Abstract
Alterations of gut microbes play a role in the pathogenesis and progression of many disorders including liver and gastrointestinal diseases. Both qualitative and quantitative changes in gut microbiota have been associated with liver disease. Intestinal dysbiosis can disrupt the integrity of the intestinal barrier leading to pathological bacterial translocation and the initiation of an inflammatory response in the liver. In order to sustain symbiosis and protect from pathological bacterial translocation, antimicrobial proteins (AMPs) such as a-defensins and C-type lectins are expressed in the gastrointestinal tract. In this review, we provide an overview of the role of AMPs in different chronic liver disease such as alcoholic steatohepatitis, non-alcoholic fatty liver disease, and cirrhosis. In addition, potential approaches to modulate the function of AMPs and prevent bacterial translocation are discussed.
Keywords: Dysbiosis, Innate immune system, Bacterial translocation, Microbiome
Introduction
The gastrointestinal tract is the largest surface area in the body and is home to a vast consortium of symbiotic bacteria that play an important role in human health and disease. Microbiota are involved in basic human biological processes, including food digestion, modulation of immune responses, regulation of epithelial development and generation of a variety of products as a result of microbial metabolic activities. These products together with host–bacteria interactions influence both normal physiology and disease susceptibility. A disruption of the symbiosis between microbiota and host is known as dysbiosis and is described in multiple chronic diseases such as obesity [1], malnutrition [2], neurological disorders [3], inflammatory bowel disease [4], diabetes mellitus [5], metabolic syndrome, atherosclerosis [6], cancer [7] and liver disease [8–10].
Nutrition, other environmental and genetic factors can independently cause changes in the gut microbiota composition, which can present as qualitative changes such as increased proportions of harmful bacteria and reduced levels of beneficial bacteria, and also quantitative changes in the total amount of bacteria (intestinal bacterial overgrowth) [10, 11]. Intestinal bacterial overgrowth can affect both the luminal compartment and mucosa-associated bacteria [12]. As a result of dysbiosis, intestinal epithelial integrity is lost, mucus-associated defense is weakened and the intestine becomes more permeable. Hence, viable bacteria or microbial products are able to migrate from the intestines to mesenteric lymph nodes or other extra-gastrointestinal organs via the bloodstream, causing disease [13]. At distant sites, these bacterial products can be recognized via toll-like receptors (TLRs). Specific TLRs recognize pathogen-associated molecular patterns associated with bacteria. TLR-2 may be activated by various membrane components of Gram-positive bacteria. TLR-4 recognizes the lipid A portion of lipopolysaccharide (LPS). TLR-5 may be activated by flagellin, and bacterial DNA activates TLR-9. Activation of TLRs on macrophages leads to a variety of inflammatory cascades that in turn cause inflammation and may represent the driving force behind disease progression [14]. In addition, upon liver injury, hepatic stellate cells undergo an activation process in which they express TLR4. Therefore, LPS and other TLR ligands may enhance fibrogenic responses via direct stellate cell activation. Lastly, as also hepatocytes express TLR-2 and TLR-4, bacterial recognition by TLRs on hepatocytes may account for cell death occurring during liver injury [15].
Besides the tightly interconnected intestinal epithelial lining, the physical barrier to separate microbiota from intestinal surface is formed by a mucus layer. This mucus is secreted by goblet cells and largely consists of mucin glycoprotein sheets. In the colon, where mucus consists of two layers, the inner mucus layer is devoid of bacteria whereas the outer is colonized [16]. Likewise, in the small intestine bacteria are kept on distance from the epithelial wall [17]. In order to sustain the mucosal barrier and protect the host against enteric pathogens, a range of host antimicrobial factors are produced in the intestinal epithelium [18]. These intestinal antimicrobial proteins (AMPs) mediate killing of bacteria by attacking the basic cell wall structures through enzymatic and non-enzymatic mechanisms. Interestingly, dysfunctional antimicrobial defense has been described in different chronic liver diseases [19–21].
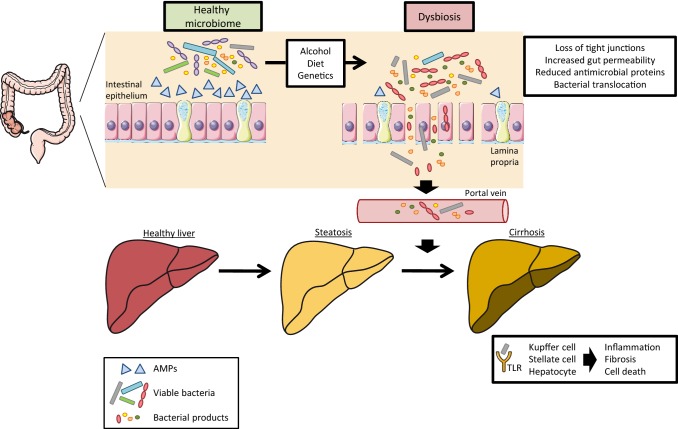
In this review, we summarize evidence supporting the contribution of bacterial translocation and the role of antimicrobial proteins in the development and progression of different chronic liver diseases such as alcoholic and non-alcoholic fatty liver disease (NAFLD), and cirrhosis (Fig. 1). Moreover, potential approaches to modulate the function of antimicrobial proteins to prevent bacterial translocation are discussed.
