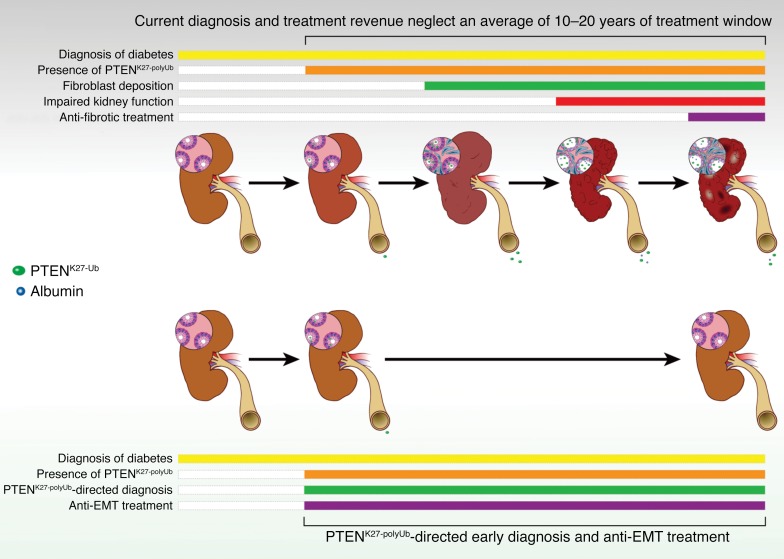
Abstract
Epithelial-mesenchymal transition (EMT) contributes significantly to interstitial matrix deposition in diabetic kidney disease (DKD). However, detection of EMT in kidney tissue is impracticable, and anti-EMT therapies have long been hindered. We reported that phosphatase and tensin homolog (PTEN) promoted transforming growth factor beta 1 (TGF-β), sonic hedgehog (SHH), connective tissue growth factor (CTGF), interleukin 6 (IL-6), and hyperglycemia-induced EMT when PTEN was modified by a MEX3C-catalyzed K27-linked polyubiquitination at lysine 80 (referred to as PTENK27-polyUb). Genetic inhibition of PTENK27-polyUb alleviated Col4a3 knockout–, folic acid–, and streptozotocin-induced (STZ-induced) kidney injury. Serum and urine PTENK27-polyUb concentrations were negatively correlated with glomerular filtration rate (GFR) for diabetic patients. Mechanistically, PTENK27-polyUb facilitated dephosphorylation and protein stabilization of TWIST, SNAI1, and YAP in renal epithelial cells, leading to enhanced EMT. We identified that a small molecule, triptolide, inhibited MEX3C-catalyzed PTENK27-polyUb and EMT of renal epithelial cells. Treatment with triptolide reduced TWIST, SNAI1, and YAP concurrently and improved kidney health in Col4a3 knockout–, folic acid–injured disease models and STZ-induced, BTBR ob/ob diabetic nephropathy models. Hence, we demonstrated the important role of PTENK27-polyUb in DKD and a promising therapeutic strategy that inhibited the progression of DKD.
Keywords: Cell Biology, Nephrology
Keywords: Chronic kidney disease, Diabetes, Proteases
Introduction
Diabetic kidney disease (DKD) affects approximately 40% of diabetic patients and is one of the major causes of morbidity and mortality (1). In addition to cardiovascular disease, which is a leading cause of death in persons with DKD, about 20%–40% of those with DKD will progress to end-stage renal disease (ESRD) (2). In addition to hyperglycemia, which is one of the main risk factors for the development of DKD (3), growth factors, including transforming growth factor beta 1 (TGF-β), interleukin 6 (IL-6), connective tissue growth factor (CTGF/CCN2), and sonic hedgehog (SHH), are key regulators of kidney fibrosis (4–8). Fibrosis is characterized by the accumulation of myofibroblasts, the collagen-depositing cells. In addition to myofibroblasts derived from bone marrow, resident fibroblasts generated by epithelial-mesenchymal transition (EMT) play an important role in the development of kidney fibrosis (9). Lineage tracing analysis demonstrates that EMT in tubular epithelial cells is necessary for the progression of kidney fibrosis (10). Rather than directly transforming into myofibroblasts, the renal epithelial cells undergo a partial EMT and dedifferentiate to secrete cytokines and chemokines that promote fibrogenesis and inflammation, which in turn promote the development of fibrosis (11). Genetic evidence indicates that master regulators of EMT, such as TWIST and SNAI1, play important roles in the progression of kidney fibrosis (12). However, the regulation of TWIST and SNAI1 under hyperglycemic conditions or other stimuli is largely unknown. Most importantly, treatment strategies that inhibit EMT and alleviate kidney disease progression and related mortality remain elusive.
Phosphatase and tensin homolog (PTEN) is a negative regulator of the protein kinase B (AKT/PKB) and mTOR (mechanistic target of rapamycin) pathways (13). Conventional knockout (KO) of Pten in mice is embryonically lethal (14). However, mice with transgenic expression of activated AKT exhibit a different spectrum of tumor development (15). Therefore, it is highly likely that PTEN exhibits biological roles other than dephosphorylation of phosphatidylinositol (3,4,5)-triphosphate (PIP3), such as acting as a protein phosphatase in vivo. Posttranslational modifications, including ubiquitination, are major regulatory mechanisms that control the protein stability, subcellular localization, and enzymatic activity of PTEN (16). The level of unmodified PTEN is dynamically regulated in kidney injury (17), suggesting that PTEN may harbor posttranslational modifications, which play important roles in kidney disease. However, the function, mechanism, and posttranslational modification of PTEN in kidney disease remain unclear.
We report that PTEN promotes TGF-β, SHH, CTGF, IL-6, and hyperglycemia-induced EMT when PTEN is modified with a K27-linked polyubiquitin chain (K27-polyUb) at lysine 80 (referred to as PTENK27-polyUb). Homozygous mice harboring the Pten K80R mutant abolished EMT and alleviated Col4a3-KO–, folic acid–, and streptozotocin-induced (STZ-induced) kidney injury. Elevated concentrations of PTENK27-polyUb were observed in blood and urine samples of diabetic patients, which is negatively correlated with estimated GFR (eGFR). Furthermore, elevated PTENK27-polyUb was detected in the kidney tissue of patients with DKD. We screened and characterized that a small molecule, triptolide, specifically binds to the RING domain of MEX3C to inhibit MEX3C-UBE2S interaction and MEX3C-catalyzed PTENK27-polyUb, leading to inhibition of EMT in renal epithelial cells. Treatment with triptolide significantly improved kidney health and extended median survival times in diabetic mouse models compared with vehicle administration. These findings suggest that PTENK27-polyUb plays important roles in the development of DKD. Targeting MEX3C-dependent PTENK27-polyUb using small molecular compounds could serve as a promising strategy for preventing the progression of renal fibrosis before diabetic patients reach ESRD.
Results
Genetic inhibition of PTENK27-polyUb reduces renal fibrosis.
We established that MEX3C catalyzes K27-linked polyubiquitination of PTEN (referred as PTENK27-polyUb) at lysine 80. K80R mutation abolishes the K27-linked polyubiquitination. PTENK27-polyUb switches the enzymatic activity of PTEN from a phosphoinositol phosphatase to a protein serine/threonine phosphatase. PTENK27-polyUb dephosphorylates TWIST1, SNAI1, and YAP1, leading to accumulation of these proteins and EMT (18). Given that EMT plays important roles in promoting organ fibrosis, we assessed whether MEX3C-PTENK27-polyUb could be involved in DKD, which eventually leads to kidney fibrosis and ESRD (19).
We generated a genetically edited mouse model that replaced lysine 80 with arginine using CRISPR/Cas9 system-mediated genomic editing (20). We detected a single-nucleotide (AAA-AGA) mutation after the founder mice were validated by Sanger sequencing. PtenK80R/K80R mice exhibit similar homeostasis development and renal function (Supplemental Figure 1, A–G; supplemental material available online with this article; https://doi.org/10.1172/JCI121987DS1). We crossed PtenK80R/K80R mice with Col4a3–/– mice to generate Col4a3+/– PtenWT/K80R, Col4a3+/– PtenK80R/K80R, Col4a3–/– PtenWT/K80R, and Col4a3–/– PtenK80R/K80R mice in a mixed genetic background of FVB/129S1 (Figure 1A). The depletion of the Col4a3 gene and Pten K80R mutant exhibited minimal effect on the body weight and organ development of young animals (Supplemental Figure 2, A–D).
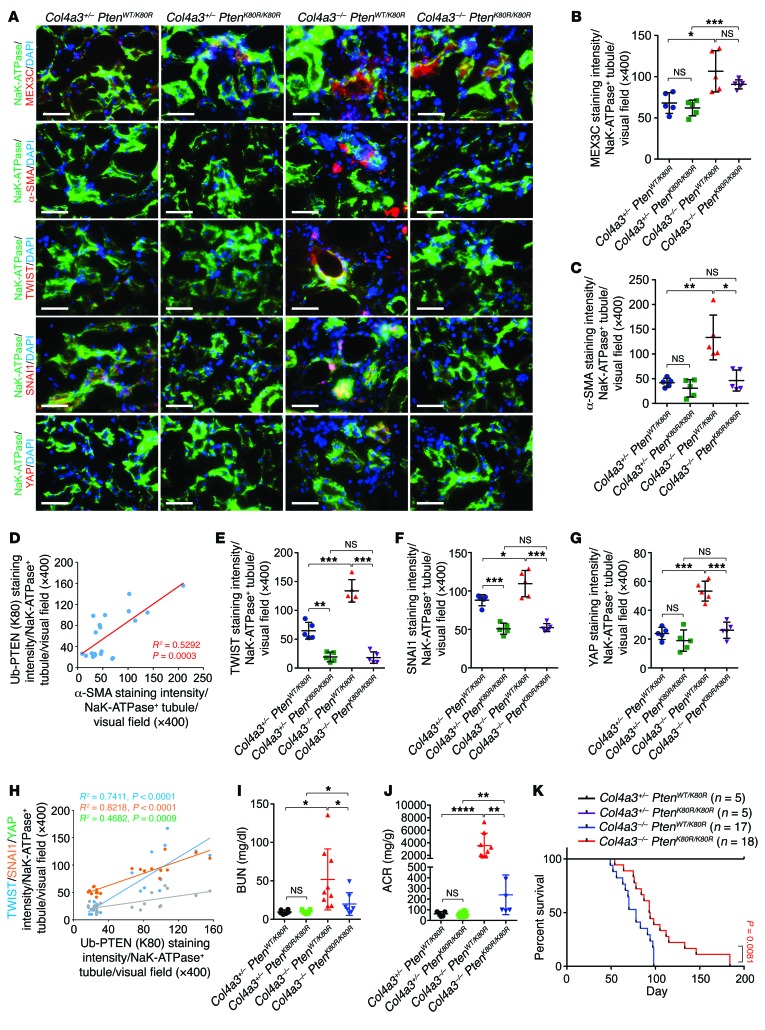
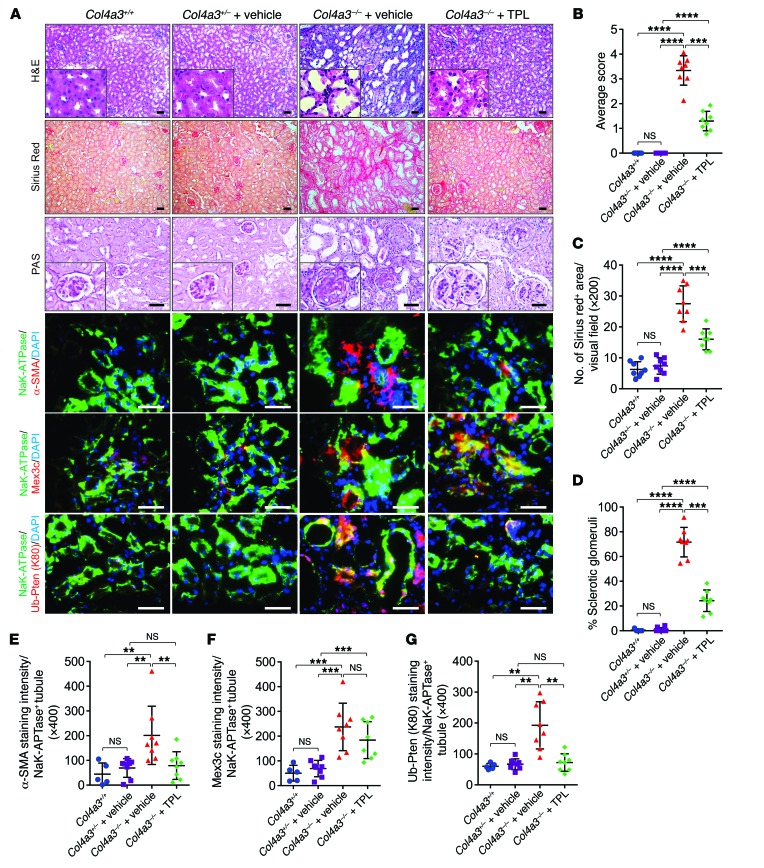
Figure 1. PtenK27-polyUb is required for renal fibrosis.
