Abstract
T-cell receptor–engineered T-cell therapy and chimeric antigen receptor T-cell therapy are 2 types of adoptive T-cell therapy that genetically modify natural T cells to treat cancers. Although chimeric antigen receptor T-cell therapy has yielded remarkable efficacy for hematological malignancies of the B-cell lineages, most solid tumors fail to respond significantly to chimeric antigen receptor T cells. T-cell receptor–engineered T-cell therapy, on the other hand, has shown unprecedented promise in treating solid tumors and has attracted growing interest. In order to create an unbiased, comprehensive, and scientific report for this fast-moving field, we carefully analyzed all 84 clinical trials using T-cell receptor–engineered T-cell therapy and downloaded from ClinicalTrials.gov updated by June 11, 2018. Informative features and trends were observed in these clinical trials. The number of trials initiated each year is increasing as expected, but an interesting pattern is observed. NY-ESO-1, as the most targeted antigen type, is the target of 31 clinical trials; melanoma is the most targeted cancer type and is the target of 33 clinical trials. Novel antigens and underrepresented cancers remain to be targeted in future studies and clinical trials. Unlike chimeric antigen receptor T-cell therapy, only about 16% of the 84 clinical trials target against hematological malignancies, consistent with T-cell receptor–engineered T-cell therapy’s high potential for solid tumors. Six pharma/biotech companies with novel T-cell receptor–engineered T-cell ideas and products were examined in this review. Multiple approaches have been utilized in these companies to increase the T-cell receptor’s affinity and efficiency and to minimize cross-reactivity. The major challenges in the development of the T-cell receptor–engineered T-cell therapy due to tumor microenvironment were also discussed here.
Keywords: adoptive T-cell therapy, TCR-T, tumor immunotherapy, tumor antigen, clinical trial
Introduction
Adoptive T-cell therapy is one potentially powerful treatment for cancer that genetically modifies natural T cells to make them tumor-specific and to improve their ability to destroy tumor cells.1 The genetically modified T cells are able to express chimeric antigen receptors (CARs) or T-cell receptors (TCRs), showing impressive results in multiple clinical trials.1 Chimeric antigen receptor generally consists of 3 parts: an ectodomain containing a single-chain fragment variant (scFv) for recognition of specific antigen, a transmembrane domain, and an endodomain including a CD3ζ chain with 3 immunoreceptor tyrosine-based activation motifs (ITAMs).2 Chimeric antigen receptor-T-cell therapy has shown excellent results against hematological malignancies, while its effect against solid tumors is unsatisfactory by comparison.3,4 This review focuses on TCR-engineered T (TCR-T) cells, which have shown greater promises against solid tumor than CAR-T cells.5 This review elucidates the basic mechanism of TCR-T-cell therapy and different types of tumor antigen targeted by TCRs, followed by a summary of information about TCR-T-cell therapy clinical trials gathered on ClinicalTrials.gov and about technologies developed and trials run by several major pharma/biotech companies. Finally, this review ends with a summary of major challenges and improvements made to this cancer therapy.
The potency of TCRs relies on their interaction with peptide-major histocompatibility complex (pMHC), complexes formed by peptide bound to MHC.6 Intracellular antigens are cut up into peptide chains and displayed by MHC molecules to form pMHCs.7 Cytoplasmic proteins to be expressed by class I MHC proteins, most of which are defective ribosomal translation products, are cleaved into peptide chains by proteolysis.8 These peptides are then bound to class I MHC proteins, which are expressed on all nucleated cells’ cell surface.7 Some cells, called antigen-presenting cells (APCs), express class II MHC proteins.9 They internalize foreign material proteins by endocytosis and cleave them into peptide chains to bind with class II MHC proteins.10 T-cell receptors from T cells, which must be matched to human leukocyte antigen (HLA) alleles of patients,5 recognize these pMHCs and cause the killing of cancer cells.11 (Human class I MHC protein is expressed from 3 gene regions: HLA-A, HLA-B, and HLA-C, and human class II MHC protein is also expressed from 3 gene regions: HLA-DR, HLA-DP, and HLA-DQ.12)
T-cell receptor is a heterodimer composed of 2 different transmembrane polypeptide chains: an α chain and a β chain, each consisting of a constant region, which anchors the chain inside the T-cell surface membrane, and a variable region, which recognizes and binds to the antigen presented by MHCs.11 The TCR complex is associated with 6 polypeptides forming 2 heterodimers, CD3γε and CD3δε, and 1 homodimer CD3 ζ, which together forms the CD3 complex.13 In total, the CD3 complex contains 10 ITAMs, which take part in T-cell activation.14 The different structures of CAR-T cells and TCR-T cells are illustrated in Figure 1. Additional costimulatory signals are also essential to the full execution of T-cell function, including CD8 on the surface of cytotoxic T cells, which binds to class I MHC complex, and CD4 on the surface of helper T cells, which binds to class II MHC complex.12 There are also other well-studied costimulatory molecules including CD28 involved in CD28: B7 engagement on APCs and 4-1BB (CD137) that upregulate antiapoptotic factors to promote T-cell survival when binding with ligands on the surface of APCs.14 Co-inhibitory molecules, such as cytotoxic T-lymphocyte antigen 4 (CTLA-4) and programmed cell death protein 1 (PD-1), are also part of the T-cell system in charge of extinguishing T-cell signaling.15
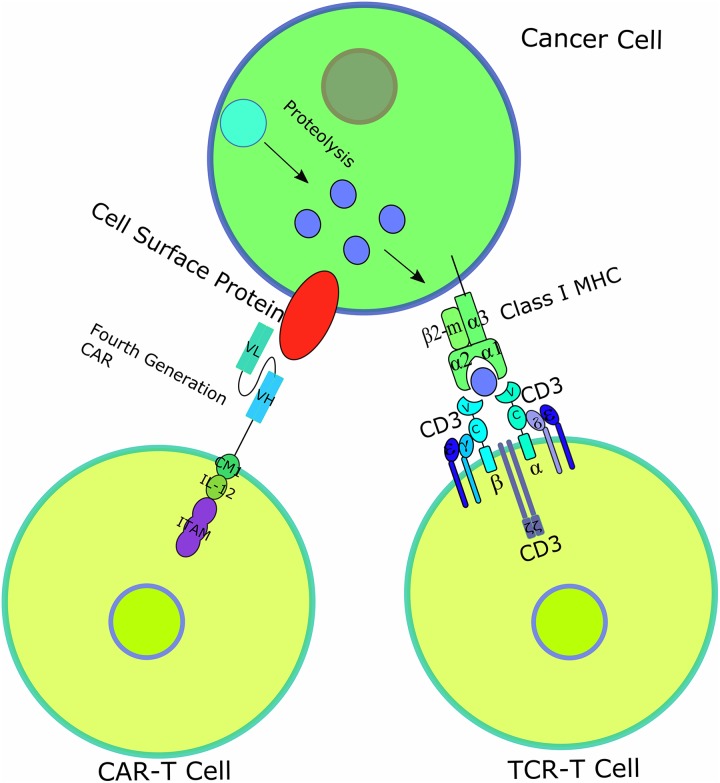
Figure 1.
Diagrams of CAR-T and TCR-T-cell therapy. CAR-T cell, which normally targets only cell surface proteins, has had 4 generations and the latest generation is shown here. All generations of CAR-T cell contain an scFv consisting of an antibody’s variable light (VL) and variable heavy (VH) chain, a transmembrane domain and a CD3ζ chain with ITAM for T-cell activation. The fourth-generation CAR further includes a costimulatory molecule (CM1), such as CD28 and B7, and an interleukin-12 (IL-12) domain for enhanced T-cell effects. T-cell receptor-engineered T cell has 1 TCR consisting of an α chain and a β chain, each containing a variable domain (v) and a constant domain (c), as well as an unlabeled transmembrane domain and 6 CD3 chains for T-cell activation. Class I MHC protein consists of an α chain with 3 domains α1, α2, α3, and a β2-microglobulin, presenting peptides derived from proteolysis of intracellular proteins. CAR indicates chimeric antigen receptor; ITAM, immunoreceptor tyrosine-based activation motif; MHC, major histocompatibility complex; scFv, single-chain fragment variant; TCR, T-cell receptor.
