Abstract
Background
Chagas disease is still prevalent in rural areas of South America. In endemic areas of Bolivia, school children are screened for the program of Chagas disease eradication of the Ministry of Health, and positive children are treated. Here, we compared the fecal, oral and skin microbiomes of children with or without Chagas disease, and before and after benznidazol treatment of infected children.
Methods
A total of 543 Bolivian children (5–14 years old) were tested for Chagas disease, and 20 positive children were treated with Benznidazole. Fecal samples and oral and skin swabs were obtained before and after treatment, together with samples from a group of 35 uninfected controls. The 16S rRNA genes were sequenced and analyzed using QIIME to determine Alpha diversity differences and community distances, and linear discriminant analyses to determine marker taxa by infection status or treatment.
Results
Twenty out of 543 children screened were seropositive for Chagas disease (3.7%) and were included in the study, together with 35 control children that were seronegative for the disease. Fecal samples, oral and skin swabs were taken at the beginning of the study and after the anti-protozoa therapy with Benznidazole to the chagasic children. Infected children had higher fecal Firmicutes (Streptococcus, Roseburia, Butyrivibrio, and Blautia), and lower Bacteroides and also showed some skin -but not oral- microbiota differences. Treatment eliminated the fecal microbiota differences from control children, increasing Dialister (class Clostridia) and members of the Enterobacteriaceae, and decreasing Prevotella and Coprococcus, with minor effects on the oral and skin bacterial diversity.
Conclusions
The results of this study show differences in the fecal microbiota associated with Chagas disease in children, and also evidence that treatment normalizes fecal microbiota (makes it more similar to that in controls), but is associated with oral and skin microbiota differences from control children. Since microbiota impacts in children, it is important to determine the effect of drugs on the children microbiota, since dysbiosis could lead to physiological effects which might be avoidable with microbiota restoration interventions.
Introduction
Chagas disease, also known as American Trypanosomaisis, is a neglected parasitic disease caused by the protozoan Trypanosoma cruzi, and transmitted to humans and animals via Triatomine insect vectors [1, 2]. T. cruzi is a stercoraria trypanosome that is deposited with the feces of the vector during a blood meal, and infection of the new host takes place when parasites penetrate through a skin lesion or by contact with mucous tissue (oral, nasal, conjunctivas); the parasite can be also transmitted by ingestion, vertical transmission or transfusion [1, 2].
It is estimated that infection with T. cruzi affects eight million people worldwide, with the majority of cases occurring in the Latin American countries where the parasite is endemic [1–4], and more recently Chagas disease has emerged in non-endemic regions such as the United States, Canada, Western Europe, Japan, and Australia, due to widespread immigration [5]. The clinical course of disease is usually divided into an acute and a chronic phase. After an incubation period of 2–3 weeks, the acute phase presents general mild and non-specific symptoms, that can include fever, inflammation at the inoculation site (chancre), unilateral palpebral edema, enlarged lymph nodes, and splenomegaly; this phase often passes unnoticed and symptoms resolve spontaneously in 4 to 8 weeks [1–3], after which most patients remain chronically infected, and approximately 30% of them develop chronic chagasic cardiomyopathy [6], where the apical aneurysm of the left ventricle constitutes the hallmark of the disease, and 10% gastrointestinal disease predominantly affecting the esophagus, colon, or both [3]. Although less frequent than cardiomyopathy, gastrointestinal Chagas disease has higher incidence in Bolivia, Chile and Paraguay than in the rest of South America, Central America and Mexico [7]. These geographical differences were thought to be the result of the genetic diversity of T. cruzi [8]. In fact, at least three major lineages (A, B and C) have been described in this parasite [9], and currently is accepted that there exist at least six discrete typing units (DTU) [10], and more recently T. cruzi isolates from bats were included as the seventh DTU. However, the analysis of circulating strains did not show significant association between DTUs and clinical manifestations of Chagas disease [1, 2, 11].