(A) Scheme of the experimental approach. (B) Representative images of H&E staining, Sirius red staining, PAS staining, and immunofluorescence staining using indicated antibodies in Col4a3+/– PtenWT/K80R, Col4a3+/– PtenK80R/K80R, Col4a3–/– PtenWT/K80R, and Col4a3–/– PtenK80R/K80R kidneys. Black circles: sclerotic glomeruli; scale bars: 100 μm. (C–F) Statistical analysis of Ub-PTEN (K80) per Na+K+-ATPase–positive tubules (C), tubular injury score (D), percentage of Sirius red–positive area (E), and percentage of sclerotic glomeruli (F) per Na+K+-ATPase–positive tubules per visual field. All error bars indicate SD; n = 5 animals and 6–8 independent fields per animal were calculated (1-way ANOVA). NS indicates P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We first demonstrated the presence of PTENK27-polyUb in fibrotic tubules using site-specific antibodies targeting PTENK27-polyUb [Ub-PTEN (K80)]. Heterozygous Col4a3+/– PtenWT/K80R kidneys exhibited low levels of PTENK27-polyUb. PTENK27-polyUb was significantly higher in Col4a3–/– PtenWT/K80R kidneys when the animals suffered from kidney fibrosis (Figure 1, B and C). The tubular areas for each measurement between experimental conditions were similar (Supplemental Figure 2E). PTENK27-polyUb was undetectable in Col4a3+/– PtenK80R/K80R and Col4a3–/– PtenK80R/K80R kidneys, which both harbor the homozygous Pten K80R mutant (Figure 1, B and C).
FVB mice are known for their susceptibility to kidney disease (21). By 10 weeks of age, no significant phenotypic abnormalities were observed in the kidneys of Col4a3+/– mice with the homozygous Pten mutant (Col4a3+/– PtenK80R/K80R) compared with littermates with the heterozygous Pten mutant (Col4a3+/– PtenWT/K80R) (Figure 1, B and D). However, Col4a3–/– PtenWT/K80R animals with a mixed FVB/129S1 genetic background exhibited severe kidney fibrosis at the same age. Kidneys of Col4a3–/– PtenWT/K80R mice exhibited fewer healthy tubules than those of Col4a3+/– PtenWT/K80R mice (Figure 1, B and D). Kidneys of Col4a3–/– mice with the homozygous Pten mutant (Col4a3–/– PtenK80R/K80R) exhibited significantly reduced injury score compared with kidneys with the heterozygous Pten mutant (Col4a3–/– PtenWT/K80R) (Figure 1, B and D). Kidneys with homozygous depletion of Col4a3 (Col4a3–/– PtenWT/K80R) showed greater accumulation of connective fibers compared with heterozygous depletion of the Col4a3 gene (Col4a3+/– PtenWT/K80R), which was significantly alleviated in the Col4a3–/– PtenK80R/K80R kidneys (Figure 1, B and E). Genetic inhibition of PTENK27-polyUb reduced glomerulosclerosis significantly (Figure 1, B and F). The staining intensities of MEX3C in Col4a3–/– PtenWT/K80R kidney tubules are higher compared with Col4a3+/– PtenWT/K80R animals (Figure 2, A and B and Supplemental Figure 2F). These data suggest that MEX3C-dependent PTENK27-polyUb plays important roles in regulating kidney fibrosis.
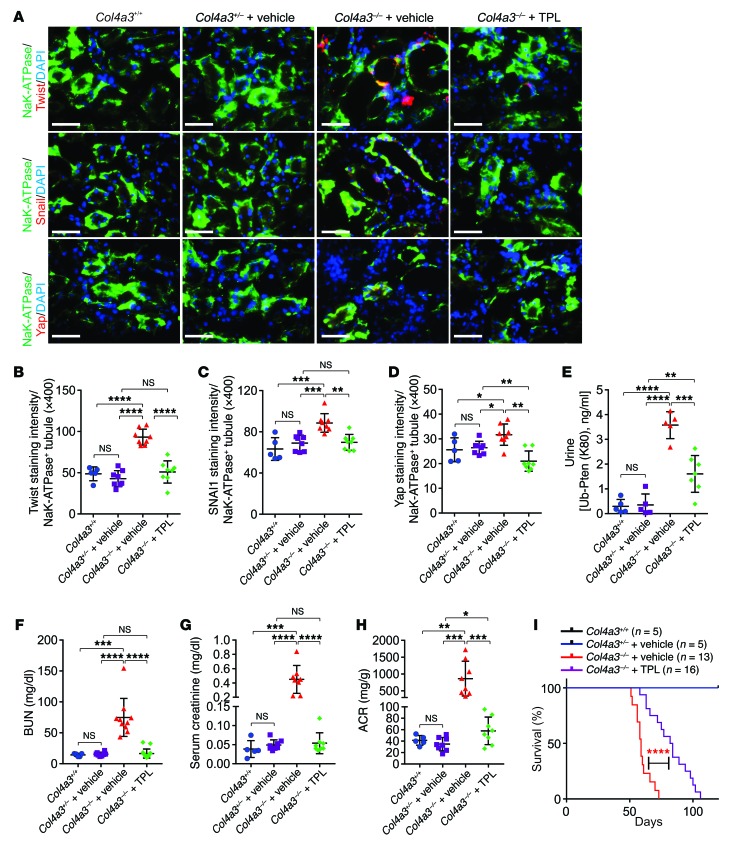
Figure 2. Pten K80R mutant reduces EMT during renal fibrosis.
(A) Representative images of immunofluorescence staining using indicated antibodies of Col4a3+/– PtenWT/K80R, Col4a3+/– PtenK80R/K80R, Col4a3–/– PtenWT/K80R, and Col4a3–/– PtenK80R/K80R kidneys. Scale bars: 100 μm. (B–C) Statistical analysis of staining intensity of MEX3C (B) and α-SMA (C) per Na+K+-ATPase–positive tubules. Error bars indicate SD; n = 5 animals and 6 independent fields per animal were calculated (1-way ANOVA). (D) Pearson correlation of the staining intensity of Ub-PTEN (K80) with α-SMA per Na+K+-ATPase+ tubule (n = 20, Pearson χ2 test). (E–G) Statistical analysis of TWIST-staining intensity (E), SNAI1-staining intensity (F), and YAP-staining intensity (G) per Na+K+-ATPase positive tubules. Error bars, SD, n = 5 animals and 6 independent fields per animal were calculated (1-way ANOVA). (H) Pearson correlation of the staining intensity of Ub-PTEN (K80) with TWIST, SNAI1, and YAP per Na+K+-ATPase+ tubule (n = 20, Pearson χ2 test). (I–J) Detection of BUN (I), or ACR (J) in blood or urine samples of Col4a3+/– PtenWT/K80R, Col4a3+/– PtenK80R/K80R, Col4a3–/– PtenWT/K80R, and Col4a3–/– PtenK80R/K80R animals. Error bars indicate SD; n = 6, 7, 9, 7 (I); n = 5, 8, 9, 6 (J) respectively (1-way ANOVA). (K) Kaplan-Meier survival analysis of Col4a3+/– PtenWT/K80R, Col4a3+/– PtenK80R/K80R, Col4a3–/– PtenWT/K80R, and Col4a3–/– PtenK80R/K80R animals (n = 5, 5, 17, and 18 respectively, log-rank test). NS indicates P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
One of the major morphological characteristics of myofibroblasts is the expression of α-smooth muscle actin (α-SMA) (22). In Col4a3–/– mice, the accumulation of α-SMA–positive staining in the kidneys of mice with the homozygous Pten mutant (Col4a3–/– PtenK80R/K80R) was less than that in mice with the heterozygous Pten mutant (Col4a3–/– PtenWT/K80R) (Figure 2, A–C and Supplemental Figure 2G). The staining intensities of PTENK27-polyUb correlated with α-SMA (Figure 2D).
Since PTENK27-polyUb stabilizes TWIST, SNAI1, and YAP based on its protein serine/threonine phosphatase activity (18), we determined the status of TWIST, SNAI1, and YAP in Col4a3+/– PtenWT/K80R, Col4a3+/– PtenK80R/K80R, Col4a3–/– PtenWT/K80R, and Col4a3–/– PtenK80R/K80R mice. TWIST and SNAI1 promote kidney fibrosis after unilateral ureteral obstruction (UUO) (9). In heterozygous Col4a3+/– PtenWT/K80R kidneys, low expression of TWIST, SNAI1, and YAP was observed; however, in Col4a3–/– mice with heterozygous Pten K80R, elevated staining intensity of TWIST, SNAI1, and YAP in Na+K+-ATPase–positive tubules was detected. TWIST, SNAI1, and YAP were abolished in PtenK80R/K80R animals regardless of the status of the Col4a3 gene (Figure 2, A, E–G and Supplemental Figure 2, H–J). The status of PTENK27-polyUb was highly correlated with the presence of TWIST, SNAI1, and YAP in mouse kidneys (Figure 2H). These data suggest that PTENK27-polyUb may facilitate EMT through stabilization of TWIST, SNAI1, and YAP in vivo.
Next, we determined the kidney function of Col4a3+/– PtenWT/K80R, Col4a3+/– PtenK80R/K80R, Col4a3–/– PtenWT/K80R, and Col4a3–/– PtenK80R/K80R mice, finding that blood urea nitrogen (BUN) and albumin-to-creatinine ratio (ACR) were similar in Col4a3+/– PtenK80R/K80R and Col4a3+/– PtenWT/K80R littermates (Figure 2, I and J). Homozygous Col4a3–/– animals with the heterozygous Pten K80R mutant exhibited significant elevation of BUN and ACR, which was significantly alleviated in Col4a3–/– PtenK80R/K80R double homozygous animals (Figure 2, I and J). Further, Col4a3–/– PtenK80R/K80R mice exhibited extended survival time compared with Col4a3–/– PtenWT/K80R animals (Figure 2K). These observations indicated that inhibiting PTENK27-polyUb may alleviate the progression of kidney fibrosis and improve kidney function.
PTENK27-polyUb is required for growth factor–induced EMT.
Using human kidney tubular epithelial HK-2 cells, we found that in addition to high glucose (25 mM), growth factors/cytokines (CTGF, IL-6, SHH, TGF-β, Cripto-1, and SDII-1) induced PTENK27-polyUb (Figure 3A). Using human HK-2 and mouse tubular epithelial MCT cells (23), we confirmed that CTGF, IL-6, SHH, and TGF-β triggered PTENK27-polyUb and reduced the level of unmodified PTEN in both cell lines (Figure 3B). Previous research demonstrates the importance of CTGF, IL-6, SHH, and TGF-β in promoting EMT, fibrosis, and inflammation associated with kidney diseases (24–27). Knocking down MEX3C using siRNAs abolished the growth factor–induced PTENK27-polyUb, validating that MEX3C acts as an E3 ligase to catalyze PTENK27-polyUb (Figure 3C).
Figure 3. PTENK27-polyUb is required for growth factors-induced EMT.
(A) Sandwich ELISA assay detection of PTENK27-polyUb using Ub-PTEN (K80) antibody in HK-2 cells treated with 25 mM glucose or indicated growth factors for 1 hour. Error bar indicates SD; n = 3 independent experiments (Student’s t test). (B–C) Immunoblotting detection using indicated antibodies of HK-2 or MCT cells transfected with indicated siRNA (C), followed by treatment with indicated human or mouse growth factors for 1 hour. Scr: scramble. (D–F) Representative images (D) and statistical analysis of vimentin-positive cells (E) and SNAI1-positive cells (F) in HK-2 cells with or without PTEN sgRNAs transducted with indicated lentivirus. The cells were subjected to vehicle or indicated growth factor treatments for 72 hours. Scale bars: 50 μm. Error bar indicates SD; n = 6 independent experiments (1-way ANOVA). NS indicates P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Mechanistically, high glucose–induced PTENK27-polyUb showed minimal effect on the subcellular localization of PTEN and protein stability (Supplemental Figure 3A). HK-2 with PTEN depletion using sgRNAs was reintroduced with the PTEN WT or K80R mutant, finding that the cellular PIP3 concentration increased in the presence of WT PTEN but not the PTEN K80R mutant (Supplemental Figure 3B). The expression of PTEN WT/mutant did not affect the enzymatic activity of PI3K (Supplemental Figure 3C). Expression of PTEN WT in PTEN-null HK-2 cells led to reduced phosphorylation of TWIST and SNAI1, suggesting that high glucose–induced PTENK27-polyUb at the K80 residue facilitates dephosphorylation of TWIST and SNAI1 (Supplemental Figure 3D). This dephosphorylation event is independent of the protein phosphatase activity of SHP2 or PP2A (Supplemental Figure 3, E and F).
We knocked out PTEN in HK-2 cells using sgRNAs and restored the expression of PTEN by expressing the WT or K80R mutant. TGF-β, SHH, IL-6, and CTGF all effectively induced the downregulation of E-cadherin and upregulation of Vimentin/Snail as mesenchymal markers (Figure 3, D–F). Depletion of PTEN abolished TGF-β, SHH, IL-6, and CTGF-induced EMT. Expression of WT PTEN restored the growth factor–induced morphology alteration, but the PTEN K80R mutant failed to do so (Figure 3, D–F).
PTENK27-polyUb is associated with DKD.
