Abstract
Immune checkpoint inhibitors and adoptive transfer of gene-engineered T cells have emerged as novel therapeutic modalities for hard-to-treat solid tumors; however, many patients are refractory to these immunotherapies, and the mechanisms underlying tumor immune resistance have not been fully elucidated. By comparing the tumor microenvironment of checkpoint inhibition–sensitive and –resistant murine solid tumors, we observed that the resistant tumors had low immunogenicity. We identified antigen presentation by CD11b+F4/80+ tumor–associated macrophages (TAMs) as a key factor correlated with immune resistance. In the resistant tumors, TAMs remained inactive and did not exert antigen-presenting activity. Targeted delivery of a long peptide antigen to TAMs by using a nano-sized hydrogel (nanogel) in the presence of a TLR agonist activated TAMs, induced their antigen-presenting activity, and thereby transformed the resistant tumors into tumors sensitive to adaptive immune responses such as adoptive transfer of tumor-specific T cell receptor–engineered T cells. These results indicate that the status and function of TAMs have a significant impact on tumor immune sensitivity and that manipulation of TAM functions would be an effective approach for improving the efficacy of immunotherapies.
Keywords: Immunology
Keywords: Cancer immunotherapy
Introduction
Accumulating preclinical and clinical evidence has shown that inhibition of immune checkpoint proteins, such as cytotoxic lymphocyte–associated protein 4 (CTLA-4), programmed cell death receptor 1 (PD-1), and programmed cell death receptor ligand 1 (PD-L1), can induce durable regression of solid tumors by countering the suppression of antitumor T cell immunity (1–5). However, as reported in non–small-cell lung cancer (NSCLC), breast cancer, pancreatic cancer, and other solid tumors, the objective response rate in this type of immunotherapy ranges from 15% to 50%, indicating that many cancer patients are still refractory to checkpoint inhibition (6, 7). In addition, certain types of tumors such as prostate tumors, sarcomas, and hepatocellular carcinomas are considered totally refractory. Many efforts to reveal the reasons for this unresponsiveness have been made in the clinical setting. For instance, in patients with melanoma, associations among CD8+ T cell infiltration, PD-L1 expression at the tumor site, and clinical response to treatment with anti–PD-1 Ab were previously reported (8, 9). In patients with NSCLC, a correlation between the efficacy of checkpoint inhibition and the number of mutations that can lead to the generation of neoantigens has been reported (10). Thus, a series of recent findings clearly indicate that the immunological environment at the tumor site influences the sensitivity of tumors to checkpoint inhibition (4, 8–13).
The mechanisms of resistance to checkpoint inhibition have also been intensively investigated in preclinical animal models by analyzing the immunological status at the tumor site in detail. For example, loss of phosphatase and tensin homolog (PTEN) function in cancer cells enhances the production of immunosuppressive cytokines and decreases T cell accumulation at the tumor site (14). PTEN loss also causes autophagy inhibition and thereby makes cancer cells resistant to T cell–mediated cytotoxicity. An in vivo genetic screen recently identified protein tyrosine phosphatase, nonreceptor type 2 (PTPN2) as an additional tumor-intrinsic key factor in immune resistance; deletion of the PTPN2 gene regulating the IFN-γ pathway sensitizes cancer cells to treatment with anti–PD-1 Ab and GVAX vaccine by increasing the response of cancer cells to IFN-γ signaling (15). Activation of the β-catenin pathway in cancer cells results in reduced C-C motif chemokine 4–dependent (CCL4-dependent) accumulation of CD103+ DCs in tumors and causes the attenuation of antigen presentation to CD8+ T cells by these DCs in the tumor-draining lymph node (16). Consequently, checkpoint inhibition is ineffective in this type of tumor because of the absence of tumor-specific CD8+ T cells. Another study in mice treated with checkpoint inhibitors revealed a different mechanism involving DCs that may greatly affect resistance: the gut microbiome. Certain bacteria, particularly Bifidobacterium or Bacteroidales, alter DC activity in the lymph nodes, thereby contributing to the improvement of tumor-specific T cell function and influencing sensitivity to checkpoint inhibition (17, 18).
Tumor-associated macrophages (TAMs) have recently attracted attention as an important mechanism for inducing immune suppression at the tumor site. Monocytes accumulating at the tumor site in a CCL2-dependent manner differentiate into TAMs (19–21). Terminal TAM differentiation is regulated by futalic acid and intracellular Notch signaling and is characterized by the loss of Ly6c expression and gain of MHC class II expression (19, 21). Differentiation into immunosuppressive M2-like MHClo TAMs was shown to be associated with hypoxia (22). IL-10 produced by TAMs negatively regulates the secretion of inflammatory cytokines (e.g., IL-12) from myeloid cells and promotes a Th2-type immune response (23). Arginase-1 is induced in immunosuppressive TAMs by IL-4, IL-10, and hypoxia and impairs T cell function by depleting arginine in the tumor microenvironment (24, 25). Nitric oxide production and PD-L1 expression by TAMs also suppress the T cell response. A series of recent studies reported more direct involvement of TAMs in tumor resistance to checkpoint inhibition. V-domain Ig suppressor of T cell activation (VISTA) expressed on TAMs serves as an additional checkpoint pathway and helps tumor cells to escape from the effect of anti–PD-1 Ab (26). Thus, important roles of TAMs in the regulation of tumor immunity have been established, making TAMs a potential therapeutic target to overcome tumor immune resistance. Some attempts to develop TAM-targeted antitumor drugs have focused on the depletion of TAMs using agents such as anti-CSF1R Ab (27), trabectedin (28), docetaxel (24−26), or clodronate liposome (CL) (29). Novel approaches to transform TAMs from the immunosuppressive M2 phenotype into the immunostimulatory M1 phenotype have also been investigated. For instance, treatment of the tumor with a PI3Kγ inhibitor was shown to switch TAMs from a M2-like phenotype to a M1-like state, leading to growth suppression of checkpoint inhibition–resistant tumors (30).
We have developed a series of nano-sized hydrogels (nanogels) to create nanomaterials for biomedical applications. In particular, cholesteryl pullulan (CHP), a pullulan polysaccharide partially hydrophobized by modification with cholesteryl groups, is well established as a highly biocompatible and efficient vaccine delivery system targeting lymph node macrophages. CHP forms nanogel particles with a diameter of less than 100 nm by self-assembly (31–33), and the CHP nanogel particle can efficiently entrap peptide antigens or protein antigens (34, 35). Although the CHP nanogel lacks known ligands for immune cells, surface charge, and immune-stimulating activity (our unpublished observations), a subcutaneously injected CHP nanogel efficiently and promptly delivered antigen to lymph node macrophages with high cross-presenting activity, thereby inducing a prominent antigen-specific T cell response (36).
In this study, we characterized the mechanism underlying tumor resistance to T cell immunity–dependent immunotherapies. By comparing in detail the immunological status at the local tumor site among checkpoint inhibition–resistant and –sensitive murine tumors, CD11b+F4/80+ TAMs were identified as a key factor closely correlated with such resistance. In the resistant tumors, TAMs were inactive and did not exert antigen-presenting activity. We then found that the intravenously injected CHP nanogel could efficiently deliver a long peptide antigen to TAMs, and also that, when accompanied by a TLR agonist such as CpG oligoDNA, TAM-targeted antigen delivery could efficiently elicit antigen presentation by TAMs. This manipulation of TAM function sensitized the resistant tumors to T cell immunity–dependent immunotherapies, in particular, adoptive transfer of tumor-specific T cell receptor–engineered (TCR-engineered) T cells. These results indicate that TAMs play a significant role in tumor immune resistance and that manipulation of TAM function represents a promising approach for enhancing the efficacy of cancer immunotherapies.
Results
Murine CMS5a fibrosarcoma serves as a preclinical model of checkpoint inhibition–resistant tumors.
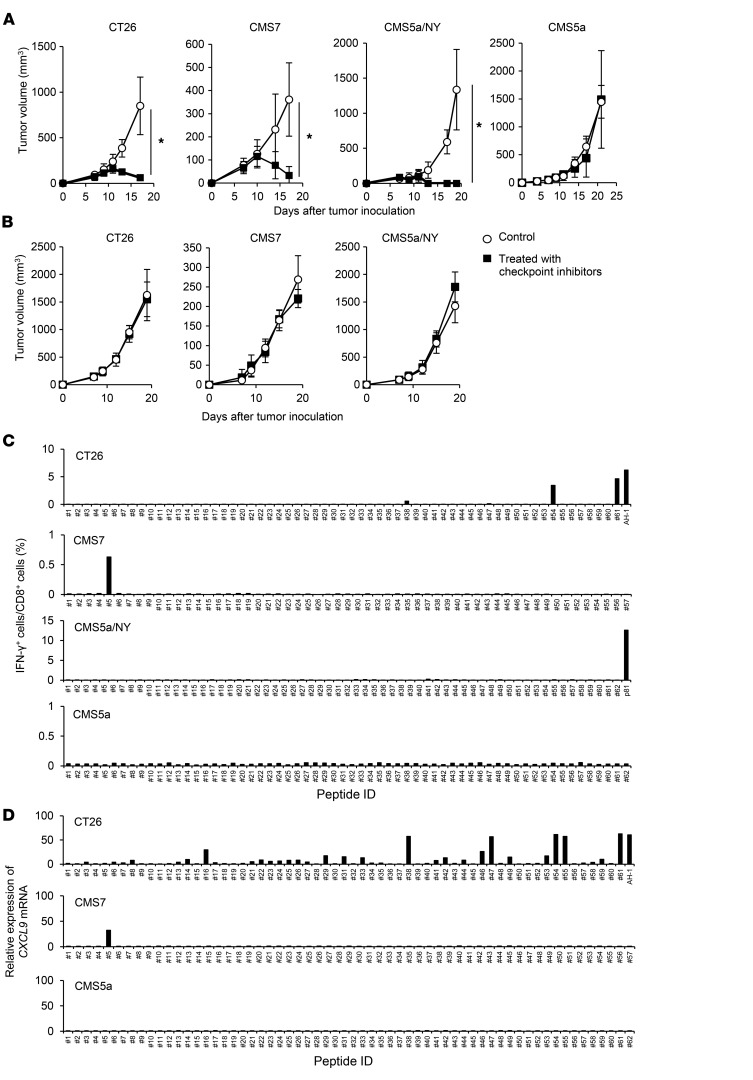
To identify preclinical murine tumor models refractory to immune checkpoint inhibition, we treated several syngeneic murine tumors subcutaneously implanted into BALB/c mice with checkpoint inhibitors, including anti–PD-1 (37, 38), anti–CTLA-4 (38–40), and anti–glucocorticoid-induced TNFR-related (anti-GITR) Abs (41, 42) on days 7, 9, and 11 after tumor inoculation. The tested tumors included colon tumor CT26, fibrosarcoma CMS7, fibrosarcoma CMS5a, and CMS5a engineered to express exogenous human NY-ESO-1 protein (CMS5a/NY) (43–46). NY-ESO-1 antigen is known to be highly immunogenic (47, 48), and its forced expression was expected to make CMS5a more immunogenic. First, we examined the effect of treating these tumors with either an anti–PD-1, anti–CTLA-4, or anti-GITR Ab. Although treatment with anti–CTLA-4 or anti-GITR Abs partially inhibited growth in CT26 and CMS5a/NY tumors, the anti–PD-1 Ab was not effective (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI97642DS1). Therefore, we decided to use a cocktail of these 3 Abs to obtain more potent antitumor activity. As a result, the growth of CT26, CMS7, and CMS5a/NY tumors was strongly retarded by this treatment, indicating that these tumors are sensitive to checkpoint inhibition (Figure 1A). In contrast, the growth of CMS5a tumors was totally unaffected, indicating that CMS5a tumors are highly refractory to checkpoint inhibition (Figure 1A). A previous report showed that the efficacy of checkpoint inhibition depends at least in part on tumor-specific cellular responses (5). Consistent with this finding, the antitumor effects of checkpoint inhibitors on the CT26, CMS7, and CMS5a/NY tumors were abolished in nude mice (Figure 1B). To further examine the contribution of T cell immunity to the effect of checkpoint inhibition, we evaluated specific T cell responses to potential neoantigens in checkpoint inhibitor–treated tumors using a combination of exome and RNA-Seq, in silico prediction of T cell epitopes, and in vitro stimulation of splenic T cells with predicted, neoantigen-derived CD8+ T cell epitope (neoepitope) peptides (Supplemental Tables 1–3), followed by measurement of IFN-γ or CXCL9 production (49). We found that the mutation burden in the tested tumors was generally comparable (Supplemental Table 4). In treated CT26 tumors, we detected specific CD8+ T cell responses to some neoepitopes and an epitope derived from endogenous murine leukemia provirus antigen (50) (Figure 1, C and D). Similarly, we detected a CD8+ T cell response to a neoepitope in treated CMS7 tumors (Figure 1C). In contrast, in the CMS5a and CMS5a/NY tumors, we observed no CD8+ T cell responses to the tested neoepitopes, including the previously reported mutated ERK2 kinase–derived 9m epitope (51) (Figure 1, C and D). However, in the CMS5a/NY tumors, a specific CD8+ T cell response to the exogenous NY-ESO-1 antigen was clearly detected (Figure 1, C and D). Duan et al. previously discovered that several neoantigens can act as tumor rejection antigens in the CMS5a tumors (52). Nevertheless, we did not detect these neoantigen-specific CD8+ T cell responses in the CMS5a tumors treated with checkpoint inhibitors (Supplemental Figure 2). Thus, no specific CD8+ T cell response was detected in the CMS5a tumor model. These results indicate that highly immunoresistant CMS5a fibrosarcomas with very low immunogenicity resemble checkpoint inhibition–resistant human tumors and may be useful as a preclinical model for studying the mechanisms of immune resistance.
