Abstract
MAPK4 is an atypical MAPK. Currently, little is known about its physiological function and involvement in diseases, including cancer. A comprehensive analysis of 8887 gene expression profiles in The Cancer Genome Atlas (TCGA) revealed that MAPK4 overexpression correlates with decreased overall survival, with particularly marked survival effects in patients with lung adenocarcinoma, bladder cancer, low-grade glioma, and thyroid carcinoma. Interestingly, human tumor MAPK4 overexpression also correlated with phosphorylation of AKT, 4E-BP1, and p70S6K, independent of the loss of PTEN or mutation of PIK3CA. This led us to examine whether MAPK4 activates the key metabolic, prosurvival, and proliferative kinase AKT and mTORC1 signaling, independent of the canonical PI3K pathway. We found that MAPK4 activated AKT via a novel, concerted mechanism independent of PI3K. Mechanistically, MAPK4 directly bound and activated AKT by phosphorylation of the activation loop at threonine 308. It also activated mTORC2 to phosphorylate AKT at serine 473 for full activation. MAPK4 overexpression induced oncogenic outcomes, including transforming prostate epithelial cells into anchorage-independent growth, and MAPK4 knockdown inhibited cancer cell proliferation, anchorage-independent growth, and xenograft growth. We concluded that MAPK4 can promote cancer by activating the AKT/mTOR signaling pathway and that targeting MAPK4 may provide a novel therapeutic approach for cancer.
Keywords: Cell Biology, Oncology
Keywords: Cancer, Oncogenes
Introduction
The AKT/mTOR signaling pathway is essential for regulating cell survival, proliferation, and metabolism. In the canonical pathway for AKT activation, PI3K catalyzes the production of phosphatidylinositol-3,4,5-triphosphate (PIP3), which binds to the pleckstrin homology (PH) domain of AKT, thereby recruiting AKT to the plasma membrane for activation (1–3). Phosphorylation of AKT at Thr308 (T308) of the activation loop by PDK1 (4) and phosphorylation of Ser473 (S473) in the hydrophobic loop are required for full AKT activation. mTOR exists in 2 distinct complexes, mTORC1 and mTORC2, which act coordinately with AKT. AKT activates mTORC1, which is an essential hub integrating extracellular stimuli and nutrient signals to regulate cell growth and metabolism (5). In contrast, mTORC2 is a major AKT S473 kinase (6). Precise control of PI3K/AKT/mTOR signaling is crucial for tissue homeostasis, and its aberrant activation leads to a number of pathological outcomes, including cancers. The PI3K/AKT/mTOR pathway has emerged as a major therapeutic target in cancer (7, 8).
The 13 human MAP kinases include the extensively studied ERK1/2 (MAPK3/1), JNK1/2/3 (MAPK8/9/10), and P38α/β/γ/δ (MAPK14/11/12/13). In contrast, only a few studies have focused on MAPK4 (ERK4), an atypical MAPK that lacks the canonical Thr-X-Tyr (TxY) activation motif for phosphorylation by the dual Ser/Thr and Tyr MAPK kinase (MAPKK) (9, 10). Currently, very little is known about the physiological functions of MAPK4 and its involvement in human diseases, including cancers.
Here we report that MAPK4 overexpression in a subset of diverse types of human cancers is associated with significantly decreased overall patient survival, and that MAPK4 overexpression in human cancer tissues correlates with AKT and mTORC1 activation. MAPK4 directly and specifically activates AKT/mTOR via an alternative pathway that is not dependent on PI3K/PDK1. MAPK4 directly binds and phosphorylates AKT at T308, and also activates mTORC2 to phosphorylate AKT at S473 for full activation. MAPK4 overexpression induces oncogenic outcomes, and inhibiting MAPK4 in cancer cells with high endogenous MAPK4 blocks cell proliferation, anchorage-independent growth, and xenograft growth. These results identify MAPK4 as a promising therapeutic target for human cancers.
Results
MAPK4 overexpression correlates with poor survival, and AKT and mTOR activation in human cancers.
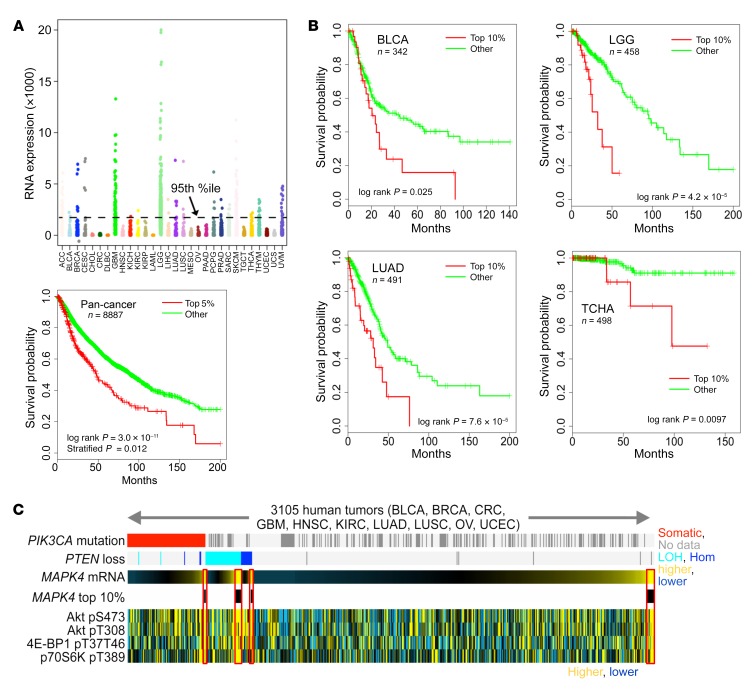
Very little is known about the physiological function of MAPK4, and previous studies have not linked MAPK4 to human cancers. A comprehensive analysis of 8887 gene expression profiles in The Cancer Genome Atlas (TCGA) (11) showed that MAPK4 is overexpressed in diverse tumor types, but typically only in a relatively small (5%–10%) subset (Figure 1A), which may account for the lack of prior interest. In this large pan-cancer panel, survival was significantly decreased in the subset of patients above the 95th percentile of MAPK4 expression (Figure 1A). Although a low number of patients and a shorter period of follow-up data limited analysis in some cases, survival was also markedly decreased in patients with MAPK4 overexpression in several different cancer types, including bladder urothelial carcinoma (BLCA), low-grade glioma (LGG), lung adenocarcinoma (LUAD), and thyroid carcinoma (THCA) (Figure 1B). These data indicate a potential tumor-promoting role of MAPK4 in human cancers.
Figure 1. Overexpression of MAPK4 in a subset of human cancers is associated with decreased overall survival and increased AKT activity.
(A) Upper panel, MAPK4 mRNA expression across 8887 tumors of various histological subtypes from The Cancer Genome Atlas (TCGA). Lower panel, Kaplan-Meier plot of overall survival in patients across all TCGA data sets, as stratified by high MAPK4 expression. P values by log-rank test and by stratified log-rank test (correcting for tumor type), as indicated. (B) Kaplan-Meier plot of overall survival in patients within specific cancer types, as stratified by high MAPK4 expression. BLCA, bladder cancer; LGG, low-grade glioma; LUAD, lung adenocarcinoma; THCA, thyroid carcinoma. P values by log-rank test. (C) Heatmaps showing the correlation of MAPK4 expression with expression of specific proteins profiled in TCGA pan-cancer datasets by Pearson’s coefficient. Significance of correlation: P < 1 × 10–17 for each protein feature.
We probed reverse-phase protein arrays (12) to identify potential growth control pathways that could be targeted by MAPK4 overexpression. Interestingly, MAPK4 overexpression in human cancers was associated with increased AKT activity. In a panel of 10 tumor types (12), loss of PTEN or PIK3CA mutation was associated with AKT phosphorylation at T308 and S473 and also with phosphorylation of its downstream mTORC1-dependent targets 4E-BP1 and p70S6K, as expected (P < 0.001, Student’s t test, for each protein, comparing tumors with PIK3CA or PTEN alteration versus other tumors). However, all 4 markers of AKT activation were also increased in patients with high MAPK4 expression but without genetic activation of the PI3 kinase pathway (Figure 1C, right end). MAPK4 expression was also increased with these markers in patients with heterozygous loss of PTEN or PIK3CA mutation (Figure 1C). These data are consistent with a potential oncogenic role of MAPK4 in activation of the key prosurvival and proliferative kinase AKT in human cancers.
MAPK4 activates AKT.
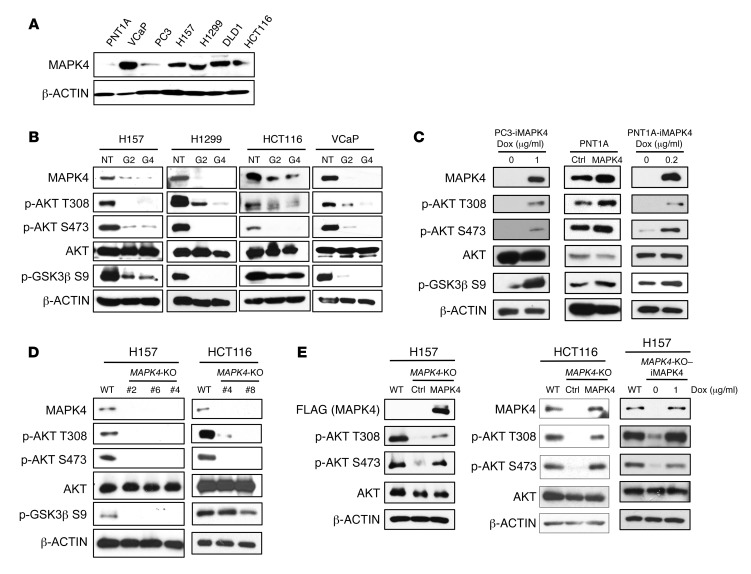
To assess MAPK4 biology in human cancers, we first surveyed its expression in a series of human cancer cell lines and found high levels of endogenous MAPK4 (MAPK4-high) in human lung cancer H157 and H1299 cells, colon cancer HCT116 and DLD1 cells, and prostate cancer VCaP cells. In contrast, prostate cancer PC3 cells and the immortalized human normal prostate epithelial PNT1A cells express lower levels of MAPK4 (MAPK4-low, Figure 2A). To investigate MAPK4 biology, we used lentiviral shRNA to knock down MAPK4 in the MAPK4-high cells. We found that MAPK4 knockdown in all 4 MAPK4-high cell lines inhibited AKT phosphorylation at both T308 and S473 and deactivated AKT, as evidenced by inhibition of GSK3β phosphorylation (Figure 2B). These results suggest that MAPK4 is indispensable for AKT phosphorylation and activation in MAPK4-high cells. We overexpressed MAPK4 in the MAPK4-low PC3 cells and PNT1A cells in a Dox-inducible manner, and also constitutively overexpressed MAPK4 in the PNT1A cells. In accordance with the shRNA loss-of-function results, MAPK4 overexpression induced AKT phosphorylation and activation in all 3 cell lines (Figure 2C).
