Abstract
Cell-free DNA (cfDNA) is a liquid biopsy marker that can carry signatures (i.e., mutations) associated with certain pathological conditions. Therefore, the extraction of cfDNA from a variety of clinical samples can be an effective and minimally invasive source of markers for disease detection and subsequent management. In the oncological diseases, circulating tumor DNA (ctDNA), a cfDNA sub-class, can carry clinically actionable mutations and coupled with next generation sequencing or other mutation detection methods provide a venue for effective in vitro diagnostics. However, cfDNA mutational analyses require high quality inputs. This necessitates extraction platforms that provide high recovery over the entire ctDNA size range (50 → 150 bp) with minimal interferences (i.e., co-extraction of genomic DNA), and high reproducibility with a simple workflow. Herein, we present a novel microfluidic solid-phase extraction device (μSPE) consisting of a plastic chip that is activated with UV/O3 to generate surface-confined carboxylic acid functionalities for the μSPE of cfDNA. The μSPE uses an immobilization buffer (IB) consisting of polyethylene glycol and salts that induce cfDNA condensation onto the activated plastic microfluidic surface. The μSPE consists of an array of micropillars to increase extraction bed load (scalable to loads >700 ng of cfDNA) and can be produced at low-cost using replication-based techniques. The entire μSPE can be fabricated in a single molding step negating the need for adding additional extraction supports to the device simplifying production and keeping device and assay cost low. The μSPE allowed for recoveries >90% of model cfDNA fragments across a range of sizes (100–700 bp) and even the ability to extract efficiently short cfDNA fragments (50 bp, >70%). In addition, the composition of the IB allowed for reducing the interference of co-extracted genomic DNA. We demonstrated the clinical utility of the μSPE by quantifying the levels of cfDNA in healthy donors and patients with non-small-cell lung and colorectal cancers. μSPE extracted cfDNA from plasma samples was also subjected to a ligase detection reaction (LDR) for determining the presence of mutations in the KRAS gene for colorectal and non-small cell lung cancer patients.
Introduction
Cell-free DNA (cfDNA) released into body fluids from both diseased and non-diseased cells was first reported by Mandel and Mëtais in 1948, but went largely unnoticed for approximately three decades.1 In the late 1970’s,2 a number of publications appeared demonstrating the correlation between high levels of cfDNA in patient plasma and tumor incidence.3–9 While in healthy individuals DNases clear the cfDNA from the blood within minutes, accumulation of a certain type of cfDNA, termed circulating tumor DNA (ctDNA), has been observed in cancer patients.1 Thus, early work in using cfDNA/ctDNA involved using the total cfDNA concentration in plasma as a disease marker.
Later, it was postulated that the length of ctDNA might be used as an indicator of its origin with ctDNA being shorter than the cfDNA originating from non-diseased cells.10,11 In terms of size, cfDNA from healthy individuals can be 200–10 000 bp while the majority of ctDNA is <150 bp.2,3
A renewed interest in cfDNA has evolved with the ability to detect mutations at a reasonable cost and throughput,4 which has partly been enabled by the advent of next generation sequencing (NGS).5 However, the quality of the sequencing data depends intimately on the quality of the input, and NGS requires mutant copy abundance >0.1%.6 As such, demands on the efficient solid phase extraction (SPE) of cfDNA from a clinical sample is important. Besides NGS, other mutation detection assays using cfDNA are also transitioning into the clinic. For example, the FDA has now approved the EGFR Cobas test from Roche for non-small cell lung cancer, which uses an allele-specific type PCR assay.7 Even for PCR-based tests, high quality cfDNA extracts are required to provide reliable results. As can be surmised from this brief discussion, the efficient extraction and subsequent mutational profiling of cfDNA can provide a minimally-invasive method for disease detection and/or management.1,8,9
As noted, cfDNA mutational analyses require highly efficient methods for the SPE of ctDNA fragments from plasma. Due to the relatively short size of ctDNA fragments (<150 bp), high inter-laboratory variability of the extraction process, and multi-step workflows requiring trained operators,10,11 there exists shortcomings associated with many existing benchtop SPE kits for cfDNA. Methods currently available for the SPE of cfDNA comprise column or bead based-assays. For example, the Qiagen kits typically use silica beads with adsorption of the plasma-borne cfDNA fragments onto the extraction beds induced by a chaotropic salt.12 Although the commercial kits can achieve reasonable recovery,13 studies have demonstrated that the variability among laboratories using these kits are high due to analytical and pre-analytical factors that typically depend on skilled operators.14–21 In addition, the workflow can require extensive sample handling and as such, prone to errors (see ESI,† Fig. S1).22 Finally, the extraction efficiency of short ctDNA fragments can be modest. It should be noted that any cfDNA/ctDNA SPE method will require plasma preprocessing before the extraction with an example being protein digestion to not only remove endogenous plasma proteins, but also release cfDNA fragments from histones.
Microfluidic devices allow for some unique operational characteristics compared to benchtop approaches making them attractive platforms for clinical use, in particular for the analysis of liquid biopsy markers, such as ctDNA. These operational characteristics include: (i) reduced processing time; (ii) closed architecture to minimize sample contamination and/or loss; (iii) conducive to automation; and (iv) the ability to allow for process integration including downstream molecular analysis. These operational characteristics can address many of the aforementioned challenges associated with existing benchtop cfDNA SPE assays.23
Microfluidic devices earmarked for the SPE of cfDNA should provide the following: (i) high specificity and recovery of ctDNA (i.e., specifically short ctDNA fragments with respect to genomic DNA (gDNA)); (ii) high loads to search for rare ctDNA fragments in patients with early stage disease; (iii) simplified workflow; and (iv) low cost per assay.8 A few publications have reported the μSPE of cfDNA using microfluidics. Lamanda et al.24 and Yang et al.10 developed devices based on electrokinetic traps, while Sonnenberg et al.25 used dielectrophoresis to isolate cfDNA. These methods could accommodate a sample input of 10–20 μL making it difficult to enrich for rare ctDNA fragments that can be a vast minority in the cfDNA population. Also, these devices required the application of an electric potential necessitating the need for patterning electrodes onto the microfluidic that can increase the cost of the assay.
Centrifugal-based microfluidics for the μSPE of cfDNA using silica beads contained within a fluidic channel was reported and provided a recovery of 75% for a model 300 bp DNA fragment.13 In this work, silica beads from a commercial kit were packed into a microchannel of the microfluidic disk and the SPE used the commercial kit’s extraction reagents. Despite nicely addressing the automation challenge by using the microfluidic to accept whole blood, isolate the plasma, and then perform the μSPE, the reported method will have implicit limitations associated with commercial kits such as poor recovery of ctDNA fragments that are typically <150 bp in length. In addition, this microfluidic SPE assay will require not only the microfluidic chip, but also silica beads from a commercial kit that must be packed into the microchannel increasing assay cost and post-fabrication assembly steps.
Previously, our group reported a microfluidic for the μSPE of gDNA and total RNA using a solid-phase reversible immobilization (SPRI) approach.26–31 In these assays, microfluidic devices fabricated in a plastic containing arrays of pillars were used as the μSPE beds. When activating the polymer surface with UV/O3 to generate -COOH surface functionalities, nucleic acids were selectively extracted through the appropriate composition of the immobilization buffer (IB). For example, to extract gDNA, we used an IB consisting of 3% PEG, 0.5 M NaCl and 63% EtOH, while total RNA was extracted by an IB containing 5% PEG, 0.4 M NaCl and 63% EtOH.
