcription factors that recognize DNA sequences within the proximity of MyoD binding sites. Indeed, our in-silico analysis of enriched binding motifs that are spatially localized near MyoD uncovered a few transcription factors that are known to be myogenic regulators, among them c-Jun [Bengal et al., 1992], Meis [Knoepfler et al., 1999], Runx1 [Wang et al., 2005] and Jdp2 [Ostrovsky et al., 2002]. Computational analysis of putative binding sites revealed that MyoD and c-Jun co-localize to 54% of myoblasts enhancers. Interestingly, our biochemical studies comparing MyoD-null myoblasts to normal myoblasts show that there is a significant decrease in the recruitment of c-Jun to myogenic enhancers in the absence of MyoD. This suggests that MyoD plays a central role in enhancer assembly via the stabilization of transcription factors that have been previously implicated in myogenic differentiation on chromatin. In addition, depletion of c-Jun in the myoblast stage caused a significant decrease in deposition of H3K27ac and H3K4me1 at myoblast enhancers that are normally bound by c-Jun, but did not affect the enhancers that do not recruit c-Jun [Blum et al., 2012]. These epigenetic studies therefore revealed the strong association of MyoD and c-Jun in their ability to activate myogenic enhancers, which is consistent with their binding interaction that was demonstrated in vitro [Bengal et al., 1992].
Previous investigations have shown that once recruited to sequence-specific DNA-motifs at gene promoters, MyoD interacts with the acetyltransferases PCAF and p300 to promote acetylation of histone H3 and histone H4 [Cao et al., 2006; Cao et al., 2010; Puri et al., 1997], and acetylation of MyoD by itself [Dilworth et al., 2004; Sartorelli et al., 1999]. Our analysis discovered that most of the condition-specific enhancers that were occupied by MyoD were also occupied by the acetyltransferase p300. In fact, the fraction of MyoD-bound enhancers that were co-occupied by p300 was found to be significantly larger than the fraction of MyoD enhancers that were deficient of this enzyme, indicating that the presence of MyoD at enhancers may confer a strong impact on the engagement of p300 to active myogenic enhancers. Further biochemical studies in MyoD−/− myoblasts displayed significantly reduced levels of H3K27ac and p300 at a selected set of MyoD-bound enhancers. Moreover, to further investigate the cause and effect relationship, we re-expressed MyoD in null myoblasts and discovered that reintroduction of MyoD in myoblasts caused a partial restoration of myoblast enhancers, as evidenced by increased PolII binding and deposition of H3Kme1 but not H3K27ac. At the same time a more complete restoration was demonstrated across myotube-specific enhancers in which the enrichment of all three enhancer-related marks was up-regulated upon introduction of MyoD. These findings suggest that there is a great importance in the timely manner in which MyoD acts to establish myogenic enhancers. Uncoordinated binding of MyoD to myogenic enhancers in a specific stage may result in poor recruitment of enhancer-activating regulators, and therefore may lead to incorrect enhancer assembly. Moreover, it is possible that the plasticity of chromatin at enhancer regions is limited to specific stages during differentiation and could become irreversibly temporally and developmentally constrained [Blum et al., 2012].
Additionally, our study investigated the role of methyltransferase Set7 at muscle enhancers. qChIP assays showed that both, Set7 recruitment and H3K4me1 deposition at MyoD-bound enhancers, sharply decreased at MyoD-bound enhancers in MyoD-null myoblasts, as compared to their wild-type counterparts – indicating that the manner in which Set7 was tethered to myogenic enhancers was MyoD-dependent [Blum et al., 2012]. These results are in good accordance with a previous report that revealed physical interactions between MyoD and Set7 at the enhancer segment of MCK, and marked the vital role of Set7 in the induction of myoblast differentiation through the control of the levels of H3K4 mono methylation [Tao et al., 2011]. Along these lines, our experimental data indicated that in addition to being recruited to MyoD-bound enhancers, Set7 was also recruited to MyoD-unbound enhancers, suggesting that other transcription factors aside from MyoD are actively involved in the recruitment of this methyltransferase to muscle enhancers.
