Abstract
p53 regulates the expression of hundreds of genes. Recent surprising observations indicate that no single protein-coding gene controls the tumor suppressor effects of p53. This raises the possibility that a subset of these genes, regulated by a p53-induced long noncoding RNA (lncRNA), could control p53’s tumor suppressor function. We propose molecular mechanisms through which lncRNAs could regulate this subset of genes and hypothesize an exciting, direct role of lncRNAs in p53’s genome stability maintenance function. Exploring these mechanisms could reveal lncRNAs as indispensable mediators of p53 and lay the foundation for understanding how other transcription factors could act via lncRNAs.
Transcription factors regulate hundreds of genes, a subset of which could mediate its effects in a given context. This Unsolved Mystery article explores mechanisms by which long noncoding RNAs might regulate such a subset downstream of p53, a well-studied transcription factor and major tumor suppressor.
Introduction
Regulation of gene expression plays a crucial role during development and disease. In a given cell, this regulation generally occurs at the level of transcription and is controlled by transcription factors that activate the expression of hundreds of genes to control multiple pathways and diverse phenotypes. In a complex disease such as cancer, is the biology of a transcription factor controlled via a single target gene, controlled via a subset of target genes, or distributed among all its targets? We propose a model according to which a transcription factor mediates its effects via a subset of its target genes. This subset is potentially selected by specific long noncoding RNAs (lncRNAs), an emerging class of regulatory RNAs greater than 200 nucleotides long [1]. This model and the underlying mechanistic scenarios that we propose are based on our current understanding of lncRNAs and recent observations on p53 [2–5], the most well-studied transcription factor and a major tumor suppressor mutated in more than half of human cancers [6–8]. Exploring this model will enhance our understanding of the molecular mechanisms by which gene expression is controlled and lead to the development of improved cancer therapies. In addition to lncRNAs, p53-mediated tumor suppression could also be regulated by p53 binding at enhancers and p53-regulated enhancer RNAs [9–12]. Here, we will focus on the role of lncRNAs in mediating the tumor suppressor effects of p53.
A lncRNA can regulate a subset of p53 targets. Why?
Recent unexpected findings have led to a new search for mechanisms responsible for p53-mediated tumor suppression. p53 has been studied for almost four decades and is known to directly bind to DNA in a sequence-specific manner to induce the expression of a myriad of genes. The outcomes of p53 activation are diverse and include cell cycle arrest, apoptosis, and senescence [13]. Although the genes that mediate these phenotypes downstream of p53 have been identified, recent puzzling observations strongly suggest that they are not sufficient on their own, for p53-mediated tumor suppression in vivo [4, 14–16]. For example, unlike p53-knockout mice that develop highly penetrant spontaneous tumors within six months of age, the triple knockout for p21, Puma, and Noxa—the critical mediators of p53-induced apoptosis, cell cycle arrest, and senescence—are not prone to spontaneous tumor development [5]. Perhaps what is even more surprising are the findings from two recent innovative functional genomics studies in which the authors found that no single protein-coding gene controls the antiproliferative effects of p53 [2, 3]. These studies utilized RNA interference (RNAi) to knock down protein-coding genes, but noncoding RNAs were not targeted. Because some lncRNAs have been shown to directly regulate the transcription of many genes [17, 18] or even an entire chromosome [19, 20], it raises the exciting possibility that lncRNAs play a major role in mediating the effects of p53 by regulating a subset of p53 targets. We propose that some p53-induced lncRNAs control the expression of a subset of genes directly or indirectly up-regulated by p53 and, consequently, p53-mediated tumor suppression (Fig 1). This regulation could be mediated by lncRNAs directly regulated by p53 and/or lncRNAs that are activated in coordination with p53 but not in a direct manner [12].
Fig 1. A p53-induced lncRNA can regulate a subset of genes in the p53-regulated transcriptome to play a major role in tumor suppression.