Fig. 1.
Steatosis (fatty liver) due to obesity (diet), alcohol consumption, other environmental or genetic factors is associated with dysbiosis, loss of intestinal tight junctions, reduced intestinal epithelial integrity, increased permeability and lower expression of antimicrobial proteins (AMPs). This allows bacterial products such as LPS (via the paracellular route) or viable bacteria (via not further detailed mechanisms) to translocate into the bloodstream and mesenteric lymph nodes. Via the portal vein, bacteria and their products reach the liver, where they promote progression to more severe stages of liver disease via recognition by Toll-like receptors (TLRs) on Kupffer cells, hepatic stellate cells and hepatocytes, leading to inflammation, fibrosis and cell death
(Figure made using Servier Medical Art; http://smart.servier.com)
Antimicrobial proteins
The surface of the mammalian intestine continuously encounters bacteria, fungi, viruses and parasites that could act as pathogens. In order to cope with these microbial challenges, a diverse collection of AMPs are produced in the intestinal epithelium and Paneth cells which rapidly kill or inactivate microorganisms. These AMPs consist of different protein families, which include defensins, cathelicidins, C-type lectins (such as the regenerating islet-derived protein (REG) family), ribonucleases (RNases, such as angiogenin 4) and S100 proteins (such as calprotectin (also known as S100A8–S100A9) and psoriasin (also known as S100A7)) [22]. Although most AMPs attack bacterial cell wall components, different AMP families use distinct molecular mechanisms to kill microorganisms [18]. This might explain that microbial resistance to multiple AMPs is rare.
The expression, secretion and activity of AMPs are under tight control of different factors. First, studies of germ-free mice have revealed that some intestinal AMPs require bacterial signals for their expression, whereas others are expressed independently of the microbiota [18]. For example, the expression of regenerating island-derived protein 3 gamma (REG3γ) is essentially absent in germ-free mice and is upregulated on colonization with a conventional microbiota [23]. Besides bacterial signals, host immune function controls the expression of these bacterially regulated AMPs. Studies in mice have shown that bacterial recognition by TLRs on intestinal epithelial cells is required for and upregulates the expression of REG3γ and REG3β [24, 25]. In addition, intestinal epithelial cell expression of REG3γ also requires interleukin 22 (IL-22), a cytokine mainly expressed by RORγt+ type 3 innate lymphoid cells (ILC3s) in the gut [26]. The production of this cytokine by ILC3s is enhanced by activation of the aryl hydrocarbon receptor (AhR) via specific bacterially derived molecules [27–29], indicating that bacteria might enhance the immune response to protect the host from harmful pathogens. Taken together, the expression and function of AMPs are the result of a symbiotic interplay between host and commensals, in which interruptions might cause disease. Here, we further describe the role of AMPs, more specifically C-type lectins, in different chronic liver diseases.
Alcoholic liver disease
Alcoholic liver disease affects several million people worldwide and can progress from hepatic fat accumulation (steatosis) and alcoholic steatohepatitis to cirrhosis and hepatocellular carcinoma [30]. Based on current understanding, multiple pathogenic factors are involved in the development of alcoholic liver disease [31, 32]. Both clinical and experimental evidence show that alcohol abuse is associated with dysbiosis. Transfer of the dysbiotic intestinal microbiota from alcoholic hepatitis patients to germ-free and conventionalized mice demonstrated that alcohol-associated dysbiosis contributes to the development of alcoholic liver disease [33]. In addition, gut barrier dysfunction and increased intestinal permeability have been implicated in alcohol-induced liver injury [34]. Mechanistically, acetaldehyde, which is a product of ethanol metabolism, and the generation of reactive oxygen species through cytochrome P450 2E1 induction might contribute to tight junction disruption leading to increased permeability during alcohol consumption [35]. In addition, intestinal inflammation is an important mediator of intestinal barrier dysfunction during alcohol consumption [11].
Bacterial translocation in alcoholic liver disease
In line with increased intestinal permeability, translocation of bacteria has been implicated in the development of alcoholic liver disease. Experimental induction of bacterial overgrowth in the small intestine alone is sufficient to result in bacterial translocation and subsequent liver disease [36]. Inversely, administration of non-absorbable antibiotics prevented bacterial overgrowth, reduced pathological bacterial translocation and ameliorated ethanol-induced steatohepatitis in rodents [37]. Moreover, plasma levels of gut-derived microbial products such as LPS and peptidoglycan are increased during alcohol administration [38, 39]. Recently, our group demonstrated that besides bacteria, also fungi contribute to alcohol-related liver disease. Increased fungal growth and translocation of fungal products to the liver activate inflammatory immune responses in the liver of ethanol-fed mice. This process is mediated via β-glucan recognition by CLEC7A on Kupffer cells. Moreover, relatively to healthy people, alcohol-dependent patients showed altered fungal signature and increased exposure and immune reactivity to fungal products in the blood, indicating fungal translocation [40]. Taken together, these data support that during alcoholic liver disease, epithelial damage in the intestine leads to pathological translocation of microbial products, thereby causing liver damage.