The only drugs proven effective against Chagas disease are Benznidazole (Bzn), formerly commercialized as Rochagan and Radanil (Roche), and Nifurtimox (Nfx), marketed as Lampit (Bayer). Both contain a nitro group linked to an imidazole or furan ring, respectively. Bzn has the best safety and efficacy profile, and is therefore the most used as first line treatment. Major limitations are the low potency of these drugs against parasites in the established chronic disease, which is the form most commonly encountered clinically, and unwanted side effects that lead to treatment discontinuation in some cases. The therapeutic benefit of Bzn in established mild to moderate Chagas disease has been under scrutiny in the Benznidazole Evaluation for Interrupting Trypanosomiasis (BENEFIT) trial [12], and up to date the mode of action of these drugs remains unknown.
It is well known the relevance of human microbiome, and how significant changes at its level (dysbiosis) can affect health, mainly as a consequence of the importance of microbial communities in immunological and biochemical functions. However the role of treatments on microbiome composition remains underexplored. In any case, it is clear that the effects of treatment should be disentangled from the effects of specific diseases on the human-associated microbiota [13], as is the case for the parasitic disease schistosomiasis, where either the parasite and the treatment produce significant differences in faecal microbiome [14]. It is noteworthy that Benznidazol has antibacterial action [15], but its effect on the microbiota remains unknown. In this study we compared the fecal, oral, and skin microbiota of children with or without Chagas, and the effect of Bnz therapy in infected children.
Materials and methods
Ethics
Ethical approval was received from the Ethical Committee of the SEDES (#001/2013), Departmental Health Service of Chuquisaca, Bolivia. Adult parents provided written informed consent on the child’s behalf, and children agreed verbally to participate. A total of 543 Bolivian children aged 5–15 years were screened for Chagas disease on a School in the Bolivian Qetchua town of Tarabuco, 63 Km SE of Sucre. A total of 20 children were seropositive for Chagas disease, and asymptomatic, and were recruited for the study and treated with Benznidazole by the local medical team (Program for Chagas Eradication, Bolivian Ministry of Health, following WHO protocols -WHO Chagas facts sheets, http://www.who.int/mediacentre/factsheets/fs340/en/-). A total of 35 Chagas disease negative children were matched as negative controls (S1 Fig).
Diagnostic, treatment and sample collection
Diagnostic of Chagas disease was performed by immunocromatography (Chagas STAT-PAK Assay) and indirect hemagglutination test (HAI Chagas Polychaco). Positive children were treated with Benznidazol (5mg/kg/day) for 60 days. Negative children were not treated. Fecal, oral and skin samples were taken pre (day 0) and post (day 60)-treatment. In non-infected children, samples used as controls were taken at days 0 (pre) and 60 (post). Dry swabs were taken from the children’s oral mucosa, skin (volar arm) and children provided stool samples in appropriate collectors. A total of 20 Chagas disease positive and 35 negative children provided samples at the time before and after positive children has their treatment. Samples were frozen at -80 Celsius until processing.
16S rRNA gene sequencing and analyses
DNA was extracted from the samples using the MoBio PowerSoil kit. The region V3-V4 of the 16S rRNA gene was amplified as described previously [16], and amplicons were purified and sequenced using the Illumina MiSeq platform. The QIIME (version 1.8.0) pipeline was used to process and analyze 16S rRNA sequences [17], estimate bacterial diversity and compare bacterial communities between the children groups. Forward and reverse reads were joined using fastq-join specifying a minimum of 10 overlapping base pairs and 20% maximum allowed differences within overlap regions. A total of 2,719,027 sequences (mean of 10,662 sequences/sample) were retained after quality filtering, using default parameters except for a Phred score cutoff of >19 and a maximum allowance of 3 N characters. Operational taxonomic units (OTUs) were picked through a closed-reference protocol with uclust [18] against the GreenGenes [19] 13_8 reference database at the 97% identity level, aligned using PyNAST [20]. Taxonomic assignments were made using RDP Classifier 2.2 [21]. Beta diversity calculations were done using unweighted UniFrac [22] and visualized through Emperor [23], R (3.2.1) and phyloSeq (1.12.2). Taxa that differentiated infected and non-infected children and microbiota before and after treatment were identified using the biomarker discovery algorithm LEfSe, version 1.0 [24] with LDA scores > 3.0. Only taxa with a minimum mean relative abundance of 0.5% were retained for further analysis. Raw sequencing reads were uploaded to the public database Qiita (Study ID # 11724) and to the European Bioinformatics Institut (EBI) database under accession number ERP113722.