SIGNALING PATHWAYS THAT CONTROL MYOGENESIS AND MAY IMPLICATED IN ENHANCER REGULATION
An accumulated body of research from the recent years has paved the way for deciphering the molecular signaling pathways controlling the differentiation of mesodermal precursor cells to myoblasts and consequently to terminally differentiated myotubes [Buckingham and Rigby, 2014; Charge and Rudnicki, 2004]. The myogenic precursor cells, which are confined to the dermomyotome (the dorsal region of the somites), are being nourished by the surrounding tissues, which transmit multiple signaling pathways aiming to specify the differentiation conditions to support a muscle formation. Ultimately, these intracellular signals are being convergent into the nucleus, where they converted by transcription factors into epigenetic signals that modulate the chromatin landscape. These regulatory pathways, including the activating Sonic hedgehog (SHH), Noggin and the Wnt, and repressive BMP4 pathways, regulate the transcriptional program dictated by Myf5 and MyoD and therefore establish the commitment of myogenic precursor cells to proceed through the muscle lineage. Myoblasts are located in the lateral portion of the demomyotome, become committed by expressing increasing levels of MyoD, and then migrate laterally to form the myotome that subsequently evolves into the skeletal musculature. Subsequently, the timed expression of myogenin and Mrf4 help to accelerate the more advanced myogenesis steps, which include myoblasts fusion, to form multinucleated elongated bundles, and formation of well aligned myoubes [Buckingham and Rigby, 2014; Charge and Rudnicki, 2004]. In addition to the four MRFs that define the myogenic program, an additional supportive network of transcriptional factors, comprised by Pax3, Pax7, Six1, and Six4, is involved in the preliminary steps of the myogenic program [Buckingham and Rigby, 2014; Liu et al., 2013]. The two factors, Pax3 and Pax7 are expressed in the dermomyotome, where Pax3 plays significant roles during embryonic myogenesis [Bober et al., 1994; Tremblay et al., 1998], and Pax7 plays a predominant role during postnatal myogeneis [Oustanina et al., 2004; Seale et al., 2000]. In the subpopulation of self-renewing myogenic stem cells, both Pax7 [Soleimani et al., 2012] and the Ploycomb Repressive Complex 2 (PRC2) [Caretti et al., 2004] play essential roles in keeping myoblasts at their proliferative state − Pax7 through the induction of proliferation genes, and PRC2 via repression of genes involved in muscle differentiation. In addition, during myoblast proliferation, two classes of histone deacetylases oppose the activity of the histone acetyltranserases p300 and PCAF. Direct interactions between HDAC1 and MyoD were shown to promote hypoacetylation of MyoD, consequently reducing its activity [Mal et al., 2001; Puri et al., 2001], while on the other hand, Class-II HDACs, such as HDAC4 and HDAC5, were shown to repress MEF2 activity and consequently reduce the expression of MEF2 target genes [Lu et al., 2000a; Lu et al., 2000b; McKinsey et al., 2000]. Other signals supporting the myoblastic cell population in its proliferative stage are obtained by the miRNA miR-31, which controls the translation of Myf5 by sequestering its transcripts inside cytoplasmatic mRNP storage granules [Crist et al., 2012].
Upon receiving myogenic-committing signals, Pax7 mediates the recruitment of the histone methyltransferase complex, Wdr5-Ash2L-MLL2, to the promoter of Myf5, resulting in increased deposition of H3K4me3 and elevated expression of Myf5 [McKinnell et al., 2008]. One of the dominant pathways that known to promote muscle differentiation is the p38 MAP kinase signaling pathway. Upon acquiring differentiation-commitment signals, p38 becomes activated and phosphorylates E47, leading to dimerization of MyoD and E47, which allows the subsequent recruitment to E-box elements found at the promoters of multiple muscle genes [Lluis et al., 2005]. Moreover, p38α/β phosphorylates the SWI/SNF component, BAF60, and via this mechanism accelerates the recruitment of this chromatin-remodeling complex to the promoters of myogenic genes [Simone et al., 2004]. In addition, blockade of p38α/β by a chemical inhibitor abrogated the interaction between the two ATPase subunits of SWI/SNF, BRM and BRG1, and MyoD [Simone et al., 2004]. An additional facet of how p38 plays a role in stimulating myogenic differentiation is demonstrated by its ability to phosphorylate MEF2A and MEF2C on their transactivation domain. Consequently, the affinity of these transcription factors to interact with MyoD increases enhancing their transcriptional activity [Puri et al., 2000; Zhao et al., 1999].
One of the well-established signaling circuits characterizing differentiated skeletal muscle cells is the loss of enzymatic activity of the PRC2-EZH2-YY1 complex, which leads to H3K27 hypomethylation, the activation of MyoD, and an increase in histone hyperacetylation at chromatin regulatory region of myogenic genes [Asp et al., 2011; Caretti et al., 2004]. Complementary studies have linked this loss in enzymatic activity to the combinatorial function of three miRNAs – miR26a [Wong and Tellam, 2008], miR29 [Wang et al., 2008a] and miR214 [Juan et al., 2009] − that become active during myogenesis and inhibit the expression of EZH2, the catalytic subunit of PRC2.
Another signaling cascade that is known to regulate myogenesis and to function in parallel to the p38 kinase pathway is the PI3K/AKT signaling pathway, which translates extracellular cues emanating by growth factors such as IGF1, into intracellular signals [Wu et al., 2000]. Importantly, in addition to dictating muscle differentiation this signaling pathway has been implicated in regulating muscle regeneration [Barton et al., 2002] and muscle cell survival [Lawlor and Rotwein, 2000].