Figure 1. Murine fibrosarcoma CMS5a is highly refractory to checkpoint inhibition and lacks a specific CD8+ T cell response.
(A) The murine tumor cell line CT26, CMS7, CMS5a/NY, or CMS5a was subcutaneously inoculated into BALB/c mice. Checkpoint inhibitors including anti–PD-1 (200 μg/mouse), anti–CTLA-4 (100 μg/mouse), and anti-GITR (200 μg/mouse) Abs (n = 8–10 mice per group) or isotype control Abs (n = 8 mice per group) were i.p. injected on days 7, 9, and 11. (B) The experiment was performed in nude mice as described in A. (C and D) Induction of a tumor-specific CD8+ T cell response by checkpoint inhibition was evaluated. BALB/c mice bearing CT26, CMS7, CMS5a/NY, or CMS5a tumors were treated with checkpoint inhibitors. Seven days after the last administration, splenocytes were isolated and restimulated with peptides of predicted neoepitopes or known tumor antigens (AH-1 in CT26 and NY-ESO-1 p81 in CMS5a/NY tumors). The frequency of stimulated CD8+ T cells was quantified by (C) intracellular IFN-γ staining (n = 4 mice per group) or (D) the fold increase in CXCL9 mRNA levels compared with DMSO. The experiments were repeated at least 2 to 4 times with similar results. *P < 0.05, by Mann-Whitney U test.
TAMs are a key factor in tumor immune resistance.
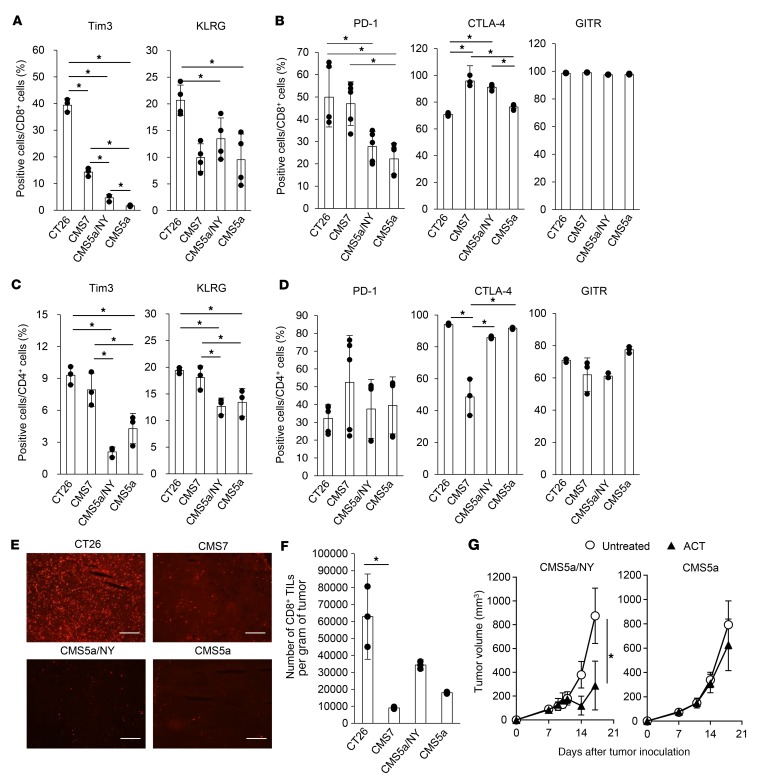
Previous studies demonstrated that the efficacy of checkpoint inhibitors correlates with the expression of PD-1 and T cell immunoglobulin and mucin domain 3 (Tim3) on tumor-infiltrating lymphocytes (TILs) prior to treatment (53). The expression of killer cell lectin-like receptor G1 (KLRG1), CTLA-4, and GITR is a useful marker of CD8+ T cell differentiation or activation (54–57). We therefore compared the expression of PD-1, Tim3, KLRG1, CTLA-4, and GITR on CD8+ TILs and CD4+ TILs from immune-sensitive and -resistant tumors. Whereas TILs frequently expressed these 5 markers at a significant level in 2 of the sensitive tumors (CT26 and CMS7), TILs in another sensitive tumor (CMS5a/NY) as well as the resistant tumor (CMS5a) expressed only CTLA-4 and GITR (Figure 2, A–D). In addition, these markers were expressed at similar levels on TILs from all 4 tumor models after treatment with checkpoint inhibitors (Supplemental Figure 3). In an immunohistochemical analysis, PD-1 expression at the tumor site was detected only in the CT26 tumors (Figure 2E). Likewise, we observed no clear trends in the number of CD8+ TILs among sensitive and resistant tumors (Figure 2F). Thus, in our system, there was no clear relationship between the efficacy of checkpoint inhibition and the activation of TILs, suggesting the presence of other mechanisms of immune resistance.
Figure 2. TIL activation and a systemic T cell response do not correlate with sensitivity to checkpoint inhibition.
(A and B) Expression of Tim3, KLRG, PD-1, CTLA-4, and GITR on CD8+ TILs from the indicated tumors was determined by flow cytometry 7 days after tumor inoculation (n = 3–4 per group). *P < 0.05, by 2-factor factorial ANOVA followed by Tukey-Kramer post hoc analysis. (C and D) The experiment was performed as in A, but on CD4+ TILs. *P < 0.05, by 2-factor factorial ANOVA followed by Tukey-Kramer post hoc analysis. The experiments were repeated 3 times with similar results. (E) PD-1 expression was also analyzed by immunohistochemistry. Scale bars: 200 μm. (F) The number of CD8+ TILs was quantified in each tumor (n = 3 per group). *P < 0.05, by 2-factor factorial ANOVA followed by Tukey-Kramer post hoc analysis. The experiments were repeated 2 times with similar results. (G) CD8+ T cells (2 × 106) isolated from DUC18 mice were infused into CMS5a/NY or CMS5a tumor–bearing mice on day 7 (shown as ACT), and the tumor size was monitored (n = 5 mice per group). *P < 0.05, by Mann-Whitney U test. The experiments were repeated 3 times with similar results.
Treated (Figure 1, C and D) and untreated (Supplemental Figure 4) CMS5a tumors lacked a spontaneous T cell response specific to neoepitopes, including the mutated ERK2-derived 9m epitope. If this is a main cause of immune resistance, adoptive cell transfer (ACT) of 9m-specific T cells might suppress the growth of CMS5a tumors. To test this possibility, T cells from DUC18 mice that were genetically engineered to express a 9m-specific TCR were infused into CMS5a tumor–bearing mice; however, the growth of CMS5a tumors was not affected (Figure 2G). In contrast, CMS5a/NY tumor–bearing mice given the same treatment showed significant inhibition of tumor growth. These results indicate that CMS5a tumors possess other mechanism(s) of resistance.