Antimicrobial defense during alcoholic liver disease
We previously described that chronic alcohol consumption suppresses REG3γ and REG3β mRNA and protein levels in murine small intestine [19, 21], and duodenal REG3γ in patients with alcohol use disorder [21]. In mice, the lowest levels of Reg3γ and Reg3β were observed in the proximal small intestine, where the bacterial overgrowth was most pronounced and luminal alcohol concentrations are highest [21]. Decreased REG3γ can be restored using prebiotics, which are associated with suppression of intestinal bacterial overgrowth. We have demonstrated that ethanol-fed Reg3γ−/− and Reg3β−/− mice have increased susceptibility to ethanol-induced liver disease, in association with increased mucosa-associated bacteria and more translocation of bacteria to the liver. In line, intestine-specific overexpression of Reg3γ protects mice against ethanol-induced liver disease by maintaining an inner mucus layer devoid of bacteria and reducing bacterial translocation [12]. How a reduced number of mucosa-associated bacteria results in lower bacterial translocation is not known. Further, mice deficient for mucin-2 production that were protected against alcohol-induced liver lesions showed increased defensin production as well as that of Reg3β and Reg3γ [19]. These data indicate that antimicrobial defense plays an important role in preventing bacterial translocation and protect against alcoholic liver disease development. Other antimicrobial molecules do not seem to be suppressed by chronic ethanol treatment [21], although a global analysis using transcriptomic or proteomic approaches should be done in future studies.
Recent findings from our laboratory suggest that Reg3γ and Reg3β suppression during alcoholic liver disease is an indirect effect of alcohol consumption. Using chronic–binge ethanol-fed mice as a model of alcoholic steatohepatitis [41], we found that dysbiosis upon ethanol consumption is associated with altered tryptophan metabolism by bacteria [42, unpublished data]. Ethanol feeding resulted in lower levels of indole-3-acetic acid, a ligand for the AhR [29, ], and reduced production of IL-22 by intestinal lamina propria ILC3. Importantly, AhR-dependent production of IL-22 regulates REG3γ and REG3β expression. Administration of non-absorbable antibiotics to ethanol-fed mice restored IL-22 production, indicating the influence of microbiota on regulating IL-22 expression [42, unpublished data]. Taken together, these data suggest that alcohol consumption changes microbial composition, thereby affecting the bacterial metabolome which alters host immunity and allows bacterial translocation.
Nevertheless, the exact mechanism of how chronic ethanol administration results in changes of the luminal intestinal microbiota composition is not fully elucidated. Undoubtedly, chronic alcohol consumption affects multiple factors in the host and more mechanistic studies are needed to fully understand how changes in the gut microbiome impact liver function during alcoholic liver disease and vice versa.
Non-alcoholic fatty liver disease
The prevalence of NAFLD is increasing worldwide and is considered to be a hepatic manifestation of the metabolic syndrome. Due to its strong association with obesity and type 2 diabetes, the pathogenesis of NAFLD and its progression to more complicated conditions have been widely accepted to be the result of multiple factors including intestinal dysbiosis [43, 44]. Similar to patients with alcoholic fatty liver disease, studies show that a shift in the gut microbiota composition correlates closely with the prevalence and progression of NAFLD. In patients with NAFLD, a decrease of some selected members of Firmicutes has been observed and obese patients with non-alcoholic steatohepatitis (NASH) had reduced Bacteroidetes compared with healthy controls [45]. Severity of NAFLD is associated with gut dysbiosis and microbial metabolome [46, 47]. These studies indicate that an alteration in the composition of the gut microbiota is closely associated with the development of NAFLD.
Different microbiota-dependent mechanisms have been suggested to contribute to NAFLD pathogenesis and progression. Ethanol-producing bacteria were proposed to be more abundant in NASH patients [48]. Further, dysbiosis may result in production and translocation of LPS and other inflammatory factors, changes in bile acid metabolism, and increased gut permeability in a subset of NAFLD patients [49]. This facilitates translocation of bacterial products into the portal circulation and activation of inflammatory processes.
Bacterial translocation in NAFLD
Studies in rodent models have shown correlations between hepatic inflammation and dysfunction of the intestinal mucosal barrier, which suggest that intestinal mucosal barrier malfunction and bacterial translocation influence the pathogenesis of NAFLD and NASH. Indeed, it has been shown that tight junction disruption in mice and NAFLD patients increases intestinal permeability and bacterial translocation to the liver through the bloodstream [50–52]. Nevertheless, data suggest that only a portion of NAFLD patients have increased intestinal permeability. It was reported that serum endotoxin levels were increased in only 42.1% (8/19) patients with NASH and a meta-analysis found that only 39.1% of patients with NAFLD (n = 128) had increased intestinal permeability [53]. Therefore, gut barrier dysfunction with subsequent translocation of microbial products might make only a small contribution to development or progression of fatty liver disease, or only in a subset of patients.