Chimeric antigen receptors, on the other hand, employ an antibody–antigen recognition machinery that consists of a scFv derived from an antibody in order to bind to antigens on the target cell’s surface, which, along with transmembrane domains and costimulatory domains, activates immune responses.16 Most proteins, however, are expressed inside cells instead of on the cell surface (only about 28% is expressed on the cell surface), making them unavailable to act as antigen for CARs.17 As a result, the variety of antigens that can be targeted by CARs is often limited; moreover, the densities of cell surface antigens vary from cell to cell.18 Traditionally, it is generally considered that CARs have higher affinity than TCRs. According to a recent study, however, by comparing between the affinity of a single-chain TCR (Vβ-linker-Vα), an analog of scFv that serves as a CAR-like receptor, and that of a CAR with the same K D, it has been shown that the full-length TCR has greater sensitivity than CAR even when CARs are expressed at higher densities and without the presence of CD8 coreceptor (which can lower the TCR affinity required by roughly 100 times and reduce the amount of pMHC required per target cell from over 30 molecules to just 1 molecule).19 This higher sensitivity can enable more rapid destruction of tumor cells but also increases the risk of “on-target, off-tumor” toxicity, as observed in multiple clinical trials.20-23 Interestingly, despite the higher sensitivity of TCRs than CARs, TCRs are found to mediate release of less amount of cytokines.24,25 Thus, the risk of cytokine release syndrome is potentially lower with TCR-T-cell therapy compared to CAR-T-cell therapy. Although CAR-T cells have shown promising results against hematological cancers, their efficacy for solid tumor treatments is less so, which might be due to the immunosuppressive microenvironment of solid tumors, which will be discussed in the later sections, as well as the availability of cancer antigens that are present at sufficient level of density to be targeted without “on-target, off-tumor” toxicity.18,26 T-cell receptor-engineered T cells, on the other hand, have shown some successes in treating both solid tumors, such as metastatic melanoma, and hematological cancers,18,27 possibly due to the fact that the latter is conferred with higher penetrating power due to the low number of copies present on the surface of tumors, which is contrasted by the large number of copies of CAR-T antigens.5
T-cell receptor-engineered T-cell therapy utilizes the modification of T cells that retain these complexes to specifically target the antigens expressed by particular tumor cells.
Types of Antigens Used in TCR-T-Cell Therapy
In practice, there are different ways of categorizing tumor antigens targeted in TCR-T-cell therapy. Generally, human tumor antigens are classified into 2 main types: shared tumor-associated antigens (shared TAAs) and unique tumor-associated antigens (unique TAAs), or tumor-specific antigens. The former includes cancer-testis (CT) antigens, overexpressed antigens, and differentiation antigens, while the latter includes neoantigens and oncoviral antigens.1,8,28 Human papillomavirus (HPV) E6 protein and HPV E7 protein belong to the category of oncoviral antigens. The diversity and complexity of tumor antigens often give rise to “on-target, off-tumor” toxicity.1 Each type of antigen has its own characteristic advantages for TCR targeting but also its particular disadvantages.
Cancer-testis antigens are expressed in various tumor types as well as in testis, during times of spermatogenesis, and placenta.8 Normally they are silent in adult tissues,29 but they are active for transcription in different tumor types.30 In theory, because many tumor types express CT antigens at a high level and normal tissues rarely express them, they are deemed attractive and safe immunotherapy targets,5 but in practice, this is not always the case. Melanoma-associated antigen (MAGE)-A gene family is a group of CT antigens: MAGE-A1, MAGE-A2, MAGE-A3, MAGE-A4, MAGE-A6, MAGE-A10, and MAGE-A12 are expressed at a frequency of more than 1/10 000.22 In an NCI MAGE-A3 trial, TCR targeted at MAGE-A3 unexpectedly cross-reacted with the related peptide, MAGE-A12, which is expressed in the brain, resulting in the death of 2 of the first 9 patients and severe mental damage of the third.23 It was found that the human brain might also express MAGE-A1, MAGE-A8, and MAGE-A9.23 In another trial conducted by Adaptimmune, the TCR targeting MAGE-A3 bound to an unrelated protein, titin, in the heart.5 These interactions between TCRs and normal tissues show the potential danger of targeting CT antigens.
Differentiation antigens are encoded by genes that express in a tissue-specific manner.28 As a result, they are shared between tumor cells and healthy cells of corresponding tissues of origin,8 unlike overexpressed antigens, which are expressed in certain, if not all, healthy tissue types.28 Most such antigens, including glycoprotein (gp100), melanoma antigen recognized by T cells (MART-1), and tyrosinase, are mainly found in melanomas and normal melanocytes.31-36 However, differentiation antigens do include carcinoembryonic antigen, which is often highly expressed in colon cancer,37 and others are found in some epithelial tissues and tumors, including prostate cancers, in which case prostate-specific antigen is identified as one type of differentiation antigen.38,39 Targeting such differentiation antigens will be likely to induce “on-target, off-tumor toxicity” on normal cells in critical organs.40 The first 2 trials of TCR-T-cell therapy against MART-1 protein used 2 different proteins that were class I MHC-restricted but that were targeting similar epitopes, DMF4 and DMF5. Although DMF5-engineered T cells proved to be more avid than DMF4-engineered ones in functional assays, expanded trials with DMF5 T cells reported autoimmune toxicity on the eye, ear, and skin not reported with DMF4 T cells.21 In one trial, such toxicity consisted of erythematous skin rash observed in 14 of 20 patients, anterior uveitis observed in 11 of 20 patients, and hearing loss observed in 10 of 20 patients.41 Lethal cardiac toxicity was also observed in 3 patients undergoing trials against MART-1.42 On one hand, to effectively eliminate solid tumors, TCR should be extremely potent in order to make any progress.5 On the other hand, when a therapeutic is highly effective, it would have the same effect upon tissues with low expression level as well as upon those with high expression level.5 Thus, the potency of TCR should be controlled in a precise range to not induce immune response up to a certain level but be potent enough to kill tumor cells after the threshold has been reached to reduce “on-target, off-tumor” toxicity.
Overexpressed antigens are antigens that are expressed in many normal tissues as well as different types of tumor.8 They are expressed at a higher level in tumor cells to reach the threshold of T-cell recognition.8 Because such antigens are also expressed, although, at a lower level, there are risks of “on-target, off-tumor” toxicity in normal tissues. For example, Wilms tumor 1 (WT1) is one kind of overexpressed antigen that is highly expressed in most acute myeloid leukemia (AML), acute lymphoid leukemia, and almost every type of solid tumor.43 It is also present in several critical tissues, such as heart tissues.44 Mesothelin, another kind of overexpressed antigen, is highly expressed in mesothelioma but is also present on mesothelial cells of several tissues, including trachea.45-47 Despite common apprehension of the “on-target, off-tumor” toxicity by targeting such antigens, reports have suggested that WT1-targeting T cells are able to differentiate between tumor cells and normal tissues.48 Using murine models, it was also suggested that enhanced-affinity TCRs targeting WT1 antigens, though surpassing the threshold for thymic selection, showed no autoimmune toxicity when transferred into wild-type mice.48 It thus could be deducted that WT1, and other overexpressed antigens, might have the ability to attack tumor cells but largely spare normal cells, making them ideal targets for TCR-T therapy.5
Neoantigens are formed by random somatic mutations specific to individual tumors and also vary when tumor cells are isolated from patients at different sites or different times.49 Targeting such antigens thus can reduce the risk of “on-target, off-tumor” toxicity but are expensive in practice. This is because the identification of neoantigens sometimes requires the sequencing of each individual tumor’s whole genome so as to identify mutated genes and to choose peptides comprising motifs predicted to be presented by HLA alleles of the patient.8 Moreover, several neoantigens have to be sequenced and targeted for the same type of tumor due to tumor heterogeneity.5,50-54 Tumor heterogeneity is divided into certain types, including interpatient tumor heterogeneity, intratumor heterogeneity, and intermetastatic heterogeneity.55 It results in the target for immunotherapies being specific to only one portion of all tumor cells, which reduces the efficacy of such therapies as well as increases the risk of metastasis.56 The nature of tumor heterogeneity means that targeting neoantigen is necessary if complete eradication of tumor is desired.
Because of the different characteristics of each individual type of tumor antigen, products from different pharma/biotech companies and research institutes have different choices regarding which category to focus on.
Statistics for Clinical Trials of TCR-T-Cell Therapy
Among the 376 clinical trials containing the keyword “TCR” published on ClinicalTrials.gov updated by June 11, 2018, 84 of these trials have been identified to employ TCR-T-cell therapy technique by manual curation. Certain patterns can be revealed from these 84 clinical trials, which are presented in Figures 2 and 3. Since June 11, when the data were gathered, to July 28, 13 more trials were included in ClinicalTrials.gov, where only 1 was related to TCR-T-cell therapy (with trial number NCT03578406) and was targeting HPV E6 antigen. The information on this new trial was not included in the figures and supplementary table.
