Zheng et al. show that levels of histone methyltransferases EHMT1/2, and of the histone methylation H3K9me2, are elevated in prefrontal cortex of a familial Alzheimer’s disease (FAD) mouse model and human postmortem tissue. Inhibition of EHMT1/2 rescues cognitive deficits by relieving H3K9me2-mediated repression of glutamate receptor transcription in FAD mice.
Keywords: Alzheimer’s disease, histone methylation, epigenetics, memory, glutamate receptor
Abstract
Epigenetic dysregulation, which leads to the alteration of gene expression in the brain, is suggested as one of the key pathophysiological bases of ageing and neurodegeneration. Here we found that, in the late-stage familial Alzheimer’s disease (FAD) mouse model, repressive histone H3 dimethylation at lysine 9 (H3K9me2) and euchromatic histone methyltransferases EHMT1 and EHMT2 were significantly elevated in the prefrontal cortex, a key cognitive region affected in Alzheimer’s disease. Elevated levels of H3K9me2 were also detected in the prefrontal cortex region of post-mortem tissues from human patients with Alzheimer’s disease. Concomitantly, H3K9me2 at glutamate receptors was increased in prefrontal cortex of aged FAD mice, which was linked to the diminished transcription, expression and function of AMPA and NMDA receptors. Treatment of FAD mice with specific EHMT1/2 inhibitors reversed histone hyper-methylation and led to the recovery of glutamate receptor expression and excitatory synaptic function in prefrontal cortex and hippocampus. Chromatin immunoprecipitation-sequencing (ChIP-seq) data indicated that FAD mice exhibited genome-wide increase of H3K9me2 enrichment at genes involved in neuronal signalling (including glutamate receptors), which was reversed by EHMT1/2 inhibition. Moreover, the impaired recognition memory, working memory, and spatial memory in aged FAD mice were rescued by the treatment with EHMT1/2 inhibitors. These results suggest that disrupted epigenetic regulation of glutamate receptor transcription underlies the synaptic and cognitive deficits in Alzheimer’s disease, and targeting histone methylation enzymes may represent a novel therapeutic strategy for this prevalent neurodegenerative disorder.
Introduction
Alzheimer’s disease, a devastating neurodegenerative disorder, afflicts a large portion of the ageing population. Despite the identification of a few Alzheimer’s disease-associated genetic risk factors, a genetic basis for the majority of this disease has not emerged, calling for the need to find alternative strategies to probe the underpinnings of Alzheimer’s disease. Recent evidence has suggested that epigenetic dysregulation, which leads to brain region-specific changes of gene expression, is one of the key pathophysiological bases of ageing and neurodegeneration (Mehler, 2008; Mitchell et al., 2014; Woldemichael et al., 2014; Lardenoije et al., 2015). However, the importance of epigenetic changes in Alzheimer’s disease from the perspectives of pathogenesis, biomarkers and potential treatment awaits to be elucidated (Millan, 2014). To complement the ongoing epigenome-wide association studies of neurodegeneration, more mechanistic investigations are required for understanding the causal role of epigenetic processes in Alzheimer’s disease (Lunnon and Mill, 2013).
One epigenetic way to control gene expression is through chromatin remodelling, a process that dynamically modifies chromatin architecture to allow or prevent transcription machinery proteins to access condensed genomic DNA (Li et al., 2007; Borrelli et al., 2008). A key process to alter chromatin architecture is the addition and removal of methyl groups to histones controlled by a plethora of histone methyltransferases and demethylases, respectively (Greer and Shi, 2012). Lysine (K) residues on histone proteins can be mono-, di- or trimethylated by specific histone modifying enzymes, which is associated with gene activation or repression (Barski et al., 2007; Heintzman et al., 2007).
