Abstract
Symbioses between insects and microbes are ubiquitous, but vary greatly in terms of function, transmission mechanism, and location in the insect. Lepidoptera (butterflies and moths) are one of the largest and most economically important insect orders; yet, in many cases, the ecology and functions of their gut microbiomes are unresolved. We used high-throughput sequencing to determine factors that influence gut microbiomes of field-collected fall armyworm (Spodoptera frugiperda) and corn earworm (Helicoverpa zea). Fall armyworm midgut bacterial communities differed from those of corn earworm collected from the same host plant species at the same site. However, corn earworm bacterial communities differed between collection sites. Subsequent experiments using fall armyworm evaluating the influence of egg source and diet indicated that that host plant had a greater impact on gut communities. We also observed differences between regurgitant (foregut) and midgut bacterial communities of the same insect host, suggesting differential colonization. Our findings indicate that host plant is a major driver shaping gut microbiota, but differences in insect physiology, gut region, and local factors can also contribute to variation in microbiomes. Additional studies are needed to assess the mechanisms that affect variation in insect microbiomes, as well as the ecological implications of this variability in caterpillars.
Introduction
Insect herbivores inhabit a diverse set of niches, and therefore face a wide variety of challenges such as nutritionally recalcitrant food sources, toxins, environmental extremes, and threats from parasites and pathogens. Insects have integrative strategies to contend with these challenges, which often include forming symbiotic associations with microbes1. Microbial associations are ubiquitous among animals, but vary along functional and ecological continua2. Compared to endosymbiotic bacteria, the evolutionary trajectories and transmission strategies of facultative gut symbionts are far more variable3,4. Extracellular symbionts can be obtained through environmental sources5, shared food resources6,7, trophallaxis3, deposition on egg surfaces, and copulation7.
The insect gut can be rich in microbial symbionts, and the associations, locations, and functions of these associates can vary considerably. Some insects possess special gut modifications or structures such as paunches, diverticula, and caeca to house symbionts8, while others lack morphological modifications. The roles of insect gut symbionts are diverse. For example, insect gut symbionts can be involved in metabolism of recalcitrant food sources9,10, provisioning of vitamins11 and nutrients12,13, and metabolism of plant allelochemicals14–16. Oral bacteria found in the regurgitant can also be involved in manipulating plant responses to herbivore feeding, thereby suppressing induction of plant defences and leading to increases in herbivore growth17,18.
Lepidoptera (butterflies and moths) is the third largest insect order with over 200,000 described species. Larval lepidopteran guts are characterised by a simple, tube-like morphology that facilitates the rapid transit of food associated with high consumption rates19. These insect guts represent extreme environments for microorganisms due to their high alkalinity (pH > 10)20–22.
Commonly, caterpillar gut microbiomes are simple and variable, usually being comprised of relatively few dominant taxa23–26, and appear to be shaped in part by dietary and environmental sources26–28. Bacterial communities have also been described from lepidopteran eggs25,29, suggesting there is potential for maternal transmission of microbiota. The roles of lepidopteran oral and gut bacteria in facilitating plant–insect interactions remain unclear, but they likely have facultative functions, particularly in relation to plant defences14,30. The sources of gut microbiota in lepidopterans have received little attention; understanding the factors that influence bacterial community composition may shed light upon symbiont–host co-adaptation and the strategies insects use to acquire their microbial partners.
The lepidopteran family Noctuidae includes many polyphagous agricultural pests that can cause significant economic losses. Spodoptera frugiperda (fall armyworm) is a highly polyphagous noctuid that is a major agricultural pest in South America, the Caribbean, and most recently in Africa31,32. In the northern United States and Canada, sporadic fall armyworm infestations occur from populations that annually migrate from southern Texas and Florida as this species cannot overwinter in the cooler northern climates33. Although fall armyworm develops faster and prefers grasses such as maize and wheat, it can also complete development on broadleaf crops such as soybean and cotton34. Corn earworm (Helicoverpa zea) is another agricultural noctuid pest that is distributed throughout the American continent35. Its major host plant is maize, although it will also feed on other crops such as tomato, cotton and soybean36.
Despite the economic importance of corn earworm and fall armyworm, little is known about the composition of their midgut and oral bacteria and how these communities are shaped by transmission from eggs and host plant feeding. We used bacterial 16S-rRNA sequencing to determine if there were differences in gut communities from different population and/or host plant sources. Specifically, our objectives were to: (i) characterise and compare the midgut bacterial community of wild-collected fall armyworm and corn earworm from different locations in Pennsylvania, (ii) determine whether egg source (wild-collected or laboratory-reared) or diet source (soybean leaves or corn silk) were more important in shaping fall armyworm midgut bacterial communities, and (iii) compare bacterial communities present in the regurgitant with those of the midgut in fall armyworm. This latter objective is of interest because specific bacteria from oral secretions (regurgitant) of lepidopterans have been shown to influence plant defences30, but whether these bacteria are a subset of those found in the midgut or comprise a unique community is unknown. To address these objectives, we used culture-independent high throughput sequencing methods. We also used traditional culture-based and 16S sequencing methods to examine bacteria present in the regurgitant of fall armyworm collected from Pennsylvania and Puerto Rico.
Results
Comparisons of midgut microbiota between fall armyworm and corn earworm
There were significant differences in the bacterial communities inhabiting the midguts of fall armyworm and corn earworm (Fig. 1a,b; Supplemental Fig. 1). Multivariate analyses revealed significant differences between fall armyworm and corn earworm microbiota based on Bray-Curtis (Fig. 1a; PerMANOVA p < 0.001) and Jaccard (Fig. 1b; PerMANOVA p < 0.001) similarity indices. In addition to the differences between these two species collected at the same location, corn earworm collected from different locations also exhibited divergent communities (Fig. 1). We found substantial differences in the bacterial midgut communities of corn earworm obtained from the two sites in Pennsylvania, despite the fact they were collected from the same host plant species. Alpha-diversity metrics also differed between lepidopteran species, with fall armyworm exhibiting marginally significantly lower Simpson (p = 0.053) and Shannon (p = 0.033) metrics than corn earworm (Supplemental Fig. 1; Supplemental Table 1). This suggests that corn earworm midgut microbiota exhibited greater levels of diversity and greater evenness.