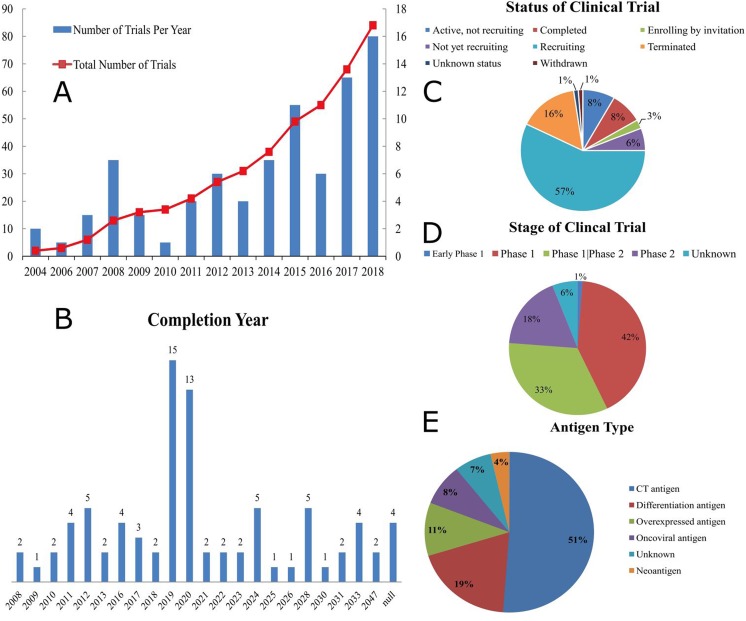
Figure 2.
Major information and trend of 84 TCR-T clinical trials. (A) Figure 2A is the number of clinical trials started each year (bar chart, based on the right-hand side y-coordinate) and the number of clinical trials that have been completed since 2004 (line graph, based on the left-hand side y-coordinate). (B) Figure 2B is the number of clinical trials that was completed or will be completed each year. (C) Figure 2C is the respective proportions of trials not yet recruiting, active but not recruiting, recruiting, enrolling by invitation, withdrawn, terminated, completed and of unknown status. (D) Figure 2D is the respective proportions of different clinical stages in the 84 trials. (E) Figure 2E is the proportion of each type of antigen in the 84 trials.
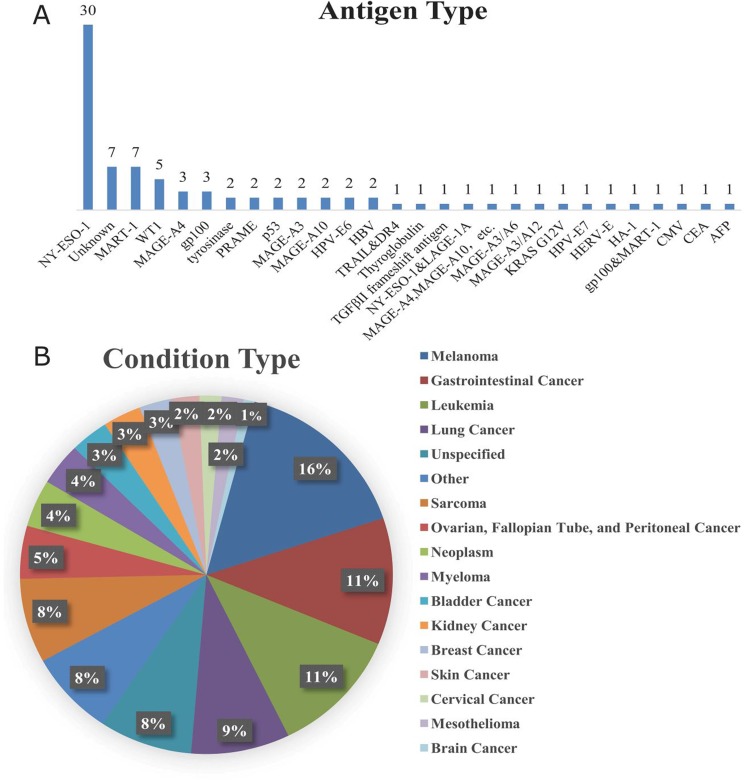
Figure 3.
Proportion of each type of antigen and cancer targeted by 84 TCR-T clinical trials. (A) Figure 3A is the proportion of each particular antigen targeted in the 84 trials. (B) A total of 210 cancers are targeted in the 84 trials collected (many trials target more than one cancer, and many types of cancer are targeted by more than one trial). Figure 3B is the proportion of each type of cancer targeted in the 84 trials.
First, as shown in Figure 2A, the start years of these clinical trials have shown a peculiar pattern. Three earliest clinical trials for TCR-T-cell therapy began in 2004. Since then, starting in 2006, the number of clinical trials started each year has shown a periodic pattern of fluctuation for every 3 or 4 years. Take the 8 years from 2006 to 2013 as an example, from 2006 to 2008, the number of clinical trials showed a steady increase and reached a peak in 2008, but the number then experienced a decrease in 2009, followed by a steady increase from 2010 to 2012 and then a decrease in 2013, and so on, but demonstrating an overall trend of increase in number of clinical trials from 2004 to 2018. This may be due to the fact that funds are periodically injected in this field, and it may require a period of 3 or 4 years to reach another peak in the number of clinical trials, whereas the demand and interest for TCR-T-cell therapy have increased in the long run due to its huge potential. As shown in Figure 2B, 28 of these 84 clinical trials are expected to be completed in 2019 and 2020. It will be the best time to evaluate the results of the TCR-T application over other immunotherapy approaches.
Second, as presented by Figure 2C, about 57% of these clinical trials (48 clinical trials) are still recruiting, whereas only less than one-quarter of these clinical trials (20 clinical trials) are either terminated or completed. This proves that TCR-T-cell therapy is still a relatively new target for research, compared to CAR-T-cell therapy, indicating that TCR-T-cell therapy may have huge potential in the future.5
Third, according to Figure 2D, about 42% of these trials (35 trials) are for phase 1 clinical trials, whereas approximately 33% of these trials (28 trials) are for both phase 1 and phase 2 clinical trials, again showing that most of the TCR-T clinical trials are at the start of development, and the application of TCR-T-cell therapy in industries or hospitals is still a long way to go.
Fourth, as illustrated by Figures 2E and 3A, more than half of the antigens targeted by the 84 clinical trials (43 trials) are CT antigens, and 31 of these 43 trials target NY-ESO-1 (30 trials target NY-ESO-1, 1 trial targets NY-ESO-1&LAGE-1A), probably because the NY-ESO-1-specific TCR-T cells have been most thoroughly examined and tested in terms of therapeutic potentials in synovial cell sarcoma, melanoma, and myeloma.40 Only 3 trials are targeting neoantigens, probably due to the fact that it requires a great deal of time and costs to perform the sequencing as well as to identify the effective TCRs for each patient and the same process needs to be repeated for each individual patient. On the other hand, if the method of sequencing and good manufacturing process can be made more efficient and cost-effective, such as applying robotics system to make sequencing faster and using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system or sleeping beauty transposons,5,57 neoantigens can be of huge potential without much competition for TCR-T industries.
Finally, as indicated by Figure 3B, the most frequently targeted cancer type is melanoma, being the target of 33 trials. This is probably because that the frequency of melanoma cases is high. Melanoma accounts for almost 92% of new cases of skin cancer and about 69% of skin cancer-related death in the United States in 2018,58 and the incidence of melanoma has been increasing at a faster rate than the majority of other cancers.59 Another reason might be that melanoma lesions are relatively accessible and that the nature of melanoma to easily adapt to tissue culture means that tissue samples and cell lines can be readily available for research purposes, making melanoma being preferred by clinical trials.59 Leukemia and gastrointestinal cancers fall as the second, being the target of 24 trials. Of the 210 cancers mentioned in 84 clinical trials, only 34 trials (approximately 16%) focuses on hematological malignancies only (leukemia: 24 trials; myeloma: 8 trials; lymphoma: 1 trial; [unspecified] hematological malignancies: 1 trial), so 84% of cancer types targeted by these 84 clinical trials belong to the category of solid tumor, possibly due to the fact that CAR-T has already well proved efficiency in treating hematological malignancies, whereas, as proven by multiple studies, its effects toward solid tumor has been proven unsuccessful as compared to TCR-T-cell therapy. Nevertheless, there have indeed been attempts to improve CAR-T cells’ performance against solid tumor because of such unsatisfactory results.60-65 Therefore, it is possible that CAR-T cells might be widely available for solid tumor treatment in the future.66,67
Major Pharma/Biotech Companies Developing TCR-T-Cell Therapy
Table 1 has presented clinical trials of several major pharma/biotech companies discussed later in this section.
Table 1.
