Abstract
The electrophoretic mobility shift assay (EMSA) is a well-established method to detect formation of complexes between proteins and nucleic acids and to determine, among other parameters, equilibrium constants for the interaction. Mixtures of protein and nucleic acid solutions of various ratios are analyzed via polyacrylamide gel electrophoresis (PAGE) under native conditions. In general, protein-nucleic acid complexes will migrate more slowly than the free nucleic acid. From the distributions of the nucleic acid components in the observed bands in individual gel lanes, quantitative parameters such as the dissociation constant (Kd) of the interaction can be measured. This unit describes a simple and rapid EMSA that relies either on precast commercial or handcast polyacrylamide gels and uses unlabeled protein and nucleic acid. Nucleic acids are instead detected with SYBR Gold stain and band intensities established with a standard gel imaging system. We used this protocol specifically to determine Kd values for complexes between the PAZ domain of Argonaute 2 (Ago2) enzyme and native and chemically modified RNA oligonucleotides. EMSA-based equilibrium constants are compared to those determined with isothermal titration calorimetry (ITC, UNIT 7.4). Advantages and limitations of this simple EMSA are discussed by comparing it to other techniques used for determination of equilibrium constants of protein-RNA interactions and a troubleshooting guide is provided.
Keywords: Ago2, electrophoresis, equilibrium constant, protein-nucleic acid interaction, RNA
INTRODUCTION
Nucleic acid-protein interactions are ubiquitous and absolutely essential in biological information transfer, including replication, transcription, repair, and RNA metabolism (Cusack et al., 2017). Gel electrophoresis using polyacrylamide gels (PAGE, UNIT 10.4) and the electrophoretic mobility shift assay (EMSA) remain important and are widely used approaches for detecting nucleic-acid protein interactions and determining the stability of individual complexes (Fried, 1989; Hellman and Fried, 2007; Chen, 2011; Alves and Cunha, 2012). This protocol details the steps for establishing the equilibrium dissociation constant Kd of an RNA-protein complex by EMSA. Solutions of RNA and protein are mixed in different ratios, and the binding reactions then separated by non-denaturing PAGE (https://tools.thermofisher.com/content/sfs/brochures/1601945-Protein-Interactions-Handbook.pdf). In general, a protein-RNA complex will migrate more slowly through the gel matrix relative to the RNA alone, thus causing a shift on the gel, e.g. (Yang et al., 1999). Individual bands can be visualized by end-labeling the RNA radioactively (32P), or using fluorescent or chemiluminescent probes in combination with a gel imager. However, in our assay, we have used non-labeled RNA together with the SYBR Gold stain (Tuma et al., 1999) for detecting and quantifying bands. From the distribution of the RNA component among the protein-RNA and RNA bands in individual lanes on the gel, one can determine the equilibrium dissociation constant Kd of the complex in a straightforward manner. An approximate value for the equilibrium constant can be obtained by finding the concentration of protein at which roughly half the nucleic acid component is bound and half remains free. A more precise way to determine Kd is to plot individual fractions of the nucleic acid bound in each reaction versus the concentration of protein and then perform a non-linear regression (Heffler et al., 2012) using free or commercially available software.
Human Argonaute 2 (Ago2) is an 859 amino acid protein that lies at the heart of the RNA-induced silencing complex (RISC) (Sheu-Gruttadauria and MacRae, 2017). The protein binds small interfering RNA (siRNA) duplexes consisting of 21mer guide (antisense) and passenger (sense) strands with 3′-terminal dinucleotide overhangs. Ago2 contains multiple domains, including the MID (binds 5′-end of guide siRNA), PIWI (harbors the endonuclease active site), PAZ (binds 3′-end of guide siRNA) and two linkage domains. The passenger strand is eventually cleaved or discarded and the target RNA loaded into Ago2 opposite the guide strand, resulting in cleavage of the former by the PIWI domain via a dual-metal ion mechanism (Watts and Corey, 2012; Wilson and Doudna, 2013). RISC contains other Ago proteins besides Ago2, i.e. Ago1, Ago3 and Ago4, and while all are associated with micro RNAs (miRNAs), only Ago2 exhibits endonuclease activity (Meister et al., 2004). Chemically modified siRNAs are widely explored as therapeutics for treatment of a range of diseases (Crooke et al., 2018; Shen and Corey 2018). ONPATTRO (Patisiran) is a modified siRNA formulated in lipids and manufactured by Alnylam Pharmaceuticals Inc. (Cambridge, MA) that was approved by the US FDA in August of 2018 for treatment of polyneuropathy of hereditary transthyretin-mediated amyloidosis in adults (Sheridan, 2017). The PAZ domain of Ago2 binds the last two nucleotides of guide siRNA and assists in separating guide and passenger siRNA (Ma et al., 2004; Elkayam et al., 2012; Schirle and MacRae, 2012) (Figure 1). The identities of the 3′-overhanging nucleotides in the siRNA guide strand and their chemical modification affect the interaction with Ago2 PAZ and silencing activity (Kandeel et al., 2014; Alagia et al., 2018). We relied on the EMSA protocol presented here to determine equilibrium dissociation constants Kd of the interactions between native and chemically modified RNAs and the human Ago2 PAZ domain.