Animal studies have implicated histone methylation in cognition. For example, trimethylation of histone H3 at lysine 4 (H3K4me3, a mark associated with gene activation) and the H3K4-specific histone methyltransferase MLL2 are required for proper long-term memory formation (Gupta et al., 2010; Kerimoglu et al., 2013). Mutations in MLL2 have been linked to Kabuki syndrome, a disorder that is accompanied by cognitive impairment (Ng et al., 2010). Dimethylation of histone H3 at lysine 9 (H3K9me2, a mark associated with gene repression) and the H3K9-specific histone methyltransferase EHMT1/2 have been linked to memory consolidation (Gupta-Agarwal et al., 2012). Postnatal neuron-specific deficiency of EHMT1/2 leads to the defects in learning and environmental adaptation (Schaefer et al., 2009), similar to key symptoms of the human 9q34 deletion syndrome (also known as Kleefstra syndrome), which is caused by haploinsufficiency of EHMT1 (Kleefstra et al., 2006, 2012). While global acquisition or loss of histone methylation marks and the ensuing alteration of many genes are implicated in ageing and cognitive functioning (Greer and Shi, 2012), the role of the diverse array of histone methylation events in Alzheimer’s disease has not been identified (Fischer, 2014; Lardenoije et al., 2015).
In this study, we sought to reveal the role of histone methylation in Alzheimer’s disease pathophysiology. We focused our studies on prefrontal cortex (PFC), a command centre for high-level ‘executive’ functions, such as working memory, planning and attention, since the significant PFC hypofunction has been observed in patients with Alzheimer’s disease (Garrido et al., 2002; Rémy et al., 2005; Maillet and Rajah, 2013). Transgenic mice carrying five familial Alzheimer’s disease (FAD) mutations on human amyloid precursor protein (APP) and human presenilin 1 (PS1, encoded by PSEN1) develop a spectrum of severe Alzheimer’s disease-like pathologies by age 5–6 months,, including amyloid plaques, synapse degeneration, learning and memory deficits (Oakley et al., 2006; Zhao et al., 2007; Chen et al., 2012; Seo et al., 2014), as well as the significant impairment of basal synaptic transmission and long-term potentiation in hippocampus and cortex (Kimura and Ohno, 2009; Chen et al., 2012; Crouzin et al., 2013; Seo et al., 2014), so these late-stage animals were used as the FAD model. Results from this study will not only define disease-specific epigenetic signatures, but also identify corresponding therapeutic strategies for Alzheimer’s disease.
Materials and methods
Animals and human post-mortem tissues
All experiments were performed with the approval of State University of New York at Buffalo Animal Care Committee. The transgenic mice carrying five familial Alzheimer’s disease mutations on human amyloid precursor protein (K670N/M671L+I716V+V717I) and human presenilin 1 (M146L+L286V), 5×FAD, were a generous gift from Dr. William E. Van Nostrand (Stony Brook University). The transgenic mice expressing the mutant (P301S) human T34 tau isoform (1N4R) were obtained from Jackson Laboratory. Genotyping were performed by PCR according to the manufacturer’s protocol (Oakley et al., 2006; Yoshiyama et al., 2007). Frozen human post-mortem tissues (Brodmann area 10) from patients with Alzheimer’s disease and normal control subjects were provided by NIH NeuroBioBank.
Western blots
Total proteins were detected by western blotting as described previously (Gu et al., 2009; Yuen et al., 2012; Deng et al., 2016). In brief, PFC was punched from brain slices (300 µm) incubated in oxygenated artificial CSF. The tissue was homogenized with 1% SDS containing proteinase inhibitor (complete, Roche) and 1 mM phenylmethylsulphonyl fluoride (PMSF), followed by centrifugation at 4°C for 10 min at 16 000 g. The supernatant was collected and denatured by boiling in 4× SDS loading buffer for 5 min. Equal amounts of proteins were subjected to 7.5% or 15% SDS-polyacrylamide gels and transferred to nitrocellulose membranes. The blots were blocked with 5% non-fat milk for 1 h at room temperature, followed by incubation overnight with the following primary antibodies: EHMT1 (Millipore, 1:500, 09878), EHMT2 (Abcam, 1:1000, ab180815), H3 (Cell Signaling, 1:500, CST4499s), GluA2 (NeuroMeb, 1:1000, 75-002), NR1 (Cell Signaling, 1:500, 5704), NR2A (Upstate, 1:1000, 07-632), NR2B (Upstate, 1:1000, 06-600), H3K9me2 (Abcam, 1:1000, ab1220), H3K27me3 (Cell Signaling, 1:500, C36B11), PSD95 (Abcam, 1:1000, ab2723), synaptophysin (Millipore, 1:2000, MAB332), MAP2 (Santa Cruz, 1:250, SC80012), tubulin (Sigma, 1:10 000, T8578), actin (Santa Cruz, 1:2000, SC1616r), tau (Thermo Fisher, 1:1000, AHB0042) and phosphorylated tau (Thermo Scientific, 1:1000, MN1020). After the incubation with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare), blots were exposed to enhanced chemiluminescence substrate (Thermo Scientific) and scanned with Bio-Rad imaging system. The densitometric value of immunoblots was measured with ImageJ software (NIH).