Conclusively, the vast majority of the signaling pathways mentioned here are ultimately affecting MyoD activity. Therefore, since MyoD was shown to be actively involved in the regulation of activated myogenic enhancers, alterations in these signaling pathways should be well correlated with changes in the activation status of myogenic enhancers. Given the profound nature of the myogenic program, it is expected that certain subtleties would exist. Future studies would have to closely examine the implications of specific signaling pathways on the activation of distinct subsets of myogenic enhancers.
ENHANCER PLACEHOLDERS AS POTENTIAL REGULATORS OF MYOGENIC LINEAGE
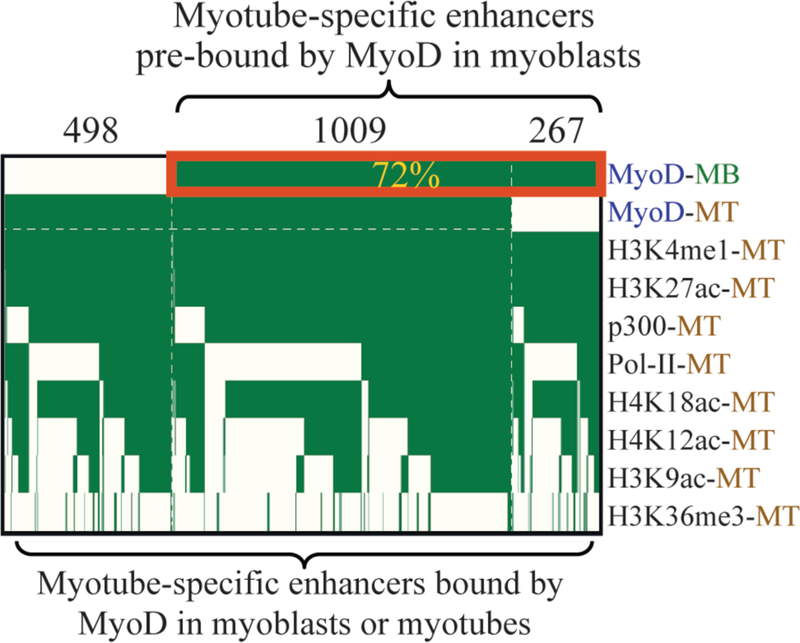
An intriguing aspect that remains largely uncharacterized is the fact that within the developing limb bud and regenerating satellite cell populations myogenic precursor cells (myoblasts) already express MyoD at high levels but are kept in an undifferentiated state prior to receiving the proper differentiating cues that induce their transformation from myoblasts to myotubes. Efforts to pinpoint a distinct molecular modification that could alter MyoD from an inactive state to operative state failed to identify such mechanism [Ludolph and Konieczny, 1995]. Interestingly, our epigenetic studies of myogenic enhancers have indicated that approximately 72% of MyoD-bound enhancers, which turn active only in myotubes, are already bound by MyoD in the myoblastic stage. Particularly, our data indicate that deposition of the marks associated with active enhancers (H3K4me1, H3K27ac, PolII and p300) occurs only after differentiation on myotube-specific enhancers, suggesting that that binding of MyoD alone to enhancers does not guarantee activation [Blum et al., 2012] (Fig. 1). Instead, these observations indicate that additional molecular signals besides MyoD are necessary for prompting the enhancer to its activate stage. These results imply that during proliferation, myotube-specific enhancers are bound by additional transcription factors that negate the action of MyoD. Therefore binding of MyoD to enhancer region may not be sufficient in order to prompt their activation, but rather the eviction of negative regulators is also required. Thus, we hypothesize that specific placeholders that bind to myogenic enhancers prior to the binding of MyoD may act as co-repressors that suppress the activity of myogenic enhancers even in face of MyoD binding [Sekiya and Zaret, 2007; Zaret and Carroll, 2011].
Figure 1: A large fraction of myotube-specific enhancer that is bound by MyoD, becomes bound by MyoD already in the proliferative stage.
Myotube-specific enhancers become active exclusively after muscle differentiation as manifested by the presence of p300, PolII, H3K4me1 and H3K27ac. Clustering of myotube-specific enhancers that are bound by MyoD before or after muscle differentiation indicates that the majority (~72%) of these condition-specific enhancers becomes bound by MyoD already in the proliferative stage and prior to the point in which they turn active, suggesting that throughout the myoblastic stage these myotube-specific enhancers could be co-bound by MyoD and another placeholder that negates the activity of MyoD and preserves these enhancers as inactive/poised. MB, myoblasts; MT, myotubes.