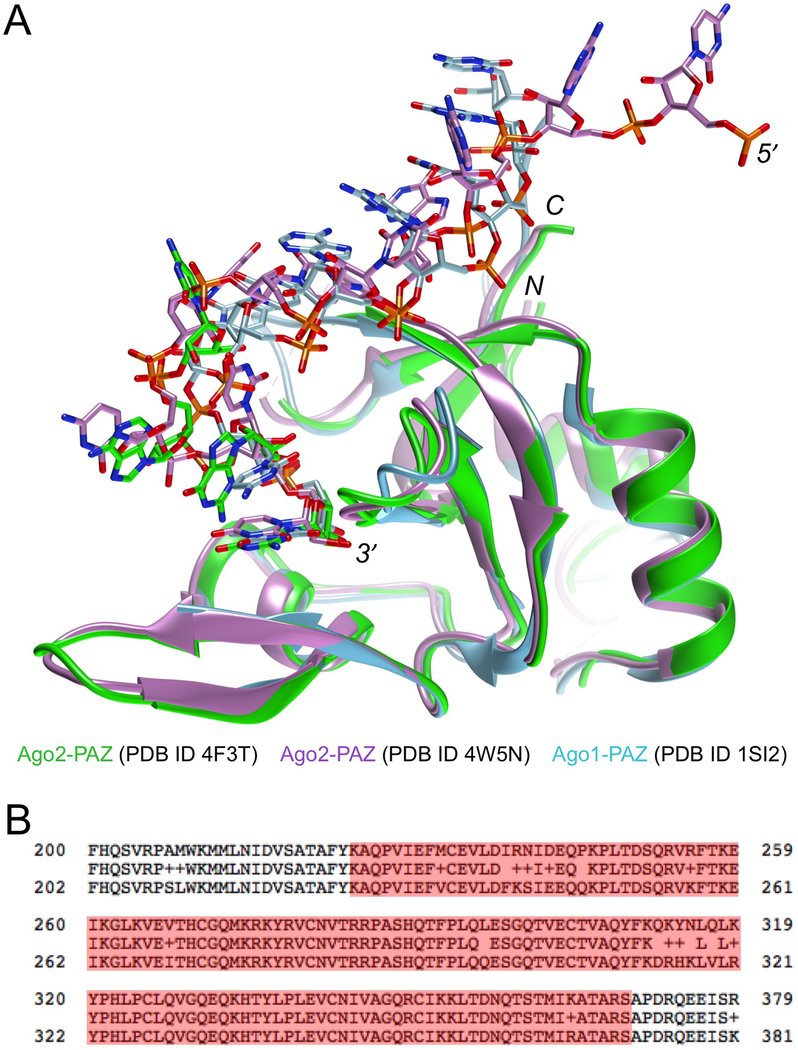
Figure 1.
Overlay of crystal structures of human Ago1- and Ago2-PAZ in complex with RNA oligonucleotides (A) and alignment of the human Ago1 (top line) and Ago2 (bottom line) sequences (B). Only a portion of the 859 amino acids is included; the consensus sequence is shown in the middle line and PAZ domain residues are highlighted: 225–369 (Ago1) and 227–371 (Ago2). The PAZ domain binds the 3′-end of guide siRNA. The models shown are from crystal structures of full-length Ago2 bound either to duplex RNA (Schirle & MacRae, 2012) or an miRNA single strand (Elkayam et al., 2012). In both cases only PAZ domain residues 226–351 were included in the illustration. In both crystal structures, only portions of the 3′-half of the RNA guide strand were visible in the electron density. The third structure depicted is of a complex between a separately expressed human Ago1 PAZ domain (residues 224–349 were resolved in the electron density) and an RNA nonamer (Ma et al., 2004). Coordinates were retrieved from the Protein Data Bank (https://www.rcsb.org/) (Berman et al., 2000) and the structural figure in panel A was generated with the program UCSF Chimera (Pettersen et al., 2004).