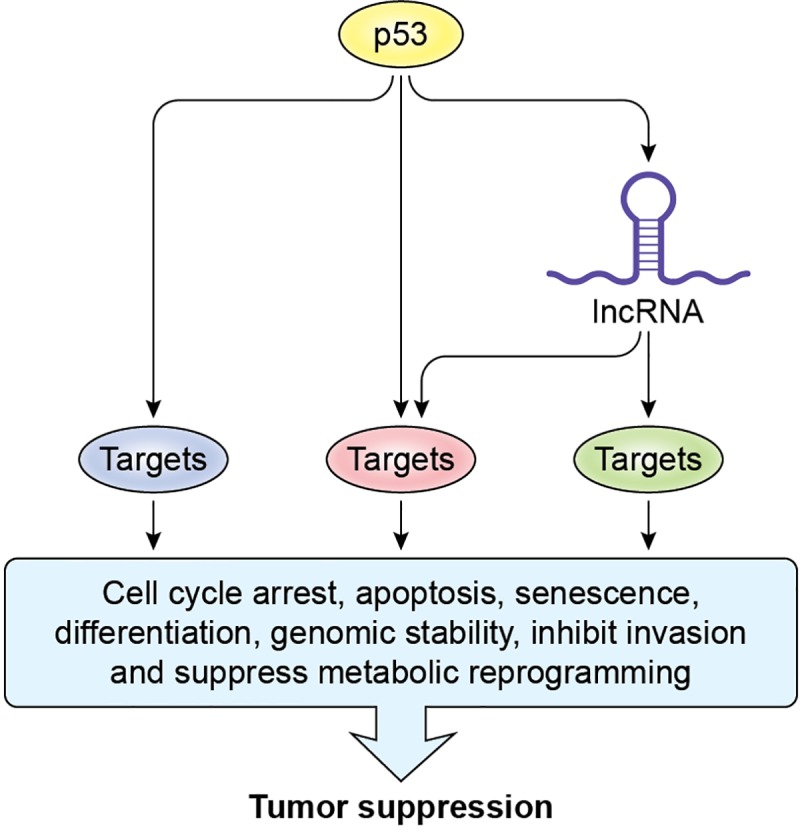
lncRNA, long noncoding RNA.
Why lncRNAs? First, although the majority of lncRNAs transcribed from the human genome have not been studied at the molecular level, some lncRNAs play a crucial role in regulating key cellular processes, including, but not limited to, cellular proliferation, metastasis, differentiation, and genomic instability [21]. Because these processes are also controlled by p53, many lncRNAs have the potential to be functionally integrated into the p53 pathway. For example, we have recently shown that the p53-induced lncRNAs PURPL [22] and PINCR [23] regulate p53 itself or the induction of a subset of p53 target genes, including BTG2, RRM2B, and GPX1, which are implicated in cell cycle arrest and apoptosis during DNA damage. In addition, the p53-induced lncRNAs NEAT1 and PINT have been recently shown to play crucial roles in p53-mediated tumor suppression [16, 24–28]. Second, because lncRNAs are typically expressed at low levels, it may be that they do not directly regulate the expression of hundreds of genes. Although this argument is very difficult to address experimentally, lincRNA-p21, a well-studied p53-induced lncRNA [29–31], is expressed at eight molecules per cell and has been proposed to lack genome-wide regulatory functions [31, 32]. Therefore, lncRNAs are more likely to regulate a subset of genes within a pathway. Third, just as p53 directly or indirectly controls the expression of genes at various stages transcriptionally and post-transcriptionally, lncRNAs have been shown to modulate gene expression at a variety of stages, often depending on their patterns of subcellular localization. Therefore, lncRNA low abundance and high functional correlation with p53 suggest that lncRNAs likely play a critical role in the p53 pathway by regulating a subset of p53 targets, which control tumor suppressor activities.
A lncRNA can regulate a subset of p53 targets by associating with DNA. Mechanisms?
The p53-regulated transcriptome consists of genes that are directly up-regulated, indirectly up-regulated, and indirectly repressed (Fig 2). The subset of genes in the p53-regulated transcriptome that mediate the tumor suppressor effects of p53 could, in principle, belong to either or all of these categories. A lncRNA could act as an activator and/or repressor of gene expression and modulate the expression of a subset of the p53-regulated transcriptome. What are the molecular mechanisms by which a lncRNA regulates these genes? Perhaps the most widely conceived function of lncRNAs is their association with chromatin to activate or repress gene expression. Therefore, the efficacy of lncRNA function lies in its specificity for target gene loci and must involve mechanisms through which it can associate with chromatin at the intended loci.