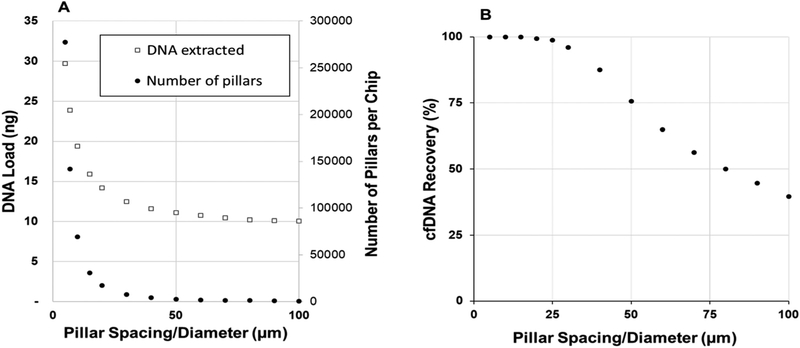
An attractive advantage of using polymer pillars is that the extraction bed is scalable in terms of target load by increasing the bed size, adding multiple beds in parallel, and/or reducing the pillar size/spacing (Fig. 1A). As the μSPE process is diffusion-controlled, decreasing the inter-pillar spacing can also increase the recovery of cfDNA due to the reduction in diffusional distances (Fig. 1B; see ESI† and Table S1 for specific details on simulation parameters). However, while the DNA load and recovery increases with an increase in the pillar density, device cost does not; the device is made from a plastic via replication using the appropriate molding master and as such, the cost of the device is increased only slightly with increased pillar density.32
Fig. 1.
(A) An estimate of the DNA load and pillar number as a function of pillar spacing and diameter (in these calculations, the pillar diameter and spacing value was equivalent for each data point). For a device with the same SPE bed dimensions, decreasing the size/spacing of the pillars increased the bed surface area, and therefore the DNA load. (B) Simulated cfDNA (122 bp) recovery versus inter-pillar spacing. Average velocity, bed length, and SPE probability were 0.6 mm s−1, 24 mm (corrected for circular pillars), and 2%, respectively. Simulations were iterated over 11 cfDNA starting positions, and if any data points did not converge, the data was fit with a fourth-order polynomial and averaged over 5–100% of the half-channel width. For more information on the simulation and parameters, see ESI† and Table S1.
Herein, we report a polymer-based microfluidic device for the μSPE of cfDNA/ctDNA from plasma. We describe the composition of the IB to allow for the efficient extraction of cfDNA/ctDNA fragments directly from plasma. We also discuss the development and fabrication of a device with the ability to process large input volumes of plasma (>5 mL) that satisfies sampling statistics to search for rare ctDNA fragments (load >700 ng of cfDNA) and can be mass-produced using either hot embossing or injection molding to reduce microfluidic device cost.32 Also, because the molding step produces the extraction bed and requires only UV/O3 activation, no beads are necessary keeping device cost low and reducing post-molding assembly steps. We will show the utility of the device for selecting model cfDNA fragments spiked into plasma over a large size range (50–700 bp) with high efficiency and using the device for selecting cfDNA/ctDNA from plasma samples secured from cancer patients (colorectal cancer, CRC and non-small cell lung cancer, NSCL). Finally, we will show the ability to detect point mutations (KRAS) within the μSPE extracted ctDNA.
Experimental
Reagents and chemicals
Genomic DNA was extracted from HT29 and LS180 cell lines that were purchased from ATCC (USA). SW620, SW480 and HTC116 cells were generously provided by Prof. Dan A. Dixon’s research group at KU Medical Center (KUMC). Plasma samples from healthy donors and cancer patients were secured from Bioreclamation (USA) and the KUMC IRB approved Biospecimen Repository Core Facility (see ESI,† Table S2 for patient information). TapeStation supplies were purchased from Agilent (USA). Polyethylene glycol (PEG) and MgCl2 were secured from Sigma-Aldrich (USA), ethanol and acetic acid came from Fisher (USA). TE buffer was purchased from G Biosciences (USA), PBS from Hyclone (USA) and nuclease free water from VWR (USA). Carboxyl magnetic particles (4.0–4.5 μm) used for protein/peptide clearing were purchased from Spherotech (USA). PEEK tubing and capillaries for connecting the device to syringe pumps for fluidic operation was secured from IDEX (USA). The cyclic olefin copolymer (COC) and polycarbonate (PC) used for the microfluidics was acquired from Knightsbridge Plastics, Inc. (USA). The release agent used for the embossing (MoldWiz® F-57-M) was from Axel Plastics (USA). The supplies for protein electrophoresis, SSO SYBR® Green Supermix, and the DNA ladder were acquired from Bio-Rad (USA). The DNA 80 bp size standards for capillary gel electrophoresis were provided by Sciex (USA). The primers for PCR, qPCR and LDR were purchased from IDT DNA (USA), and the ligase and proteinase K enzymes as well as the OneTaq® Quick-Load® 2X Master Mix with Standard Buffer used in PCR were secured from New England Bio-labs (USA). Two commercial kits – Plasma/Serum Cell-Free Circulating DNA Purification Mini Kit from Norgen (USA) and QIAamp® MiniElute ccfDNA Mini Kit from Qiagen (USA) for cfDNA isolation were compared to the μSPE for extracting different model DNA fragment sizes. Qiagen also supplied the QIAquick® PCR purification kit for PCR product purification.
Device fabrication
The mold masters for the 1-bed and 3-bed microfluidic devices were fabricated in brass using high precision micromachining.28 The 24-bed device mold master was fabricated using a lithography technique due to the size/spacing of the pillars (~5 μm) and is discussed in detail in the ESI.† Irrespective of device architecture, the polymer replicas of the mold master were produced using hot embossing either into PC or COC by utilizing a Precision Press model P3H-15-PLX (Wabash MPI, USA). Prior to embossing, polymer wafers were rinsed with 2-propanol (Sigma) and deionized (DI) water (>17.9 MΩ) from an E-pure water purification system (Barnstead, USA) and subsequently dried in an oven at 65 °C overnight. Embossing was performed at 175 psi for 4 min and at 162 °C and 185 °C for PC and COC, respectively. Solution and buffer reservoirs (1 mm in diameter) were drilled into the 5 mm thick plastic chips. Before assembly, the cover plate and fluidic substrate were sonicated in 10% Micro-90 for 15 min, rinsed with isopropanol and DI water, dried at 65 °C, and UV/O3-modified for 30 min (22 mW cm−2) in a homebuilt UV chamber equipped with a quartz, low-pressure Hg lamp to create surface confined carboxy groups. COC chips were thermally fusion bonded at 131 °C for 90 min. PC chips were bonded at 149 °C for 25 min (Heratherm oven, ThermoFischer, USA). Prior to assembly, non-contact profilometry (VK-X250, Keyence, Japan) was employed to assess the quality and dimensions of the molds and devices in different stages of fabrication.
cfDNA quantification
μSPE optimization was evaluated with the following cfDNA models: 122 and 290 bp amplicons generated from PCR of the KRAS gene secured from gDNA of cell lines (HT29). To obtain the amplicons, PCR cycles consisted of an initial denaturation step at 94 °C for 3 min followed by 40 cycles of the following: 94 °C for 30 s, 55 °C for 15 s, 72 °C for 30 s, with a final extension at 72 °C for 3 min. Purified amplicons, spiked into the plasma of healthy donors or into PBS. After extraction, quantitative PCR (qPCR) was used to assess the amount of cfDNA in the extract. Two μL of the extract was used in 10 μL of the PCR mixture with 0.25 μM reverse and forward primers. For primer sequences see ESI,† Table S3. Amplification curves are presented in the ESI† (Fig. S2). qPCR quantification of the cfDNA isolated from clinical samples was performed with reference to the expression of 18S and GAPDH housekeeping genes.33 The correction for cfDNA target availability was done according to the procedure described by Horlitz et al.34 The ESI† presents a detailed protocol.