BASIC PROTOCOL
ELECTROPHORETIC MOBILITY SHIFT ASSAY (EMSA) USING LABEL-FREE, NATIVE AND CHEMICALLY MODIFIED OLIGO-RIBONUCLEOTIDES AND HUMAN ARGONAUTE 2 PAZ DOMAIN PROTEIN (AGO2-PAZ) TO DETERMINE EQUILIBRIUM DISSOCIATION CONSTANTS Kd OF PROTEIN-RNA COMPLEXES
The following procedures outline the mixing of RNA and protein solutions of defined concentrations in various ratios to establish the binding reactions, the preparation of polyacrylamide gels to run EMSAs for analyzing protein-RNA binding, staining and imaging of gels to measure the intensities of individual bands and the distribution of RNA between the free and protein-bound forms, and the determination of Kd values using standard software to compute non-linear regressions. However, neither the synthesis of RNA oligonucleotides nor the expression and purification of the Ago2 PAZ domain are described in detail. Briefly, RNAs were provided by Alnylam Pharmaceuticals Inc. (Cambridge, MA; native oligoribonucleotides) or AM Biotechnologies LLC (Houston, TX; 2′-OMe/3′-phosphorodithioate-(PS2; both non-bridging phosphate oxygen atoms replaced with sulfur) oligoribonucleotide, here referred to as MS2-modified RNA). Native RNAs were synthesized by standard or adapted solid phase phosphoramidite synthesis, purified (UNIT 10.3) by high performance liquid chromatography (HPLC, UNIT 10.5) and desalted (UNIT 10.7). Identities and purities of all oligonucleotides were confirmed by electrospray ionization mass spectrometry (ESI-MS) and ion-exchange HPLC (IEX-HPLC), respectively. The MS2-modified RNA was synthesized and purified as reported in (Yang et al., 2017; UNIT 4.77). An expression system for human Ago2 PAZ (amino acids 227–371, Figure 1B) was generated by gene synthesis (GenScript Inc.) and subcloning into the pHD116 plasmid. The plasmid was transformed into E. coli BL21 (DE3) Gold cells using standard procedures. After expression the protein was first purified by Ni affinity chromatography and, following cleavage of the His6 tag by PreScission protease (GE Healthcare), by ion exchange and gel filtration chromatography. The identity of the Ago2 PAZ domain was established by tryptic digestion in combination with MS analysis.
Materials
Human Ago2 PAZ domain protein (180 μM in 5 mM HEPES, 100 mM KCl, 10 mM DTT)
RNA(s) of interest (500 nM); see Figure 2 for sequences.
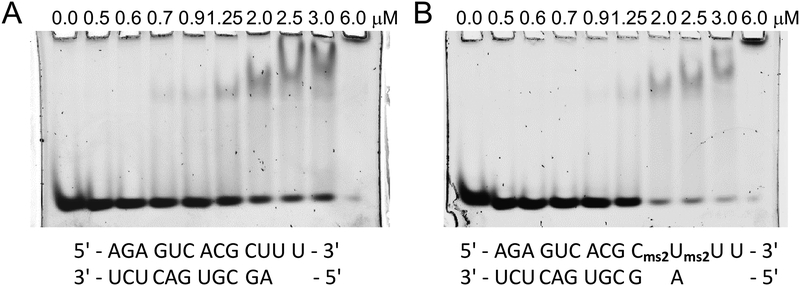
Figure 2.
Gel images of binding reactions with various ratios between Ago2 PAZ and (A) native RNA and (B) MS2-modified RNA. RNA sequences are shown below the gels, protein concentrations are shown above individual lanes, and the RNA concentration was 250 nM.
NaCl (Acros, CAS number: 7647-14-5)
EDTA (Sigma, CAS number: 60-00-4)
DTT (Sigma, CAS number: 3483-12-3)
Tris base (RPI Corp., CAS number: 77-86-1)
Boric acid (RPI Corp., CAS number: 10043-35-3)
APS (Biorad catalog number: 161–0700)
Bromophenol blue dye
10,000x Sybr Gold (Invitrogen catalog number: S11494)
Nuclease free water (Promega P119E-C)
Agarose (see recipes)
Glycerol (RPI, CAS number: 56-81-5)
Ficoll (25mg/ml) (Sigma, CAS number: 26873-85-8)
Gel Materials
30% acrylamide/bis solution 37.5:1 (Bio-Rad catalog number: 161–0158)
Sterile water
Tetramethylethylenediamine (TEMED, Bio-Rad catalog number: 161–0800)
10% alkaline phosphatase buffer (APS, see recipes)
5x Tris/borate/EDTA buffer (TBE, see recipes)
10x binding buffer
5x TBE buffer (1 L)
10% APS (1 mL)
5% polyacrylamide gel solution (see Reagents and Solutions)
Microcentrifuge tubes
Equipment
1.5 mm Mini gel plates
Gel electrophoresis chamber, e.g. Bio-Rad Mini-PROTEAN Tetra Cell System
Black cassettes
Shaker
Microcentrifuge tubes
ChemiDoc MP imaging system (Bio-Rad)
Make 5% native TBE polyacrylamide gel by first assembling the two 1.5 mm-thick glass gel plates (i.e. with clips) and set into the gel apparatus.
-
Then seal the bottom of the gel plates with 1% agarose with a 1.0 mL micropipette.
Make sure to pipette agarose on both sides of the gel plates to seal them as well.
Prepare 10 mL of 5% (0.5x TBE) polyacrylamide gel solution (see reagents and solutions).
Using a 1.0 mL micropipette, inject the polyacrylamide solution into the gel cast. Stick the comb into the assembled gel sandwich. Wait 1 hour to let the gel polymerize.
Prepare electrophoresis running buffer by diluting 100 mL of 5x TBE buffer with 900 mL of sterile water to create 1 L of 0.5x TBE.
Pre-run the polyacrylamide gel by submerging the cast gel in the running buffer from step 5 and run at 45V-60V for 1 hour or until the current is stable.