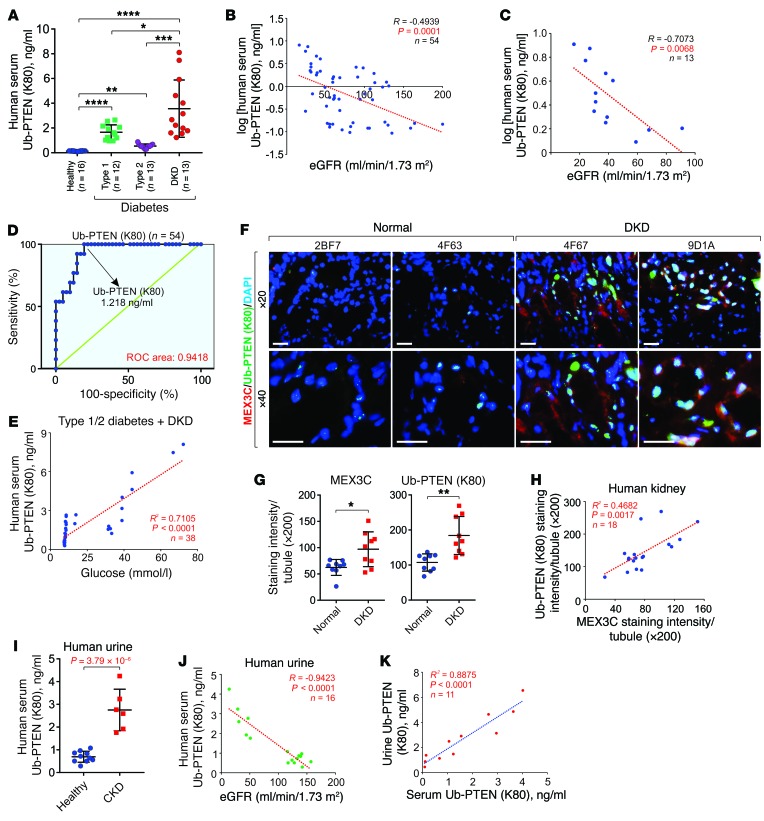
We measured the PTENK27-polyUb concentration in 54 human serum samples using a modification-specific antibody (Figure 4A, Table 1, and Supplemental Table 1). Low concentrations of PTENK27-polyUb were detected in the serum samples of healthy kidney donors compared with serum from patients with either type 1 or type 2 diabetes (Figure 4A). Serum from patients with DKD contained even higher concentrations of PTENK27-polyUb than the diabetic patients (Figure 4A). The presence of serum PTENK27-polyUb concentration is negatively correlated with estimated GFR (eGFR) (Figure 4, B and C). The sensitivity and specificity of PTENK27-polyUb as a biomarker of DKD are 92.31% and 85.37%, respectively, with a receiver operating characteristic (ROC) area of 0.9418 (Figure 4D). Serum PTENK27-polyUb is also correlated with the blood glucose levels of type 1/2 diabetic and DKD patients but not the age of donors (Figure 4E and Supplemental Figure 4A).
Figure 4. PTENK27-polyUb is correlated with DKD.
(A) Presence of PTENK27-polyUb in human serum samples from healthy donors, type 1 diabetic patients, type 2 diabetic patients, or patients with DKD (n = 16, 12, 13, 13 serum samples, respectively; 1-way ANOVA). (B) Pearson’s correlation of serum PTENK27-polyUb with eGFR (n = 54 serum, Pearson χ2 test). (C) Pearson’s correlation of serum PTENK27-polyUb with eGFR of DKD patients (n = 13 serum, Pearson χ2 test). (D) ROC curves for analytes in serum PTENK27-polyUb for the diagnosis of DKD patients. (E) Pearson’s correlation of serum PTENK27-polyUb with blood glucose level in patients with type 1/2 diabetes and DKD (n = 38 serum, Pearson χ2 test). (F–G) Representative images (F) and statistical analysis of immunofluorescence staining of MEX3C (G, left) and PTENK27-polyUb (G, right) and in kidney tissues of healthy donors and DKD patients. Error bar indicates SD of staining intensities of 6 random fields per tissue (n = 9 and 9, respectively; Student’s t test). Scale bars: 100 μm. (H) Pearson’s correlation of MEX3C and PTENK27-polyUb in human kidney tissues (n = 18, Pearson χ2 test). (I) Measurement of PTENK27-polyUb concentration in human urine samples from healthy donors or DKD patients (n=10 and 6 urine samples), Error bars, SD, Student’s t-test. (J) Pearson’s correlation of urine PTENK27-polyUb with eGFR of healthy and DKD patients (n=16 urine samples, Pearson χ2 test). (K) Measurement of PTENK27-polyUb concentration in paired serum and urine samples of healthy donors, type 2 diabetes patients, and DKD patients (n = 11, Pearson χ2 test). NS indicates P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Table 1. Basic characteristics of the cohort of human serum samples used in the present study.

Of the mex3 RNA binding family members, MEX3C is the member most abundantly expressed in the kidneys (Supplemental Figure 4B). Therefore, we determined the status of PTENK27-polyUb and protein levels of MEX3C in human kidney tubules of healthy kidney donors and patients with DKD. Protein levels of MEX3C and the status of PTENK27-polyUb were both elevated in glomeruli and tubules from human DKD tissues compared with normal kidneys (Figure 4, F and G and Supplemental Figure 4, C and D). The levels of MEX3C and PTENK27-polyUb were correlated in human kidney tissues (Figure 4H).
We then determined the presence of PTENK27-polyUb in human urine samples from healthy kidney donors or patients with DKD. PTENK27-polyUb concentrations were significantly higher in urine samples from DKD patients than from healthy donors (Figure 4I). PTENK27-polyUb levels in the urine were negatively correlated with eGFR (Figure 4J). We further determined the concentration of PTENK27-polyUb in paired serum and urine samples of healthy donors, diabetic patients, and DKD patients (Supplemental Table 1), finding that serum PTENK27-polyUb concentration is faithfully correlated with urine concentration of PTENK27-polyUb (Figure 4K). Furthermore, in this cohort, serum and urine concentrations of PTENK27-polyUb were elevated in type 2 diabetic patients compared with healthy donors. The status of serum and urine concentrations of PTENK27-polyUb was further increased in DKD patients compared with type 2 diabetic patients (Supplemental Figure 4, E and F). Our data suggest that PTENK27-polyUb detected in DKD serum and urine specimens correlates with the presence of DKD.
Characterization of triptolide as EMT inhibitor.
Although EMT is a key factor in promoting kidney fibrosis, anti-EMT therapy has long been hindered because the master regulators of EMT, including TWIST and SNAI1, are transcription factors (28). Hence, we aimed to identify potential small molecule inhibitors that target MEX3C, which we hypothesized may reduce the progression of kidney fibrosis. We established a screening platform that took advantage of a Ub-PTEN (K80) antibody and high-throughput ELISA assay to detect the in vitro formation of this poly-ubiquitin chain in the presence of E1, E2, MEX3C, PTEN, and ubiquitin (Figure 5A). We assembled a validated library of 2027 bioactive compounds, comprised of FDA approved drugs, known inhibitors, and natural compounds (catalog Z87653, Selleck Chemicals). The screening revealed that a natural compound, triptolide (TPL), exhibited effective inhibition of the enzymatic activity of MEX3C in vitro, potentially serving as a potent E3 inhibitor (Figure 5A).
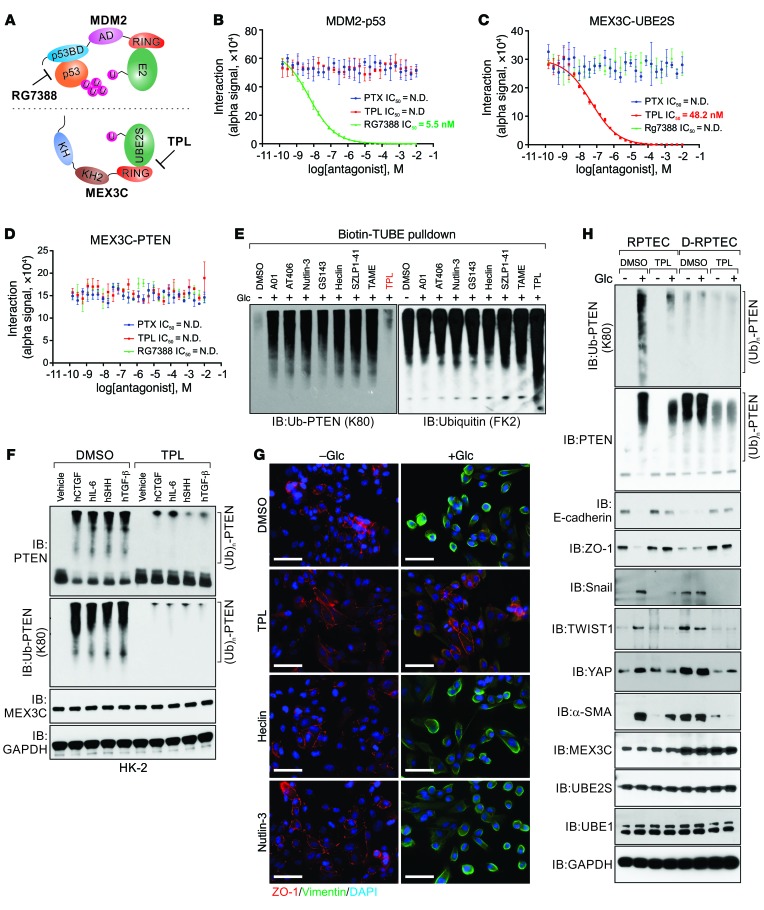
Figure 5. Characterization of triptolide as a MEX3C inhibitor.
(A) Top panel: flow chart of sandwich ELISA using His6-TUBE and Ub-PTEN (K80) antibody screening customized compound library and the generation of PTENK27-polyUb was detected by OD450. Bottom panel: The log2 of relative fold change of polyUb chain formation activity was shown. (B) Illustration of Bio-Linker and Bio-TPL. (C) Streptavidin pulldown using Bio-Linker or Bio-TPL and Col4a3+/+ or Col4a3–/– kidneys. The Bio-TPL–binding proteins were excised (blue box) and subjected to LC-MS analysis. (D) Protein scores of protein identification of LC-MS. (E) Competition curve determination of KD value of TPL and indicated recombinant proteins with 2-serial fold dilution. Error bars indicate SD; n = 3 independent experiments. (F) Competition curve determination of IC50 value of TPL on the enzymatic activity of indicated E3 ligases. The IC50 values of TPL against each E3 ligase are shown. Error bars indicate SD; n = 3 independent experiments. (G) In vitro ubiquitination assay using indicated recombinant proteins with or without TPL followed by immunoblotting detection (ERCC3/MEX3C molar ratio 10:1). (H–I) Streptavidin pulldown using Bio-TPL and indicated recombinant proteins of MEX3C followed by immunoblotting detection using anti-FLAG antibody.
To demonstrate the target of TPL, we synthesized biotinylated-TPL (Bio-TPL) (Figure 5B) and performed a chemical pulldown using Col4a3+/+ or Col4a3–/– kidneys (Figure 5C and Supplemental Figure 5A). The protein targets pulled down by Bio-TPL were subjected to liquid chromatography–mass spectrometry (LC-MS) for protein identification (Figure 5C, blue boxes, and Supplemental Table 2). TPL associated with MEX3C, LIN-41, ERCC3, TRIM63, PSD10, and DCT91, by which the interaction and inhibitory effect of TPL against ERCC3 and DCTP1 was previously reported (29, 30), validating our chemical pulldown (Figure 5D). We determined the binding affinity between Bio-TPL and recombinant MEX3C, TRIM63, and TRIM71 (LIN-41), which are all potential E3 ligases, finding that TPL exhibits robust interaction with MEX3C with a KD value of 27.5 nM (Figure 5E). Using the representative E3 ligase of Ring-finger E3 ligase (TRIM63, MDM2, RNF4, and MEX3C), U-box E3 ligase (UBE3A), and HECT family of E3 ligase (ITCH) (31), TPL showed effective inhibition of MEX3C with an IC50 value of 48 nM (Figure 5F). The MEX3C-dependent PTENK27-polyUb was abolished upon TPL treatment in cell-free assays, by which the inhibitory effect of TPL was neutralized by a TPL-binding protein, ERCC3 (XPB) (ERCC3/MEX3C molar ratio 10:1) (Figure 5G).
MEX3C has 2 K homology (KH) RNA binding domains and a C-terminal RING type zinc finger domain. Deletion mutants (ΔZNF) abolished the interaction between Bio-TPL and MEX3C, suggesting that TPL targets the RING-type zinc finger domain of MEX3C (Figure 5H and Supplemental Figure 5B). The conserved cysteine and histidine residues in the RING type zinc finger domain are essential in mediating domain folding and association with E2 ubiquitin-conjugating enzymes (32). MEX3C C806S and H625A mutants impaired the MEX3C-TPL interaction (Figure 5I and Supplemental Figure 5C), further validating TPL association with the RING-type zinc finger domain of MEX3C.
