Microcephaly is a genetically heterogeneous disorder. Through molecular genetic studies, Perez et al. demonstrate that homozygous loss-of-function mutations in C7orf43 (now termed microtubule-associated protein 11, MAP11) cause microcephaly in humans and zebrafish. Functional analyses show that MAP11 associates with α-tubulin in mitotic spindles and influences cell proliferation.
Keywords: C7orf43, MAP11, microcephaly, microtubule-associated protein, CRISPR/Cas9 zebrafish
Abstract
Microtubule associated protein 11 (MAP11, previously termed C7orf43) encodes a highly conserved protein whose function is unknown. Through genome-wide linkage analysis combined with whole exome sequencing, we demonstrate that human autosomal recessive primary microcephaly is caused by a truncating mutation in MAP11. Moreover, homozygous MAP11-orthologue CRISPR/Cas9 knock-out zebrafish presented with microcephaly and decreased neuronal proliferation, recapitulating the human phenotype. We demonstrate that MAP11 is ubiquitously transcribed with high levels in brain and cerebellum. Immunofluorescence and co-immunoprecipitation studies in SH-SY5Y cells showed that MAP11 associates with mitotic spindles, co-localizing and physically associating with α-tubulin during mitosis. MAP11 expression precedes α-tubulin in gap formation of cell abscission at the midbody and is co-localized with PLK1, a key regulator of cytokinesis, at the edges of microtubule extensions of daughter cells post cytokinesis abscission, implicating a role in mitotic spindle dynamics and in regulation of cell abscission during cytokinesis. Finally, lentiviral-mediated silencing of MAP11 diminished SH-SY5Y cell viability, reducing proliferation rather than affecting apoptosis. Thus, MAP11 encodes a microtubule-associated protein that plays a role in spindle dynamics and cell division, in which mutations cause microcephaly in humans and zebrafish.
Introduction
Primary hereditary microcephaly is a neurodevelopmental disorder characterized by reduction in brain volume, with an occipital-frontal circumference ≤−2 standard deviations (SD) that of the age and sex-matched population at birth. This genetically heterogeneous disorder leads to intellectual disability with variable severities (Woods and Parker, 2013). To date, mutations in 20 different genes have been shown to cause autosomal recessive primary microcephaly (MCPH; OMIM phenotypic series: PS251200). Humans have a larger and more complex neocortex compared to other mammals because of a substantially greater neuronal proliferative capacity and differentiation of neuronal stem cells at early stages of cortical development (Kriegstein et al., 2006; Ostrem et al., 2017). Thus, the proper maintenance of the progenitor pool, correct spatiotemporal transition of progenitors’ symmetric/asymmetric divisions and differentiation dynamics are crucial to normal neocortex development. It is thus not surprising that most hereditary microcephaly genes have been implicated predominantly in critical cellular processes of mitosis: mitotic spindle assembly and structure, centrosome and centriole function, DNA repair and damage response during cell cycle, as well as kinetochore-associated functions, chromatin remodeling complexes, cleavage furrow and midbody regulation (Gilmore and Walsh, 2013; Kadir et al., 2016; Moawia et al., 2017; Shohayeb et al., 2017; Létard et al., 2018).
We now show that autosomal recessive primary microcephaly in both humans and zebrafish can be caused by loss of function truncating mutations in a previously unstudied gene, the chromosome 7 open reading frame 43 (C7orf43) gene (which we now name microtubule associated protein 11; MAP11), and demonstrate that the encoded protein binds α-tubulin during mitosis, affecting cell proliferation.
Materials and methods
Subjects and clinical phenotyping
DNA samples were obtained from all available samples following informed consent and approval of the Soroka Medical Center Internal Review Board (IRB). Clinical phenotyping was determined by an experienced paediatric neurologist and geneticist. Patient’s photographs are presented following legal guardian consent and written approval.
Homozygosity mapping
Genome-wide linkage analysis using all available DNA samples (Fig. 1A) was performed using Affymetrix GeneChip Human Mapping 500 K Set Nsp (Affymetrix) as described previously (Perez et al., 2014). Homozygosity mapping analysis was carried out using the open online software HomozygosityMapper (http://www.homozygositymapper.org/) (Seelow et al., 2009).
Figure 1.
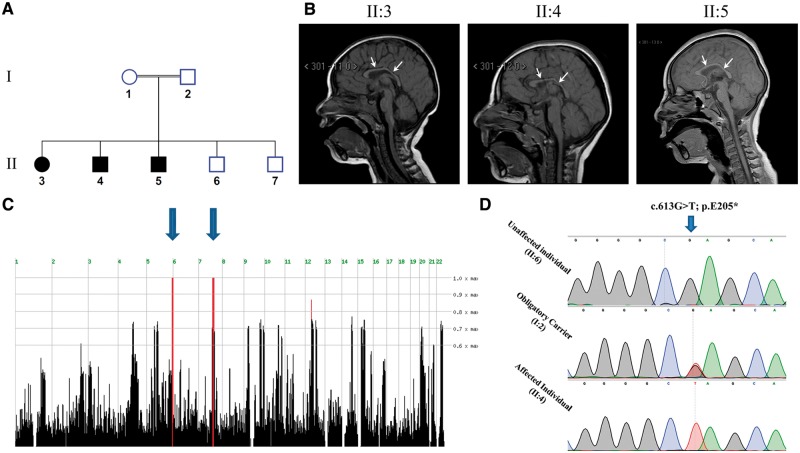
The disease phenotype and MAP11 mutation. (A) Pedigree of the consanguineous Bedouin kindred studied showing three affected siblings. (B) Patients’ brain MRI showing microcephaly and decreased white matter. White arrows are indicating hypoplasia of corpus callosum. (C) Homozygosity-Mapper plot showing two homozygous loci, on chromosome 5 and 7, shared by all affected individuals. (D) Sanger sequencing of the c.613G>T mutation in an unaffected, obligatory carrier and affected individuals.
DNA sequence analysis
Whole-exome sequencing was performed as previously described (Perez et al., 2016). Data were analysed using QIAGEN’s Ingenuity® Variant Analysis™ software (www.qiagen.com/ingenuity, QIAGEN Redwood City) as described previously (Perez et al., 2016). Briefly, variants observed with an allele frequency ≥0.5% in the 1000 genomes project, NHLBI ESP exomes (All) or the Allele Frequency Community were excluded. In addition, variants that appeared in a homozygous state in our in-house whole exome sequencing database of 350 Bedouin control samples were also excluded. We focused on variants which were predicted to have a deleterious effect upon protein coding sequences (e.g. frameshift, in-frame indel, stop codon change, missense or predicted to disrupt splicing by MaxEnt Scan) and variants which were experimentally observed to be associated with a phenotype according to the Human Gene Mutation Database (HGMD) (Stenson et al., 2003). Of those variants, we selected only homozygous variants which were located within homozygous loci shared by all affected individuals (per genome wide homozygosity mapping analysis). Validation and segregation analysis of the MAP11 (C7orf43) mutation was done via Sanger, using sequencing primers: Forward 5′-GCTCAGCCATCTGTCTTTCC-3′; Reverse 5′-AGCCACTTTCCAGTGTGACC-3′.