Antimicrobial defense during NAFLD
Similar to chronic ethanol consumption, animal models of diet-induced obesity indicate a downregulation of intestinal Reg3γ [54]. Recently, we explored the role of Reg3 lectins in the development of NASH. To induce NASH, mice deficient for REG3β or REG3γ were fed a Western-style fast-food diet (rich in saturated fat, cholesterol and fructose) for 20 weeks. Loss of REG3β or REG3γ did not cause more severe liver disease than in their WT littermates, despite elevated endotoxemia in Reg3γ-deficient mice. In addition, intestinal overexpression of REG3γ did not protect mice against NASH development [55]. Overall, these results indicate that loss of REG3β or REG3γ is insufficient to aggravate diet-induced obesity and NAFLD.
Vitamin D insufficiency, which has been associated with metabolic syndrome and NAFLD, has been found to be associated with loss of Paneth cell defensins, which may consequently lead to intestinal dysbiosis and endotoxemia. Moreover, oral administration of human alpha-defensin 5 rebalanced gut microbiota and resolved hepatic steatosis in mice [56]. Further, cathelicidin, another antimicrobial peptide, suppresses lipid accumulation and hepatic steatosis via the inhibition of the CD36 receptor [57]. Recently, the expression of cathelicidin-related antimicrobial peptide (Cramp), the only member of cathelicidin antimicrobial peptide family in mice, was found to be decreased by alcohol exposure to mice [58]. More studies investigating the function of different antimicrobial proteins during fatty liver disease development and progression are needed. Nevertheless, current data support the notion that bacterial translocation might not be as important during NAFLD as observed in patients with alcohol use disorder and alcohol-induced liver disease.
Cirrhosis
For most chronic liver diseases, cirrhosis is the common end-stage histologic distortion, characterized by the presence of regenerative nodules that causes portal hypertension. This in turn induces bacterial overgrowth by altering intestinal motility [59]. The gut–liver axis is well studied during cirrhosis and complications such as hepatic encephalopathy, spontaneous bacterial peritonitis and variceal bleeding are the result of pathological translocation of bacteria or their products into the blood of cirrhotic patients [60–63]. Evidence from both animal and patient studies indicates a loss of epithelial tight junctions during cirrhosis. Patients or mouse models of liver cirrhosis showed reduced intestinal expression of zonula occludens-1, occludin and claudin-1 compared to controls, and these changes were more evident in cases of decompensated or more advanced stage of disease [64]. Moreover, intestinal permeability is enhanced via increased production of lipid peroxidation products such as malondialdehyde in the intestine, as described in patients and rats with cirrhosis [65, 66].
Bacterial translocation in cirrhosis
In patients with cirrhosis, changes in gastrointestinal barrier and dysbiosis increase the rate of bacterial translocation [62]. Indeed, cirrhotic patients have increased levels of LPS and bacterial DNA in the portal circulation compared to healthy controls, with increasing amounts as the liver function worsens [67, 68]. Whereas the rate and degree of translocating bacterial products are higher in early cirrhosis compared to healthy conditions, pathological translocation of viable bacteria occurs in the decompensated stage of the disease. During decompensated cirrhosis, a further increase in intestinal permeability could be triggered by intestinal inflammation and may contribute to enhanced translocation of viable bacteria [69, 70]. Living bacteria appear to translocate via the transcellular route (transcytosis), whereas microbial products migrate via disrupted tight junctions using the paracellular route and further enhance the local and systemic inflammatory response [71]. Positive cultures from mesenteric lymph nodes are found in about 50–60% of rats with CCl4-induced cirrhosis and in 30% of cirrhotic patients [72, 73], supporting that pathological bacterial translocation plays an important role in disease progression during cirrhosis.
Antimicrobial defense during cirrhosis
Besides intestinal immune cell damage, several studies point to deficiencies in the production of intestinal antimicrobial peptides during cirrhosis. Cirrhotic rats with ascites and translocation of viable bacteria to mesenteric lymph nodes produce lower levels of defensins molecules compared with cirrhotic rats without bacterial translocation [20]. The transcription factor farnesoid X receptor (FXR), which is the nuclear receptor for conjugated bile acids, plays a crucial role in preserving intestinal epithelial integrity by increasing antimicrobial peptide production and secretion [74]. Treatment of cirrhotic rats with obeticholic acid, a potent agonist of FXR, significantly reduced bacterial translocation from the intestine to the blood via upregulation of antimicrobial proteins angiogenin-1 and α-5 defensin, as well as tight junction proteins, and reduced liver fibrosis [75]. Further, liver cirrhosis in advanced stages is frequently associated with malnutrition [76], which has deleterious effects on gut mucosal integrity and antimicrobial peptides [77]. Data on the expression of intestinal AMPs in cirrhosis are scarce. Mucosal expression of several different AMPs was not altered in the ileum or colon of patients with cirrhosis as compared with healthy controls [78]. Taken together, experimental data in animal models underline the concept of intestinal antimicrobial deficiency in cirrhosis. Measurements of AMPs in more and larger patient cohorts should be undertaken.