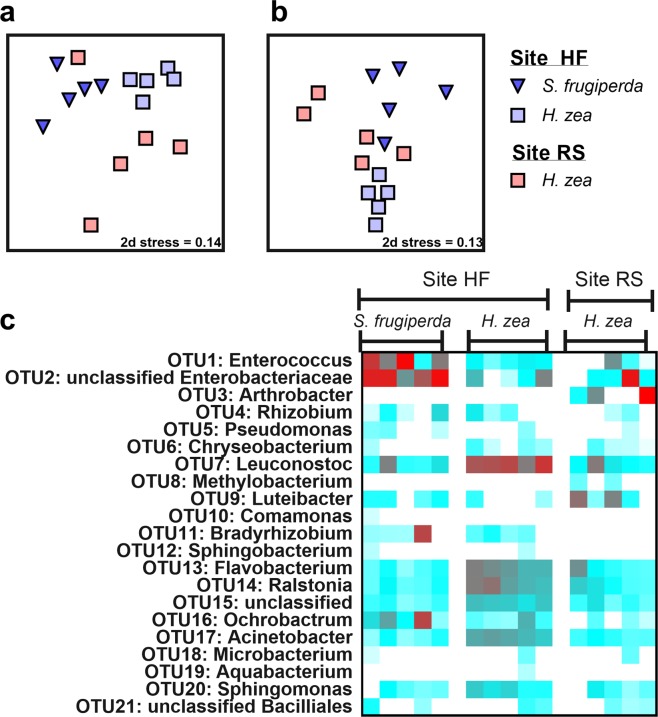
Figure 1.
Differences between fall armyworm (Spodoptera frugiperda) and corn earworm (Helicoverpa zea) midgut bacterial communities collected from sweet corn ears in Pennsylvania. Corn earworm was collected from Site HF (Harner Farm) and Site RS (Rock Springs), while fall armyworm was collected only from Site HF. Non-metric multidimensional scaling (nMDS) plots were constructed using Bray-Curtis (a) and Jaccard (b) dissimilarities. Stress values indicate a good fit in two dimensions. Heat maps (c) show relative abundance (log2 transformed) of OTUs (operational taxonomic units). Red boxes indicate greater relative abundances, blue boxes correspond to lower values, and white boxes correspond to no detection of that particular OTU.
Heat maps of midgut microbiota indicated that conspecific individuals exhibited a high degree of variability in terms of both membership and abundance (Fig. 1c). Among the most enriched OTUs (operational taxonomic units), fall armyworm had the greatest relative abundances of Enterococcus and unclassified Enterobacteriaceae. Corn earworm from the HF population had lower relative abundances of Enterococcus and unclassified Enterobacteriaceae, but were enriched for Leuconostoc. Corn earworm from the RF site were highly variable as to which OTUs were present in high relative abundances, and had fewer Leuconostoc than corn earworm from the HF site.
Influence of egg population source and plant diet on fall armyworm midgut bacterial communities
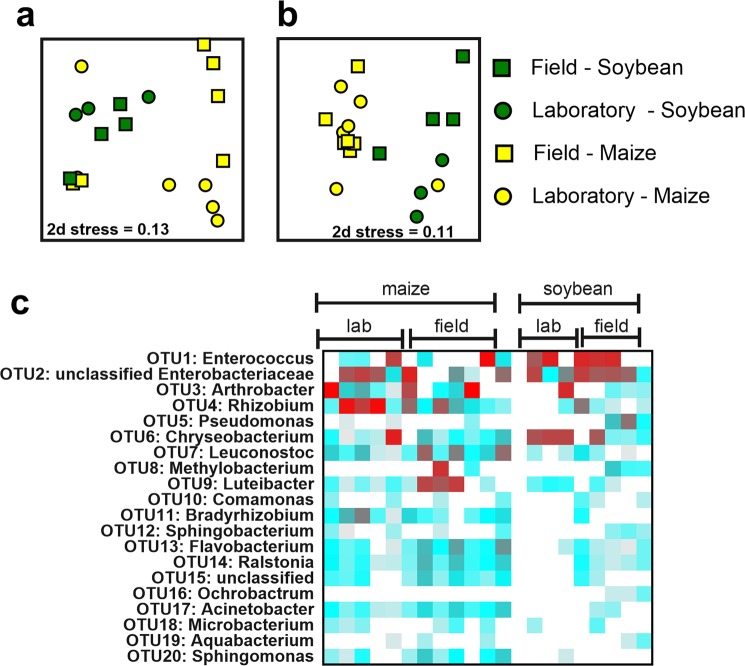
Multivariate analyses revealed there was a significant impact of host plant on midgut bacterial communities in terms of both Bray-Curtis (Fig. 2a) and Jaccard (Fig. 2b) similarities (Table 1). There was no impact of egg population source or its interaction with host plant on midgut bacterial communities (Fig. 2a,b; Table 1). At both the order and individual OTU levels, there was substantial variation in abundance of bacterial taxa between individual fall armyworm larvae within treatments (Fig. 2c; Supplemental Fig. 2). In general larvae feeding on maize had high relative abundances of OTUs corresponding to unclassified Enterobacteriaceae, Rhizobium, and Chryseobacterium. Soybean-fed larvae generally had higher relative abundances of Enterococcus, unclassified Enterobacteriaceae, Chryseobacterium, and Pseudomonas (Fig. 2c).
Figure 2.
Influence of diet (maize or soybean) and egg population source (field-collected or laboratory) on fall armyworm (Spodoptera frugiperda) midgut bacterial communities. Non-metric multidimensional scaling (nMDS) plots were constructed using Bray-Curtis (a) and Jaccard (b) dissimilarities. Stress values indicate a good fit in two dimensions. Heat maps (c) show relative abundance (log2 transformed) of OTUs (operational taxonomic units). Red boxes indicate greater relative abundances, blue boxes correspond to lower values, and white boxes correspond to no detection of that particular OTU.
Table 1.
PERMANOVA output assessing differences between fall armyworm (Spodoptera frugiperda) midgut bacterial communities between egg population source (field or laboratory) and host plant (maize or soybean).
| Source | df | SS | MS | Pseudo-F | p-value |
|---|---|---|---|---|---|
| Bray-Curtis Similarity | |||||
| Plant | 1, 19 | 8057 | 8057 | 4.15 | 0.029 |
| Population | 1, 19 | 3407 | 3407 | 1.75 | 0.156 |
| Plant*Population | 1, 19 | 1447 | 1447 | 0.74 | 0.512 |
| Jaccard Similarity | |||||
| Plant | 1, 19 | 8025 | 8025 | 2.07 | 0.002 |
| Population | 1, 19 | 4381 | 4381 | 1.13 | 0.181 |
| Plant*Population | 1, 19 | 4167 | 4167 | 1.07 | 0.297 |
PERMANOVAs were generated using 999 permutations, and the individual insect was included in the model as a random effect. Bold values indicate significant (p < 0.05) differences.
