Abstract
Myroides odoratimimus is an important nosocomial pathogen. Management of M. odoratimimus infection is difficult owing to the multidrug resistance and the unknown pathogenesis mechanisms. Based on our previous genomic sequencing data of M. odoratimimus PR63039 (isolated from a patient with the urinary tract infection), in this study, we further performed comparative genomic analysis for 10 selected Myroides strains. Our results showed that these Myroides genome contexts were very similar and phylogenetically related. Various prophages were identified in the four clinical isolate genomes, which possibly contributed to the genome evolution among the Myroides strains. CRISPR elements were only detected in the two clinical (PR63039 and CCUG10230) isolates and two environmental (CCUG12700 and H1bi) strains. With more stringent cutoff parameters in CARD analysis, the four clinical M. odoratimimus contained roughly equal antibiotic resistance genes, indicating their similar antibiotic resistance profiles. The three clinical (CCUG10230, CCUG12901, CIP101113) and three environmental (CCUG12700, L41, H1bi) M. odoratimimus strains were speculated to carry the indistinguishable virulent factors (VFs), which may involve in the similar pathogenesis mechanism. Moreover, some VFs might confer to the high capacity of dissemination, attacking tissue cells and induction of autoimmune complications. Our results facilitate the research of antibiotic resistance and the development of therapeutic regimens for the M. odoratimimus infections.
Keywords: antibiotic resistance genes, comparative genomics, Myroides odoratimimus, virulence factors
1. INTRODUCTION
Myroides odoratimimus is a gram‐negative and opportunistic pathogen. It causes a variety of serious infections mainly reported in China (summarized by Hu et al., 2016) and outbreak of urinary tract infection (Ktari et al., 2012). Recently, the increasing infections emerged in patients with the recurrent calcaneal ulcer (Pompilio et al., 2017), fulminant erysipelas and sepsis (Willems, Muller, Verhaegen, Saegeman, & Desmet, 2017), bacteremia (Belloir, Billy, Hentgen, Fille, & Barrans, 2016), or prosthesis joint infection (Jover‐Sáenz, Pérez‐Villar, & Barcenilla‐Gaite, 2016).
M. odoratimimus infections are life‐threatening due to its multidrug resistance and unknown pathogenicity (as summarized in Hu et al., 2016). In our previous report (Hu et al., 2017), to some extent, we correlated the phenotype in antibiotic susceptibilities and infectivity of M. odoratimimus with the genomic findings of a variety of resistance genes, virulence factor (VF) genes. To better accomplish these purposes and verify the possibility of infection source of environmental strains, here, we further performed comparative genomic analysis of 10 Myroides strains, including four clinically pathogenic (PR63039, CCUG10230, CCUG12901, CIP101113), four environmental (CCUG 3837 CCUG 12700 H1bi L41), and two human‐associated Myroides isolates (CCUG39352, Myroides sp. A21) by focusing on their antibiotic resistance and pathogenesis mechanisms.
2. MATERIALS AND METHODS
2.1. Genome sequences
In the NCBI Genome RefSeq Assembly Database, only nine genomic sequences of M. odoratimimus were found (Table 1). They included four clinically pathogenic strains, a human‐associated strain, and four environmental isolates. Only PR63039 genome (Hu et al., 2017) was complete. Strain PR63039 (Hu et al., 2017) and CCUG12901 were isolated from the urine of patients with postinjury urinary tract infection. CCUG10230 and CIP101113 were isolated from skin wounds. Human‐associated strain CCUG39352 was collected and sequenced by Shandong University. M. odoratimimus H1 bi, L41, CCUG 12700, and CCUG 3837 are environmental isolates. For the phylogenetic tree analysis of Myroides genomes, another human‐associated strain Myroides sp. A21 (CP010327) (Burghartz et al., 2015) with highly homologous 16S rRNA gene sequence to strain PR63039 (coverage 100%, identity 100%, 1,388 bp) (GenBank No. KR349266) was also included. Myroides sp. A21 was isolated from the urethral catheter of a patient without symptoms of a urinary tract infection, had extensive drug resistance; its full genomic sequence was available.
Table 1.
General genomic characteristics of 10 Myroides strains
| Sources | Strain | Site of isolation | Type | Assembly No. | Level | Scaffold | Size (Mb) | GC (%) | Gene | Protein | rRNA | tRNA | Other RNA | Pseudo gene |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinically pathogenic M. odoratimimus | PR63039 | Urine | GCA_001481655.1 | 2 | 4.46 | 34.14 | 4,084 | 3,840 | 27 | 105 | 4 | 108 | ||
| Chr | NZ_CP013690.1 | Complete | 4.37 | 34.20 | 3,988 | 3,745 | 27 | 105 | 4 | 107 | ||||
| Plsm | NZ_CP013691.1 | 0.09 | 31.30 | 96 | 95 | ‐ | ‐ | ‐ | 1 | |||||
| CCUG 10230 | Skin wound | Chr | GCA_000242075.2 | Scaffold | 7 | 4.03 | 34.10 | 3,610 | 3,458 | 5 | 76 | 4 | 67 | |
| CCUG 12901 | Urogenital tract | Chr | GCA_000242095.1 | Scaffold | 3 | 4.07 | 34.20 | 3,653 | 3,505 | 9 | 75 | 4 | 60 | |
| CIP 101113 | Skin wound | Chr | GCA_000242135.1 | Scaffold | 3 | 4.14 | 34.10 | 3,673 | 3,516 | 8 | 71 | 4 | 74 | |
| Environmental M. odoratimimus | CCUG 3837 | N/A | Chr | GCA_000297855.1 | Scaffold | 4 | 4.14 | 34.40 | 3,611 | 3,459 | 11 | 72 | 4 | 65 |
| CCUG 12700 | N/A | Chr | GCA_000413415.1 | Scaffold | 7 | 4.04 | 34.30 | 3,581 | 3,423 | 7 | 87 | 4 | 60 | |
| H1bi | Carnivorous plant Phytotelma | Chr | GCA_000633375.1 | Contig | 183 | 3.88 | 34.00 | 3,549 | 3,260 | 4 | 85 | 4 | 196 | |
| L41 | Lake water | Chr | GCA_000812825.1 | Contig | 168 | 4.16 | 33.60 | 3,802 | 3,599 | 82 | 4 | 114 | ||
| Human‐associated Myroides | CCUG 39352 | Wound | Chr | GCA_001485415.1 | Contig | 65 | 4.24 | 33.90 | 3,789 | 3,650 | 74 | ‐ | ‐ | |
| Myroides sp. A21 | Urethral catheter | Chr | GCA_000807225.1 | Complete | 1 | 4.16 | 34.10 | 3,750 | 3,554 | 24 | 101 | 4 | 67 |
Chr, chromosome; Plsm, plasmid; N/A, not available.
2.2. Softwares and databases used for comparative genomics analysis
The analyses of whole‐genome phylogenetic tree, circular genome mapping, insertion sequence elements (IS), multiple genome alignment, prophage, CRISPR, antibiotic resistance genes, and VF genes in the M. odoratimimus genomes were performed with the softwares and databases listed in Table 2.
Table 2.
The softwares and databases used for comparative analysis
| Analysis | Software/database | References | Clinically pathogenic strains | Environmental strains | Human‐associated strains | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PR63039 | CCUG 10230 | CCUG 12901 | CIP 101113 | CCUG 12700 | L41 | H1bi | CCUG 3837 | CCUG 39352 | Myroides sp. A21 | |||
| Sequence level | Complete | Scaffold | Scaffold | Scaffold | Scaffold | Contig | Contig | Scaffold | Contig | Complete | ||
| Phylogenetic tree | REALPHY | Bertels, Silander, Pachkov, Rainey, & van Nimwegen, 2014; | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CG | CG viewer | Grant & Stothard, 2008; | Yes | Yes | Yes | Yes | ||||||
| ISs | IS Finder | Siguier, Perochon, Lestrade, Mahillon, & Chandler, 2006; | Yes (43) | Yes (169) | Yes (117) | Yes (104) | ||||||
| Synteny | Progressive mauve | Darling et al., 2010; | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |||
| Prophage | PHAST | Zhou, Liang, Lynch, Dennis, & Wishart, 2011; | Yes (2 incomplete) | Yes (2) | Yes (5) | Yes (1) | ||||||
| CRISPR | CRISPR Finder | Bland et al., 2007; | Yes (3) | Yes (4) | Yes (ND) | Yes (ND) | Yes (1) | Yes (ND) | Yes (1) | |||
| Antibiotic resistance genes | CARD Resistance Gene identifier | Mcarthur et al., 2013; Jia et al., 2017; | Yes | Yes | Yes | Yes | ||||||
| Virulence factors | VFDB protein Set B database | Chen et al., 2012, 2016 | Yes | Yes | Yes | Yes | Yes | Yes | ||||
CG, Circular genome maps (Genomic variants); ISs, insertion sequence elements; Yes, analyzed; (): number of the predicted; ND, not detected.