Protein conservation
Protein multiple sequence alignment (MSA) was performed using Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/) (Larkin et al., 2007). RefSeq sequence accession numbers for Homo sapiens, Pan troglodytes, Bos taurus, Mus musculus, Canis lupus familiaris, Xenopus tropicalis and Danio rerio of C7orf43 orthologues: NP_060745.3, JAA25474.1, XP_002698258.1, NP_694801.2, XP_849734.1, NP_00112 1523.1 and XP_001339329.2, respectively.
Lentiviral shRNA mediated MAP11 (C7orf43) silencing
Two different shRNA oligos were designed to hybridize and silence coding regions of the human MAP11 transcript: MAP11_shRNA (A): 5′-GATCCTTGTGATGACCGCTTCTT GTAAGTCcttcctgtcagaGACTTACAAGAAGCGGTCATCAC AATTTTTG-3′; MAP11_shRNA (B): 5′-GATCCGAACCC GTCAAGTAGCACCGTGCCActtcctgtcagaTGGCACGGTG CTACTTGACGGGTTCTTTTTG-3′. Lower case letters represent the shRNA hairpin loop. Oligonucleotides were annealed and ligated into BamHI/EcoRI site of pGreenPuro™ lentiviral vector (System Biosciences). Lentivirus production and transduction of SH-SY5Y cells was conducted following manufacturer’s guidelines. Three different infection events were done for each silencing oligo. MAP11 silencing was assessed via real-time quantitative polymerase chain reaction (RT-qPCR). Results represent the mean of all three.
MAP11 (C7orf43) transcription analysis
MAP11 transcription levels were assayed via RT-PCR and RT-qPCR. For MAP11 semi-quantitative (RT-PCR) transcription level assay in different normal human tissues, a panel of cDNA samples was prepared from total RNA derived of 21 normal human tissues (Clontech Laboratories), using Verso cDNA kit (Thermo Scientific™). Two sets of PCR primers were designed to amplify cDNA rather than genomic DNA of either human MAP11 (C7orf43) transcript variant 1 only (NM_018275.4) or all known MAP11 transcript isoforms (per RefSeq database). Primers used to amplify a segment of MAP11 unique to transcript variant 1 (366 bp amplicon): Forward 5′-CTGCAGCCCCCTTCTCAC-3′; Reverse 5′-AGCACGGTCAAGTGTTTTCCA-3′; primers used to amplify a segment of MAP11 shared by all transcript isoforms (238 bp amplicon): Forward 5′-AGTCCCCTGTTCGGACCTAC-3′; Reverse 5′-AAGGCCACACTGAAGGTGAG-3′ Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as housekeeping gene. Primers used for GAPDH amplification (452 bp amplicon): Forward 5′-ACCACAGTCCATGCCATCAC-3′; Reverse 5′-TCCACCACCCTGTTGCTGT-3′. Annealing temperature used was 60°C, the extension time was set for 30 s and the PCR reaction repeats was of 35 cycles for all reactions. PCR products from each reaction were mixed together (equal amounts) and run on 2% agarose gel electrophoresis. Gel was visualized by a ChemiDoc™ MP imaging system (Bio-Rad). For RT-qPCR analyses of MAP11, total RNA was extracted from either MAP11 silenced SH-SY5Y cells or patients’ and controls’ lymphocytes using GENzolTM Tri RNA Pure Kit (Geneaid Biotech Ltd.) according to manufacturer’s instructions. Single stranded cDNA libraries were prepared as described above. RT-qPCR was done using a FastStart universal SYBR® Green master reaction master mix (Roche). Analysis was performed using Rotor-Gene RG-3000 machine (Corbett Research). Primers used for RT-qPCR of MAP11 (111 bp amplicon): Forward 5′-TCAACAACCTTGGCTTTTCC-3′; Reverse 5′-GCAGCTTCAGTTTCATGTGC-3′. MAP11 transcript levels were normalized to GAPDH housekeeping gene. Primers used for GAPDH (87 bp amplicon): Forward 5′-TCGACAGTCAGC CGCATC-3′ ; Reverse 5′-CCGTTGACTCCGACCTTC-3′.
Cell viability assay
Cell viability assay (XTT) was done according to manufacturer’s instructions (Cell Proliferation Kit, 20–300–1000; Biological industries). Briefly, MAP11-silenced and shRNA scrambled control SH-SY5Y cells (5000 cells/well) were seeded on a corning 96-well flat transparent plate in 100 µl Dulbecco’s modified Eagle medium (DMEM) containing 10% foetal bovine serum (FBS). Wells containing complete medium without cells were used as control blanks. Twenty-four hours post seeding, cells were supplemented with XTT and activation reagent (50 µl per well) and incubated for 3 h. Then, absorbance of the samples was measured against a background control using an Infinite® 200 PRO TECAN reader spectrophotometer (ELISA reader) at 500 nm wavelength. To measure reference absorbance (eliminating non-specific readings), measurements were done at 690 nm wavelength and the average absorbance of the blank control wells was subtracted from that of the assay wells. All experiments were done five times; results represent the mean of all five.
Confocal imaging
Endogenous MAP11 protein was detected using rabbit anti-human MAP11 (C7orf43) antibody (HPA019359; Sigma-Aldrich), which was validated by western blot of SH-SY5Y MAP11 overexpressing cells (Supplementary Fig. 1). Initially, SH-SY5Y cells were grown to ∼70% confluence (seeded on coverslips) in 12-well plates. Cells were washed twice with PBST (PBS + 0.05% Tween 20), fixed in 4% paraformaldehyde (PFA) for 20 min, permeabilized and blocked using Triton™ X-100 (0.5% v/v) in an antibody diluent reagent (E09–300; GBI Labs). For immunostainings of MAP11 and α-tubulin, cells were incubated with primary rabbit anti-MAP11 antibody for 1 h, washed twice with PBST and then incubated with a primary mouse monoclonal anti-α-tubulin antibody (T6199; Sigma-Aldrich) for 1 h. Cells were then washed twice with PBST and incubated with secondary goat anti-rabbit IgG Alexa Fluor® 594 (ab150080; Abcam) and donkey anti-mouse IgG Alexa Fluor® 488 (ab150105; Abcam) for 1 h. For immunostainings of MAP11 and PLK1, cells were incubated with primary rabbit anti-MAP11 (C7orf43) antibody for 1 h, washed twice with PBST and then incubated with a primary mouse anti-PLK1 antibody (05–844; Milipore) for 1 h. Cells were then washed twice with PBST and incubated with secondary Dnk anti-rabbit Alexa Fluor® 488 (ab150065; Abcam) and Dnk anti-mouse Alexa Fluor® 647 (ab150107; Abcam). Post antibody incubation, cells were washed twice with PBST and mounted using Vectashield containing DAPI (H-1200; Vector Laboratories). All antibodies were used at 1:500 dilutions. Subcellular localization was visualized using an Olympus confocal microscope (×60 objective). Confocal images were recorded under identical conditions. Excitation was performed with a 488 nm (for Alexa Fluor® 488), 405 nm (for DAPI), 594 nm (for Alexa Fluor® 594) and 647 nm (for Alexa Fluor® 647) laser filtered accordingly. Co-localization of MAP11 and α-tubulin was assayed by calculating Pearson’s correlation coefficient using image J software.