Alpha-diversity metrics indicated that, in general, gut bacterial communities of larvae fed on soybean were more diverse than those fed on maize (Supplemental Fig. 3, Supplemental Table 2). Midguts from insects reared from field-collected and laboratory (Benzon) egg sources generally had the same levels of OTU richness. However maize-fed larvae from the field population had higher bacterial gut diversity than those from the laboratory population (Supplemental Fig. 3).
Differences in fall armyworm bacterial communities in midgut and regurgitant
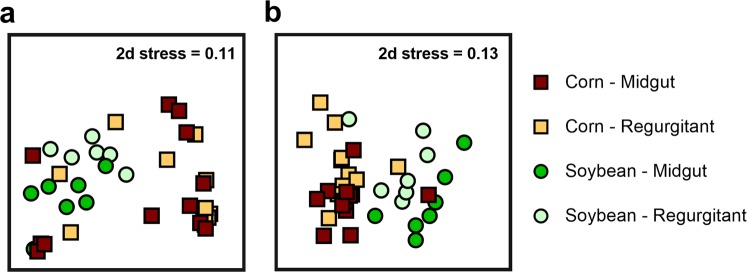
We collected paired samples corresponding to fall armyworm regurgitant and midguts to determine if there were differences between the microbiota that reside in the foregut versus the midgut. There were substantial differences in the communities present, and like in prior analyses, there was a major impact of host plant on the midgut bacterial communities using both Bray-Curtis (Fig. 3a) and Jaccard (Fig. 3b) similarities (Table 2). In addition to these differences between host plants, there was a substantial impact of whether the community of origin was from the midgut or regurgitant (Table 2; p = 0.013). However, we observed no interaction between the host plant and the physical origin of the bacterial community. These trends were consistent for both Bray-Curtis and Jaccard metrics.
Figure 3.
Influence of sample type (midgut or regurgitant) and diet (maize or soybean) on fall armyworm (Spodoptera frugiperda) bacterial communities. Non-metric multidimensional scaling (nMDS) plots were constructed using Bray-Curtis (a) and Jaccard (b) similarities. Stress values indicate a good fit in two dimensions.
Table 2.
PERMANOVA output assessing differences between fall armyworm (Spodoptera frugiperda) bacterial communities between sample type (midgut or regurgitant) and host plant (maize or soybean).
| Source | df | SS | MS | Pseudo-F | p-value |
|---|---|---|---|---|---|
| Bray-Curtis Similarity | |||||
| Sample type | 1, 39 | 7191 | 7191 | 3.82 | 0.013 |
| Plant | 1, 39 | 16257 | 16257 | 8.64 | 0.001 |
| Sample type*Plant | 1, 39 | 2075 | 2075 | 1.10 | 0.331 |
| Jaccard Similarity | |||||
| Sample type | 1, 39 | 4772 | 4772 | 1.22 | 0.020 |
| Plant | 1, 39 | 11526 | 11526 | 2.95 | 0.001 |
| Sample type*Plant | 1, 39 | 4222 | 4222 | 1.08 | 0.169 |
PERMANOVAs were generated using 999 permutations, and the individual insect was included in the model as a random effect. Bold values indicate significant (p < 0.05) differences.
Using Bray-Curtis similarities, we used a two-way SIMPER analysis to identify the major OTUs that contributed to the differences between host plant and regurgitant, and then conducted paired statistical tests. While host plant had a significant impact on the relative abundance of several of the OTUs that inhabited the midgut and regurgitant (Table 3), the effects of regurgitant were far more limited. Only Enterococcus exhibited significant differences between midgut and regurgitant from fall armyworm feeding on maize (p = 0.045) and soybean (p = 0.001).
Table 3.
Pairwise comparisons of abundances of individual bacterial OTUs (operational taxonomic units) between host plant (soybean or maize) and sample type (midgut or regurgitant) in fall armyworm (Spodoptera frugiperda). Bold values indicate significant (p < 0.05) differences. OTUs were selected using a SIMPER analysis.
| OTU | Maize | Soybean | Plant | Sample type (maize) | Sample type (soybean) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Midgut | Regurgitant | Midgut | Regurgitant | W | p-value | W | p-value | W | p-value | |
| OTU001: Enterococcus | 27.48 ± 10.8 | 9.82 ± 6.2 | 32.94 ± 5.95 | 15.43 ± 2.69 | 427 | 0.0055 | 185 | 0.0449 | 98 | 0.0006 |
| OTU002: Enterobacteriaceae | 39.93 ± 10.9 | 55.15 ± 9.46 | 22.25 ± 5.52 | 27.74 ± 9.76 | 253 | 0.0387 | 131 | 0.2913 | 59 | 0.3823 |
| OTU003: Arthrobacter | 5.33 ± 4.66 | 4.14 ± 3.1 | 24.49 ± 4.97 | 34.01 ± 7.78 | 455 | 0.0003 | 159.5 | 0.5968 | 50 | 0.065 |
| OTU004: Rhizobium | 6.87 ± 3.44 | 2.31 ± 0.85 | 0.91 ± 0.36 | 1.24 ± 0.63 | 348 | 0.5944 | 143 | 0.7125 | 57 | 0.2786 |
| OTU005: Pseudomonas | 0.97 ± 0.4 | 1.44 ± 0.58 | 0.07 ± 0.03 | 0.09 ± 0.05 | 192 | <0.0001 | 140 | 0.5899 | 65 | 0.798 |
| OTU008: Methylobacterium | 1.46 ± 0.64 | 1.73 ± 0.81 | 0.04 ± 0.02 | 0.03 ± 0.01 | 222 | 0.0028 | 155.5 | 0.7679 | 65 | 0.798 |
| OTU011: Bradyrhizobium | 0.98 ± 0.38 | 0.91 ± 0.42 | 0.04 ± 0.03 | 0.01 ± 0 | 191 | <0.0001 | 158 | 0.6707 | 61.5 | 0.5169 |
| OTU015: Unclassified | 3.85 ± 2.42 | 1.25 ± 0.47 | 0.02 ± 0.01 | 0.01 ± 0.01 | 181.5 | <0.0001 | 157 | 0.7122 | 58 | 0.2821 |
| OTU016: Ochrobactrum | 1.43 ± 0.79 | 0.5 ± 0.16 | 0.76 ± 0.33 | 1.15 ± 0.61 | 328 | 1.000 | 154 | 0.8426 | 59 | 0.3667 |