We should mention that, for identifying the resistance genes using CARD Resistance Gene Identifier (RGI) software (Jia et al., 2017; Mcarthur et al., 2013), we performed BLASTp search (collaborated with Beijing Novogene Bioinformatics Technology Co., Ltd, BNNT) of the protein sequences of M. odoratimimus (downloaded from RefSeq assembly) against the CARD reference sequences, and more stringent parameters were set up [Query ID, Chromosome, Gene start, Gene end, Direction, ARO ID, ARO name, Category, Query length, Query start, Query end, Subject length, Subject start, Subject end, Gap, Mismatch length, Match length, Bit score, E value ≤ 1e‐30, Identity (%), Query coverage, Subject coverage]. The stringency of extracting antibiotic resistance genes from the primary output was improved by setting the cutoff parameters (Protein identity [>50%], Query coverage [>50%], and Subject coverage [>50%]).
The genes coding for the virulence factors was predicted by performing BLAST search (collaborated with Beijing Novogene Bioinformatics Technology Co., Ltd) of the protein sequences of M. odoratimimus against the VFDB protein Set B database (Chen, Xiong, Sun, Yang, & Jin, 2012; Chen, Zheng, Liu, Yang, & Jin, 2016). The stringent parameters were set up (Gene ID, VFDB internal ID, VF ID, VF name, Genes, Characteristics, Structure features, Functions, Mechanisms, Descriptions, Query length, Query start, Query end, Subject length, Subject start, Subject end, Match length, Mismatch length, Gap, Identity, E value, Bit score, Query coverage, Subject coverage). The cutoff parameters for extracting VF genes from the primary outputs were same as the extracting resistance genes as the above.
3. RESULTS
3.1. The basic genome statistics of 10 Myroides genomes
The general features of 10 Myroides genomes, including four clinically pathogenic M. odoratimimus (PR63039, CCUG12901, CCUG10230, CIP101113), four environmental (CCUG 12700, L41, H1bi, CCUG 3837), and two human‐associated Myroides strains (CCUG 39352, Myroides sp. A21) were similar (summarized in Table 1). Their GC contents were approximately 34%. The sizes of the genomes varied from 3.88 to 4.46 Mb. The numbers of genes, proteins, and tRNAs in PR63039 genome were larger than that of the other nine genomes. We should mention that eight Myroides genomes were incomplete, and the sequencing of the plasmids in PR63039 strains were not completed even its chromosome was fully sequenced (Figure 1 in Hu et al., 2016, 2017).
3.2. Phylogenetic analysis of 10 Myroides genomes
Whole‐genome phylogenetic tree of the 10 Myroides genomes was created (Figure 1). It showed that PR63039 formed a different clade from Myroides sp. A21 even although they had highly homologous 16S rRNA gene sequence (100% identity). The clinically pathogenic strain CCUG 10230 and the environmental strain CCUG 39352 were closest. Strain CCUG3837, CIP101113, and CCUG12700 formed a clade, while strain H1bi was located alone. It seemed that strain CCUG3837, CIP101113, and CCUG12700 were phylogenetically closer and H1bi might have a different origin compared to the above three strains.
Figure 1.

Whole‐genome phylogenetic tree of 10 Myroides isolates. This Whole‐genome phylogenetic tree was produced by REALPHY with the default parameters. Strain L41 was used as the root
Collectively, these 10 Myroides strains were related phylogenetically, indicating that there might similarly evolve to some pathogenesis traits. However, further study to illuminate the evolution pathway is warranted.
3.3. Genomic variants among four clinically pathogenic M. odoratimimus strains
We compared the genomes of three clinically pathogenic M. odoratimimus strains (CCUG12901, CCUG10230, CIP101113) with the clinically pathogenic PR63039 genome as the reference (Table 2). Many highly variable regions were found (Figure 2). Specifically, the following regions on the above three genomes were absent or had low identity with our strain PR63039: from 150 to 250 kb, 700 to 780 kb, 1,650 to 1,700 kb, 2,300 to 2,450 kb, 2,680 to 2,720 kb, 3,370 to 3,530 kb, 3,720 to 3,800 kb, 4,110 to 4,200 kb, 4,250 to 4,270 kb, and 4,350 to 4,560 kb. Interestingly, the region from 1,650 to 1,700 kb was predicted to be located in one prophage locus of CCUG‐12901 genome. Circular map of the genome comparisons indicated that there were a number of conserved or diverged genome segments among the genomes of these four clinically pathogenic M. odoratimimus.
Figure 2.
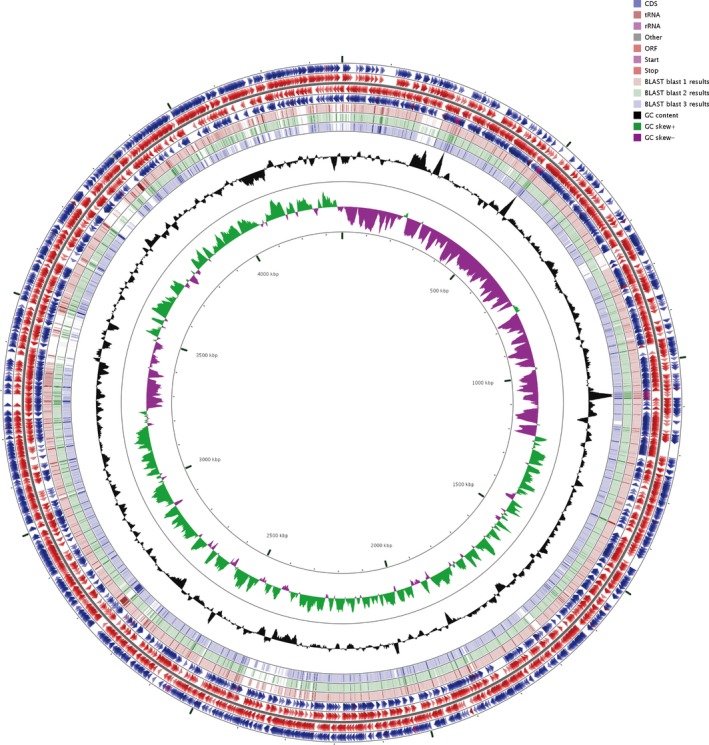
Genomic comparisons in four clinically pathogenic M. odoratimimus strains. Blast 1: CCUG10230. Blast 2: M. CCUG12901. Blast 3: CIP101113. The contents of the feature rings (starting with the outermost ring) are as follows: Ring 1: forward strand features the read from the primary sequence of PR63039. Rings 2: forward strand ORFs from the primary sequence of PR63039. Rings 3: reverse strand ORFs from the primary sequence of PR63039. Ring 4: reverse strand features read from the primary sequence of PR63039. Ring 5 (vermilion): blast 1. Ring 6 (green): blast 2. Ring 7 (blue): blast 3. Ring 8 (black): the G + C content. Ring 9 (green and purple): the GC skew value
In addition, the abovementioned variable regions were partially accompanied by several insertion elements, which might assist the integration of resistance‐ and pathogenesis‐related genes and facilitate the transfer of drug resistance and pathogenic genes among M. odoratimimus strains. Furthermore, IS elements may enhance drug resistance and virulence by promoting gene expression (Heritier, Poirel, & Nordmann, 2006; Higgins, Dammhayn, Hackel, & Seifert, 2010).
3.4. Synteny analysis among four clinically pathogenic and three environmental M. odoratimimus strains
Genome alignments can identify evolutionary traits. To study the genome synteny and rearrangements in four clinically pathogenic (PR63039, CCUG10230, CCUG12901, and CIP101113) and three environmental (CCUG12700, L41, and H1bi) M. odoratimimus bacteria (Table 2), the genome alignment software progressive MAUVE (Darling, Mau, & Perna, 2010) was used Figure (S1). The synteny between the PR63039 genome and Myroides sp. A21 was approximately 83.7% (Hu et al., 2017). The genome arrangement of these four clinically pathogenic isolates mimics each other. Similarly, the genome context and arrangement in the three environmental strains exhibited great similarity. However, the genome synteny between the clinically pathogenic and environmental isolates was relatively low.
The alignment of the four genomes of clinically pathogenic isolates showed that their genome rearrangements were similar although there were inversions in some regions. Moreover, the chromosomal alignments of CCUG12901 and CIP101113 were nearly identical with large segments of high similarity. There were some white areas not aligned well because they might contain elements specific to a particular genome.