Modulation of antimicrobial proteins as treatment for liver disease
Since bacterial translocation and reduced expression of certain antimicrobial proteins in the gut are observed in rodents and humans with liver disease, designing a strategy to increase intestinal concentrations of antimicrobial proteins or their production by intestinal epithelial cells might be developed to prevent liver disease.
Antibiotics
Non-absorbable antibiotics have a beneficial effect on NASH [79] and alcoholic liver disease [37], and are commonly used to treat patients with cirrhosis [80]. Although the use of antibiotics seems to have a beneficial effect by reducing bacterial overgrowth and preventing bacterial translocation, no evidence supports that this is achieved by increased expression of antimicrobial proteins. Nevertheless, recent data from our group indicate that non-absorbable antibiotics can restore IL-22 production by ILC3 s during ethanol diet [42, unpublished data]. Therefore, changes in the microbial metabolome, the composition of microbiota, or host immunity due to antibiotics might affect antimicrobial responses indirectly.
Probiotics and prebiotics
Probiotics regulate antimicrobial defense. For instance, probiotic Escherichia coli Nissle 1917 and a variety of other probiotics such as lactobacilli strongly induced the expression of human beta-defensin-2 in epithelial cell lines [81]. In addition, probiotic lactobacilli strains are not only able to upregulate enterocyte human beta-defensin 2 (hBD-2) production in vitro [82]; some species, such as Lactobacillus lactis, have been demonstrated to be resistant to the antimicrobial effects of this defensin [83]. Apart from the induction of AMP, probiotics might affect cytokine-producing innate cells in the mucosa (e.g., IL-22) that can increase the expression of Reg3 lectins. As such, probiotic Lactobacillus reuteri was found to produce AhR ligands from tryptophan metabolism, thereby enhancing IL-22 production and mucosal defense [29]. This host–commensal interplay is a prime example of how beneficial bacteria might enhance the immune response to protect the host from pathogens.
Prebiotics are complex short-chain saccharides that cannot be digested by host pancreatic and brush-border enzymes, but can be selectively used and fermented by the commensal microbiota. They stimulate probiotic bacteria such as lactobacilli and bifidobacteria [84]. Interestingly, we found that decreased REG3γ level during ethanol-induced liver disease can be partly restored using prebiotics. Adding fructooligosaccharides to ethanol-fed mice reduced ethanol-induced steatohepatitis and intestinal bacterial overgrowth by partial restoration of REG3γ [21]. Current evidence supporting a beneficial effect of prebiotics for NAFLD and cirrhosis is lacking. More studies and larger clinical trials are needed to support the use of pre- and probiotics in different chronic liver diseases.
Conclusion
In conclusion, several human and mouse studies have demonstrated that intestinal barrier dysfunction, bacterial translocation and a deficiency in various antimicrobial proteins are implicated in the development of chronic liver disease. We are gaining increased insight into the close relationship between the gut and the liver evoked by dysbiosis. The evaluation of the gut–liver axis and the intervention of the relationships between antimicrobial peptides and bacteria might aid the development of treatment and prevention for liver disease patients. Finally, besides bacteria, the intestinal microbiota also includes eukaryotic viruses [85], bacteriophages [86], and eukaryotic organisms such as fungi [87]. However, studies were mainly focused on the interaction between AMPs and bacteria. Therefore, studies to explore the influence of AMPs on other microbial communities will be interesting. In that way, more insights in the communication between host and microbiome will be made which may provide new strategies for improving health and disease management.
Abbreviations
- AhR
Aryl hydrocarbon receptor
- AMPs
Antimicrobial proteins
- FXR
Farnesoid X receptor
- IL-22
Interleukin 22
- ILC3
Innate lymphoid cells type 3
- LPS
Lipopolysaccharide
- NAFLD
Non-alcoholic fatty liver disease
- NASH
Non-alcoholic steatohepatitis
- Reg
Regenerating island-derived protein
- TLR
Toll-like receptor
Funding
This manuscript was supported by an Erwin Schrodinger Fellowship (J4063) from the Austrian Science Fund (to T. Hendrikx), and NIH Grants R01 AA020703, R01 AA24726, U01 AA021856 and by Award number I01BX002213 from the Biomedical Laboratory Research and Development Service of the VA Office of Research and Development (to B. Schnabl).