Cloning and co-immunoprecipitation
A custom gene synthesis plasmid, Puc57-Amp (GENEWIZ, Inc.), with MAP11 (C7orf43) reference sequence (derived from GenBank; NM_018275.4) followed by FLAG sequence (5′-GACTACAAAGACGATGACGACAAG-3′), was obtained. The MAP11 fused to FLAG sequence was subsequently cloned into the expression pCDNA3.1 (−) plasmid. HEK-293T cells were seeded on 10 cm tissue culture dishes with 10% FBS containing DMEM medium. Cells were transfected with MAP11-FLAG-fused overexpressing construct or mock pCDNA3.1 (−) plasmids (8 µg in 10 cm culture dish) using Lipofectamine 2000® (Thermo Fisher Scientific) per manufacturer’s instructions. After 48-h incubation, cells were lysed with 1.5 ml non-denaturing lysis buffer (50 mM Tris HCl, pH 7.4, with 150 mM NaCl, 1 mM EDTA, 1% Triton™), incubated for 30 min at 4°C and sonicated (50% amplitude for 20 s, five times). Immunoprecipitation of cell lysates was done using ANTI-FLAG M2 Magnetic Beads (Sigma-Aldrich) following manufacturer’s instructions. Bead elution was done using protein sample buffer (for 4×, final concentration: 40% glycerol, 240 mM Tris/HCl pH 6.8, 8% SDS, 0.04% bromophenol blue, 5% beta-mercaptoethanol). Post elution, lysates were heated (95°C) for 10 min and centrifuged (14 000 rpm) for 5 min to remove residual cell debris. The supernatant was loaded onto 10% polyacrylamide gel. Following electrophoresis (120 V, 1.5 h), proteins were transferred to nitrocellulose membranes (1 h, 400 mA) using a wet transfer apparatus. The nitrocellulose membranes were blocked by incubation in TTBS (0.02 M Tris, pH 7.5, 0.15 M NaCl, 0.9 mM Tween 20; Bio Lab) containing 5% skimmed milk for 1 h at room temperature. The blocked membranes were then incubated for 1 h with primary mouse monoclonal ANTI-FLAG® M2 antibody (F1804; Sigma-Aldrich) at room temperature in a 1:1000 dilution. Following incubation with primary antibody, membranes were washed three times with TTBS for 5 min each, and then incubated with a secondary goat anti-mouse IgG-HRP conjugated antibody (Santa Cruz Biotechnology Inc.; Sc-2005) for 1 h (room temperature, 1:10 000 dilution), washed three times in TTBS and visualized using an enhanced chemiluminescence detection kit (SuperSignal™ West Pico chemiluminescent substrate; Thermo Scientific) in ChemiDoc™ MP imaging system (Bio-Rad). Membranes were then washed three times in TTBS, incubated with a mouse anti-α-tubulin antibody for 1 h, washed three times with TTBS and incubated again with a goat anti-mouse IgG-HRP-conjugated antibody (Santa Cruz Biotechnology; Sc-2005) for 1 h at room temperature, washed and visualized.
Cell proliferation assay
Proliferation assays were performed using Pacific Blue™ anti-mouse Ki-67 antibody (BioLegend) according to manufacturer’s instructions. Briefly, MAP11 shRNA silenced and control shRNA SH-SY5Y cells were assayed. Each line assay was repeated three times with cells from different lentiviral infection events. Approximately 0.5 × 106 cells were seeded on a 6-well plate. Twelve hours later, cells were harvested (0.05% trypsin) washed twice with PBS and fixed using cold 70% ethanol (added drop by drop to the cell pellet while vortexing) and incubated for 1 h at −20°C. Cells were then washed three times with FACS buffer (25 mM HEPES, 1 mM EDTA, 1% FBS and 0.1% sodium azide in PBS) and incubated with the Pacific Blue™ anti-mouse Ki-67 antibody at room temperature in the dark for 30 min. Cells were washed twice with FACS buffer prior to analysis. Single-cell analyses were performed with a 13-channel flow cytometry analyzer (CytoFLEX, configuration B5-R3-V5; Beckman Coulter). Flow cytometry results were analysed with FlowJo (FlowJo, LLC) focusing on the percentage GFP-positive single cells expressing Ki-67 (Supplementary Fig. 2).
Generating map11 (c7orf43) specific sgRNAs
Single guide RNAs (sgRNAs) targeting map11 (ZFIN: si:dkey-3k24.5) for CRISPR/Cas9-mediated knock-out, was done following a published protocol (Gagnon et al., 2014) that uses a cloning-independent system. Briefly, two different sgRNAs were designed to hybridize near the truncating mutation of the zebrafish MAP11 orthologue. The sgRNAs were designed using CHOPCHOP online (http://chopchop.cbu.uib.no/) (Montague et al., 2014; Labun et al., 2016). For each sgRNA, we used a 60-base oligonucleotide containing a T7 promoter for in vitro transcription, a 20-base spacer region specific to the target site (‘gene-specific oligo’) and an overlap region that anneals to an 80-base constant oligonucleotide (constituting the RNA scaffold recruiting the Cas9 endonuclease). The selected gRNAs sequences: map11_gRNA1: 5′-TAATACGACTCACTATAAGATGGATCAGTGCTGCTGGGTTTTAGAGCTAGAAATAGCAAG-3′; map11_gRNA2: 5′-TAATACGACTCACTATAGTTAAACGGGAGCTCGCAAGGTTTTAGAGCTAGAAATAGCAAG-3′. The constant 80-base oligo used: 5′-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3′. Gene-specific oligos were annealed in 10 mM Tris-Hcl, pH 8.5 buffer in a PCR machine, ramping temperature down from 95°C to 4°C in a stepwise manner. Gaps of annealed oligos were then filled-in using T4 DNA polymerase (New England Biolabs) and run on agarose gel electrophoresis to ensure correct annealing. Products were purified using a standard PCR clean-up column. RNAs from both clones were in vitro transcribed using MEGAscript® T7/SP6 kit according to manufacturer’s instructions (Ambion). Post transcription, RNA was purified in an RNA column clean-up kit (RNAeasy® mini kit; Qiagen). There are two main approaches to achieve CRISPR/Cas9 mediated knockout. One approach is to generate a double strand break that will be endogenously repaired via the highly efficient, yet error prone, non-homologous end joining (NHEJ) repair system. A less efficient yet less error-prone approach is to insert a single stranded oligo donor (ssODN) as a template to encourage double strand break repair through homology directed repair (HDR) pathway. Due to the nature of the MAP11 mutation (truncating), we aimed to increase knockout probability by using the NHEJ and HDR mechanisms in parallel: injecting the sgRNA and Cas9 protein alone (for NHEJ) or by adding a ssODN sequence encoding a stop cassette (for HDR). The ssODN was designed to contain stop codons in all three reading frames. The stop cassette contains two 20-base homology arms flanking the predicted Cas9-mediated breaking point. These homology arms surround the stop cassette sequence. The stop cassette sequence used: Stop Cassette of sgRNA1: 5′-TTACCAAACATTGTCCACCAGTCATGGCGTTTAAACCTTAATTAAGCTGTTAGGCAGCACTGATCCATCTGGC-3′; Stop Cassette of sgRNA2: 5′-ACGCACACACACTTCCTCTTGTCATGGCGTTTAAACCTTAATTAAGCTGTTGTAGGCGAGCTCCCGTTTAACACT-3′.