Fig 2. The subset of genes in the p53-regulated transcriptome that a p53-induced lncRNA could regulate.
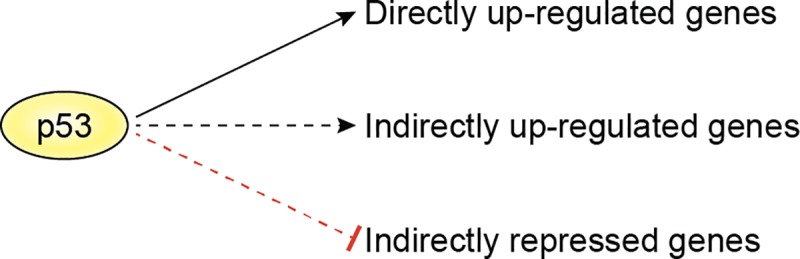
lncRNA, long noncoding RNA.
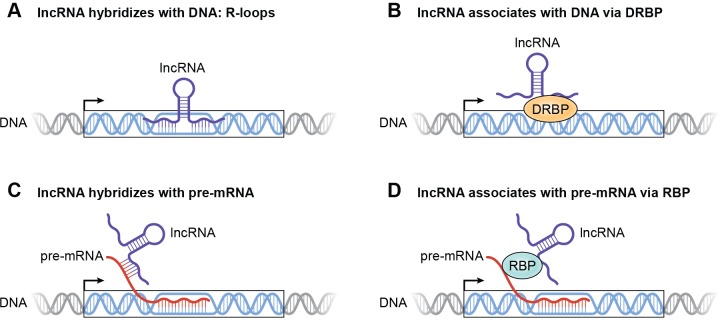
One mechanism through which this could be achieved is via direct association of the lncRNA with chromatin through the formation of RNA-DNA hybrids (R-loops) at regulatory DNA sequences of a subset of p53 targets (Fig 3A). R-loops are thermodynamically favorable and thus represent a plausible mechanism to facilitate direct, sequence-specific binding of the lncRNA at target gene loci [33]. These interactions could occur at regulatory regions of a subset of p53 targets that are hot spots for R-loop formation, including promoters, 1 to 2 kb downstream of the transcription start site or near the polyadenylation sequence at the 3ʹ end of the target gene. Formation of such R-loops by lncRNAs may be aided by sequence elements that promote R-loop formation, such as G-rich sequences in the lncRNA and negatively supercoiled DNA. Alternatively, RNA helicases bound to a lncRNA could facilitate the conversion of intramolecular Guanine-Cytosine (GC)-rich sequences in the lncRNA to single-stranded regions thereby increasing the affinity of the lncRNA for the target DNA. In addition, a lncRNA can be recruited to chromatin by its interaction with DNA- and RNA-binding proteins (DRBPs), a class of proteins that can bind to DNA as well as RNA [34]. In this case, the DNA sequence recognized by the lncRNA will be determined by the specific binding motifs in the DRBP (Fig 3B).
Fig 3. Mechanisms by which a p53-induced lncRNA could modulate gene expression by associating with chromatin at regulatory regions.
(A) lncRNA directly associates with DNA by forming R-loops. (B) lncRNA indirectly associates with DNA via a DRBP. (C) lncRNA indirectly associates with DNA by binding to pre-mRNA. (D) lncRNA associates with DNA indirectly by binding to pre-mRNA via an RBP. DRBP, DNA- and RNA-binding protein; lncRNA, long noncoding RNA; R-loop, RNA-DNA hybrid; RBP, RNA-binding protein.