Prepare 100 μL of RNA stock (500 nM) per gel in nuclease free water and dilute PAZ protein with 1x binding buffer to create two protein stock solutions, one of 10 μM concentration and the other of 40 μM concentration.
Allow the stock solutions prepared in the previous step to warm to room temperature and then prepare nine human Ago2 PAZ protein and RNA binding mixtures in separate microcentrifuge tubes according to Table 1. Create control sample by mixing 10 μL of RNA and 10 μL of bromophenol bue dye. Incubate at room temperature for 30 minutes.
After the incubation is complete, add 2 μL of ficoll solution to each tube, including the control.
Load 20 μL of each sample into a well with the gel; the control sample should be loaded into the first lane.
Run the gel at 40–70V, 6–15mA for 1 hour or until the samples are roughly 2/3 down the gel (Figure 2).
Add 5 μL of 10,000x Sybr Gold to 50 mL of 0.5x TBE buffer to create 50 mL of 1x Sybr Gold solution.
-
Stain each gel by submerging it in 50 mL of 1x Sybr gold in a black cassette for 30 minutes.
Note: the Sybr Gold stain does not bind to the Ago2 PAZ domain.
Rinse the gels with deionized water (optional).
Image gel using ChemiDoc MP Imaging System with a Blot/UV/Stain-Free Sample Tray (http://www.bio-rad.com/en-us/product/chemidoc-mp-imaging-system?ID=NINJ8ZE8Z) (Figure 2).
Calculate the binding affinity between the RNA and the protein by analyzing the gel images with ImageJ. See the anticipated results section below for more details.
Table 1.
Sample Volumes and Concentrations of PAZ and RNA needed for ESMA.
| Protein concentration (nM) | PAZ protein added (μL) | 1x binding buffer added (μL) | RNA(s) of interest added (μL) |
|---|---|---|---|
| 500 | 1.0 (10 μM stock) | 9.0 | 10.0 (500 nM stock) |
| 600 | 1.2 | 8.8 | 10.0 |
| 700 | 1.4 | 8.6 | 10.0 |
| 900 | 1.6 | 8.2 | 10.0 |
| 1250 | 2.5 | 7.5 | 10.0 |
| 2000 | 4.0 | 6.0 | 10.0 |
| 2500 | 5.0 | 5.0 | 10.0 |
| 3000 | 6.0 | 4.0 | 10.0 |
| 6000 | 3.0 (40 μM stock) | 7.0 | 10.0 |
Reagents and Solutions
- 10x binding buffer
- 0.1 mM HEPES
- 1.5 M NaCl
- 30 mM EDTA
- 10 mM DTT
- 1x binding buffer (10 mL)
- 10x binding buffer: 1 mL
- 50% glycerol: 2 mL
- Sterile water: 7 mL
- 5x TBE buffer (1L)
- 54 g Tris base
- 27.5 g boric acid
- 20 mL of 0.5M EDTA pH 8.0
- 10% APS (1 mL)
- 0.1 g APS
- 1 mL of sterile water
- 1% agarose
- 0.5 g of agarose
- 50 mL of 0.5x TBE buffer
- 5% polyacrylamide gel solution
- Sterile H2O: 7.27 mL
- 30% acrylamide: 1.66 mL
- 5x TBE: 1.00 mL
- 10% APS: 70 mL
- TEMED: 3.5 mL
COMMENTARY
Background Information
There are multiple techniques for determining the equilibrium dissociation constant Kd of a protein-nucleic acid binding interaction besides the EMSA (Fried, 1989; Hellman and Fried, 2007; Chen, 2011; Alves and Cunha, 2012; Heffler et al., 2012). These include the filter-binding assay (Woodbury and von Hippel, 1983), fluorescence intensity (Teplova et al., 2000), chemical shift differences using 1H-15N heteronuclear single quantum coherence (HSQC) NMR titration experiments (Frank et al., 2012), surface plasmon resonance (SPR; McDonnell, 2001; Ma et al., 2004), acoustic measurements (Cooper and Whalen, 2005; Godber et al., 2005), biolayer interferometry (BLI; Lou et al., 2016; UNIT 7.25), microscale thermophoresis (MST; Mueller et al., 2017), and isothermal titration calorimetry (ITC; Rozners et al., 2015; UNIT 7.4), among others.