CRISPR/Cas 9 zebrafish mutants
To generate CRISPR/Cas9-mediated zebrafish knockouts, we directly injected the sgRNA with recombinant Cas9 protein (kindly provided by Prof. Gil Levkowitz, Weizmann institute) with or without the ssODN stop cassette into one-cell stage zebrafish embryos. The injected solution contained 300 ng sgRNA, 600 ng Cas9 protein, 3 µM stop cassette and phenol red dye in a single mix. Embryos were grown for 3–4 months post injection and then genotyped by dorsal fin clipping, DNA extraction and PCR amplification of the targeted MAP11 homologue double strand break region. Amplicons were subcloned into pGEM®-T-Easy vectors and Sanger-sequenced. Selected knockout (KO) F0 zebrafish were grown and bred with wild-type zebrafish to generate non-chimeric F1 KO heterozygotes. Heterozygote F1 offspring were then bred and F2 offspring underwent double-blinded head to body surface area measurements at day 28 post-fertilization (prior to genotyping), using Discovery. V12 microscope (Zeiss) and ImageJ software. Post analysis, F2 zebrafish were sacrificed and amplification of the targeted MAP11 homologue double strand break region was done using tissue direct PCR amplification kit (Terra™ PCR Direct Polymerase Mix; Clontech). As the two mutations in our zebrafish lines abrogated a BbvI restriction site, amplicon digestion with BbvI restriction enzyme (New England Biolabs) followed by gel electrophoresis was performed to screen for mutant, wild-type and heterozygous carriers of the two different map11 KO mutations. Primers used for mutation screenings: F: 5′-AAACCTTCAGAACCTA AGGACTGTT-3′; R: 5′-TTTGCGATAGTGAATTTGCTG-3′.
Zebrafish whole mount cell death detection by TUNEL assay
Terminal deoxynucleotidyl transferase (TdT) nick-end labelling (TUNEL) assay was performed for map11-KO line according to manufacturer’s instructions (Roche; 12156792910). Briefly, chorions of 24 h-post-fertilization (hpf) zebrafish embryos were removed using gentle forceps and fixed with 4% PFA over-night at 4°C. Embryos were washed three times for 10 min each in PBST (0.5% Triton™ X-100 in PBS) for permeabilization and blocked for 1 h at room temperature in blocking solution [0.5% Triton™ X-100, 10% foetal calf serum (FCS), 1% dimethyl sulphoxide (DMSO) in PBS]. Embryos were then labelled using TUNEL reaction mixture (labelling solution with terminal transferase enzyme). Embryos served as positive controls were incubated with 5 U/ml of recombinant DNase I in 50 mM Tris-HCl, pH 7.5, 1 mg/ml bovine serum albumin) for 10 min at room temperature prior to labelling. Embryos served as negative control were incubated without terminal transferase enzyme. After incubation, embryos were washed in PBST and visualized with fluorescent Discovery.V12 microscope (Zeiss). Image analysis was done using ImageJ. Fluorescent intensity was measured by integrated density of whole embryos to calculate relative apoptosis.
Zebrafish whole mount proliferation assay
Zebrafish proliferation was measured by anti-phospho-Histone H3 (PH3) staining. Briefly, chorions of 24 hpf zebrafish embryos were removed using gentle forceps and fixed with 4% PFA overnight at 4°C. Embryos were then washed twice in PBS and incubated for 7 min in −20°C acetone. Acetone was removed and embryos were washed in double-distilled water followed by two washes in PBST (0.1% Tween in PBS) for 5 min. Embryos were blocked for 2 h at room temperature in blocking solution (10% FCS, 1% DMSO in PBST). After blocking, embryos were incubated in blocking solution with a 1:750 dilution of rabbit p-Histone H3 antibody (Cell Signaling Technology; #9701) overnight at 4°C. Post antibody incubation, embryos were washed 4 × 5 min in PBST and incubated for 2 h at room temperature with a 1:300 dilution of donkey anti-rabbit HRP conjugated antibody (Abcam; ab97085). Embryos were washed 4 × 15 min in PBST and incubated for 10 min in 3,3′-diaminobenzidine (DAB) solution (D4418; Sigma-Aldrich). After incubation, embryos were washed three times in PBST. Images were captured by Discovery.V12 microscope (Zeiss) and analyzed for proliferation capacity by measuring pixel integrated density of zebrafish heads using ImageJ software.
Data availability
The raw data that support the findings of this study are available from the corresponding author, upon reasonable request.
Results
Disease phenotype
Three affected individuals of consanguineous Bedouin Israeli kindred presented with apparently autosomal recessive primary microcephaly without dysmorphic features (Fig. 1A, B and Supplementary Fig. 3). All affected individuals were born at term with normal birth weights following uneventful pregnancies. Occipital-frontal circumferences were below −2 SD at birth, with microcephaly progressing later in life (Table 1). Global developmental delay leading to intellectual disability was evident (with no regression), most notably affecting verbal and expressive skills, capable of expressing only three words at the ages of 3–4 years. All patients were incontinent at 4 years of age and had severe attention deficit hyperactivity disorder (ADHD). Ophthalmological and hearing tests were normal, as were karyotypes, chromosomal microarrays and biochemical and metabolic work-up. In all patients, except for microcephaly, brain MRI demonstrated no gross abnormal findings aside from reduction of white matter and hypoplasia (without dysplasia) of corpus callosum, similar to apple core corpus callosal abnormalities (Hanna et al., 2011). One patient (Patient II:3) had also tethered spinal cord and lipoma of filum terminale (Fig. 1B and Table 1). Patients were within normal height and weight compared to age and sex-matched control population at birth. Later in life (age 18–31 months), patients presented some delay in growth (height ≤3rd centile; Table 1) but reached normal height by the age of 4 years (data not shown). Weight was within normal range throughout development, except for Patient II:3 at the age of 18 months (7.6 kg; <3rd centile) and 30 months (Table 1); however, that patient’s weight also normalized by the age of 44 months (14 kg; 25th centile). Unaffected siblings (Subjects II:6 and II:7) had normal occipital-frontal circumference at birth (35 cm, >25th centile and 34 cm, ≥15th centile, respectively) as well as later on in life (data not shown).
Table 1.