Clinical Trials by Major Pharma/Biotech Companies.
| Sponsors/Collaborators | NCT Trial Number | Target Antigen | Agent | Targeted Tumor Type | Stage |
|---|---|---|---|---|---|
| Medigene | NCT03503968 | PRAME | MDG1011 | Unknown | Phase I/II |
| Bellicum Pharmaceuticals | NCT02743611 | PRAME | BPX-701 | AML, myelodysplastic syndrome | Phase I |
| Kite Pharma | NCT03139370 | MAGE-A3/A6 | KITE-718 | Unknown | Phase I |
| Immatics | NCT03247309 | Unknown | IMA-201 | Head and neck squamous cell carcinoma, squamous cell NSCLC | Phase I |
| Adaptimmune | NCT02592577 | MAGE-A10 | MAGE-A10c796T | NSCLC | Phase I |
| Adaptimmune | NCT01343043 | NY-ESO-1 | NY-ESO-1c259T | Synovial sarcoma | Phase I/II |
| Adaptimmune | NCT01892293 | NY-ESO-1 | NY-ESO-1c259T | Multiple myeloma | Phase I/II |
| Adaptimmune | NCT01352286 | NY-ESO-1 | NY-ESO-1c259T | Multiple myeloma | Phase I/II |
| Adaptimmune | NCT01567891 | NY-ESO-1 | NY-ESO-1c259T | Ovarian cancer | Phase I/II |
| Adaptimmune | NCT01350401 | NY-ESO-1 | NY-ESO-1c259T | Melanoma | Phase I/II |
| Adaptimmune | NCT02588612 | NY-ESO-1 | NY-ESO-1c259T | NSCLC | Phase I/II |
| Adaptimmune | NCT03132792 | AFP | AFPc332T | Hepatocellular cancer | Phase I |
| Adaptimmune | NCT03168438 | NY-ESO-1 | NY-ESO-1c259T | Refractory multiple myeloma | Phase I |
| Adaptimmune | NCT02989064 | MAGE-A10 | MAGE-A10c796T | Urinary bladder cancer, head and neck cancer, melanoma | Phase I |
Abbreviations: AFP, α-fetoprotein; AML, acute myeloid leukemia; MAGE, melanoma-associated antigen; NSCLC, non-small cell lung carcinoma; PRAME, preferentially expressed antigen in melanoma.
Adaptimmune (Oxfordshire, United Kingdom) modifies a patient’s own CD4+ and CD8+ cells’ TCRs to express specific peptide enhanced affinity receptors (SPEARs) to increase the binding affinity of natural TCRs, overcoming their low affinities as a result of negative thymic selection during maturation of T cells in the thymus.68-70 Specific peptide enhanced affinity receptors are made by modifying the hypervariable complementarity-determining regions of native TCRs and selecting for TCRs with high affinity and lack of alloreactivity.71-73 The main targets for Adaptimmune’s SPEARs are CT antigens, including MAGE-A4, MAGE-A10, and NY-ESO-1.74 It is also collaborating with GlaxoSmithKline (Brentford, United Kingdom) to target preferentially expressed antigen in melanoma (PRAME), another CT antigen.75 These antigens are either not processed into peptides or processed minimally on normal adult tissues.5 Adaptimmune is also developing SPEAR for α-fetoprotein (AFP), a glycoprotein highly expressed in fetal serum but is of low content in adult blood, mainly due to mature hepatocytes’ inability to produce AFP.74,76 α-fetoprotein is expressed in high levels in hepatocellular carcinoma, which Adaptimmune is trying to cure with AFP TCRs. Targeting these antigens should reduce the risk of “on-target, off-tumor” toxicity. In addition, it has developed a proprietary preclinical screening program consisting of molecular analysis to systematically identify peptides similar to the target cancer peptide inside human bodies to eliminate further cross-reactivity. It has developed human cell testing to determine whether the SPEARs bind to samples of normal cells and whether they are effective in whole blood samples as well.70
Kite Pharma is targeting MAGE-A3/MAGE-A6, using KITE-718 (NCT trial number: NCT03139370), as well as neoantigens.5,77 It has strengthened its TCR product platform in 2015 through its acquisition of T-cell Factory B.V. (TCF), a privately held Dutch company, and renamed it as Kite Pharma EU, thus acquiring its proprietary TCR-GENErator discovery platform in order for rapid, high-throughput identification of TCRs, including those that target neoantigens.78 It mainly incorporates T cells that induce apoptosis of cancer cells when the engineered TCRs bind to tumor antigens.79
Medigene employs viral vector–mediated transfer to infuse genes that code for specific TCRs into both CD4+ and CD8+ T cells.80 Its TCR-T platform delivers TCR-T cells recognizing various tumor antigens, including common antigens shared by tumors and neoantigens specific to each individual patient.80 Since March 2018, the company has been engaging in its first clinical trial study: a phase I/II clinical trial with MDG1011, its TCR-T therapy targeting PRAME, for the treatment of various types of hematological malignancies, including multiple myeloma (MM), AML, and myelodysplastic syndrome80 (NCT trial number: NCT03503968). Meanwhile, it is collaborating with Charité Hospital and the Max-Delbrück-Center for an investigator-initiated TCR study that equips patients suffering from relapsed or refractory MM with TCR-T cells recognizing MAGE-A1 antigen in Germany.80
TCR2 Therapeutics has a unique TRuC (which stands for “T-cell receptor fusion construct”, Cambridge, Massachusetts) platform for recruiting TCRs without the need of HLA matching.81 This platform conjugates tumor antigen binder, such as scFv, single-domain antibody, and antigen-binding fragment, to the CD3 γ chain of TCR.81 This construct enables engineered TCRs to target surface antigens, which do not require HLA matching, overcoming the major limitation of TCR-T-cell therapies, but retains the complete TCR system consisting of TCR and CD3 complex.81 Another advantage of the TRuC platform is that it is flexible and can take variable forms, one of which is the dual-target TruC platform, where 2 tumor antigen binders are conjugated to a single TRuC-T cell product to overcome antigen escape mechanisms common in some tumor types.81 Finally, each TRuC-T cell product can be engineered to carry built-in accessories or modulators including T-cell enhancers, up/down control genes, and stroma remodelers, to activate T cells or inactivate tumor cells81 (tissue-resident stromal cells are known to be actively participating in tumor onset and/or evolution82). This innovation, therefore, provides a more intricate machinery for T cells to more effectively kill tumor cells but at the same time evade “on-target, off-tumor” toxicity in theory, while its practical application still requires clinical trials to assess.
Immatics company has developed its ACTengine approach that genetically engineers a patient’s autologous T cells to express exogenous TCR upon lentiviral transduction.83 Highly specific exogenous TCRs with optimal affinity are identified from natural, human T-cell repertoire through proprietary high-throughput screening (HTS), similar to what Kite Pharma and Medigene are doing.83 By this approach, it has developed 3 products: IMA-201, IMA-202, which have entered the clinical phase I/II stage, and IMA-203, in collaboration with MD Anderson Cancer Center.84 It has also invented bispecific TCR molecules that could be easily synthesized in mammalian host cells and that contain 2 domains: a T-cell recruiting antibody domain that targets immunomodulating T-cell surface proteins, such as CD3, and a specific TCR domain that targets and binds to tumor antigens presented by class I MHC complexes.85 This design has the benefit of activating T cells to attack tumor cells regardless of their intrinsic specificity.85 Thus, it is like an indirect recruitment of T cells against tumor cells, which can also prevent “on-target, off-tumor” toxicity because no enhanced exogenous TCRs are required. Nevertheless, it also requires several clinical trials to determine whether this design is actually effective in actual human bodies.