The choice to use one method versus another may depend on the amounts of material available, the particular environment in which the experiment is to be conducted (e.g. solution, chip, microfluidics, cell, etc.), whether labeling can be accomplished relatively easily and/or cheaply, or whether equipment that is needed with some of the above techniques is accessible to the investigator. Thus, EMSAs, spectroscopic approaches and SPR require less material than filter-binding assays and ITC. An important consideration concerns the Kd limit, that is the upper end of the stability range for the complex between protein and nucleic acid of interest at which a particular technique still affords precise data. In this regard, EMSA, SPR and BLI allow reliable measurements of interactions with Kd values of as low as 10−12 M (Jing and Bowser, 2011). This contrasts with the Kd limit for ITC that is around 10−8 to 10−9 M. However, ITC, despite this limitation and the need for relatively large amounts of material, offers the benefits of a solution environment and label-free binding partners; it is also the method of choice to establish precise thermodynamic parameters of a binding interaction (ΔH, ΔS and ΔG; UNIT 7.4). Most of the alternative approaches that yield the Kd of an interaction require labeling of either protein or nucleic acid, e.g. 15N (protein; NMR), 32P (DNA or RNA; filter-binding assay, EMSA), biotinylation (DNA or RNA; SPR, BLI UNIT 7.25), or fluorescent probes (protein or nucleic acid; MST, EMSA) (Jiang and Egli, 2011; UNIT 7.15).
We opted to use EMSA with label-free oligoribonucleotides to determine the Kd of the interaction between human Ago2 PAZ and RNA. Advantages of this approach are speed, low cost and the small amounts of material needed. A familiar limitation lies in the environment, i.e. a gel matrix and not solution, that can potentially affect the binding reaction. The complex between PAZ domain and RNA is known to be of a 1:1 stoichiometry (Figure 1A), but published data reveal somewhat divergent values for the stability of the complex. Thus, the Kd values for the interactions between Ago2 PAZ and mono- (e.g. UMP) and dinucleotides (e.g. UpUMP) varied between 10 and 60 μM based on ITC (Kandeel et al., 2014). In contrast, the Kd of the interaction between Ago1 PAZ and an 11mer/13mer RNA duplex with a 3′-terminal dinucleotide overhang (Figure 2A) was reported to be 2.2 nM as determined by SPR (Ma et al. 2004; Ago1 and Ago2 PAZ have highly similar sequences, Figure 1B). In order to verify the EMSA-based Kd values, we therefore decided to also conduct independent ITC experiments. The outcomes of the two approaches are discussed in the section ‘Anticipated Results’.
Critical Parameters and Troubleshooting
We tested two types of polyacrylamide gels and varied their percentages.
TBE polyacrylamide gel. We started from 15% and then continuously lowered the percentage to 12%, 10%, 8%, 5%, and finally to 3.5%, as gels of higher % polyacrylamide prevented the complex from getting into and migrating through the gel. However, a 3.5% gel turned out to be too frail to handle and it also showed no significant improvement compared to the 5% gel in terms of the migration of the complex. Thus, a 5% gel was judged to be optimal in terms of structural integrity and migration of the complex, even at higher concentrations of PAZ protein.
Tris-glycine polyacrylamide gel. We assayed binding reactions on 4%−20%, 12%, 10%, and 7.5% gels. Altough the complex band migrated through the gel, increasing the protein:RNA ratio did not result in reduced and increased intensity of the RNA and complex band, respectively and such gels do apparently not constitute a suitable environment for the PAZ-RNA complex.
We also examined the effects of varying the pH, e.g. 1% TBE pH 8.0 and 0.5% TBE pH 7.6, and the addition of detergents (sodium cholate 0.04%, 1/10 CMC, and 0.008% Triton X-100, ½ CMC). However, detergents proved to be detrimental to the complex. Thus, a 0.5x TBE buffer for both gel and running buffer yielded much better results. We did not use Mg2+ or Ca2+ in the buffer and neither was used for annealing the RNAs. We don’t anticipate that the presence of these ions would have a significant effect. Moreover, neither ion is present in or near the binding pocket of the PAZ domain.
We observed that gel bands representing the PAZ-RNA complex tended to smear and that migration was limited at the highest ratio between protein and RNA. This issue may become exacerbated by the use of higher concentrations of protein and RNA owing to the lack of labels on the latter. However, the RNA bands were typically quite sharp, allowing reasonably precise density measurements even in cases where migration of the complex was limited or bands representing the complex were spread out.
For PAGE experiments one can of course use either pre- or handcast gels. We relied on commercial precast gels in some cases but also poured our own gels. There are various systems on the market that facilitate handpouring gels, e.g. Sure Cast Gel Handcast System offered by ThermoFisher (https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-gel-electrophoresis/protein-gels/surecast-gel-handcast-system.html).
Anticipated Results
To analyze the gel image obtained, the NIH program ImageJ was used (Schneider et al., 2012). The program can be downloaded from the NIH website for free (https://imagej.nih.gov/ij). The image shown in Figure 2 was first inverted and then the raw integrated density (RID) of each band was measured with ImageJ. ImageJ may not have the RID as a default setting. If that is the case, go to the “Analyze” tab, click “Set Measurements”, and then check the “Integrated Density” box and click “OK”. The results were then transferred into an Excel spreadsheet to perform the following calculations. Each RID was subtracted from the lowest RID, with the lowest RID being subtracted from itself resulting in zero. These values are the relative RIDs. Each relative RID was divided by the largest relative RID and then multiplied by 100, with the largest relative RID being divided by itself and then mutltiplied by 100, resulting in 100. These are the percentage RIDs. Finally, each percentage RID was subtracted from 100, resulting in the percentage of binding, i.e. complex formation, for each protein concentration.