E3 ligase inhibitors block E2-E3 interactions and E3-substrate interactions (33). As an example, an MDM2 inhibitor, RG7388, abolishes MDM2-p53 interaction (34) (Figure 6A). We performed an alpha assay to evaluate MEX3C-PTEN and MEX3C-UBE2S interactions in the presence of TPL. The MDM2-p53 interaction in the presence of RG7388 (34) was included as a positive control and paclitaxel (PTX) (35) was included as negative control (Figure 6, B–D). Consistent with a previous report, our data indicates that RG7388 effectively inhibits MDM2-p53 interaction with a KD value of 5.5 nM (34). By contrast, PTX and TPL exhibit minimal effects on MDM2-p53 interaction (Figure 6B). The interaction between MEX3C and PTEN was not significantly affected in the presence of PTX, TPL, or RG7388 (Figure 6D). However, the MEX3C-UBE2S (E3-E2) association was abolished in the presence of TPL but not the other compounds we tested (Figure 6C). Taken together, our data indicate that TPL interferes with the UBE2S-MEX3C interaction, potentially serving as an E3 ligase inhibitor.
Figure 6. Triptolide inhibits PTENK27-polyUb and EMT.
(A) Graphic illustration of the molecular mechanisms of E3 ligase inhibitors. (B–D) Competition curve determination of KD value of PTX, TPL, and RG7388 against interactions of MDM2-p53 (B), MEX3C-UBE2S (C), and MEX3C-PTEN (D). Error bars indicate SD; n = 3 independent experiments. (E) HK-2 cells treated with indicated inhibitors with glucose stimulation (5 or 25 mM, 1 hour) were subjected to Biotin-TUBE pulldown and immunoblotting detection using indicated antibodies. (F) HK-2 cells treated with indicated growth factors (1 hour) were subjected to immunoblotting using indicated antibodies. (G) Immunofluorescence staining using indicated antibodies were performed using RPTECs in the presence of glucose with indicated treatment for 48 hours. Scale bars: 50 μm. (H) Immunoblotting detection using indicated antibodies in RPTECs or D-RPTECs treated with DMSO or TPL (1 μM, 2 hours), with glucose 5 mM or 25 mM for 1 hour.
In HK-2 cells, treatment with TPL, but not the other inhibitors we tested, reduced glucose-induced PTENK27-polyUb with minimal effect on the total ubiquitination of PTEN (Figure 6E). Pretreatment with TPL also effectively inhibited the CTGF-, IL-6–, SHH-, and TGF-β–induced PTENK27-polyUb (Figure 6F). Human renal tubule epithelial cells (RPTECs) exhibited expression of ZO-1 and low expression of vimentin and cell-cell contact (Figure 6G). In the presence of high glucose (25 mM) or TGF-β, RPTECs exhibited decreased expression of ZO-1, increased expression of vimentin, and loss of cell-cell contact, which were reversed by pretreatment with TPL but not with the other E3 ligase inhibitors we tested (Figure 6G and Supplemental Figure 5D).
By taking advantage of the paired primary cell lines, RPTECs, and diabetic human renal tubule epithelial cells (D-RPTECs), we found that protein levels of MEX3C were higher in D-RPTECs than in RPTECs (Figure 6H). High glucose stimulation induced PTENK27-polyUb, decreased unmodified PTEN, upregulated α-SMA, SNAI1, TWIST, and YAP, and downregulated E-cadherin/ZO-1 in RPTECs. D-RPTECs exhibited elevated PTENK27-polyUb, reduced unmodified PTEN, and augmented α-SMA and SNAI1 without glucose stimulation (Figure 6H). Treatment with TPL inhibited PTENK27-polyUb in both cell types as well as the expression of α-SMA and SNAI1 without affecting the protein level of MEX3C (Figure 6H). Biotin-TUBE (tandem ubiquitin binding entities) pulldown indicated that TPL specifically inhibited PTENK27-polyUb in glucose-treated RPTECs and D-RPTECs, with minimal effect on the global ubiquitination status (Supplemental Figure 5E).
Triptolide alleviates kidney fibrosis.
We determined the efficiency of TPL in alleviating kidney fibrosis using the Col4a3–/– model with a pure 129S1 genetic background (36). We included 4 groups: Col4a3 WT mice (Col4a3+/+), Col4a3 heterozygous mice treated with vehicle (Col4a3+/– + vehicle), Col4a3 homozygous mice treated with vehicle (Col4a3–/– + vehicle), and Col4a3 homozygous mice treated with TPL (Col4a3–/– + TPL) (Figure 7A). The animals were treated with vehicle or TPL, given orally daily and dosed 0.1 mg/kg, starting from 3 weeks of age and continuing until death. At 8 weeks of age, Col4a3–/– + vehicle animals showed reduced body weight and kidney weight compared with the Col4a3+/– + vehicle group, while TPL-treated animals showed improved kidney and body weight compared with the vehicle group (Supplemental Figure 6, A–C). The histology and liver function of the experimental animals were minimally affected (Supplemental Figure 6, D and E). Kidneys of Col4a3–/– animals treated with TPL exhibited improved tubular health, decreased sclerotic glomeruli, reduced myofibroblast deposition, lower degrees of α-SMA deposition, and reduced PTENK27-polyUb without a significant effect on the MEX3C protein level in tubules (Figure 7, B–G). Treating Col4a3–/– mice with TPL significantly reduced the protein levels of TWIST, SANI1, and YAP in kidney tubules (Figure 8, A–D and Supplemental Figure 6, F–H).
Figure 7. Triptolide alleviates kidney fibrosis.
(A) Representative images of H&E staining, Sirius red staining, PAS staining, immunofluorescence staining of α-SMA, MEX3C, and Ub-PTEN (K80) per Na+K+-ATPase+ tubule. Scale bars: 100 μm. (B–G) Statistical analysis of tubular injury score (B), percentage of Sirius red staining area (C), percentage of sclerotic glomeruli (D), staining intensity of α-SMA (E), MEX3C (F) and Ub-PTEN (K80) (G) per Na+K+-ATPase+ tubules of Col4a3+/+, Col4a3+/– + vehicle, Col4a3–/– + vehicle, and Col4a3–/– + TPL kidneys. Error bars indicate SD; n = 5, 8, 8, and 8 animals; 6 independent fields per animal were calculated (1-way ANOVA). NS indicates P > 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Figure 8. Triptolide improves renal function of fibrotic kidneys.
(A–D) Representative images (A) and statistical analysis of immunofluorescence staining of TWIST (B), SNAI1 (C) and YAP (D) per Na+K+-ATPase+ tubules of Col4a3+/+, Col4a3+/– + vehicle, Col4a3–/– + vehicle, and Col4a3–/– + TPL kidneys. Error bars indicate SD; n = 5, 8, 8, 8 animals; 6 independent fields per animal were calculated (1-way ANOVA). Scale bars: 100 μm. (E) Measurement of PTENK27-polyUb in urine of Col4a3+/+, Col4a3+/– + vehicle, Col4a3–/– + vehicle, and Col4a3–/– + TPL animals. Error bars indicate SD; n = 5, 6, 5, 7 animals, respectively (1-way ANOVA). (F) Measurement of BUN of Col4a3+/+ mice (8 weeks), Col4a3+/– + vehicle mice (8 weeks), Col4a3–/– + vehicle mice (8 weeks), and Col4a3–/– + TPL mice. Error bar indicates SD; n = 5, 8, 10, 8 animals, respectively (1-way ANOVA). (G–H) Measurement of serum creatinine using LC-MS (G) or ACR (H) of Col4a3+/+, Col4a3+/– + vehicle, Col4a3–/– + vehicle, and Col4a3–/– + TPL mice by age of 8 weeks. Error bar indicates SD; n = 5, 8, 8, 8 animals, respectively (1-way ANOVA). (I) Survival curves of Col4a3+/+, Col4a3+/– + vehicle, Col4a3–/– + vehicle, and Col4a3–/– + TPL animals (n = 5, 5, 13, and 16 animals, respectively, log-rank test). NS indicates P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Increased urine concentration of PTENK27-polyUb was detected in Col4a3–/– animal urine compared with the vehicle treatment (Figure 8E). TPL treatment resulted in reduced urine PTENK27-polyUb levels, which is consistent with the pathology and expression of EMT markers in these tissues (Figure 8E). Furthermore, while Col4a3–/– mice with vehicle treatment exhibited elevated serum BUN, creatinine, and ACR at 8 weeks of age, these characteristics were improved in Col4a3–/– mice with TPL treatment (Figure 8, F–H). TPL treatment significantly improved survival times, with a median survival time of 83 days compared with 59 days for the vehicle treatment group (Figure 8I).
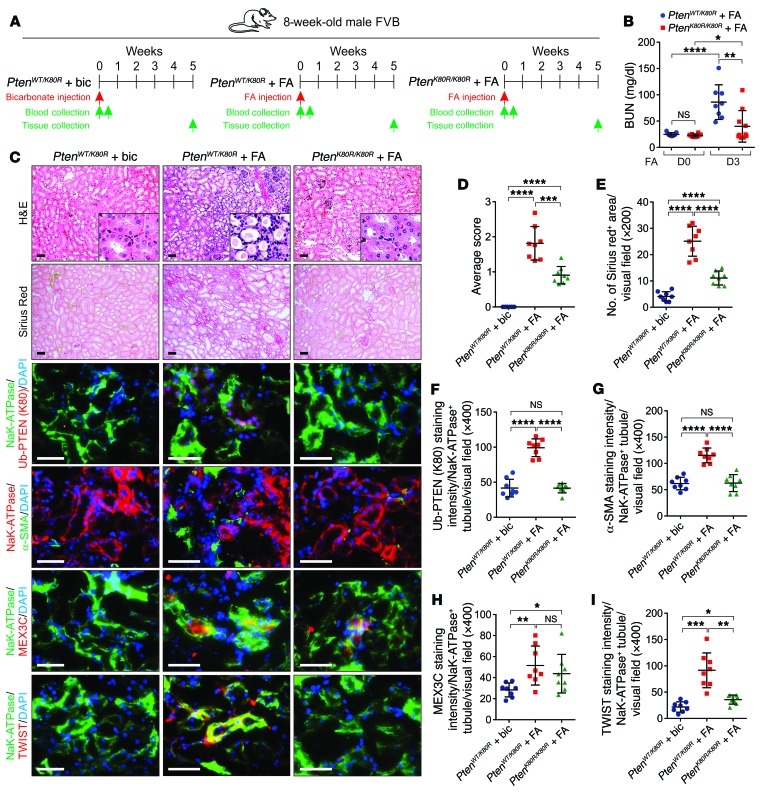
We determined the effect of PTENK27-polyUb in folic acid–induced (FA-induced) chronic progressive kidney fibrosis (37) (Figure 9A). Three days after FA injection, the serum BUN concentrations of PtenWT/K80R animals were significantly higher than in the PtenK80R/K80R animals (Figure 9B). Compared with bicarbonate only, FA injection induced interstitial fibrosis 5 weeks after injection. We detected altered tubular morphology, excessive collagen deposition, upregulation of PtenK27-polyUb and MEX3C, and tubular accumulation of TWIST, SNAI1, and YAP proteins (Figure 9, C–I and Supplemental Figure 8, A–E). Genetic inhibition of PtenK27-polyUb (PtenK80R/K80R) reduced FA-induced kidney fibrosis compared with the PtenWT/K80R animals (Figure 9, C–I and Supplemental Figure 7, A–H). Whether EMT plays important roles in the acute injury remains to be investigated. It is likely that the FA-induced damage during the acute phase triggers EMT of tubular epithelial cells (38). Consistent with this hypothesis, mitochondrial-derived ROS potentially induces EMT during FA-induced acute renal injury (39, 40). The mesenchymal-like cells triggered by EMT release multiple cytokines/chemokines (41), which further recruit inflammatory cells for kidney damage and consequently elevate the level of BUN.
Figure 9. Inhibition of PtenK27-polyUb reduces FA-induced renal fibrosis.
(A) Scheme of the experimental approach. (B) Measurement of BUN of PtenWT/K80R + FA mice (8 weeks) and PtenK80R/K80R + FA mice (8 weeks), before and 3 days after FA injection. Error bars indicate SD; n = 8, 8, 8, 8 animals, respectively (1-way ANOVA). (C) Representative images of H&E staining, Sirius red staining and immunofluorescence staining using indicated antibodies in PtenWT/K80R with biocarbonate (bic), PtenWT/K80R with FA, and PtenK80R/K80R with FA injection kidneys. Scale bars: 100 μm. (D–I) Statistical analysis of tubular injury score (D), percentage of Sirius red–positive area (E), Ub-PTEN (K80) (F), staining intensity of α-SMA (G), MEX3C (H), and TWIST (I) per Na+K+-ATPase–positive tubule per visual field. Error bars indicate SD; n = 8 animals; 6 independent fields per animal were calculated (1-way ANOVA). NS indicates P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Next, we treated the WT C57BL/6J mice with FA, followed by oral administration of vehicle or TPL 1 week after the FA injection until the end of the experiment (Figure 10A). TPL treatment resulted in improved tubular morphology, reduced collagen deposition, and decreased PtenK27-polyUb status in mouse kidneys (Figure 10, B–F and Supplemental Figure 9, A–I). Compared with vehicle administration, TPL treatment significantly reduced the expression of TWIST, SNAI1, and YAP (Figure 10, G and H, and Supplemental Figure 10, A–E).