Results
We performed fecal microbiome analyses on 55 children, 20 that resulted Chagas positive, and 35 age matched controls. Positive children (but not control population) were treated with benznidazol.
Fecal, oral and skin microbiome and Chagas disease
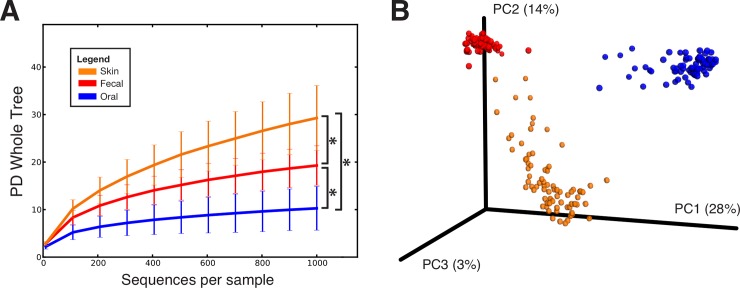
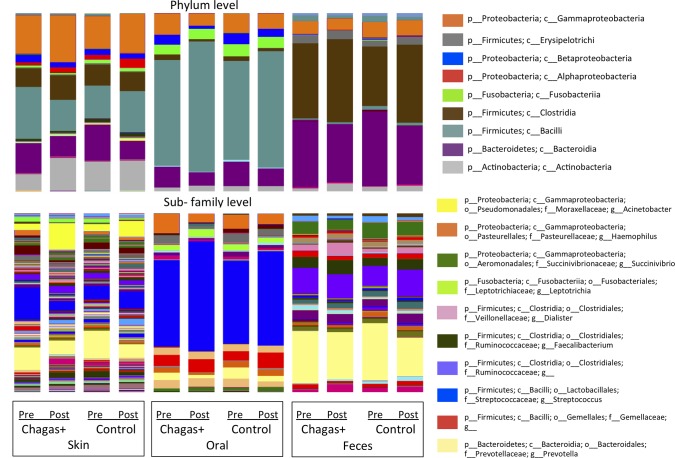
The microbiota differed by body site, as expected (Fig 1). Rarefaction curves of bacterial species diversity by body site, showed that the skin had the highest diversity, followed by fecal and oral. There were no significant differences by gender. The most abundant arm skin taxa were Prevotella (15.2%) and Streptococcus (9.7%; Fig 2). The fecal microbiota was dominated by Prevotella (30.2%), Ruminococcaceae (9.8%) and Succinivibrio (7.1%) (Fig 2). The oral microbiota was dominated by Streptococcus (45%), Prevotella (7.2%), Haemophilus (6.8%), and members of the family Gemellaceae (5%), and there were no differences by infection status (Fig 2).
Fig 1. Alpha and Beta diversity of combined fecal, oral, and skin samples from all individuals.
(A) Rarefaction curves using Faith’s Phylogenetic Diversity (PD Whole Tree) metric at 1000 sequences per sample (* p<0.01, t-test) (B) PCoA plot using the unweighted UniFrac metric shows that samples cluster first by body site.
Fig 2. Relative bacterial taxa abundances in body sites of children with and without Chagas disease both before and after the treatment time point.
Taxa are shown at the (A) phylum level and (B) the lowest classified level, and the labels below the plots indicate infection status (Chagas+ or control) as well as the time point (“Pre” = before treatment, “Post” = post-treatment). A taxa name ending in “g__” indicates that the OTU was not identified at the genus level. Sequences were rarefied at 1000 sequences/sample.