Figure 2. MAPK4 activates AKT.
(A) Western blots on MAPK4 expression in various human cancer cell lines and PNT1A, an immortalized human prostate epithelial cell line. (B) Two distinct shRNAs targeting MAPK4 (G2 and G4 as described in Methods) were used to knock down MAPK4 expression in H157, H1299, HCT116, and VCaP cells. Western blots were used to detect AKT phosphorylation and activation (phosphorylation of GSK3β). (C) MAPK4 was overexpressed in PC3 and PNT1A cells. Western blots were used to detect AKT phosphorylation and activation. (D) CRISPR/Cas9 technology was used to knockout MAPK4 in H157 cells (clones 2, 4, 6) and HCT116 cells (clones 4 and 8). Western blots were used to compare AKT phosphorylation and activation among these cells. (E) MAPK4 was ectopically expressed in the H157 MAPK4-KO cells (clone 2) and HCT116 MAPK4-KO cells (clone 4). Both constitutive and Dox-induced expression systems were used to express MAPK4 in the H157 MAPK4-KO cells. Western blots were used to detect AKT phosphorylation and activation. Ctrl: control; NT: nontargeting control; iMAPK4: Dox-inducible overexpression of MAPK4. Data are representative of at least 3 independent experiments.
To confirm and extend these results, we used CRISPR/Cas9 to generate MAPK4 null cells (13, 14). As expected, knockout of MAPK4 in both H157 cells and HCT116 cells greatly inhibited AKT phosphorylation and activation (Figure 2D). Finally, we confirmed that constitutive and/or Dox-inducible overexpression of MAPK4 rescued AKT phosphorylation in the MAPK4-KO H157 cells and HCT116 cells (Figure 2E). Altogether, these data support a crucial role of MAPK4 in inducing AKT phosphorylation and activation.
MAPK4 phosphorylates and activates AKT independent of the PI3K pathway.
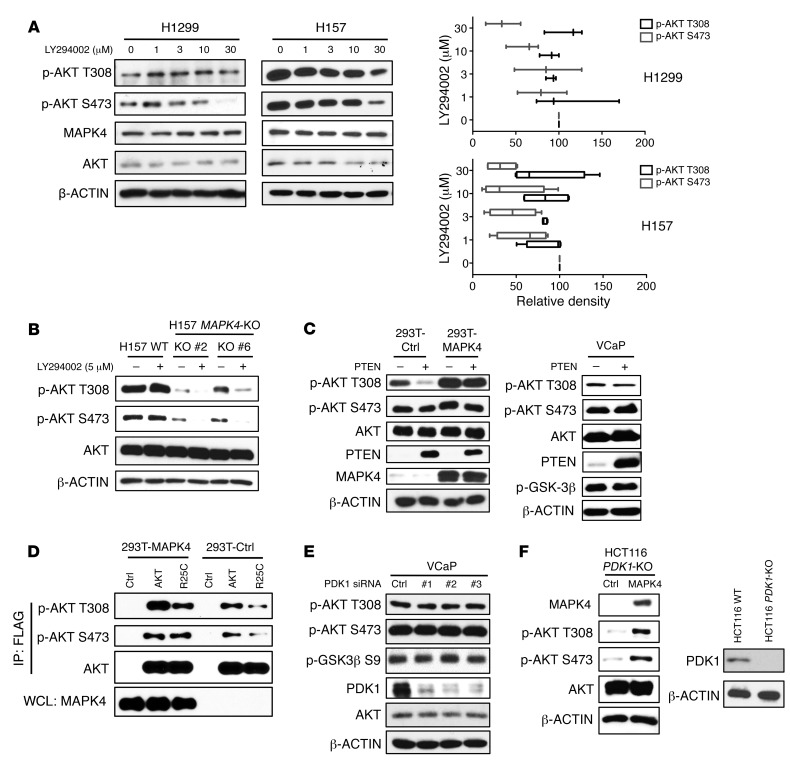
The PI3K/PDK1 pathway is thought to be essential for AKT activation via direct PDK1 phosphorylation at AKT residue T308. We used 4 different approaches to determine whether MAPK4 activation of AKT is dependent on PI3K/PDK1. First, we tested the impact of the PI3K inhibitor LY294002 on MAPK4-induced AKT phosphorylation and activation. The MAPK4-high H157 and H1299 cells are resistant to LY294002 treatments (Figure 3A). In contrast, LY294002 largely abolished AKT phosphorylation at T308 and S473 in the MAPK4-KO H157 cells (Figure 3B). We next tested whether overexpression of the PI3K pathway inhibitor PTEN affects AKT phosphorylation in cells overexpressing MAPK4. While PTEN greatly inhibited AKT phosphorylation at T308 in the control HEK293T cells, it had minimal effect on T308 phosphorylation in HEK293T cells overexpressing MAPK4 and in the MAPK4-high VCaP cells (Figure 3C). We also used the R25C mutation within the AKT PH domain, which disrupts PIP3 binding (15) and prevents AKT1R25C mutant from being efficiently recruited to the plasma membrane for PI3K/PDK1-induced phosphorylation and activation. AKT1R25C phosphorylation is significantly increased in HEK293T-MAPK4 cells, indicating that MAPK4 phosphorylation of AKT1R25C is not dependent on PI3K/PDK1 (Figure 3D). However, this MAPK4-induced phosphorylation of AKT1R25C appears to be weaker than that of WT AKT1, which is induced by both PI3K/PDK1 and MAPK4. Finally, we asked whether manipulation of PDK1 protein levels affects MAPK4-associated AKT phosphorylation. Knockdown of PDK1 in the MAPK4-high VCaP cells did not alter AKT phosphorylation or activity (Figure 3E), whereas MAPK4 overexpression induced significant AKT phosphorylation in HCT116 PDK1-KO cells (Figure 3F). Altogether, we conclude that MAPK4 induction of AKT phosphorylation and activation is independent of the PI3K/PDK1 pathway.
Figure 3. MAPK4 activates AKT independent of PI3K/PDK1.
(A) H1299 and H157 cells with high endogenous MAPK4 were treated with increasing doses of PI3K inhibitor LY294002 for 24 hours. Western blots were used to detect AKT phosphorylation. The phosphorylation of AKT at T308 and S473 was analyzed using ImageJ (33). The data are from 3 (for H1299 cells) and 4 (for H157 cells) independent studies. Box and whisker plots indicate average value with the box extending from the 25th to 75th percentiles and the whisker extending from Min to Max. (B) H157 and the H157 MAPK4-KO cells (KO clone 2 and KO clone 6) were treated with 5 μM LY294002 for 2 hours. Western blots were used to detect AKT phosphorylation. (C) PTEN was ectopically overexpressed in HEK293T cells overexpressing control (293T-Ctrl) or MAPK4 (293T-MAPK4), as well as in VCaP cells. Western blots were used to detect AKT phosphorylation and confirm PTEN overexpression. (D) The FLAG-tagged AKT1 and AKT1R25C mutants were overexpressed in HEK293T-Ctrl and HEK293T-MAPK4 cells. After immunoprecipitation using anti-FLAG M2 affinity gel, phosphorylation of the overexpressed AKT1 was detected with Western blots. WCL: whole-cell lysate. (E) Three independent siRNAs against PDK1 were used to knock down PDK1 expression in VCaP cells. Western blots were used to detect AKT phosphorylation and confirm knockdown of PDK1. (F) Left, MAPK4 was ectopically overexpressed in the HCT116 PDK1-KO cells. Western blots were used to detect AKT phosphorylation. Right, Western blots were used to confirm loss of PDK1 expression in the HCT116 PDK1-KO cells. Ctrl: control. Data are representative of at least 3 independent experiments.
MAPK4 directly interacts with AKT and phosphorylates AKT at T308.