Through these mechanisms of lncRNA association with chromatin, a lncRNA will be able to interact with regulatory regions of p53 targets in a sequence-specific manner. Where could these regulatory regions be in the genome? These regions could be in enhancers, promoters, insulators, or the gene body of a subset of p53 targets. In this way, a lncRNA may recruit transcriptional regulators to aid in transcription, thus increasing the expression of certain p53 target genes. A lncRNA may also increase target gene expression by facilitating chromatin looping. In this case, the lncRNA can interact with enhancer regions to increase the transcriptional activity of p53 targets by altering chromatin structure. For example, PINCR likely plays a role in the formation of chromatin loops between the promoters and enhancers of the p53 targets BTG2, GPX1, and RRM2B [23]. Additionally, lncRNAs may influence histone modifications at regulatory sites to repress or induce gene expression. This regulation can be achieved through lncRNA interactions with chromatin-modifying enzymes. lncRNAs could also facilitate association of RNA-binding proteins (RBPs) with chromatin to regulate the expression of a subset of p53 targets. For example, the lncRNA XIST has been shown to bind to several RBPs to achieve X-chromosome inactivation [35].
A lncRNA can regulate a subset of p53 targets by associating with pre-mRNAs. How?
A lncRNA can indirectly associate with chromatin via interactions with pre-mRNAs of a subset of p53 targets. RNA–RNA interactions are extremely stable, indicating that this can be a significant mechanism through which lncRNAs associate with chromatin. These interactions may be direct, involving sequence-specific binding of the lncRNA to the pre-mRNA (Fig 3C). Alternatively, this pre-mRNA–facilitated association of the lncRNA with chromatin could occur indirectly via RBPs that recognize specific motifs in the pre-mRNA and the lncRNA (Fig 3D). For example, MALAT1, a nuclear-speckle–localized lncRNA, has been shown to interact indirectly with pre-mRNAs through RBPs [36]. These direct or indirect RNA–RNA interactions will allow for sequence-specific control of a subset of p53 target genes at the post-transcriptional level, resulting in facilitating or inhibiting mRNA splicing and/or processing by a p53-induced lncRNA.
A lncRNA could regulate a subset of p53 targets beyond interactions with chromatin. How?
The mechanisms of associating with chromatin outlined above may represent the most commonly utilized mode by which nuclear-retained lncRNAs dictate a subset of p53 targets. However, lncRNAs that localize to the cytoplasm also have the potential to exert similar functions. Some cytoplasmic lncRNAs are known to promote either mRNA stability or degradation [37]. A p53-induced lncRNA could bind sites targeted by the mRNA degradation machinery to increase the stability of mRNAs in the p53-regulated transcriptome. Alternatively, the lncRNA may recruit degradation machinery and bind to such mRNAs to decrease stability. These lncRNA–mRNA interactions could also occur indirectly via RBPs that provide sequence specificity. For example, mRNAs of specific p53 targets may be destabilized by lncRNAs under normal conditions but stabilized by lncRNAs in response to p53 activation. Additionally, lncRNAs could promote or inhibit the translation of a subset of p53-regulated mRNAs on polysomes. For example, the human lncRNA RoR has been shown to repress p53 mRNA translation [38]. Finally, a lncRNA that localizes to the cytoplasm may alter mRNA levels by acting as competing endogenous RNA (ceRNA) [39]. In this scenario, the lncRNA may prevent interaction of a microRNA (miRNA) with its target mRNAs by outcompeting mRNAs for shared miRNAs.
An important point to consider with each of these mechanistic scenarios is the copy number per cell of the lncRNA that is being considered. The p53-induced lncRNA and the target miRNA or protein should be present in stoichiometric amounts to substantially affect the expression of the target gene. Perhaps the best example of a cytoplasmic lncRNA in which the issue of stoichiometry was carefully considered is NORAD, a very abundant lncRNA that sequesters Pumilio proteins to maintain genome stability [40].
Direct regulation of DNA repair by a p53-induced lncRNA. How?
One of the hallmarks of cancer is DNA double-strand breaks (DSBs), which, if not repaired accurately, lead to mutations and chromosomal rearrangements [41]. DSBs are generally repaired by homologous recombination (HR) or nonhomologous end joining (NHEJ). The preferred substrate for HR is the sister chromatid, but this substrate is only available during the S and G2 phases of the cell cycle. Therefore, during G0/G1, NHEJ can be used to join broken DNA ends without the use of a template DNA, but this is error prone. Alternatively, to repair DNA by HR in G0/G1 phase of the cell cycle, a cell can potentially use endogenous RNA as a substrate for DNA synthesis. Although direct involvement of RNA in DNA repair was unanticipated and considered a rare mechanism, there is now mounting evidence that endogenous RNAs can serve as a template for DSB repair [42–47]. We propose that, in addition to the known roles of some lncRNAs—such as DDSR1 and damage-induced lncRNAs (dilncRNAs) in DNA repair pathways [48, 49], and NORAD in the maintenance of genome stability [40, 50]—specific antisense lncRNAs play direct roles in DSB repair by base-pairing to the damaged DNA.