Clinical characteristics of patients
| MAP11 genomic mutation coordinates | NC_000007.13:g.99755280G>T | ||
| MAP11 mutation cDNA nomenclature | NM_018275.4; c.613G>T | ||
| MAP11 mutation protein nomenclature | NP_060745.3; p.(E205*) | ||
| Patient ID | II:3 | II:4 | II:5 |
| Sex | Female | Male | Male |
| Present age, years | 6 | 7 | 5 |
| Birth weight, g | 2940 | 3080 | 2599 |
| Weight, kg / age measured, months / centile | 10.8 / 30 / 3rd | 12 / 31 / ≥10th | 10.8 / 18 / 25th |
| Head circumference at birth, cm / centile | 32 / < 5th | 32.5 / ≤ 5th | 32.5 / ≤ 5th |
| Head circumference, cm / age measured, months / SD below mean | 39 / 18 / −6 SD | 42 / 30 / −6 SD | 41 / 23 / −5 SD |
| Height, cm / age measured, months / centile | 72 / 18 /< 3rd | 84 / 31 / 3rd | 80 / 25 /< 3rd |
| Primary microcephaly | + | + | + |
| Developmental delay | + | + | + |
| Intellectual disability | + | + | + |
| MRI | Microcephaly; thin corpus callosum | Microcephaly; thin corpus callosum | Microcephaly; thin corpus callosum |
| Other findings | Tethered spinal cord; lipoma of filum terminale | − | − |
Patient IDs correspond with pedigrees in Fig. 1. + = positive; − = negative.
Genetic studies
Genome-wide homozygosity mapping, testing all individuals of the kindred studied (Fig. 1A), identified two homozygous segments shared by affected individuals: a ∼5.9 Mb homozygous segment on chromosome 5 between SNPs rs930875 and rs6894609 and a ∼12.4 Mb homozygous segment on chromosome 7 between SNPs rs17165823 and rs2215815 (Fig. 1C). Whole-exome sequencing (WES) data of Individual II:4 (Fig. 1A) were filtered for normal variants as described in the materials and methods section. Exons and their boundaries not well covered (<10 reads) in the WES data underwent Sanger sequencing. Following the filtering described above, no homozygous variants were found within the homozygous chromosome 5 locus (Fig. 1C). However, within the 12.4 Mb homozygous chromosome 7 locus, we identified a single homozygous deleterious mutation, c.613G>T, p.(E205*) in chromosome 7 open reading frame 43 (C7orf43, now termed MAP11) as the only likely disease-causing mutation: The MAP11 mutation, validated by Sanger sequencing, segregated in the kindred as expected for recessive heredity (Fig. 1D). Screening of 300 ethnically matched controls identified no carriers and no homozygous mutants (data not shown). The mutation has not been previously reported in the genome Aggregation Database (gnomAD; http://gnomad.broadinstitute.org/) (Lek et al., 2016). Only eight MAP11 (C7orf43) loss-of-function variants have been reported in the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/), none of which in a homozygous state. The MAP11 mutation is located within transcript variant 1 (NM_018275.4) that encodes a 580 amino acid protein which is highly conserved in all vertebrates (Supplementary Fig. 4). MAP11 is ubiquitously expressed, with high levels in brain tissues (Supplementary Fig. 5) yet has no identifiable predicted domains. Per RefSeq database, an additional transcript variant of MAP11 (NM_001303470.1), encoding a 311 amino acid protein, lacks the mutated exon. The expression levels of this transcript within the human brain and the predicted effect of the mutation in its 5′ UTR is unknown. RT-qPCR analysis of affected and unaffected lymphocytes showed that the c.613G>T mutation is not likely to trigger nonsense mediated mRNA decay of the MAP11 truncated transcript (Supplementary Fig. 6). Moreover, western blot analysis of HEK293T cells overexpressing the mutated form of MAP11 fused to a FLAG sequence, presented minor levels of the MAP11 truncated protein at the predicted molecular weight, suggesting that an aberrant form of MAP11 is being generated (Supplementary Fig. 7). Thus, the MAP11, c.613G>T mutation is predicted to cause a premature stop codon of the mature encoded protein after 205 aa, generating an aberrant protein by deleting almost two thirds of its C’-terminus (Supplementary Figs 4, 6 and 7).
MAP11 tissue expression
Analysis of MAP11 expression in various normal human tissues by RT-PCR demonstrated that it is ubiquitous and highly transcribed in all tissues examined with elevated transcription in brain, cerebellum, testis and whole blood. This MAP11 expression pattern is in line with GTEx consortium data (Lonsdale et al., 2013) (Supplementary Fig. 5).
MAP11 localization and interaction
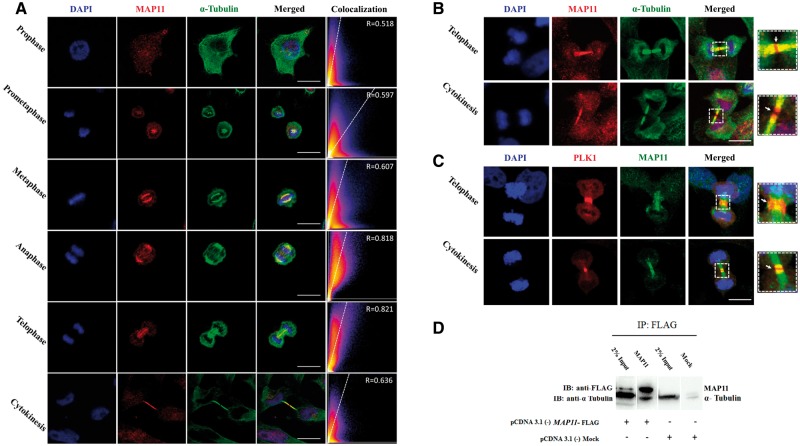
To determine the intra-cellular localization of MAP11, we initially visualized the endogenous MAP11 protein in SH-SY5Y cells. We found that MAP11 is localized to undetermined cytoskeletal associated vesicles. Further investigation of dividing cells revealed that MAP11 is enriched in mitosis, especially during early metaphase, anaphase and telophase (Fig. 2A). Co-staining of MAP11 and α-tubulin showed co-localization of both, mainly during anaphase and telophase (Fig. 2A). In telophase cytokinesis, we found that α-tubulin precedes the microtubule gap formation at the midbody leading to cell abscission while MAP11 is still continuous and undisrupted (Fig. 2B), suggesting that MAP11 has a role in cytokinesis and cell abscission. Immunostainings of endogenous MAP11 and PLK1 (a key regulator of cytokinesis, pivotal in midbody assembly and furrow cleavage formation) (Barr et al., 2004; Archambault and Glover, 2009; Hu et al., 2012; Hasegawa et al., 2013) revealed intriguing cellular dynamics of MAP11 during telophase-cytokinesis stages (Fig. 2C). MAP11 is surrounded by PLK1 at the spindle midzone during telophase with partial co-localization (Fig. 2C). During cytokinesis, PLK1 shifts to the centre of the cell midbody and is required for the formation and maintenance of the midbody. Moreover, PLK1 is known to be required for formation and ingression of the cleavage furrow and plays a role in cellular abscission. During abscission, MAP11 and PLK1 appeared to co-localize at the edges of each daughter cell microtubule extension post abscission (Fig. 2C). In contrast, MAP11 did not co-localize with PLK1 during metaphase, and was not seen at the centrosomes at any stage during mitosis (data not shown). Co-immunoprecipitation analysis demonstrated that MAP11 physically associates with α-tubulin and that α-tubulin could be co-precipitated with MAP11 in SH-SY5Y overexpressing lysates (Fig. 2D). Thus, MAP11 is associated with microtubule spindles through α-tubulin interaction in mitosis and is co-localized with PLK1 during cytokinesis and cell abscission.