Overall, the four clinically pathogenic strain genomes were similar although the genome synteny in the latter three (CCUG10230, CCUG12901, and CIP101113) was more related than that in PR63039.
3.5. Prophages in four clinically pathogenic M. odoratimimus strains
All the four clinically pathogenic isolates genomes contained incomplete prophage elements (Figure 3). In our strain PR 63039, two incomplete prophages were identified (Hu et al., 2017). CIP101113 contained one prophage with 54 CDs extending from 985,588 bp to 1,032,415 bp (46.8 kb). CCUG10230 was predicted to carry two prophages (9 CDs, 9.2 kb and 12 CDs, respectively). CCUG12901 had five prophages. The region length was 9.2, 9.7, 9.9, 10.1, and 9 kb, respectively, and the number of CDs was 9, 8, 7, 6, 6, respectively. CIP101113 contained only one prophage, but it was larger and more complete than any other prophages. It consisted of hypothetical proteins, phage‐like proteins, attachment sites, tail shafts, and proteases. Among these predicted prophages, attachment sites and proteases only existed in CIP101113 prophage. In bacterial genomes, integrases are useful markers for mobile DNA elements, such as prophages, integrative plasmids, and pathogenicity islands (Liu et al., 2015). However, no integrase was identified in these predicted prophages.
Figure 3.
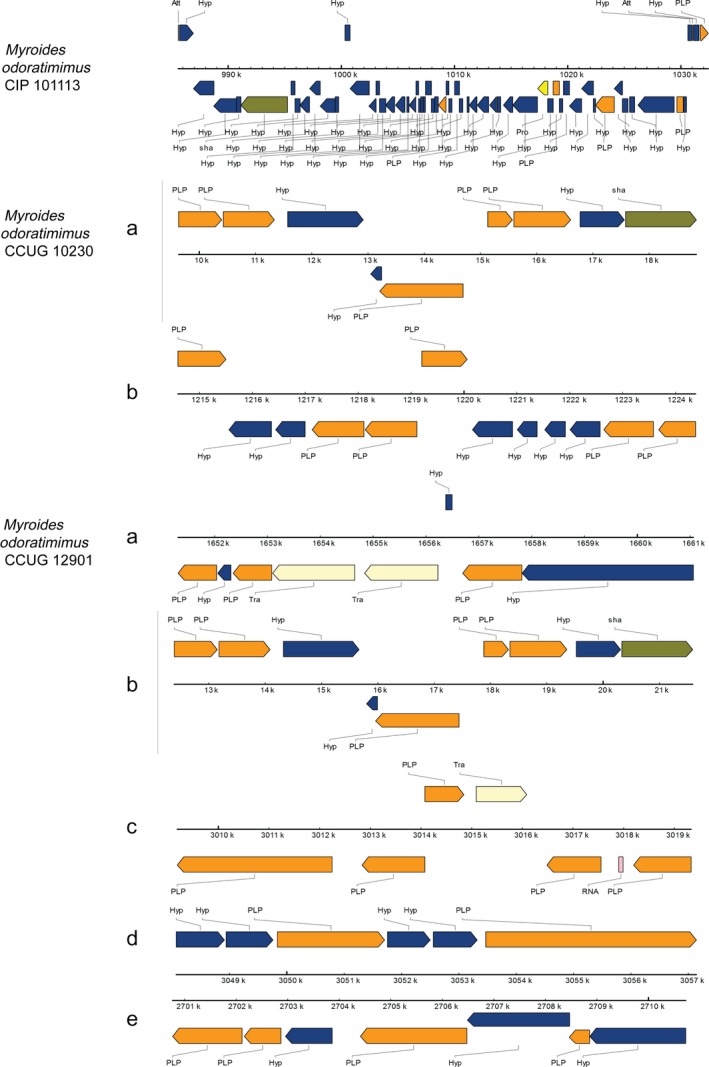
Prophage regions and predicted elements in three clinically pathogenic M. odoratimimus strains. Different colored rectangles indicated different phage elements. Att, attachment site; Hyp, hypothetical protein; PLP, Other phage‐like protein; sha, Tail shaft; Pro, protease; Tra, transposase
3.6. CRISPR prediction in the genomes of four clinically pathogenic and three environmental M. odoratimimus strains
CRISPR is well known to contribute to the antibiotic resistance and prevent the foreign virulence genes from invading into pathogens. It may be involved in the bacterial evolution, regulation of virulence gene expression, and the enhancement of pathogenicity (Hatoum‐Aslan & Marraffini, 2014). Particularly, in the pathogen, CRISPR is able to edit genome and modulate gene functions as an adaptive immune system (Arras et al., 2016; Sontheimer & Barrangou, 2015). Comparative analysis of the seven strain genomes (PR63039, CCUG10230, CCUG12901, CIP101113, CCUG12700, L41, and H1bi) showed that PR63039 genome contained three types of CRISPRs (Table S1); CCUG10230 genome contained four types of CRISPRs. CCUG12700 and H1bi contained only one CRISPR. However, no CRISPRs were identified in the genomes of CCUG12901, CIP101113, and L41 (Table S1).
3.7. Comparative analysis of antibiotic resistance genes in the genomes of four clinically pathogenic M. odoratimimus strains
With CARD RGI software (Jia et al., 2017; Mcarthur et al., 2013), all the genomes of four clinically pathogenic M. odoratimimus strains PR63039, CCUG10230, CCUG12901, and CIP101113 were predicted to contain a number of genes related to antibiotic resistance, including the β‐lactam resistance gene, fluoroquinolone resistance gene, antibiotic target replacement protein, antibiotic inactivation enzyme, triclosan resistance gene, diaminopyrimidine resistance gene, phenicol resistance gene, elfamycin resistance gene, and efflux pumps conferring antibiotic resistance (Table 3).
Table 3.
The predicted resistance genes in the genomes of four clinically pathogenic M. odoratimimus strains
| Category | PR63039 | CCUG12901 | CCUG10230 | CIP101113 |
|---|---|---|---|---|
| Efflux pump complex or subunit conferring antibiotic resistance | abeS | abeS | abeS | abeS |
| KPC‐2 | KPC‐2 | KPC‐2 | KPC‐2 | |
| ‐ | mefC | ‐ | ‐ | |
| msrB | msrB | msrB | msrB | |
| qacH | qacH | qacH | qacH | |
| rosA | rosA | rosA | rosA | |
| Determinant of elfamycin resistance | basS | basS | basS | basS |
| LpxC | LpxC | LpxC | LpxC | |
| SPM‐1 | SPM‐1 | SPM‐1 | SPM‐1 | |
| Determinant of phenicol resistance | catB2 | ‐ | ‐ | ‐ |
| catB6 | ‐ | ‐ | ‐ | |
| catB7 | ‐ | ‐ | catB7 | |
| catB8 | ‐ | ‐ | ‐ | |
| catB9 | ‐ | ‐ | ‐ | |
| catB10 | ‐ | ‐ | ‐ | |
| ‐ | ‐ | catII | catI | |
| ‐ | ‐ | catIII | ‐ | |
| Determinant of diaminopyrimidine resistance | dfrE | dfrE | dfrE | dfrE |
| Determinant of triclosan resistance | MexR | MexR | MexR | MexR |
| Antibiotic inactivation enzyme | catB3 | ‐ | ‐ | ‐ |
| ‐ | catII | catII | ||
| ‐ | mphD | ‐ | ‐ | |
| ‐ | mphG | ‐ | ‐ | |
| OXA‐78 | OXA‐78 | OXA‐78 | OXA‐78 | |
| tetX | ||||
| VIM‐2 | VIM‐2 | VIM‐2 | VIM‐2 | |
| Antibiotic target replacement protein | sul1 | ‐ | ‐ | ‐ |
| sul2 | ‐ | ‐ | ‐ | |
| sul3 | ‐ | ‐ | ‐ | |
| Determinant of fluoroquinolone resistance | oqxB | oqxB | oqxB | oqxB |
| rpsJ | rpsJ | rpsJ | rpsJ | |
| tet(35) | tet(35) | tet(35) | tet(35) | |
| tetB(48) | tetB(48) | tetB(48) | tetB(48) | |
| Determinant of beta‐lactam resistance | OXA‐209 | ‐ | ‐ | ‐ |
| OXA‐347 | ‐ | ‐ | ‐ | |
| TLA‐3 | TLA‐3 | TLA‐3 | TLA‐3 |
‐not predicted
Overall, more resistance genes were predicted in the fully sequenced clinically pathogenic PR63039 genome than in the other three partially sequenced strains, such as, cat gene variant catB2, catB3, catB6, catB7, catB8, catB9, and catB10, tetracycline resistance gene tetX, sulfonamide resistance gene sul1, sul2, and sul3, and β‐lactam resistance gene OXA‐209, OXA‐347. Moreover, in PR63039 genome, the resistance genes tetX, cat, OXA‐347, and OXA‐209 were clustered in an approximately 6 kb region, called MY63039‐RR (Hu et al., 2017). No similar resistance gene cluster could be identified in the genomes of other three clinically pathogenic M. odoratimimus strains.