Figure 10. Triptolide reduces FA-induced kidney fibrosis.
(A) Scheme of the experimental approach. C57BL/6J mice were treated with vehicle or FA, followed with vehicle or TPL treatment. (B) Representative images of H&E staining, Sirius red staining, and immunofluorescence staining using indicated antibodies in vehicle, FA and FA + TPL kidneys. Scale bars: 100 μm. (C–H) Statistical analysis of tubular injury score (C), percentage of Sirius red–positive area (D), Ub-PTEN (K80) (E), staining intensity of α-SMA (F), MEX3C (G), and TWIST (H) per Na+K+-ATPase–positive tubule per visual field. Error bars indicate SD; n = 8 animals; 6 independent fields per animal were calculated (1-way ANOVA). NS indicates P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Inhibition of PtenK27-polyUb attenuates STZ-induced nephropathy.
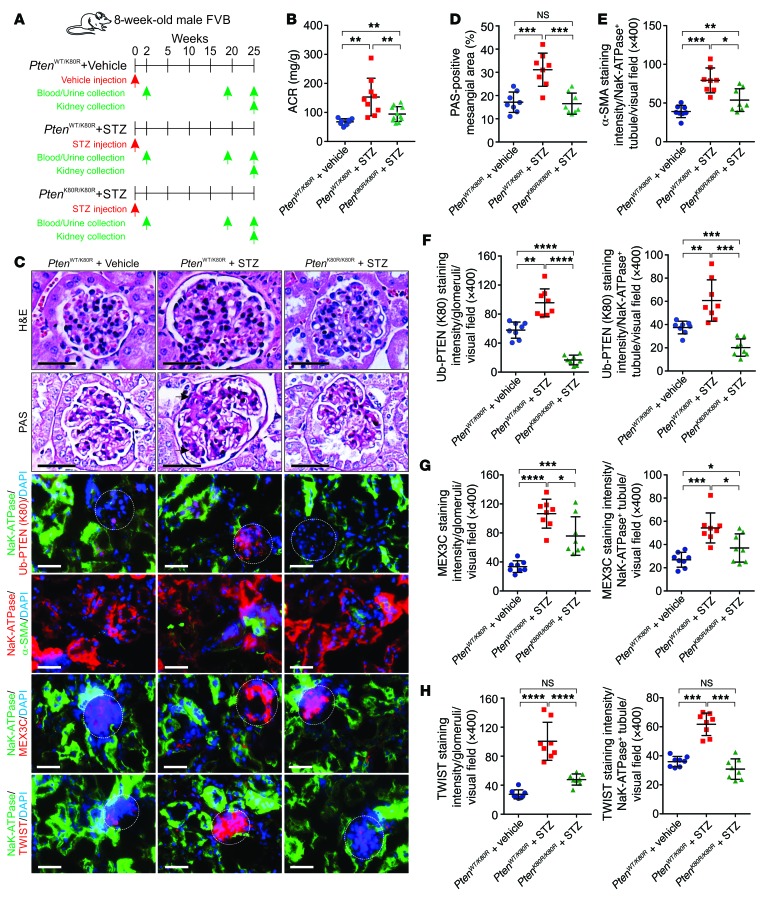
Streptozotocin-induced (STZ-induced) diabetic mouse models mimic type 1 diabetes by disrupting the insulin-producing pancreatic beta cells (42). To address the hypothesis that genetic inhibition of PtenK27-polyUb may reduce STZ-induced kidney injury, we included 3 groups of animals: PtenWT/K80R with vehicle injection, PtenWT/K80R with STZ injection, and PtenK80R/K80R with STZ injection (Figure 11A). The plasma glucose concentrations of these animals were determined 2 weeks after STZ injection and every 2 weeks until the end of the experiment. PtenWT/K80R and PtenK80R/K80R animals that were subjected to the STZ injection exhibited similar levels of hyperglycemia (Supplemental Figure 11A). PtenK80R/K80R animals showed significantly reduced ACR, improved body weight, and reduced kidney weight compared with PtenWT/K80R littermates upon STZ injection (Figure 11B and Supplemental Figure 11, B–G), suggesting that genetic inhibition of PtenK27-polyUb may improve kidney function in hyperglycemia. PtenWT/K80R kidneys exhibited expanded mesangial area, collagen deposition, elevated PtenK27-polyUb, and expression of TWIST1 25 weeks after STZ injection, which were attenuated by genetic inhibition of PtenK27-polyUb (Figure 11, C–H and Supplemental Figure 11, H–K). Therefore, our data suggested that PtenK27-polyUb is an important contributor to the development of DKD.
Figure 11. Inhibition of PtenK27-polyUb alleviates STZ-induced diabetic nephropathy.
(A) Scheme of the experimental approach. Three groups of animals were compared: PtenWT/K80R with vehicle injection, PtenWT/K80R with STZ injection, and PtenK80R/K80R with STZ injection. (B) Measurement of ACR of PtenWT/K80R + vehicle injection, PtenWT/K80R + STZ and PtenK80R/K80R + STZ animals. Error bar indicates SD; n = 8, 9, 8 animals, respectively (1-way ANOVA). (C) Representative images of H&E staining, PAS staining, and immunofluorescence staining using indicated antibodies in PtenWT/K80R + vehicle injection; PtenWT/K80R + STZ and PtenK80R/K80R + STZ kidneys. Scale bars: 50 μm (black) and 100 μm (white). White circle: glomerular; black arrow: expanded mesangial area. (D–H) Statistical analysis of PAS-positive mesangial area (D), staining intensity of α-SMA (E), staining of Ub-PTEN (K80) (F), MEX3C (G), and TWIST (H) per Na+K+-ATPase–negative glomerular (left panels) or –positive tubules (right panels) per visual field. Error bars indicate SD; n = 8 animals; 6 independent fields per animal were calculated (1-way ANOVA). NS indicates P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To assess the effect of the EMT inhibitor TPL on DKD, we treated male C57BL/6J mice with or without STZ-induced diabetes (Figure 12A). Two weeks after injection, the presence of PtenK27-polyUb in mouse urine (Figure 12B) indicated the occurrence of EMT in the diabetic kidneys, which provided rationale for TPL treatment. Hyperglycemic animals were subjected to vehicle or TPL treatment 3 weeks after STZ injection. Although blood glucose concentrations were not significantly affected by TPL treatment, body weight, ACR, and BUN concentration in the animals treated with TPL were significantly improved compared with the vehicle treatment group (Figure 12, C and D and Supplemental Figure 12, A–F). Animals treated with TPL exhibited reduced glomerular tuft, decreased mesangial area, lowered PtenK27-polyUb, and inhibited expression of mesenchymal markers (Figure 12, E–J and Supplemental Figure 12, G–J).
Figure 12. Triptolide prevents diabetic nephropathy.
(A) Scheme of the experimental approach. C57BL/6J mice were treated with vehicle or STZ, followed with vehicle or TPL treatment. (B–D) Measurement of urine PtenK27-polyUb (B), BUN (C), or ACR (D) of vehicle, STZ and STZ + TPL animals 2 and 25 weeks after injection. Error bars indicate SD; n = 8, 8, 12, 10, 12, 12 animals (B–C) or n = 8, 10, 12 animals (D), respectively (1-way ANOVA). (E) Representative images of H&E staining, PAS staining, and immunofluorescence staining using indicated antibodies in vehicle, STZ, and STZ + TPL kidneys. Scale bars: 50 μm (black) or 100 μm (white). White circle: glomerular; black arrow: expanded mesangial area. (F–J) Statistical analysis of PAS-positive mesangial area (F), staining intensity of α-SMA (G), staining of Ub-PTEN (K80) (H), MEX3C (I), and TWIST1 (J) per Na+K+-ATPase–negative glomerular (left panels) or –positive tubules (right panels) per visual field. Error bars indicate SD; n = 8 animals; 6 independent fields per animal were calculated (1-way ANOVA). NS indicates P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
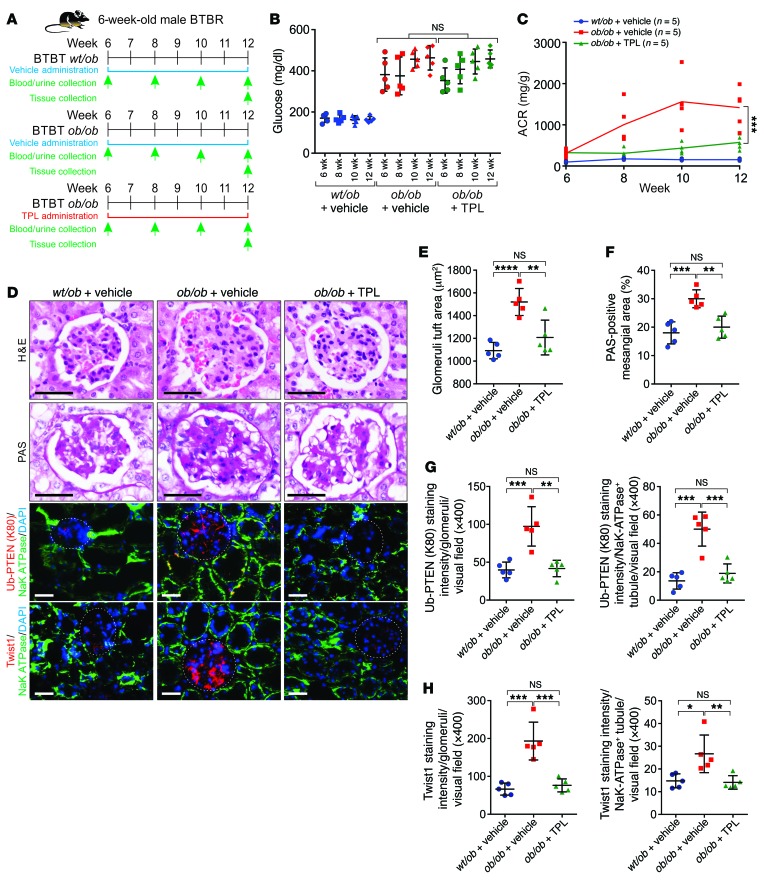
We further determined the efficacy of TPL in BTBR ob/ob-induced nephropathy. BTBR ob/ob mice exhibit increased albuminuria and glomerular mesangial matrix accumulation but marginal interstitial fibrosis (43). We treated BTBR ob/ob mice with TPL from 6 weeks old to 12 weeks old. The body weight, blood glucose, and ACR were monitored biweekly (Figure 13A). Treatment with TPL did not affect the blood glucose level or body weight of BTBR ob/ob mice. However, it significantly reduced proteinuria as indicated by reduced ACR, kidney weight, and BUN, but showed minimal effect on serum creatinine concentration and liver functions (Figure 13, B and C and Supplemental Figure 13, A–E). At 12 weeks of age, the animals were sacrificed for histological analysis. BTBR ob/ob mice exhibited enlarged glomerular tuft area and expansion of the glomerular mesangial matrix (Figure 13, D–H). BTBR ob/ob animals treated with TPL showed reduced glomerular tuft and a decreased mesangial matrix compared with the vehicle group (Figure 13, D–H). BTBR ob/ob kidneys increased PTENK27-polyUb and the status of Twist1 protein in both glomerular and tubular areas, which was efficiently inhibited upon TPL treatment (Figure 13, D–H and Supplemental Figure 13, F–G). Our findings indicated that the administration of TPL significantly improved glomerulosclerosis under diabetic conditions compared with the vehicle group.
Figure 13. Pharmaceutical inhibition of PtenK27-polyUb alleviates diabetic nephropathy.
(A) Scheme of the experimental approach. Three groups of animals were compared: BTBR ob/wt + vehicle, BTBR ob/ob + vehicle, and BTBR ob/ob + TPL injection. (B–C) Measurement of blood glucose (B) or ACR (C) of BTBR ob/wt + vehicle, BTBR ob/ob + vehicle, and BTBR ob/ob + TPL animals at 6, 8, 10, and 12 weeks of age (n = 5 animals per group). Error bar indicates SD (1-way ANOVA). (D) Representative images of H&E staining, PAS staining, and immunofluorescence staining using indicated antibodies in BTBR ob/wt + vehicle, BTBR ob/ob + vehicle, and BTBR ob/ob + TPL kidneys. Scale bars: 50 μm (black) and 100 μm (white). White circle: glomerular. (E–H) Statistical analysis of glomeruli tuft area (E), PAS-positive mesangial area (F), staining intensity of Ub-PTEN (K80) (G), and TWIST (H) per Na+K+-ATPase–negative glomerular (left panels) or –positive tubules (right panels) per visual field. Error bars indicate SD; n = 5 animals; 6 independent fields per animal were calculated (1-way ANOVA). NS indicates P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Discussion
About 40% of diabetic patients develop DKD, and the resultant kidney damage often leads to kidney failure, which requires dialysis or a kidney transplant. Once renal structural damage occurs, it is exceedingly difficult to reverse the damage that has already taken place. Our findings suggest that although the level of the unmodified form of Pten is reduced, the level of PtenK27-polyUb is significantly increased with kidney tubular injury in mice. Once modified, PtenK27-polyUb achieves adequate protein serine/threonine phosphatase activity to remove the phospho-groups of TWIST, SNAI1, and YAP. As a consequence, these EMT master regulators are stabilized to activate the EMT program. By taking advantage of CRISPR technology, we generated a mouse model harboring a single nucleotide mutation of Pten, PtenK80R/K80R, to genetically inhibit PtenK27-polyUb and convert the modified Pten to unmodified Pten. We reasoned that unmodified PTEN (EMT-preventing) and PtenK27-polyUb (EMT-promoting) are dynamically regulated in kidney disease, and knockout of Pten leads to a combined phenotype. On the contrary, PtenK80R/K80R genetically inhibited the EMT-promoting role of PTEN, leading to alleviated kidney damage and improved mouse survival. The proliferation of fibroblasts also contributes significantly to the progression of kidney fibrosis. Genetic or pharmaceutical inhibition of PTENK27-polyUb showed minimal effect on the proliferation of kidney resident fibroblasts.