| OTU018: Microbacterium | 0.48 ± 0.25 | 1.25 ± 0.7 | 0.56 ± 0.17 | 0.7 ± 0.31 | 364.5 | 0.3207 | 128 | 0.2184 | 62 | 0.5737 |
| OTU033: Bacillus | 0.02 ± 0.02 | 0.29 ± 0.2 | 2.1 ± 1.56 | 2.11 ± 1.39 | 468.5 | <0.0001 | 116.5 | 0.3205 | 63 | 0.6454 |
16S sequencing of individual isolates from Puerto Rico and Pennsylvania
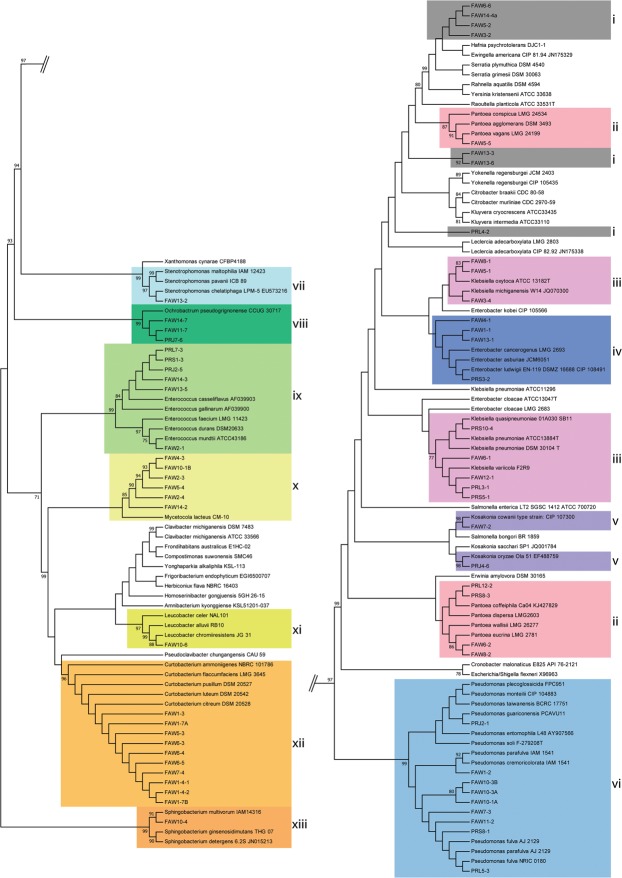
Culture-dependent sequencing of bacteria isolated from the regurgitant of fall armyworm collected in both Puerto Rico and Pennsylvania support the findings of the community analysis data reported above. We found that bacterial taxa in the Enterobacteriaceae, Pseudomonadaceae and Enterococcaceae and Microbacteriaceae were commonly identified based on sequencing of the V4 region of the bacterial 16S gene (Fig. 4). Common bacterial genera that were isolated include Pantoea (Fig. 4, clade ii), Klebsiella (Fig. 4, clade iii), Enterobacter (Fig. 4, clade iv), Pseudomonas (Fig. 4, clade v), Ochrobactrum (Fig. 4, clade vii), Enterococcus (Fig. 4, clade viii), Mycetocola (Fig. 4, clade x) and Curtobacterium (Fig. 4, clade xi). In some cases, the first six of these genera were detected in larvae collected in both Pennsylvania and Puerto Rico.
Figure 4.
Phylogenetic tree of near full-length 16S rRNA gene sequences from bacterial isolates cultured from fall armyworm (Spodoptera frugiperda) regurgitant collected in Pennsylvania, United States (indicated by FAW prefix) Puerto Rico (indicated by PR prefix), and selected type strains from the RDP database. The phylogenetic tree was constructed using the Maximum Likelihood method based on the Jukes-Cantor model in MEGA7. The tree with the highest log likelihood (−5718.26) is shown. Bootstrap values with values of 70 or greater are shown. Annotations correspond to clades that are identified as likely the following genera (i) unresolved (ii) Pantoea (iii) Klebsiella (iv) Enterobacter (v) Kosakonia (vi) Pseudomonas (vii) Stenotrophomonas (viii) Ochrobactrum (ix) Enterococcus (x) Mycetocola (xi) Leucobacter (xii) Curtobacterium and (xiii) Sphingobacterium.
Discussion
Bacterial gut communities of two lepidopteran species, fall armyworm and corn earworm, were variable and influenced by multiple factors. Bacterial communities were distinct depending on insect species and collection location. We also found that host plant was much more important in shaping midgut bacterial communities in fall armyworm than the population source of the eggs. Midgut and regurgitant bacterial composition of fall armyworm were also distinct, suggesting that the physiology of different gut regions may affect bacterial composition or abundance.
We found high variability in gut bacterial composition and abundance between individuals of the same species, even from those feeding on the same food source. This is consistent with reports of several other lepidopteran species that possess microbial gut assemblages that differ between individuals29,37,38. The high variability in lepidopteran gut bacterial assemblages and the apparent lack of a resident microbiota has fuelled speculation that gut associates lack functional importance in lepidopterans38–41. These suggestions likely stem from the limited research into functional roles of gut bacteria in Lepidoptera. However, demonstrated roles of lepidopteran oral and gut bacteria include suppression30 and detoxification14 of plant defences. Moreover, these associates can mediate other ecological associations, such as interactions with entomopathogens42–44. Future studies manipulating the presence/absence and assemblage of gut bacterial associates in controlled studies are needed to address functions of these associates and the consequences of variations in community composition/abundance.
Although we observed high intraspecific variation in bacterial gut composition and structure, we also found that the two lepidopteran species had distinct midgut bacterial communities despite feeding on the same host plant species at the same location. This suggests that acquisition of bacteria from the environment is not a completely stochastic process and that bacterial communities are not merely a reflection of the host plant microbiome. There are likely gut physiological mechanisms that select for certain taxa, which may help shape bacterial composition in these different species. Competition among taxa may also play a role and it is likely that microbial community composition is dynamic. The ability of insect guts to filter certain taxa from the wider environmental pool of bacteria has been observed in several systems including cockroaches45, bean bugs46–48 and bumble bees49. There is little known about the mechanisms involved in bacterial acquisition and establishment in lepidopteran guts, but variation in physiochemical properties and competition among taxa are likely important for shaping these communities.