Evaluation of the size distribution of the μSPE cfDNA fragments
Size distributions of cfDNA μSPE extracted fragments was analyzed using the electrophoresis TapeStation (Agilent, USA). The sample preparation and data analysis were carried out according to the supplier’s suggested protocol.
Mutation detection
Most commonly observed KRAS mutations35 in CRC and NSCLC are present in codons 12 and 13 (Fig. S3†) and were detected using PCR/LDR36 and the extract from the μSPE device containing cfDNA/ctDNA. PCR used the same conditions as that described to produce the 122 and 290 bp control fragments. PCR/LDRs were performed using a commercial thermocycler (Eppendorf, USA). The LDR of the PCR amplicons was performed with 4 nM discriminating and common-dye labeled primers (ESI,† Table S4) and 4 units of Taq ligase enzyme. The LDR mix was preheated to 94 °C for 2 min followed by 25 cycles using the following temperatures: 94 °C for 30 s; 54 °C for 4 min. Products were separated and quantified by capillary gel electrophoresis (CEQ8000, Beckman, USA), using a 50 cm, 8 capillary array filled with POP6® gel. After denaturation at 92 °C for 2 min, the samples were injected electrokinetically for 45 s at 2.5 kV (50 V cm−1) and separated at 8.5 kV (170 V cm−1) for 25 min. Size markers (20 and 80 nt) were co-injected with the sample.
Results and discussion
Condensation and μSPE of cfDNA using a plastic microfluidic chip with pillars
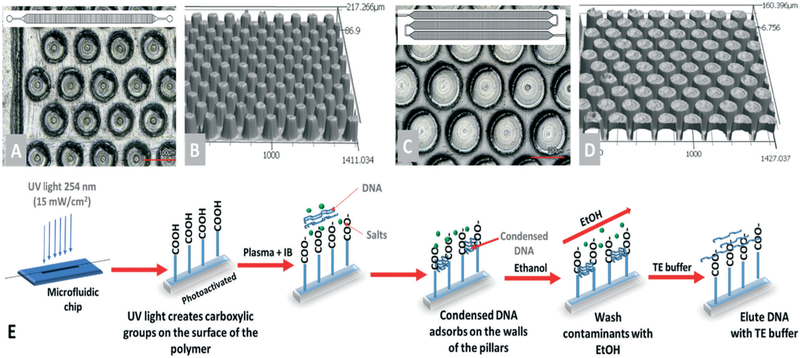
The μSPE of cfDNA reported herein relies on a mechanism known as polymer and salt-induced condensation (psi-condensation) of DNA.37 The neutralized charge of DNA due to the presence of a salt in the IB allows for DNA to condense onto a negatively charged surface.37 Also required is a neutral polymer, such as PEG, to assist in the condensation due to a volume exclusion mechanism and in some cases, an organic solvent such as ethanol (EtOH). The methodology is similar to SPRI31,37 in that surface-confined carboxylate groups are used for the selective extraction of DNA. The process of DNA condensation is reversible; in low ionic strength buffer or water, DNA recovers its negative charge and desorbs from the surface quantitatively. The amount of DNA that condenses onto the carboxylated surface depends on the density of the carboxylate groups. The size of DNA that condenses onto the surface depends on the concentration of PEG used in the IB as well as the salt composition and EtOH concentration.
To demonstrate that the cfDNA μSPE was not affected by microfluidic scaling issues (pillar spacing and size as well a bed size), devices were designed with different bed sizes, bed numbers, and pillar spacing/size (Fig. 2). We used three different configurations with each possessing different pillar sizes, inter-pillar spacing, extraction bed length, and number of extraction beds. For the device shown in Fig. 2A and B, the length of the extraction bed was 24 mm, width was 1.4 mm, and the depth was 94 ± 4 μm with the bed consisting of 3604 pillars that were 61 ± 2 μm diameter, a spacing of 34 ± 2 μm, and surface area of 1.2 cm2. The bed capacity or load, assuming extraction of 122 bp fragments, was calculated to be 7 ng (see Table 1). The device shown in Fig. 2C and D possessed 3 extraction beds connected in series with a 5.6× larger surface area compared to the device in Fig. 2A (1-bed device). Each bed was 34 mm long, 1.7 mm wide and 91 ± 1 μm deep. This device (15 202 pillars) had a load of 36 ng (122 bp DNA). Due to the relatively large size of the pillars and pillar spacing, we could fabricate these mold masters using high precision micromilling and use these mold masters for either hot embossing or injection molding.
Fig. 2.
Microfluidic devices used for μSPE of cfDNA. (A) Top view of the pillars in the 1-bed device in which the bed length was 24 mm, and bed width was 1.4 mm. (B) Profile image of a section of the pillared channel of the 1-bed chip. (C) View of the 3-bed design: 34 mm bed length with a 1.7 mm width. (D) A profile image of the pillars in the 3-bed device. (E) General workflow for the cfDNA extraction from the plasma inside the microfluidic device. Prior to injection, samples were digested with proteinase K, protein/peptide clearance with magnetic particles, and then mixed with IB. In the device, the condensed DNA adsorbs onto the μSPE surfaces, washed using 70% EtOH, and then dried before elution with a low ionic strength buffer.
Table 1.
Design characteristic and most relevant figures of the devices presented in this paper
| Chip characteristic | 1-Bed | 3-Bed | 24-Bed |
|---|---|---|---|
| Bed disposition | Single | Series | Parallel |
| Pillar size (μm) | 61 ± 2 | 108 ± 6 | 5.8 ± 0.5 |
| Pillar spacing (μm) | 34 ± 2 | 21 ± 5 | 6.9 ± 0.5 |
| Pillar height (μm) | 94 ± 4 | 91 ± 1 | 11.6 ± 0.4 |
| Bed length (mm) | 24 | 34 | 40 |
| Bed width (mm) | 1.4 | 1.7 | 2.0 |
| Number of pillars | 3604 | 15 202 | 1.4 M |
| Surface area (cm2) | 1.22 | 6.80 | 125 |
| Bed-capacity (ng) 122 bp cfDNA model | 7 | 36 | 717 |
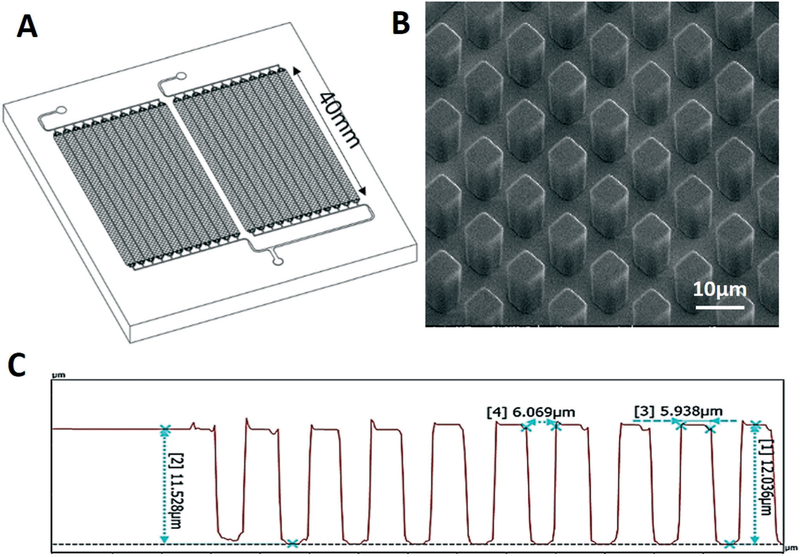
To improve the DNA recovery and mass load of the μSPE, and based on the calculations shown in Fig. 1, we designed a third configuration, which was comprised of 24-beds positioned in parallel (Fig. 3A). Each bed was 40 mm long and populated with 5.8 ± 0.5 μm diamond-shaped pillars that were spaced by 6.9 ± 0.5 μm with a height of 11.6 ± 0.4 μm (Fig. 3B and C and Table 1). This device had a cfDNA load of 717 ng based on the size of a 122 bp fragment and can process ~5 mL of plasma assuming a ~150 ng mL−1 concentration of cfDNA for a cancer patient (Table 1). Due to the pillar size and spacing, a Ni/Co mold master was made using a lithographic process (see ESI† and Fig. S4) for the 24-bed configuration.27
Fig. 3.