Real-time RT-PCR
Prefrontal cortex was dissected quickly from the mouse brain, followed by liquid nitrogen grinding, and cold TRIzol® reagent (Invitrogen) extracting to get total RNA. Then, the cDNA was obtained using iScript™ cDNA Synthesis Kit (Bio-Rad) from the tissue mRNA and real-time PCR was performed using iQ™ SYBR® Green Supermix (Bio-Rad CFX96 Touch system) according to the manufacturer’s instruction. Fold changes in the target gene expression in FAD mice was determined by normalized CT value [2−Δ(ΔCT), ΔCT = CTtarget − CTGAPDH, and Δ(ΔCT) = ΔCTFAD − ΔCTWT]; Gapdh was used as housekeeping gene; CT = threshold cycle, which is defined as the cycle number getting detectable fluorescence signal. A PCR mixture of 20 µl per well (96-well thin-wall PCR plate, Bio-Rad) was amplified according to the following cycling condition: 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 60 s. Quantitative real-time PCR was performed in double reactions and repeated at least three times. The sequences of primers were as follows: Ehmt1: 5′- CATAGCAAAAGCAGACA CAA-3′; 5′-ACTTTCCAAGGTTTCCTTTC-3′; Ehmt2: 5′-GC TCCACCTGTCTACATCAT-3′; 5′-GCAGATGTTTTCCTCA TTGT-3′; GluA2 (Gria2): 5′-AGCCTATGAGATCTGGAT GT-3′, 5′-GAGAGAGATCTTGGCGAAAT-3′; NR1 (Grin1): 5′-CATCGGACTTCAGCTAATCA-3′, 5′-GTCCCCATCCT CATTGAATT-3′; NR2A (Grin2a): 5′-GGCTACAGAGACTTCATCAG-3′, 5′-ATCCAGAAGAAATCGTAGCC-3′; NR2B (Grin2b): 5′-TTAACAACTCCGTACCTGTG-3’, 5’-TGGAACTTCTTGTCACTCAG-3′; Gapdh: 5′-GACAACTCACTCAAGATTGTCAG-3′, 5′-ATGGCATGGACTGTGGTCATGAG-3′.
Quantitative chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed as we previously described (Wei et al., 2016; Qin et al., 2018). In brief, PFC slices (1.5 mm diameter, eight punches per mouse) were cross-linked with 1% formaldehyde for 12 min at room temperature. Glycine (125 mM) was added to quench the cross-link, followed by two washes with cold PBS-containing proteinase inhibitors and PMSF. Then SDS lysis buffer containing proteinase inhibitors and PMSF was used to homogenize the tissue. The chromatin fragments (∼200–500 bp in length) were obtained by sonication (Fisher Scientific Sonic Dismembrator Model 300) at 28% power with 12 rounds of 15 s pulse (30 s pause between pulses) at 4°C while the samples were immersed in an ice-water bath. After centrifugation at 16 000 g for 10 min at 4°C, sheared chromatin was collected and incubated with antibody against H3K9me2 (1:100, Abcam, ab1220) or RNA polymerase II (1:100, Millipore, 05-623) overnight at 4°C with gentle rotation, and mouse IgG was used as the negative control. Immunocomplexes were precipitated by salmon sperm DNA-protein A agarose beads, followed by low-salt, high-salt, LiCl, and Tris-EDTA buffer sequential washes. The protein-DNA cross- linking was reversed by incubation with 4 µl (per 100 µl complex) 5 M NaCl at 65°C for 4 h with rotation, and DNA fragments were purified with phenol/chloroform/isoamyl alcohol (American Bioanalytical) followed by acid ethanol precipitation. Input DNA was recovered from 50 µl total sheared chromatin.