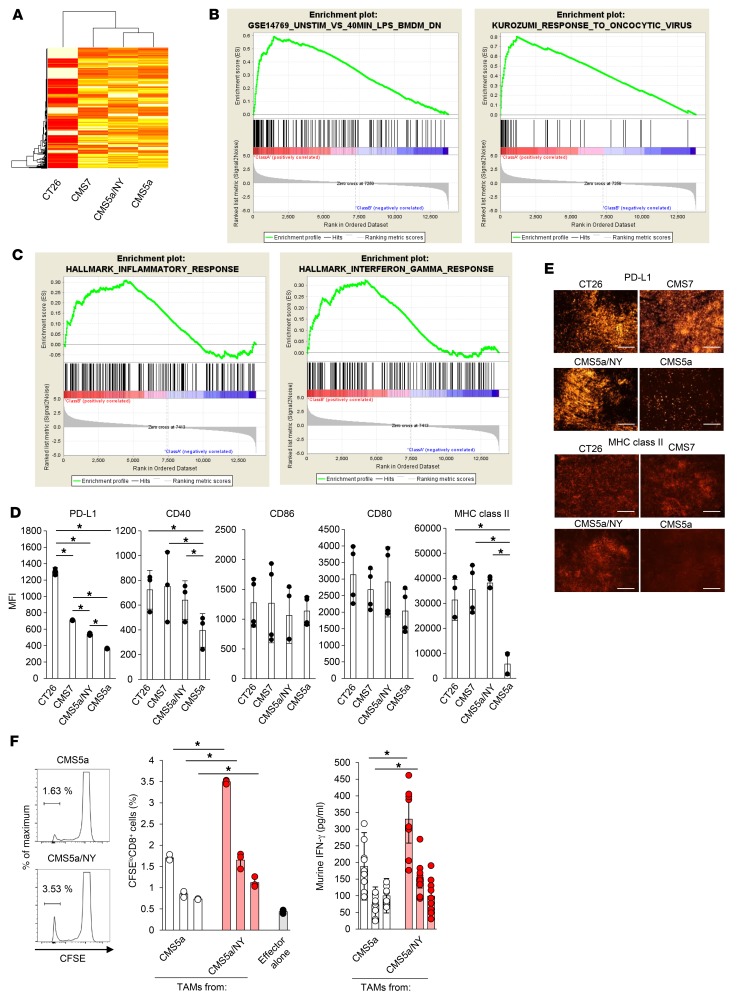
To obtain more clues on the mechanism(s) of resistance, we analyzed whole gene expression at the tumor site. mRNAs isolated from in vivo CMS5a, CMS5a/NY, CMS7, and CT26 tumors were subjected to microarray analysis. Gene set enrichment analysis (GSEA) using database-curated gene sets revealed that in the CMS5a tumors, the expression of genes induced in LPS-treated bone marrow–derived macrophages (Gene Expression Omnibus [GEO] database: GSE14769) (58) or in oncolytic virus–treated tumors (59) was different from that in the sensitive tumors (CT26, CMS7, and CMS5a/NY) (Figure 3, A and B). Intratumoral injection of oncolytic virus is known to induce inflammatory responses, including macrophage activation. We therefore subjected mRNA of TAMs isolated from in vivo resistant and sensitive tumors to microarray analysis. GSEA identified significant differences (FDR <0.05) in the expression of genes related to inflammatory responses (60) and IFN-γ responses (60) between the sensitive tumors and the resistant tumors, indicating that TAMs in the sensitive tumors have a highly inflammatory phenotype (Figure 3C). On the basis of these results, we hypothesized that there may be a difference in TAM status and/or function between resistant and sensitive tumors. Indeed, TAMs in the resistant CMS5a tumors expressed PD-L1, CD40, and MHC class II at considerably lower levels than those in the sensitive tumors (CT26, CMS7, and CMS5a/NY) (Figure 3, D and E), indicating that TAMs in the resistant tumors remain inactive. We also compared the antigen-presenting activity of TAMs between immune-resistant CMS5a and sensitive CMS5a/NY tumors. TAMs were isolated from in vivo tumors and cocultured with CD8+ T cells from 9m TCR–transgenic DUC18 mice as responder cells (61). We found that TAMs from the resistant CMS5a tumors did not stimulate DUC18 CD8+ T cells, whereas those from the sensitive CMS5a/NY tumors clearly induced proliferation and IFN-γ production in antigen-specific CD8+ T cells (Figure 3F). Consistent with this observation, TAMs from the sensitive CT26 tumors also stimulated antigen-specific T cells (Supplemental Figure 5). These data indicate that activation of and antigen presentation by TAMs are closely correlated with tumor sensitivity to checkpoint inhibition.
Figure 3. TAMs remain inactive in the resistant CMS5a tumor.
(A) Tumor tissue from CT26-, CMS7-, CMS5a/NY-, or CMS5a tumor–bearing mice was collected 7 days after inoculation and subjected to microarray analysis. (B) Subsequent GSEA identified the downregulation of 2 indicated gene sets in resistant CMS5a tumors. (C) TAMs from the CT26-, CMS7-, CMS5a/NY-, or CMS5a tumor–bearing mice were collected, and total RNA from these TAMs was subjected to microarray analysis. Subsequent GSEA identified the downregulation of 2 indicated gene sets in TAMs from CMS5a tumors. (D) TAMs were isolated from each tumor 7 days after inoculation and were tested by flow cytometry for the expression of PD-L1, CD40, CD86, CD80, and MHC class II (n = 6 tumors per group). MFI, mean fluorescence intensity. *P < 0.05, by 2-factor factorial ANOVA followed by Tukey-Kramer post hoc analysis. The experiments were repeated 3 times with similar results. (E) PD-L1 and MHC class II expression was also analyzed by immunohistochemistry. Scale bars: 200 μm. (F) TAMs isolated from CMS5a tumor–bearing mice on day 7 were cocultured for 72 hours as antigen-presenting cells with 9m-specific DUC18 CD8+ T cells as responder cells. Antigen-dependent proliferation of DUC18 CD8+ T cells and production of IFN-γ were measured by CFSE dilution assay (n = 3–4 per group) or ELISA (n = 8–9 per group), respectively. Histograms show representative data, and the numbers shown in the histograms indicate the percentage of proliferating cells. *P < 0.05, by 2-tailed Student’s t test. Data represent the mean ± SD. The experiments were repeated 2 times with similar results.
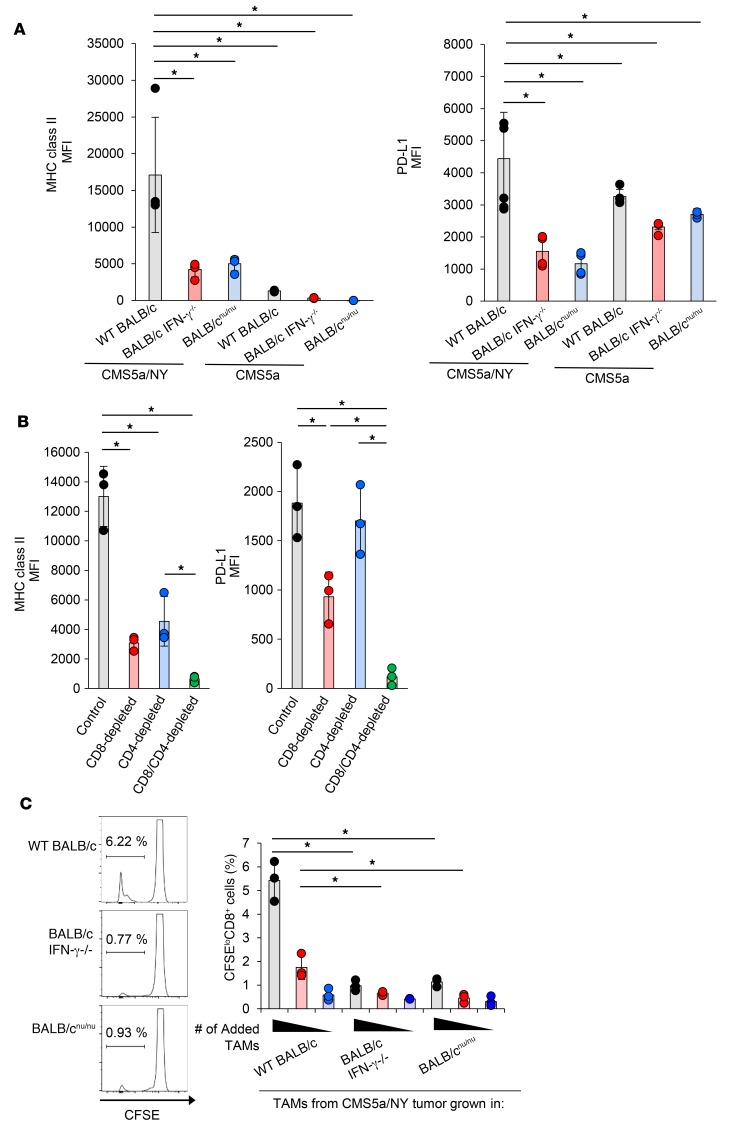
Inflammation at the tumor site has been reported to affect the activation of TAMs and antigen presentation by these cells (62). To understand the mechanisms contributing to TAM activation in sensitive tumors, we examined TAM activation in sensitive CMS5a/NY tumors implanted into IFN-γ–knockout mice or nude mice. As a result, upregulation of PD-L1 and MHC class II on TAMs from these mice was abrogated (Figure 4A). Again, we observed activation of TAMs in the sensitive CMS5a/NY tumor in terms of MHC class II and PD-L1 upregulation as compared with that in the resistant CMS5a tumors (Figure 4A). We found that depletion of CD8+ T cells or CD4+ T cells in the CMS5a/NY tumor–bearing mice also hampered TAM activation (Figure 4B). Furthermore, the antigen-presenting ability of TAMs was abrogated in the CMS5a/NY tumors implanted into IFN-γ–knockout mice and nude mice (Figure 4C). Finally, given that resistant tumors lack specific CD8+ T cell responses, the presence of highly immunogenic tumor antigens in sensitive tumors stimulated CD8+ TILs (and CD4+ TILs), leading to TAM activation by T cell–produced inflammatory molecules such as IFN-γ and substantially affecting the sensitivity of tumors to immune attack.
Figure 4. TAM activation in sensitive tumors is dependent on IFN-γ signaling and T cells.
(A) CMS5a/NY or CMS5a cells were subcutaneously inoculated into WT BALB/c mice (gray, n = 3), BALB/c IFN-γ–/– mice (blue, n = 3), or BALB/cnu/nu mice (red, n = 3). Seven days after inoculation, TAMs were isolated and analyzed for PD-L1 and MHC class II expression. Histograms show representative data. (B) The experiment was performed as described in A, but in CD8+ T cell–depleted (red, n = 3), CD4+ T cell–depleted (blue, n = 3), or CD8+ T cell– and CD4+ T cell–depleted (green, n = 3) mice. (C) TAMs were sorted from CMS5a/NY tumors grown in WT BALB/c mice (n = 3), BALB/c IFN-γ–/– mice (n = 3), or BALB/cnu/nu mice (n = 3) and were cocultured for 72 hours as antigen-presenting cells (1 × 105, 0.25 × 105, or 0.125 × 105 cells/well, respectively) with DUC18 CD8+ T cells as responder cells. Antigen-dependent proliferation of DUC18 CD8+ T cells was measured using a CFSE dilution assay. Histograms show representative data, and the numbers shown in the histograms indicate the percentage of proliferating CD8+ T cells. Data represent the mean ± SD. *P < 0.05, by 2-factor factorial ANOVA followed by Tukey–Kramer post hoc analysis. The experiments were repeated at least 2 to 4 times with similar results.
Nanogel-based antigen delivery with a TLR agonist induces antigen presentation by TAMs.