(A) Schematic of the 24-bed device fabricated to scale up the extraction load of cfDNA. (B) SEM images of post arrays. (C) Profilometry analysis of the device embossed in PC.
We evaluated PC and COC as possible plastics for the μSPE chip because of our success in using PC as a substrate for the extraction of gDNA and total RNA using SPRI.26–28 COC was tested because of the high efficiency of creating -COOH groups on its surface.38,39 While PC or COC can be injection molded using a molding master, PC is cheaper and we found that it was more robust for embossing, especially the 24-bed device. It can withstand larger variations in temperatures and pressure during molding compared to COC. Also, it does not create sticking friction with the NiCo molding master. This is a critical advantage when molding pillars with ~5 μm features (see ESI† and Fig. S5). However, COC can be readily injection molded.40
We evaluated both COC and PC as the substrate for the μSPE device using the optimized workflow and IB composition as we will discuss below. When cfDNA was extracted from the same plasma samples spiked with 122 bp fragments, the recovery was not significantly different (p = 0.028) for COC or PC devices. Therefore, either material can be used for the μSPE device with the choice predicated on substrate cost and/or moldability.
cfDNA μSPE workflow
The schematic illustrating the workflow for cfDNA μSPE is shown in Fig. 2E. In previous work from our laboratory, we demonstrated the extraction of gDNA and total RNA using 3–5% PEG with monovalent salts that efficiently condensed these nucleic acids onto -COOH microfluidic surfaces.26–28 In order to extract short ctDNA fragments (<150 bp) in particular with high efficiency from plasma, we sought to investigate different IB cocktails, for example the use of higher concentrations of PEG. However, high concentrations of PEG can cause the precipitation of plasma proteins and/or peptides.41,42 To mitigate this, digestion of plasma proteins and peptide clearance before mixing with the IB was performed. We should note that even the commercial kits for the SPE of cfDNA do a protein digestion step as well as peptide clearance before the SPE process (see Fig. S1†).
After pre-processing of the plasma, which included digestion with proteinase K, the sample was mixed with the IB (1 : 3) and hydrodynamically pumped through the μSPE device (2 μL min−1 for the 1-bed device and 0.8 μL min−1 for the 3-bed device). Devices were then rinsed with 70% EtOH to remove salts and other interfering components, but not the cfDNA that is adsorbed onto the surface of the chip. The microfluidic is then air-dried and the cfDNA eluted from the chip quantitatively using water or low ionic strength buffer. As can be seen from the workflow chart shown in Fig. S1† for the commercial kits, there are basically 11 processing steps (starting with plasma input), and the μSPE chip workflow utilized 6 processing steps with no centrifugation required for plasma as the input.
Plasma protein digestion
The enzymatic digestion of endogenous plasma proteins and histones was necessary to produce peptides that are less susceptible to the exclusion effect induced by PEG as well as release cfDNA fragments from histones. We evaluated the use of proteinase K, trypsin and pepsin. At fixed time intervals during the enzymatic reaction, aliquots of the plasma and proteolytic enzyme mixture were collected and mixed with the IB. The formation of a protein precipitate – visible as a white cloud (ESI,† Fig. S6A) – was deemed as an indicator of incomplete digestion. When no visual precipitate was observed, the level of digestion was also assessed using SDS-PAGE (ESI,† Fig. S6B).
Trypsin cleaves peptide bonds predominately at the carboxyl side of lysine or arginine residues and produces peptides with an average size of 700–1500 Da. Pepsin cleaves amide bonds at the N-terminal side43 of aromatic amino acids and produces peptide sizes similar to trypsin, however, more hydrophobic in nature. Proteinase K is considered a non-specific protease, with the predominant site of cleavage being the peptide bond adjacent to the carboxyl group of aliphatic and aromatic amino acids with blocked alpha amino groups and typically results in complete digestion of proteins to dipeptides.44 Upon tryptic and pepsin digestion and mixing with the IB, we observed the formation of a white precipitate, suggestive of pepsin and trypsin producing too large of peptide fragments to avoid precipitation with high concentrations of PEG (ESI,† Fig. S6A). When proteinase K was used, no precipitation was detected upon addition of the IB. Gel electrophoresis was used to demonstrate that most of the original proteins were digested (ESI,† Fig. S6B).
After digestion of plasma proteins with proteinase K and mixed with the IB and hydrodynamically shuttled through the μSPE chip, Coomassie blue was used to stain for proteins remaining within the chip after washing the chip with water (see ESI,† Fig. S6C). Only small amounts of peptide/proteins were observed when proteinase K digestion was used. We note that without endogenous protein digestion, the recovery of cfDNA models (122 and 200 bp fragments seeded into normal plasma) was significantly reduced. Therefore, as with all commercial cfDNA SPE kits using silica and/or magnetic particles, digestion with proteinase K is necessary to prevent co-extraction of intact proteins to the SPE bed and also, to release cfDNA from histones (see below).
IB composition for optimal cfDNA μSPE
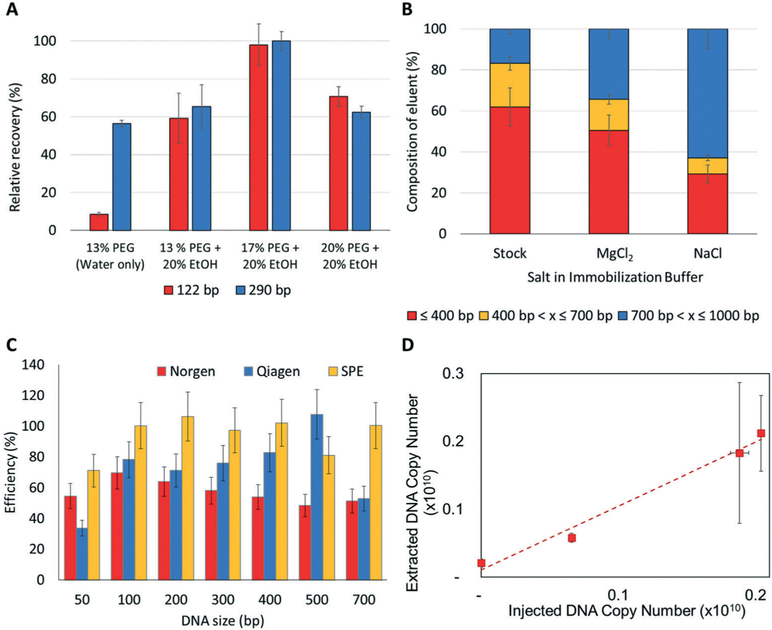
To isolate fragments from 50 bp to 1000 bp in length, which should cover the majority of ctDNA fragments in plasma, we evaluated the following components of the IB: (i) PEG concentration; (ii) cation source; and (iii) EtOH concentration (Fig. 4A and B). Each buffer component was evaluated using cfDNA models (122 bp and 290 bp fragments) that were spiked into the plasma of healthy donors with the recovery quantitatively determined using qPCR and the appropriate primers that flanked the sequence of the test templates (see Fig. S2† for qPCR profiles). These models were designed to have no homology with any sequence within the human genome.