Bellicum Pharmaceutical, on the other hand, does not provide another alternative for enhancing the affinity or potency of TCRs against cancer cells. Instead, its novel technology is to prevent occasions when engineered T cells demonstrate side effects in patients’ body. It develops a switch technology for TCR-T cell therapy called CaspaCIDe. This technology involves transforming T cells with genes that code for caspase-9 and chemically induced dimerization (CID) protein with specially designed domain allowing for binding of rimiducid.86 When a patient is experiencing severe side effect due to “on-target, off-tumor” toxicity, rimiducid is introduced and dimerizes CID proteins. The dimerization of CID proteins initiates a signaling cascade that activates caspase-9 and then caspase-3, which eventually leads to apoptosis of T cells to alleviate side effects.86
Obstacles and Breakthroughs
One everlasting challenge for TCRs is their binding affinity to tumor antigens. Several methods have been proposed, including those platforms applied by companies mentioned above. Medigene, in collaboration with other institutes, has proposed a humanized mouse model for identification of affinity-enhanced TCRs with higher affinity and more substantial therapeutic effect compared to human-derived TCRs.87 As a matter of fact, the affinity of TCRs should be controlled as well, not just because of cross-reactivity but also because of self-reactivity among transduced T cells via fratricide, making such enhanced T cells not viable in culture, which is the case when the affinity of TCRs for NY-ESO-1 is enhanced to K D = 26 pM.88 The affinity of TCRs is also directly related to their “on-target, off-tumor” toxicity toward normal body cells. Intratumoral injections of TCR-T cells, a potential solution to this problem, enables high concentrations and bioavailability of T cells to be reached locally while the actual systemic exposure of normal body cells to these infused T cells can remain (very) low.89 Another approach would be to add an inhibitory CAR molecule to the TCR-T cell, in which a separate CAR containing an scFv targeting an antigen on normal tissue but not on tumor tissue is fused to an inhibitory cytoplasmic domain, for example, the immune checkpoint molecule PD-1.18 This approach can enable TCR-T cells to distinguish between tumor cells and normal cells with the same antigen. Moreover, there have been proposals of inducing the apoptosis of engineered T cells: iCas9, where caspase 9 gene is modified to be inducible upon the addition of a small molecule is a potential ideal suicide switch for T cells.90 Finally, another approach is to transduce engineered T cells with a gene for modified human CYP4B1 enzyme, which leads to bioactivation of the protoxin 4-ipomeanol and induces T-cell killing.91
T-cell receptors also have trouble eradicating metastatic tumors because of the immunosuppressive microenvironment of tumors. Tumor tissue inhibits T-cell trafficking toward tissues by limiting expression in tumor endothelial cells of T cell-specific adhesion molecules, such as intercellular adhesion molecule 1, costimulatory ligands, or shutting down T-cell-specific chemoattractants.92,93 Tumor cells hinder T-cell migration by cancer-associated fibroblasts and extracellular matrix components.94 Certain molecules derived from tumor cells, including vascular endothelial growth factor (VEGF), interleukin 10 (IL-10), and prostaglandin E2, which cooperate to induce expression of FAS-ligand and thus can mediate the apoptosis of FAS-positive CD8 effector T cells.95 The second barrier to T-cell-mediated killing of tumor cells is suppressed T-cell activation. T cell will generally encounter hypoxia, which, when sustained, often leads to T-cell evasion as well as tumor progression: all mammalian cells that divide rapidly require high glucose uptake to sustain their proliferation.96 As a result, tumor cells, stromal cells, and immune cells must undergo fierce competition against the limited glucose in the natural environment.96 However, tumor cells can drive higher expression of the glucose transporter GLUT1 under situations of hypoxia, maintaining a high metabolic rate and proliferation, and outcompete T cells, reducing their antitumor activity.96 Moreover, tumor cells often increase the expression of co-inhibitory ligands (checkpoint inhibitors), including PD-1 ligand 1 (PD-L1) and PD-1 ligand 2 (PD-L2), as well as reduce the expression of B7 proteins that produces costimulatory signals when bind to CD28 on T cells.94 Cytotoxic T-lymphocyte antigen-4, a homolog of CD28 but have greater binding affinities than CD28 and is expressed mainly by activated T cells, prevents further activation of T cells when binding to ligand B7 on APCs.40,97,98 The PD-1, another inhibitory molecule belonging to the immunoglobulin superfamily, induces apoptosis of antigen-specific T cells and reduces apoptosis of regulatory T cells when binding to PD-L1.98-100 Moreover, engagement of PD-1 by PD-L2 can drastically inhibit TCR-mediated proliferation and cytokine production by helper T cells.101 There might also be an insufficient amount of chemokine receptors, such as CXC chemokine receptor type-3 (CXCR3), in tumor cells to attract T cells, and tumors may induce enhanced necrosis.96,102,103
Fortunately, TCRs could be modified to improve T-cell trafficking and activation. One method is to engineer T cells with genes coding for receptors for chemokines expressed by tumors to improve T-cell trafficking. One study demonstrated the effective induction of interferon-γ secretion by T cells transduced with genes that encoded CXCR2, receptors for growth-regulated oncogene α (CXCL1), which is expressed by a range of tumor cell lines.92 Another method, which has been proved successful on 5 different types of vascularized tumors using CAR-T cells, is to engineer T cells with CARs targeted against VEGFR-2 protein, which is overexpressed in tumor endothelial cells.104 As a result, TCRs could be engineered to express such receptors as well as to improve T-cell trafficking. For T-cell activation, one approach is to incorporate a signal switch to the T cells that reverses the suppressive signal when binding to tumor chemokines into an activation signal that increases T-cell proliferation, such as a chimeric chemokine receptor by fusing a IL-7 receptor endodomain to a IL-4 receptor exodomain, which showed striking antitumor benefits against EBV-transformed B-cell tumors.105
Multiple immune checkpoint blockade inhibitors were developed to enhance the efficacy of immunotherapy against poorly responding tumors. Cytotoxic T-lymphocyte antigen-4 blockade and PD-1 blockade were proved to be effective in enhancing T-cell activation.97,106-108 Cytotoxic T-lymphocyte antigen-4 blocking antibody MDX-CTLA-4, which is now called ipilimumab, is commonly used as immune checkpoint blockade and is used in 2 of the 84 identified clinical trials.98 Programmed cell death protein-1 inhibitors and PD-L1 inhibitors are also common among clinical trials aiming at the treatment of cancers; moreover, 2 PD-1 inhibitors, nivolumab (which is used in 1 of the 84 trials) and pembrolizumab (which is used in 2 of the 84 trials), and 1 PD-L1 inhibitor, atezolizumab, has been approved by Food and Drug Administration in the treatment of certain cancer types.109 Another method involves the removal of genes in T cells coding for co-inhibitory-ligand-binding molecules using gene-editing technologies. For example, transcription activator-like effector nuclease (TALEN)-mediated PD-1 gene inactivation in tumor-reactive CD4+ and CD8+ T cells (which was transferred adoptively) has shown to increase T cells’ resistance to PD-1-mediated cell death in tumor tissues.110
Another challenge, especially for neoantigen-specific TCRs, is to efficiently identify such special antigens. As applied by companies like Kite Pharma, HTS-IR technology at both bulk and single-cell levels, including computational methods like TraCeR and single-cell TCRseq for reconstructing TCRs and identifying immunogenic neoantigens have provided useful tools for analyses of the diversity, dynamics, and clonality of T cells.111 Although its cost might be lowered by future advancement in gene-sequencing and bioinformatics analysis, it is still a relatively expensive method for now.111,112
Other improvements have also been attempted, including the knockdown of endogenous TCRs by nuclease genome editing using zinc finger, TALEN, or CRISPR/Cas9 system, for more efficient recognition and higher level of expression of exogenous TCRs. Targeting multiple antigens simultaneously, such as the adoptive transfer of 2 populations of CD8+ T cells targeting gp100 and ovalbumin, in one trial resulted in delayed recurrence of B16-OVA melanoma.42,113 With such improvements, TCR-T-cell therapy is likely to be more effective in eradicating tumors, but the lowering of cost is likely to be another issue for patients with cancer.1,49,114
Overall, TCR-T-cell therapy has been evolving rapidly and has become a promising strategy against various types of cancer, especially against solid tumors. Our study did not require an ethical board approval, because it did not contain human or animal trials.
Supplemental Material
Supplementary_Table-The_Emerging_World_of_TCR-T_Cell_Trials_Against_Cancer_A_Systematic_Review_(1) for The Emerging World of TCR-T Cell Trials Against Cancer: A Systematic Review by Jianxiang Zhang, and Lingyu Wang in Technology in Cancer Research & Treatment
Acknowledgments
The authors would like to thank Rui Chen, Liuyang Wang, and Mengyang Chong for their feedback and advice.