RNA-mediated DNA repair reverses the central dogma in which DNA is synthesized by RNA. These repair mechanisms, therefore, could rely on RNA interacting with a reverse transcriptase (RT). Although this interaction is well established in the context of retroviruses, retrotransposons, and during telomere synthesis, reverse transcription is likely not limited to these scenarios. Nearly half of the noncoding human genome consists of repetitive elements [51], and over 75% of lncRNA sequences contain elements derived from retrotransposons [52]. Therefore, the majority of lncRNAs contain sequences that are known to interact with RT in the context of DSB repair. One could imagine that genes that play more significant roles in cellular pathways may contain antisense lncRNAs to preserve their integrity. Taken together, this evidence leads to the hypothesis that lncRNAs play crucial roles in “guarding the genome” by functioning in DSB repair pathways.
p53 is known to play a crucial role in maintaining genomic stability [53]. A recent report that utilized in vivo RNAi screening on select p53 targets provided strong evidence that in some contexts the genome stability function of p53 is regulated by the DNA repair gene Mlh1, and the regulation of the DNA repair process plays a very important role in p53-mediated tumor suppression [54]. Most lncRNAs were not included in the RNAi library. This raises the following question. In addition to the regulation of the DNA repair function of p53 via Mlh1, can a p53-induced lncRNA also play a direct role in DSB repair? How could this occur, and what could be the advantages over other mechanisms? p53 is the guardian of the genome [13, 55, 56]. It accomplishes its genome stability maintenance function by a variety of well-established mechanisms, including direct transcriptional regulation of specific genes that control cell cycle arrest, apoptosis, senescence, and DNA repair. For example, upon DNA damage, induction of cell cycle arrest by p53 ensures that DNA replication is turned off, thereby allowing more time for the damaged DNA to be repaired. If the DNA damage is too severe to be repaired, p53 kills the cell by inducing apoptosis. In cancer cells that express dysfunctional p53 or have no p53, these processes are disrupted leading to increased genome instability.
For the regulation of these processes via a protein-coding gene, the p53 target gene(s) would have to be transcriptionally induced by p53, and the corresponding mRNA would then be exported to the cytoplasm and translated. The effector protein may need to be post-translationally modified and, in some cases, imported to the nucleus. We propose that a p53-induced lncRNA could provide a faster and more efficient mechanism of fixing the damaged DNA by directly facilitating lncRNA-mediated DNA repair in the nucleus.
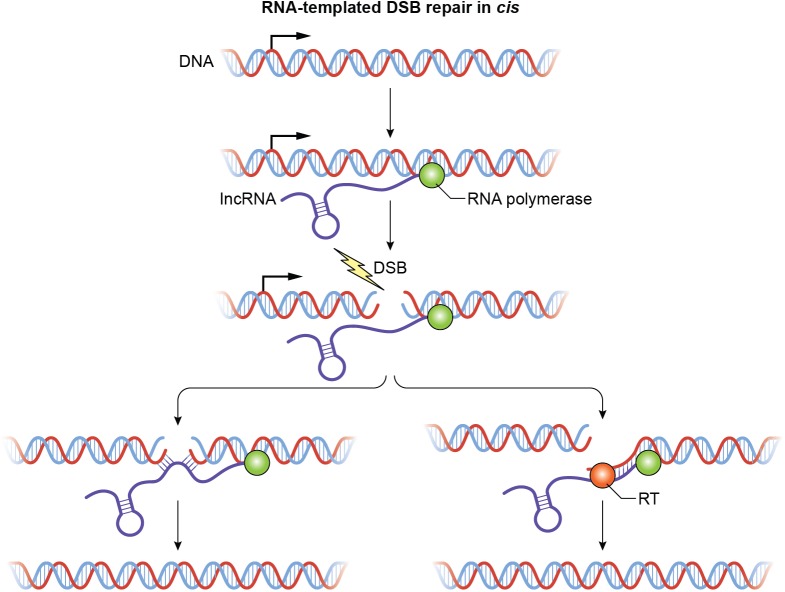
How could this occur? The repair may be templated or nontemplated and will utilize a lncRNA transcript homologous to the DNA flanking the DSB. This transcript could be an antisense lncRNA transcribed from the damaged locus prior to DNA damage or from the undamaged homologous allele in response to DNA damage. An exciting possibility is that the antisense lncRNA could pair with homologous DNA at the site of a DSB. In cis, a lncRNA could base-pair with both sides of the DSB and facilitate end joining (Fig 4). This lncRNA–DNA interaction at the site of the break would occur by forming R-loops. HR machinery has been shown to modulate R-loop formation in the context of genomic instability [57]. Additionally, RNA–DNA hybrids have been shown to promote HR [58]. In trans, an antisense lncRNA or a lncRNA that shows partial complementarity to the regions near the damaged DNA may associate with chromatin at the site of the DSB through a DRBP or directly by forming R-loops. Although both scenarios are possible, lncRNA repair in cis would be preferred because a transcript from the same site as the DSB would have greater repair frequency.