Host plant (maize or soybean) played a much bigger role in influencing the composition of midgut bacteria in fall armyworm than egg source (Fig. 2). These results are comparable to studies on other folivores showing that host plant can shape gut bacterial communities, including Spodoptera littoralis24, Helicoverpa spp.24,26,37, Lymantria dispar23,28, and Leptinotarsa decemlineata50. There are pre- and post-digestive interactions between the plant and microbe that may affect these communities. For example, differences in leaf surface, wax composition, availability of sugars, and interactions with other bacterial species can alter the bacterial community composition present on plant leaves51, therefore altering the composition and quantify of bacteria the insect may encounter. There are also chemical interactions occurring in the gut that may mediate these interactions; for example, foliar concentrations of plant secondary compounds can affect bacterial composition in insect guts52. To what degree fall armyworm gut bacterial composition reflects that of their host plant was not investigated in this study, but is likely influenced by a combination of phyllosphere bacteria and the chemical composition of the plant tissues.
We also found that corn earworm midgut bacterial communities from two sites in Pennsylvania were distinct, even though the host plant (sweet corn) was the same at both sites, likely due to differences in phyllosphere bacteria inhabiting the host plants at each site. Several studies investigating factors that influence phyllosphere bacteria found that geographical location of plants altered bacterial community composition53,54.
Although fall armyworm individuals possessed different overall bacterial communities in paired regurgitant and midgut samples, pairwise comparisons revealed that this was primarily driven by a single Enterococcus OTU. There are several physiological and morphological differences between the midgut and foregut that may differentially impact microbial gut associates. The foregut is covered in a cuticular lining while the midgut cells are protected by the peritrophic membrane lining the midgut wall. Additionally, the pH of the lepidopteran alimentary canal changes across its length; the midgut is highly alkaline, while the foregut is usually closer to neutral55,56. These differences may provide differential binding and colonization affinities for microbiota, resulting in greater abundances of certain microbial taxa. The foregut may also pose greater disturbances to bacterial communities, because the cuticular lining is replaced during each moult. It is unclear if and how microbes recolonise the foregut after moulting, although in S. littoralis fluorescently-labelled bacteria were observed in the foregut throughout different instars57.
We observed relatively few bacterial taxa associated with the regurgitant of fall armyworm. A recent study of the “regurgitome” of Mexican bean beetle (Epilachna varivestis) revealed a highly diverse microbiome with 1230 bacterial species in 577 genera58. In our study we observed less diversity and richness; culture-independent sequencing detected about 100 OTUs in larvae fed on maize and soybean. In both plant species, Enterococcus and an unclassified Enterobacteriaceae OTU comprised the majority of the taxa in our study. These results paired well with our culture-dependent survey. Culture-dependent 16S sequencing of bacteria from regurgitant of fall armyworm larvae collected in Pennsylvania and Puerto Rico showed that isolates belonged to genera including Pantoea, Enterobacter, Klebsiella, Pseudomonas, Curtobacterium, Mycetocola, and Enterococcus. These genera are commonly associated with many different insect groups59, and have documented roles in mediating fall armyworm–plant interactions30.
Based on the phylogenetic tree constructed from sequencing near-full length 16S-rRNA, we found some bacterial isolates could not be reliably identified (Fig. 4i) and several genera including Pantoea, Klebsiella, Enterobacter, and Kosakonia were not monophyletic. The 16S rRNA gene is widely used to study bacterial ecology, but can have limitations distinguishing closely related taxa due to high sequence similarities60. The Enterobacteriaceae tends to have poor resolution using hypervariable regions of the 16S rRNA gene, including the broadly used V4 region61,62. It is likely that the unclassified Enterobacteriaceae OTU in our culture-independent analyses actually contains several bacterial genera, and thus underestimated the diversity in our samples.
This study contributes to our understanding of the factors that influence gut microbiomes in two lepidopteran insects and offers insight into the composition of bacterial communities in fall armyworm regurgitant, which are likely to be relevant for mediating plant–insect interactions. We show that variation in insect host, gut region, and host plant can affect the composition of bacterial gut communities of lepidopteran larvae. Further research will be required to identify the factors that alter bacterial communities at each of these levels of scale.
Methods
Insect and plant sources
Plant and insect material were collected from two sites in Centre County, PA in September 2016: a local farm in State College, PA (HF) and the Pennsylvania State University Russell E. Larson Research Farm (RS). These two sites are separated by approximately 11 km (Supplemental Table 3). We obtained fall armyworm from HF, and corn earworm from HF and RS. Insects were collected from the ears of sweet corn (Zea mays). At the time of insect collection, we also collected soybean (Glycine max) foliage and maize silk and stored them at 4 °C. Fall armyworm from HF were used for culture-based sequencing of bacteria in regurgitant and bacterial community analysis of regurgitant and midguts. Corn earworm collected from HF and RS were used for bacterial community analysis of midguts.
Fall armyworm larvae were also collected from three sites in southern Puerto Rico in February 2017, which included two local farms (PRL and PRS) and the Juana Diez Experimental Research Station (PRJ) (Supplemental Table 3). Fall armyworm from the Puerto Rico sites were used for culture-based analysis of regurgitant. Upon removal from the field, larvae were placed in individual plastic cups with maize silk from the plant they were collected from for food, until sample collection. For larvae from Site HF and RS, regurgitant and midgut samples were collected the following day. For larvae collected in Puerto Rico, due to shipping time back to the United States, regurgitant was collected three days after larval collection.
A laboratory colony of fall armyworm was obtained from Benzon Research (Carlisle, PA), and maintained at Pennsylvania State University. To determine the contributions of host plant and egg source on gut bacterial composition in fall armyworm we used egg masses produced by 1) the Benzon laboratory colony and 2) field-collected fall armyworm. The field-collected eggs were produced by the moths that developed from caterpillars originally collected from Site HF on September 7, 2016. These caterpillars were final instars when collected and were fed exclusively on maize silk collected from the same site until they pupated, with the aim of maintaining their natural gut microbiota. Upon hatching, neonate larvae were placed in individual cups with either soybean leaves (FS Hisoy HS33A14-98SB132B) or maize silk (var. Providence), from plant material collected from Site RS. Larvae were fed ad libitum and leaves were replaced every 2–3 days until larvae reached the final instar. Regurgitant and midgut collections were conducted on the second day of the final instar.