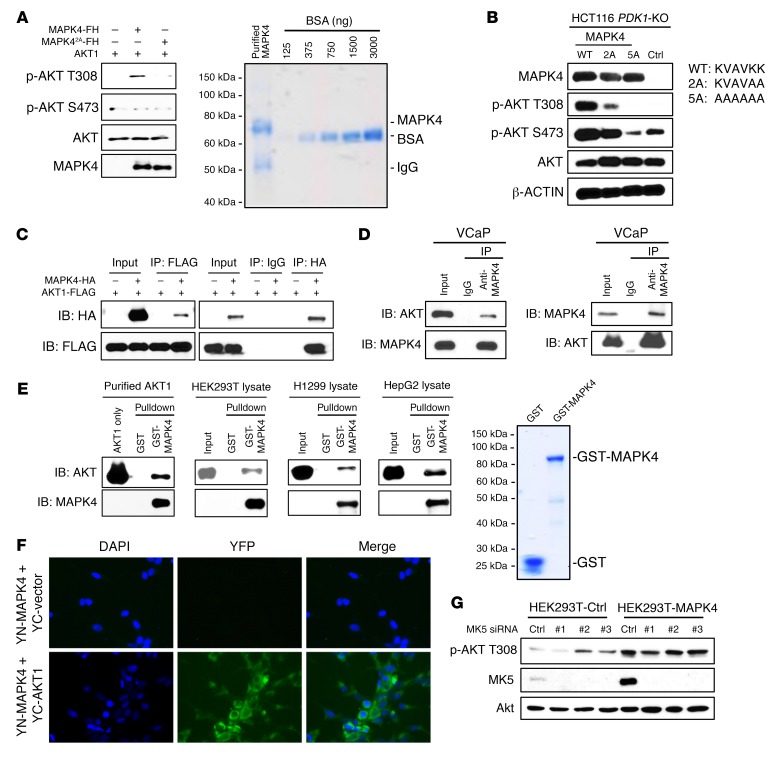
The results noted above suggest that MAPK4 directly binds and phosphorylates AKT. To test this, we overexpressed FLAG/His-tagged MAPK4 in PNT1A cells and purified it using Ni-NTA affinity column followed by immunopurification using anti-FLAG antibody–conjugated beads. This purified MAPK4 (with calculated molecular weight of 69.6 kDa) contained 1 major band around 70 kDa (Coomassie blue staining, Figure 4A). We then performed in vitro kinase assays as previously described (6) using commercially available purified AKT1 protein as substrate. MAPK4 strongly phosphorylated AKT1 on T308 but not S473 in these in vitro kinase assays, identifying MAPK4 as a novel AKT T308 kinase (Figure 4A). The VAIK motif in protein kinases interacts with ATP to promote phosphate transfer to substrate (16). In MAPK4, a variant KVAVKK motif at positions 45–50 is predicted to engage ATP. To create a kinase-dead MAPK4, we first made a MAPK42A mutant with KVAVAA in place of the KVAVKK motif. In vitro kinase assay revealed that AKT T308 kinase activity was greatly inhibited in this MAPK42A mutant (Figure 4A); however, kinase activity was still evident when MAPK42A was expressed in HCT116 PDK1-KO cells (Figure 4B). To further block kinase activity, we made a MAPK45A mutant that carries AAAAAA in place of the KVAVKK motif. This MAPK45A mutant no longer exhibited AKT T308 kinase activity when expressed in HCT116 PDK1-KO cells (Figure 4B). These results support the role of MAPK4 as a novel AKT T308 kinase.
Figure 4. MAPK4 interacts with AKT and is a novel T308 kinase.
(A) FLAG/His-tagged MAPK4 and the MAPK42A mutant were overexpressed in PNT1A cells and purified using Ni-NTA column followed by immunoprecipitation using anti-FLAG M2 affinity gel. In vitro kinase assay was performed using a commercially available recombinant AKT1. AKT phosphorylation was detected by Western blots. MAPK42A carries KVAVAA in place of the KVAVKK ATP binding motif. Coomassie blue staining reveals that the purified FLAG/His-tagged MAPK4 (with a calculated molecular weight of 69.6 kDa) contains one major band around 70 kDa. F/H: 2× FLAG and 10× His tag. (B) MAPK4 and the MAPK42A and MAPK45A mutants were overexpressed in HCT116 PDK1-KO cells. AKT phosphorylation was detected by Western blots. MAPK45A carries AAAAAA in place of the KVAVKK ATP binding motif. Coimmunoprecipitation (co-IP) assays reveal (C) ectopically overexpressed AKT1 binding to ectopically overexpressed MAPK4 in HEK293T cells, and (D) endogenous AKT1 binding to endogenous MAPK4 in VCaP cells. (E) Pull-down assay on the binding between purified GST-MAPK4 protein and purified AKT1 as well as endogenous AKT in the HEK293T, H1299, and HepG2 cell lysates. GST was used as control. GST-MAPK4 and GST were overexpressed in E. coli and purified using the glutathione sepharose beads. Coomassie blue staining confirms major bands of around 90 kDa and 26 kDa in the purified GST-MAPK4 and GST, respectively. (F) BiFC assay for AKT association with MAPK4 in cytoplasm. Hela cells were cotransfected with YN-MAPK4 and YC-AKT1 or YC control vectors. Forty-eight hours later, the cells were fixed, counterstained with DAPI, and imaged for YFP fluorescence to indicate MAPK4-AKT1 interaction. Original magnification: ×400. (G) Three different MK5 siRNAs were transfected into control and MAPK4-overexpressing HEK293T cells. Western blots were used to confirm MK5 knockdown and compare AKT phosphorylation. Ctrl: control. Data are representative of at least 3 independent experiments.
To further define this role, we investigated MAPK4-AKT interaction. We expressed FLAG-tagged AKT1 in HEK293T cells overexpressing a HA-tagged MAPK4, and performed immunoprecipitation using anti-FLAG antibody–conjugated beads followed by Western blot for HA, and reciprocal immunoprecipitation using anti-HA antibody followed by Western blot for FLAG. In both cases, we readily detected the coimmunoprecipitation of MAPK4 and AKT1 (Figure 4C). To confirm the physiological interaction between MAPK4 and AKT, we immunoprecipitated endogenous MAPK4 from VCaP cell lysate followed by Western blot for AKT, and reciprocally immunoprecipitated AKT followed by Western blot for MAPK4. Again, we readily detected the interaction of endogenous MAPK4 and AKT (Figure 4D).
The direct interaction was confirmed by the ability of overexpressed and purified GST-MAPK4 fusion protein, but not GST, to pull down commercially available purified AKT1 protein (Figure 4E). We also performed pull-down assays using HEK293T, H1299, and HepG2 cell lysates and demonstrated that GST-MAPK4 protein can pull down the endogenous AKT in these cell lysates (Figure 4E).
Bimolecular florescence complementation (BiFC) enables visualization of protein-protein interactions and subcellular locations in live cells (17). Accordingly, we cotransfected Hela cells with a YN-MAPK4 construct encoding MAPK4 fused to the N-terminal fragment of YFP, along with a YC-AKT1 construct encoding AKT1 fused to the C-terminal fragment of YFP. Observation of a positive cytoplasmic signal in cells coexpressing YN-MAPK4 and YC-AKT1, but not in the YN-MAPK4 and YC vector control cotransfected cells, provided additional confirmation of the MAPK4-AKT1 interaction (Figure 4F).
Although these results strongly support the direct activation of AKT by MAPK4, they do not rule out a potential role of its only known substrate, MK5 (9, 10). To test this, we used siRNA to knock down MK5 expression in HEK293T control and MAPK4-overexpressing cells. MK5 knockdown did not significantly affect AKT activation, indicating that MK5 is not essential for this pathway (Figure 4G).
The molecular basis for MAPK4-AKT interaction.
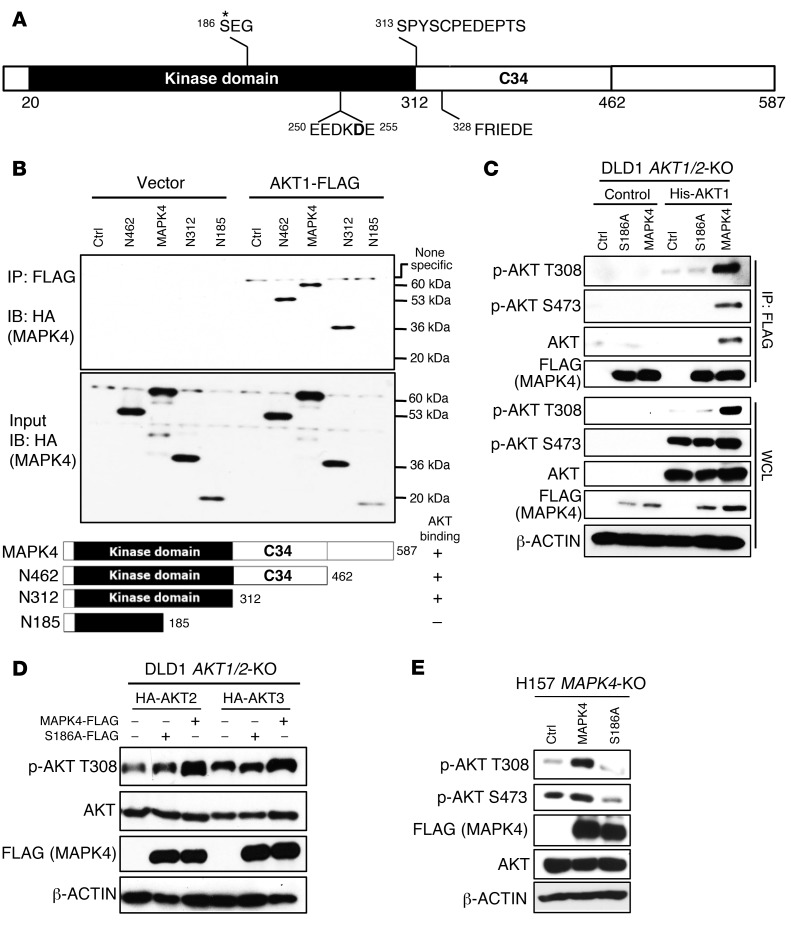
MAPK4 contains an N-terminal kinase domain, a C34 conserved region shared between MAPK4 and MAPK6/Erk3, and a C-terminal tail (Figure 5A). To determine the key region critical for MAPK4 interaction with AKT, we performed coimmunoprecipitation of FLAG-tagged AKT1 with C-terminally truncated HA-tagged MAPK4 proteins overexpressed in HEK293T cells. We found that the C-terminal fragment of the kinase domain (N186-312) is essential for MAPK4 binding to AKT1 (Figure 5B).
Figure 5. The molecular basis for MAPK4 binding to AKT, part 1.