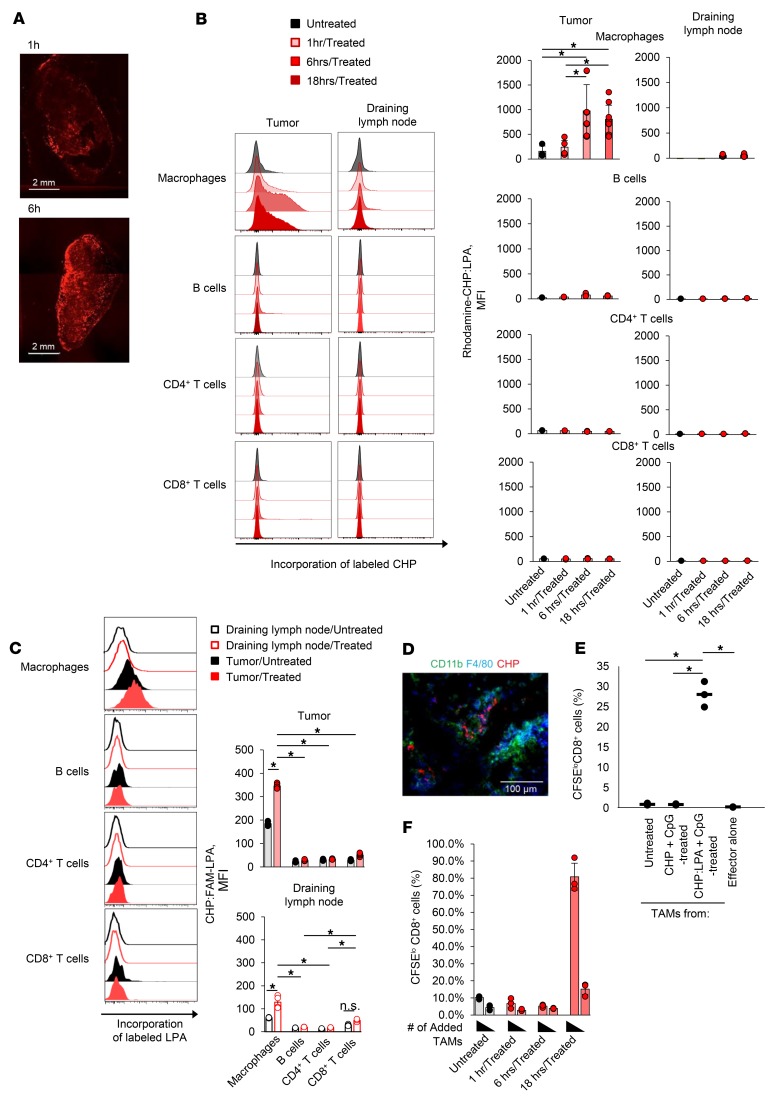
Given that the lack of antigen presentation by inactive TAMs is closely associated with immune resistance, we speculated that manipulation of TAMs to activate antigen presentation might improve tumor sensitivity. Inactive TAMs in resistant tumors may still have the potential to present antigens (Supplemental Figure 6). To deliver tumor antigens to TAMs in vivo, we used a novel antigen delivery system, cholesteryl pullulan (CHP) nanogel. CHP is a pullulan polysaccharide chemically modified with cholesteryl groups and spontaneously forms a nanogel with a diameter of 20 to 50 nm via hydrophobic interactions among the cholesteryl groups in an aqueous solution (Supplemental Figure 7). CHP nanogel is an inert monodisperse nanoparticle with no electric charge (Supplemental Figure 7) and no ligands for phagocytic receptors on its surface. When subcutaneously injected, the CHP nanogel promptly accumulates in the draining lymph node and is efficiently incorporated into lymph node medullary macrophages in a highly selective manner (36). It is also known that intravenously injected nanoparticles can penetrate into tumor tissue by an enhanced permeability and retention (EPR) effect and/or stochastic eruptions (63–65). Considering this information, we speculated that after intravenous injection, a complex of CHP and tumor antigen might migrate into the tumor and might be efficiently engulfed by TAMs. We therefore prepared a complex (CHP:LPA) of a fluorescently labeled CHP nanogel and a synthetic long peptide antigen (LPA) containing the 9m epitope and tested the effects of its intravenous injection into CMS5a tumor–bearing mice. As early as 1 hour after injection, we observed accumulation of CHP:LPA in the whole tumor, however CHP:LPA was not yet incorporated into TAMs or any other immune cells at that point (Figure 5, A–C). At 6 and 18 hours after injection, we noted cellular uptake of CHP:LPA in TAMs but not in T cells or B cells at the tumor site (Figure 5, B and C). Incorporation of the CHP nanogel into CD11b+F4/80+ TAMs was also confirmed by immunohistochemistry (Figure 5D). Next, we investigated whether targeted antigen delivery induced antigen-presenting activity in TAMs. The CMS5a tumor–bearing mice were intravenously injected with the CHP:LPA complex and a TLR9 agonist, CpG oligoDNA (ODN), and TAMs were then isolated from the tumors. We evaluated the antigen-presenting activity of these cells by coculturing them with DUC18 CD8+ T cells. TAMs from the treated mice showed potent antigen-presenting activity, whereas those from the untreated or CHP (without LPA) plus CpG ODN–treated mice did not (Figure 5E). It was not until 18 hours after the injection of CHP:LPA plus CpG ODN that we detected antigen-presenting activity in TAMs from the treated mice (Figure 5F). Interestingly, although the CHP:LPA complex was also incorporated into macrophages in other tissues (e.g., liver, lung, and spleen), macrophages in these normal tissues did not stimulate specific CD8+ T cells (Supplemental Figure 8, A and B). In DCs in the tumor-draining lymph nodes of the treated mice, we detected no CHP:LPA incorporation or subsequent antigen presentation (Supplemental Figure 8C). Histopathological analysis showed no toxicological changes caused by the CHP:LPA complex itself in the tissues in which CHP:LPA incorporation was detected, although some changes derived from CpG ODN were found in the livers and spleens of mice treated with either CpG ODN or CHP:LPA plus CpG ODN (Supplemental Figure 9). Thus, the CHP nanogel delivery system enabled the efficient and safe delivery of antigens to TAMs, thereby eliciting antigen presentation in these cells.
Figure 5. Targeted antigen delivery using CHP nanogel improves the antigen-presenting activity of TAMs.
(A) CMS5a tumor–bearing BALB/c mice were intravenously injected with rhodamine-CHP:LPA, and 1 hour or 6 hours later, tumors were removed. The distribution of rhodamine-CHP in the tumor was observed using confocal laser microscopy. Scale bars: 2 mm. (B) CMS5a tumor–bearing BALB/c mice were intravenously injected with the rhodamine-CHP:LPA complex, and 1 hour, 6 hours, or 18 hours later, immune cells including CD11b+F4/80+ macrophages, B cells, CD8+ T cells, and CD4+ T cells in the tumor or the tumor-draining lymph node were isolated (n = 4 per group). The uptake of labeled CHP:LPA in these cells was measured by flow cytometry. Histograms show representative data for CHP:LPA incorporation. *P < 0.05, by2-factor factorial ANOVA followed by Tukey-Kramer post hoc analysis.(C) CMS5a tumor–bearing BALB/c mice were intravenously injected with the CHP:FAM-LPA complex, and 18 hours later, the uptake of CHP:FAM-LPA in these cells was measured using flow cytometry (n = 3 mice per group). Histograms show representative data for CHP:LPA incorporation. *P < 0.05, by 2-factor factorial ANOVA followed by Tukey-Kramer post hoc analysis. The experiments were repeated 3 times with similar results. (D) CMS5a tumor–bearing BALB/c mice were treated as in A, and 6 hours later, cryosections of tumor were prepared. Incorporation of rhodamine-CHP:LPA into CD11b+F4/80+ TAMs was observed by immunohistochemistry. Scale bar: 100 μm. (E and F) The complex of CHP with 9m-containing LPA (50 μg) was intravenously injected with CpG ODN (50 μg) into BALB/c mice. After 1 hour, 6 hours, and 18 hours, TAMs were isolated and cocultured for 72 hours as antigen-presenting cells with DUC18 CD8+ T cells as responder cells. Antigen-dependent proliferation of DUC18 CD8+ T cells was measured using a CFSE dilution assay (n = 3 per group). Histograms show representative data, and the numbers shown in the histograms indicate the percentage of proliferating CD8+ T cells. Data represent the mean ± SD. *P < 0.05, by 2-factor factorial ANOVA followed by Tukey-Kramer post hoc analysis. The experiments were repeated 2 times with similar results.
Induction of antigen presentation by TAMs overcomes tumor immune resistance.
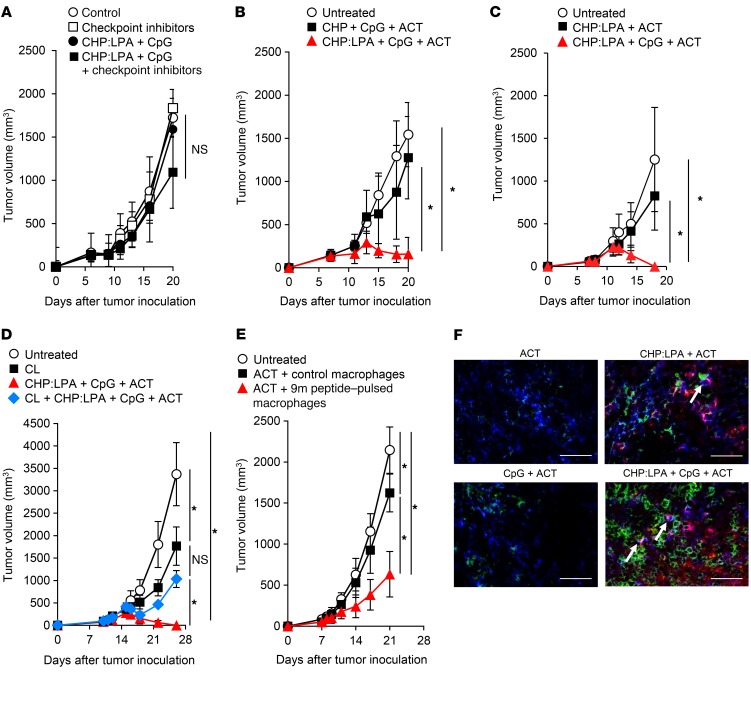
We next investigated whether the CHP:LPA-induced antigen presentation in TAMs would transform tumors from being immune resistant to immune sensitive. Intravenous administration of the CHP:LPA and CpG ODN was nearly ineffective in the CMS5a tumors, even when combined with immune checkpoint inhibitors (Figure 6A). This result was likely due to a lack of tumor-specific CD8+ T cell response induction during treatment in the nonimmunogenic CMS5a tumor model, as intravenous administration of the CHP:LPA and CpG ODN alone was not accompanied by de novo induction of specific T cells (Supplemental Figure 10). To establish a tumor-specific CD8+ T cell response in the CMS5a tumor–bearing mice, we next infused 9m epitope–specific DUC18 CD8+ T cells into the mice after intravenous injection of CHP:LPA plus CpG ODN. As a result, injection of CHP:LPA with CpG ODN on days 7 and 11 followed by ACT of DUC18 CD8+ T cells on days 8 and 12 substantially slowed the growth of CMS5a tumors, leading to rejection of these hard-to-treat, highly immune-resistant tumors (Figure 6, B and C). The therapeutic effect was decreased or abolished when 1 of the 3 components was omitted (Figure 6, A−C). CpG ODN was found to support TAM activation (Supplemental Figure 11). We also observed a similar synergistic effect of CHP:LPA and engineered T cell transfer in the presence of poly-IC RNA instead of CpG ODN (Supplemental Figure 12). These results suggest that intravenous administration of CHP:LPA and a TLR agonist transforms solid immune-resistant tumors into immune-sensitive tumors; furthermore, when combined with engineered T cell transfer, it can eradicate immune checkpoint inhibition–resistant tumors.
Figure 6. Induction of antigen presentation by TAMs improves tumor immune sensitivity.