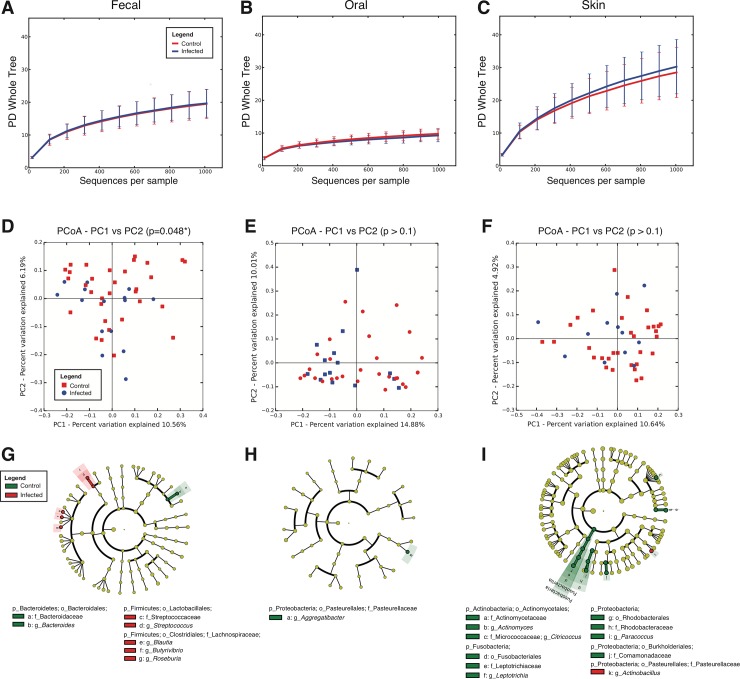
After screening, a total of 20 children seropositive for Chagas disease were studied, whereas 35 samples from seronegative children were used as control population. Although this group was not treated with benznidazol, samples were taken at days 0 and 60, and named "pre" and "post" respectively. We then compared infected and non-infected children using Faith’s Phylogenetic Diversity (PD_whole_tree), and no differences were observed in Alpha diversity (Fig 3A–3C). However, when comparing Beta diversities using the UniFrac distance marginal differences were evidenced in fecal communities (Fig 3D–3F; p = 0.048, Permanova test). Infected children showed increased fecal Streptococcus, Blautia, Butyrivibrio and Roseburia and lower fecal Bacteroides (Fig 3G). Also, infected children presented increased skin Actinobacillus, and decreased Actinobacteria (Actinomyces and Citricoccus), Leptotrichia, Paracoccus, and Comamonadaceae, and no differences in oral bacteria were found between infected children and controls (Fig 3H and 3I).
Fig 3. Effect of Chagas disease infection status on Alpha and Beta diversity of the microbiota in different body sites.
(A-C) Rarefaction curves for fecal, oral, and skin samples infected (respectively 15, 15 and 12 samples) and uninfected children (respectively 32, 29 and 32 samples) at the initial time point, using Faith’s Phylogenetic Diversity (PD Whole Tree) metric at 1000 sequences per sample. (D-F). PCoA plots for fecal, oral, and skin samples using the unweighted UniFrac metric. (G-I) Taxa that best differentiate between Chagas disease seropositive and seronegative (control) children using LEfSe (LDA score cutoff of 3.0 and a minimum mean abundance of 0.1%).