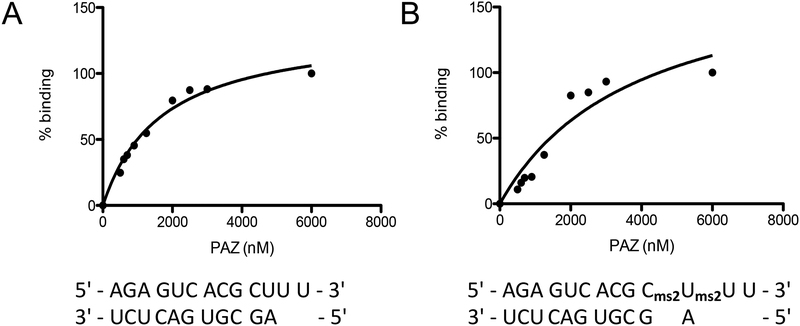
These results then served as input for the program GraphPad Prism (version 5.00 for Mac; GraphPad Software, La Jolla CA; https://www.graphpad.com/). Select the option that plots a single “Y” value for each data point and press create. Enter the protein concentrations in the x-column and enter the percent binding in the y-column. Label both columns. Next, click on the results tab and select nonlinear regression (curve fit) under “XY analyses”. Then select the equation that is labeled “one site – specific binding”, which computes the Kd value (Figure 3).
Figure 3.
Non-linear regressions for the PAZ complex with (A) native RNA, and (B) modified RNA based on the gels depicted in Figure 2.
The Kd values obtained for the PAZ complexes with native and modified RNA are shown in Table 2.
Table 2:
Kd values obtained for PAZ complexes with native and modified RNA.
| RNA | Kd [μM], nonlinear fit |
|---|---|
| 5′ - AGA GUC ACG CUU U -
3′ 3′ - UCU CAG UGC GA - 5′ |
1.7 |
| 5′ - AGA GUC ACG
Cms2Ums2U U - 3′ 3′ - UCU CAG UGC G A - 5′ |
3.9 |
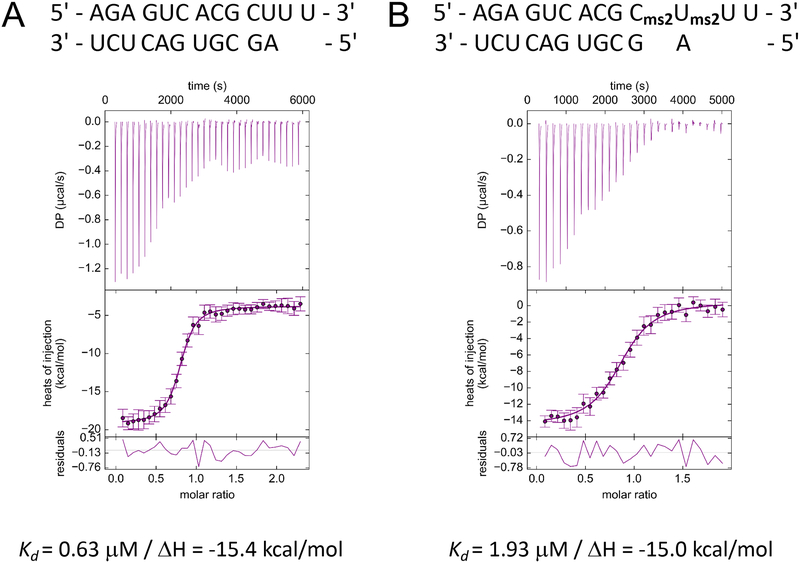
Our EMSA is relatively simple, cheap and quick. But does it also provide reasonably accurate data, keeping in mind the usual limitations of a gel-based assay? We decided to compare the EMSA-based data to Kd values obtained from isothermal titration calorimetry (ITC) experiments. These Kds were 0.6 μM (PAZ complex with native RNA; Figure 4A) and 1.9 μM (PAZ complex with modified RNA; Figure 4B) and therefore quite similar to the EMSA data. Typically, one would carry out the EMSA in triplicate and then report average values.
Figure 4.
Results of isothermal titration calorimetry experiments for the PAZ complex with (A) native RNA and (B) modified RNA. The experiments were conducted under the following conditions: 56 μM Ago2 PAZ and 600 μM RNA in a buffer composed of 5 mM HEPES pH 7.5, 150 mM KCl, 10% glycerol and10 mM DTT. RNA was added to the protein solution by 28 to 33 1.2 μL injections at 25°C. Note the 1:1 stoichiometry of the complexes, consistent with the structural models depicted in Figure 1A.