CMS5a cells were subcutaneously inoculated into BALB/c mice. The complex of CHP with 9m epitope–containing LPA (50 μg) was intravenously injected with CpG ODN (50 μg) into CMS5a tumor–bearing BALB/c mice on days 7 and 11. (A) Checkpoint inhibitors including anti–PD-1 (200 μg/mouse), anti–CTLA-4 (100 μg/mouse), and anti-GITR (200 μg/mouse) Abs or isotype Abs were intraperitoneally injected on days 7, 9, and 11, or (B–F) naive CD8+ cells isolated from DUC18 mice were infused on days 8 and 12 into the same mice. Tumor size was then monitored. (D) CLs (200 μl) were intravenously injected into CMS5a tumor–bearing mice on days 4 and 8, and other reagents were administered as in B. (E) CMS5a tumors were inoculated into BALB/c mice on day 0. Control or 9m peptide–pulsed bone marrow–derived macrophages were directly injected into the CMS5a tumor on days 6 and 10. DUC18 CD8+ cells were infused on days 7 and 11 (n = 8–10 mice per group). *P < 0.05, by Steel-Dwass test. Experiments in A–E were performed at least 2 to 3 times with similar results. (F) Rhodamine-labeled CHP:LPA complex and CpG ODN were intravenously injected into CMS5a tumor–bearing BALB/c mice on day 7, followed by infusion of DUC18/CD90.1 mouse-derived CD8+ cells on day 8. Cryosections of the treated tumor were prepared and stained with anti-CD90.1 mAb (green). TAMs engulfing the labeled CHP:LPA complex are shown in red; blue indicates F4/80+ TAMs. Colocalization of CHP:LPA-ingested TAMs and specific CD8+ T cells is indicated by arrows. Scale bars: 50 μm. The experiments were repeated 2 times with similar results.
To investigate the significance of TAMs in the efficacy of this combination immunotherapy, macrophages in the CMS5a tumor and spleen, but not in the lymph nodes, were depleted by intravenous injection of CL (36, 66, 67) (Supplemental Figure 13). We found that CL-mediated TAM depletion prior to combination immunotherapy treatment severely limited tumor growth inhibition (Figure 6D). Tumor growth was comparable between the mice treated with CL alone and those treated with CL and CHP:LPA plus CpG ODN. In contrast, bone marrow–derived macrophages treated in vitro with the 9m epitope peptide and CpG ODN that were intratumorally injected into CMS5a tumor–bearing mice resulted in improved efficacy of ACT (Figure 6E). Immunohistochemical analysis revealed the colocalization of CHP:LPA-ingested TAMs and specific CD8+ T cells at the tumor site (Figure 6F). Thus, TAMs were essential for CHP:LPA-enhanced efficacy of ACT.
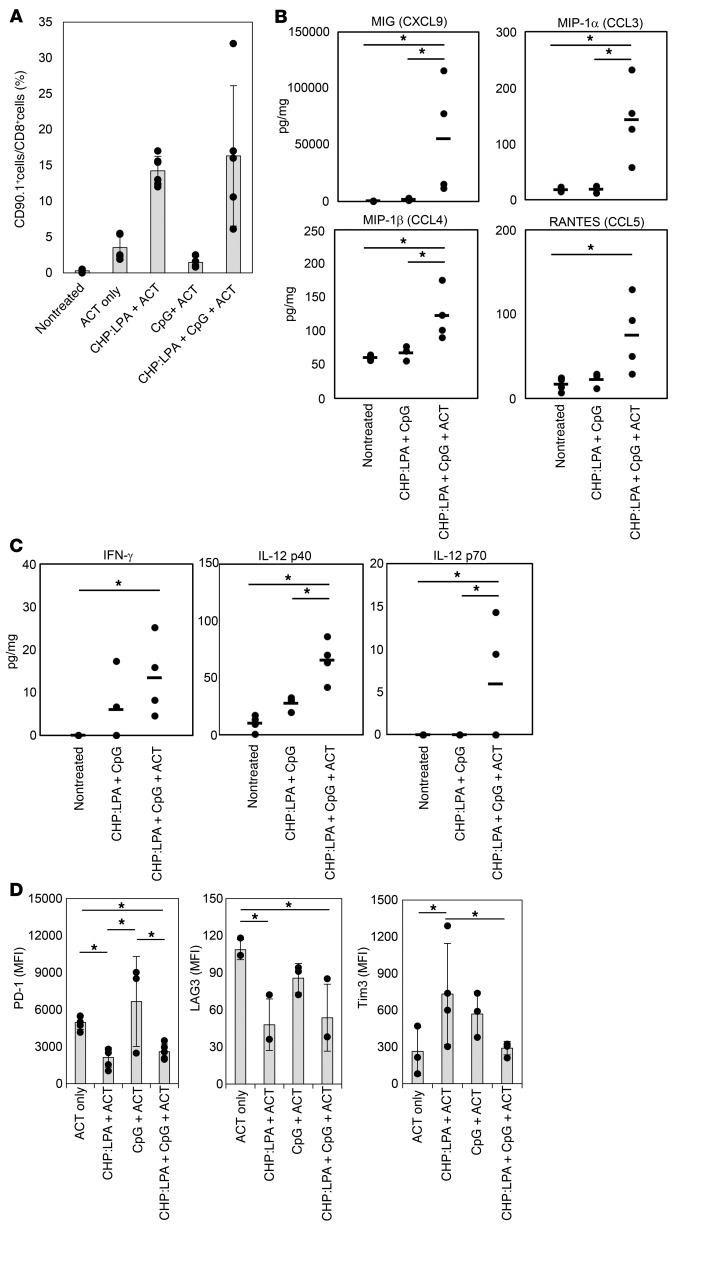
Three days after ACT, we assessed the changes in the local tumor site caused by TAM-targeted antigen delivery followed by ACT. We found that intravenous injection of CHP:LPA clearly enhanced the accumulation of infused 9m-specific CD8+ T cells at the tumor site (Figure 7A). Consistent with this finding, the levels of chemokines known to recruit T cells such as MIG (also known as CXCL9), MIP-1a (also known as CCL3), and RANTES (also known as CCL5) at the tumor site were increased by treatment (Figure 7B). The levels of Th1-type cytokines such as IFN-γ and IL-12 were also increased (Figure 7C). These results indicate that antigen presentation by TAMs leads to the establishment of Th1 status in the tumor microenvironment and thereby efficiently enhances specific CD8+ T cell infiltration. Interestingly, expression of PD-1 in tumor-infiltrating 9m-specific CD8+ T cells in the combination therapy group was markedly lower than that in the ACT group or CpG ODN plus ACT group (Figure 7D). A similar result was also obtained in endogenous tumor-infiltrating specific CD8+ T cells in a B16 tumor–bearing mouse model (38) after treatment with CHP:LPA plus CpG ODN (Supplemental Figure 14). We detected an increased amount of the transcription factor T-bet, a repressor of PD-1 expression (68), in 9m-specific CD8+ T cells that had infiltrated the CMS5a tumor after intravenous injection of CHP:LPA (Supplemental Figure 15), indicating that appropriate antigen presentation at the tumor site prevents PD-1 induction on CD8+ T cells via upregulation of T-bet (68).
Figure 7. Induction of antigen presentation by TAMs enhances the accumulation of specific CD8+ T cells at the local tumor site.
(A) The complex of CHP with 9m epitope–containing LPA (50 μg) was intravenously injected with CpG ODN (50 μg) into CMS5a tumor–bearing BALB/c mice (n = 2 mice per group) on day 7. Naive CD8+ cells isolated from DUC18/CD90.1 mice were infused into the same mice on day 8. The tumors were removed from the mice on day 11, and the frequency of CD90.1+CD8+ T cells in the tumors was determined by flow cytometry. Data from 3 experiments were pooled and represent the mean ± SD. (B and C) CMS5a tumor–bearing BALB/c mice (n = 3–4 per group) were treated as in A, and the tumors were isolated on day 11. The concentration of chemokines and cytokines in the tumors was measured by Bio-Plex assay. Circles in the graph indicate the values of the individual tumors, and bars show the mean. *P < 0.05, by 2-factor factorial ANOVA followed by Tukey-Kramer post hoc analysis. (D) Expression of immune checkpoint molecules on CD90.1+CD8+ T cells infiltrating the tumors isolated on day 11 was analyzed by flow cytometry (n = 3–4 per group). Data represent the mean ± SD. *P < 0.05, by 2-factor factorial ANOVA followed by Tukey-Kramer post hoc analysis. The experiments were repeated at least 2 to 3 times with similar results.
Taken together, our data indicate that manipulation of TAMs to activate antigen presentation at the tumor site transforms resistant tumors into T cell immunity–sensitive tumors. In addition, in combination with ACT, TAM-targeted antigen delivery can lead to the cure of highly immune-resistant tumors.
Discussion
The present work demonstrates that TAMs are one of the key factors determining tumor immune sensitivity. In immune-resistant tumors, TAMs remained inactive and did not serve as antigen-presenting cells. However, in the presence of TLR stimulation, nanogel-mediated selective antigen delivery elicited antigen-presenting activity in TAMs. Consequently, resistant tumors became sensitive to T cell–mediated immune attack. Because we did not observe TAM activation in the absence of IFN-γ or T cells, we concluded that the interaction between TAMs and T cells was critical for this phenomenon). Analysis of the local tumor site after treatment confirmed the increase in tumor-infiltrating specific CD8+ T cells, inflammatory cytokines, and T cell–recruiting chemokines, also indicating that the interplay between activated antigen-presenting TAMs and specific T cells is vital for enhancement of the therapeutic activity of treatment. Consistent with our results, a previous study reported that infusion of IL-12–producing CD8+ T cells induced inflammation at the tumor site and therefore the acquisition of antigen-presenting ability in myeloid cells, resulting in enhanced IFN-γ cellular responses (62).
The molecular mechanism(s) underlying the treatment-induced functional activation of TAMs is of great interest. We sought to identify transcription factor(s) responsible for the treatment-induced antigen presentation activity of TAMs. We focused on some transcription factors related to M1/M2 polarization of macrophages and, using microarray analysis, compared mRNA expression levels between treated and nontreated CMS5a tumors or between sensitive CMS5a/NY tumors and the resistant CMS5a tumors (Supplemental Table 5). We detected no significant differences in levels of the tested transcription factors, implying that other molecular mechanisms such as posttranslational modification of these transcription factors and/or epigenetic regulation of other macrophage-associated genes are involved in the phenomenon. To understand the mechanisms and to obtain more clues regarding TAM-targeted cancer immunotherapy, we conducted a detailed analysis of the cellular and molecular events that occur in TAMs after treatment with CHP:LPA plus CpG ODN.
All 3 components of our combination immunotherapy, i.e., the CHP:LPA complex, the TLR agonist, and the tumor-specific, receptor-engineered T cells, played indispensable roles in the efficacy of treatment. Intravenously injected CHP:LPA successfully induced antigen presentation by TAMs. To our knowledge, this is the first report of selective antigen delivery to antigen-presenting cells localizing at tumor tissues. Interestingly, this modification improved tumor sensitivity to T cell–mediated immunity, thereby rendering this approach different from conventional lymph node–targeted delivery of antigens and TLR ligands to reinforce antitumor T cell–mediated immunity (36, 69). A TLR agonist was also essential in the present combination immunotherapy, as macrophages that engulf antigens but are inactive cannot efficiently present antigens to T cells. Infused tumor-specific CD8+ T cells were necessary for tumor regression.