Figure 2.
MAP11 is localized to mitotic spindles and interacts with α-tubulin. (A) Immunofluorescence microscopy of MAP11 in SH-SY5Y cells. Confocal imaging showing endogenous MAP11 co-localizing with α-tubulin during mitosis. Scale bar = 20 µm. Co-localization is highly significant in anaphase and telophase. Graphs in the right panel show Pearson’s correlation coefficient score (denoted by R) between MAP11 and α-tubulin for each image (0 < R < 1; A higher R-score indicates better co-localization between MAP11 and α-tubulin). (B) Immunofluorescence microscopy of MAP11 in SH-SY5Y cells focusing on telophase and cytokinesis. At early cytokinesis, α-tubulin precedes MAP11 in the gap formation followed by cell abscission at the midbody. At late cytokinesis, MAP11 fluorescence projections are sticking out of the microtubule α-tubulin. Right: Enlarged image of the region of interest. White arrow marks the midbody and cell abscission point. Scale bar = 20 µm. (C) MAP11 is co-localized with PLK1 at the edges of each daughter cell’s microtubule extension post cytokinesis abscission. Right: Enlarged image of the region of interest; white arrow marks the midbody and cell abscission point. Scale bar = 20 µm. (D) Co-immunoprecipitation of MAP11 and α-tubulin proteins from SH-SY5Y cells lysates overexpressing MAP11 fused to a FLAG sequence. Immunoprecipitation of cell lysates was performed using anti-FLAG magnetic beads. Immunoblotting was done using primary anti-FLAG and anti-α-tubulin antibodies followed by appropriate secondary HRP-conjugated antibodies.
MAP11 knock-down and proliferation assay
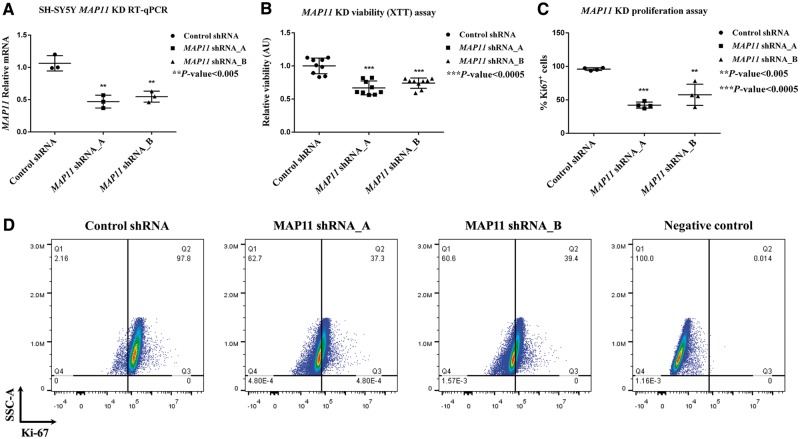
To characterize MAP11 function, we silenced its expression in SH-SY5Y cells. The efficacy of lentiviral shRNA-mediated MAP11 silencing was measured by RT-qPCR: MAP11 mRNA levels were silenced to ∼40% for shRNA_A and to 58% for shRNA_B. Both were statistically significant (P < 0.005 for mRNA levels of shRNA_A; P < 0.05 for mRNA levels of shRNA_B) (Fig. 3A). Cell viability assay (XTT) of MAP11-silenced SH-SY5Y cells showed statistically significant (P < 0.0005) lower viability for both shRNA_A and shRNA_B (Fig. 3B) compared to shRNA control cells. We speculated that this might be explained by either increase of apoptosis or decrease of proliferation capacity of MAP11 knock-down cells. Indeed, proliferation assays demonstrated that MAP11 silenced cells were less proliferative (measured by ratios of Ki-67 positive cells in FACS analysis) compared to shRNA control cells (Fig. 3C and D). In contrast, no marked differences in apoptosis rates (Annexin V assay) were identified between silenced and control cells (Supplementary Fig. 8A–C).
Figure 3.
Cell viability and proliferation capacity of MAP11 knock-down SH-SY5Y cells. (A) RT-qPCR results of MAP11, showing the degree of lentiviral-mediated silencing of SH-SY5Y cells. (B) Viability assay, demonstrating low viability of MAP11 knock-down cells compared to shRNA controls. (C) A graph summarizing proliferation assay (measured by Ki-67 positive cells via FACS), showing reduced proliferation of MAP11-silenced cells. (D) Fluorescent activated cell sorting results of MAP11 knock-down cells compared to shRNA control and negative controls.
Zebrafish MAP11 orthologue gene knock-out using CRISPR/Cas9
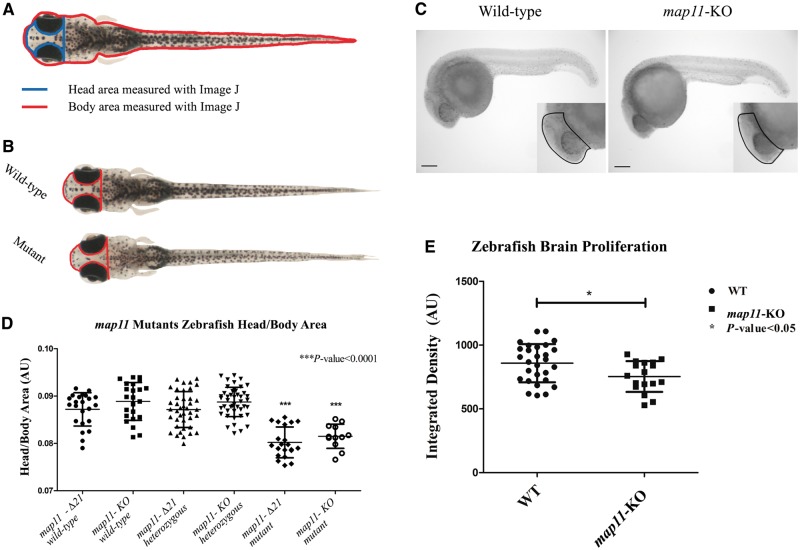
To determine the in vivo function of MAP11, we generated zebrafish knockout model of MAP11 homologous gene using CRISPR/Cas9. We injected sgRNA and Cas9 protein, with or without the ssODN oligo as a template for HDR, into one-cell-stage embryos (F0) that yielded many mutant lines (∼10% of injected embryos presented a genetic variation; data not shown). At 3–4 months post injection, the genetic editing of the MAP11 orthologue gene was validated by genomic DNA extraction, amplicon subcloning into pGEM®- T Easy plasmid and Sanger sequencing. One knockout line (map11-KO) and one mutant (map11-Δ21) line were selected for further analysis. The map11-KO line is a result of HDR (injected with ssODN stop cassette), presenting a 34-nucleotide insertion of the ssODN stop cassette and a 2 nucleotide deletion at the 5′ end of the insertion point, causing frameshift and a premature stop codon at position 308 of the MAP11 orthologue (Supplementary Fig. 9A and C). The map11-Δ21 line is a result of NHEJ repair (injected without ssODN), presenting a 22-nucleotide insertion mutation followed by a single nucleotide deletion at the 5′ end of the insertion point, causing an in-frame seven-amino-acid insertion mutation in the zebrafish MAP11 orthologue (Supplementary Fig. 9B and D). Notably, in the map11-KO line, genomic editing via CRISPR/Cas9 generated additional truncating codons downstream of the mutations reported above (data not shown). By breeding F1 heterozygotes, we generated F2 zebrafish offspring that showed normal Mendelian inherent ratios in both map11-KO and map11-Δ21 lines (data not shown). Next, we measured head to body area ratios at 28 days post-fertilization as described in the ‘Materials and methods’ section (Fig. 4A); while head to body ratios of F2map11-KO and map11-Δ21 heterozygotes were similar to those of control wild-types, head to body ratios of both homozygous mutants were significantly lower than those of wild-type or heterozygous controls (Fig. 4). Notably, map11-KO head to body ratios were slightly higher than those of map11-Δ21. These ratios were maintained also in wild-types and heterozygous carriers and likely stem from the genetic nature of their founders. Thus, the smaller head size observed in the two knockout zebrafish lines recapitulates the autosomal recessive inheritance and the human microcephaly phenotype (Fig. 3C). We also noticed that some degree of the observed smaller head size might be attributed to an anterior-posterior axis shortening of zebrafish map11 mutants (Supplementary Fig. 10). We investigated brain cell proliferation of map11-KO zebrafish embryos further, and found that knockout embryos had decreased brain cell proliferation rate at 24 hpf (measured by phosphohistone-H3 staining) (Fig. 4D and E) but with no change in total cellular apoptosis (measured by TUNEL assay, Supplementary Fig. 11), when compare to wild-type embryos. Thus, map11 loss-of-function likely reduces neuronal proliferation and leads to smaller head size in the two mutated zebrafish lines, recapitulating the human microcephaly phenotype (Fig. 4).
Figure 4.