Among CCUG12901, CIP101113, and CCUG10230, the identified resistance genes were almost similar. By comparing with PR63039 genome, CCUG12901 genome contained mefC (efflux pump conferring antibiotic resistance) and antibiotic inactivation enzyme (mphD and mphG), but lacked the resistance gene cat. The gene catII, catIII in CCUG10230, and catI in CIP101113 were also not predicted in PR63039 genome.
3.8. Virulence factors in the genomes of three clinically pathogenic and three environmental M. odoratimimus strains
With the help of VFDB protein Set B database (Chen et al., 2012, 2016) and more stringent cutoff parameters, we obtained the VF genes in the genome of three clinically pathogenic (CCUG.12901, CCUG.10230, CIP.101113) and three environmental (H1bi, L41, CCUG.12700) M. odoratimimus isolates (Table 4).
Table 4.
The VFs predicted in three clinically pathogenic and three environmental M. odoratimimus genomes
| Classification | Definition | Genes coding for virulence factors | |||||
|---|---|---|---|---|---|---|---|
| Clinically pathogenic M. odoratimimus | Environmental M. odoratimimus | ||||||
| CCUG. 12901 | CCUG. 10230 | CIP. 101113 | H1bi | L41 | CCUG. 12700 | ||
| Capsular polysaccharide | UDP‐N‐acetyl‐D‐galactosamine 6‐dehydrogenase | capL (hasB2) | capL | ‐ | capL | ‐ | capL |
| UDP‐glucose 4‐epimerase | galE | galE | galE | galE | galE | galE | |
| Bifunctional UDP‐N‐acetylglucosamine pyrophosphorylase/glucosamine‐1‐phosphate N‐acetyltransferase | glmU | glmU | glmU | glmU | glmU | ‐ | |
| UDP‐glucose 6‐dehydrogenase | ugd | ugd | ugd | ‐ | ugd | ‐ | |
| Capsule sialic acid | UDP‐N‐acetylglucosamine 2‐epimerase | wbjD/wecB | wbjD/wecB | wbjD/wecB | wbjD/wecB | ‐ | wbjD/wecB |
| Cell wall Peptidoglycan | Undecaprenyl diphosphate synthase | uppS | uppS | uppS | uppS | uppS | uppS |
| Glucose‐1‐phosphate thymidylytransferase | rmlA | rmlA | rmlA | rmlA | rmlA | rmlA | |
| dTDP‐glucose 4,6‐dehydratase | rmlB | rmlB | rmlB | rmlB | ‐ | ‐ | |
| UDP‐N‐acetyl‐D‐mannosaminuronic acid dehydrogenase | wecC | wecC | wecC | ‐ | ‐ | wecC | |
| Intracellular survival factors | Catalase katA | katA | katA | katA | katA | katA | katA |
| ATP‐dependent Clp protease proteolytic subunit | clpP | clpP | clpP | clpP | clpP | clpP | |
| Elongation factor Tu | EF‐Tu | EF‐Tu | EF‐Tu | EF‐Tu | EF‐Tu | EF‐Tu | |
| Superoxide dismutase | sodB | SodB | sodB | sodB | sodB | sodB | |
| Molecular chaperones | CT396 molecular chaperone DnaK | DnaK | DnaK | DnaK | DnaK | DnaK | DnaK |
| 60k heat‐shock protein HtpB | Hsp60 | Hsp60 | Hsp60 | Hsp60 | Hsp60 | Hsp60 | |
| Urease | Urease | ureA | ureA | ureA | ureA | ureA | ureA |
| Urease | ureB | ureB | ureB | ureB | ureB | ureB | |
| Urease/hydrogenase‐associated predicted GTPase | ureG | ureG | ureG | ureG | ureG | ureG | |
| Acinetobactin | ABC‐type enterochelin transport system, ATPase component | bauE | bauE | bauE | bauE | ‐ | bauE |
| Streptococcal enolase | Streptococcal enolase | eno | eno | eno | eno | eno | eno |
| Pantothenate synthesis | Aspartate 1‐decarboxylase | panD | panD | panD | panD | panD | panD |
| Heme biosynthesis | Porphobilinogen synthase | hemB | hemB | hemB | hemB | hemB | hemB |
| glutamate‐1‐semialdehyde aminotransferase | hemL | hemL | hemL | hemL | hemL | hemL | |
| Acyl carrier protein | acpXL | acpXL | acpXL | acpXL | acpXL | acpXL | |
| T4SS effectors | Trans‐2‐enoyl‐CoA reductase (no unique name) | + | + | + | + | + | + |
‐not predicted; +,predicted; bold/italic, were discussed.
Overall, all these M. odoratimimus genomes had similar VF profiles with a little difference. The VFs included capsule/capsular polysaccharide (GalE, GlmU, wbjD/wecB, ugd, uppS, RmlA, RmlB, capL, wecC), intracellular survival and invasion factors (katA, clpP, EF‐Tu, sodB), molecular chaperone (hsp60, DnaK), urease (ureA, ureB, ureG), acinetobactin (bauE), Streptococcal enolase (eno), heme biosynthesis (hemB, hemL), acyl carrier protein (acpXL), and T4SS effectors (Trans‐2‐enoyl‐CoA reductase).
4. DISCUSSION
4.1. Genomic evolution, variants, synteny of M. odoratimimus
To some contents, the 10 Myroides genomes had similar general features. However, the genomes of four clinically pathogenic M. odoratimimus strains (PR63039, CCUG12901, CCUG10230, and CIP101113) contained highly variable regions. The genome arrangement, rearrangements of the four clinically pathogenic isolates, and the three environmental strains were similar, respectively. These data implied that they might be evolutionarily related. All four clinically pathogenic isolate genomes contained prophage elements. CRISPRs were not always identified in all the genomes of four clinically pathogenic and three environmental M. odoratimimus strains. Complete genomic sequencing of these M. odoratimimus strains and their plasmids are indispensable for confirming these analyses.
4.2. Resistance genes in clinically pathogenic M. odoratimimus
All these four clinically pathogenic strains (PR63039, CCUG12901, CCUG10230, CIP101113) contained a number of antibiotic resistance genes. PR63039 genome might have several possibly unique resistance genes, including catB2, catB3, catB6, catB7, catB8, catB9, catB10, tetX, OXA‐209, OXA‐347, sul1, sul2, and sul3. Some were already discussed to be involved in drug resistance in our previous report (Hu et al., 2017). Here, we mainly discuss mph and cat genes (catB2, catB3, catB6, catB8) with clear functions in antibiotic resistance described by literatures.
The mphD and mphG genes provide a high level of resistance to 14‐ and 15‐membered‐ring macrolides via coding for a macrolide 2′‐phosphotransferase (https://card.mcmaster.ca/). Cat (chloramphenicol acetyltransferase gene) has many variants in a variety of bacteria, such as Staphylococcus aureus, Staphylococcus haemolyticus, Enterococcus faecium, and Bacillus clausii (Bruckner & Matzura, 1985; Galopin, Cattoir, & Leclercq, 2009; Grady & Hayes, 2003; Schwarz & Cardoso, 1991). All the above identified catB genes (as well catI, catII, catIII) are plasmid, chromosome, or integron‐mediated cat variants (https://card.mcmaster.ca/). The cat variants usually participate in the composition of gene cassette or integron, and confer to the ability of antibiotic resistance. For instance, catB2, aacC4, and aadA1 form the gene cassettes aacC4‐aadA1‐catB2, which confers multidrug and broad‐spectrum cephalosporin resistance in Salmonella clinical isolates (Villa et al., 2002). CatB3, one member of gene cassette aacA7‐catB3‐aadB‐oxa2‐orfD, can be mobilized by the integron‐encoded DNA integrase and plays a role in chloramphenicol resistance of plasmid pBWH301 (Bunny, Hall, & Stokes, 1995; Houang, Chu, Lo, Chu, & Cheng, 2003). CatB6, a chloramphenicol acetyltransferase‐encoding allele of the catB family inserted in integron In31, functions to decrease the in vitro antibiotic susceptibilities of Pseudomonas aeruginosa strains (Laraki et al., 1999). CatB8 consists of a resistance gene cassette aacA4‐catB8‐aadA1 which is prevalent in many clinical antibiotic‐resistant bacteria, such as carbapenem‐resistant Klebsiella pneumoniae (Ou, Li, Li, & Yu, 2017) and carbapenem‐resistant Acinetobacter baumannii (Farshadzadeh et al., 2015; Lin, Liou, Tu, Yeh, & Lan, 2013).