Culture-based sequencing of regurgitant bacteria
Regurgitant was collected by gently squeezing larvae with soft forceps until they regurgitated. The regurgitation droplet was collected directly from the caterpillar’s oral cavity using a sterile pipette tip30. One µl of regurgitant was diluted with sterile Milli-Q water. Serial dilutions were plated on 2xYT media and the number of colony-forming units (CFUs) per ml of regurgitant was quantified. Colonies that displayed unique morphology were subcultured and these pure cultures were stored as glycerol stocks at −80 °C. Pure cultures were prepared from glycerol stocks on solid media for 48 h. DNA was extracted using the CTAB protocol63 and PCR was performed on extracted DNA using the 16S primers 27F (5′ AGAGTTTGATYMTGGCTCAG 3′) and 1392R (5′ ACGGGCGGTGTGTRC 3′) with GoTaq® Green Master Mix (Promega, Madison, WI, USA). The PCR conditions were as follows: initial denaturation at 95 °C for 2 min; followed by 30 cycles of 95 °C for 1 min, 50 °C for 45 s, 72 °C for 1 min; then 72 °C for 5 min. PCR products were purified using Exo-SAP-it (Affymetrix. Santa Clara, CA) following manufacturer’s instructions. Sanger Sequencing of purified DNA products was performed at the Penn State Genomics Core Facility, University Park, PA.
Contigs for bacterial sequences were assembled and trimmed using SeqMan Pro (DNASTAR, Madison, WI). Consensus sequences were used to search the Ribosomal Database Project database for bacterial type strains with similar (0.80–1.00 match) sequences64,65. The Muscle algorithm within MEGA7 software66 was used to align the bacterial type strains and samples. The aligned sequences were then used to construct an unrooted phylogenetic tree using the Maximum Likelihood method based on the Jukes-Cantor model67 in MEGA7.
Sample collection and DNA extraction
Regurgitant was collected from larvae as described above. Approximately 20 µl of regurgitant was collected from each larva and stored at −80 °C until needed. After regurgitant collection, caterpillars were starved for 2–3 h, surface sterilised in 10% Coverage Plus NPD (Steris, Mentor, OH, USA), and rinsed twice in sterile molecular grade water. Midguts were dissected under sterile conditions and stored at −80 °C until DNA extraction. DNA extractions were performed using the Quick-DNA™ Fecal/Soil Microbe Microprep Kit (Zymo Research, Irvine, CA, USA) according to manufacturer’s instructions.
Generation and sequencing of 16S amplicons
Primers used for bacterial V4 16S-rRNA amplification were 515F and 806R68. Amplicons were generated in 25 μL volumes using Phusion Hi-Fidelity Polymerase (New England BioLabs, Ipswich, MA, USA) containing 0.5 μM of forward and reverse primers and 25 ng of template DNA. Reaction conditions for 16S amplification were: 94 °C 3 min, 30 cycles of 94 °C for 45 sec, 50 °C for 60 sec, and 72 °C for 90 sec, followed by a final extension of 72 °C for 10 min. Indices and Illumina sequencing adapters were added to amplicon pools with 5 additional cycles of PCR. Barcoded products were pooled and sequenced on an Illumina MiSeq using 2 × 300 bp paired end reads. Generation of amplicon pools and sequencing of products was completed by the PSU Hershey Genomics Facility.
Processing of sequencing data
Bacterial amplicons were processed and analyzed using mothur v. 1.3769. The recommended workflow was modified such that the mothur command ‘pcr.seqs’ was implemented with pdiffs = 2 after the command ‘make.contigs’ to remove primer sequences from the reads. Bacterial operational taxonomic units (OTUs) were picked at 97% similarity and used for subsequent 3000 analyses. We used mothur to conduct OTU subsampling to OTUs. Taxonomies were determined using a mothur-formatted version of the Ribosomal Database Project (v. 9). Prior to subsequent analysis, samples were evaluated against a negative sequencing control, and two OTUs were removed from subsequent analyses.
Statistical analyses
Analyses of the bacterial communities were conducted using non-metric multidimensional scaling (nMDS) ordination and permutation-based multivariate analysis of variance (PerMANOVA). Heat maps were generated using the 20 most abundant OTUs by performing a log2 [x] transformation. We used Bray-Curtis similarities generated from standardised data that incorporated relative abundance of OTUs to assess community structure; Jaccard similarities were generated using subsampled data incorporating presence/absence to assess composition. PerMANOVA and nMDS was conducted in PRIMER-E (v. 7.0) using these metrics. PERMANOVA was also run in PRIMER-E using 999 iterations of the model. First, we assessed if there were differences between corn earworm and fall armyworm collected from the field using a one-way analysis. Then, we evaluated if insect egg source and plant sources have impacts on fall armyworm gut communities using a two-way analysis. Finally, we determined if there were differences between gut and regurgitant communities, and if there was an impact of host plant using a two-way analysis. Following this last test, we used similarity percentages (SIMPER) analysis to identify the OTUs that drive the differences, and then conducted non-parametric Wilcoxon rank sum analysis on the relative abundances.
Supplementary information
Acknowledgements
We appreciate the constructive comments from two anonymous reviewers. We thank Dr. Shelby Fleischer, Dr. Rob Meagher and Dr. Ermita Hernandez for field assistance in PR, Dan Harner of Harner Farm, Pine Grove Mills, PA for access to Site HF, and Penn State College of Agricultural Sciences Tag Along Fund for facilitating travel to PR. Funding was provided by USDA-NIFA Grant 2015-67013-2328 to KH, USDA-NIFA Grant 2017-67013-26596 to GWF and KH, USDA-NIFA PEN0476 and Accession #1004464 to KH and GWF, and USDA-NIFA Fellowship 2018-67012-27979 to CJM.
Author Contributions
A.G.J., C.J.M., G.W.F., and K.H. conceived of the overall question, A.G.J. and C.J.M. designed the experiments, A.G.J. collected samples, A.G.J. and C.J.M. performed experiments and statistical analyses, A.G.J., C.J.M., and K.H. interpreted the data, A.G.J. wrote the initial draft of the manuscript, with feedback from C.J.M., G.W.F., and, K.H. All authors reviewed the manuscript.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. Short read sequences have been submitted to NCBI SRA under accession number PRJNA507591. Sanger sequences of individual bacterial isolates have been submitted to NCBI Genbank Under Submission Number SUB4856027.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39163-9.