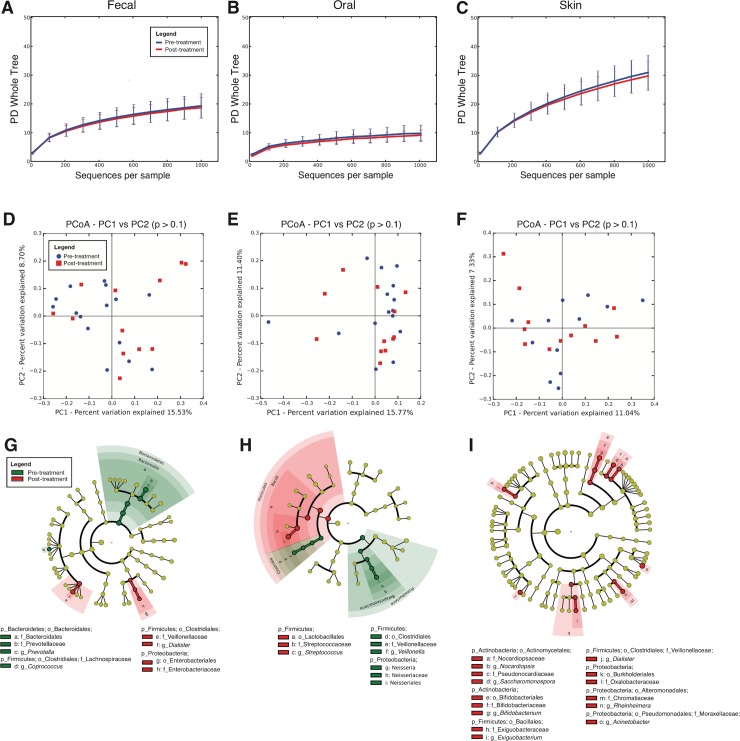
Effect of treatment on the microbiota of infected children
We then compared the microbiota of infected children before and after the treatment with Bzn, and neither Alpha nor Beta diversity changed significantly (Fig 4A–4F and S2 Fig), whereas Beta diversity differed in the fecal, oral and skin microbiota. After the treatment, the fecal microbiota had reduced Prevotella and Coprococcus (Clostridia), and increased Dialister and Enterobacteriaceae (Fig 4G); the oral microbiota decreased Veillonella and Neisseria, and increased Streptococcus (Fig 4H); and the skin had increased Bifidobacterium, Saccharomonospora and Nocardiopsis, and increased Firmicutes (Exiguobacterium and Dialister) and Proteobacteria (Acinetobacter, Rheinheimera, and Oxalobacteraceae) (Fig 4I). Children in the control group did not change significantly their microbiota Alpha or Beta diversities over time, and some populations changed in different ways to those in treated children (S3 and S4 Figs). Their decreased oral Shannon diversity index and also decreased over time (S2 Fig).
Fig 4. Effect of treatment on the microbiota of infected children.
(A-C) Rarefaction curves for fecal, oral, and skin samples of infected children pre-treatment (respectively 15, 12 and 12 samples) and post treatment (respectively 12in each body site), using Faith’s Phylogenetic Diversity (PD Whole Tree), rarefied at 1000 sequences per sample (D-F) PCoA plots for fecal, oral, and skin samples using the unweighted UniFrac metric. (G-I) Taxa that best differentiate between children before and after treatment using LEfSe (LDA score cutoff of 3.0 and a minimum mean abundance of 0.1%).
Discussion
Chagas disease constitutes a major concern for public health in Latin America, and more recently has emerged in non-endemic regions such as the Western Europe, United States, Canada, Japan and Australia, due to widespread immigration. The nitroimidazole Bnz constitutes the first line treatment, and its efficacy has been demonstrated in children [25]. Moreover, recently the U.S. Food and Drug Administration (FDA) granted accelerated approval to Bnz for use in 2 to 12 year-old children with Chagas disease, which constituted the first drug approved in the United States for the treatment of this disease. Although its mode of action remains unclear, its pleiotropic effects are well known and can be explained by the large amount of their derived metabolites that form adducts with functionally relevant biomolecules [26]. In this work we found differences in the microbiota of children infected with T. cruzi, and also differences associated with treatment. The differences in fecal microbiota associated to Chagas disease in children disappear with the treatment, suggesting that the effect of the infection on the gut microbiota might be more important than the effect of the treatment. Despite the age and diet effect on the variations in the fecal microbiota, there were statistically significant variation between children that were Chagas positive or not, and of Chagas infected and treated children in relation to their microbiota before treatment. The immune response against T. cruzi might be responsible, at least in part, of these changes, but this deserves further study.
We found differences in fecal, skin and oral microbiota associated to the Bnz treatment, which could be a direct consequence of the antibacterial activity of Bnz. In fact, its biotransformation produces several covalent thiol adducts resulting in the depletion of glutathione, trypanothion and cysteine in the parasite, with the consecutive increase in sensitivity to oxidants [26] and, in addition, Bnz covalently interacts with nucleic acids, proteins and lipids [27], which may also account for its activity on microbiota composition. Remarkably, Bnz is a pro-drug without activity and its oral ingestion not necessarily causes antibacterial effects. By the contrary, Bnz needs to be activated and in T. cruzi the nitroreductases NTR I and TcOYE are the main responsible for this activation [28–29]. Interestingly, both nitroreductases belong to highly conserved families in prokaryotes, and even a bacterial origin through horizontal transfer has been postulated for TcOYE [29]. We therefore postulate that activation of Bnz could account immediately after ingestion by bacterial nitroreductases, although it deserves to be studied in detail.