TAMs, as well as myeloid-derived suppressor cells, have been generally recognized as suppressor cells that work locally at the tumor site, and inhibition or depletion of TAMs is therefore expected to enhance antitumor immune responses. However, several studies have recently proposed the modulation of tumor-associated myeloid cell function as a novel, effective therapeutic strategy (23, 30, 70, 71). Because macrophage-produced IL-10 inhibits the tumor-inhibitory functions of tumor-associated DCs (TADCs), treatment with an anti–IL-10R Ab enhances IL-12 production from TADCs and induces potent CD8+ T cell–dependent antitumor responses (23). By affecting TAMs, IL-12 also augments cellular immune responses, inducing tumor regression accompanied by TADC activation. Vaccination with LPA activates TAMs and promotes their secretion of inflammatory cytokines, thereby suppressing tumor growth (70). The treatment of tumors with a PI3Kγ inhibitor was shown to switch TAMs from a M2-like phenotype to a M1-like state, leading to growth suppression of checkpoint inhibition–resistant tumors (30). Our study revealed for the first time to our knowledge the importance of antigen presentation by TAMs. Recent findings along with those presented here clearly suggest that the manipulation of TAM function could provide a promising strategy to potentiate the efficacy of immunotherapy.
CHP nanogel particles have a diameter of approximately 40 nm (36), which is small enough to pass through the walls of lymph capillary and tumor vessels. In addition, CHP is an immunologically stealthy biomaterial that has no immune-stimulating activity, surface charge, or known ligands. Our previous study demonstrated that when subcutaneously injected, the CHP nanogel efficiently and selectively delivers LPA to medullary macrophages in the draining lymph node; in the presence of TLR stimulation, these cells acquire the ability to present antigens to specific CD8+ T cells (36). In the current study, we found that the intravenously injected CHP:LPA complex preferentially accumulated in tumors rather than lymph nodes, possibly through an EPR effect and/or stochastic eruptions (63–65). It is also likely that the inertness of nanogel helps it to avoid capture by the reticuloendothelial system and/or the extracellular matrix. This indicates that the injection route greatly affects the in vivo distribution and action of nanogel-complexed antigen. Our early cancer vaccine products containing a protein antigen and CHP nanogel have been evaluated clinically, and their safety has been confirmed, indicating that the CHP nanogel is highly biocompatible and that TAM-targeted delivery using the CHP nanogel would be feasible in the clinical setting (72–74).
Many efforts have been made to overcome tumor resistance to checkpoint inhibition (75–77). These studies have used several strategies that involve the combination of checkpoint inhibitors with other reagents. For instance, Zamarin et al. reported that in immune-resistant B16 melanoma–bearing mice, the specific T cell–inducing and therapeutic effects of anti–CTLA-4 Ab and oncolytic Newcastle disease virus (NDV) were greater than those with NDV alone (77). In contrast, our combination immunotherapy eradicated checkpoint inhibition–resistant tumors without the use of checkpoint inhibitors. These 2 therapies differ in their dependency on checkpoint inhibition, which can be caused by the differential expression of checkpoint molecules on CD8+ T cells, a main effector of tumor killing. B16 melanoma cells killed by NDV probably secrete highly immunogenic antigens, and T cells recognizing these antigens express PD-1 and CTLA-4 (38). In contrast, the DUC18 CD8+ T cells used for adoptive transfer in our study did not express checkpoint molecules, because they were naive. Although further elucidation is necessary, to our knowledge, the current work successfully identified a combination immunotherapy that can cure checkpoint inhibition–resistant tumors without the use of checkpoint inhibitors.
To fully understand the mode of action of our combination immunotherapy, further studies are needed to identify the specific population of TAMs that play a vital role in this immunotherapy. Moreover, we did not investigate whether this effect occurred in other tumors with low immunogenicity, because we could not identify another tumor model that was similar to CMS5a fibrosarcoma in terms of low immunogenicity and checkpoint inhibition resistance. An analysis of human tumor samples would also be important to clarify the relationship between inactive TAMs and the sensitivity to checkpoint inhibition. Nevertheless, we believe our TAM-targeted combination immunotherapy addresses an important issue in current oncology — that of eradicating tumors that are completely resistant to checkpoint inhibition — and provides a new opportunity to treat checkpoint inhibition–resistant human tumors.
Methods
Abs and other reagents.
Fluorescently labeled mAbs were purchased from BioLegend. These included: anti-CD8 (clone 53-6.7); anti-Ly6C (clone HK1.4); anti-CD4 (clone RM4-5); anti-CD45RB/B220 (clone RM4-5); anti-CD11b (clone M1/70); anti-CD11c (clone N418); anti-F4/80 (clone BM8); anti-CD80 (clone 16-10A1); anti-CD86 (clone GL-1); anti-CD274 (clone 10F.9G2); anti-CD40 (clone 3/23); anti–I-A/I-E (clone M5/114); anti-KLRG1 (clone 2F1/KLRG1); anti-Tim3 (clone RMT3-23); anti-CD45 (clone 30-F11); anti–PD-1 (clone 29F.1A12); anti–IFN-γ (clone XMG1.2); anti-CD16/32 (clone 2.4G2); anti–Gr-1 (clone RB6-8C5); anti–T-bet (clone 4B10); purified rat IgG2a, κ isotype control Ab (clone RTK2758); purified rat IgG2b, κ isotype control Ab (clone RTK4530); and mouse IgG2b, κ isotype control Ab (clone MPC11). Anti–mouse PD-1 (clone RMP1–14); anti–mouse CTLA-4 (clone 9D9); and anti–mouse GITR (clone DTA-1) mAbs were produced in-house from hybridomas and purified using a protein G column. CpG ODN 1668 (5′-TCCATGACGTTCCTGATGCT-3′) was synthesized by Hokkaido System Science. Chemically synthesized LPAs were obtained from Bio-Synthesis Inc. CLs and control liposomes were purchased from FormuMax Scientific.
Fabrication of CHP nanogel and the LPA complex.
CHP powder was provided by Nippon Oil and Fat. The LPA consisting of 38–amino acid residues (SNPARYEFLYYYYYYQYIHSANVLYYYYYYRGPESRLL) contained 3 CD8+ T cell epitopes, including MAGE-A4 p265-273 (SNPARYEFL) (48), 9m (QYIHSANVL) (51), and NY-ESO-1 p81-88 (RGPESRLL) (46), and another LPA consisting of 38–amino acid residues (SVYDFFVWLYYYYYYTWHRYHLLYYYYYYEGSRNQDWL) contained 3 CD8+ T cell epitopes, including TRP2 p180-188 (SVYDFFVWL), TRP1 p222-229 (TWHRYHLL), and gp100 p25-33 (EGSRNQDWL); these LPAs were synthesized by Bio-Synthesis Inc. Oligotyrosine (YYYYYY) was included for efficient antigen processing. For analysis of antigen incorporation, the LPA was labeled with FAM. CHP and LPA were dissolved in PBS containing 6 M urea and dimethyl sulfoxide, respectively. Both solutions were combined and gently mixed at 4°C, followed by dialysis against PBS to facilitate the formation of a complex between LPA and CHP by removing urea. The obtained CHP:LPA nanogel complex solution was stored at 4°C until use.
Mice.
Female BALB/c and BALB/cnu/nu mice were obtained from SLC Japan and used at 6 to 12 weeks of age. DUC18 mice, transgenic for TCRα/β-reactive with a Kd-restricted 9m epitope, were established as previously described (61). DUC18 mice and CD90.1-congenic mice were mated to generate DUC18/CD90.1 mice. Mice on a BALB/c background deficient in IFN-γ (IFN-γ–/–) were purchased from Taconic. All mice were maintained at the Animal Center of Mie University.
Tumors.
CMS5a is a subclone derived from CMS5 that expresses the mutated ERK2 antigen containing the 9m epitope. CMS5a/NY stably expresses the human cancer/testis antigen NY-ESO-1 (46). The colon epithelial tumor cell line CT26 (43, 44) was purchased from the American Type Culture Collection (ATCC). CT26, CMS7 (45), CMS5a, and CMS5a/NY cells were cultured in RPMI 1640 medium containing 10% FBS. The melanoma tumor cell line B16 was purchased from the ATCC. B16 cells were cultured in DMEM containing 10% FBS. Mice were inoculated subcutaneously into the hind flank with 1 × 106 cells of each tumor cell line and monitored 3 times a week. Tumor volume was calculated using the following formula: tumor volume = 0.5 × length (mm) × (width in mm)2.
Administration of the CHP:LPA complex, the TLR agonist, and DUC18 CD8+ T cells.
To deplete macrophages, CL solution (200 μl/mouse) was intravenously injected into mice 24 hours prior to immunization. A CHP:LPA complex (50 μg LPA/mouse) was intravenously injected into mice with or without CpG ODN (50 μg/mouse). One day after immunization, 2 × 106 CD8+ T cells prepared from DUC18 mice were infused.
Flow cytometric analysis of immune cells.
Cell suspensions prepared from tumor, spleen, and other tissues were stained for surface markers using mAbs at appropriate concentrations (4 μg/ml) in PBS containing 2% FBS for 15 minutes at 4°C and analyzed on a FACSCanto II system (BD Biosciences). For intracellular cytokine staining, draining lymph node cells were incubated with the antigen peptide for 1 hour at 37°C and then incubated for an additional 6 hours with GolgiPlug (BD Biosciences). These cells were stained for surface markers. After permeabilization and fixation using a Cytofix/Cytoperm Kit (BD Biosciences), cells were stained with Abs. After washing, cells were analyzed on a FACSCanto II using FlowJo software (Tree Star).
Reverse transcription quantitative PCR.
Total RNA from cultured cells was reverse transcribed to first-strand cDNA (cDNA) using a QuantiTect Reverse Transcription Kit (QIAGEN). Reverse transcription quantitative PCR was performed using the StepOnePlus Real-Time PCR System (Applied Biosystems) according to the manufacturer’s protocol. The primers and probe for CXCL9 were selected from the ABI TaqMan Gene Expression Assay catalog. CXCL9 expression was normalized to that of GAPDH. Fold change was determined by the ΔΔCt method. Each experiment was performed in triplicate.
Tracking of injected antigen.
A fluorescently labeled complex of CHP nanogel and LPA was intravenously or subcutaneously injected into tumor-bearing mice. The inguinal lymph node or tumor was harvested 16 hours after the injection, homogenized, and then filtered through a nylon mesh. The resultant cell suspension was analyzed by flow cytometry to determine incorporation of labeled CHP or LPA and expression of CD45, CD11b, CD11c, F4/80, CD8, CD4, and B220.
Ex vivo antigen presentation.
Antigen presentation by TAMs was evaluated ex vivo by measuring antigen-specific proliferation of CD8+ T cells. CHP:LPA and CpG ODN were first intravenously injected into tumor-bearing mice. The tumor was resected 18 hours after injection. To isolate TAMs, the total cell suspension prepared from the tumor was mixed with anti-CD11b microbeads (Miltenyi Biotec) and separated by positive selection on a magnetic bead column. CD11b+ cells were then stained with APC-labeled anti–Gr-1, PE-labeled anti-CD11b, PerCP-Cy5.5–labeled Ly6C, PE/Cy7-labeled F4/80, and APC/Cy7-labeled anti-CD45 mAbs, and then sorted using a FACSAria (BD Biosciences). To isolate DCs, the total cell suspension prepared from the lymph node was mixed with anti-CD11c microbeads (Miltenyi Biotec) and separated by positive selection on a magnetic bead column. Isolated cells were cocultured with 2.5 × 105 DUC18 T cells prelabeled with a CFSE dye for 72 hours. T cell proliferation was determined by quantifying CFSE dilution on a FACSCanto II system (BD Biosciences). IFN-γ production in the culture medium at 72 hours was quantified using a mouse IFN-γ ELISA kit (BD Biosciences).