Thus far, studies to identify specific transcription factors that act as placeholders in muscle have not been carried out. Nevertheless, previously, our survey to identify such factors led us to two candidates that have the potential of acting as placeholders in muscle cells − Msc (also known as musculin or MyoR (myogenic repressor)), and Stat1. Msc is a basic helix-loop-helix factor known to be a marker of undifferentiated muscle precursor cells [von Scheven et al., 2006]. This factor functions as a lineage-restricted transcriptional repressor of myogenesis since (i) throughout the skeletal muscle lineage it is specifically expressed between days 10.5 and 16.5 of mouse embryogenesis, and at the onset of myogenesis its expression level drops; and (ii) in culture conditions Msc is expressed in undifferentiated myoblasts and its expression declines during differentiation [Lu et al., 1999]. Luciferase reporter assays in 10T1/2 fibroblasts showed that wild-type Msc blocks MyoD from activating transcription. These experiments further indicated that Msc-mediated transcriptional inhibition that was not relieved by supplementing ectopic E proteins, suggesting that sequestration of E proteins is not the major mechanism by which Msc suppresses MyoD [Lu et al., 1999]. Conversely, a mutated form of Msc that lacks the basic DNA-binding domain but that can heterodimerize with E2A, only modestly reduced transcriptional activation by MyoD, indicating that the major mechanism for repression by Msc involves DNA binding [Lu et al., 1999]. Importantly, mobility shift assays in the C2C12 skeletal muscle cells indicated that Msc binds to an E-box elements located in the Ckm (muscle creatine kinase) enhancer [Lu et al., 1999]. Additional support for the role of Msc as a negative regulator of myogenic enhancers was provided by ChIP experiments that were employed with isolated cells from the branchial arch of 10.5 dpc embryos that give rise to muscles of mastication. These studies showed that at this developmental stage, Msc binds to the enhancer regions of both Myf5 and MyoD located approximately 1kb and 5.2kb upstream of their TSSs, respectively [Moncaut et al., 2012].
Recent studies have pointed to a plausible role for Msc in Rhabdomyosarcoma (RMS), a neoplasm composed of skeletal myoblast-like cells, which is the most common soft tissue sarcoma in children. Ample evidence suggests that an unbalanced regulation of various inhibitory transcription factors that typically suppress MyoD activity is keeping RMS cells trapped in a proliferative state that is similar to that of myobalst cells. Importantly, RMS cells are characterized by the expression of myogenic specification genes, such as MyoD and Myf5, however they fail to progress properly through the myogenic lineage. Interestingly, knockdown of Msc in the presence of retroviral expression of MyoD in RMS RD cells resulted in 10-fold higher levels of Ckm (muscle creatine kinase) expression [Yang et al., 2009]. In addition, supportive evidences point to the suppressive effect that Msc can imposes on miR-206, a muscle-specific microRNA that is known to promote muscle differentiation [Kim et al., 2006]. Recent studies have shown that Msc suppresses the activation of miR-206 by binding to a critical E-box, located ~400bp upstream of its promoter, which is required for its transcriptional induction by MyoD [Macquarrie et al., 2012]. ChIP-seq analysis of Msc and MyoD in undifferentiated RD cells revealed their distinct peaks over two adjacent E-boxes, indicating that MyoD binds an E-box domain located adjacently to another E-box, which bound by Msc. Thus, these results further suggest that Msc may play an important role in preventing the transcriptional activation of the DNA-bound MyoD. The fact that Msc binds in proximity to the promoter of miR-206 could suggests that it plays an active role as a placeholder in both promoter and enhancer regions.
An additional candidate that may play a role as a myogenic placeholder is Stat1. Several studies have shown that the signaling pathway Jak1–Stat1–Stat3 is required for myoblast proliferation and for preventing premature differentiation [Jang et al., 2011; Sun et al., 2007; Trenerry et al., 2011; Xiao et al., 2011]. Specific knockdown of either Stat1 or its transducer Jak1 in both immortalized and primary myoblasts caused a dramatic reduction in cell proliferation and induction of precocious myogenic differentiation [Sun et al., 2007]. Interestingly, among all the Stat encoding genes that are known to be transcribed in C2C12 cells (i.e., Stat1, 2, 3, 5A, and 5B), only the knockdown of Stat1 led to an early induction of myogenin, an effect reminiscent of Jak1 knockdown. Interestingly, our motif binding studies have shown that Stat1 binding site were enriched at myotube-specific enhancers, including those that are bound by MyoD exclusively after differentiation (Blum et al., 2012). The idea that Stat1 may play an active role as a placeholder at enhancers is reinforced by genome-wide mapping studies of Stat1 performed in human HeLa S3 cells that indicated that approximately 25% of Stat1 binding occurs in intergenic regions that could be flanked by enhancers [Robertson et al., 2007]. Notably, Stat1 was not enriched at myoblast-specific enhancers, suggesting that Stat1 may be involved in regulation of the myotube enhancers activity prior to the period where they become active and bind MyoD [Blum et al., 2012].