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Tilg H. Obesity, metabolic syndrome, and microbiota: multiple interactions. J Clin Gastroenterol. 2010;44(Suppl 1):S16–S18. doi: 10.1097/MCG.0b013e3181dd8b64. [DOI] [PubMed] [Google Scholar]
- 2.Kau AL, Ahern PP, Griffin NW, et al. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsiao EY, McBride SW, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut. 2014;63:1513–1521. doi: 10.1136/gutjnl-2014-306928. [DOI] [PubMed] [Google Scholar]
- 6.Gerardi V, Del Zompo F, D’Aversa F, et al. The relationship between gut microbiota and cardiovascular diseases. G Ital Cardiol (Rome). 2016;17:11-4. Il legame tra il microbiota intestinale e le patologie cardiovascolari. [DOI] [PubMed]
- 7.Yoshimoto S, Loo TM, Atarashi K, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 8.Schnabl B. The microbiome and the liver. Gastroenterol Hepatol (N Y). 2014;10:519–521. [PMC free article] [PubMed] [Google Scholar]
- 9.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tripathi A, Debelius J, Brenner DA, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15:397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandl K, Schnabl B. Is intestinal inflammation linking dysbiosis to gut barrier dysfunction during liver disease? Expert Rev Gastroenterol Hepatol. 2015;9:1069–1076. doi: 10.1586/17474124.2015.1057122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Fouts DE, Starkel P, et al. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe. 2016;19:227–239. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouts DE, Torralba M, Nelson KE, et al. Bacterial translocation and changes in the intestinal microbiome in mouse models of liver disease. J Hepatol. 2012;56:1283–1292. doi: 10.1016/j.jhep.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mencin A, Kluwe J, Schwabe RF. Toll-like receptors as targets in chronic liver diseases. Gut. 2009;58:704–720. doi: 10.1136/gut.2008.156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwabe RF, Seki E, Brenner DA. Toll-like receptor signaling in the liver. Gastroenterology. 2006;130:1886–1900. doi: 10.1053/j.gastro.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 16.Johansson M, Phillipson M, Petersson J, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaishnava S, Yamamoto M, Severson KM, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee S, Hooper LV. Antimicrobial defense of the intestine. Immunity. 2015;42:28–39. doi: 10.1016/j.immuni.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 19.Hartmann P, Chen P, Wang HJ, et al. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology. 2013;58:108–119. doi: 10.1002/hep.26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teltschik Z, Wiest R, Beisner J, et al. Intestinal bacterial translocation in rats with cirrhosis is related to compromised Paneth cell antimicrobial host defense. Hepatology. 2012;55:1154–1163. doi: 10.1002/hep.24789. [DOI] [PubMed] [Google Scholar]
- 21.Yan AW, Fouts DE, Brandl J, et al. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53:96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okumura R, Takeda K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp Mol Med. 2017;49:e338. doi: 10.1038/emm.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cash HL, Whitham CV, Behrendt CL, et al. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandl K, Plitas G, Schnabl B, et al. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaishnava S, Behrendt CL, Ismail AS, et al. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 27.Lamas B, Richard ML, Leducq V, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JS, Cella M, McDonald KG, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2011;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zelante T, Iannitti RG, Cunha C, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Rehm J, Samokhvalov AV, Shield KD. Global burden of alcoholic liver diseases. J Hepatol. 2013;59:160–168. doi: 10.1016/j.jhep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Szabo G. Gut-liver axis in alcoholic liver disease. Gastroenterology. 2015;148:30–36. doi: 10.1053/j.gastro.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starkel P, Schnabl B. Bidirectional Communication between liver and gut during alcoholic liver disease. Semin Liver Dis. 2016;36:331–339. doi: 10.1055/s-0036-1593882. [DOI] [PubMed] [Google Scholar]
- 33.Llopis M, Cassard AM, Wrzosek L, et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut. 2016;65:830–839. doi: 10.1136/gutjnl-2015-310585. [DOI] [PubMed] [Google Scholar]
- 34.Starkel P, Leclercq S, de Timary P, et al. Intestinal dysbiosis and permeability: the yin and yang in alcohol dependence and alcoholic liver disease. Clin Sci (Lond). 2018;132:199–212. doi: 10.1042/CS20171055. [DOI] [PubMed] [Google Scholar]
- 35.Elamin EE, Masclee AA, Dekker J, et al. Ethanol metabolism and its effects on the intestinal epithelial barrier. Nutr Rev. 2013;71:483–499. doi: 10.1111/nure.12027. [DOI] [PubMed] [Google Scholar]
- 36.Lichtman SN, Sartor RB, Keku J, et al. Hepatic inflammation in rats with experimental small intestinal bacterial overgrowth. Gastroenterology. 1990;98:414–423. doi: 10.1016/0016-5085(90)90833-M. [DOI] [PubMed] [Google Scholar]