Fig. 4.
(A) Effect of PEG concentration on the recovery of 122 bp (red) and 290 bp (blue) DNA fragments. Values represent the concentration of PEG after mixing with sample. All samples tested contained 10 mM MgCl2. (B) Effect of the salt type on the recovery of cfDNA. The stock consisted of different size ranges of oligonucleotides seeded into PBS. The bar graphs show the relative contribution of each fragment size range to the isolate using MgCl2 or NaCl. (C) μSPE performed with a 100 bp ladder as cfDNA model that was spiked into PBS. (D) Recovery of 122 bp cfDNA model as a function of input copy number spiked into a plasma from a normal donor (slope of the curve is 0.92 ± 0.12).
High PEG concentrations strengthen the exclusion effect by promoting the condensation of smaller DNA molecules but also increases the viscosity of the solution that can reduce the diffusion rate of even small DNA fragments to the μSPE surface, therefore reducing recovery. As shown in Fig. 4A, the maximum recovery of the cfDNA models was achieved when using 17% PEG with the recovery decreasing when using higher concentrations of PEG. We speculate that the loss of efficiency at high PEG concentrations (>20%) arises from the increased viscosity of the solution reducing the diffusion rate of cfDNA fragments to the surface. Therefore, we opted to use 17% PEG in the IB. This compares sharply to our previous reports of using PEG-based IB for gDNA and total RNA, which required 3–5% PEG.
Traditionally, Na+ salts are the source of cations used in psi-condensation.45 The cation neutralizes the charge on the DNA phosphate backbone assisting in the condensation of DNA. For cfDNA fragments, we evaluated an alternative cation, Mg2+. Besides neutralization, Mg2+ also forms bridges between DNA molecules.46 This is hypothesized to be particularly important for small DNA fragments; Cheng et al.47 demonstrated that Mg2+ produces higher condensation forces compared to monovalent salts. It also requires a lower concentration of the ion for efficient condensation; 10 mM for Mg2+ vs. 1 M for Na+, which is advantageous when EtOH and large concentrations of PEG are employed. We therefore compared the efficiency of μSPE using an IB containing NaCl, which has been used in our previous SPRI for gDNA,28 or MgCl2 along with the effect of salt concentration on the efficiency of μSPE for cfDNA. For these experiments, a stock solution with fragments ranging from 50–1000 bp was prepared with a different concentration of each fragment (see Fig. 4B for size distribution of the stock solution). As can be seen in Fig. 4B following μSPE, MgCl2 did a better job of maintaining the relative size distribution of the stock sample compared to NaCl due primarily to its better efficiency in extracting DNA fragments <400 bp. NaCl showed a preference for the extraction of the larger (>700 bp) DNA fragments. Changes in MgCl2 concentration (10 to 50 mM) had no effect on the cfDNA recovery (data not shown). Because plasma samples contain EDTA (blood drawn into EDTA tubes), it is important to ensure that Mg2+ is not completely chelated by the EDTA. Therefore, additional amounts of Mg2+ to make the final concentration >10 mM was added to compensate for chelation by EDTA found in the plasma sample.
EtOH affects the solubility of DNA and its conformation during condensation.48 Paithankar and Prasad62 concluded that EtOH is more efficient than PEG in inducing condensation of DNA fragments <150 bp, which is relevant as ctDNA fragments are within this size range. We observed that the highest cfDNA recoveries for the 122 and 200 bp fragments were achieved with 20–25% of EtOH in the IB, similar to what was reported by Arscott et al.63 In summary, for the model cfDNA fragments used in these studies, we found that the optimal IB composition to maximize recovery of cfDNA and ctDNA fragments should consist of 17% PEG, >10 mM MgCl2, and 20% EtOH.
For elution of the cfDNA from the μSPE nuclease free water, 10 mM Tris, and 1 mM EDTA was employed; no significant differences were observed in cfDNA elution with other aqueous buffers, such as PCR buffers (data not shown). Tween 20 was added to the elution solution at a concentration of 0.05% to improve the wettability of the chip surface and to prevent air from being trapped between pillars.
Comparison of μSPE to commercial cfDNA kits
Using a DNA ladder consisting of 50–700 bp double-stranded DNA fragments, we compared the efficiency of μSPE with two commercial kits, the Norgen and Qiagen cfDNA isolation kits. In these experiments, the DNA ladder was spiked into PBS so that no interfering components, such as peptides and/or proteins, were present and we could monitor exclusively recovery of the cfDNA targets. As shown in Fig. 4C, the μSPE showed recoveries that were >90% across the cfDNA size range of 100–700 bp. The results for the commercial kits are comparable to literature values for these kits and ranged from 50–70% for the Norgen kit and 53–100% for the Qiagen kit over the 100–700 bp size range.18 In terms of the recovery of the shortest fragment tested (50 bp), the μSPE method gave significantly higher recovery compared to the commercial kits. We note that the SPE of these short fragments is important as they can be a large size sub-population within the total cfDNA population for certain cancer diseases.49
We also spiked plasma from healthy donors with the 122 bp fragment bearing KRAS mutations (PCR from SW620, SW480 or LS180 cells, which carry KRAS mutations) to mimic ctDNA found in cancer patients. Using the conditions optimized for our μSPE devices, 92 ± 12% of the DNA spiked into the plasma sample was recovered (Fig. 4D). When the 122 bp DNA fragment was spiked into plasma and recovered, we observed an RSD of ~22% for samples processed over a 6 month period of time. The gDNA was found to be recovered at a significantly lower efficiency (0.01%). We also compared the recovery of the model 122 bp fragment bearing KRAS mutations seeded into plasma for the 3-bed device and noted no significant differences in terms of recovery compared to the 1-bed device.