(A) The domain architecture of human MAPK4. MAPK4 contains an N-terminal kinase domain (aa20 to aa312), a conserved C34 (aa313 to aa462) motif shared between MAPK4 (Erk4) and MAPK6 (Erk3), and a C-terminal tail. Asterisk indicates phosphorylation site. (B) HEK293T cells were transfected with FLAG-tagged AKT1 and HA-tagged WT or C-terminally truncated MAPK4 including N462, N312, and N185. Immunoprecipitation was performed using anti-FLAG M2 affinity gel followed by Western blots using anti-HA antibody. The kinase domain of MAPK4 binds to AKT1 and the aa186–aa312 fragment is essential for this interaction. (C) His-tagged AKT1 along with FLAG-tagged MAPK4 and MAPK4S186A mutant were reconstructed into the DLD1 AKT1/2-KO cells that lack all 3 isoforms of AKT. Immunoprecipitation was performed using anti-FLAG M2 affinity gel followed by Western blots for phosphorylation of AKT and total AKT. WCL: whole-cell lysate. (D) HA-tagged AKT2 and AKT3 were reconstructed into the DLD1 AKT1/2-KO cells along FLAG-tagged MAPK4 or MAPK4S186A mutant. Western blots using Phospho-AKT T308 antibody were used to detect the phosphorylation of AKT2 at T309 and AKT3 at T305, respectively. (E) FLAG-tagged MAPK4 and MAPK4S186A mutant were transduced into the H157 MAPK4-KO cells. Western blots were used to determine AKT phosphorylation. Ctrl: control. Data are representative of at least 3 independent experiments.
Conventional MAP kinases share a conserved TxY motif in the activation loop that is the target for MAPKK-induced serine/threonine and tyrosine dual phosphorylation. In MAPK4 and MAPK6/Erk3, this is replaced by SEG. The N186-312 fragment that is required for AKT interaction contains this S186. To assess whether S186 is critical for regulating MAPK4 activation of AKT, we introduced His-tagged AKT1 and FLAG-tagged WT MAPK4 or the S186A mutant MAPK4 (MAPK4S186A) into the DLD1 AKT1/2-KO cells that lack expression of all 3 isoforms of AKT (18). We found that MAPK4, but not MAPK4S186A, binds to and phosphorylates AKT1 (Figure 5C). This suggests a crucial role of S186 in promoting MAPK4 binding to and phosphorylating AKT1. In accord with this, MAPK4 but not MAPK4S186A phosphorylates AKT2 and AKT3, the other 2 isoforms of AKT, when overexpressed in DLD1 AKT1/2-KO cells (Figure 5D). We further confirmed the crucial role of S186 in MAPK4 phosphorylation of AKT by demonstrating that MAPK4, but not MAPK4S186A, rescues AKT phosphorylation in H157 MAPK4-KO cells (Figure 5E).
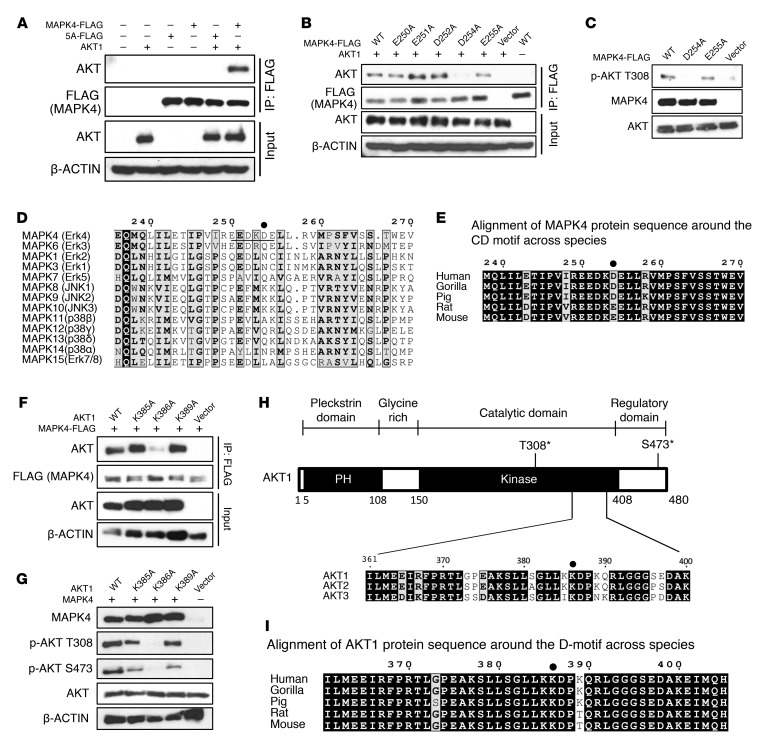
The common docking (CD) motif provides the major docking site for MAPK family members to bind to their partners including substrates (19). MAPK4 contains a conserved CD motif SPYSCPEDEPTS (20); however, it is dispensable for MAPK4 binding to its substrate MK5 (21). Instead, MAPK4 uses a unique FRIEDE-motif to bind to MK5 (21). Interestingly, N186-312 contains neither of these 2 motifs, raising the question of how MAPK4 binds to AKT. The CD motif consists of a cluster of D/E negatively charged amino acids, which binds to the positively charged MAPK docking motif (D-motif, also known as the kinase interaction motif [KIM]) in the partner protein (19). Interestingly, the N186-312 segment contains another cluster of D/E-enriched amino acids (EEDKDE) at positions 250–255. Therefore, we asked whether this EEDKDE motif functions as an AKT docking site. We made mutant FLAG-tagged MAPK4 proteins containing 5 single D/E to A mutations within this EEDKDE motif as well as mutation of all 5 D/E residues to A. We expressed these FLAG-tagged MAPK4 proteins and AKT1 in HEK293T cells and performed immunoprecipitation using anti-FLAG antibody–conjugated beads followed by Western blot for AKT. We found that mutation of all 5 D/E residues largely blocked MAPK4-AKT interaction (Figure 6A), but among the single mutants only D254A strongly decreased interaction (Figure 6B). In accordance with this, overexpression of the MAPK4D254A mutant in HCT116 MAPK4-KO cells did not rescue AKT T308 kinase activity (Figure 6C). These data suggest that the newly identified EEDKDE motif functions as a CD motif. This motif, especially D254, provides the key docking site for AKT binding. It is interesting that this EEDKDE motif is highly conserved in MAPK4 proteins across species, but is completely absent in most other MAP kinases (Figure 6, D and E). In MAPK6/Erk3, a variant of this motif is present, but D254, the key residue essential for MAPK4-AKT binding, is replaced by Q (Figure 6D). Altogether, this specific CD motif may account for the unique AKT activating activity of MAPK4.
Figure 6. The molecular basis for MAPK4 binding to AKT, part 2.
(A) The FLAG-tagged MAPK4 and MAPK4E/D5A (5A-FLAG) mutant were transduced into HEK293T cells with or without cotransfection of AKT1. Immunoprecipitation was performed using anti-FLAG M2 affinity gel followed by Western blots using anti-AKT antibody. The MAPK4E/D5A (5A-FLAG) mutant carries AAAKAA in place of the potential CD motif EEDKDE. (B) FLAG-tagged WT MAPK4 and the MAPK4E250A, MAPK4E251A, MAPK4D252A, MAPK4D254A, MAPK4E255A mutants were transfected into HEK293T cells together with AKT1. Immunoprecipitation was performed using anti-FLAG M2 affinity gel followed by Western blots using anti-AKT antibody. (C) FLAG-tagged WT MAPK4 (WT) and the MAPK4D254A, MAPK4E255A mutants were transfected into the HCT116 MAPK4-KO cells. Phosphorylation of AKT was detected with Western blots. Data are representative of at least 3 independent experiments. (D) Sequence alignments of 13 human MAP kinases using T-Coffee and ESPript (34, 35). (E) Alignment of MAPK4 protein sequence around the CD motif across species. (F) WT AKT1 and the AKT1K385A, AKT1K386A, and AKT1K389A mutants were transduced into HCT116 cells together with FLAG-tagged MAPK4. Immunoprecipitation was performed using anti-FLAG M2 affinity gel followed by Western blots using anti-AKT antibody. (G) WT AKT1 and the AKT1K385A, AKT1K386A, and AKT1K389A mutants were transfected into the HCT116 PDK1-KO cells. Phosphorylation of AKT was detected with Western blots. Data are representative of at least 3 independent experiments. (H) The domain architecture of human AKT1 and the sequence alignment of human AKT1, AKT2, and AKT3 proteins. (I) Alignment of AKT1 protein sequence around the D-motif across species.
The AKT1 segment KKDPKQR (positions 385–391) has the potential to be the corresponding positively charged D-motif. To examine whether this KKDPKQR motif provides the potential MAPK4 binding site, we overexpressed mutant AKT1 proteins containing single K/R to A mutations along with FLAG-tagged MAPK4 in the HCT116 cells. We then performed immunoprecipitation using anti-FLAG antibody–conjugated beads followed by Western blot for AKT. Only the AKT1 K386A mutant showed decreased MAPK4 binding (Figure 6F and data not shown). In accordance with this, the K386A mutation largely blocked MAPK4-induced AKT phosphorylation (Figure 6G). It is interesting that K386 in AKT1 is highly conserved among all 3 AKT isoforms and across species (Figure 6, H and I). This conservation may account for MAPK4 activating all 3 AKT isoforms.
mTORC2 activation is indispensable for MAPK4-induced AKT S473 phosphorylation.