- 37.Chen P, Starkel P, Turner JR, et al. Dysbiosis-induced intestinal inflammation activates tumor necrosis factor receptor I and mediates alcoholic liver disease in mice. Hepatology. 2015;61:883–894. doi: 10.1002/hep.27489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parlesak A, Schafer C, Schutz T, et al. Increased intestinal permeability to macromolecules and endotoxemia in patients with chronic alcohol abuse in different stages of alcohol-induced liver disease. J Hepatol. 2000;32:742–747. doi: 10.1016/S0168-8278(00)80242-1. [DOI] [PubMed] [Google Scholar]
- 39.Tabata T, Tani T, Endo Y, et al. Bacterial translocation and peptidoglycan translocation by acute ethanol administration. J Gastroenterol. 2002;37:726–731. doi: 10.1007/s005350200118. [DOI] [PubMed] [Google Scholar]
- 40.Yang AM, Inamine T, Hochrath K, et al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. 2017;127:2829–2841. doi: 10.1172/JCI90562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertola A, Mathews S, Ki SH, et al. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8:627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hendrikx T, Duan Y, Wang Y, et al. Bacteria engineered to produce IL22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. 2018 (unpublished data). [DOI] [PMC free article] [PubMed]
- 43.Byrne CD, Targher G. NAFLD: a multisystem disease. J Hepatol. 2015;62:S47–S64. doi: 10.1016/j.jhep.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Boursier J, Diehl AM. Nonalcoholic fatty liver disease and the gut microbiome. Clin Liver Dis. 2016;20:263–275. doi: 10.1016/j.cld.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Brandl K, Schnabl B. Intestinal microbiota and nonalcoholic steatohepatitis. Curr Opin Gastroenterol. 2017;33:128–133. doi: 10.1097/MOG.0000000000000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loomba R, Seguritan V, Li W, et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 2017;25(1054–62):e5. doi: 10.1016/j.cmet.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 49.Leung C, Rivera L, Furness JB, et al. The role of the gut microbiota in NAFLD. Nat Rev Gastroenterol Hepatol. 2016;13:412–425. doi: 10.1038/nrgastro.2016.85. [DOI] [PubMed] [Google Scholar]
- 50.Miele L, Valenza V, La Torre G, et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. 2009;49:1877–1887. doi: 10.1002/hep.22848. [DOI] [PubMed] [Google Scholar]
- 51.Rahman K, Desai C, Iyer SS, et al. Loss of junctional adhesion molecule a promotes severe steatohepatitis in mice on a diet high in saturated fat, fructose, and cholesterol. Gastroenterology. 2016;151(733–46):e12. doi: 10.1053/j.gastro.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luther J, Garber JJ, Khalili H, et al. Hepatic injury in nonalcoholic steatohepatitis contributes to altered intestinal permeability. Cell Mol Gastroenterol Hepatol. 2015;1:222–232. doi: 10.1016/j.jcmgh.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan J, Baker SS, Liu W, et al. Endotoxemia unrequired in the pathogenesis of pediatric nonalcoholic steatohepatitis. J Gastroenterol Hepatol. 2014;29:1292–1298. doi: 10.1111/jgh.12510. [DOI] [PubMed] [Google Scholar]
- 54.Everard A, Lazarevic V, Gaia N, et al. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014;8:2116–2130. doi: 10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bluemel S, Wang L, Martino C, et al. The role of intestinal C-type regenerating Islet derived-3 lectins for nonalcoholic steatohepatitis. Hepatol Commun. 2018;2:393–406. doi: 10.1002/hep4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su D, Nie Y, Zhu A, et al. Vitamin D signaling through induction of paneth cell defensins maintains gut microbiota and improves metabolic disorders and hepatic steatosis in animal models. Front Physiol. 2016;7:498. doi: 10.3389/fphys.2016.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoang-Yen Tran D, Hoang-Ngoc Tran D, Mattai SA, et al. Cathelicidin suppresses lipid accumulation and hepatic steatosis by inhibition of the CD36 receptor. Int J Obes (Lond). 2016;40:1424–1434. doi: 10.1038/ijo.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao T, Zhao C, Li F, et al. Intestinal HIF-1alpha deletion exacerbates alcoholic liver disease by inducing intestinal dysbiosis and barrier dysfunction. J Hepatol. 2018;69:886–895. doi: 10.1016/j.jhep.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gomez-Hurtado I, Such J, Sanz Y, et al. Gut microbiota-related complications in cirrhosis. World J Gastroenterol. 2014;20:15624–15631. doi: 10.3748/wjg.v20.i42.15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alexopoulou A, Agiasotelli D, Vasilieva LE, et al. Bacterial translocation markers in liver cirrhosis. Ann Gastroenterol. 2017;30:486–497. doi: 10.20524/aog.2017.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giannelli V, Di Gregorio V, Iebba V, et al. Microbiota and the gut-liver axis: bacterial translocation, inflammation and infection in cirrhosis. World J Gastroenterol. 2014;20:16795–16810. doi: 10.3748/wjg.v20.i45.16795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ponziani FR, Zocco MA, Cerrito L, et al. Bacterial translocation in patients with liver cirrhosis: physiology, clinical consequences, and practical implications. Expert Rev Gastroenterol Hepatol. 2018;12:641–656. doi: 10.1080/17474124.2018.1481747. [DOI] [PubMed] [Google Scholar]