In the future it would be important to closely examine whether Msc and Stat1 are indeed playing a role as placeholders of myogenic enhancers. It would be especially interesting to identify new candidate factors that may act as placeholders of myotube-specific enhancers since perturbation of such proteins due to mutation or deletion may result in abrogation of myotube enhancer assembly. Consequently, myotube-specific enhancers are likely to remain in their inactive/poised stage and therefore the induction of key myogenic genes would be delayed or completely abolished, thus locking myoblast cells in their proliferative state. To identify new candidates, we propose that to clone the genomic regions of myotube-specific enhancers into a reporter plasmid. Expression of this plasmid exclusively under non-differentiating conditions (in myoblasts) should result in a lack of activity due to the expression of inhibitory placeholders, which block the activation of myotube-specific enhancers. Loss of the placeholder through genetic targeting would relive the inhibitory effect on the enhancer construct and then should lead to promiscuous expression of the reporter plasmid. Such a myotube specific enhancer-derived reporter could be further utilized in the context of high-throughput RNAi screening, and under appropriate conditions and proper controls, lead to the identification of new genes that prevent the early activation of myotube-specific enhancers. Further genome-wide mapping studies would help to determine the binding patterns such placeholder candidates and would assist in investigating the manner in which placeholders may restrain the transcriptional activity of MyoD and possibly of other myogenic regulatory factors. Identification and functional exploration of transcription factors that behave as placeholders during myoblast proliferation will remarkably advance our perception of the mechanistic layers that govern myogenic differentiation.
CONCLUSION
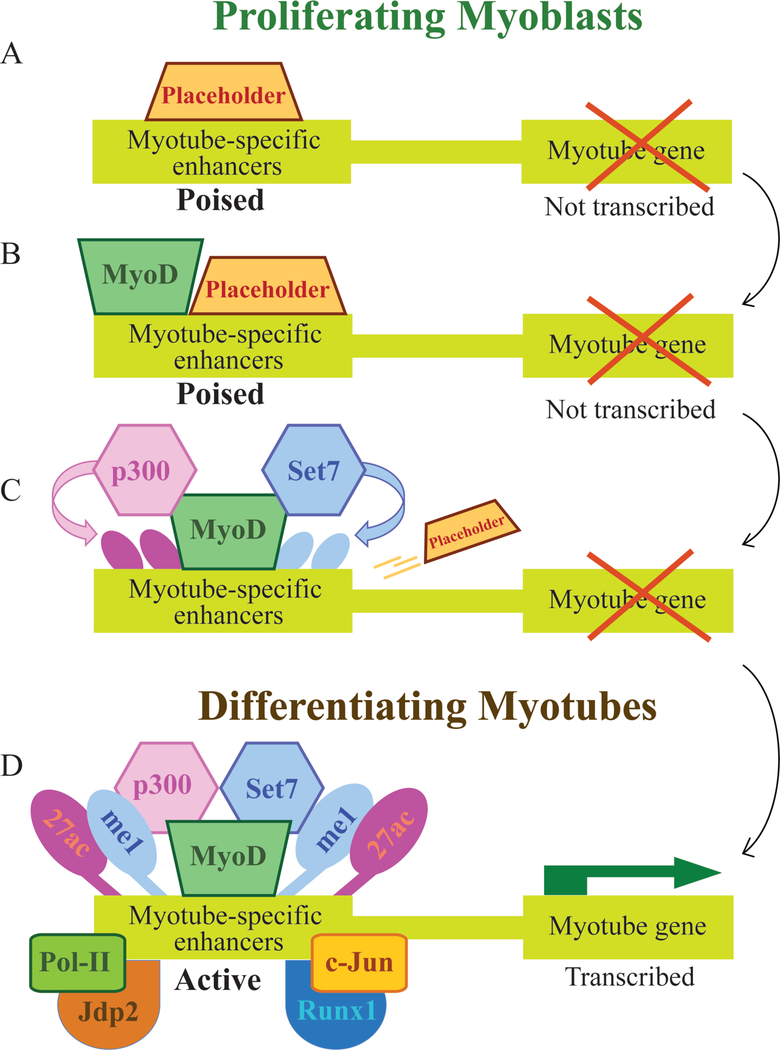
Our current studies indicate that while MyoD occupies multiple promoters that induce the transcription of genes vital for establishing the myogenic fate, an additional new role for this master regulator is recognized due to its enhancer binding capacity. MyoD is implicated as a mediator of numerous chromatin modifying enzymes, among them p300 and Set7, that upon their recruitment to myogenic enhancers facilitate the deposition of histone marks that promote enhancer activation (Fig. 2). Our observations support enhancer-associated interaction between MyoD and multiple transcription factors such as c-Jun. The recruitment of such factors seems to be reduced in the absence of MyoD and therefore dependent on its presence. However, we find that in some cases the presence of MyoD alone is not sufficient for enhancer activation. We suggest that specific placeholders co-localized with MyoD to enhancer regions and may contribute directly to its functional status.
Figure 2: A hypothetical model for the involvement of placeholder in regulation of myotube-specific enhancer assembly and activation.