Fig 4. Regulation of DSB repair in cis by a p53-induced antisense lncRNA.
The antisense lncRNA binds DNA at either side of the break and facilitates end joining or acts as a template for resynthesis of damaged DNA. The RNA polymerase that transcribes the antisense lncRNA is shown as a green circle; the RT for synthesis of cDNA using the lncRNA as template is shown as an orange circle. DSB, double-strand break; lncRNA, long noncoding RNA; RT, reverse transcriptase.
In addition to facilitating end joining, an antisense lncRNA could also act as a template for DNA repair, allowing for extension of one of the free DNA ends using a currently unidentified RT (Fig 4). A major advantage of lncRNA-mediated DSB repair could be that only a single molecule of the antisense lncRNA will be required. Our proposed models of lncRNA-mediated DNA repair could be exciting, prevalent, and largely unexplored mechanisms that may unmask the functions of hundreds of currently functionally uncharacterized lncRNAs.
Conclusions and beyond
We propose that a subset of p53 targets regulated by a lncRNA are required for effective tumor suppression. This hypothesis is supported by the low abundance of lncRNAs and their functional correlation with p53. As described above, lncRNAs may control this subset of p53 targets through direct association with chromatin or indirect association with chromatin via nascent pre-mRNA; these interactions may be facilitated by RBPs or DRBPs. Moreover, lncRNAs could modulate p53 targets at the level of translation via interactions with miRNAs or mRNAs on polysomes. Through these mechanisms, lncRNAs may be necessary for effective p53 tumor suppression through regulation of the expression of a subset of p53 targets. Our proposed model on a potential role of antisense p53-regulated lncRNAs in DNA DSB repair, in addition to the known role of Mlh1 in DNA repair downstream of p53 [54], is exciting to explore given the fact that p53 is the guardian of the genome. It is likely that these roles of lncRNAs may be broadly applicable to other transcription factors, accounting for the wide array of lncRNA cellular functions and a significant number of lncRNA genes in the human genome. Therefore, lncRNAs may represent integral regulators of transcription factor pathways and may be necessary for carrying out the observed functions of such transcription factors to play critical roles in development and disease.
Acknowledgments
We thank K.V. Prasanth and Myriam Gorospe for their comments on this manuscript. We also wish to thank the NIH Medical Arts section for help generating figures. We apologize to those whose work we were unable to cite due to space limitations.