We could not further correlate these predicted antibiotic resistance gene profiles to the antibiotic susceptibility of the other three clinically pathogenic strains (CCUG12901, CCUG10230, CIP101113) due to the lack of these related data. However, the conservation of the antibiotic resistance genes among the clinically pathogenic M. odoratimimus strains indicated that these predicted antibiotic resistance gene profiles potentially provide the guidance for treating M. odoratimimus infections later.
4.3. Pathogenicity of clinically pathogenic and environmental M. odoratimimus indicated by the predicted virulence factors
The VFs in M. odoratimimus were identified using VFDB protein Set B database which curates the experiment‐verified and predicted virulence factor genes (Table 4). The finding of similar VFs in both clinically pathogenic and environmental M. odoratimimus genomes might explain the pathogenicity of the three clinically pathogenic M. odoratimimus isolates and indicate that the three environmental M. odoratimimus isolates are also potentially pathogenic. We only discuss the experimentally verified VF genes (in bold/italic, Table 4).
bauE encodes the ferric siderophore ABC transporter/ATP‐binding protein BauE with a high‐affinity iron‐chelating capacity, belonging to acinetobactin. In a mouse sepsis model, the expression of a fully active acinetobactin‐mediated iron uptake apparatus by Acinetobacter baumannii was verified to be vital for the bacteria to establish infection and kill mouse, by competing with host cells for iron (Gaddy et al., 2012). Thus, bauE should be a survival factor of M. odoratimimus during infection.
Capsular LPS is the predominant virulence determinant in many gram‐negative and ‐positive bacteria (García & López, 1997). It inhibits complement‐mediated lysis, phagocytosis, and immune recognition in host (Rowe & Huntley, 2015). Several genes involved in capsular LPS biosynthesis pathway were found in the M. odoratimimus genomes, such as GalE, GlmU, ugd, wbjD/wecB, uppS, RmlA, RmlB. UDP‐sugar 4‐epimerase (GalE) plays an essential role in LPS synthesis (Beerens, Soetaert, & Desmet, 2015; Fry et al., 2000) by catalyzing the interconversion of UDP‐galactose and UDP‐glucose (Fry et al., 2000), is a critical virulence factor in many gram‐negative bacteria (Beerens et al., 2015; Fry et al., 2000). N‐acetylglucosamine‐1‐phosphateuridyltransferase/glucosamine‐1‐phosphate‐acetyltransferase (GlmU) is involved in the synthesis of peptidoglycan and LPS in Gram‐negative and ‐positive bacteria (Sharma & Khan, 2017). The bifunctional enzyme UDP‐N‐acetylglucosamine 2‐epimerase/ManNAc kinase (encoded by wbjD/wecB, also known as siaA or neuC) catalyzes biosynthesis of Escherichia coli K1 capsule, an alpha‐2,8‐linked polymer of sialic acid, and is a vital meningitis virulence factor for this pathogen (Vann et al., 2004). Both mammals (Chou, Hinderlich, Reutter, & Tanner, 2003) and bacteria (Murkin, Chou, Wakarchuk, & Tanner, 2004) produce this bifunctional enzyme. Both source of this enzyme can catalyze the conversion of UDP‐GlcNAc into ManNAc and UDP, the first two steps in the sialic acid biosynthesis in mammals (Chou et al., 2003) and the first step of sialic acid (N‐acetylneuraminic acid) biosynthesis in bacteria (Murkin et al., 2004). UDP‐glucose 6‐dehydrogenase (UGD) has an indispensable role in hyaluronic acid capsule production and pathogenicity in Group A Streptococcus (Cole et al., 2012), is required for bacterial growth inside macrophages (Mouslim & Groisman, 2003). Undecaprenyl diphosphate synthase (UPPS) is involved in cell wall biosynthesis (peptidoglycan and wall teichoic acid synthesis) by catalyzing the synthesis of a polyisoprenoid, becoming an attractive antibacterial drug target (Farha et al., 2015). Glucose‐1‐phosphate thymidylyltransferase (RmlA) is vital for bacteria survival (Mansuri et al., 2016). It participates in L‐rhamnose synthesis (Alphey et al., 2013; Mansuri et al., 2016), a critical linker of peptidoglycan and arabinogalacton in bacterial cell wall (Mansuri et al., 2016), by catalyzing the generation of dTDP‐D‐glucose and pyrophosphate (PPi) (Alphey et al., 2013; Mansuri et al., 2016). dTDP‐D‐glucose 4,6‐dehydratase (RmlB) is also involved in L‐rhamnose biosynthesis, by catalyzing the conversion of dTDP‐D‐glucose into dTDP‐4‐keto‐6‐deoxy‐D‐glucose in cell wall (Allard et al., 2002). Bacteria with truncated LPS molecules due to the L‐rhamnose synthesis failure could not prevent clearance by the host cells and become avirulent (Allard et al., 2002). The presence of capsular LPS biosynthesis genes in six M. odoratimimus indicates their infectivity. The presence of these LPS biosynthesis genes in both the three clinically pathogenic and the three environmental M. odoratimimus isolates is in concert with the fact that M. odoratimimus is gram‐negative and confer it this bacterium with in infectivity.
The presence of bacterial intracellular survival factors katA, clpP, EF‐Tu, and sodB in the six M. odoratimimus genomes suggested that this bacterium might be able to survive within host cells, increase the antibiotic therapy difficulty and thus explain the reported high death rate of M. odoratimimus infections (summarized in Hu et al., 2016). Catalase katA is a critical virulence factor for Campylobacter jejuni, a facultatively intracellular microbe and the principal pathogen of human gastroenteritis. Its resistance ability to the bacteriocidal activity from host cell‐produced hydrogen peroxide and intramacrophage persistence/growth is dependent on this catalase (Day, Sajecki, Pitts, & Joens, 2000). ClpP, a highly conserved protease in prokaryotes and eukaryotes, is involved in the rapid adaption capacity during infection for Listeria monocytogenes, another facultative intracellular pathogen (Gaillot, Bregenholt, Jaubert, Di Santo, & Berche, 2001; Gaillot, Pellegrini, Bregenholt, Nair, & Berche, 2000). By interacting with host surface nucleolin, the bacterial surface EF‐Tu (elongation factor Tu), a GTP‐binding protein involved in protein translation in Francisella tularensis, a highly infectious intracellular gram‐negative bacterium, plays a crucial role in its invasion to host tissues (Barel et al., 2008). It is also an adhesion/invasion factor secreted by microbes during infection by bacteria (like Helicobacter pylori) (Chiu, Wang, Tsai, Lei, & Liao, 2017) and fungi (Marcos et al., 2016) through binding (Mycobacterium avium subsp. paratuberculosis) with fibronectin on host cells (Viale et al., 2014). Superoxide dismutases (SODs) protect the bacteria from oxidative damage by converting endogenously generated superoxide radicals into hydrogen peroxide and oxygen, are indispensable for intraphagocytic viability for pathogenic bacteria (Dhar, Gupta, & Virdi, 2013), SodB is required for colonization of Helicobacter pylori in the stomach (Tsugawa et al., 2015).
The presence of bacterial DnaK (known as Hsp70 in eukaryotes) and Hsp60 in M. odoratimimus imply that the autoimmunological response might be complicated by the infection. Heat‐shock proteins are ubiquitous proteins with high homology between eukaryotes and prokaryotes. Bacterial DnaK is crucial bacterial virulence factor. Both host Hsp70 and bacterial DnaK mediate bacterial attachment to host cells. After infection, bacterial DnaK switches on bacterial survival processes and arouses autoimmune sequelae (Ghazaei, 2017). Hsp60 is also involved in Clostridium difficile attachment to host cells (Hennequin et al., 2001), and the strong proinflammatory reaction (IL8) of monocytic cells induced by Helicobacter pylori (Lin et al., 2005). Chlamydia pneumonia Hsp60 can help to spread Chlamydial infection of blood monocytes to vascular wall cells (Rupp et al., 2005), and increase the pathogenesis and severity of Chlamydia infection‐correlated atherosclerosis because of sequence homology between bacterial and human Hsp60 (mitochondria in endothelial cells) and subsequent induction of a strong autologous humoral and cellular immune responses (Kalayoglu et al., 2000; Mehta et al., 2005).