An earlier publication had reported a 2.2 nM Kd for the Ago1 PAZ complex with the same native RNA construct that was employed here and using SPR (Ma et al., 2004). However, we believe that this may be an overestimation of the tightness of binding between PAZ domain and RNA. For one, Ago1 and Ago2 PAZ have similar sequences (Figure 1B) and it is unlikely that the former should exceed the latter in terms of its affinity for RNA by three orders of magnitude. In addition, the buried surface (binding interface, calculated with the program PRince (Barik et al., 2012); http://www.facweb.iitkgp.ac.in/~rbahadur/prince/home.html) in a complex between thrombin and an anti-thrombin RNA aptamer (Long et al., 2008; PDB ID 3DD2) that exhibits a Kd of 1 nM (Abeydeera et al., 2016), protein 719 Å2 and RNA 790 Å2, is much larger than that between Ago2 PAZ and the 3′-end of an RNA (complex with PDB ID 4F3T, Figure 1A): protein 344 Å2 and RNA 465 Å2. However, it is of note that complexes can exhibit dramatically different stabilities as a consequence of a key interaction that leaves the buried surface essentially unchanged. Thus, replacement of a single phosphate (PO2) by a phosphorodithioate (PS2) moiety in RNA aptamers was reported to trigger a dramatic change in the dissociation constant of the complexes with their respective targets from ca. 1 nM to ca. 1 pM (anti-thrombin and anti-VEGF aptamers; Abeydeera et al., 2016). Finally, the Kd values for complexes between Piwi protein PAZ domains and various RNAs based on ITC were recently reported to be between 2 and 34 μM (Tian et al., 2011).
Time Considerations
The entire experiment, from pouring the gels to imaging and calculating the Kd value takes around 6 hours. Pouring, prepping, waiting for the gel to polymerize, and pre-running it can be accomplished in around 2 hours. Doing the calculations to prepare the individual samples, preparing them, loading the samples and running the gel takes 2–3 hours. Finally, gel imaging, measuring band densities with ImageJ and obtaining the Kd value using Prism took another hour. With one gel apparatus, two gels can be run simultaneously and thus Kd values for two complexes can be obtained on the same day.
Acknowledgements
We are grateful to Alnylam Pharmaceuticals Inc. for synthesis of native RNAs and AM Biotechnologies LLC (Dr. Xianbin Yang) for synthesis of the MS2-modified oligo. We would like to thank Drs. Muthiah Manoharan, Martin Maier, Klaus Charisse, Alexander Kel’in, Mark Schlegel, Vasant Jadhav and Ivan Zlatev (Alnylam Pharmaceuticals) as well as Drs. David Cortez, Huzefa Dungrawala, F. Peter Guengerich and Carl Sedgeman (Vanderbilt University) for helpful discussions. ITC data were acquired by Dr. Jia Ma at the Biophysical Analysis Laboratory, Purdue University, West Lafayette, IN. Supported by Alnylam Pharmaceuticals and the US NIH (grant R01 GM071461).
Literature Cited
- Abeydeera ND, Egli M, Cox N, Mercier K, Conde JN, Pallan PS, Mizurini DM, Sierant M, Hibti F-E, Hassell T, Wang T, Liu F-W, Liu H-M, Martinez C, Sood AK, Lybrand TP, Frydman C, Monteiro RQ, Gomer RH, Nawrot B, and Yang X 2016. Evoking picomolar binding in RNA by a single phosphorodithioate linkage. Nucleic Acids Res 44:8052–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagia A, Jorge AF, Aviñó A, Cova TFGG, Crehuet R, Grijalvo S, Pais AACC, and Eritja R 2018. Exploring PAZ/3′-overhang interaction to improve siRNA specificity. A combined experimental and modeling study. Chem. Sci 9:2074–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves C, and Cunha C 2012. Electrophoretic mobility shift assay: analyzing protein-nucleic acid interactions In: Gel Electrophoresis - Advanced Techniques, Magdeldin S (Ed.), pp. 205–228; ISBN: 978-953-51-0457-5. [Google Scholar]
- Barik A, Mishra A, and Bahadur RP 2012. PRince: a web server for structural and physicochemical analysis of Protein-RNA interface. Nucleic Acids Res 40:W440–W444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, and Bourne PE 2000. The Protein Data Bank. Nucleic Acids Res 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R 2011. A general EMSA (gel-shift) protocol. Bio Protoc 1:e24. [Google Scholar]
- Cooper MA and Whalen C 2005. Profiling molecular interactions using label-free acoustic screening. Drug Discov. Today Technol 2:241–245. [DOI] [PubMed] [Google Scholar]
- Crooke ST, Witztum JL, Bennett CF, and Baker BF 2018. RNA-targeted therapeutics. Cell Metab 227:714–739. [DOI] [PubMed] [Google Scholar]
- Cusack S, Müller C, Orengo C and Thornton J (Edts.) 2017. Protein-nucleic acid interactions. Catalysis and regulation. Curr. Opin. Struct. Biol 47, 1–176. [DOI] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, and Joshua-Tor L 2012. The structure of human Argonaute-2 in complex with miR-20a. Cell 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Hauver J, Sonenberg N, and Bushan N 2012. Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J 31:3588–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried MG 1989. Measurement of protein-DNA interaction parameters by electrophoresis mobility shift assay. Electrophoresis 10:366–376. [DOI] [PubMed] [Google Scholar]