Abbreviations
- AFP
α-fetoprotein
- AML
acute myeloid leukemia
- APC
antigen-presenting cell
- CAR
chimeric antigen receptor
- CD
cluster of differentiation
- CID
chemically induced dimerization
- CRISPR
clustered regularly interspaced short palindromic repeats
- CT
cancer-testis
- CTLA-4
cytotoxic T-lymphocyte antigen 4
- CXCR3
CXC chemokine receptor type-3
- gp100
glycoprotein 100
- HLA
human leukocyte antigen
- HPV
human papillomavirus
- HTS
high-throughput screening
- IL
interleukin
- ITAM
immunoreceptor tyrosine-based activation motif
- MAGE
melanoma-associated antigen
- MART-1
melanoma antigen recognized by T cells
- MHC
major histocompatibility complex
- MM
multiple myeloma
- NSCLC
non-small cell lung carcinoma
- PD-1
programmed cell death protein 1
- PD-L1
programmed cell death ligand-1
- pMHC
peptide-major histocompatibility complex
- PRAME
preferentially expressed antigen in melanoma
- scFv
single-chain fragment variant
- SPEAR
specific peptide enhanced affinity receptor
- TAA
tumor-associated antigen
- TALEN
transcription activator-like effector nuclease
- TCR
T-cell receptor
- TCR-T
T-cell receptor-engineered T
- TRuC
T-cell receptor fusion construct
- VEGF
vascular endothelial growth factor
- VEGFR
VEGF receptor
- WT1
Wilms tumor 1
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Jianxiang Zhang  https://orcid.org/0000-0001-5119-4157
https://orcid.org/0000-0001-5119-4157
Lingyu Wang, PhD  https://orcid.org/0000-0002-2736-7588
https://orcid.org/0000-0002-2736-7588
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Mo Z, Du P, Wang G, Wang Y. The multi-purpose tool of tumor immunotherapy : gene-engineered T cells. J Cancer. 2017;8(9):1690–1703. doi:10.7150/jca.18681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang C, Liu J, Zhong JF, Zhang X. Engineering CAR-T cells. Biomark Res. 2017;5(1):22 doi:10.1186/s40364-017-0102-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu S, Li A, Liu Q, et al. Chimeric antigen receptor T cells: a novel therapy for solid tumors. J Hematol Oncol. 2017;10(1):78 doi:10.1186/s13045-017-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kato D, Yaguchi T, Iwata T, et al. Prospects for personalized combination immunotherapy for solid tumors based on adoptive cell therapies and immune checkpoint blockade therapies. Jpn J Clin Immunol. 2017;40(1):68–77. doi:10.2177/jsci.40.68. [DOI] [PubMed] [Google Scholar]
- 5. Garber K. Driving T-cell immunotherapy to solid tumors. Nat Biotechnol. 2018;36(3):215–219. doi:10.1038/nbt.4090. [DOI] [PubMed] [Google Scholar]
- 6. Kass I, Buckle AM, Borg NA. Understanding the structural dynamics of T cell receptor – peptide-bound major histocompatibility complex interactions. Trends Immunol. 2014:1–9. doi:10.1016/j.it.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 7. Hewitt EW, Biology M. The MHC class I antigen presentation pathway : strategies for viral immune evasion. Immunology. 2003;110(2):163–169. doi:10.1046/j.1365-2567.2003.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buonaguro L, Petrizzo A, Tornesello ML, Buonaguro FM. Translating tumor antigens into cancer vaccines. Clin Vaccine Immunol. 2011;18(1):23–34. doi:10.1128/CVI.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mantegazza AR, Magalhaes JG, Amigorena S, Marks MS. Presentation of phagocytosed antigens by MHC class I and II Adriana. Traffic. 2014;14(2):135–152. doi:10.1111/tra.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berger AC, Paul A, Berger AC, Roche PA. MHC class II transport at a glance. J Cell Sci. 2009;2009:1–4. doi:10.1242/jcs.035089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Golubovskaya V, Berahovich R, Xu S, Harto H, Wu L. Major highlights of the CAR-TCR Summit, Boston, 2016. Anticancer Agents Med Chem. 2017;17(10):1344–1350. doi:10.2174/1871520617666170110151900. [DOI] [PubMed] [Google Scholar]
- 12. Wieczorek M, Abualrous ET, Sticht J, et al. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front Immunol. 2017;8(MAR):1–16. doi:10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun ZYJ, Kim KS, Wagner G, Reinherz EL. Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3εγ heterodimer. Cell. 2001;105(7):913–923. doi:10.1016/S0092-8674(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 14. Medsker B, Forno E, Simhan H, Juan C, Sciences R. Adoptive T cell therapies: a comparison of T cell receptors and chimeric antigen receptors. Trends Pharmacol Sci. 2016;70(12):773–779. doi:10.1016/j.tips.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pardoll DM. The blockade of immune slide checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi:10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maus MV, June CH, Hospital G. Making better chimeric antigen receptors for adoptive T-cell therapy. 2017;22(8):1875–1884. doi:10.1158/1078-0432.CCR-15-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uhlen M, Fagerberg L, Hallstrom BM, et al. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419–1-1260419–9 doi:10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 18. Harris DT, Kranz DM. Adoptive T cell therapies: a comparison of T cell receptors and chimeric antigen receptors. Trends Pharmacol Sci. 2016;37(3):220–230. doi:10.1016/j.tips.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stone JD, Harris DT, Soto CM, et al. A novel T cell receptor single-chain signaling complex mediates antigen-specific T cell activity and tumor control. Cancer Immunol Immunother. 2014;63(11):1163–1176. doi:10.1007/s00262-014-1586-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang JC. Toxicities associated with adoptive T-cell transfer for cancer. Cancer J (Sudbury, Mass). 2015;21(6):506–509. doi:10.1097/PPO.0000000000000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Borbulevych OY, Santhanagopolan SM, Hossain M, Baker BM. TCRs used in cancer gene therapy cross-react with MART-1/Melan-A tumor antigens via distinct mechanisms. J Immunol. 2011;187(5):2453–2463. doi:10.4049/jimmunol.1101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elroisse JD, Urquin CL, Laen EDEP, Errano AS, Ethe BL. Quantitative evaluation of the expression of MAGE genes in tumors by limiting dilution of cDNA libraries. Int J Cancer. 1999;83(5):664–669. doi:10.1002/(SICI)1097-0215(19991126)83:5<664:: AID-IJC16>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 23. TCR A, Morgan RA, Chinnasamy N, et al. Cancer regression and neurologic toxicity following anti-MAGE- A3 TCR gene therapy Richard. J Immunother. 2014;36(2):133–151. doi:10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harris DT, Hager MV, Smith SN, et al. Comparison of T cell activities mediated by human TCRs and CARs that use the same recognition domains. J Immunol (Baltimore, Md : 1950). 2018;200(3):1088–1100. doi:10.4049/jimmunol.1700236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu Y, Yang Z, Horan LH, et al. A novel antibody-TCR (AbTCR) platform combines Fab-based antigen recognition with gamma/delta-TCR signaling to facilitate T-cell cytotoxicity with low cytokine release. Cell Discov. 2018;4(1):62 doi:10.1038/s41421-018-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rabinovich GA, Gabrilovich D, Sotomayor EM. Immunosuppressive strategies that are mediated by tumor cells. Annu Rev Immunol. 2007;25(1):267–296. doi:10.1146/annurev.immunol.25.022106.141609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson LA, Morgan RA, Dudley ME, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114(3):535–546. doi:10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vigneron N. Human tumor antigens and cancer immunotherapy. Biomed Res Int. 2015:17 doi:10.1155/2015/948501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smet C, Lurquin C, Bruggen P, Plaen E, Brasseur F, Boon T. Sequence and expression pattern of the human MAGE2 gene. Immunogenetics. 1994;39(2):121–129. doi:10.1007/BF00188615. [DOI] [PubMed] [Google Scholar]
- 30. Smet CDE, Lurquin C, Lethe B, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19(11):7327–7335. doi:10.1128/MCB.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bakker BABH, Schreurs MWJ, Boer AJD, et al. Melanocyte lineage-specific antigen gpl00 is recognized by melanoma-derived tumor-infiltrating lymphocytes. J Exp Med. 1994;179(3):1005–1009. doi:10.1084/jem.179.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kawakami BY, Eliyahu S, Sakaguchi K, et al. Identification of the immunodominant peptides of the MART-1 human melanoma antigen recognized by the majority of HLA-A2-restricted tumor infiltrating lymphocytes. J Exp Med. 1994;180(1):1–6. doi:10.1084/jem.180.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salgaller ML, Afshar A, Marincola FM, Rivoltini L, Kawakami Y, Rosenberg SA. Recognition of multiple epitopes in the human melanoma antigen gp100 by peripheral blood lymphocytes stimulated in vitro with synthetic peptides. Cancer Res. 1995;55(21):4972–4979. [PubMed] [Google Scholar]