In vitro generation of mouse bone marrow–derived macrophages.
Bone marrow cells were isolated from femurs of 7- to 10-week-old mice. Cells were cultured in 20 ng/ml M-CSF (Peprotech) in RPMI 1640 containing 10% FBS. After 7 days, macrophages were stimulated with 10 μg/ml CpG ODN for 12 hours and pulsed with 10 μg/ml 9m peptide (QYIHSANVL) for 1 hour.
Immunohistochemistry.
Cryosections were prepared from tumors collected 16 hours after infusion of DUC18 CD8+ T cells. OCT-embedded cryosections were stained with fluorescent dye–conjugated anti–PD-1, anti–PD-L1, or anti–MHC class II mAbs and observed under a fluorescence microscope (BX53, Olympus).
Microarray analysis of tumors.
RNA was extracted from tumors or TAMs collected on day 7, and RNA quality was assessed using spectrophotometry and an Agilent Bioanalyzer 2100 (Agilent Technologies). Total RNA was amplified and labeled using the Ovation Pico WTA System and Encore Biotin Module (NuGEN). Prepared cDNA was hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 arrays and scanned using a GeneChip Scanner 3000 7G (Affymetrix). Microarray data are available in the DNA Data Bank of Japan (DDBJ) (http://trace.ddbj.nig.ac.jp/DRASearch/) under accession numbers E-GEAD-289 and E-GEAD-290.
Measurement of neoepitope-specific CD8+ T cell responses.
DNA and total RNA were extracted from in vitro cultured CT26, CMS7, and CMS5a cells. DNA was also extracted from the tails of BALB/c mice. Exome capture was performed using the SureSelectXT Mouse Exon Kit (Agilent Technologies). Exome capture libraries were then sequenced by Riken Genesis on a HiSeq 2000 (Illumina) using an Illumina V3 Kit, resulting in 20 million (2 × 150 bp) exome reads from each library. RNA samples were used to generate RNA-Seq libraries. RNA-Seq was performed by Hokkaido System Science on a HiSeq 2000, resulting in 30 million (2 × 100 bp) RNA fragments from each library.
Whole-exome sequence reads were aligned to the mm9 reference sequence using the Burrows-Wheeler Aligner (BWA) (78). Somatic single nucleotide variants (SNVs) were detected using Fisher’s exact test to compare read counts of normal and tumor samples. Variants with a P value of less than 10–10 were considered statistically significant. From the identified SNVs, we extracted only nonsynonymous SNVs. We also aligned the RNA-Seq reads to the mm9 genome with TopHat using the Bowtie aligner (79). We next analyzed the RNA-Seq data to confirm that RNAs carrying the detected SNVs were actually transcribed. Among the aligned RNA sequence reads, candidate variants in the whole-exome sequence were compared using Fisher’s exact test. Variants with a P value of less than 10–10 were considered to be transcribed SNVs. Whole-exosome sequencing data and RNA-Seq data are available in the DDBJ (accession nos. E-GEAD-291 and E-GEAD-292).
To prepare potential tumor-specific epitopes able to bind to murine MHC class I, we used the Immune Epitope Database (IEDB) and Analysis Resource (http://www.iedb.org/). All candidate epitopes (8–10 mers) containing missense mutations were analyzed for their binding affinities for H-2Kd, H-2Dd, or H-2Ld molecule according to the IEDB-recommended method (consensus) consisting of the artificial neural network (ANN), the stabilized matrix method, and the scoring matrices derived from combinatorial peptide library (Comblib_Sidney2008) algorithms (80). We prepared 61, 57, and 62 different candidate neoepitopes for CT26, CMS7, and CMS5a, respectively, on the basis of those with a percentile rank of 0.6 or lower (Supplemental Tables 1−3).
Measurement of cytokines and chemokines in tumors.
Tumors were harvested from CMS5a tumor–bearing mice, immediately frozen in liquid N2, and stored at –80°C. The tumors were then subjected to preparation of tumor lysate using the Bio-Plex Cell Lysis Kit (Bio-Rad Laboratories) according to the manufacturer’s instruction. The protein concentration of the tumor lysate was determined using the DC Protein Assay Kit (Bio-Rad Laboratories). The concentration of cytokines and chemokines was measured using Bio-Plex Pro Mouse Cytokine GI 23-Plex and GII 9-Plex panels (Bio-Rad Laboratories).
Retrovirus infection.
Whole splenic cells (1.5 × 107 cells/5 ml) from BALB/c mice were stimulated with immobilized anti-CD3 (1 μg/ml; 145-2C11) and soluble anti-CD28 (1 μg/ml; 37.51). One day after stimulation, 5 × 105 cells were transduced with the viral vector encoding TCRs against AH-1 using the RetroNectin-bound virus infection method (81). On day 5, the cells were harvested and used for experiments. Recombinant human IL-2 (Novartis) at 60 IU/ml was added during culturing.
Dynamic light scattering.
Dynamic light-scattering measurement was performed using a Zetasizer Nano ZS (Malvern Instruments Ltd.) at 633 nm and a 173° detection angle at 25°C. The measured autocorrelation function was analyzed using the cumulant method. The hydrodynamic diameter (DH) of the samples was calculated using the Stokes-Einstein equation. The ζ-potential of the CHP:LPA complex was also measured using the Zetasizer Nano ZS at a 90° detection angle at 25°C in PBS.
Transmission electron microscopy.
The sample was applied to a carbon-coated grid, and the grid was stained with TI Blue (Nisshin EM), dried, and subjected to transmission electron microscopic analysis (HT7700, Hitachi) at an accelerating voltage of 100 kV.
H&E staining.
Paraffin-embedded sections of mouse tissues fixed with 10% buffered formalin were subjected to H&E staining.
Statistics.
Data obtained in the in vivo tumor growth experiments were analyzed using the Mann-Whitney U test or the Steel-Dwass test. The remaining data were calculated using the 2-tailed Student’s t test, Dunnett’s test, or a 2-factor factorial ANOVA followed by Tukey-Kramer post hoc analysis. To compare the multiple groups in microarray analyses, the FDR was calculated. A P value or FDR of less than 0.05 was considered statistically significant.
Study approval.
The experimental protocols were approved by the Ethics Review Committee for Animal Experimentation of Mie University. All animal studies were performed with the approval of the IACUCs of Mie University and the University of Shizuoka.
Author contributions
DM, NH, and HS designed the study and wrote, reviewed, and revised the manuscript. DM, TH, KF, IT, YM, and KO performed animal and immunological experiments. NS and YT performed immunohistochemical analysis. NH and TH designed and prepared the CHP:LPA complex. DM, NS, TH, and NH analyzed and interpreted data. DM, RY, MK, and SI performed bioinformatics analysis on neoantigens. YT and MG performed chemical analysis of CHP:LPA. HY and SS provided key materials. KA, AA, HI, and SM provided key discussions.
Supplementary Material
Acknowledgments
We thank T. Takahashi (Comprehensive Health Science Center) for helpful discussion and L. Wang and K. Mori (Mie University) for technical assistance. This study was partially supported by the Japan Society for the Promotion of Science KAKENHI (grant nos. 15K18443 and 16H06313) and Exploratory Research for Advanced Technology of the JST (JST ERATO).
Version 1. 01/10/2019
In-Press Preview
Version 2. 02/11/2019
Electronic publication
Version 3. 03/01/2019
Print issue publication
Footnotes
Conflict of interest: NH is CEO of United Immunity, Co., Ltd.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(3):1278–1294.https://doi.org/10.1172/JCI97642.
Contributor Information
Daisuke Muraoka, Email: dmuraoka@nagasaki-u.ac.jp.
Naohiro Seo, Email: seo-naohiro@clin.medic.mie-u.ac.jp.
Tae Hayashi, Email: madhura884@yahoo.co.jp.
Yoshiro Tahara, Email: tahara.yoshiro.181@m.kyushu-u.ac.jp.
Keisuke Fujii, Email: minamikaze805@gmail.com.
Isao Tawara, Email: itawara@clin.medic.mie-u.ac.jp.
Yoshihiro Miyahara, Email: miyaou35@yahoo.co.jp.
Kana Okamori, Email: bluerose.seiyuki@gmail.com.
Hideo Yagita, Email: hyagita@juntendo.ac.jp.
Seiya Imoto, Email: imoto@ims.u-tokyo.ac.jp.
Rui Yamaguchi, Email: ruiy@ims.u-tokyo.ac.jp.
Mitsuhiro Komura, Email: komura@ims.u-tokyo.ac.jp.
Satoru Miyano, Email: miyano@ims.u-tokyo.ac.jp.
Masahiro Goto, Email: m-goto@mail.cstm.kyushu-u.ac.jp.
Shin-ichi Sawada, Email: sawada@bio.polym.kyoto-u.ac.jp.
Akira Asai, Email: aasai@u-shizuoka-ken.ac.jp.
Hiroaki Ikeda, Email: hikeda@nagasaki-u.ac.jp.
Kazunari Akiyoshi, Email: akiyoshi@bio.polym.kyoto-u.ac.jp.
Naozumi Harada, Email: naozumi.harada@unitedimmunity.co.jp.
Hiroshi Shiku, Email: shiku@clin.medic.mie-u.ac.jp.