The presence of ureA, ureB, ureG in M. odoratimimus implies that this bacterium might be pathogenic in human stomach. Urease is a principal virulence factor of human gastric bacterium Helicobacter pylori (Stingl et al., 2008). It has oligomeric Ni2+‐containing heterodimer of UreA and UreB subunits involved in converting gastric juice urea into NH3 in bacterial periplasm which maintains an optimal pH, inner membrane potential and proton motive force, being critical for colonization within the human stomach (Sachs, Weeks, Melchers, & Scott, 2003). Urease activity needs an assembly of a lysine‐carbamate functional group with two Ni2+ ions facilitated partially by GTP hydrolysis by UreG (Martin‐Diaconescu, Bellucci, Musiani, Ciurli, & Maroney, 2012; Zambelli, Turano, Musiani, Neyroz, & Ciurli, 2009).
The surface enolase (eno) of bacteria, a glycolytic pathway enzyme, can bind human plasminogen and convert it into active plasmin (Cork et al., 2009) to facilitate bacterial adherence to host cells and destruction of host tissues through plasmin degrading intercellular junctions and extracellular matrix components (Attali, Durmort, Vernet, & Di Guilmi, 2008), like cellulitis (Bachmeyer et al., 2008), necrotizing fasciitis (Crum‐Cianflone, Matson, & Ballon‐Landa, 2014), and make the bacterial infection life‐threatening (Cork et al., 2009). The containing eno in M. odoratimimus genome might explain the high death rate of patients infected by M. odoratimimus (as summarized in Hu et al., 2016).
In brief, M. odoratimimus not only possesses common virulence factors, like using bauE gene to compete the iron with host, general LPS synthesis genes, adherence factors (DnaK, Hsp60), but also can survive intracellularly (katA, clpP, EF‐Tu, and sodB), even in human stomach (ureA, ureB, ureG), but also disseminate easily, destroy human tissues, induce autoimmune diseases. So, the M. odoratimimus is a life‐threatening pathogen as reported (summarized in Hu et al., 2016).
5. CONCLUSION
The genomic analysis demonstrated that these M. odoratimimus isolates are closely related. Our analyses provided some insights in bacterial pathogenicity and antibiotic resistance mechanisms of M. odoratimimus and contribute to future development of the therapeutic regimens in M. odoratimimus infections.
COMPLIANCE WITH ETHICAL STANDARDS
This article does not contain any studies with human participants or animals performed by any of the authors.
CONFLICT OF INTEREST
None declared.
Supporting information
ACKNOWLEDGMENT
This work was supported by the Fong Shu Fook Tong and Fong Yun Wah Foundations (14X30127), the Technology Planning Projects of Quanzhou Social Development Fields (2014Z24), and the Natural Science Fund Project of Fujian Province (2015J01514).
Hu S, Cao L, Wu Y, et al. Comparative genomic analysis of Myroides odoratimimus isolates. MicrobiologyOpen. 2019;8:e634 10.1002/mbo3.634
Contributor Information
Qiujing Wang, Email: qiujingwang1@126.com.
Desong Ming, Email: mds6430@126.com.
Shicheng Chen, Email: shicheng@msu.edu.
Mingxi Wang, Email: mxwang@hqu.edu.cn.
REFERENCES
- Allard, S. T. , Beis, K. , Giraud, M. F. , Hegeman, A. D. , Gross, J. W. , Wilmouth, R. C. , … Naismith, J. H. (2002). Toward a structural understanding of the dehydratase mechanism. Structure, 10, 81–92. 10.1016/S0969-2126(01)00694-3 [DOI] [PubMed] [Google Scholar]
- Alphey, M. S. , Pirrie, L. , Torrie, L. S. , Boulkeroua, W. A. , Gardiner, M. , Sarkar, A. , … Naismith, J. H. (2013). Allosteric competitive inhibitors of the glucose‐1‐phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa . ACS Chemical Biology, 8, 387–396. 10.1021/cb300426u [DOI] [PubMed] [Google Scholar]
- Arras, S. D. , Chua, S. M. , Wizrah, M. S. , Faint, J. A. , Yap, A. S. , & Fraser, J. A. (2016). Targeted genome editing via CRISPR in the pathogen Cryptococcus neoformans . PLoS ONE, 11, e0164322 10.1371/journal.pone.0164322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attali, C. , Durmort, C. , Vernet, T. , & Di Guilmi, A. M. (2008). The interaction of Streptococcus pneumoniae with plasmin mediates transmigration across endothelial and epithelial monolayers by intercellular junction cleavage. Infection and Immunity, 76, 5350–5356. 10.1128/IAI.00184-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmeyer, C. , Entressengle, H. , Khosrotehrani, K. , Goldman, G. , Delisle, F. , Arlet, G. , & Grateau, G. (2008). Cellulitis due to Myroides odoratimimus in a patient with alcoholic cirrhosis. Clinical and Experimental Dermatology, 33, 97–98. [DOI] [PubMed] [Google Scholar]
- Barel, M. , Hovanessian, A. G. , Meibom, K. , Briand, J. P. , Dupuis, M. , & Charbit, A. (2008). A novel receptor‐ligand pathway for entry of Francisella tularensis in monocyte‐like THP‐1 cells: Interaction between surface nucleolin and bacterial elongation factor Tu. BMC Microbiology, 8, 145 10.1186/1471-2180-8-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens, K. , Soetaert, W. , & Desmet, T. (2015). UDP‐hexose 4‐epimerases: A view on structure, mechanism and substrate specificity. Carbohydrate Research, 414, 8–14. 10.1016/j.carres.2015.06.006 [DOI] [PubMed] [Google Scholar]
- Belloir, L. , Billy, P. A. , Hentgen, C. , Fille, A. , & Barrans, A. (2016). Myroides odoratimimus bacteremia. Medecine at Maladies Infectieuses, 46, 396–397.[Article in French] 10.1016/j.medmal.2016.05.003 [DOI] [PubMed] [Google Scholar]
- Bertels, F. , Silander, O. K. , Pachkov, M. , Rainey, P. B. , & van Nimwegen, E. (2014). Automated reconstruction of whole‐genome phylogenies from short‐sequence reads. Molecular Biology and Evolution, 31, 1077–1088. 10.1093/molbev/msu088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland, C. , Ramsey, T. L. , Sabree, F. , Lowe, M. , Brown, K. , Kyrpides, N. C. , & Hugenholtz, P. (2007). CRISPR recognition tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics, 8, 209 10.1186/1471-2105-8-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner, R. , & Matzura, H. (1985). Regulation of the inducible chloramphenicol acetyltransferase gene of the Staphylococcus aureus plasmid pUB112. The EMBO Journal, 4, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunny, K. L. , Hall, R. M. , & Stokes, H. W. (1995). New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrobial Agents and Chemotherapy, 39, 686–693. 10.1128/AAC.39.3.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghartz, M. , Bunk, B. , Sproer, C. , Voget, S. , Daniel, R. , Overmann, J. , … Jahn, M. (2015). Complete genome sequence of the urethral catheter isolate Myroides sp. A21. Genome Announcements, 3, e00068–15. 10.1128/genomeA.00068-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Xiong, Z. , Sun, L. , Yang, J. , & Jin, Q. (2012). VFDB 2012 update: Toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Research, 40, D641–D645. 10.1093/nar/gkr989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Zheng, D. , Liu, B. , Yang, J. , & Jin, Q. (2016). VFDB 2016: Hierarchical and refined dataset for big data analysis‐10 years on. Nucleic Acids Research, 44, D694–D697. 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, K. H. , Wang, L. H. , Tsai, T. T. , Lei, H. Y. , & Liao, P. C. (2017). Secretomic analysis of host‐pathogen interactions reveals that elongation factor‐Tu is a potential adherence factor of Helicobacter pylori during pathogenesis. Journal of Proteome Research, 16, 264–273. 10.1021/acs.jproteome.6b00584 [DOI] [PubMed] [Google Scholar]
- Chou, W. K. , Hinderlich, S. , Reutter, W. , & Tanner, M. E. (2003). Sialic acid biosynthesis: Stereochemistry and mechanism of the reaction catalyzed by the mammalian UDP‐N‐acetylglucosamine 2‐epimerase. Journal of the American Chemical Society, 125, 2455–2461. 10.1021/ja021309g [DOI] [PubMed] [Google Scholar]