The core signaling MEX3C-catalyzed PTENK27-polyUb axis is triggered by multiple stimuli, including TGF-β, CTGF, SHH, IL-6, and hyperglycemia. It is noteworthy that TGF-β, CTGF, SHH, IL-6, and hyperglycemia had all been linked to kidney fibrosis, but the underlying mechanisms remained unknown. The MEX3C-PTENK27-polyUb axis regulates the downstream protein targets of TWIST, SNAI1, and YAP concurrently. TWIST, SNAI1, and YAP all contribute to kidney fibrosis. Hence, targeting TWIST or SNAI1 alone would not maximally inhibit EMT due to potential compensation by the other pathways. Targeting TWIST, SNAI1, and YAP simultaneously with individual inhibitors is also not feasible given the likelihood for potential toxicity. Genetic or pharmaceutical inhibition of MEX3C-PTENK27-polyUb, however, reduced the protein levels of TWIST, SNAI1, and YAP simultaneously. The detection of PTENK27-polyUb concentrations in blood and urine can serve as a risk assessment marker that suggests when the administration of TPL would be beneficial for patients with DKD.
Triptolide, a natural compound originally extracted from the traditional Chinese medicine Tripterygium wilfordii, has been used to treat autoimmune diseases and chronic kidney diseases via PTEN-dependent and independent mechanisms (44, 45). However, the direct target of TPL has remained elusive. TPL has been reported to inhibit XPB (ERCC3) with a IC50 value of 109 nM ± 8 nM (29); undetermined for NF-κB in vitro (46); undetermined for PC2 in vitro (47); undetermined for ADAM10 (48); and undetermined for the autosomal dominant polycystic kidney disease (ADPKD) model (44). The interaction affinity between triptolide and TAB1 is between 9.7 μM–10.1 μM (49). Our research demonstrates that TPL specifically binds with MEX3C with a KD value of 27.5 nM, suggesting that TPL exhibits a strong binding affinity toward MEX3C in fibrotic kidneys. We further indicated that TPL directly associates with the RING domain of MEX3C, inhibiting the interaction between MEX3C and UBE2S with a IC50 value of 48.2 nM, situating TPL as a promising E3 ligase inhibitor. Clinical trials using TPL for ADPKD indicated no noticeable changes in laboratory parameters indicative of possible toxicity and no sign of bone marrow suppression or liver toxicity (50). The toxicity and solubility of TPL could be improved using the nanoparticle coating method (51), pretreatment with verapamil (52) or CYP3A (53), or development of TPL analogues (54). Hence, we demonstrate that DKD patients with high serum/urine PTENK27-polyUb could benefit from TPL-based treatment options to inhibit progression of renal fibrosis.
Methods
Animal studies.
PtenK80R/K80R mice were generated by replacing Pten lysine 80 with arginine using CRISPR/Cas system–mediated genomic editing technology. A single-nucleotide (AAA-AGA) mutation of founder mice was validated by TaqMan PCR and Sanger sequencing. Col4a3-KO (Col4a3–/–) mice were provided by Raghu Kalluri, MD Anderson Cancer Center (Houston, TX). Genotyping was carried out by PCR. Col4a3–/– mice were bred with PtenK80R/K80R mice to generate 4 different groups of mice in a mixed 129S1 and FVB genetic background: Col4a3+/– PtenWT/K80R, Col4a3+/– PtenK80R/K80R, Col4a3–/– PtenWT/K80R, and Col4a3–/– PtenK80R/K80R. We first crossed Col4a3–/– in 129S1 and PtenK80R/K80R in FVB background animals to achieve Col4a3+/– PtenWT/K80R animals in the first generation of 129S1/FVB mixed genetic background. These animals were self-crossed to achieve Col4a3+/– PtenWT/K80R, Col4a3+/– PtenK80R/K80R, Col4a3–/– PtenWT/K80R, and Col4a3–/– PtenK80R/K80R animals in the second generation of 129S1/FVB mixed genetic background. Male and female littermates were used for the experiments. The sample size was chosen on the basis of our previous experience and is indicated for each experiment in the figure legends.
For TPL treatment, Col4a3–/– mice were bred in a 129S1 genetic background to reduce individual differences. Heterozygous mice from the same litter were used as controls. TPL was dissolved in 30% PEG400, 0.5% Tween 80, and 5% propylene glycol to prepare a stock concentration of 0.01 mg/ml. The treatment group mice received daily gavages with TPL (0.1 mg/kg/day), while control group was administered the same amount of vehicle. All the treatments were initiated in 3-week-old mice and terminated when the mice reached the end of disease.
To induce renal fibrosis, a single dose of folic acid (250 mg/kg body weight) in 0.3 M NaHCO3 or vehicle alone was intraperitoneally administered to 8-week-old adult mice as described (55). For the TPL treatment studies, the animals were administrated TPL or the vehicle 1 week after FA injection until end of experiment. Blood was collected before and 3 days after FA injection for BUN measurement. Mice were euthanized at 34 days after FA injection, and the kidneys were collected for histopathological studies.
Induction of diabetes by STZ was performed as previously described (56). Briefly, mice with the indicated genetic background (8 weeks old) were given 5 consecutive daily intraperitoneal injections of STZ (40 mg/kg body weight in 0.1 M citrate buffer). The mice receiving the injection were administered 5% sucrose water within 24 hours to prevent hypoglycemic shock. Blood glucose levels were examined before the administration of STZ, after 24 hours, by end of the second week, and every 2 weeks until the end of experiments using an AimStrip Plus Blood Glucose Meter (Germaine Laboratories). Mice with blood glucose levels greater than 300 mg/dl were considered diabetic. When blood glucose levels exceeded 25 mM (450 mg/dl), diabetic mice were given 0.4U of insulin every other day to prevent weight loss while maintaining hyperglycemia (13.9 mM–25 mM) (42). For the TPL treatment studies, the diabetic animals were administered TPL or the vehicle 3 weeks after STZ injection until the end of the experiments.
Six-week-old male BTBR ob/ob mice were purchased from Jackson Laboratories and divided into 3 groups: BTBR wt/ob (n = 5), BTBR ob/ob (n = 5), and BTBR ob/ob treated with 0.1 mg/kg/day TPL (n = 5). The treatment group mice received daily gavages with TPL starting from 6 weeks old, and lasting for 6 weeks until 12 weeks old, while the control group was administered the same amount of vehicle. All animal studies were reviewed and approved by the MD Anderson Cancer Center Institutional Animal Care and Use Committee. No animals were excluded from the analysis.
Blood and urine analysis.
Mouse serum was collected by cardiac puncture in anesthetized mice. Metabolic cages were used to collect 24-hour urine from mice. BUN concentrations were measured via colorimetric assay (Arbor Assays). Alanine transaminase (ALT) and aspartate aminotransferase (AST) activity were tested by Alanine Transaminase Colorimetric Activity Assay Kit (Cayman Chemical) and EnzyChrom Aspartate Transaminase Assay Kit (BioAssay Systems). Urinary ACR was determined using mouse albumin ELISA kit (Molecular Innovations) and QuantiChrom Creatinine Assay Kit (BioAssay Systems), per the manufacturers’ protocols. The absolute concentration of serum creatinine was determined by Metabolomics Core, Baylor College of Medicine (57).
Cell culture and treatments.
Human tubule epithelial cell HK-2 cells were purchased from ATCC and maintained in DMEM supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2 (vol/vol). Mouse tubule epithelial MCT cells were provided by Raghu Kalluri, MD Anderson Cancer Center (Houston, TX) and the Characterized Cell Line Core Facility (MD Anderson Cancer Center), and were maintained in DMEM supplemented with 10% FBS at 37°C in 5% CO2 (vol/vol). RPTECs and D-RPTECs were purchased from Lonza and cultured in respective media (REGM) basal medium with REGM BulletKit (Lonza) at 37°C in 5% CO2 (vol/vol).
For glucose treatment, cells were washed and refreshed with glucose-free DMEM with 10% dialyzed FBS. Eighteen hours later, cells were treated with d-glucose (5 mM or 25 mM; Sigma-Aldrich) for the indicated time in various experiments. For growth factor treatment, cells were serum starved for 24 hours followed by treatment with Angiotensin II (100 ng/ml), BMP2 (100 ng/ml), CCL18 (100 ng/ml), CoCl2 (250 μM), Cripto-1 (100 ng/ml), hCTGF (10 ng/ml), rCTGF (10 ng/ml), EGF (10 ng/ml), FGF2 (10 ng/ml), HGF (20 ng/ml), IGF-1 (50 ng/ml), IL-1 (10 ng/ml), hIL-6 (10 ng/ml), mIL-6 (10 ng/ml), IL-8 (10 ng/ml), MMP2 (100 ng/ml), OSM (50 ng/ml), PDGF (10 ng/ml), PGE2 (10 ng/ml), SDII-1, hSHH (10 ng/ml), mSHH (10 ng/ml), hTGF-β (10 ng/ml), mTGF-β (10 ng/ml), TNF-α (10 ng/ml), and Wnt3α (50 ng/ml) (Peprotech) for 30 minutes or 72 hours, as indicated in the figure legends. For inhibitor treatment, cells were serum starved for 24 hours followed by treatment with 1 μM A01 (Tocris), AT406 (Sellekchem), Nutlin-3 (Tocris), GS143 (Tocris), Heclin (Tocris), SZLP1-41 (Tocris), TAME (Tocris), and triptolide (Sellekchem) for 2 hours or 72 hours, as indicated.
Antibodies, constructs, recombinant proteins, and oligonucleotides.
Antibodies, constructs, recombinant proteins, and oligonucleotides were purchased from a variety of manufacturers, as follows. Anti-PTEN (D4.3) rabbit mAb (catalog 9188), anti-YAP (D8H1X) rabbit mAb (catalog 14074), anti-SNAIL (L70G2) mouse mAb (catalog 3895), anti–ZO-1 (D6L1E) rabbit mAb (catalog 13663), anti-UBE1 rabbit antibody (catalog 4890), anti–His-Tag rabbit antibody (catalog 2365), anti-GST (26H1) mouse mAb (catalog 2624) were from Cell Signaling Technology. Anti-sodium potassium ATPase rabbit mAb (Alexa Fluor 488, catalog ab197713), anti–E cadherin (M168) mouse mAb (catalog ab76055), anti-vimentin rabbit pAb (catalog ab92547), anti–N cadherin rabbit pAb (catalog ab18203) were from Abcam. Anti-α-SMA (1A4) mouse mAb (catalog A5228), anti-UBE2S (2G7) mouse antibody (catalog SAB1405042), anti-FLAG M1 mouse mAb (catalog F3040) were from Sigma-Aldrich. Anti-GAPDH (6C5) mouse mAb (catalog sc-32233) and anti-TWIST (H81) rabbit pAb (catalog sc-15393) were from Santa Cruz Biotechnology. Anti-MEX3C rabbit pAb (catalog NBP1-76341) was from Novus Biologicals. Anti-ubiquitin TUBE1 (catalog UM201) and anti-ubiquitin (catalog FK2) were from LifeSensors. Anti–E cadherin (NCH-38) mouse mAb (catalog M3612) was from Dako. Anti–ZO-1 monoclonal antibody (catalog ZO1-1A12, Alexa Fluor 594) was from Thermo Fisher Scientific. Anti–Ub-PTEN(K80) was raised against synthetic bunching peptides AERHYDTAK(GGRLRLV)FN based on the human PTEN sequence flanking lysine 80, which was custom generated by YenZym Antibodies, LLC, and used in immunoblotting, immunoprecipitation, immunofluorescence, and immunohistochemistry experiments.
The full-length of MEX3C and PTEN mammalian expression vectors were obtained from OriGene. Bacterial expression vectors for His6-tagged MEX3C (WT and mutants) were subcloned into pET-28a vector. His6-tagged PTEN (WT and mutants) were constructed into the pET-DEST42 vectors (Life Technologies). The point or domain deletion mutants were generated from the WT sequence using QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). Plasmid transfections were performed using Lipofectamine3000 (Life Technologies) according to the manufacturer’s instruction.