As mentioned, Bnz has several undesirable side effects, being the most notorious skin manifestations like hypersensitivity and dermatitis with cutaneous eruptions, which have been classically attributed to a direct effect on skin [28]. However the results found here on skin microbiota can also explain, at least in part, these skin effects. Finally, we cannot exclude the possibility that host response to treatment, because of its high toxicity, can also affect microbiota. Future studies, including metatranscriptomics and metabolomics, should add important mechanistic information on the effect of infection and of treatment on the microbiome and host immune responses.
In summary, human Chagas disease was traditionally analyzed as a binomial host-parasite interaction, and we studied here a third variable that is the affection on host microbiota both through the parasite by itself, and through the effects of Bnz. Considering this, together with previous reports in animal models showing that early impacts on the microbiota lead to physiological effects [30–33], understanding the effects of child infections and treatment drugs on the microbiota emerges as a new target to optimize Chagas disease treatment strategies.
Supporting information
(PDF)
(PDF)
Rarefaction curves for fecal, oral, and skin samples at 1000 sequences per sample are shown, comparing (A-C) infected and control groups, (D-F) pre- and post-treatment groups, and (G-I) days 0 (blue) and 60 (red). Oral diversity significantly (*p<0.05, nonparametric t-test) decreased at the post-treatment time point in both control and infected groups).
(PDF)
Rarefaction curves for fecal, oral, and skin samples based on the number of unique OTUs at 1000 sequences per sample are shown, comparing (A-C) Chagas infected and control groups, (D-F) pre- and post-treatment groups, and (G-I) days 0 (blue) and 60 (red). No significant (p < 0.05) differences between groups were found.
(PDF)
(A-C) Rarefaction curves for fecal, oral, and skin samples using Faith’s Phylogenetic Diversity (PD Whole Tree) metric at 1000 sequences per sample. (D-F) PCoA plots for fecal, oral, and skin samples using the unweighted UniFrac metric. (G-I) Taxa that best differentiate between children at the time points before (n = 32 for fecal and skin, and 29 for oral) and after treatment (n = 22 for fecal and skin, 21 for oral) found using using LEfSe using an LDA score cutoff of 3.0 and a minimum mean abundance of 0.1%.
(PDF)
Acknowledgments
We thank the support of the Hospital Ricardo Bacherer of Municipio Tarabuco, and the School Rosalia Voa de Antezana, in Tarabuco, Bolivia. We also thank Gail Ackermann for assistance during the sequencing data submission to public databases.
Data Availability
Raw sequencing reads were uploaded to the public database Qiita (Study ID # 11724) and to the European Bioinformatics Institut (EBI) database under accession number ERP113722.