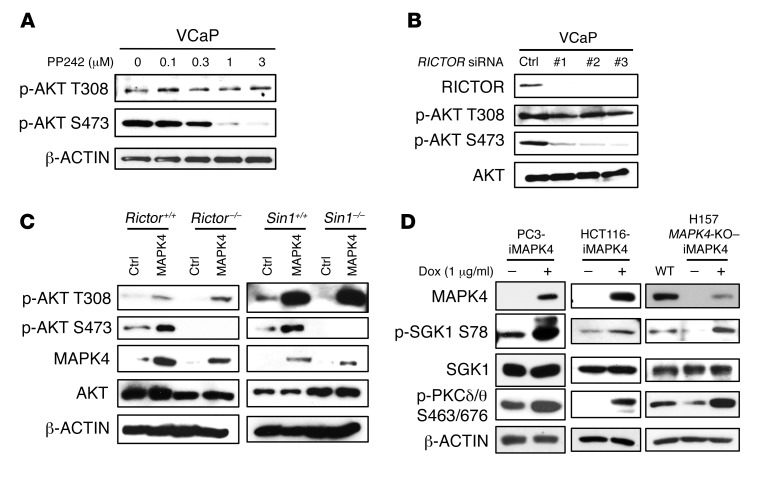
MAPK4 induces AKT phosphorylation at both T308 and S473 in cells, but in the in vitro kinase assay it acts mainly as an AKT T308 kinase. mTORC2, consisting of mTOR, RICTOR, SIN1, and mLST8/GβL, is considered the major AKT S473 kinase (6). To test whether mTORC2 is required for MAPK4-dependent phosphorylation of AKT at Ser473, we first treated the MAPK4-high VCaP cells with PP242, an mTOR kinase–specific inhibitor (22). PP242 dose-dependently inhibited AKT S473 phosphorylation, whereas T308 phosphorylation was less sensitive, indicating that mTOR kinase activity is required for MAPK4-induced S473 phosphorylation (Figure 7A). Consistent with this, knockdown of RICTOR, a key unique component of the mTORC2 complex, significantly inhibited AKT phosphorylation at S473 but not T308 in VCaP cells (Figure 7B). Most importantly, while overexpression of MAPK4 induced AKT phosphorylation at both T308 and S473 in WT MEFs, it strongly induced phosphorylation of AKT at T308, but not S473, in Rictor or Sin1 knockout MEFs that lack a functional mTORC2 complex (Figure 7C). Finally, MAPK4 induced the phosphorylation of mTORC2 substrates, including SGK1 and PKCδ/θ, in PC3 cells and HCT116 cells (Figure 7D). These results indicate that MAPK4 activates mTORC2 (Figure 7D). Consistent with this, phosphorylation of SGK1 and PKCδ/θ was greatly repressed in the H157 MAPK4-KO cells, and such phosphorylation was largely rescued by the ectopic overexpression of MAPK4 (Figure 7D). Altogether, these results suggest that MAPK4 is an upstream effector of mTORC2 and activates mTORC2 for AKT phosphorylation at S473.
Figure 7. mTORC2 mediates MAPK4-induced AKT phosphorylation at S473.
(A) VCaP cells were treated with increasing doses of the mTOR-specific inhibitor PP242 for 2 hours. AKT phosphorylation was detected with Western blots. (B) Three independent siRNAs were transfected into VCaP cells for knockdown of RICTOR. Phosphorylation of AKT was detected with Western blots. (C) MAPK4 was transduced into the Rictor–/–, Rictor+/+, Sin1–/–, and Sin1+/+ MEFs. Western blots were used to determine AKT phosphorylation and confirm MAPK4 expression. Ctrl: control. (D) MAPK4 was ectopically expressed in PC3, HCT116, and the H157 MAPK4-KO cells (clone 6) in a Dox-inducible manner. Western blots were used to detect the phosphorylation of SGK1 and PKCδ/θ. iMAPK4: Dox-inducible overexpression of MAPK4. Data are representative of at least 3 independent experiments.
MAPK4 promotes tumor growth.
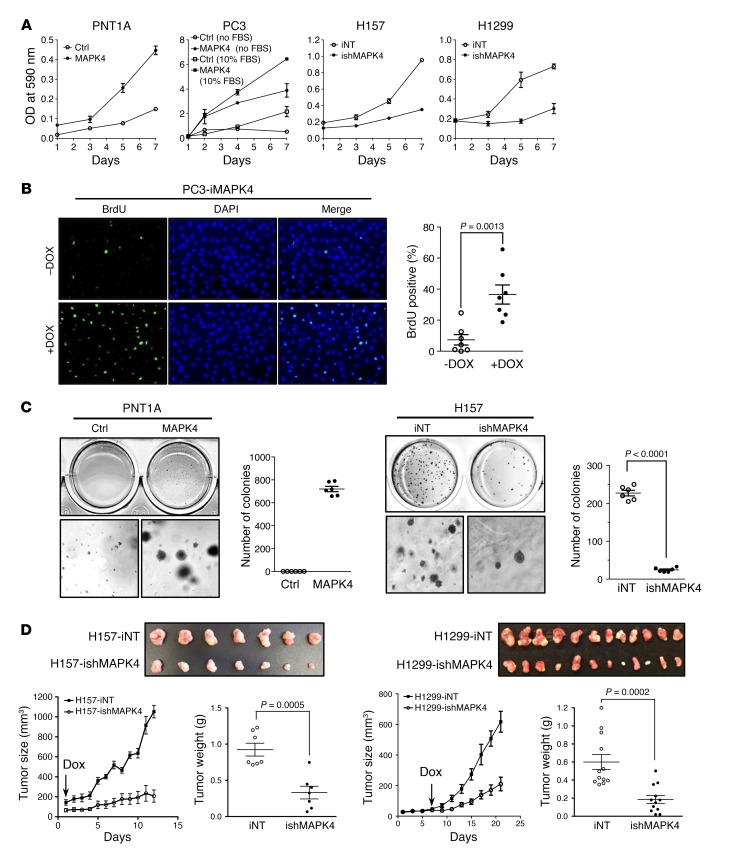
We next determined the impact of MAPK4 overexpression on cell proliferation. As expected from the activation of the AKT/mTOR signaling pathway, MAPK4 overexpression in the MAPK4-low PNT1A and PC3 cells significantly enhanced cell proliferation (Figure 8A). Notably, MAPK4 overexpression also potently promoted PC3 cell proliferation in the absence of serum (Figure 8A). Increased BrdU incorporation confirmed the enhanced proliferation of PC3 cells with Dox-induced MAPK4 overexpression (Figure 8B). Interestingly, MAPK4 overexpression also transformed the PNT1A cells into anchorage-independent growth, implicating MAPK4 as a direct oncogene (Figure 8C). In accordance with this, knockdown of MAPK4 in the MAPK4-high H157 and H1299 cells greatly inhibited cell proliferation and anchorage-independent growth in vitro, and attenuated the growth of xenograft tumors in severe combined immunodeficient (SCID) mice (Figure 8, A, C, and D, and data not shown). Finally, knockdown of MAPK4 in the H157 xenograft tumors correlated with reduced AKT phosphorylation, decreased proliferation, and increased apoptosis (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI97712DS1). Altogether, these results support the tumor-promoting activity of MAPK4.
Figure 8. MAPK4 induces oncogenic outcomes.
(A) MTT assays comparing the proliferation of the control (Ctrl) and MAPK4-overexpressing PNT1A cells, the control and MAPK4-overexpresssing PC3 cells cultured in 10% FBS (open and filled squares) and in serum-free media (open and filled circles), the H157 cells with 4 μg/ml Dox-induced knockdown of MAPK4 (ishMAPK4) and control (iNT), and the H1299 cells with 4 μg/ml Dox-induced knockdown of MAPK4 and control. Data are mean ± SD. (B) BrdU incorporation assay comparing the cell proliferation of the PC3-iMAPK4 cells with (+) and without (–) Dox induction. Data are mean ± SEM. P value by unpaired 2-tailed Student’s t test. (C) Soft agar assays comparing the anchorage-independent growth of the control and MAPK4-overexpressing PNT1A cells (left panels) and the H157 cells with 4 μg/ml Dox-induced knockdown of MAPK4 and control (right panels). Higher magnifications are shown in the bottom panels. Original magnification: ×50. Also shown are the quantification of colony numbers. Data are mean ± SEM. P value by unpaired 2-tailed Student’s t test. (D) Dox-inducible knockdown of MAPK4 significantly delays tumor progression of H157 and H1299 xenografts in SCID mice. A quantity of 1 × 106 iNT or ishMAPK4 cells were s.c. injected into the lateral flanks of SCID mice (iNT: left side; ishMAPK4: right side). Mice began receiving 2 mg/ml Dox and 10% sucrose in drinking water as indicated by the arrow. Tumors were harvested as indicated. Ctrl: control; iNT: Dox-inducible nontargeting control; ishMAPK4: Dox-inducible knockdown of MAPK4. Data are mean ± SEM. P values by unpaired 2-tailed Student’s t test. Data are representative of at least 3 independent experiments.
AKT activation is crucial for the tumor-promoting activity of MAPK4.
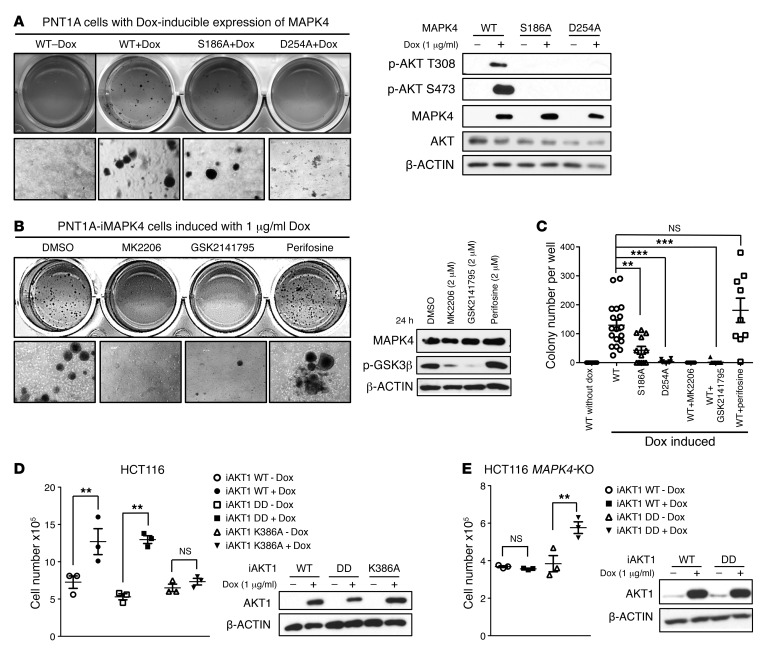
Since AKT is a key oncogenic kinase, we next investigated the functional significance of AKT activation as a mediator of the oncogenic and tumor-promoting activity of MAPK4. Whereas MAPK4 overexpression potently transformed the PNT1A cells into anchorage-independent growth, the S186A mutant significantly reduced and the D254A mutant largely blocked the PNT1A-transforming activity of MAPK4 (Figure 9, A and C). These data indicate that binding and activation of AKT is crucial for the oncogenic activity of MAPK4. In accordance with this, treatments using AKT kinase inhibitors MK2206 and GSK2141795 largely abolished MAPK4-induced, anchorage-independent growth of PNT1A cells (Figure 9, B and C). In contrast, treatments using perifosine, an AKT inhibitor blocking AKT recruitment to the cell membrane for PI3K/PDK1-mediated phosphorylation, neither repressed the MAPK4-induced AKT activation (phosphorylation of GSK3β) nor blocked MAPK4 activities in transforming PNT1A cells into anchorage-independent growth (Figure 9, B and C).