- Cole, J. N. , Aziz, R. K. , Kuipers, K. , Timmer, A. M. , Nizet, V. , & van Sorge, N. M. (2012). A conserved UDP‐glucose dehydrogenase encoded outside the hasABC operon contributes to capsule biogenesis in group A Streptococcus . Journal of Bacteriology, 194, 6154–6161. 10.1128/JB.01317-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cork, A. J. , Jergic, S. , Hammerschmidt, S. , Kobe, B. , Pancholi, V. , Benesch, J. L. , … Walker, M. J. (2009). Defining the structural basis of human plasminogen binding by streptococcal surface enolase. The Journal of Biological Chemistry, 284, 17129–17137. 10.1074/jbc.M109.004317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum‐Cianflone, N. F. , Matson, R. W. , & Ballon‐Landa, G. (2014). Fatal case of necrotizing fasciitis due to Myroides odoratus . Infection, 42, 931–935. 10.1007/s15010-014-0626-0 [DOI] [PubMed] [Google Scholar]
- Darling, A. E. , Mau, B. , & Perna, N. T. (2010). progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE, 5, e11147 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, W. A. Jr , Sajecki, J. L. , Pitts, T. M. , & Joens, L. A. (2000). Role of catalase in Campylobacter jejuni intracellular survival. Infection and Immunity, 68, 6337–6345. 10.1128/IAI.68.11.6337-6345.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar, M. S. , Gupta, V. , & Virdi, J. S. (2013). Detection, distribution and characterization of novel superoxide dismutases from Yersinia enterocolitica, Biovar 1A. PLoS ONE, 8, e63919 10.1371/journal.pone.0063919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farha, M. A. , Czarny, T. L. , Myers, C. L. , Worrall, L. J. , French, S. , Conrady, D. G. , … Brown, E. D. (2015). Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase. Proceedings of the National Academy of Sciences of the United States of America, 112, 11048–11053. 10.1073/pnas.1511751112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farshadzadeh, Z. , Hashemi, F. B. , Rahimi, S. , Pourakbari, B. , Esmaeili, D. , … Bahador, A. (2015). Wide distribution of carbapenem resistant Acinetobacter baumannii in burns patients in Iran. Frontier Microbiology, 6, 1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, B. N. , Feng, S. , Chen, Y. Y. , Newell, D. G. , Coloe, P. J. , & Korolik, V. (2000). The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infection and Immunity, 68, 2594–2601. 10.1128/IAI.68.5.2594-2601.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddy, J. A. , Arivett, B. A. , Mcconnell, M. J. , Lopez‐Rojas, R. , Pachon, J. , & Actis, L. A. (2012). Role of acinetobactin‐mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infection and Immunity, 80, 1015–1024. 10.1128/IAI.06279-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillot, O. , Bregenholt, S. , Jaubert, F. , Di Santo, J. P. , & Berche, P. (2001). Stress‐induced ClpP serine protease of Listeria monocytogenes is essential for induction of listeriolysin O‐dependent protective immunity. Infection and Immunity, 69, 4938–4943. 10.1128/IAI.69.8.4938-4943.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillot, O. , Pellegrini, E. , Bregenholt, S. , Nair, S. , & Berche, P. (2000). The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes . Molecular Microbiology, 35, 1286–1294. 10.1046/j.1365-2958.2000.01773.x [DOI] [PubMed] [Google Scholar]
- Galopin, S. , Cattoir, V. , & Leclercq, R. (2009). A chromosomal chloramphenicol acetyltransferase determinant from a probiotic strain of Bacillus clausii . FEMS Microbiology Letters, 296, 185–189. 10.1111/j.1574-6968.2009.01633.x [DOI] [PubMed] [Google Scholar]
- García, E. , & López, R. (1997). Molecular biology of the capsular genes of Streptococcus pneumoniae . FEMS Microbiology Letter, 49, 1–10. 10.1016/S0378-1097(97)00026-8 [DOI] [PubMed] [Google Scholar]
- Ghazaei, C. (2017). Role and mechanism of the Hsp70 molecular chaperone machines in bacterial pathogens. Journal of Medical Microbiology, 66, 259–265. 10.1099/jmm.0.000429 [DOI] [PubMed] [Google Scholar]
- Grady, R. , & Hayes, F. (2003). Axe‐Txe, a broad‐spectrum proteic toxin‐antitoxin system specified by a multidrug‐resistant, clinical isolate of Enterococcus faecium . Molecular Microbiology, 47, 1419–1432. 10.1046/j.1365-2958.2003.03387.x [DOI] [PubMed] [Google Scholar]
- Grant, J. R. , & Stothard, P. (2008). The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Research, 36, W181–W184. 10.1093/nar/gkn179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum‐Aslan, A. , & Marraffini, L. A. (2014). Impact of CRISPR immunity on the emergence and virulence of bacterial pathogens. Current Opinion in Microbiology, 17, 82–90. 10.1016/j.mib.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennequin, C. , Porcheray, F. , Waligora‐Dupriet, A. , Collignon, A. , Barc, M. , Bourlioux, P. , & Karjalainen, T. (2001). GroEL (Hsp60) of Clostridium difficile is involved in cell adherence. Microbiology, 147(Pt 1), 87–96. 10.1099/00221287-147-1-87 [DOI] [PubMed] [Google Scholar]
- Heritier, C. , Poirel, L. , & Nordmann, P. (2006). Cephalosporinase over‐expression resulting from insertion of ISAba1 in Acinetobacter baumannii . Clinical Microbiology and Infection, 12, 123–130. 10.1111/j.1469-0691.2005.01320.x [DOI] [PubMed] [Google Scholar]
- Higgins, P. G. , Dammhayn, C. , Hackel, M. , & Seifert, H. (2010). Global spread of carbapenem‐resistant Acinetobacter baumannii. Journal of Antimicrobial Chemotherapy, 65, 233–238. 10.1093/jac/dkp428 [DOI] [PubMed] [Google Scholar]
- Houang, E. T. , Chu, Y. W. , Lo, W. S. , Chu, K. Y. , & Cheng, A. F. (2003). Epidemiology of rifampin ADP‐ribosyltransferase (arr‐2) and metallo‐beta‐lactamase (blaIMP‐4) gene cassettes in class 1 integrons in Acinetobacter strains isolated from blood cultures in 1997 to 2000. Antimicrobial Agents and Chemotherapy, 47, 1382–1390. 10.1128/AAC.47.4.1382-1390.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, S. , Jiang, T. , Zhou, Y. , Ming, D. , Gao, H. , & Wang, M. (2017). Genomic analysis of the multi‐drug‐resistant clinical isolate Myroides odoratimimus PR63039. Molecular Genetics and Genomics, 292, 133–144. 10.1007/s00438-016-1261-5 [DOI] [PubMed] [Google Scholar]
- Hu, S. H. , Yuan, S. X. , Qu, H. , Jiang, T. , Zhou, Y. J. , Wang, M. X. , & Ming, D. S. (2016). Antibiotic resistance mechanisms of Myroides sp . Journal of Zhejiang University‐Science B, 17, 188–199. 10.1631/jzus.B1500068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, B. , Raphenya, A. R. , Alcock, B. , Waglechner, N. , Guo, P. , Tsang, K. K. , … McArthur, A. G. (2017). CARD 2017: Expansion and model‐centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Research, 45(D1), D566–D573. 10.1093/nar/gkw1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover‐Sáenz, A. , Pérez‐Villar, F. , & Barcenilla‐Gaite, F. (2016). Severe sepsis caused by infected prosthesis joint due to Myroides odoratimimus . Medicina Clinica (Barc), 147, 276–277. [Article in Spanish] 10.1016/j.medcli.2016.04.011 [DOI] [PubMed] [Google Scholar]
- Kalayoglu, M. V. , Indrawati, Morrison, R. P. , Morrison, S. G. , Yuan, Y. , & Byrne, G. I. (2000). Chlamydial virulence determinants in atherogenesis: The role of Chlamydial lipopolysaccharide and heat shock protein 60 in macrophage‐lipoprotein interactions. Journal of Infectious Diseases, 181(Suppl 3), S483–S489. 10.1086/315619 [DOI] [PubMed] [Google Scholar]
- Ktari, S. , Mnif, B. , Koubaa, M. , Mahjoubi, F. , Ben Jemaa, M. , Mhiri, M. N. , & Hammami, A. (2012). Nosocomial outbreak of Myroides odoratimimus urinary tract infection in a Tunisian hospital. The Journal of Hospital Infection, 80, 77–81. 10.1016/j.jhin.2011.09.010 [DOI] [PubMed] [Google Scholar]