(A) A putative placeholder (pioneering factor) is bound to myotube-specific enhancer during the myoblastic stage and maintains the enhancer in its poised state. (B) During the proliferative stage MyoD becomes co-bound to the poised enhancer conjointly with the placeholder that negates the activity of MyoD and retains the enhancer as inactive. (C) Upon termination of the myoblastic stage and concomitantly with accumulating differentiation signals, enhancer-bound placeholder becomes inactive or evicts the enhancer, enabling MyoD to become active and to facilitate the recruitment of histone modifiers that reshape the poised enhancer and convert it to active. (D) In its active state, deposited with H3K4me1 and H3K27ac, the activated enhancer becomes occupied by transcription factors that co-bind in conjunction with MyoD and stimulate the enhancer activity.
ACKNOWLEDGMENTS
I thank Dr. J. Cheng for critical reading of the manuscript.
REFERENCES
- Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339:1074–7. [DOI] [PubMed] [Google Scholar]
- Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, Bowman C, Kluger Y, Dynlacht BD. 2011. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci U S A 108:E149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J, Olson L, Schaffner W. 1983. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33:729–40. [DOI] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. 1981. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27:299–308. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–37. [DOI] [PubMed] [Google Scholar]
- Barton ER, Morris L, Musaro A, Rosenthal N, Sweeney HL. 2002. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J Cell Biol 157:137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengal E, Ransone L, Scharfmann R, Dwarki VJ, Tapscott SJ, Weintraub H, Verma IM. 1992. Functional antagonism between c-Jun and MyoD proteins: a direct physical association. Cell 68:507–19. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Ramskold D, Zaouter C, Klum S, Sandberg R, Muhr J. 2011. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev 25:2453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell 14:465–77. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–26. [DOI] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. 2005. An initial blueprint for myogenic differentiation. Genes Dev 19:553–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R, Vethantham V, Bowman C, Rudnicki M, Dynlacht BD. 2012. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev 26:2763–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bober E, Franz T, Arnold HH, Gruss P, Tremblay P. 1994. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development 120:603–12. [DOI] [PubMed] [Google Scholar]
- Bogdanovic O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen SJ, Veenstra GJ, Gomez-Skarmeta JL. 2012. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res 22:2043–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R 2014. The role of chromatin and epigenetics in the polyphenisms of ant castes. Brief Funct Genomics. 13:235–245. [DOI] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44:148–56. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell 132:311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Rigby PW. 2014. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell 28:225–38. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144:327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49:825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. 2006. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J 25:502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, Gentleman RC, Tapscott SJ. 2010. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 18:662–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. 2004. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18:2627–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal JJ, Keith A, Rigby PW. 2008. Global transcriptional regulation of the locus encoding the skeletal muscle determination genes Mrf4 and Myf5. Genes Dev 22:265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charge SB, Rudnicki MA. 2004. Cellular and molecular regulation of muscle regeneration. Physiol Rev 84:209–38. [DOI] [PubMed] [Google Scholar]
- Cheng J, Blum R, Bowman C, Hu D, Shilatifard A, Shen S, Dynlacht BD. 2014. A Role for H3K4 Monomethylation in Gene Repression and Partitioning of Chromatin Readers. Mol Cell 53:979–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. 2012. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res 22:490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, Zhou D, Luo S, Vasicek TJ, Daly MJ, Wolfsberg TG, Collins FS. 2006. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res 16:123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107:21931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist CG, Montarras D, Buckingham M. 2012. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell 11:118–26. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51:987–1000. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol 25:3997–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8:e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth FJ, Seaver KJ, Fishburn AL, Htet SL, Tapscott SJ. 2004. In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proc Natl Acad Sci U S A 101:11593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisuya M, Yamamoto T, Nakajima M, Nishida E. 2008. Ripples from neighbouring transcription. Nat Cell Biol 10:1106–13. [DOI] [PubMed] [Google Scholar]
- Farajollahi S, Maas S. 2010. Molecular diversity through RNA editing: a balancing act. Trends Genet 26:221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcales SV, Albini S, Giordani L, Malecova B, Cignolo L, Chernov A, Coutinho P, Saccone V, Consalvi S, Williams R, Wang K, Wu Z, Baranovskaya S, Miller A, Dilworth FJ, Puri PL. 2012. Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J 31:301–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. 1997. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev 11:436–50. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, Natoli G. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32:317–28. [DOI] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. 2007. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 17:877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhamer DJ, Brunk BP, Faerman A, King A, Shani M, Emerson CP Jr. 1995. Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development 121:637–49. [DOI] [PubMed] [Google Scholar]
- Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. 2008. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A 105:2946–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39:311–8. [DOI] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. 2012. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev 26:2604–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. 2013. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol 33:4745–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YN, Lee IJ, Park MC, Baik EJ. 2011. Role of JAK3 in myogenic differentiation. Cell Signal 24:742–9. [DOI] [PubMed] [Google Scholar]
- Jeong KW, Kim K, Situ AJ, Ulmer TS, An W, Stallcup MR. 2011. Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat Struct Mol Biol 18:1358–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 30:249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Wold BJ, Hauschka SD. 1989. Muscle creatine kinase sequence elements regulating skeletal and cardiac muscle expression in transgenic mice. Mol Cell Biol 9:3393–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. 2009. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell 36:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK. 2013. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 51:310–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. 2012. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe Y, Wang YX, McKinnell IW, Bedford MT, Rudnicki MA. 2012. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell 11:333–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Schedl P. 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol 12:2424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G. 2007. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res 35:125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. 2006. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 174:677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465:182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Bergstrom DA, Uetsuki T, Dac-Korytko I, Sun YH, Wright WE, Tapscott SJ, Kamps MP. 1999. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with pbx-Meis1/Prep1. Nucleic Acids Res 27:3752–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. 2007. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res 17:691–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. 2011. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell 44:410–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. 2013. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor MA, Rotwein P. 2000. Insulin-like growth factor-mediated muscle cell survival: central roles for Akt and cyclin-dependent kinase inhibitor p21. Mol Cell Biol 20:8983–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, Ge K. 2013. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2:e01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Haslinger A, Karin M, Tjian R. 1987. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature 325:368–72. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chakroun I, Yang D, Horner E, Liang J, Aziz A, Chu A, De Repentigny Y, Dilworth FJ, Kothary R, Blais A. 2013. Six1 regulates MyoD expression in adult muscle progenitor cells. PLoS One 8:e67762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluis F, Ballestar E, Suelves M, Esteller M, Munoz-Canoves P. 2005. E47 phosphorylation by p38 MAPK promotes MyoD/E47 association and muscle-specific gene transcription. EMBO J 24:974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Nicol RL, Olson EN. 2000a. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci U S A 97:4070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Zhang CL, Olson EN. 2000b. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell 6:233–44. [DOI] [PubMed] [Google Scholar]
- Lu J, Webb R, Richardson JA, Olson EN. 1999. MyoR: a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proc Natl Acad Sci U S A 96:552–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludolph DC, Konieczny SF. 1995. Transcription factor families: muscling in on the myogenic program. FASEB J 9:1595–604. [DOI] [PubMed] [Google Scholar]
- Macquarrie KL, Yao Z, Young JM, Cao Y, Tapscott SJ. 2012. miR-206 integrates multiple components of differentiation pathways to control the transition from growth to differentiation in rhabdomyosarcoma cells. Skelet Muscle 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. 2001. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J 20:1739–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L, Waskiewicz AJ, Paul B, Cao Y, Tyler A, Moens CB, Tapscott SJ. 2007. Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development 134:3371–82. [DOI] [PubMed] [Google Scholar]
- May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Afzal V, Simpson PC, Rubin EM, Black BL, Bristow J, Pennacchio LA, Visel A. 2012. Large-scale discovery of enhancers from human heart tissue. Nat Genet 44:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, Rudnicki MA. 2008. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol 10:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. 2010. Comparative epigenomic analysis of murine and human adipogenesis. Cell 143:156–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncaut N, Cross JW, Siligan C, Keith A, Taylor K, Rigby PW, Carvajal JJ. 2012. Musculin and TCF21 coordinate the maintenance of myogenic regulatory factor expression levels during mouse craniofacial development. Development 139:958–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncaut N, Rigby PW, Carvajal JJ. 2013. Dial M(RF) for myogenesis. FEBS J 280:3980–90. [DOI] [PubMed] [Google Scholar]
- Moreau P, Hen R, Wasylyk B, Everett R, Gaub MP, Chambon P. 1981. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res 9:6047–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtha M, Tokcaer-Keskin Z, Tang Z, Strino F, Chen X, Wang Y, Xi X, Basilico C, Brown S, Bonneau R, Kluger Y, Dailey L. 2014. FIREWACh: high-throughput functional detection of transcriptional regulatory modules in mammalian cells. Nat Methods 11:559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, Nabeshima Y. 1993. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 364:532–5. [DOI] [PubMed] [Google Scholar]
- Narlikar L, Sakabe NJ, Blanski AA, Arimura FE, Westlund JM, Nobrega MA, Ovcharenko I. 2010. Genome-wide discovery of human heart enhancers. Genome Res 20:381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Andrau JC. 2012. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet 46:1–19. [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG. 2011. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12:283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrovsky O, Bengal E, Aronheim A. 2002. Induction of terminal differentiation by the c-Jun dimerization protein JDP2 in C2 myoblasts and rhabdomyosarcoma cells. J Biol Chem 277:40043–54. [DOI] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. 2013. Latent enhancers activated by stimulation in differentiated cells. Cell 152:157–71. [DOI] [PubMed] [Google Scholar]