The following recombinant proteins were used in this study. GST-UBE1, GST-UBE2D3, UBE2S, ubiquitin, MDM2, RNF4, UBE3A, and ITCH (BostonBiochem); USP2 and Biotin-TUBE1 (LifeSensors); GST-PTEN, GST-MEX3C, GST-ERCC3 (Novus Biologicals); GST-TRIM63 (Millipore); LIN-41 (Abcam); GST-PSMD10 (LSBio); GST-DCTP1 (Proteintech). Recombinant His6-MEX3C and His6-PTEN proteins were expressed in E. coli strain BL21-CodonPlus (DE3)–RIPL (Agilent Technologies) and purified using HisPur Cobalt Spin Columns (Life Technologies).
Nontargeting control siRNAs (D-001320) and ON-TARGETplus SMARTpool siRNA targeting MEX3C (L-006989) obtained from GE Healthcare Dharmacon were used in this study. Control CRISPR/Cas9 plasmid (catalog sc-418922) and PTEN double nickase plasmids (catalog sc-400103-NIC, catalog sc-400103-NIC-2) were purchased from Santa Cruz Biotechnology.
Biotinylated linker (Bio-Linker) and triptolide (Bio-TPL) were custom synthesized from KareBay Biochem Inc. with over 90% purity as validated by vendor.
Cell lysis, immunoprecipitation, immunoblotting, and streptavidin pulldown.
Cells were homogenized in 1× RIPA buffer (EMD Millipore) as previously described. Supernatants were analyzed for immunoprecipitation with the indicated antibodies and the immunoprecipitated proteins were subjected to immunoblotting (58).
Chemical pulldown and mass spectrometry.
Chemical pulldown followed by mass spectrometry were performed using custom synthesized Bio-TPL and control compound Bio-Linker from KareBay Biochem Inc. Fresh kidneys were collected from 8-week-old male Col4a3+/+ and Col4a3–/– animals. Tissues were minced to prepare single-cell suspensions. They were washed 3–4 times with ice-cold PBS. The cell lysates were freshly prepared as previously described (59). The biotinylated compound-protein complexes were washed sequentially and eluted by NuPAGE LDS Sample Buffer (Thermal Fisher Scientific), and boiled for 10 minutes. The eluted protein were analyzed in SDS-PAGE and virtualized using Pierce Silver Stain for Mass Spectrometry Kit (Thermal Fisher Scientific). Protein bands were excised, extracted, alkylated, and digested with immobilized trypsin (Promega) for MS analysis at MD Anderson Cancer Center Proteomics Facility. The matched peptide queries are listed in Supplemental Table 2. The Multidimensional Protein Identification Technology (MudPIT) scoring for each identified protein is shown.
Measurement of PTENK27-polyUb.
Concentration of total K27-polyubiquitinated PTEN was measured with the UbiQuant ELISA kit (LifeSensors) from serum or urine samples following the manufacturer’s protocol with minor modification using anti-Ub-PTEN (K80) as primary antibody. Chemiluminescence was measured on an EnSpire Multimode Plate Reader (PerkinElmer) and quantified by MaterPlex ReaderFit software (Hitachi Solutions America, Ltd.)
Histology and histopathology.
Kidneys were fixed in 10% neutral buffered formalin and embedded in paraffin. Tissue sections (5 μm) were used for hematoxylin and eosin (H&E) staining and Sirius red staining. PicroSirius red stain (Abcam) was used according to the manufacturer’s instructions. The extent of renal injury and fibrosis was estimated using morphometric assessment of tubular damage and interstitial fibrosis. Tubular injury was scored semiquantitatively by a blinded pathologist who examined at least 10 cortical fields (×200 magnification) of PAS-stained sections (n = 8). Tubular injury was defined as tubular dilation, tubular atrophy, tubular cast formation, vacuolization, degeneration, and sloughing off of tubular epithelial cells or loss of the brush border and thickening of the tubular basement membrane. The tubules were evaluated according to the following scoring system: 0 = no tubular injury; 1 = 10% or fewer tubules injured; 2 = 11%–25% tubules injured; 3 = 26%–50% tubules injured; 4 = 51%–74% tubules injured; and 5 = 75% or more tubules injured. The average value of random histological fields for each animal (5–8 animals per group as indicated in the figure legends) were used for comparison between experimental groups.
For the analysis of the interstitial fibrosis, eight ×200 visual fields were randomly selected for Picrosirius red–stained kidney sections and number of Sirius red–positive area was manually assessed by a grid intersection analysis using Adobe Photoshop. The average value of 8 random histological fields for each animal (5 to 8 animals per group as indicated in figure legend) were used for comparison between experimental groups. Representative images were acquired with Zeiss Axioskop2 plus microscope and analyzed with ProgRes Capture Pro software. The slides were scanned on the Automated Cellular Image System III (Dako, Agilent) for quantification by digital image analysis.
Kidney tissues fixed with paraformaldehyde were sectioned and stained with periodic acid–Schiff (PAS). Light microscopic analysis was performed by surveying the entire cortical area of a PAS-stained slide containing 100–150 glomeruli per section. PAS-stained kidney tissue was graded blindly depending on the amount of mesangial matrix. Percentage of PAS-positive mesangial area was quantified as previously described (60). Briefly, 20 PAS-stained, nonoverlapping glomeruli from each mouse were examined on a digital microscope screen grid containing 667 (29 × 23) points. The number of grid points that hit pink or red mesangial matrix deposition were divided by the total number of points in the glomerulus to obtain the percentage of mesangial matrix deposition (mesangial matrix index) in a given glomerulus.
The tuft area of each individual glomerular profile was measured using a traditional stereologic point-counting method and an orthogonal grid system (each grid square measured 2.5 × 2.5 cm). Glomerular profiles that were closer than one glomerular diameter to the edge of the sections were excluded from the study. All glomerular profiles within the more central parts of the samples were measured and averaged as the average glomeruli tuft area (100–150 glomeruli per animal, n = 5 animals per experimental condition were measured).
Immunofluorescence.
Frozen sections of kidneys embedded in optimum cutting temperature (OCT, Sakura Finetek) medium, and 5-μm frozen sections were blocked for 1 hour with 5% normal goat serum and immunostained using antibodies against Na+K+-ATPase (Abcam, EP1845Y, 1:200), α-SMA-Cy3 (Sigma-Aldrich, C6198, 1:200), MEX3C (Santa Cruz, D-10, 1:100), Ub-PTEN (K80, 1:200), YAP (CST, D8H1X, 1:400), Twist (Santa Cruz, Twist2C1a, 1:100), Snail (CST, C15D3, 1:100), ZO-1 (ZO1-1A12, Alexa Fluor 594, 1:200), E-cadherin (ab76055, 1:400), vimentin (ab92547, 1:400), and N-cadherin (ab18203, 1:400). Nuclei were stained with Vectashield Antifade Mounting Medium with DAPI (Vector Laboratories) and imaged with confocal microscope (Zeiss). The slides were scanned on the Automated Cellular Image System III (Dako, Agilent) for quantification by digital image analysis. The staining intensity and measurement area (×400) for 6 random glomerular or Na+K+-ATPase–positive tubules per animal per channel were scored by ImageJ. The average intensity of 6 random microscopic fields per channel for each animal (5–8 animals per group as indicated in the figure legends) were used for comparison between experimental groups. The measurement area per tubule for each experimental condition is shown in corresponding Supplemental Figures.
For EMT analysis, HK-2 and RPTECs were fixed with 4% paraformaldehyde at room temperature for 15 minutes and stained as previously described (61) with primary antibodies for 1 hour. Cells were then incubated with anti-Alexa Fluor 488 (Thermal Fisher Scientific, 1:500) or anti-AlexaFluor 594 (Thermal Fisher Scientific, 1:500) for 1 hour if necessary, followed by nuclear staining using Vectashield Antifade Mounting Medium with DAPI. Images were acquired with Zeiss Axioskop2 plus microscope and analyzed with ProgRes Capture Pro software. The slides were scanned on the Automated Cellular Image System III (Dako, Agilent) for quantification by digital image analysis.
Screening of small molecule inhibitors targeting MEX3C.
The small molecular inhibitor screening were performed as previously described (58) using Ub-PTEN (K80) antibody. A validated library of 2027 bioactive compounds (Selleck Chemicals) was tested. The polyUb formation of PTEN was detected by incubation with Ub-PTENK80 antibody. Background-subtracted average absorbance of each tested compounds was normalized to DMSO-treated samples and the log2 fold change was plotted.
Determination of KD value using alpha assay and in vitro ubiquitination assay.
Alpha binding assay was used to determine KD for interactions as previously described (62), using streptavidin donor beads and anti-His6 AlphaLISA acceptor beads or anti-His6 AlphaLISA donor beads and anti-GST AlphaLISA acceptor beads as pair (PerkinElmer). The KD was determined by a competition experiment in which unlabeled TPL was titrated in serial of 2-fold dilution from 10 μM to 0.06 nM. The competitive inhibition curve were calculated based on alpha signal readings by fitting to a “log (inhibitor) versus response-variable slope (4 parameters)” model respectively (GraphPad Prism 6 software).
E3LITE MEX3C ubiquitin ligase assay.
The ubiquitin ligase activity was determined using the E3LITE Customizable Ubiquitin Ligase Kit from LifeSensors as instructed by the manufacturer. The IC50 value of TPL was determined by a serial of 2-fold dilution from 10 μM to 0.06 nM in triplicate. The competitive inhibition curve were calculated by fitting to a nonlinear regression “log (inhibitor) versus response-variable slope (4 parameters)” model (GraphPad Prism 6 software). In vitro ubiquitination assays were performed using an ubiquitin protein conjugation kit as manufactory instructed (BostonBiochem).
Statistics.
The experiment was set up to use 3–8 samples/repeats per experiment/group/condition to detect a 2-fold difference with power of 80% and at the significance level of 0.05 by a 2-sided test for significant studies. For immunofluorescence and immunohistochemical staining, the representative images are shown. Each of these experiments was independently repeated for 3–5 times. Results are reported as mean ± SEM or SD of at least 3 independent experiments. Each exact n value is indicated in the corresponding figure legend. Comparisons were performed using unpaired 2-tailed Student’s t test or 1-way ANOVA (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001) as indicated in individual figures. Pearson χ2 test was implemented for statistical analyses of the correlation between markers. Kaplan-Meier survival curves were compared using the log rank test with GraphPad Prism (GraphPad Software). A P value of less than 0.05 was considered statistically significant.
Study approval.
All human subject studies were approved by the Institutional Review Board of the University of Texas, MD Anderson Cancer Center. Human serum samples from healthy donors and patients with diabetic kidney disease (DKD), human urine samples from healthy donors and kidney disease patients, and paired serum/urine samples from the same patients were purchased from ReproCELL. Human serum samples of type 1 and type 2 diabetic patients and fresh frozen human kidney tissues within normal limit or diabetic nephropathy were purchased from ProteoGenex, Inc. All tissue samples were collected in compliance with informed consent policy. Clinical information is summarized in Supplemental Table 1. All animal experiments were performed in accordance with protocol approved by the Institutional Animal Care and Use Committee of the MD Anderson Cancer Center.
Author contributions
YL and QH devised and performed most experiments. YL and C. Li performed animal treatment. YL and QH performed biochemistry studies. HH and TKN helped with animal breeding. NP, CRA, and DHH performed mass spectrometry measurement and analysis. Clinical specimens were ascertained and processed by LY. The histological staining and corresponding analysis were performed by KL and YL. PKP and SDE assisted with manuscript drafting. YX and LH performed bioinformatics analysis. MCH and FRD contributed to discussion and data interpretation. LY and C. Lin initiated and supervised the project and wrote the manuscript with input from all authors.
Supplementary Material
Acknowledgments
The authors thank Raghu Kalluri for providing mouse tubule epithelial MCT cells and the Col4a3–/– mouse model. We thank D. Aten for assistance with figure presentation. The Proteomics and Metabolomics Facility was supported in part by grants from the Cancer Prevention and Research Institute of Texas (CPRIT) (RP130397) and the NIH (1S10OD012304-01). This project was partially supported by the Metabolomics Core at Baylor College of Medicine with funding from the NIH (P30 CA125123), CPRIT Proteomics and Metabolomics Core Facility (to NP) (RP170005), and the Dan L. Duncan Cancer Center, American Cancer Society Award 127430-RSG-15-105-01-CNE (to NP) and NIH R01CA220297 and R01CA216426 (to NP). This work was also supported by grants from NIH R00DK094981, NCI R01CA218025-01 and R01CA231011, CPRIT 150094 and 180259, DOD BC180196 (to C. Lin), and NIH R00CA166527, NCI R01 CA218036-01, CPRIT R1218, and DOD BC151465, and the Andrew Sabin Family Fellowship (to LY).
Version 1. 02/11/2019
Electronic publication
Version 2. 03/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(3):1129–1151.https://doi.org/10.1172/JCI121987.
Contributor Information
Yajuan Li, Email: YLi43@mdanderson.org.
Qingsong Hu, Email: QHu1@mdanderson.org.
Chunlai Li, Email: CLi10@mdanderson.org.
Ke Liang, Email: kliang@mdanderson.org.
Yu Xiang, Email: Yu.Xiang@uth.tmc.edu.
Heidi Hsiao, Email: Heidi.h.hsiao@gmail.com.