Quantification of ctDNA in cancer patients’ plasma samples
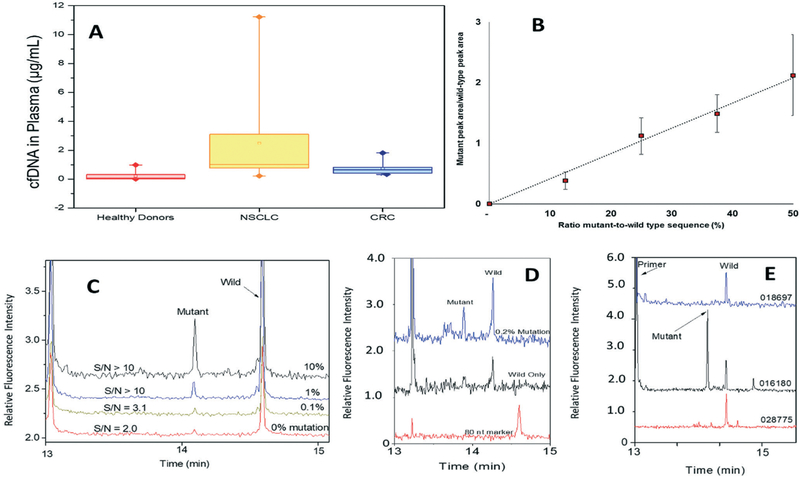
The cfDNA from plasma collected from patients with CRC and NSCLC as well as healthy donors was extracted using μSPE. Extracted cfDNA was quantified using qPCR and the 18S and GAPDH housekeeping genes according to the method of Horlitz et al.34 (see ESI†). The cfDNA average/median concentrations in plasma from healthy donors, NSCLC, and CRC were 0.065 μg mL−1 (range 0.020–0.980 μg mL−1), 1.0 μg mL−1 (range 0.222–9.3 μg mL−1) and 0.534 μg mL−1 (0.335–1.8 μg mL−1), respectively (see Fig. 5A). cfDNA mass differences between healthy donors and cancer patients was significant (healthy controls vs. NSCLC, p = 4 × 10−6; healthy controls vs. CRC, p = 3 × 10−4; Mann Whitney U-Wilcox test). These values agree with other reports.14,50–52
Fig. 5.
(A) cfDNA level detected in the plasma of healthy donors and patients diagnosed with CRC and NSCLC. DNA quantification was done using the 18S housekeeping gene as the reference according to the procedure described in the Experimental section and ESI.† Samples from 6 healthy donors, 3 NSCLC and 5 CRC patients were processed (≥3 replica each). (B) Peak area ratio for detected mutant and wild-type ctDNA/cfDNA targets as observed by capillary gel electrophoresis. (C) Electropherograms for detection of the G12V mutation present at different frequencies in the plasma sample. (D) Separation of LDR products for samples containing different levels of G12D mutant ctDNA model. (E) Electropherograms of LDR products following G12S KRAS mutation analysis of clinical samples (see sample IDs in the figure and in Table S2†).
Identification of KRAS mutations in the ctDNA extracted from plasma using the μSPE
Results from clinical trials have demonstrated that cancer patients with the wild-type KRAS genes respond better to antiepidermal growth factor receptor (EGFR) monoclonal antibody therapies than patients with mutated KRAS. Because ~40% of CRCs are driven by mutations in the RAS family of genes,53 clinical guidelines recommend that RAS mutational testing should be performed for all patients being considered for anti-EGFR mAb therapy.54 In NSCLC, testing for KRAS mutations are recommended following diagnosis to eliminate the need to probe for EGFR alterations, which are mutually exclusive with KRAS.55 Additionally, ASCO 2018 guidelines state that it is appropriate to include KRAS testing either initially or when routine EGFR, ALK, BRAF, and ROS1 testing is negative.55 ASCO also provides input on the role of cfDNA in patients with NSCLC, suggesting that in clinical settings in which tissue is limited and/or insufficient for molecular testing, physicians may use cfDNA to identify actionable mutations.55 However, reports have demonstrated that the correlation between tumor and ctDNA mutation status is 70%.56,57 Therefore, we used our μSPE chip for the extraction of cfDNA from plasma samples and tested samples for KRAS mutations; a list of the mutations tested can be found in Table S4 and Fig. S3 (see ESI†).
We performed a Ligase Detection Reaction (LDR) to test for KRAS mutations in μSPE extracted cfDNA.36 LDR has the ability to identify mutations in low copy numbers even in high abundance of wild-type copies, such as would be the case for ctDNA co-extracted within the entire cfDNA population. We evaluated the sensitivity of the LDR assay by spiking controlled levels of mutated KRAS amplicons into the plasma of healthy donors, which initially tested negative for KRAS mutations. The cfDNA/ctDNA isolated from plasma were recovered using μSPE and analyzed by LDR with the results shown in Fig. 5B and C. The products of LDR were separated by capillary gel electrophoresis to determine which mutations were present. The ratio of peak areas for mutant and wild-type cfDNA fragments showed a linear relationship with the concentration of mutant fragments in the mix. Additionally, the mutated cfDNA was detected at 0.1% for the G12V (Fig. 5C) and 0.2% for the G12D mutant-to-wild-type ratios (Fig. 5D).
We performed LDR mutation detection assays on the cfDNA/ctDNA μSPE extracted from clinical plasma samples that were banked in our Biorepository and stored at −80 °C (see Table 2 for summary of results). PCR/LDR identified KRAS G12D mutations in the ctDNA from NSCLC sample #028775 (tissue was not tested). Also, G12S/G13D mutations in sample #016180 from the cfDNA were detected while only the G13D was detected in the tumor tissue (Fig. 5E). This observation is not surprising given that concordance between ctDNA mutational status and that from the primary tumor is ~70% as noted above. This 70% concordance can arise from the fact that the mutational status of metastatic sites and the primary tumor can be different and that ctDNA represents contributions from both the primary tumor and metastatic sites. In the case of CRC patient #021000, we did not detect the G12V mutation identified in the tumor tissue. It has also been documented that during chemotherapy, KRAS mutations can be acquired,58 and as such, tumor tissue can possess a different mutational status compared to the cfDNA due to different sampling times (before versus after chemotherapy). Another NSCLC patient was negative for mutated KRAS, which was in concordance with the KRAS mutational status of the tumor tissue.
Table 2.
cfDNA mutational status in clinical samples
| Sample ID | Cancer type | Mutation in tumor tissue | cfDNA mutation |
|---|---|---|---|
| μSPE | |||
| 028775 | NSCLC | Not tested | G12D |
| 016180 | NSCLC | G13D | G12S/G13D |
| 018697 | NSCLC | Negative | Negative |
| 021000 | CRC | G12V | Negative |
| 021019 | CRC | Negative | Negative |
| 022282 | CRC | Negative | Negative |
| 022327 | CRC | Negative | Negative |
| 021957 | CRC | Negative | Negative |
Conclusions
A μSPE assay and device for the efficient extraction of cfDNA and ctDNA from plasma was reported herein. The μSPE provided high recovery of cfDNA/ctDNA fragments that was found to be >90% over the size range of 100–700 bp and a recovery higher than 70% of very short DNA fragments (50 bp). In addition, the extraction efficiency of interfering gDNA was low(0.01%). Comparison of the recovery of the μSPE to commercial kits also showed comparable efficiency, except for the smallest DNA fragment tested (50 bp), with the μSPE providing significantly better recovery. This will be important in providing a better representation of the entire ctDNA population, which can produce on average smaller DNA fragments compared to the cfDNA fragments produced from non-diseased cells.
An additional advantage of the μSPE compared to the commercial benchtop kits was the smaller number of processing steps (11 steps for commercial kits and 6 for μSPE). This observation will reduce the potential errors associated with sample handling and improve assay reproducibility (we are currently engaged in large scale testing to compare assay reproducibility of the commercial kits versus the μSPE).
The μSPE uses an extraction bed made from a thermoplastic via replication with the extraction bed populated with micropillars that can permit high-scale production of devices at low cost; the device can be injection molded at <$2 per part with ~1000 devices produced per day per injection molding machine based upon the injection molding cycle time. In addition, post-molding device assembly and preparation steps are simple requiring only fusion bonding a cover plate to the molded device and UV/O3 activation of the plastic surface to produce -COOH groups for the SPE of cfDNA. Thus, no beads or additional reagents are required to be inserted into the μSPE device following molding, keeping the consumable price low.