Funding Statement
We acknowledge the support of the C&D Fund for Microbial Anthropology, and the Emch Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Clayton J. Chagas disease 101. Nature. 2010;465(7301):S4–5. Epub 2010/06/24. 10.1038/nature09220 [DOI] [PubMed] [Google Scholar]
- 2.Rassi A Jr., Rassi A, Marin-Neto JA. Chagas disease. Lancet. 2010;375(9723):1388–402. 10.1016/S0140-6736(10)60061-X . [DOI] [PubMed] [Google Scholar]
- 3.Bern C. Chagas' Disease. N Engl J Med. 2015;373(19):1882 Epub 2015/11/05. 10.1056/NEJMc1510996 . [DOI] [PubMed] [Google Scholar]
- 4.Bonney KM. Chagas disease in the 21st century: a public health success or an emerging threat? Parasite. 2014;21:11 10.1051/parasite/2014012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coura JR, Vinas PA. Chagas disease: a new worldwide challenge. Nature. 2010;465(7301):S6–7. Epub 2010/06/24. doi: nature09221. 10.1038/nature09221 [DOI] [PubMed] [Google Scholar]
- 6.Lewis MD, Kelly JM. Putting Infection Dynamics at the Heart of Chagas Disease. Trends Parasitol. 2016;32(11):899–911. Epub 2016/10/31. doi: S1471-4922(16)30126-X [pii] 10.1016/j.pt.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.da Silveira AB, Lemos EM, Adad SJ, Correa-Oliveira R, Furness JB, D'Avila Reis D. Megacolon in Chagas disease: a study of inflammatory cells, enteric nerves, and glial cells. Hum Pathol. 2007;38(8):1256–64. 10.1016/j.humpath.2007.01.020 . [DOI] [PubMed] [Google Scholar]
- 8.Virreira M, Serrano G, Maldonado L, Svoboda M. Trypanosoma cruzi: typing of genotype (sub)lineages in megacolon samples from bolivian patients. Acta Trop. 2006;100(3):252–5. 10.1016/j.actatropica.2006.11.005 . [DOI] [PubMed] [Google Scholar]
- 9.Robello C, Gamarro F, Castanys S, Alvarez-Valin F. Evolutionary relationships in Trypanosoma cruzi: molecular phylogenetics supports the existence of a new major lineage of strains. Gene. 2000;246(1–2):331–8. Epub 2000/04/18. . [DOI] [PubMed] [Google Scholar]
- 10.Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernandes O, et al. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104(7):1051–4. Epub 2009/12/23. . [DOI] [PubMed] [Google Scholar]
- 11.del Puerto R, Nishizawa JE, Kikuchi M, Iihoshi N, Roca Y, Avilas C, et al. Lineage analysis of circulating Trypanosoma cruzi parasites and their association with clinical forms of Chagas disease in Bolivia. PLoS Negl Trop Dis. 2010;4(5):e687 Epub 2010/05/27. 10.1371/journal.pntd.0000687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marin-Neto JA, Rassi A Jr., Avezum A Jr., Mattos AC, Rassi A, Morillo CA, et al. The BENEFIT trial: testing the hypothesis that trypanocidal therapy is beneficial for patients with chronic Chagas heart disease. Mem Inst Oswaldo Cruz. 2009;104 Suppl 1:319–24. Epub 2009/09/24. . [DOI] [PubMed] [Google Scholar]
- 13.Devkota S. MICROBIOME. Prescription drugs obscure microbiome analyses. Science. 2016. January 29;351(6272):452–3. 10.1126/science.aaf1353 [DOI] [PubMed] [Google Scholar]
- 14.Kay GL, Millard A, Sergeant MJ, Midzi N, Gwisai R, Mduluza T, et al. (2015) Differences in the Faecal Microbiome in Schistosoma haematobium Infected Children vs. Uninfected Children. PLoS Negl Trop Dis 9(6): e0003861 10.1371/journal.pntd.0003861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hof H. Antibacterial activities of the antiparasitic drugs nifurtimox and benznidazole. Antimicrob Agents Chemother. 1989;33(3):404–5. Epub 1989/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller NT, Shin H, Pizoni A, Werlang IC, Matte U, Goldani MZ, et al. Birth mode-dependent association between pre-pregnancy maternal weight status and the neonatal intestinal microbiome. Sci Rep. 2016;6:23133 10.1038/srep23133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. Epub 2010/04/13. doi: nmeth.f.303. 10.1038/nmeth.f.303 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. Epub 2010/08/17. doi: btq461. 10.1093/bioinformatics/btq461 . [DOI] [PubMed] [Google Scholar]