Figure 9. AKT activation is crucial for mediating MAPK4 tumor-promoting activity.
(A) Soft agar assays comparing the anchorage-independent growth of PNT1A cells with Dox-induced expression of WT MAPK4 (WT+Dox), MAPK4S186A (S186A+Dox), and MAPK4D254A mutant (D254A+Dox). The PNT1A-iMAPK4 cells without Dox induction (WT-Dox) were used as control. Higher magnifications are shown in the bottom panels. Original magnification: ×50 (A and B). Western blots were used to confirm comparable levels of Dox-induced MAPK4 expression, and for detection of AKT phosphorylation. (B) Soft agar assays comparing the anchorage-independent growth of the Dox-induced PNT1A-iMAPK4 cells under different treatments as indicated. Western blots were used to assess AKT activation status (phosphorylation of GSK3β) in these cells. (C) Quantification of colony numbers in the soft agar assays in A and B. The group labeled WT contains both the WT+Dox group in A and the DMSO group in B. The q values were calculated and used to account for the FDR in the multiple comparisons (32). No test was applied to the WT+MK2206 group because no colonies were found. Data are mean ± SEM. **q < 0.01, ***q < 0.001. (D) Cell counting comparing the proliferation of HCT116 cells with (+) and without (–) Dox-induced expression of WT AKT1, and the T308D/S473D (DD) and K386A mutants. Western blots were used to confirm comparable levels of induced AKT1 expression. (E) Cell counting comparing the proliferation of HCT116 MAPK4-KO (KO, clone 4) cells with (+) and without (–) Dox-induced expression of WT AKT1 and the T308D/S473D (DD) mutant AKT1. Data are mean ± SEM. P values determined by unpaired 2-tailed Student’s t test. **P < 0.01. Western blots were used to confirm comparable levels of induced AKT1 expression. Data are representative of at least 3 independent experiments.
Since we identified K386 of AKT1 as the key site for AKT-MAPK4 interaction, we examined whether the AKT1K386A mutant loses growth-promoting activity when overexpressed in the MAPK4-high HCT116 cells. Indeed, while both WT AKT1 and a constitutively active AKT1T308D/S473D (DD) mutant promoted HCT116 cell growth, the AKT1K386A mutant exhibited minimal activity (Figure 9D). These data further support MAPK4 activation of AKT as a key pathway for promoting tumor cell growth. In accordance with this, overexpression of the constitutively active AKT1T308D/S473D (DD) mutant but not the WT AKT1 significantly enhanced the growth of the HCT116 MAPK4-KO cells (Figure 9E). Altogether, these results show that AKT activation is crucial for the oncogenic activity of MAPK4.
Discussion
We concluded that MAPK4 provides a PI3K independent pathway for AKT activation. MAPK4 directly phosphorylates AKT T308 and functions as a novel mTORC2 activator to phosphorylate S473. In cell-based models, MAPK4 overexpression is associated with AKT activation, increased proliferation, and xenograft growth, and this association is strongly validated by the AKT activation and decreased patient survival in the small subsets of diverse types of human tumors that overexpress MAPK4. Decreased MAPK4 activity is associated with decreases in both AKT activity and proliferation in cell-based models.
The structure of the MAPK4 protein has not yet been solved. However, our data suggest that the segments containing D254 on MAPK4 and K386 on AKT1 provide a matching pair of electrostatic interactions that underlie MAPK4-AKT binding. In accordance with this, the MAPK4D254A mutant cannot effectively phosphorylate WT AKT1 (Figure 6C) and conversely, WT MAPK4 cannot effectively phosphorylate the AKT1K386A mutant (Figure 6G). It is interesting that the newly identified EEDKDE motif is specific to MAPK4, which may account for the unique ability of MAPK4 to activate AKT.
It has been well documented that the activating phosphorylation of the TxY (TDY) motif of Erk2 induces conformation changes, Erk2 dimerization, and nuclear translocation (23, 24). We found that S186 in the corresponding SEG motif is required for MAPK4 binding and phosphorylation of AKT. Further detailed studies will be required to determine whether MAPK4 S186 phosphorylation also induces allosteric activation and/or dimerization to allow the EEDKDE motif to efficiently bind AKT.
By activating the oncogenic AKT/mTOR signaling pathway, MAPK4 emerges as a novel oncogene. Its oncogenic activity is supported by its ability to transform PNT1A cells into anchorage-independent growth, and to promote cancer cell proliferation and xenograft tumor growth (Figure 8). It is interesting that the PI3K-independent MAPK4 pathway is essential for AKT activation in the MAPK4-high cancer cells. In turn, our results confirm that AKT activation is essential for the growth-promoting effects of MAPK4. This MAPK4-addiction phenotype (25) identifies targeting MAPK4 as an exciting new therapeutic avenue.
Although only a relatively small subset of human cancers express high levels of MAPK4, this MAPK4 overexpression is observed in different cancer types, including low-grade glioma (LGG), glioblastoma (GBM), skin cutaneous melanoma (SKCM), adrenocortical carcinoma (ACC), and to a lesser extent in lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), breast invasive carcinoma (BRCA), thyroid carcinoma (THCA), and bladder urothelial carcinoma (BLCA), among other cancers (Figure 1A). MAPK4 expression in BLCA, LGG, LUAD, and THCA correlates with survival (Figure 1B), indicating that MAPK4 is a novel prognosis marker. Although survival effects were not observed in GBM, SKCM, and ACC, perhaps due to the low number of overexpressing tumors, our results indicate that targeting MAPK4 provides a novel therapeutic strategy to inhibit the oncogenic AKT/mTOR pathway in diverse cancer types.
The PI3K pathway is the target of many ongoing clinical trials. Our results indicate that MAPK4-high cells can evade this pathway and have intrinsic resistance to PI3K inhibitors. Indeed, the MAPK4-high H157 and H1299 cells are resistant to LY294002 treatment (Figure 3A). It will be interesting to determine whether MAPK4-high status is correlated with loss of response to PI3K inhibition in broad clinical trials. It will also be interesting to investigate whether MAPK4 expression and/or activity is induced in tumors that gain resistance to PI3K inhibitors, and whether attacking MAPK4 sensitizes those tumors again to PI3K inhibition.
Methods
Reagents and antibodies.
Recombinant His-AKT1 (inactive, catalog 14-279) was purchased from Upstate/Millipore. Anti-HA (catalog sc-805) and anti-AKT1 (catalog sc-1618) antibodies and Protein A-Agarose (catalog sc-2001) were purchased from Santa Cruz Biotechnology. EZview Red ANTI-FLAG M2 Affinity Gel (catalog F2426), PP242 (catalog P0037), LY294002 (catalog L9908), MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), and anti-β ACTIN antibody were purchased from MilliporeSigma. Antibodies including those against p-AKT T308 (catalog 4056), p-AKT S473 (catalog 9271 and 3787), AKT (catalog 2920), p-GSK3β S9 (catalog 9558), PDK1 (catalog 3062), RICTOR (catalog 2114), PTEN (catalog 9188), p-SGK1 S78 (catalog 5599), SGK1 (catalog 12103), p-PKCδ/θ S643/676 (catalog 9376), CD31 (catalog 77699), and Ki67 (catalog 12202) were from Cell Signaling Technology. Anti-MAPK4 antibodies AP7298b and TA505872 were purchased from Abgent and Origene, respectively.
Plasmids.
MAPK4 cDNA was PCR amplified from the cDNA prepared from LNCaP cells and cloned into the pBMN-IRES-GFP vector between the BamHI and NotI sites. An HA tag and a FLAG/His tag (2 times FLAG and 10 times His) were synthesized and added to the C-terminal end of MAPK4. The tagged and WT MAPK4 was then cloned into the pCDH-CMV-MCS-EF1α-RFP+Puro vector (CD516B-2, System Biosciences) to generate lentiviral vector for MAPK4 overexpression. The pCDNA3.1-PTEN and pCDNA6.1-AKT1-HA, AKT2-HA, and AKT3-HA vectors were provided by Hui-Kuan Lin at Wake Forest University, Winston-Salem, North Carolina. LentiCRISPR v2 (Addgene plasmid 52961) was a gift from Feng Zhang at MIT, Cambridge, Massachusetts (26). GIPZ lentiviral shRNA constructs were purchased from Open Biosystem (Thermo Fisher Scientific). The pInducer10 and pInducer20 were provided by Thomas Westbrook at Baylor College of Medicine, Houston, Texas (27). For inducible overexpression of the target gene, we reengineered the pInducer20 to pInducer20-YF by replacing the vector sequence between attR1 and attR2 with a synthesized multiple cloning site, ACTAGTGCTAGCACGCGTTGATCAGTTAACGTCGACCTCGAG. This allowed the subcloning gene of interest into the vector independent of the Gateway system (Thermo Fisher Scientific). For inducible overexpression of MAPK4, MAPK4 cDNA was cloned into the pInducer20-YF vector between MluI and SalI. For inducible knockdown, the shRNA coding sequence was released from the GIPZ vector and cloned into the pInducer10 vector between MluI and XhoI. The YN and YC vectors for BiFC assay were provided by Xin-Hua Feng at Baylor College of Medicine, Houston, Texas. MAPK4 and AKT1 were cloned into YN and YC vectors, respectively. The obtained YN-MAPK4 and YC-AKT1 vectors were used in the BiFC assay as described in detail below.