In addition, we demonstrated that the device could be scaled-up in terms of DNA load (7 → 717 ng) by simply changing the extraction bed number (1, 3 or 24), size and pillar density. Because we are using replication techniques to produce the chip, the cost of the chip consumable is relatively independent of scale-up. For example, the number of posts for the 1-bed device was 3604 while that for the 24-bed device it was 1.4 M pillars. The injection molding price per device is the same with the only difference being that associated with generating the optical mask required to pattern the resist onto the Si wafer for defining the features on the molding master.
We performed the SPE of cfDNA/ctDNA from clinical plasma samples collected from CRC and NSCLC patients. Using our μSPE device, we found that the plasma from NSCLC patients had on average higher concentrations of cfDNA than CRC patients consistent with previous literature reports. From the extracted cfDNA, we were able to perform a mutation detection assay of KRAS genes and found similar mutations in the primary tumor tissue with some disconcordance, which is not surprising given the difference in biological sourcing of the cfDNA (primary tumor tissue versus plasma).
While in the current work, we reported only the μSPE on chip to allow us to explicitly test the utility of the assay for cfDNA, our future work will build on technologies that we already reported59,60 in terms of integrating other processing steps to the μSPE, such as plasma isolation, proteinase K digestion, and protein/peptide clearance. In addition, we will seek to integrate the molecular analysis to the chip as well to provide a viable platform for fully automated cfDNA analyses.61
Supplementary Material
Acknowledgements
The authors acknowledge financial support from the National Institute of Biomedical Imaging and Bioengineering P-41 Center grant P41EB020594. Research reported in this publication was also supported by the National Cancer Institute Cancer Center Support Grant P30CA168524 and used the Biospecimen Shared Resource at KUMC (Kansas City, KS). Authors are grateful to the Ralph Adams Microfabrication Facilities. We thank Dr. Mateusz L. Hupert for valuable advice and help on mold fabrication and Prof. Dan A. Dixon’s research group for donation of cell lines. De-identified clinical samples were provided from the KU Cancer Center’s Biospecimen Repository Core Facility (BRCF) along with detailed clinical outcomes and phenotypes. Blood-plasma specimens were obtained from individuals enrolled under the repository’s IRB approved protocol (HSC #5929) and following U.S. common rule. Once the patient provides written, informed consent in accordance with the BRCF IRB protocol, a blood sample is collected, centrifuged, the plasma withdrawn and then banked. The plasma sample is de-identified to the user and transported directly to the laboratory for evaluation.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c8lc00716k
Conflicts of interest
Authors do not have any conflict of interest.
References
- 1.Fleischhacker M and Schmidt B, Nat. Med, 2008, 14, 914–915. [DOI] [PubMed] [Google Scholar]
- 2.Mouliere F, Robert B, Arnau Peyrotte E, Del Rio M, Ychou M, Molina F, Gongora C and Thierry AR, PLoS One, 2011, 6, e23418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP and Shendure J, PLoS Genet., 2016, 12, e1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siravegna G, Marsoni S, Siena S and Bardelli A, Nat. Rev. Clin. Oncol, 2017, 14, 531. [DOI] [PubMed] [Google Scholar]
- 5.Bennett CW, Berchem G, Kim YJ and El-Khoury V, Oncotarget, 2015, 7, 71013–71035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos CDM, Jackson JM, Witek MA and Soper SA, Cancer J., 2018, 24, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malapelle U, Sirera R, Jantus-Lewintre E, Reclusa P, Calabuig-Fariñas S, Blasco A, Pisapia P, Rolfo C and Camps C, Expert Rev. Mol. Diagn, 2017, 17, 209–215. [DOI] [PubMed] [Google Scholar]
- 8.Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, Gillet B, Gongora C, Dechelotte P, Robert B, Del Rio M, Lamy P-J, Bibeau F, Nouaille M, Loriot V, Jarrousse A-S, Molina F, Mathonnet M, Pezet D and Ychou M, Nat. Med, 2014, 20, 430–435. [DOI] [PubMed] [Google Scholar]
- 9.Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O’Connell A, Feeney N, Mach SL, Jänne PA and Oxnard GR, JAMA Oncol., 2016, 2, 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Selvaganapathy PR, Gould TJ, Dwivedi DJ, Liu D, Fox-Robichaud AE and Liaw PC, Lab Chip, 2015, 15, 3925–3933. [DOI] [PubMed] [Google Scholar]
- 11.Pao E, Renier C, Lemaire C, Che J, Matsumoto Di Carlo M, Triboulet M, Srivinas S, Jeffrey SS, Kulkarni RP, Rettig M, Sollier E and Di Carlo D, Cancer Res., 2016, 76, 4967–4967. [Google Scholar]
- 12.Qiagen, QIAamp® Circulating Nucleic Acid Handbook, Qiagen, 3rd edn, 2013. [Google Scholar]
- 13.Kim CJ, Park J, Sunkara V, Kim TH, Lee Y, Lee K, Kim MH and Cho YK, Lab Chip, 2018, 18, 1320–1329. [DOI] [PubMed] [Google Scholar]
- 14.van Ginkel JH, van den Broek DA, van Kuik J, Linders D, de Weger R, Willems SM and Huibers MMH, Cancer Med., 2017, 6, 2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Messaoudi S, Rolet F, Mouliere F and Thierry AR, Clin. Chim. Acta, 2013, 424, 222–230. [DOI] [PubMed] [Google Scholar]
- 16.Bronkhorst AJ, Aucamp J and Pretorius PJ, Clin. Chim. Acta, 2015, 450, 243–253. [DOI] [PubMed] [Google Scholar]
- 17.Fleischhacker M, Schmidt B, Weickmann S, Fersching DMI, Leszinski GS, Siegele B, Stoetzer OJ, Nagel D and Holdenrieder S, Clin. Chim. Acta, 2011, 412, 2085–2088. [DOI] [PubMed] [Google Scholar]
- 18.Devonshire AS, Whale AS, Gutteridge A, Jones G, Cowen S, Foy CA and Huggett JF, Anal. Bioanal. Chem, 2014, 406, 6499–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pérez-Barrios C, Nieto-Alcolado I, Torrente M, Jiménez-Sánchez C, Calvo V, Gutierrez-Sanz L, Palka M, Donoso-Navarro E, Provencio M and Romero A, Transl. Lung Cancer Res, 2016, 5, 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorber L, Zwaenepoel K, Deschoolmeester V, Roeyen G, Lardon F, Rolfo C and Pauwels P, J. Mol. Diagn, 2017, 19, 162–168. [DOI] [PubMed] [Google Scholar]
- 21.Mauger F, Dulary C, Daviaud C, Deleuze JF and Tost J, Anal. Bioanal. Chem, 2015, 407, 6873–6878. [DOI] [PubMed] [Google Scholar]
- 22.Malentacchi F, Pizzamiglio S, Verderio P, Pazzagli M, Orlando C, Ciniselli CM, Günther K and Gelmini S, Clin. Chem. Lab. Med, 2015, 53, 1935–1942. [DOI] [PubMed] [Google Scholar]
- 23.Dittrich PS and Manz A, Nat. Rev. Drug Discovery, 2006, 5, 210–218. [DOI] [PubMed] [Google Scholar]
- 24.Lamanda A, Lu Y, Gill N and Wong PK, presented in part at the 2015 Transducers - 2015 18th International Conference on Solid-State Sensors, Actuators and Microsystems, TRANSDUCERS 2015, Anchorage, USA, 2015. [Google Scholar]
- 25.Sonnenberg A, Marciniak JY, McCanna J, Krishnan R, Rassenti L, Kipps TJ and Heller MJ, Electrophoresis, 2013, 34, 1076–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witek MA, Hupert ML, Park DSW, Fears K, Murphy MC and Soper SA, Anal. Chem, 2008, 80, 3483–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park DS, Hupert ML, Witek MA, You BH, Datta P, Guy J, Lee JB, Soper SA, Nikitopoulos DE and Murphy MC, Biomed. Microdevices, 2008, 10, 21–33. [DOI] [PubMed] [Google Scholar]
- 28.Witek MA, Llopis SD, Wheatley A, McCarley RL and Soper SA, Nucleic Acids Res., 2006, 34, e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, Vaidya B, Patel AB, Ford SM, McCarley RL and Soper SA, Anal. Chem, 2003, 75, 2975–2984. [DOI] [PubMed] [Google Scholar]
- 30.Deangelis MM, Wang DG and Hawkins TL, Nucleic Acids Res., 1995, 23, 4742–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkins TL, O’Connor-Morin T, Roy A and Santillan C, Nucleic Acids Res., 1994, 22, 4543–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker H, Lab Chip, 2009, 9, 2759–2762. [DOI] [PubMed] [Google Scholar]
- 33.Gong B, Xue J, Yu J, Li H, Hu H, Yen H, Hu J, Dong Q and Chen F, Oncol. Lett, 2012, 3, 897–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horlitz M, Lucas A and Sprenger-Haussels M, PLoS One, 2009, 4, e7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.COSMIC, COSMIC - Catalogue of Somatic Mutations in Cancer, https://cancer.sanger.ac.uk/cosmic, Accessed 01-Mai-2018, 2018.