References
- 1.Douglas AE. Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 2015;60:17–34. doi: 10.1146/annurev-ento-010814-020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pontes MH, Dale C. Culture and manipulation of insect facultative symbionts. Trends Microbiol. 2006;14:406–412. doi: 10.1016/j.tim.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Salem H, Florez L, Gerardo N, Kaltenpoth M. An out-of-body experience: the extracellular dimension for the transmission of mutualistic bacteria in insects. Proc. R. Soc. B Biol. Sci. 2015;282:20142957. doi: 10.1098/rspb.2014.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mason, C. J., Jones, A. G. & Felton, G. W. Co-option of microbial associates by insects and their impact on plant–folivore interactions. Plant Cell Environ. 0 (2018). [DOI] [PubMed]
- 5.Kikuchi Y, Hosokawa T, Fukatsu T. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 2007;73:4308–4316. doi: 10.1128/AEM.00067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caspi-Fluger A, et al. Horizontal transmission of the insect symbiont Rickettsia is plant-mediated. Proc. R. Soc. B Biol. Sci. 2012;279:1791. doi: 10.1098/rspb.2011.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonella E, et al. Horizontal transmission of the symbiotic bacterium Asaia sp. in the leafhopper Scaphoideus titanus Ball (Hemiptera: Cicadellidae) BMC Microbiol. 2012;12:S4. doi: 10.1186/1471-2180-12-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dillon RJ, Dillon VM. The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 9.Kaufman MG, Klug MJ. The contribution of hindgut bacteria to dietary carbohydrate utilization by crickets (Orthoptera: Gryllidae) Comp. Biochem. Physiol. A Physiol. 1991;98:117–123. doi: 10.1016/0300-9629(91)90588-4. [DOI] [Google Scholar]
- 10.Salem H, et al. Drastic genome reduction in an herbivore’s pectinolytic symbiont. Cell. 2017;171:1520–1531.e13. doi: 10.1016/j.cell.2017.10.029. [DOI] [PubMed] [Google Scholar]
- 11.Salem H, et al. Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc. R. Soc. B Biol. Sci. 2014;281:20141838. doi: 10.1098/rspb.2014.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scully ED, et al. Functional genomics and microbiome profiling of the Asian longhorned beetle (Anoplophora glabripennis) reveal insights into the digestive physiology and nutritional ecology of wood feeding beetles. BMC Genomics. 2014;15:1096. doi: 10.1186/1471-2164-15-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, et al. Herbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome. Nat. Commun. 2018;9:964. doi: 10.1038/s41467-018-03357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason CJ, Couture JJ, Raffa KF. Plant-associated bacteria degrade defense chemicals and reduce their adverse effects on an insect defoliator. Oecologia. 2014;175:901–910. doi: 10.1007/s00442-014-2950-6. [DOI] [PubMed] [Google Scholar]
- 15.Ceja-Navarro JA, et al. Gut microbiota mediate caffeine detoxification in the primary insect pest of coffee. Nat. Commun. 2015;6:7618. doi: 10.1038/ncomms8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welte CU, et al. Plasmids from the gut microbiome of cabbage root fly larvae encode SaxA that catalyses the conversion of the plant toxin 2-phenylethyl isothiocyanate. Environ. Microbiol. 2016;18:1379–1390. doi: 10.1111/1462-2920.12997. [DOI] [PubMed] [Google Scholar]
- 17.Chung SH, et al. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. 2013;110:15728–15733. doi: 10.1073/pnas.1308867110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, et al. Herbivore oral secreted bacteria trigger distinct defense responses in preferred and non-preferred host plants. J. Chem. Ecol. 2016;42:463–474. doi: 10.1007/s10886-016-0712-0. [DOI] [PubMed] [Google Scholar]
- 19.Dow, J. A. Insect midgut function. in Advances in Insect Physiology (eds Evans, P. D. & Wigglesworth, V. B.) 19, 187–328 (Academic Press, 1987).
- 20.Harrison JF. Insect acid-base physiology. Annu. Rev. Entomol. 2001;46:221–250. doi: 10.1146/annurev.ento.46.1.221. [DOI] [PubMed] [Google Scholar]
- 21.Appel HM, Martin MM. Gut redox conditions in herbivorous lepidopteran larvae. J. Chem. Ecol. 1990;16:3277–3290. doi: 10.1007/BF00982098. [DOI] [PubMed] [Google Scholar]
- 22.Johnson KS, Felton GW. Physiological and dietary influences on midgut redox conditions in generalist lepidopteran larvae. J. Insect Physiol. 1996;42:191–198. doi: 10.1016/0022-1910(95)00096-8. [DOI] [Google Scholar]
- 23.Broderick NA, Raffa KF, Goodman RM, Handelsman J. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 2004;70:293–300. doi: 10.1128/AEM.70.1.293-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang X, et al. Complexity and variability of gut commensal microbiota in polyphagous lepidopteran larvae. Plos One. 2012;7:e36978. doi: 10.1371/journal.pone.0036978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen B, et al. Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 2016;6:29505. doi: 10.1038/srep29505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang H, et al. Microbial communities in the larval midgut of laboratory and field populations of cotton bollworm (Helicoverpa armigera) Can. J. Microbiol. 2006;52:1085–1092. doi: 10.1139/w06-064. [DOI] [PubMed] [Google Scholar]
- 27.Chaturvedi S, Rego A, Lucas LK, Gompert Z. Sources of variation in the gut microbial community of Lycaeides melissa caterpillars. Sci. Rep. 2017;7:11335. doi: 10.1038/s41598-017-11781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mason CJ, Raffa KF. Acquisition and structuring of midgut bacterial communities in gypsy moth (Lepidoptera: Erebidae) larvae. Environ. Entomol. 2014;43:595–604. doi: 10.1603/EN14031. [DOI] [PubMed] [Google Scholar]
- 29.Staudacher H, et al. Variability of bacterial communities in the moth Heliothis virescens indicates transient association with the host. Plos One. 2016;11:e0154514. doi: 10.1371/journal.pone.0154514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Acevedo, F. E. et al. Fall armyworm-associated gut bacteria modulate plant defense responses. Mol. Plant. Microbe Interact (2017). [DOI] [PubMed]
- 31.Stokstad E. New crop pest takes Africa at lightning speed. Science. 2017;356:473. doi: 10.1126/science.356.6337.473. [DOI] [PubMed] [Google Scholar]
- 32.Goergen G, Kumar PL, Sankung SB, Togola A, Tamò M. First report of outbreaks of the fall armyworm Spodoptera frugiperda (J E Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in west and central Africa. Plos One. 2016;11:e0165632. doi: 10.1371/journal.pone.0165632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagoshi RN, Meagher RL, Hay-Roe M. Inferring the annual migration patterns of fall armyworm (Lepidoptera: Noctuidae) in the United States from mitochondrial haplotypes. Ecol. Evol. 2012;2:1458–1467. doi: 10.1002/ece3.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva DMda, et al. Biology and nutrition of Spodoptera frugiperda (Lepidoptera: Noctuidae) fed on different food sources. Sci. Agric. 2017;74:18–31. doi: 10.1590/1678-992x-2015-0160. [DOI] [Google Scholar]
- 35.Bentivenha JPF, et al. Battle in the New World: Helicoverpa armigera versus Helicoverpa zea (Lepidoptera: Noctuidae) Plos One. 2016;11:e0167182. doi: 10.1371/journal.pone.0167182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reisig D, Suits R, Burrack H, Bacheler J, Dunphy JE. Does florivory by Helicoverpa zea (Lepidoptera: Noctuidae) cause yield loss in soybeans? J. Econ. Entomol. 2017;110:464–470. doi: 10.1093/jee/tow312. [DOI] [PubMed] [Google Scholar]
- 37.Priya NG, Ojha A, Kajla MK, Raj A, Rajagopal R. Host plant induced variation in gut bacteria of Helicoverpa armigera. PLoS One. 2012;7:e30768. doi: 10.1371/journal.pone.0030768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammer TJ, Janzen DH, Hallwachs W, Jaffe SL, Fierer N. Caterpillars lack a resident gut microbiome. Proc. Natl. Acad. Sci. USA. 2017;114:9641–9646. doi: 10.1073/pnas.1707186114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Appel, H. M. The chewing herbivore gut lumen: physicochemical conditions and their impact on plant nutrients, allelochemicals and insect pathogens. In Insect-Plant Interactions (ed. Bernays, E. A.) 5, 209–221 (CRC Press, 1994).