Abbreviations
- ceRNA
competing endogenous RNA
- dilncRNA
damaged-induced lncRNA
- DRBP
DNA- and RNA-binding protein
- DSB
double-strand break
- GC
Guanine-Cytosine
- HR
homologous recombination
- lncRNA
long noncoding RNA
- miRNA
microRNA
- NHEJ
nonhomologous end joining
- RBP
RNA-binding protein
- R-loop
RNA-DNA hybrid
- RNAi
RNA interference
- RT
reverse transcriptase
Funding Statement
E.D., S.B.L., and A.L were supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (Z01 Number: ZIA BC 011646). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22(9):1775–89. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, et al. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2(4):202–6. 10.1038/nchembio774 . [DOI] [PubMed] [Google Scholar]
- 3.Andrysik Z, Galbraith MD, Guarnieri AL, Zaccara S, Sullivan KD, Pandey A, et al. Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity. Genome research. 2017;27(10):1645–57. 10.1101/gr.220533.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149(6):1269–83. 10.1016/j.cell.2012.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valente LJ, Gray DH, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell reports. 2013;3(5):1339–45. Epub 2013/05/15. 10.1016/j.celrep.2013.04.012 . [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–10. Epub 2000/12/01. 10.1038/35042675 . [DOI] [PubMed] [Google Scholar]
- 7.Vousden KH, Lane DP. p53 in health and disease. Nature reviews Molecular cell biology. 2007;8(4):275–83. Epub 2007/03/24. 10.1038/nrm2147 . [DOI] [PubMed] [Google Scholar]
- 8.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. 10.1038/nature12912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014;3:e02200 10.7554/eLife.02200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sammons MA, Zhu J, Drake AM, Berger SL. TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome research. 2015;25(2):179–88. 10.1101/gr.181883.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Younger ST, Rinn JL. p53 regulates enhancer accessibility and activity in response to DNA damage. Nucleic acids research. 2017;45(17):9889–900. 10.1093/nar/gkx577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Younger ST, Kenzelmann-Broz D, Jung H, Attardi LD, Rinn JL. Integrative genomic analysis reveals widespread enhancer regulation by p53 in response to DNA damage. Nucleic acids research. 2015;43(9):4447–62. 10.1093/nar/gkv284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–6. Epub 1992/07/02. 10.1038/358015a0 . [DOI] [PubMed] [Google Scholar]
- 14.Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145(4):571–83. 10.1016/j.cell.2011.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang D, Brady CA, Johnson TM, Lee EY, Park EJ, Scott MP, et al. Full p53 transcriptional activation potential is dispensable for tumor suppression in diverse lineages. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(41):17123–8. Epub 2011/10/05. 10.1073/pnas.1111245108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mello SS, Sinow C, Raj N, Mazur PK, Bieging-Rolett K, Broz DK, et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes & development. 2017;31(11):1095–108. 10.1101/gad.284661.116 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Molecular cell. 2014;55(5):791–802. 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular cell. 2010;39(6):925–38. 10.1016/j.molcel.2010.08.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Rocha ST, Heard E. Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nature structural & molecular biology. 2017;24(3):197–204. 10.1038/nsmb.3370 . [DOI] [PubMed] [Google Scholar]
- 20.Cerase A, Pintacuda G, Tattermusch A, Avner P. Xist localization and function: new insights from multiple levels. Genome biology. 2015;16:166 10.1186/s13059-015-0733-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell. 2016;29(4):452–63. 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li XL, Subramanian M, Jones MF, Chaudhary R, Singh DK, Zong X, et al. Long Noncoding RNA PURPL Suppresses Basal p53 Levels and Promotes Tumorigenicity in Colorectal Cancer. Cell reports. 2017;20(10):2408–23. 10.1016/j.celrep.2017.08.041 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhary R, Gryder B, Woods WS, Subramanian M, Jones MF, Li XL, et al. Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3. eLife. 2017;6 10.7554/eLife.23244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marin-Bejar O, Mas AM, Gonzalez J, Martinez D, Athie A, Morales X, et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome biology. 2017;18(1):202 10.1186/s13059-017-1331-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mello SS, Attardi LD. Neat-en-ing up our understanding of p53 pathways in tumor suppression. Cell cycle. 2018;17(13):1527–35. 10.1080/15384101.2018.1464835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome biology. 2013;14(9):R104 10.1186/gb-2013-14-9-r104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinn JL, Huarte M. To repress or not to repress: this is the guardian's question. Trends in cell biology. 2011;21(6):344–53. 10.1016/j.tcb.2011.04.002 . [DOI] [PubMed] [Google Scholar]