- 34. Wang RF, Appella E, Kawakami Y, Kang X, Rosenberg SA. Identification of TRP-2 as a human tumor antigen recognized by cytotoxic T lymphocytes. J Exp Med. 1996;184(6):2207–2216. doi:10.1084/jem.184.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parkhurst MR, Fitzgerald EB, Southwood S, et al. Identification of a shared HLA-A*0201-restricted T-cell epitope from the melanoma antigen tyrosinase-related protein 2 (TRP2). Cancer Res. 1998;58(21):4895–4901. [PubMed] [Google Scholar]
- 36. Kawakami Y, Robbins PF, Wang X, et al. Identification of new melanoma epitopes on melanosomal proteins recognized by tumor infiltrating T lymphocytes restricted by HLA-A1, -A2, and -A3 alleles. J Immunol. 1998;161(12):6985–6992. http://cancerres.aacrjournals.org/content/58/21/4895.long. [PubMed] [Google Scholar]
- 37. Tsang KY, Zaremba S, Nieroda CA, Zhu MZ, Hamilton JM, Schlom J. Generation of human cytotoxic T cells specific for human carcinoembryonic antigen epitopes from patients immunized with recombinant vaccinia-CEA vaccine. J Natl Cancer Inst. 1995;87(13):982–990. doi:10.1093/jnci/87.13.982. [DOI] [PubMed] [Google Scholar]
- 38. Corman JM, Sercarz EE, Nanda NK. Recognition of prostate-specific antigenic peptide determinants by human CD4 and CD8 T cells. Clin Exp Immunol. 1998;114(2):166–172. doi:10.1046/j.1365-2249.1998.00678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Correale P, Walmsley K, Nieroda C, et al. In vitro generation of human cytotoxic T lymphocytes specific for peptides derived from prostate-specific antigen. J Natl Cancer Inst. 1997;89(4):293–300. doi:10.1093/jnci/89.4.293. [DOI] [PubMed] [Google Scholar]
- 40. Ping Y, Liu C, Zhang Y. T-cell receptor-engineered T cells for cancer treatment: current status and future directions. Protein Cell. 2018;9(3):254–266. doi:10.1007/s13238-016-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van den Berg JH, Gomez-Eerland R, van de Wiel B, et al. Case report of a fatal serious adverse event upon administration of T cells transduced with a MART-1-specific T-cell receptor. Mol Ther. 2015;23(9):1541–1550. doi:10.1038/mt.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kunert A, Straetemans T, Govers C, et al. TCR-engineered T cells meet new challenges to treat solid tumors: choice of antigen, T cell fitness, and sensitization of tumor milieu. Front Immunol. 2013;4:363 doi:10.3389/fimmu.2013.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sugiyama H. WT1 (Wilms’ tumor gene 1): biology and cancer immunotherapy. Jpn J Clin Oncol. 2010;40(5):377–387. doi:10.1093/jjco/hyp194. [DOI] [PubMed] [Google Scholar]
- 44. Duim SN, Kurakula K, Goumans MJ, Kruithof BPT. Cardiac endothelial cells express Wilms’ tumor-1. Wt1 expression in the developing, adult and infarcted heart. J Mol Cell Cardiol. 2015;81:127–135. doi:10.1016/j.yjmcc.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 45. O’Hara M, Stashwick C, Haas AR, Tanyi JL. Mesothelin as a target for chimeric antigen receptor-modified T cells as anticancer therapy. Immunotherapy. 2016;8(4):449–460. doi:10.2217/imt.16.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jirsova Katerina K, Neuwirth A, Kalasova S, Vesela V, Merjava S. Mesothelial proteins are expressed in the human cornea. Exp Eye Res. 2010;91(5):623–629. doi:10.1016/j.exer.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 47. Villena-Vargas J, Adusumilli PS. Mesothelin-targeted immunotherapies for malignant pleural mesothelioma. Ann Cardiothorac Surg. 2012;1(4):466–471. doi:10.3978/j.issn.2225-319X.2012.10.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmitt TM, Aggen DH, Stromnes IM, et al. Enhanced-affinity murine T-cell receptors for tumor/self-antigens can be safe in gene therapy despite surpassing the threshold for thymic selection. Blood. 2013;122(3):348–356. doi:10.1182/blood-2013-01-478164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ye B, Stary CM, Gao Q, et al. Genetically modified T-cell-based adoptive immunotherapy in hematological malignancies. J Immunol Res. 2017;2017:5210459 doi:10.1155/2017/5210459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Radtke I, Mullighan CG, Ishii M, et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci U S A. 2009;106(31):12944–12949. doi:10.1073/pnas.0903142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Junnila S, Kokkola A, Karjalainen-Lindsberg ML, Puolakkainen P, Monni O. Genome-wide gene copy number and expression analysis of primary gastric tumors and gastric cancer cell lines. BMC Cancer. 2010;10(1):73 doi:10.1186/1471-2407-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jones S, Zhang X, Parsons WD, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi:10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS One. 2010;5(2):e8918 doi:10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi:10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 55. Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational implications of tumor heterogeneity. Clin Cancer Res. 2015;21(6):1258–1266. doi:10.1158/1078-0432.CCR-14-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang H, Ye ZL, Yuan ZG, Luo ZQ, Jin HJ, Qian QJ. New strategies for the treatment of solid tumors with CAR-T cells. Int J Biol Sci. 2016;12(6):718–729. doi:10.7150/ijbs.14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Deniger DC, Pasetto A, Tran E, et al. Stable, nonviral expression of mutated tumor neoantigen-specific T-cell receptors using the sleeping beauty transposon/transposase system. Mol Ther. 2016;24(6):1078–1089. doi:10.1038/mt.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 59. Maio M. Melanoma as a model tumour for immuno-oncology. Ann Oncol. 2012;23(suppl 8):viii10–viii14. doi:10.1093/annonc/mds257. [DOI] [PubMed] [Google Scholar]
- 60. Lamers CHJ, Sleijfer S, Vulto AG, et al. Treatment of metastatic renal cell carcinoma with autologous T-lymphocytes genetically retargeted against carbonic anhydrase IX: first clinical experience. J Clin Oncol. 2006;24(13):e20–e22. doi:10.1200/JCO.2006.05.9964. [DOI] [PubMed] [Google Scholar]
- 61. Lamers CH, Sleijfer S, van Steenbergen S, et al. Treatment of metastatic renal cell carcinoma with CAIX CAR-engineered T cells: clinical evaluation and management of on-target toxicity. Mol Ther. 2013;21(4):904–912. doi:10.1038/mt.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Park JR, Digiusto DL, Slovak M, et al. Adoptive transfer of chimeric antigen receptor re-directed cytolytic T lymphocyte clones in patients with neuroblastoma. Mol Ther. 2007;15(4):825–833. doi:10.1038/sj.mt.6300104. [DOI] [PubMed] [Google Scholar]
- 63. McCormack E, Adams KJ, Hassan NJ, et al. Bi-specific TCR-Anti CD3 redirected T-cell targeting of NY-ESO-1- and LAGE-1-positive tumors. Cancer Immunol Immunother. 2013;62(4):773–785. doi: 10.1007/s00262-012-1384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Parkhurst MR, Yang JC, Langan RC, et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol Ther. 2011;19(3):620–626. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kakarla S, Gottschalk S. CAR T cells for solid tumors: armed and ready to go? Cancer J (Sudbury, Mass). 2014;20(2):151–155. doi:10.1097/PPO.0000000000000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Perna SK, De Angelis B, Pagliara D, et al. Interleukin 15 provides relief to CTLs from regulatory T cell-mediated inhibition: implications for adoptive T cell-based therapies for lymphoma. Clin Cancer Res. 2013;19(1):106–117. doi:10.1158/1078-0432.CCR-12-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kerkar SP, Leonardi AJ, van Panhuys N, et al. Collapse of the tumor stroma is triggered by IL-12 induction of fas. Mol Ther. 2013;21(7):1369–1377. doi:10.1038/mt.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Overview: Adaptimmune. Adaptimmune. 2018. https://www.adaptimmune.com/technology/overview. Accessed July 17, 2018.
- 69. Manufacturing: Adaptimmune. Adaptimmune. 2018. https://www.adaptimmune.com/technology/manufacturing. Accessed July 17, 2018.
- 70. Our Technology Platform: Adaptimmune. Adaptimmune. 2018. https://www.adaptimmune.com/technology/our-technology-platform. Accessed July 17, 2018.
- 71. Adaptimmune. Our Engineered T-Cell Therapies: Adaptimmune. Adaptimmune. 2018. https://www.adaptimmune.com/technology/our-engineered-t-cell-therapies. Accessed December 2, 2018.
- 72. Seeking Alpha. Adaptimmune (ADAP) Presents At 10th Annual Biotech Showcase 2018 - Slideshow - Adaptimmune Therapeutics plc (NASDAQ: ADAP) | Seeking Alpha. Seeking Alpha. 2018. https://seekingalpha.com/article/4136007-adaptimmune-adap-presents-10th-annual-biotech-showcase-2018-slideshow. Accessed December 2, 2018.
- 73. European Biotechnology. Adaptimmune raises US$100 m – European Biotechnology. European Biotechnology. 2018. https://european-biotechnology.com/up-to-date/latest-news/news/adaptimmune-raises-us100m.html. Accessed December 2, 2018.
- 74. Pipeline: Adaptimmune. Adaptimmune. 2018. https://www.adaptimmune.com/pipeline/overview. Accessed July 17, 2018.
- 75. Adaptimmune confirms GSK Nomination of Second Adaptimmune Target under Strategic Multi-Target Collaboration. Adaptimmune confirms GSK Nomination of Second Adaptimmune Target under Strategic Multi-Target Collaboration. 2017. http://ir.adaptimmune.com/phoenix.zhtml?c=253991&p=irol-newsArticle&ID=2234983. Accessed July 17, 2018.