- 19.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–72. 10.1128/AEM.03006-05 PubMed PMID: WOS:000238961000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26(2):266–7. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology. 2007;73(16):5261–7. 10.1128/AEM.00062-07 PubMed PMID: ISI:000248825900024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–35. 10.1128/AEM.71.12.8228-8235.2005 PubMed PMID: WOS:000234417600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vazquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2(1):16 10.1186/2047-217X-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biology. 2011;12(6). doi: Artn R60 10.1186/Gb-2011-12-6-R60 PubMed PMID: WOS:000296646600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Andrade AL, Zicker F, de Oliveira RM, Almeida Silva S, Luquetti A, Travassos LR, Almeida IC, de Andrade SS, de Andrade JG, Martelli CM. Randomised trial of efficacy of benznidazole in treatment of early Trypanosoma cruzi infection. Lancet. 1996. November 23;348(9039):1407–13. . [DOI] [PubMed] [Google Scholar]
- 26.Trochine A, Creek DJ, Faral-Tello P, Barrett MP, Robello C. Benznidazole biotransformation and multiple targets in Trypanosoma cruzi revealed by metabolomics. PLoS Negl Trop Dis. 2014;8(5):e2844 Epub 2014/05/24. 10.1371/journal.pntd.0002844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diaz de Toranzo EG, Castro JA, Franke de Cazzulo BM, Cazzulo JJ. Interaction of benznidazole reactive metabolites with nuclear and kinetoplastic DNA, proteins and lipids from Trypanosoma cruzi. Experientia. 1988;44(10):880–1. Epub 1988/10/15. [DOI] [PubMed] [Google Scholar]
- 28.Wilkinson SR, Bot C, Kelly JM, Hall BS. Trypanocidal activity of nitroaromatic prodrugs: current treatments and future perspectives. Curr Top Med Chem. 2011;11(16):2072–84. Review. . [DOI] [PubMed] [Google Scholar]
- 29.Díaz-Viraqué F, Chiribao ML, Trochine A, González-Herrera F, Castillo C, Liempi A, Kemmerling U, Maya JD, Robello C. Old Yellow Enzyme from Trypanosoma cruzi Exhibits In Vivo Prostaglandin F2α Synthase Activity and Has a Key Role in Parasite Infection and Drug Susceptibility. Frontiers in Immunology 2018, 9 10.3389/fimmu.2018.00456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez KA 2nd, Devlin JC, Lacher CR, Yin Y, Cai Y, Wang J, et al. Increased weight gain by C-section: Functional significance of the primordial microbiome. Sci Adv. 2017;3(10):eaao1874 Epub 2017/10/14. 10.1126/sciadv.aao1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz VE, Battaglia T, Kurtz ZD, Bijnens L, Ou A, Engstrand I, et al. A single early-in-life macrolide course has lasting effects on murine microbial network topology and immunity. Nat Commun. 2017;8(1):518 Epub 2017/09/13. 10.1038/s41467-017-00531-6 PubMed Central PMCID: PMCPMC5593929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blaser MJ. The theory of disappearing microbiota and the epidemics of chronic diseases. Nat Rev Immunol. 2017;17(8):461–3. Epub 2017/07/28. 10.1038/nri.2017.77 . [DOI] [PubMed] [Google Scholar]
- 33.Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun. 2015;6:7486 10.1038/ncomms8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
Rarefaction curves for fecal, oral, and skin samples at 1000 sequences per sample are shown, comparing (A-C) infected and control groups, (D-F) pre- and post-treatment groups, and (G-I) days 0 (blue) and 60 (red). Oral diversity significantly (*p<0.05, nonparametric t-test) decreased at the post-treatment time point in both control and infected groups).
(PDF)
Rarefaction curves for fecal, oral, and skin samples based on the number of unique OTUs at 1000 sequences per sample are shown, comparing (A-C) Chagas infected and control groups, (D-F) pre- and post-treatment groups, and (G-I) days 0 (blue) and 60 (red). No significant (p < 0.05) differences between groups were found.
(PDF)
(A-C) Rarefaction curves for fecal, oral, and skin samples using Faith’s Phylogenetic Diversity (PD Whole Tree) metric at 1000 sequences per sample. (D-F) PCoA plots for fecal, oral, and skin samples using the unweighted UniFrac metric. (G-I) Taxa that best differentiate between children at the time points before (n = 32 for fecal and skin, and 29 for oral) and after treatment (n = 22 for fecal and skin, 21 for oral) found using using LEfSe using an LDA score cutoff of 3.0 and a minimum mean abundance of 0.1%.
(PDF)
Data Availability Statement
Raw sequencing reads were uploaded to the public database Qiita (Study ID # 11724) and to the European Bioinformatics Institut (EBI) database under accession number ERP113722.