- 28.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–61. 10.1093/hmg/ddq353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–19. Epub 2010/08/03. 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groff AF, Sanchez-Gomez DB, Soruco MM, Gerhardinger C, Barutcu AR, Li E, et al. In Vivo Characterization of Linc-p21 Reveals Functional cis-Regulatory DNA Elements. Cell reports. 2016;16(8):2178–86. 10.1016/j.celrep.2016.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Molecular cell. 2014;54(5):777–90. 10.1016/j.molcel.2014.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172(3):393–407. 10.1016/j.cell.2018.01.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chedin F. Nascent Connections: R-Loops and Chromatin Patterning. Trends in genetics: TIG. 2016;32(12):828–38. 10.1016/j.tig.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hudson WH, Ortlund EA. The structure, function and evolution of proteins that bind DNA and RNA. Nature reviews Molecular cell biology. 2014;15(11):749–60. 10.1038/nrm3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161(2):404–16. Epub 2015/04/07. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159(1):188–99. Epub 2014/09/27. 10.1016/j.cell.2014.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Lucas BA, Maquat LE. New gene expression pipelines gush lncRNAs. Genome biology. 2013;14(5):117 10.1186/gb-2013-14-5-117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell research. 2013;23(3):340–50. 10.1038/cr.2012.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–8. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, et al. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164(1–2):69–80. 10.1016/j.cell.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nature genetics. 2001;27(3):247–54. 10.1038/85798 . [DOI] [PubMed] [Google Scholar]
- 42.Meers C, Keskin H, Storici F. DNA repair by RNA: Templated, or not templated, that is the question. DNA Repair (Amst). 2016;44:17–21. 10.1016/j.dnarep.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447(7142):338–41. 10.1038/nature05720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storici F, Tichon AE. RNA takes over control of DNA break repair. Nature cell biology. 2017;19(12):1382–4. 10.1038/ncb3645 . [DOI] [PubMed] [Google Scholar]
- 45.Michelini F, Jalihal AP, Francia S, Meers C, Neeb ZT, Rossiello F, et al. From "Cellular" RNA to "Smart" RNA: Multiple Roles of RNA in Genome Stability and Beyond. Chem Rev. 2018;118(8):4365–403. 10.1021/acs.chemrev.7b00487 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, et al. Transcript-RNA-templated DNA recombination and repair. Nature. 2014;515(7527):436–9. 10.1038/nature13682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keskin H, Meers C, Storici F. Transcript RNA supports precise repair of its own DNA gene. RNA biology. 2016;13(2):157–65. 10.1080/15476286.2015.1116676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma V, Khurana S, Kubben N, Abdelmohsen K, Oberdoerffer P, Gorospe M, et al. A BRCA1-interacting lncRNA regulates homologous recombination. EMBO reports. 2015;16(11):1520–34. 10.15252/embr.201540437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michelini F, Pitchiaya S, Vitelli V, Sharma S, Gioia U, Pessina F, et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nature cell biology. 2017;19(12):1400–11. 10.1038/ncb3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 2018;561(7721):132–6. 10.1038/s41586-018-0453-z . [DOI] [PubMed] [Google Scholar]
- 51.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nature reviews Genetics. 2011;13(1):36–46. 10.1038/nrg3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ganesh S, Svoboda P. Retrotransposon-associated long non-coding RNAs in mice and men. Pflugers Arch. 2016;468(6):1049–60. 10.1007/s00424-016-1818-5 . [DOI] [PubMed] [Google Scholar]
- 53.Williams AB, Schumacher B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb Perspect Med. 2016;6(5). 10.1101/cshperspect.a026070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janic A, Valente LJ, Wakefield MJ, Di Stefano L, Milla L, Wilcox S, et al. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat Med. 2018;24(7):947–53. 10.1038/s41591-018-0043-5 . [DOI] [PubMed] [Google Scholar]
- 55.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nature reviews Cancer. 2009;9(10):749–58. 10.1038/nrc2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carvajal LA, Manfredi JJ. Another fork in the road—life or death decisions by the tumour suppressor p53. EMBO reports. 2013;14(5):414–21. 10.1038/embor.2013.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. eLife. 2013;2:e00505 10.7554/eLife.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, et al. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nature structural & molecular biology. 2013;20(10):1199–205. 10.1038/nsmb.2662 . [DOI] [PubMed] [Google Scholar]