Cell culture, transfection, lentivirus, and retrovirus infection.
WT, PDK1–/–, and AKT1/2–/– HCT116 and DLD1 cells were provided by Bert Vogelstein at Johns Hopkins University, Baltimore, Maryland (18). MEF Rictor+/+ and MEF Rictor–/– cells were obtained from Mark Magnuson at Vanderbilt University, Nashville, Tennessee (28). MEF Sin1+/+ and MEF Sin1–/– cells were provided by Bing Su at Yale University, New Haven, Connecticut (29). PNT1A cells were acquired from the European Collection of Authenticated Cell Cultures (ECACC). Other cell lines were obtained from the American Type Culture Collection (ATCC).
GenMute siRNA and LipoD293 DNA transfection reagents (SignaGen Laboratories) were used for siRNA and DNA transfection. For lentiviral-mediated gene delivery, lentiviral-based expression constructs were transfected into HEK293T cells together with the packaging mix in the Trans-lentiviral packaging kit (TLP5914, Thermo Fisher Scientific). Viruses in the conditioned media were harvested 48–96 hours after the transfection and filtered (0.22 μm). The cells were infected with the viral medium for 3 days before starting drug selection. The established stabled cell lines were then expanded for further assays. Retroviruses were packaged and used to infect cells following the similar protocol as we previously described (30).
We established a set of cell lines including HEK293T, PC3, PNT1A, MEF Rictor+/+, MEF Rictor–/–, MEF Sin1+/+, MEF Sin1–/–, HCT116 PDK1–/–, and DLD1 AKT1/2–/– cells with constitutive or doxycycline-inducible (Dox-inducible) overexpression of MAPK4 (HA, FLAG/His, or no tag) by lentiviral- and retroviral-mediated gene delivery. GIPZ and pInducer10 were used for lentiviral-mediated stable and Dox-inducible knockdown of MAPK4 in VCaP, H157, H1299, and HCT116 cells (shRNA NT, G2, and G4 for noninducible, and shRNA iNT, iG2, and iG4 for inducible knockdown of MAPK4). The inducible cells were treated with up to 4 μg/ml Dox for at least 3 days to obtain significant knockdown of MAPK4.
Generation of knockout cell lines.
The coding sequence of sgRNA (ACTTCACTGTTCACTTCAGGGAG) was cloned into the lentiCRISPR-v2 vector. Lentiviral packaging vectors pMD2.G and psPAX2 along with sgRNA-expressing vector were transfected into HEK293T cells using the LipoD293 transfection reagent (SignaGen). Media was changed 24 hours after transfection. The virus-containing supernatant was collected 72 hours after transfection and passed through a 0.22-μm filter. H157 and HCT116 cells in 12-well tissue culture plates were infected in media containing 500 μl virus-containing supernatant and 200 μl fresh culture medium. Virus-containing medium was removed 24 hours after infection and cells were selected with 1–2 μg/ml puromycin for 3 days. After selection, single cells were sorted out by serial dilution in a 96-well plate. Single clones were grown for 2 weeks and validated for loss of MAPK4 via genomic sequencing and immunoblotting.
Immunoprecipitation and affinity purification.
Whole-cell extracts were prepared using immunoprecipitation buffer (20 mM Tris HCl, pH 7.5, 10 mM MgCl2, 2 mM EGTA, 10% glycerol, 137 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 1 mM DTT, phosphatase, and protease inhibitors). Cell lysate was agitated at 4°C for 1 hour and then centrifuged at 4°C for 10 minutes at 13,800 g. The supernatant was precleaned using protein A– or protein G–conjugated beads agitated for 30 minutes at 4°C. Antibodies and IgG as negative control were added and agitated overnight at 4°C. Protein A or G beads were then added and agitated at 4°C for 1 hour. Immunoprecipitates were washed 3 times with 1× PBS and then denatured in 1× SDS sample buffer with 5% β-mercaptoethanol and subjected to SDS-PAGE and immunoblotting analysis. For immunoprecipitation using FLAG antibody, EZview Red ANTI-FLAG M2 Affinity Gel (MilliporeSigma) was washed with RIPA buffer (100 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl, pH 8.0, 1% Triton X-100 with 100× protease inhibitors cocktail solution from GenDEPOT [P3100-005] and the phosphatase inhibitors NaF [10 mM] and Na3VO4 [20 mM]) 3 times and then applied into the cell lysate. After overnight incubation, the beads were washed with 1× PBS for 3 times before Western blots.
Western blot.
Protein samples for Western blot assays were prepared in RIPA buffer. Cell lysate was applied to a brief sonication before protein quantification using a bicinchoninic acid (BCA) protein assay kit (Pierce). An equal amount (5–20 μg) of protein was used in Western blot analysis.
GST pull-down assay.
MAPK4 cDNA was PCR amplified and cloned into the pGEX vector (GE Healthcare) between EcoRI (5′) and SalI (3′) to generate the GST-MAPK4 fusion. The GST-MAPK4 fusion protein and GST protein were overexpressed in E. coli and purified using glutathione sepharose beads following the manufacturer’s instruction (Amersham Biosciences). The purified proteins were run out on SDS gel, and their quality and concentration were estimated by Coomassie blue staining by comparing to serially diluted BSA standard. Recombinant AKT1 was precleared with 10–15 μl glutathione sepharose beads (50%) in 1 ml GST-binding buffer (50 mM Tris-HCl, pH 7.5, 120 mM NaCl, 2 mM EDTA, and 0.1% NP-40) at 4°C for 1 hour. For the GST pull-down assay, 500 ng GST or GST-MAPK4 protein conjugated to glutathione sepharose beads was incubated with 200 ng recombinant AKT1 in GST-binding buffer at 4°C for 16 hours, followed by washing 3 times for 10 minutes each with GST-washing buffer at 4°C (25 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, and 1% Triton X-100). The samples, including an input of 2 ng recombinant AKT1, were subjected to Western blotting using anti-AKT antibody.
Purification of recombinant MAPK4.
The recombinant FLAG/His-tagged MAPK4 protein was purified as follows. The cell lysate was prepared from PNT1A cells overexpressing MAPK4-FLAG/His using native lysis buffer (50 mM sodium phosphate, 500 mM NaCl, 10 mM imidazole, pH 8.0, 0.5% Triton X-100). Recombinant MAPK4-FLAG/His protein was first purified using the native Ni-NTA agarose column (Novagen, catalog 70666). After washing 3 times using native washing buffer (50 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, pH 8.0), the MAPK4 protein was eluted using native elution buffer (50 mM sodium phosphate, 500 mM NaCl, 250 mM imidazole, pH 8.0). The recombinant MAPK4-FLAG/His protein was then further immunopurified using the EZview Red ANTI-FLAG M2 Affinity Gel (MilliporeSigma). The cell lysate from control PNT1A cells was prepared following the same procedure and used as vehicle control.
In vitro kinase assay.
Recombinant AKT1 (Upstate, catalog 14-276) was incubated with the purified recombinant MAPK4 protein (M2-agarose bond) or vehicle control (as described above) in the in vitro kinase buffer (25 mM HEPES, 100 mM potassium acetate, 1 mM MgCl2, 500 μM ATP, pH 7.5) at 37°C for 1 hour. The reaction was stopped by adding equal volume of 2× SDS loading dye and boiling at 100°C for 3 minutes. The samples were then subjected to Western blotting for AKT phosphorylation at T308 and S473.
Bimolecular fluorescence complementation assay.
To directly visualize MAPK4 and AKT interaction in live cells, we performed bimolecular fluorescence complementation (BiFC) as previously described (31). For BiFC assay, the reporter protein (YFP) is split into the N-terminal (YN, aa 1–154) and the C-terminal (YC, aa 155–238) fragments. Each fragment is then fused with 1 of the 2 interacting proteins to be tested. Protein-protein interaction will bring the N- and C-terminal fragments together to allow reforming a functional YFP for fluorescent signal under the fluorescence microscope. Accordingly, we constructed YN-MAPK4 and YC-AKT1 expression plasmids to fuse MAPK4 and AKT1 with the N- and C-terminal fragments of YFP in pRK5 vector, respectively. We then transfected YN-MAPK4 and YC-AKT1 into Hela cells, and used transfection of YN-MAPK4 and YC-vector as a negative control.
MTT assay.
MTT substrate (5 mg/ml) dissolved in 1× PBS was added to the cell cultures and incubated at 37°C for 4 hours. The purple crystals were then dissolved in isopropanol with 0.1% Triton X-100. Absorbance was read at 590 nm. The cells with inducible knockdown of MAPK4, including H157-iNT (inducible nontargeting control), H157-ishMAPK4 (inducible knockdown of MAPK4), H1299-iNT, and H1299-ishMAPK4 cells, were first treated with 4 μg/ml Dox for 3 days before setting up MTT assay, and 4 μg/ml Dox was used throughout the assay.
BrdU incorporation assay.
PC3 cells were seeded at 1 × 105 cells/well on a cleaned coverslip in 12-well plates. Cells were labeled 24–48 hours later with 10 μM BrdU in fresh media for 3 hours at 37°C, fixed with 2% formaldehyde in PBS (pH 7.4) for 15 minutes, and then penetrated using 0.2% Triton X-100 in PBS for 5 minutes. After DNA denaturation in 2 M HCl for 30 minutes, the cells were washed 3 times with PBS and incubated in the immunofluorescence buffer (0.75 g glycine per 100 ml PBS) for 30 minutes at room temperature (RT). The cells were then stained using anti-BrdU antibody (MilliporeSigma, B2531, 1:50) at RT for 1 hour, followed by staining using the goat anti-mouse IgG(H+L) DyLight 488 antibody (Invitrogen, catalog 35502, 1:500) at RT in the dark for 1 hour. After DAPI counterstaining, the coverslips were mounted onto microscope slides for imaging under a fluorescence microscope.
Soft agar colony formation assay.