- Godber B, Thompson KS, Rehak M, Uludag Y, Kelling S, Sleptsov A, Frogley M, Wiehler K, Whalen C, and Cooper MA 2005. Direct quantification of analyte concentration by resonant acoustic profiling. Clin. Chem 51:1962–1972. [DOI] [PubMed] [Google Scholar]
- Heffler MA, Walters RD and Kugel JF 2012. Using electrophoretic mobility shift assays to measure equilibrium dissociation constants: GAL4-p53 binding DNA as a model system. Biochem. Mol. Biol. Educ 40:383–387. [DOI] [PubMed] [Google Scholar]
- Hellman LM, and Fried MG 2007. Electrophoretic mobility shift assays (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc 2:1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, and Egli M 2011. Use of chromophoric ligands to visually screen co-crystals of putative protein-nucleic acid complexes. Curr. Protoc. Nucleic Acid Chem 46:7.15.1–7.15.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing M, and Bowser M 2011. Methods for measuring aptamer-protein equilibria: A review. Anal. Chim. Acta 686:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M, Al-Taher A, Nakashima R, Sakaguchi T, Kandeel A, Nagaya Y, Kitamura Y, and Kitade Y 2014. Bioenergetics and gene silencing approaches for unraveling nucleotide recognition by the human EIF2C2/Ago2 PAZ domain. PLoS ONE 9, e94538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Long MB, White RR, and Sullenger BA 2008. Crystal structure of an RNA aptamer bound to thrombin. RNA 14:2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou X, Egli M, and Yang X 2016. Determining functional aptamer-protein interaction by biolayer interferometry. Curr. Protoc. Nucleic Acid Chem 67:7.25.1–7.25.15. [DOI] [PubMed] [Google Scholar]
- Ma J-B, Ye K, and Patel DJ 2004. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429:318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell JM 2001. Surface plasmon resonance: Towards an understanding of the mechanisms of biological molecular recognition. Curr. Opin. Chem. Biol 5:572–577. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A Dorsett Y, Teng G, and Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15:185–197. [DOI] [PubMed] [Google Scholar]
- Mueller AM, Breitsprecher D, Duhr S, Baaske P, Schubert T, and Längst G 2017. MicroScale Thermophoresis: A rapid and precise method to quantify protein-nucleic acid interactions in solution In: Kaufmann M, Klinger C, and Savelsbergh A (eds) Functional Genomics. Meth. Mol. Biol, 1654:151–164. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE 2004. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comp. Chem 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Rozners E, Pilch DS, and Egli M 2015. Calorimetry of nucleic acids. Curr. Protoc. Nucleic Acid Chem 63:7.4.1–7.4.12. [DOI] [PubMed] [Google Scholar]
- Schirle NT, and MacRae IJ 2012. The crystal structure of human Argonaute2. Science 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Meth 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, and Corey DR 2018. Chemistry, mechanism and clinical status of antisense oligo-nucleotides and duplex RNAs. Nucleic Acids Res 46:1584–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C 2017. With Alnylam’s amyloidosis success, RNAi approval hopes soar. Nat. Biotechnol 35, 995–997. [DOI] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J, and MacRae IJ 2017. Structural foundations of RNA silencing by Argonaute. J. Mol. Biol 429:2619–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Tereshko V, Sanishvili R, Joachimiak A, Bushueva T, Anderson WF, and Egli M 2000. The structure of the yrdC gene product from E. coli reveals a new fold and suggests a role in RNA-binding. Protein Sci 9:2557–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ma J-B, and Patel DJ 2011. Structural basis for piRNA 2′-O-methylated 3′-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains. Proc. Natl. Acad. Sci. USA 108:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma RS, Beaudet MP, Jin X, Jones LJ, Cheung CY, Yue S, and Singer VL 1999. Characterization of SYBR Gold nucleic acid gel stain: a dye optimized for use with 300-nm ultraviolet transilluminators. Anal. Biochem 268:278–288. [DOI] [PubMed] [Google Scholar]
- Watts JK, and Corey DR 2012. Gene silencing by siRNAs and antisense oligonucleotides in the laboratory and the clinic. J. Pathol 226:365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, and Doudna JA 2013. Molecular mechanisms of RNA interference. Annu. Rev. Biophys 42:217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury CP Jr. and von Hippel PH 1983. On the determination of deoxyribonucleic acid-protein interaction parameters using the nitrocellulose filter-binding assay. Biochemistry 22:4730–4737. [DOI] [PubMed] [Google Scholar]
- Yang X, Fennewald S, Luxon BA, Aronson J, Herzog NK, and Gorenstein DG 1999. Aptamers containing thymidine 3′-O-phosphorodithioates: Synthesis and binding to nuclear factor-κB. Bioorg. Med. Chem. Lett 9:3357–3362. [DOI] [PubMed] [Google Scholar]
- Yang X 2017. Solid-phase synthesis of RNA analogs containing phosphorodithioate linkages. Curr. Protoc. Nucleic Acid Chem 70:4.77.1–4.77.13. [DOI] [PMC free article] [PubMed] [Google Scholar]