- 76. Wang X, Wang Q. Alpha-fetoprotein and hepatocellular carcinoma immunity. Can J Gastroenterol Hepatol. 2018;2018:9049252 doi:10.1155/2018/9049252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pipeline | Kite Pharma. Kite Pharma. 2018. https://www.kitepharma.com/our-research/pipeline/. Accessed July 17, 2018.
- 78. Kite Pharma Strengthens Its T Cell Receptor (TCR) Cancer Gene Therapy Platform Through Acquisition of T-Cell Factory B.V. (TCF(TM)) (NASDAQ: KITE). Kite Pharma. 2015. http://ir.kitepharma.com/releasedetail.cfm?releaseid=901985. Accessed July 17, 2018.
- 79. Technology | Kite Pharma. 2018. https://www.kitepharma.com/our-research/technology/. Accessed July 5, 2018.
- 80. Medigene – Technologies – TCR-T Platform. Medigene. 2018. https://www.medigene.com/technologies/platforms/tcr-t-platform/. Accessed July 17, 2018.
- 81. Our Approach — TCR2 Therapeutics. TCR2 Therapeutics. 2018. http://www.tcr2.com/innovation/. Accessed July 17, 2018.
- 82. Otranto M, Sarrazy V, Bonté F, Hinz B, Gabbiani G, Desmoulière A. The role of the myofibroblast in tumor stroma remodeling. Cell Adh Migr. 2012;6(3):203–219. doi:10.4161/cam.20377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. ACT – Immatics. Immatics. 2018. https://immatics.com/act.html. Accessed July 24, 2018.
- 84. Pipeline – Immatics. Immatics. 2018. https://immatics.com/product-pipeline.html. Accessed July 24, 2018.
- 85. Bispecific TCRs – Immatics. Immatics. 2018. https://immatics.com/tcr-bispecifics.html. Accessed July 17, 2018.
- 86. Technology – Bellicum Pharmaceuticals, Inc. Bellicum Pharmaceuticals. 2018. http://www.bellicum.com/technology/. Accessed July 17, 2018.
- 87. Obenaus M, Leitão C, Leisegang M, et al. Identification of human T-cell receptors with optimal affinity to cancer antigens using antigen-negative humanized mice. Nat Biotechnol. 2015;33(4):402–407. doi:10.1038/nbt.3147. [DOI] [PubMed] [Google Scholar]
- 88. Tan MP, Gerry AB, Brewer JE, et al. T cell receptor binding affinity governs the functional profile of cancer-specific CD8 + T cells. Clin Exp Immunol. 2015;180(2):255–270. doi:10.1111/cei.12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Marabelle A, Kohrt H, Caux C, Levy R. Intratumoral Immunization: a new paradigm for cancer therapy. Clin Cancer Res. 2014;20(7):1747–1756. doi:10.1158/1078-0432.CCR-13-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Di Stasi A, Tey SK, Dotti G, et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N Engl J Med. 2011;365(18):1673–1683. doi:10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Foust KD, Kaspar BK. Identification of amino acid determinants in CYP4B1 for optimal catalytic processing of 4-ipomeanol. Biochem J. 2015;1(465):103–114. doi:10.1042/BJ20140813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kershaw MH, Wang G, Westwood JA, et al. Redirecting migration of T Cells to chemokine secreted from tumors by genetic modification with CXCR2. Human Gene Therapy. 2002;13(16):1971–1980. doi:10.1089/10430340260355374. [DOI] [PubMed] [Google Scholar]
- 93. Ritchie DS, Neeson PJ, Khot A, et al. Persistence and efficacy of second generation CAR T cell against the LeY antigen in acute myeloid leukemia. Mol Ther. 2013;21(11):2122–2129. doi:10.1038/mt.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kunert A, Debets R. Engineering T cells for adoptive therapy: outsmarting the tumor. Curr Opin Immunol. 2018;51:133–139. doi:10.1016/j.coi.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 95. Motz GT, Santoro SP, Wang LP, et al. Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat Med. 2014;20(6):607–615. doi:10.1038/nm.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Anderson KG, Stromnes IM, Greenberg PD. Obstacles posed by the tumor microenvironment to T cell activity: a case for synergistic therapies. Cancer Cell. 2017;31(3):311–325. doi:10.1016/j.ccell.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cartier SF, Chen ZY, Walder GJ, Sleppy CR, Castleman AW. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271(5256):1734–1736. doi:10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 98. Haanen JBAG, Robert C. Immune checkpoint inhibitors. Prog Tumor Res. 2015;42:55–66. doi:10.1159/000437178. [DOI] [PubMed] [Google Scholar]
- 99. Loise MF, Sage PT, Sharpe AH. PD-1 pathway in tolerance and autoimmunity. Autoimmunity. 2011;236:219–242. doi:10.1111/j.1600-065X.2010.00923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Fife BT, Pauken KE. The role of the PD-1 pathway in autoimmunity and peripheral tolerance. Ann N Y Acad Sci. 2011;1217(1):45–59. doi:10.1111/j.1749-6632.2010.05919.x. [DOI] [PubMed] [Google Scholar]
- 101. Latchman Y, Wood CR, Chernova T, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2(3):261–268. doi:10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 102. Harlin H, Meng Y, Peterson AC, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Can res. 2009;69(7):3077–3085. doi:10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mulligan AM, Raitman I, Feeley L, et al. Tumoral lymphocytic infiltration and expression of the chemokine CXCL10 in breast cancers from the Ontario Familial Breast Cancer Registry. Clin Cancer Res. 2013;19(2):336–346. doi:10.1158/1078-0432.CCR-11-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Huang J, Liang J, Tang Q, et al. An active murine-human chimeric Fab antibody derived from Escherichia coli, potential therapy against over-expressing VEGFR2 solid tumors. Appl Microbiol Biotechnol. 2011;91(5):1341–1351. doi:10.1007/s00253-011-3335-y. [DOI] [PubMed] [Google Scholar]
- 105. Leen AM, Sukumaran S, Watanabe N, et al. Reversal of tumor immune inhibition using a chimeric cytokine receptor. Mol Ther. 2014;22(6):1211–1220. doi:10.1038/MT.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kwon ED, Hurwitz AA, Foster BA, et al. Manipulation of T cell costimulatory and inhibitory signals for immunotherapy of prostate cancer. Proc Natl Acad Sci U S A. 1997;94(15):8099–8103. http://www.ncbi.nlm.nih.gov/pubmed/9223321. Accessed November 25, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Freeman GJ, Long AJ, Iwai Y, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192(7):1027–1034. http://www.ncbi.nlm.nih.gov/pubmed/11015443. Accessed November 25, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Iwai Y, Terawaki S, Honjo T. PD-1 blockade inhibits hematogenous spread of poorly immunogenic tumor cells by enhanced recruitment of effector T cells. International Immunol. 2004;17(2):133–144. doi:10.1093/intimm/dxh194. [DOI] [PubMed] [Google Scholar]
- 109. Alsaab HO, Sau S, Alzhrani R, et al. PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: mechanism, combinations, and clinical outcome. Front Pharmacol. 2017;8:1–15. doi:10.3389/fphar.2017.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Menger L, Sledzinska A, Bergerhoff K, et al. TALEN-mediated inactivation of PD-1 in tumor-reactive lymphocytes promotes intratumoral T-cell persistence and rejection of established tumors. Cancer Res. 2016;76(8):2087–2093. doi:10.1158/0008-5472.CAN-15-3352. [DOI] [PubMed] [Google Scholar]
- 111. Ye B, Smerin D, Gao Q, Kang C, Xiong X. High-throughput sequencing of the immune repertoire in oncology: applications for clinical diagnosis, monitoring, and immunotherapies. Cancer Lett. 2018;416:42–56. doi:10.1016/j.canlet.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 112. Orkin SH, Reilly P. Paying for future success in gene therapy. Science. 2016;352(6289):1059–1061. doi:10.1126/science.aaf4770. [DOI] [PubMed] [Google Scholar]
- 113. Kaluza KM, Kottke T, Diaz RM, Rommelfanger D, Thompson J, Vile R. Adoptive transfer of cytotoxic T lymphocytes targeting two different antigens limits antigen loss and tumor escape. Human Gene Therapy. 2012;23(10):1054–1064. doi:10.1089/hum.2012.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Morris EC, Stauss HJ. Optimizing T-cell receptor gene therapy for hematologic malignancies. Blood. 2016;127(26):3305–3312. doi:10.1182/blood-2015-11-629071.BLOOD. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_Table-The_Emerging_World_of_TCR-T_Cell_Trials_Against_Cancer_A_Systematic_Review_(1) for The Emerging World of TCR-T Cell Trials Against Cancer: A Systematic Review by Jianxiang Zhang, and Lingyu Wang in Technology in Cancer Research & Treatment