- Laraki, N. , Galleni, M. , Thamm, I. , Riccio, M. L. , Amicosante, G. , Frere, J. M. , & Rossolini, G. M. (1999). Structure of In31, a blaIMP‐containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrobial Agents and Chemotherapy, 43, 890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M. F. , Liou, M. L. , Tu, C. C. , Yeh, H. W. , & Lan, C. Y. (2013). Molecular epidemiology of integron‐associated antimicrobial gene cassettes in the clinical isolates of Acinetobacter baumannii from northern Taiwan. Annals of Laboratory Medicine, 33, 242–247. 10.3343/alm.2013.33.4.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. N. , Ayada, K. , Zhao, Y. , Yokota, K. , Takenaka, R. , Okada, H. , … Oguma, K. (2005). Helicobacter pylori heat‐shock protein 60 induces production of the pro‐inflammatory cytokine IL8 in monocytic cells. Journal of Medical Microbiology, 54(Pt 3), 225–233. 10.1099/jmm.0.45871-0 [DOI] [PubMed] [Google Scholar]
- Liu, C. J. , Wang, R. , Gong, F. M. , Liu, X. F. , Zheng, H. J. , Luo, Y. Y. , & Li, X. R. (2015). Complete genome sequences and comparative genome analysis of Lactobacillus plantarum strain 5‐2 isolated from fermented soybean. Genomics, 106, 404–411. 10.1016/j.ygeno.2015.07.007 [DOI] [PubMed] [Google Scholar]
- Mansuri, R. , Ansari, M. Y. , Singh, J. , Rana, S. , Sinha, S. , Sahoo, G. C. , … Das, P. (2016). Computational elucidation of structural basis for ligand binding with Mycobacterium tuberculosis glucose‐1‐phosphate thymidylyltransferase (RmlA). Current Pharmaceutical Biotechnology, 17, 1089–1099. 10.2174/1389201017666160909155959 [DOI] [PubMed] [Google Scholar]
- Marcos, C. M. , de Oliveira, H. C. , da Silva, J. F. , Assato, P. A. , Yamazaki, D. S. , da Silva, R. A. , & Fusco‐Almeida, A.M . (2016). Identification and characterisation of elongation factor Tu, a novel protein involved in Paracoccidioides brasiliensis‐host interaction. FEMS Yeast Research, 16, fow079 10.1093/femsyr/fow079 [DOI] [PubMed] [Google Scholar]
- Martin‐Diaconescu, V. , Bellucci, M. , Musiani, F. , Ciurli, S. , & Maroney, M. J. (2012). Unraveling the Helicobacter pylori UreG zinc binding site using X‐ray absorption spectroscopy (XAS) and structural modeling. Journal of Biological Inorganic Chemistry, 17, 353–361. 10.1007/s00775-011-0857-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcarthur, A. G. , Waglechner, N. , Nizam, F. , Yan, A. , Azad, M. A. , Baylay, A. J. , … Wright, G. D. (2013). The comprehensive antibiotic resistance database. Antimicrobial Agents and Chemotherapy, 57, 3348–3357. 10.1128/AAC.00419-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, T. A. , Greenman, J. , Ettelaie, C. , Venkatasubramaniam, A. , Chetter, I. C. , & McCollum, P. T. (2005). Heat shock proteins in vascular disease—a review. European Journal of Vascular & Endovascular Surgery, 29, 395–402. 10.1016/j.ejvs.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Mouslim, C. , & Groisman, E. A. (2003). Control of the Salmonella ugd gene by three‐two‐component regulatory systems. Molecular Microbiology, 47, 335–344. 10.1046/j.1365-2958.2003.03318.x [DOI] [PubMed] [Google Scholar]
- Murkin, A. S. , Chou, W. K. , Wakarchuk, W. W. , & Tanner, M. E. (2004). Identification and mechanism of a bacterial hydrolyzing UDP‐N‐acetylglucosamine 2‐epimerase. Biochemistry, 43, 14290–14298. 10.1021/bi048606d [DOI] [PubMed] [Google Scholar]
- Ou, Q. , Li, W. , Li, B. , & Yu, C. (2017). Prevalence of carbapenem‐resistant Klebsiella pneumoniae (CRKP) and the distribution of Class 1 Integron in their strains isolated from a hospital in Central China. Chinese Medical Sciences Journal, 32, 107–112. [DOI] [PubMed] [Google Scholar]
- Pompilio, A. , Galardi, G. , Gherardi, G. , Verginelli, F. , Geminiani, C. , Pilloni, A. P. , & Bonaventura, G. D. (2017). Infection of recurrent calcaneal ulcer caused by a biofilm‐producer Myroides odoratimimus strain. Folia Microbiologica (Praha), 2017, 203–207. 10.1007/s12223-017-0552-5 [DOI] [PubMed] [Google Scholar]
- Rowe, H. M. , & Huntley, J. F. (2015). From the Outside‐In: The Francisella tularensis envelope and virulence. Frontiers in Cellular and Infection Microbiology, 5, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp, J. , Koch, M. , van Zandbergen, G. , Solbach, W. , Brandt, E. , & Maass, M. (2005). Transmission of Chlamydia pneumoniae infection from blood monocytes to vascular cells in a novel transendothelial migration model. FEMS Microbiology Letters, 242, 203–208. 10.1016/j.femsle.2004.11.010 [DOI] [PubMed] [Google Scholar]
- Sachs, G. , Weeks, D. L. , Melchers, K. , & Scott, D. R. (2003). The gastric biology of Helicobacter pylori. Annual Review of Physiology, 65, 349–369. 10.1146/annurev.physiol.65.092101.142156 [DOI] [PubMed] [Google Scholar]
- Schwarz, S. , & Cardoso, M. (1991). Molecular cloning, purification, and properties of a plasmid‐encoded chloramphenicol acetyltransferase from Staphylococcus haemolyticus . Antimicrobial Agents and Chemotherapy, 35, 1277–1283. 10.1128/AAC.35.7.1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, R. , & Khan, I. A. (2017). Mechanism and potential inhibitors of GlmU: A novel target for antimicrobial drug discovery. Current Drug Targets, 18, 1587–1597. [DOI] [PubMed] [Google Scholar]
- Siguier, P. , Perochon, J. , Lestrade, L. , Mahillon, J. , & Chandler, M. (2006). ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Research, 34, D32–D36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer, E. J. , & Barrangou, R. (2015). The bacterial origins of the CRISPR genome‐editing revolution. Human Gene Therapy, 26, 413–424. 10.1089/hum.2015.091 [DOI] [PubMed] [Google Scholar]
- Stingl, K. , Schauer, K. , Ecobichon, C. , Labigne, A. , Lenormand, P. , Rousselle, J. C. , & de Reuse, H. (2008). In vivo interactome of Helicobacter pylori urease revealed by tandem affinity purification. Molecular & Cellular Proteomics, 7, 2429–2441. 10.1074/mcp.M800160-MCP200 [DOI] [PubMed] [Google Scholar]
- Tsugawa, H. , Mori, H. , Matsuzaki, J. , Masaoka, T. , Hirayama, T. , Nagasawa, H. , & Suzuki, H. (2015). Nordihydroguaiaretic acid disrupts the antioxidant ability of Helicobacter pylori through the repression of SodB activity in vitro. BioMed Research International, 2015, 734548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann, W. F. , Daines, D. A. , Murkin, A. S. , Tanner, M. E. , Chaffin, D. O. , Rubens, C. E. , … Silver, R. P. (2004). The NeuC protein of Escherichia coli K1 is a UDP N‐acetylglucosamine 2‐epimerase. Journal of Bacteriology, 186, 706–712. 10.1128/JB.186.3.706-712.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale, M. N. , Echeverria‐Valencia, G. , Romasanta, P. , Mon, M. L. , Fernandez, M. , Malchiodi, E. , … Santangelo, M. L. (2014). Description of a novel adhesin of Mycobacterium avium subsp. paratuberculosis. BioMed Research International, 2014, 729618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa, L. , Mammina, C. , Miriagou, V. , Tzouvelekis, L. S. , Tassios, P. T. , Nastasi, A. , & Carattoli, A. (2002). Multidrug and broad‐spectrum cephalosporin resistance among Salmonella enterica serotype enteritidis clinical isolates in southern Italy. Journal of Clinical Microbiology, 40, 2662–2665. 10.1128/JCM.40.7.2662-2665.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems, P. , Muller, J. , Verhaegen, J. , Saegeman, V. , & Desmet, S. (2017). How to treat a fulminant erysipelas and sepsis caused by Myroides odoratimimus: Case report and literature review. Acta Clinica Belgica, 72, 331–335. 10.1080/17843286.2016.1245173 [DOI] [PubMed] [Google Scholar]
- Zambelli, B. , Turano, P. , Musiani, F. , Neyroz, P. , & Ciurli, S. (2009). Zn2+‐linked dimerization of UreG from Helicobacter pylori, a chaperone involved in nickel trafficking and urease activation. Proteins, 74, 222–239. 10.1002/prot.22205 [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Liang, Y. , Lynch, K. H. , Dennis, J. J. , & Wishart, D. S. (2011). PHAST: A fast phage search tool. Nucleic Acids Research, 39, W347–W352. 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