- 40.Visôtto LE, Oliveira MGA, Guedes RNC, Ribon AOB, Good-God PIV. Contribution of gut bacteria to digestion and development of the velvetbean caterpillar, Anticarsia gemmatalis. J. Insect Physiol. 2009;55:185–191. doi: 10.1016/j.jinsphys.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 41.Douglas, A. E. Microbial brokers of insect-plant interactions. in Proceedings of the 8th International Symposium on Insect-Plant Relationships (eds. Menken, S. B. J., Visser, J. H. & Harrewijn, P.) 329–336 (Springer Netherlands, 1992).
- 42.Caccia S, et al. Midgut microbiota and host immunocompetence underlie Bacillus thuringiensis killing mechanism. Proc. Natl. Acad. Sci. 2016;113:9486. doi: 10.1073/pnas.1521741113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broderick NA, Raffa KF, Handelsman J. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc. Natl. Acad. Sci. 2006;103:15196. doi: 10.1073/pnas.0604865103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakubowska AK, Vogel H, Herrero S. Increase in gut microbiota after immune suppression in baculovirus-infected larvae. Plos Pathog. 2013;9:e1003379. doi: 10.1371/journal.ppat.1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikaelyan A, Thompson CL, Hofer MJ, Brune A. Deterministic assembly of complex bacterial communities in guts of germ-free cockroaches. Appl. Environ. Microbiol. 2016;82:1256–1263. doi: 10.1128/AEM.03700-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byeon JH, et al. A specific cathepsin-L-like protease purified from an insect midgut shows antibacterial activity against gut symbiotic bacteria. Dev. Comp. Immunol. 2015;53:79–84. doi: 10.1016/j.dci.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Futahashi R, et al. Gene expression in gut symbiotic organ of stinkbug affected by extracellular bacterial symbiont. Plos One. 2013;8:e64557. doi: 10.1371/journal.pone.0064557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohbayashi T, et al. Insect’s intestinal organ for symbiont sorting. Proc. Natl. Acad. Sci. USA. 2015;112:E5179–E5188. doi: 10.1073/pnas.1511454112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Näpflin K, Schmid-Hempel P. Host effects on microbiota community assembly. J. Anim. Ecol. 2017;87:331–340. doi: 10.1111/1365-2656.12768. [DOI] [PubMed] [Google Scholar]
- 50.Chung SH, et al. Host plant species determines symbiotic bacterial community mediating suppression of plant defenses. Sci. Rep. 2017;7:39690. doi: 10.1038/srep39690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindow SE, Brandl MT. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003;69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mason CJ, Rubert-Nason KF, Lindroth RL, Raffa KF. Aspen defense chemicals influence midgut bacterial community composition of gypsy moth. J. Chem. Ecol. 2015;41:75–84. doi: 10.1007/s10886-014-0530-1. [DOI] [PubMed] [Google Scholar]
- 53.Knief C, Ramette A, Frances L, Alonso-Blanco C, Vorholt JA. Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. Isme J. 2010;4:719. doi: 10.1038/ismej.2010.9. [DOI] [PubMed] [Google Scholar]
- 54.Rastogi G, et al. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012;6:1812–1822. doi: 10.1038/ismej.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Appel HM, Maines LW. The influence of host plant on gut conditions of gypsy moth (Lymantria dispar) caterpillars. J. Insect Physiol. 1995;41:241–246. doi: 10.1016/0022-1910(94)00106-Q. [DOI] [Google Scholar]
- 56.Dow JA. pH gradients in the lepidopteran midgut. J. Exp. Biol. 1992;172:355. doi: 10.1242/jeb.172.1.355. [DOI] [PubMed] [Google Scholar]
- 57.Teh B-S, Apel J, Shao Y, Boland W. Colonization of the intestinal tract of the polyphagous pest Spodoptera littoralis with the GFP-tagged indigenous gut bacterium Enterococcus mundtii. Front. Microbiol. 2016;7:928. doi: 10.3389/fmicb.2016.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gedling CR, Smith CM, LeMoine CMR, Cassone BJ. The Mexican bean beetle (Epilachna varivestis) regurgitome and insights into beetle-borne virus specificity. Plos One. 2018;13:e0192003. doi: 10.1371/journal.pone.0192003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engel P, Moran NA. The gut microbiota of insects – diversity in structure and function. FEMS Microbiol. Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 60.Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. Plos One. 2013;8:e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods. 2007;69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jovel J, et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 2016;7:459. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen WP, Kuo TT. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 1993;21:2260. doi: 10.1093/nar/21.9.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cole, R. J. et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. D633–D642, 10.1093/nar/gkt1244 (2014). [DOI] [PMC free article] [PubMed]
- 66.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jukes, T. H. & Cantor, C. R. Evolution of protein molecules. In Mammalian Protein Metabolism 21–132 (Academic Press, 1969).
- 68.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. Short read sequences have been submitted to NCBI SRA under accession number PRJNA507591. Sanger sequences of individual bacterial isolates have been submitted to NCBI Genbank Under Submission Number SUB4856027.