Cells (1 × 105) were suspended in culture media containing 0.4% low-melting agarose at 37°C and then plated onto supporting gel (0.8% agarose) in a 12-well plate. A quantity of 4 μg/ml Dox was used for inducible knockdown of MAPK4 in the H157-ishMAPK4 cells, and 1 μg/ml Dox was used for inducible expression of MAPK4 in the PNT1A-iMAPK4 cells. Dox at the same concentration (as stated in previous sentence) was also added into the control wells and maintained throughout the study. Colony numbers were counted after inoculation for 3–4 weeks.
Xenograft tumor models.
Male SCID/beige mice, 8–10 weeks old, were purchased from Envigo and used for xenograft studies. Animals were housed in a pathogen-free facility and all animal studies were performed under a protocol approved by the IACUC at the Baylor College of Medicine. H157 or H1299 cells (1 × 106 ) with Dox-inducible knockdown of MAPK4 (ishMAPK4; iNT as control) cells were s.c. injected into the lateral flanks of SCID mice (iNT: left side; ishMAPK4: right side). Mice began receiving 2 mg/ml Dox and 10% sucrose in drinking water for inducible knockdown of MAPK4 in the xenografts. Tumors were monitored every 2 days after the injection. Tumors were harvested as indicated and weighed. Tumor size was calculated as vol = 0.52 × abc (a, b, and c are the maximum length of each dimension of the tumor). Average tumor weight was compared between groups for statistical relevance using the unpaired Student’s t test. P less than 0.05 was considered statistically significant.
Histology and immunohistochemistry.
Tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Sections (5 μm) were mounted onto slides for H&E staining and immunohistochemistry. Immunostaining was performed following a general protocol published previously (30). Antigen retrieval, which consisted of boiling slides in 10 mM citrate buffer (pH 6.0) for 20 minutes, was used for immunohistochemistry staining for detection of p-AKT S473, Ki67, and CD31.
TUNEL assay.
TUNEL assay was performed on paraffin-embedded tumor tissue sections (5 μm) using the In Situ Cell Death Detection Kit, Fluorescein (MilliporeSigma, catalog 11684795910) following the manufacturer’s instructions.
siRNAs and shRNAs for knockdown.
Synthetic siRNAs targeting PDK1 and RICTOR were obtained from Invitrogen as follows, and siRNA against luciferase (siLuc) was used as negative control: siRNA PDK1-1, GGUCAGGCAGCAACAUAGAGCAGUA; PDK1-2, CGAACUCCUUUGAACUGGACUUACA; PDK1-3, GAACUUCGACCAGAGGCCAAGAAUU; siRNA RICTOR-1, UGCAGCAUCGCUUCUUUGCCGGUAU; RICTOR-2, CCAAACAAGGCUGUGAUAUUCUAAA; RICTOR-3, CGAGCCAGUGCAGCCUUGAACUGUU; siRNA MK5-1, GCACCGGCACUUUACAGAGAAGCAA; MK5-2, GCACCAAGCCAAAGGACAGUGUCUA; MK5-3, CCGAGAUGUGAUUGCUCAGUGUAUU; siLuc, GCACUCUGAUUGACAAAUACGAUUU.
Lentiviral shRNA expression constructs were obtained from Open Biosystem (Thermo Fisher Scientific) with the following targeting sequences: pGIPZ-shMAPK4-G2, GGGTTGGTAACAAAGTGGT; pGIPZ-shMAPK4-G4, CGGGAGGAAGACAAGGACG.
Analysis of human tumor molecular data sets.
For pan-cancer analyses, we collected molecular data on 8887 tumors of various histological subtypes (ACC project, n = 79; BLCA, n = 408; BRCA, n = 1094; CESC, n = 304; COAD/READ, n = 625; DLBC, n = 28; GBM, n = 161; HNSC, n = 520; KICH, n = 66; KIRC, n = 533; KIRP, n = 290; LAML, n = 173; LGG, n = 516; LIHC, n = 371; LUAD, n = 515; LUSC, n = 501; OV, n = 263; PAAD, n = 178; PCPG, n = 179; PRAD, n = 497; SKCM, n = 468; THCA, n = 501; UCEC, n = 545; UCS, n = 57) from The Cancer Genome Atlas (TCGA) (11), for which RNA-seq data (v2 platform) were available. A subset of the 8887 TCGA pan-cancer set also had survival data and data on other molecular platforms, as indicated (Figure 1, A–C). TCGA reverse-phase protein array (RPPA) data were from Akbani et al. (12). Survival data were current as of February 11, 2015; patient survival was capped at 200 months. Pearson’s correlation among features was computed using log-transformed expression values.
Statistics.
The statistical relevance in the cell-culture studies and xenograft tumor studies was analyzed using the unpaired 2-tailed Student’s t test. When multiple comparisons were made, q values were calculated and used to account for the false discovery rate (FDR) (32). The statistical relevance of patient survival was analyzed by the log-rank test and the stratified log-rank test (correcting for tumor type). P (q) less than 0.05 was considered significant.
Study approval.
All animal studies were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Author contributions
WW, TS, BD, and FY designed the experiments. WW, TS, BD, CJC, DDM, and FY wrote and revised the manuscript. WW, TS, YM, WZ, QS, and HZ performed the experiments. WW, TS, and YZ did the protein sequence alignments. WW, TS, BD, CJC, DDM, and FY analyzed the data.
Supplementary Material
Acknowledgments
We thank Dr. Jeffrey M. Rosen for critical reading the manuscript, Dr. Zhandong Liu for analyzing data, and Fengjun Chen for technical support. This research was supported by grants from the Department of Defense Congressionally Directed Medical Research Programs (W81XWH-07-1-0200, W81XWH-09-1-0651, W81XWH-13-1-0162, W81XWH-13-1-0163, W81XWH-17-1-0043 to FY), the Cancer Prevention Research Institute of Texas (RP130651 to FY), and the National Institutes of Health (P30 CA125123 to CJC). We also acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine and the MAPK4 as a Novel Therapeutic Target for Human Cancers Cancer Program.
Version 1. 01/28/2019
Electronic publication
Version 2. 03/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(3):1015–1029.https://doi.org/10.1172/JCI97712.
Contributor Information
Wei Wang, Email: ww6@bcm.edu.
Tao Shen, Email: tshen@bcm.edu.
Bingning Dong, Email: bd148049@bcm.edu.
Chad J. Creighton, Email: creighto@bcm.edu.
Yanling Meng, Email: yanling.meng@bcm.edu.
Wolong Zhou, Email: wolongz@bcm.edu.
Qing Shi, Email: henryshi87@hotmail.com.
Hao Zhou, Email: zhou_hao120@163.com.
Yinjie Zhang, Email: yinjiezhang86@gmail.com.
David D. Moore, Email: moore@bcm.edu.
Feng Yang, Email: fyang@bcm.edu.
References
- 1.Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315(6016):239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 2.Auger KR, Serunian LA, Soltoff SP, Libby P, Cantley LC. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 1989;57(1):167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- 3.Franke TF, et al. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81(5):727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 4.Alessi DR, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7(4):261–269. doi: 10.1016/S0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 5.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 7.Klempner SJ, Myers AP, Cantley LC. What a tangled web we weave: emerging resistance mechanisms to inhibition of the phosphoinositide 3-kinase pathway. Cancer Discov. 2013;3(12):1345–1354. doi: 10.1158/2159-8290.CD-13-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13(2):140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aberg E, et al. Regulation of MAPK-activated protein kinase 5 activity and subcellular localization by the atypical MAPK ERK4/MAPK4. J Biol Chem. 2006;281(46):35499–35510. doi: 10.1074/jbc.M606225200. [DOI] [PubMed] [Google Scholar]
- 10.Kant S, Schumacher S, Singh MK, Kispert A, Kotlyarov A, Gaestel M. Characterization of the atypical MAPK ERK4 and its activation of the MAPK-activated protein kinase MK5. J Biol Chem. 2006;281(46):35511–35519. doi: 10.1074/jbc.M606693200. [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akbani R, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275(5300):665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- 16.Langeberg LK, Scott JD. Signalling scaffolds and local organization of cellular behaviour. Nat Rev Mol Cell Biol. 2015;16(4):232–244. doi: 10.1038/nrm3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9(4):789–798. doi: 10.1016/S1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Ericson K, et al. Genetic inactivation of AKT1, AKT2, and PDPK1 in human colorectal cancer cells clarifies their roles in tumor growth regulation. Proc Natl Acad Sci U S A. 2010;107(6):2598–2603. doi: 10.1073/pnas.0914018107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2(2):110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Liu J, Zhang C. Evolutionary history of the vertebrate mitogen activated protein kinases family. PLoS One. 2011;6(10):e26999. doi: 10.1371/journal.pone.0026999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aberg E, Torgersen KM, Johansen B, Keyse SM, Perander M, Seternes OM. Docking of PRAK/MK5 to the atypical MAPKs ERK3 and ERK4 defines a novel MAPK interaction motif. J Biol Chem. 2009;284(29):19392–19401. doi: 10.1074/jbc.M109.023283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7(2):e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90(5):859–869. doi: 10.1016/S0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 24.Khokhlatchev AV, et al. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93(4):605–615. doi: 10.1016/S0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- 25.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297(5578):63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 26.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A. 2011;108(9):3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiota C, Woo JT, Lindner J, Shelton KD, Magnuson MA. Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev Cell. 2006;11(4):583–589. doi: 10.1016/j.devcel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Jacinto E, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127(1):125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 30.Yang F, Tuxhorn JA, Ressler SJ, McAlhany SJ, Dang TD, Rowley DR. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res. 2005;65(19):8887–8895. doi: 10.1158/0008-5472.CAN-05-1702. [DOI] [PubMed] [Google Scholar]
- 31.Shen T, et al. Specific control of BMP signaling and mesenchymal differentiation by cytoplasmic phosphatase PPM1H. Cell Res. 2014;24(6):727–741. doi: 10.1038/cr.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Tommaso P, et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39(Web Server issue):W13–W17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.