- 36.Khanna M, Park P, Zirvi M, Cao W, Picon A, Day J, Paty P and Barany F, Oncogene, 1999, 18, 27–38. [DOI] [PubMed] [Google Scholar]
- 37.Rittich B and Spanova A, J. Sep. Sci, 2013, 36, 2472–2485. [DOI] [PubMed] [Google Scholar]
- 38.Jackson JM, Witek MA, Hupert ML, Brady C, Pullagurla S, Kamande J, Aufforth RD, Tignanelli CJ, Torphy RJ, Yeh JJ and Soper SA, Lab Chip, 2014, 14, 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Neil CE, Jackson JM, Shim SH and Soper SA, Anal. Chem, 2016, 88, 3686–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C-S, Chen S-C, Liao W-H, Chien R-D and Lin S-H, Int. Commun. Heat Mass Transfer, 2010, 37, 1290–1294. [Google Scholar]
- 41.Ingham KC, Methods Enzymol., 1990, 182, 301–306. [DOI] [PubMed] [Google Scholar]
- 42.Sim SL, He T, Tscheliessnig A, Mueller M, Tan RB and Jungbauer A, J. Biotechnol, 2012, 157, 315–319. [DOI] [PubMed] [Google Scholar]
- 43.Lopez-Ferrer D, Petritis K, Robinson EW, Hixson KK, Tian Z, Lee JH, Lee SW, Tolic N, Weitz KK, Belov ME, Smith RD and Pasa-Tolic L, Mol. Cell. Proteomics, 2011, 10, M110 001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu CC, MacCoss MJ, Howell KE and Yates JR 3rd, Nat. Biotechnol, 2003, 21, 532–538. [DOI] [PubMed] [Google Scholar]
- 45.Rocha MS, Cavalcante AG, Silva R and Ramos EB, J. Phys. Chem. B, 2014, 118, 4832–4839. [DOI] [PubMed] [Google Scholar]
- 46.Teif VB and Bohinc K, Prog. Biophys. Mol. Biol, 2011, 105, 208–222. [DOI] [PubMed] [Google Scholar]
- 47.Cheng C, Jia J-L and Ran S-Y, Soft Matter, 2015, 11, 3927–3935. [DOI] [PubMed] [Google Scholar]
- 48.Roy KB, Antony T, Saxena A and Bohidar HB, J. Phys. Chem. B, 1999, 103, 5117–5121. [Google Scholar]
- 49.Perakis S, Auer M, Belic J and Heitzer E, Adv. Clin. Chem, 2017, 80, 73–153. [DOI] [PubMed] [Google Scholar]
- 50.Wu TL, Zhang D, Chia JH, Tsao KC, Sun CF and Wu JT, Clin. Chim. Acta, 2002, 321, 77–87. [DOI] [PubMed] [Google Scholar]
- 51.Schwarzenbach H, Stoehlmacher J, Pantel K and Goekkurt E, Ann. N. Y. Acad. Sci, 2008, 1137, 190–196. [DOI] [PubMed] [Google Scholar]
- 52.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW Jr., Alizadeh AA and Diehn M, Nat. Med, 2014, 20, 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fleming M, Ravula S, Tatishchev SF and Wang HL, J. Gastrointest. Oncol, 2012, 3, 153–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allegra CJ, Rumble RB, Hamilton SR, Mangu PB, Roach N, Hantel A and Schilsky RL, J. Clin. Oncol, 2016, 34, 179–185. [DOI] [PubMed] [Google Scholar]
- 55.Kalemkerian GP, Narula N, Kennedy EB, Biermann WA, Donington J, Leighl NB, Lew M, Pantelas J, Ramalingam SS, Reck M, Saqi A, Simoff M, Singh N and Sundaram B, J. Clin. Oncol, 2018, 36, 911–919. [DOI] [PubMed] [Google Scholar]
- 56.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, Anagnostou V, Fiksel J, Cristiano S, Papp E, Speir S, Reinert T, Orntoft M.-b. W., Woodward BD, Murphy D, Parpart-li S, Riley D, Nesselbush M, Sengamalay N, Georgiadis A, Li QK, Madsen MR, Mortensen FV, Huiskens J, Punt C, Grieken NV, Fijneman R, Meijer G, Husain H, Scharpf RB, D. LA Jr, Jones S, Angiuoli S, Ørntoft T, Nielsen HJ, Andersen CL, Velculescu VE, van Grieken N, Fijneman R, Meijer G, Husain H, Scharpf RB, Diaz LA, Jones S, Angiuoli S, Ørntoft, Nielsen HJ, Andersen CL and Velculescu VE, Sci. Transl. Med, 2017, 2415, eaan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih l M, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N and Diaz LA Jr., Sci. Transl. Med, 2014, 6, 224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SKN, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih LM, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N and Diaz LA, Sci. Transl. Med, 2014, 6, 224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashimoto M, Barany F and Soper SA, Biosens. Bioelectron, 2006, 21, 1915–1923. [DOI] [PubMed] [Google Scholar]
- 60.Lee J, Soper SA and Murray KK, Analyst, 2009, 134, 2426–2433. [DOI] [PubMed] [Google Scholar]
- 61.Wang H, Chen HW, Hupert ML, Chen PC, Datta P, Pittman TL, Goettert J, Murphy MC, Williams D, Barany F and Soper SA, Angew. Chem., Int. Ed, 2012, 51, 4349–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paithankar KR and Prasad KSN, Nucleic Acids Res, 1991, 19(6), 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arscott PG, Ma C, Wenner JR and Bloomfield VA, Biopolymers, 1995, 36(3), 345–364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.