CRISPR/Cas9-mediated MAP11-orthologue zebrafish mutants recapitulate the human microcephaly phenotype. (A) Head to body area measurements of CRISPR/Cas9 MAP11 orthologue mutants. Zebrafish head and body surface area were measured at age 28 days post fertilization using ImageJ software. Head to body area ratios are plotted. (B) Wild-type and homozygous map11-Δ21 mutant zebrafish (28 days post fertilization) demonstrating same body area surface. A red line highlights the head surface area of each larva showing smaller head of map11-Δ21 mutants compared to wild-types. (C) Images of 24 hpf zebrafish embryos stained for phospho-Histone H3. Images of map11-KO embryos show decreased proliferating cells within brain regions compared to wild-types (black dots). Scale bar = 0.5 mm. (D) Head-to-body ratios of map11-KO and map11-Δ21homozygous mutants. P-values represent comparison of each mutant (map11-Δ21 and map11-KO) to its corresponding wild-type control. P-values were calculated using an unpaired two-sided student t-test. (E) A graph showing greater proliferation capacity of wild-type (n = 28) compared to map11- KO (n = 17) zebrafish brains (measured by integrated density of the region of interest).
Discussion
The 508-amino-acid mature encoded MAP11 protein is highly conserved in all vertebrates (Supplementary Fig. 4). It has no identifiable domains and there are no publications regarding its function. Analysis of the expression pattern of MAP11 in various normal human tissues by RT-PCR demonstrated that it is ubiquitous, yet highly transcribed in brain, cerebellum, testis and whole blood, in line with GTEx portal data (Supplementary Fig. 5A and B).
The autosomal recessive primary microcephaly presented by our patients is characterized by global developmental delay, severe intellectual disability with speech impairment, ADHD and no dysmorphic features (Fig. 1A, B and Table 1). Aside from the microcephaly, reduced white matter and thin corpus callosum (Fig. 1B), no structural abnormalities were evident in brain MRI of affected individuals. One patient (Patient II:3) had tethered spinal cord and lipoma of filum terminale, a finding that is not necessarily associated with the MAP11 mutation, as the consanguinity of the parents might have caused another, unrelated, phenotype. Through a combination of homozygosity mapping and whole exome sequencing, we showed that this neurodevelopmental disorder is caused by a truncating mutation in MAP11 (Fig. 1C and D). RT-qPCR analysis of patient-derived lymphocytes, indicates that the mutated MAP11 transcript is not being degraded through nonsense mediated mRNA decay (Supplementary Fig. 6). Furthermore, western blot analysis of HEK293T overexpressing a mutant MAP11 fused to a FLAG sequence demonstrated that a truncated MAP11 protein is being generated (Supplementary Fig. 7), though the function of this truncated protein is yet to be elucidated. Immunofluorescence analysis of SH-SY5Y cells demonstrated that MAP11 is enriched during mitosis and localized to mitotic spindles especially during early metaphase, anaphase, telophase and cytokinesis (Fig. 2A). Co-immunostaining of MAP11 and α-tubulin, demonstrated remarkable co-localization as could be appreciated by the Pearson’s correlation coefficient scores (Fig. 2A). Further confocal and co-immunoprecipitation analyses showed that MAP11 is a microtubule-associated protein (MAP) that associates with α-tubulin at the cell midzone, and is likely involved in spindle microtubule dynamics and regulation during mitosis (Fig. 2A–C). Whether the interaction of MAP11 and α-tubulin is direct or mediated by other proteins is yet to be elucidated. Interestingly, MAP11 expression precedes α-tubulin in the gap formation of cell abscission at the midbody and is co-localized with PLK1 at the edges of each daughter cell’s microtubule extension post cytokinesis abscission (Fig. 2B and C). This suggests that MAP11 plays a role in microtubule and spindle dynamics during mitosis and might also be involved in the regulation of cell abscission during cytokinesis. This is not surprising, as many other microcephaly-causing genes are involved in mitotic spindle organization, cleavage furrow and midbody regulation (Woods et al., 2005; Thornton and Woods, 2009; Faheem et al., 2015; Ito and Goshima, 2015; Moawia et al., 2017; Létard et al., 2018). As primary microcephaly is usually a result of disturbance in cell-cycle regulation and progression of neural progenitor lineages or neural stem cells, we hypothesized that MAP11 might be involved in cell survival. Indeed, through shRNA-mediated silencing of MAP11 in SH-SY5Y cells, we showed that cells with reduced MAP11 expression have lower viability rates (XTT assay) compared to controls (Fig. 3A and B). We speculated that this could stem from either increase in apoptosis or decrease in proliferation capacity of MAP11 knock-down cells. By fluorescent activated cell sorting (FACS) analysis we demonstrated that the difference shown in the viability assay is due to lower proliferation rates rather than apoptosis (Fig. 3B–D, Supplementary Figs 2 and 8). To validate our genetic studies further, we conducted an in vivo zebrafish experiment; using CRISPR/Cas9 genome editing, we generated two zebrafish MAP11 knockout lines. By analysing head to body area ratios, we demonstrated that homozygous mutant zebrafish had smaller head size compared to heterozygous carriers and wild-types. Thus, the zebrafish mutant phenotype recapitulated the human microcephaly phenotype. In addition to an overall head size decrease, we also found shortening of the anterior-posterior axis of mutant zebrafish heads (Supplementary Fig. 10). In accordance to the proliferation and apoptosis data of MAP11 SH-SY5Y knock-down cells, we found no difference in cellular apoptosis levels (whole body) between wild-type and map11-KO zebrafish (Supplementary Fig. 11). Interestingly, we demonstrate that map11-KO zebrafish have decreased brain cellular proliferation compared to wild-types, indicating that map11 is important for neuronal proliferation at early zebrafish neurodevelopment. This is in-line with one of the key molecular features underlying primary microcephaly: low proliferation capacity of neural stem cells causing a depleted progenitor pool that is unable to give rise to sufficient number of neurons to populate the growing neocortex (Fish et al., 2008; Thornton and Woods, 2009; Gilmore and Walsh, 2013; Paridaen and Huttner, 2014; Kadir et al., 2016).