- Oustanina S, Hause G, Braun T. 2004. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J 23:3430–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, Helin K. 2010. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res 38:4958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar S, Poulin F, Shoukry M, Afzal V, Rubin EM, Couronne O, Pennacchio LA. 2006. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res 16:855–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Iezzi S, Stiegler P, Chen TT, Schiltz RL, Muscat GE, Giordano A, Kedes L, Wang JY, Sartorelli V. 2001. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol Cell 8:885–97. [DOI] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, Levrero M. 1997. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell 1:35–45. [DOI] [PubMed] [Google Scholar]
- Puri PL, Wu Z, Zhang P, Wood LD, Bhakta KS, Han J, Feramisco JR, Karin M, Wang JY. 2000. Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev 14:574–84. [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470:279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4:651–7. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell 4:725–34. [DOI] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell 102:777–86. [DOI] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS. 2007. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell 28:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, Salbert G, Eeckhoute J. 2011. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res 21:555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15:272–86. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15:1034–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10:697–708. [DOI] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. 2004. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet 36:738–43. [DOI] [PubMed] [Google Scholar]
- Soleimani VD, Punch VG, Kawabe Y, Jones AE, Palidwor GA, Porter CJ, Cross JW, Carvajal JJ, Kockx CE, van IWF, Perkins TJ, Rigby PW, Grosveld F, Rudnicki MA. 2012. Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Dev Cell 22:1208–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13:613–26. [DOI] [PubMed] [Google Scholar]
- Sun L, Ma K, Wang H, Xiao F, Gao Y, Zhang W, Wang K, Gao X, Ip N, Wu Z. 2007. JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts. J Cell Biol 179:129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Neppl RL, Huang ZP, Chen J, Tang RH, Cao R, Zhang Y, Jin SW, Wang DZ. 2011. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J Cell Biol 194:551–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. 2009. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136:3131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay P, Dietrich S, Mericskay M, Schubert FR, Li Z, Paulin D. 1998. A crucial role for Pax3 in the development of the hypaxial musculature and the long-range migration of muscle precursors. Dev Biol 203:49–61. [DOI] [PubMed] [Google Scholar]
- Trenerry MK, Della Gatta PA, Cameron-Smith D. 2011. JAK/STAT signaling and human in vitro myogenesis. BMC Physiol 11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O’Shea JJ. 2012. STATs shape the active enhancer landscape of T cell populations. Cell 151:981–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457:854–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, Hansen DV, Nord AS, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, Kaplan T, Kriegstein AR, Rubin EM, Ovcharenko I, Pennacchio LA, Rubenstein JL. 2013. A high-resolution enhancer atlas of the developing telencephalon. Cell 152:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Scheven G, Bothe I, Ahmed MU, Alvares LE, Dietrich S. 2006. Protein and genomic organisation of vertebrate MyoR and Capsulin genes and their expression during avian development. Gene Expr Patterns 6:383–93. [DOI] [PubMed] [Google Scholar]
- Wakabayashi-Takai E, Noguchi S, Ozawa E. 2001. Identification of myogenesis-dependent transcriptional enhancers in promoter region of mouse gamma-sarcoglycan gene. Eur J Biochem 268:948–57. [DOI] [PubMed] [Google Scholar]
- Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. 2008a. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14:369–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Blagden C, Fan J, Nowak SJ, Taniuchi I, Littman DR, Burden SJ. 2005. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev 19:1715–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. 2008b. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Lockshon D, Lassar A. 1990. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci U S A 87:5623–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. 1989. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A 86:5434–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth BM, Donoghue M, Engert JC, Berglund EB, Rosenthal N. 1991. Paired MyoD-binding sites regulate myosin light chain gene expression. Proc Natl Acad Sci U S A 88:1242–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CF, Tellam RL. 2008. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem 283:9836–43. [DOI] [PubMed] [Google Scholar]
- Wu Z, Woodring PJ, Bhakta KS, Tamura K, Wen F, Feramisco JR, Karin M, Wang JY, Puri PL. 2000. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol Cell Biol 20:3951–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Wang H, Fu X, Li Y, Ma K, Sun L, Gao X, Wu Z. 2011. Oncostatin M inhibits myoblast differentiation and regulates muscle regeneration. Cell Res 21:350–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. 2009. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev 23:2824–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, MacQuarrie KL, Analau E, Tyler AE, Dilworth FJ, Cao Y, Diede SJ, Tapscott SJ. 2009. MyoD and E-protein heterodimers switch rhabdomyosarcoma cells from an arrested myoblast phase to a differentiated state. Genes Dev 23:694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee SP, Rigby PW. 1993. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev 7:1277–89. [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309:626–30. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25:2227–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, New L, Kravchenko VV, Kato Y, Gram H, di Padova F, Olson EN, Ulevitch RJ, Han J. 1999. Regulation of the MEF2 family of transcription factors by p38. Mol Cell Biol 19:21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]