Nagireddy Putluri, Email: putluri@bcm.edu.
David H. Hawke, Email: dhawke@mdanderson.org.
Leng Han, Email: Leng.Han@uth.tmc.edu.
Mien-Chie Hung, Email: mhung@mdanderson.org.
Farhad R. Danesh, Email: danesh@bcm.edu.
Liuqing Yang, Email: lyang7@mdanderson.org.
Chunru Lin, Email: clin2@mdanderson.org.
References
- 1.Shahbazian H, Rezaii I. Diabetic kidney disease; review of the current knowledge. J Renal Inj Prev. 2013;2(2):73–80. doi: 10.12861/jrip.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuttle KR, et al. Diabetic kidney disease: a report from an ADA Consensus Conference. Am J Kidney Dis. 2014;64(4):510–533. doi: 10.1053/j.ajkd.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Gross JL, de Azevedo MJ, Silveiro SP, Canani LH, Caramori ML, Zelmanovitz T. Diabetic nephropathy: diagnosis, prevention, and treatment. Diabetes Care. 2005;28(1):164–176. doi: 10.2337/diacare.28.1.164. [DOI] [PubMed] [Google Scholar]
- 4.Calcutt NA, et al. Therapeutic efficacy of sonic hedgehog protein in experimental diabetic neuropathy. J Clin Invest. 2003;111(4):507–514. doi: 10.1172/JCI15792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldfarb S, Ziyadeh FN. TGF-beta: a crucial component of the pathogenesis of diabetic nephropathy. Trans Am Clin Climatol Assoc. 2001;112:27–32; discussion 33. [PMC free article] [PubMed] [Google Scholar]
- 6.Navarro-González JF, Mora-Fernández C. The role of inflammatory cytokines in diabetic nephropathy. J Am Soc Nephrol. 2008;19(3):433–442. doi: 10.1681/ASN.2007091048. [DOI] [PubMed] [Google Scholar]
- 7.Roestenberg P, et al. Temporal expression profile and distribution pattern indicate a role of connective tissue growth factor (CTGF/CCN-2) in diabetic nephropathy in mice. Am J Physiol Renal Physiol. 2006;290(6):F1344–F1354. doi: 10.1152/ajprenal.00174.2005. [DOI] [PubMed] [Google Scholar]
- 8.Edeling M, Ragi G, Huang S, Pavenstädt H, Susztak K. Developmental signalling pathways in renal fibrosis: the roles of Notch, Wnt and Hedgehog. Nat Rev Nephrol. 2016;12(7):426–439. doi: 10.1038/nrneph.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lovisa S, et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med. 2015;21(9):998–1009. doi: 10.1038/nm.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeBleu VS, et al. Origin and function of myofibroblasts in kidney fibrosis. Nat Med. 2013;19(8):1047–1053. doi: 10.1038/nm.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carew RM, Wang B, Kantharidis P. The role of EMT in renal fibrosis. Cell Tissue Res. 2012;347(1):103–116. doi: 10.1007/s00441-011-1227-1. [DOI] [PubMed] [Google Scholar]
- 12.Grande MT, et al. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat Med. 2015;21(9):989–997. doi: 10.1038/nm.3901. [DOI] [PubMed] [Google Scholar]
- 13.Lee JO, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99(3):323–334. doi: 10.1016/S0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 14.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19(4):348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 15.Blanco-Aparicio C, Renner O, Leal JF, Carnero A. PTEN, more than the AKT pathway. Carcinogenesis. 2007;28(7):1379–1386. doi: 10.1093/carcin/bgm052. [DOI] [PubMed] [Google Scholar]
- 16.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13(5):283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 17.Lan R, et al. PTEN loss defines a TGF-β-induced tubule phenotype of failed differentiation and JNK signaling during renal fibrosis. Am J Physiol Renal Physiol. 2012;302(9):F1210–F1223. doi: 10.1152/ajprenal.00660.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Q, et al. LncRNAs-directed PTEN enzymatic switch governs epithelial-mesenchymal transition. Cell Res. doi: 10.1038/s41422-018-0134-3. [published online ahead of print January 10, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karihaloo A. Anti-fibrosis therapy and diabetic nephropathy. Curr Diab Rep. 2012;12(4):414–422. doi: 10.1007/s11892-012-0290-7. [DOI] [PubMed] [Google Scholar]
- 20.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchio-Yamada K, Monobe Y, Akagi K, Yamamoto Y, Ogura A, Manabe N. Tensin2-deficient mice on FVB/N background develop severe glomerular disease. J Vet Med Sci. 2016;78(5):811–818. doi: 10.1292/jvms.15-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ina K, Kitamura H, Tatsukawa S, Fujikura Y. Significance of α-SMA in myofibroblasts emerging in renal tubulointerstitial fibrosis. Histol Histopathol. 2011;26(7):855–866. doi: 10.14670/HH-26.855. [DOI] [PubMed] [Google Scholar]
- 23.Haverty TP, et al. Characterization of a renal tubular epithelial cell line which secretes the autologous target antigen of autoimmune experimental interstitial nephritis. J Cell Biol. 1988;107(4):1359–1368. doi: 10.1083/jcb.107.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen XM, Qi W, Pollock CA. CTGF and chronic kidney fibrosis. Front Biosci (Schol Ed) 2009;1:132–141. doi: 10.2741/s13. [DOI] [PubMed] [Google Scholar]
- 25.Fielding CA, et al. Interleukin-6 signaling drives fibrosis in unresolved inflammation. Immunity. 2014;40(1):40–50. doi: 10.1016/j.immuni.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding H, et al. Sonic hedgehog signaling mediates epithelial-mesenchymal communication and promotes renal fibrosis. J Am Soc Nephrol. 2012;23(5):801–813. doi: 10.1681/ASN.2011060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-β: the master regulator of fibrosis. Nat Rev Nephrol. 2016;12(6):325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 28.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112(12):1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Titov DV, et al. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol. 2011;7(3):182–188. doi: 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corson TW, Cavga H, Aberle N, Crews CM. Triptolide directly inhibits dCTP pyrophosphatase. Chembiochem. 2011;12(11):1767–1773. doi: 10.1002/cbic.201100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morreale FE, Walden H. Types of ubiquitin ligases. Cell. 2016;165(1):248–248.e1. doi: 10.1016/j.cell.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta. 2014;1843(1):47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov. 2014;13(12):889–903. doi: 10.1038/nrd4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding Q, et al. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem. 2013;56(14):5979–5983. doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- 35.Horwitz SB. Taxol (paclitaxel): mechanisms of action. Ann Oncol. 1994;5(Suppl 6):S3–S6. [PubMed] [Google Scholar]
- 36.Gross O, et al. Preemptive ramipril therapy delays renal failure and reduces renal fibrosis in COL4A3-knockout mice with Alport syndrome. Kidney Int. 2003;63(2):438–446. doi: 10.1046/j.1523-1755.2003.00779.x. [DOI] [PubMed] [Google Scholar]
- 37.Fink M, Henry M, Tange JD. Experimental folic acid nephropathy. Pathology. 1987;19(2):143–149. doi: 10.3109/00313028709077125. [DOI] [PubMed] [Google Scholar]
- 38.Jiang YS, Jiang T, Huang B, Chen PS, Ouyang J. Epithelial-mesenchymal transition of renal tubules: divergent processes of repairing in acute or chronic injury? Med Hypotheses. 2013;81(1):73–75. doi: 10.1016/j.mehy.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 39.Hallman MA, Zhuang S, Schnellmann RG. Regulation of dedifferentiation and redifferentiation in renal proximal tubular cells by the epidermal growth factor receptor. J Pharmacol Exp Ther. 2008;325(2):520–528. doi: 10.1124/jpet.107.134031. [DOI] [PubMed] [Google Scholar]
- 40.Yuan Y, et al. Mitochondrial dysfunction accounts for aldosterone-induced epithelial-to-mesenchymal transition of renal proximal tubular epithelial cells. Free Radic Biol Med. 2012;53(1):30–43. doi: 10.1016/j.freeradbiomed.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 41.Allison SJ. Fibrosis: Targeting EMT to reverse renal fibrosis. Nat Rev Nephrol. 2015;11(10):565. doi: 10.1038/nrneph.2015.133. [DOI] [PubMed] [Google Scholar]
- 42.Tesch GH, Allen TJ. Rodent models of streptozotocin-induced diabetic nephropathy. Nephrology (Carlton) 2007;12(3):261–266. doi: 10.1111/j.1440-1797.2007.00796.x. [DOI] [PubMed] [Google Scholar]
- 43.Hudkins KL, et al. BTBR Ob/Ob mutant mice model progressive diabetic nephropathy. J Am Soc Nephrol. 2010;21(9):1533–1542. doi: 10.1681/ASN.2009121290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leuenroth SJ, Bencivenga N, Igarashi P, Somlo S, Crews CM. Triptolide reduces cystogenesis in a model of ADPKD. J Am Soc Nephrol. 2008;19(9):1659–1662. doi: 10.1681/ASN.2008030259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li XY, et al. Triptolide restores autophagy to alleviate diabetic renal fibrosis through the miR-141-3p/PTEN/Akt/mTOR pathway. Mol Ther Nucleic Acids. 2017;9:48–56. doi: 10.1016/j.omtn.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao G, Vaszar LT, Qiu D, Shi L, Kao PN. Anti-inflammatory effects of triptolide in human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2000;279(5):L958–L966. doi: 10.1152/ajplung.2000.279.5.L958. [DOI] [PubMed] [Google Scholar]
- 47.Leuenroth SJ, et al. Triptolide is a traditional Chinese medicine-derived inhibitor of polycystic kidney disease. Proc Natl Acad Sci U S A. 2007;104(11):4389–4394. doi: 10.1073/pnas.0700499104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soundararajan R, Sayat R, Robertson GS, Marignani PA. Triptolide: An inhibitor of a disintegrin and metalloproteinase 10 (ADAM10) in cancer cells. Cancer Biol Ther. 2009;8(21):2054–2062. doi: 10.4161/cbt.8.21.9803. [DOI] [PubMed] [Google Scholar]
- 49.Lu Y, et al. TAB1: a target of triptolide in macrophages. Chem Biol. 2014;21(2):246–256. doi: 10.1016/j.chembiol.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 50.Chen D, et al. Triptolide-containing formulation in patients with autosomal dominant polycystic kidney disease and proteinuria: an uncontrolled trial. Am J Kidney Dis. 2014;63(6):1070–1072. doi: 10.1053/j.ajkd.2014.01.418. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, et al. Fabrication of a triptolide-loaded and poly-γ-glutamic acid-based amphiphilic nanoparticle for the treatment of rheumatoid arthritis. Int J Nanomedicine. 2018;13:2051–2064. doi: 10.2147/IJN.S151233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, et al. Influence of verapamil on pharmacokinetics of triptolide in rats. Eur J Drug Metab Pharmacokinet. 2016;41(4):449–456. doi: 10.1007/s13318-015-0275-4. [DOI] [PubMed] [Google Scholar]
- 53.Shen G, Zhuang X, Xiao W, Kong L, Tan Y, Li H. Role of CYP3A in regulating hepatic clearance and hepatotoxicity of triptolide in rat liver microsomes and sandwich-cultured hepatocytes. Food Chem Toxicol. 2014;71:90–96. doi: 10.1016/j.fct.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 54.Aoyagi Y, et al. Semisynthesis of C-ring modified triptolide analogues and their cytotoxic activities. Bioorg Med Chem Lett. 2006;16(7):1947–1949. doi: 10.1016/j.bmcl.2005.12.098. [DOI] [PubMed] [Google Scholar]
- 55.Doi K, et al. Attenuation of folic acid-induced renal inflammatory injury in platelet-activating factor receptor-deficient mice. Am J Pathol. 2006;168(5):1413–1424. doi: 10.2353/ajpath.2006.050634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leiter EH. Multiple low-dose streptozotocin-induced hyperglycemia and insulitis in C57BL mice: influence of inbred background, sex, and thymus. Proc Natl Acad Sci U S A. 1982;79(2):630–634. doi: 10.1073/pnas.79.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vantaku V, et al. Expression of ganglioside GD2, reprogram the lipid metabolism and EMT phenotype in bladder cancer. Oncotarget. 2017;8(56):95620–95631. doi: 10.18632/oncotarget.21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li C, et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19(2):106–119. doi: 10.1038/ncb3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xing Z, Lin C, Yang L. LncRNA pulldown combined with mass spectrometry to identify the novel LncRNA-associated proteins. Methods Mol Biol. 2016;1402:1–9. doi: 10.1007/978-1-4939-3378-5_1. [DOI] [PubMed] [Google Scholar]
- 60.Sugimoto H, Grahovac G, Zeisberg M, Kalluri R. Renal fibrosis and glomerulosclerosis in a new mouse model of diabetic nephropathy and its regression by bone morphogenic protein-7 and advanced glycation end product inhibitors. Diabetes. 2007;56(7):1825–1833. doi: 10.2337/db06-1226. [DOI] [PubMed] [Google Scholar]
- 61.Lin A, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18(2):213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin A, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol. 2017;19(3):238–251. doi: 10.1038/ncb3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.