References
- 1.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480(7378):480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callahan MK, Wolchok JD. At the bedside: CTLA-4- and PD-1-blocking antibodies in cancer immunotherapy. J Leukoc Biol. 2013;94(1):41–53. doi: 10.1189/jlb.1212631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol. 2014;32(10):1020–1030. doi: 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8(328):328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwai Y, Hamanishi J, Chamoto K, Honjo T. Cancer immunotherapies targeting the PD-1 signaling pathway. J Biomed Sci. 2017;24(1):26. doi: 10.1186/s12929-017-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powles T, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 10.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taube JM, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4(127):127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larkin J, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015;373(1):23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng W, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016;6(2):202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manguso RT, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547(7664):413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 17.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran E, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franklin RA, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344(6186):921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159(6):1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colegio OR, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513(7519):559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laoui D, et al. Tumor hypoxia does not drive differentiation of tumor-associated macrophages but rather fine-tunes the M2-like macrophage population. Cancer Res. 2014;74(1):24–30. doi: 10.1158/0008-5472.CAN-13-1196. [DOI] [PubMed] [Google Scholar]
- 23.Ruffell B, et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26(5):623–637. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doedens AL, et al. Macrophage expression of hypoxia-inducible factor-1 alpha suppresses T-cell function and promotes tumor progression. Cancer Res. 2010;70(19):7465–7475. doi: 10.1158/0008-5472.CAN-10-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hesse M, et al. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: granulomatous pathology is shaped by the pattern of L-arginine metabolism. J Immunol. 2001;167(11):6533–6544. doi: 10.4049/jimmunol.167.11.6533. [DOI] [PubMed] [Google Scholar]
- 26.Gao J, et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat Med. 2017;23(5):551–555. doi: 10.1038/nm.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeNardo DG, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1(1):54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Germano G, et al. Role of macrophage targeting in the antitumor activity of trabectedin. Cancer Cell. 2013;23(2):249–262. doi: 10.1016/j.ccr.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Predina J, et al. Changes in the local tumor microenvironment in recurrent cancers may explain the failure of vaccines after surgery. Proc Natl Acad Sci USA. 2013;110(5):E415–E424. doi: 10.1073/pnas.1211850110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaneda MM, et al. PI3Kγ is a molecular switch that controls immune suppression. Nature. 2016;539(7629):437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitano S, et al. HER2-specific T-cell immune responses in patients vaccinated with truncated HER2 protein complexed with nanogels of cholesteryl pullulan. Clin Cancer Res. 2006;12(24):7397–7405. doi: 10.1158/1078-0432.CCR-06-1546. [DOI] [PubMed] [Google Scholar]
- 32.Gu XG, et al. A novel hydrophobized polysaccharide/oncoprotein complex vaccine induces in vitro and in vivo cellular and humoral immune responses against HER2-expressing murine sarcomas. Cancer Res. 1998;58(15):3385–3390. [PubMed] [Google Scholar]
- 33.Akiyoshi K, et al. Self-assembled hydrogel nanoparticle of cholesterol-bearing pullulan as a carrier of protein drugs: complexation and stabilization of insulin. J Control Release. 1998;54(3):313–320. doi: 10.1016/S0168-3659(98)00017-0. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki Y, Akiyoshi K. Nanogel engineering for new nanobiomaterials: from chaperoning engineering to biomedical applications. Chem Rec. 2010;10(6):366–376. doi: 10.1002/tcr.201000008. [DOI] [PubMed] [Google Scholar]
- 35.Tahara Y, Akiyoshi K. Current advances in self-assembled nanogel delivery systems for immunotherapy. Adv Drug Deliv Rev. 2015;95:65–76. doi: 10.1016/j.addr.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Muraoka D, et al. Nanogel-based immunologically stealth vaccine targets macrophages in the medulla of lymph node and induces potent antitumor immunity. ACS Nano. 2014;8(9):9209–9218. doi: 10.1021/nn502975r. [DOI] [PubMed] [Google Scholar]
- 37.Lu L, Xu X, Zhang B, Zhang R, Ji H, Wang X. Combined PD-1 blockade and GITR triggering induce a potent antitumor immunity in murine cancer models and synergizes with chemotherapeutic drugs. J Transl Med. 2014;12:36. doi: 10.1186/1479-5876-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A. 2010;107(9):4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krummel MF, Allison JP. CTLA-4 engagement inhibits IL-2 accumulation and cell cycle progression upon activation of resting T cells. J Exp Med. 1996;183(6):2533–2540. doi: 10.1084/jem.183.6.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson TR, et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med. 2013;210(9):1695–1710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ko K, et al. Treatment of advanced tumors with agonistic anti-GITR mAb and its effects on tumor-infiltrating Foxp3+CD25+CD4+ regulatory T cells. J Exp Med. 2005;202(7):885–891. doi: 10.1084/jem.20050940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bulliard Y, et al. Activating Fc γ receptors contribute to the antitumor activities of immunoregulatory receptor-targeting antibodies. J Exp Med. 2013;210(9):1685–1693. doi: 10.1084/jem.20130573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corbett TH, Griswold DP, Roberts BJ, Peckham JC, Schabel FM. Tumor induction relationships in development of transplantable cancers of the colon in mice for chemotherapy assays, with a note on carcinogen structure. Cancer Res. 1975;35(9):2434–2439. [PubMed] [Google Scholar]
- 44.Griswold DP, Corbett TH. A colon tumor model for anticancer agent evaluation. Cancer. 1975;36(6 Suppl):2441–2444. doi: 10.1002/1097-0142(197512)36:6<2441::aid-cncr2820360627>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 45.DeLeo AB, Shiku H, Takahashi T, John M, Old LJ. Cell surface antigens of chemically induced sarcomas of the mouse. I. Murine leukemia virus-related antigens and alloantigens on cultured fibroblasts and sarcoma cells: description of a unique antigen on BALB/c Meth A sarcoma. J Exp Med. 1977;146(3):720–734. doi: 10.1084/jem.146.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muraoka D, et al. Peptide vaccine induces enhanced tumor growth associated with apoptosis induction in CD8+ T cells. J Immunol. 2010;185(6):3768–3776. doi: 10.4049/jimmunol.0903649. [DOI] [PubMed] [Google Scholar]
- 47.Gnjatic S, et al. NY-ESO-1: review of an immunogenic tumor antigen. Adv Cancer Res. 2006;95:1–30. doi: 10.1016/S0065-230X(06)95001-5. [DOI] [PubMed] [Google Scholar]
- 48.Muraoka D, et al. Establishment of animal models to analyze the kinetics and distribution of human tumor antigen-specific CD8(+) T cells. Vaccine. 2013;31(17):2110–2118. doi: 10.1016/j.vaccine.2013.02.056. [DOI] [PubMed] [Google Scholar]
- 49.Fujii K, et al. Identification of an immunogenic neo-epitope encoded by mouse sarcoma using CXCR3 ligand mRNAs as sensors. Oncoimmunology. 2017;6(5):e1306617. doi: 10.1080/2162402X.2017.1306617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang AY, et al. The immunodominant major histocompatibility complex class I-restricted antigen of a murine colon tumor derives from an endogenous retroviral gene product. Proc Natl Acad Sci U S A. 1996;93(18):9730–9735. doi: 10.1073/pnas.93.18.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ikeda H, et al. Mutated mitogen-activated protein kinase: a tumor rejection antigen of mouse sarcoma. Proc Natl Acad Sci U S A. 1997;94(12):6375–6379. doi: 10.1073/pnas.94.12.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duan F, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med. 2014;211(11):2231–2248. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207(10):2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eikawa S, Nishida M, Mizukami S, Yamazaki C, Nakayama E, Udono H. Immune-mediated antitumor effect by type 2 diabetes drug, metformin. Proc Natl Acad Sci U S A. 2015;112(6):1809–1814. doi: 10.1073/pnas.1417636112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27(2):281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oosterwegel MA, Greenwald RJ, Mandelbrot DA, Lorsbach RB, Sharpe AH. CTLA-4 and T cell activation. Curr Opin Immunol. 1999;11(3):294–300. doi: 10.1016/S0952-7915(99)80047-8. [DOI] [PubMed] [Google Scholar]
- 57.Nocentini G, Riccardi C. GITR: a modulator of immune response and inflammation. Adv Exp Med Biol. 2009;647:156–173. doi: 10.1007/978-0-387-89520-8_11. [DOI] [PubMed] [Google Scholar]
- 58.Litvak V, et al. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10(4):437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurozumi K, et al. Effect of tumor microenvironment modulation on the efficacy of oncolytic virus therapy. J Natl Cancer Inst. 2007;99(23):1768–1781. doi: 10.1093/jnci/djm229. [DOI] [PubMed] [Google Scholar]
- 60.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanson HL, et al. Eradication of established tumors by CD8+ T cell adoptive immunotherapy. Immunity. 2000;13(2):265–276. doi: 10.1016/S1074-7613(00)00026-1. [DOI] [PubMed] [Google Scholar]
- 62.Kerkar SP, et al. IL-12 triggers a programmatic change in dysfunctional myeloid-derived cells within mouse tumors. J Clin Invest. 2011;121(12):4746–4757. doi: 10.1172/JCI58814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bae YH, Park K. Targeted drug delivery to tumors: myths, reality and possibility. J Control Release. 2011;153(3):198–205. doi: 10.1016/j.jconrel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greish K. Enhanced permeability and retention (EPR) effect for anticancer nanomedicine drug targeting. Methods Mol Biol. 2010;624:25–37. doi: 10.1007/978-1-60761-609-2_3. [DOI] [PubMed] [Google Scholar]
- 65.Matsumoto Y, et al. Vascular bursts enhance permeability of tumour blood vessels and improve nanoparticle delivery. Nat Nanotechnol. 2016;11(6):533–538. doi: 10.1038/nnano.2015.342. [DOI] [PubMed] [Google Scholar]
- 66.Schliehe C, et al. CD8– dendritic cells and macrophages cross-present poly(D,L-lactate-co-glycolate) acid microsphere-encapsulated antigen in vivo. J Immunol. 2011;187(5):2112–2121. doi: 10.4049/jimmunol.1002084. [DOI] [PubMed] [Google Scholar]
- 67.Barral P, et al. CD169(+) macrophages present lipid antigens to mediate early activation of iNKT cells in lymph nodes. Nat Immunol. 2010;11(4):303–312. doi: 10.1038/ni.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kao C, et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat Immunol. 2011;12(7):663–671. doi: 10.1038/ni.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim WG, et al. Covalent Conjugation of Small-Molecule Adjuvants to Nanoparticles Induces Robust Cytotoxic T Cell Responses via DC Activation. Bioconjug Chem. 2016;27(9):2007–2013. doi: 10.1021/acs.bioconjchem.6b00277. [DOI] [PubMed] [Google Scholar]
- 70.van der Sluis TC, et al. Therapeutic peptide vaccine-induced CD8 T cells strongly modulate intratumoral macrophages required for tumor regression. Cancer Immunol Res. 2015;3(9):1042–1051. doi: 10.1158/2326-6066.CIR-15-0052. [DOI] [PubMed] [Google Scholar]
- 71.Mills CD, Lenz LL, Harris RA. A Breakthrough: macrophage-directed cancer immunotherapy. Cancer Res. 2016;76(3):513–516. doi: 10.1158/0008-5472.CAN-15-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miyauchi K, et al. Clinical relevance of antigen spreading pattern induced by CHP-MAGE-A4 cancer vaccination. Immunotherapy. 2016;8(5):527–540. doi: 10.2217/imt-2016-0007. [DOI] [PubMed] [Google Scholar]
- 73.Kageyama S, et al. Dose-dependent effects of NY-ESO-1 protein vaccine complexed with cholesteryl pullulan (CHP-NY-ESO-1) on immune responses and survival benefits of esophageal cancer patients. J Transl Med. 2013;11:246. doi: 10.1186/1479-5876-11-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aoki M, et al. Antibody responses against NY-ESO-1 and HER2 antigens in patients vaccinated with combinations of cholesteryl pullulan (CHP)-NY-ESO-1 and CHP-HER2 with OK-432. Vaccine. 2009;27(49):6854–6861. doi: 10.1016/j.vaccine.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 75.Kim K, et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc Natl Acad Sci U S A. 2014;111(32):11774–11779. doi: 10.1073/pnas.1410626111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fu J, et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7(283):283ra52. doi: 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zamarin D, et al. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci Transl Med. 2014;6(226):226ra32. doi: 10.1126/scitranslmed.3008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moutaftsi M, et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol. 2006;24(7):817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 81.Wang L, et al. Efficient tumor regression by adoptively transferred CEA-specific CAR-T cells associated with symptoms of mild cytokine release syndrome. Oncoimmunology. 2016;5(9):e1211218. doi: 10.1080/2162402X.2016.1211218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.