- 63.Tuomisto S, Pessi T, Collin P, et al. Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol. 2014;14:40. doi: 10.1186/1471-230X-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Assimakopoulos SF, Tsamandas AC, Tsiaoussis GI, et al. Altered intestinal tight junctions’ expression in patients with liver cirrhosis: a pathogenetic mechanism of intestinal hyperpermeability. Eur J Clin Invest. 2012;42:439–446. doi: 10.1111/j.1365-2362.2011.02609.x. [DOI] [PubMed] [Google Scholar]
- 65.Chiva M, Soriano G, Rochat I, et al. Effect of Lactobacillus johnsonii La1 and antioxidants on intestinal flora and bacterial translocation in rats with experimental cirrhosis. J Hepatol. 2002;37:456–462. doi: 10.1016/S0168-8278(02)00142-3. [DOI] [PubMed] [Google Scholar]
- 66.Assimakopoulos SF, Tsamandas AC, Tsiaoussis GI, et al. Intestinal mucosal proliferation, apoptosis and oxidative stress in patients with liver cirrhosis. Ann Hepatol. 2013;12:301–307. [PubMed] [Google Scholar]
- 67.Bellot P, Garcia-Pagan JC, Frances R, et al. Bacterial DNA translocation is associated with systemic circulatory abnormalities and intrahepatic endothelial dysfunction in patients with cirrhosis. Hepatology. 2010;52:2044–2052. doi: 10.1002/hep.23918. [DOI] [PubMed] [Google Scholar]
- 68.Chan CC, Hwang SJ, Lee FY, et al. Prognostic value of plasma endotoxin levels in patients with cirrhosis. Scand J Gastroenterol. 1997;32:942–946. doi: 10.3109/00365529709011206. [DOI] [PubMed] [Google Scholar]
- 69.Du Plessis J, Vanheel H, Janssen CE, et al. Activated intestinal macrophages in patients with cirrhosis release NO and IL-6 that may disrupt intestinal barrier function. J Hepatol. 2013;58:1125–1132. doi: 10.1016/j.jhep.2013.01.038. [DOI] [PubMed] [Google Scholar]
- 70.Saitoh O, Sugi K, Lojima K, et al. Increased prevalence of intestinal inflammation in patients with liver cirrhosis. World J Gastroenterol. 1999;5:391–396. doi: 10.3748/wjg.v5.i5.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiest R, Lawson M, Geuking M. Pathological bacterial translocation in liver cirrhosis. J Hepatol. 2014;60:197–209. doi: 10.1016/j.jhep.2013.07.044. [DOI] [PubMed] [Google Scholar]
- 72.Cirera I, Bauer TM, Navasa M, et al. Bacterial translocation of enteric organisms in patients with cirrhosis. J Hepatol. 2001;34:32–37. doi: 10.1016/S0168-8278(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 73.Yeh DC, Wu CC, Ho WM, et al. Bacterial translocation after cirrhotic liver resection: a clinical investigation of 181 patients. J Surg Res. 2003;111:209–214. doi: 10.1016/S0022-4804(03)00112-4. [DOI] [PubMed] [Google Scholar]
- 74.Inagaki T, Moschetta A, Lee YK, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ubeda M, Lario M, Munoz L, et al. Obeticholic acid reduces bacterial translocation and inhibits intestinal inflammation in cirrhotic rats. J Hepatol. 2016;64:1049–1057. doi: 10.1016/j.jhep.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 76.Cheung K, Lee SS, Raman M. Prevalence and mechanisms of malnutrition in patients with advanced liver disease, and nutrition management strategies. Clin Gastroenterol Hepatol. 2012;10:117–125. doi: 10.1016/j.cgh.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 77.Hodin CM, Lenaerts K, Grootjans J, et al. Starvation compromises Paneth cells. Am J Pathol. 2011;179:2885–2893. doi: 10.1016/j.ajpath.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaltsa G, Bamias G, Siakavellas SI, et al. Systemic levels of human beta-defensin 1 are elevated in patients with cirrhosis. Ann Gastroenterol. 2016;29:63–70. [PMC free article] [PubMed] [Google Scholar]
- 79.Gangarapu V, Ince AT, Baysal B, et al. Efficacy of rifaximin on circulating endotoxins and cytokines in patients with nonalcoholic fatty liver disease. Eur J Gastroenterol Hepatol. 2015;27:840–845. doi: 10.1097/MEG.0000000000000348. [DOI] [PubMed] [Google Scholar]
- 80.Bajaj JS. Review article: potential mechanisms of action of rifaximin in the management of hepatic encephalopathy and other complications of cirrhosis. Aliment Pharmacol Ther. 2016;43(Suppl 1):11–26. doi: 10.1111/apt.13435. [DOI] [PubMed] [Google Scholar]
- 81.Wehkamp J, Harder J, Wehkamp K, et al. NF-kappaB- and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: a novel effect of a probiotic bacterium. Infect Immun. 2004;72:5750–5758. doi: 10.1128/IAI.72.10.5750-5758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schlee M, Harder J, Koten B, et al. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clin Exp Immunol. 2008;151:528–535. doi: 10.1111/j.1365-2249.2007.03587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hugo AA, Tymczyszyn EE, Gomez-Zavaglia A, et al. Effect of human defensins on lactobacilli and liposomes. J Appl Microbiol. 2012;113:1491–1497. doi: 10.1111/j.1365-2672.2012.05433.x. [DOI] [PubMed] [Google Scholar]
- 84.Kolida S, Gibson GR. Prebiotic capacity of inulin-type fructans. J Nutr. 2007;137:2503S–2506S. doi: 10.1093/jn/137.11.2503S. [DOI] [PubMed] [Google Scholar]
- 85.Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reyes A, Haynes M, Hanson N, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]