Overall, through in vitro and in vivo studies, we demonstrate that the orphan protein C7orf43, now termed MAP11, associates with mitotic spindles, and is both co-localized to and physically associates with α-tubulin during mitosis, affecting cell proliferation. We demonstrate that MAP11 expression precedes that of α-tubulin in the gap formation of cell abscission at the midbody and is co-localized with PLK1 at the edges of each daughter cell’s microtubule extensions post cytokinesis abscission. Our findings suggest a likely role of MAP11 in spindle dynamics during mitosis and in regulation of cell abscission in cytokinesis. Finally, we show that similar to other genes involved in mitotic spindle organization, cleavage furrow and midbody regulation, MAP11 loss of function mutations cause microcephaly in both humans and zebrafish through reduction of cell proliferation during early neurodevelopment.
Funding
Funding for this research was provided by the Legacy Heritage Bio-Medical Program of the Israel Science Foundation (1798/16) awarded to O.S.B. and the Bi-National Science Foundation (2013288), NARSAD (22893), CURE epilepsy foundation, and Marie Curie integration grant (630849) awarded to R.Y.B. This study was also supported by the National Knowledge Center for Rare/Orphan Diseases sponsored by the Israel Ministry of Science, Technology and Space.
Competing interests
The authors report no competing interests.
Supplementary Material
Glossary
Abbreviations
- RT-qPCR
real-time quantitative polymerase chain reaction
- ssODN
single stranded oligo donor
References
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol 2009; 10: 265–75. [DOI] [PubMed] [Google Scholar]
- Barr FA, Silljé HHW, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol 2004; 5: 429–40. [DOI] [PubMed] [Google Scholar]
- Faheem M, Naseer MI, Rasool M, Chaudhary AG, Kumosani TA, Ilyas AM, et al. Molecular genetics of human primary microcephaly: an overview. BMC Med Genomics 2015; 8: S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Dehay C, Kennedy H, Huttner WB. Making bigger brains-the evolution of neural-progenitor-cell division. J Cell Sci 2008; 121: 2783–93. [DOI] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Ahkmetova L, Pauli A, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 2014; 9: e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore EC, Walsh CA. Genetic causes of microcephaly and lessons for neuronal development. Wiley Interdiscip Rev Dev Biol 2013; 2: 461–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna RM, Marsh SE, Swistun D, Al-Gazali L, Zaki MS, Abdel-Salam GM, et al. Distinguishing 3 classes of corpus callosal abnormalities in consanguineous families. Neurology 2011; 76: 373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H, Hyodo T, Asano E, Ito S, Maeda M, Kuribayashi H, et al. The role of PLK1-phosphorylated SVIL in myosin II activation and cytokinetic furrowing. J Cell Sci 2013; 126: 3627–37. [DOI] [PubMed] [Google Scholar]
- Hu C-K, Coughlin M, Mitchison TJ. Midbody assembly and its regulation during cytokinesis. Mol Biol Cell 2012; 23: 1024–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Goshima G. Microcephaly protein Asp focuses the minus ends of spindle microtubules at the pole and within the spindle. J Cell Biol 2015; 211: 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadir R, Harel T, Markus B, Perez Y, Bakhrat A, Cohen I, et al. ALFY-controlled DVL3 autophagy regulates Wnt signaling, determining human brain size. PLoS Genet 2016; 12: e1005919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci 2006; 7: 883–90. [DOI] [PubMed] [Google Scholar]
- Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res 2016; 44: W272–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23: 2947–48. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Létard P, Drunat S, Vial Y, Duerinckx S, Ernault A, Amram D, et al. Autosomal recessive primary microcephaly due to ASPM mutations: an update. Hum Mutat 2018; 39: 319–32. [DOI] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The genotype-tissue expression (GTEx) project. Nat Genet 2013; 45: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moawia A, Shaheen R, Rasool S, Waseem SS, Ewida N, Budde B, et al. Mutations of KIF14 cause primary microcephaly by impairing cytokinesis. Ann Neurol 2017; 82: 562–77. [DOI] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 2014; 42: 401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem B, Di Lullo E, Kriegstein A. oRGs and mitotic somal translocation-a role in development and disease. Curr Opin Neurobiol 2017;42:61–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen JT, Huttner WB. Neurogenesis during development of the vertebrate central nervous system. EMBO Rep 2014;15:351–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Y, Gradstein L, Flusser H, Markus B, Cohen I, Langer Y, et al. Isolated foveal hypoplasia with secondary nystagmus and low vision is associated with a homozygous SLC38A8 mutation. Eur J Hum Genet 2014; 22: 703–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Y, Kadir R, Volodarsky M, Noyman I, Flusser H, Shorer Z, et al. UNC80 mutation causes a syndrome of hypotonia, severe intellectual disability, dyskinesia and dysmorphism, similar to that caused by mutations in its interacting cation channel NALCN. J Med Genet 2016; 53: 397–402. [DOI] [PubMed] [Google Scholar]
- Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper - An interactive approach to homozygosity mapping. Nucleic Acids Res 2009; 37: 593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohayeb B, Lim NR, Ho U, Xu Z, Dottori M, Quinn L, et al. The role of WD40-repeat protein 62 (MCPH2) in brain growth: diverse molecular and cellular mechanisms required for cortical development. Mol Neurobiol 2017; 62: 1–16. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NST, et al. Human Gene Mutation Database (HGMD): 2003 Update. Hum Mutat 2003; 21: 577–81. [DOI] [PubMed] [Google Scholar]
- Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends Genet. 2009; 25: 501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CG, Bond J, Enard W. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet 2005; 76: 717–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CG, Parker A. Investigating microcephaly. Arch Dis Child 2013; 98: 707–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data that support the findings of this study are available from the corresponding author, upon reasonable request.