Both regular and real-time PCR were performed to amplify 100–200 bp fragments around the transcription start sites region of mouse Gria2/GluA2, Grin2b/NR2B and Gapdh. Primer sets were as follows: Gria2/GluA2, 5′-CATGCAC TTTGGGCCATCAG-3′, 5′-ATACCTGCGTGGCAAGTCTC-3′; Grin2b/NR2B, 5′- GAGAGGCACACGGTATTGCT-3′, 5′- TCCCGTTCCCTGAACTTTGT-3′; Gapdh, 5′- CTCA CGTCCCAACTCTCCAC-3′, 5′- GAATTGGAGGAGGCTCA GAGG-3′. Quantification of ChIP signals was calculated as per cent input. For regular PCR, AccuPrime™ GC-Rich DNA polymerase (Invitrogen, 12337-016) was used to amplify the target sequence. Cycling conditions were as follows: 95°C for 3 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 30 s, and final extension at 72°C for 10 min.
Electrophysiological recordings in slices
Standard whole-cell voltage-clamp recordings were used to measure synaptic currents in layer V medial PFC pyramidal neurons as we described previously (Yuen et al., 2012; Duffney et al., 2015; Wei et al., 2016). Mice were sacrificed after inhaling halothane (Sigma). Brains were immediately removed, iced and cut into 300 µm slices by a Vibratome (Leica VP1000S, Leica Microsystems Inc.). Slices were then incubated in artificial CSF (in mM: 130 NaCl, 26 NaHCO3, 3 KCl, 5 MgCl2, 1.25 NaH2PO4, 1 CaCl2, 10 glucose, pH 7.4, 300 mOsm) for 1–6 h at room temperature (20–21°C) bubbling with 95% O2, 5% CO2. Slices were positioned in a perfusion chamber attached to the fixed stage of an Olympus upright microscope and submerged in continuously flowing oxygenated artificial CSF. Bicuculline (10 µM) and CNQX (25 µM) were added in N-methyl d-aspartate receptor (NMDAR) excitatory postsynaptic current (EPSC) recordings. Bicuculline and D-APV (25 µM) were added in AMPA receptor (AMPAR) EPSC recordings.
Patch electrodes contained internal solution (in mM): 130 Cs-methanesulphonate, 10 CsCl, 4 NaCl, 10 HEPES, 1 MgCl2, 5 EGTA, 2.2 QX-314, 12 phosphocreatine, 5 MgATP, 0.2 Na2GTP, 0.1 leupeptin, pH 7.2–7.3 and 265–270 mOsm. Cells were visualized with a 40× water-immersion lens and illuminated with near infrared light and the image was detected with an infrared-sensitive CCD camera. A Multiclamp 700A amplifier was used for these recordings. Tight seals (2–10 GΩ) from visualized neurons were obtained by applying negative pressure. With additional suction, the membrane was disrupted into the whole-cell configuration. Evoked EPSCs were generated with a pulse from a stimulation isolation unit controlled by a S48 pulse generator (Astro Med). A bipolar stimulating electrode (FHC) was placed ∼100 µm from the neuron under recording. Membrane potential was maintained at −70 mV for AMPAR-EPSC recordings. For NMDAR-EPSC, the cell (clamped at −70 mV) was depolarized to +60 mV for 3 s before stimulation to fully relieve the voltage-dependent Mg2+ block. To obtain the input-output responses, EPSC was elicited by a series of pulses with different stimulation intensities delivered at 0.033 Hz. To minimize experimental variations between cells, the following criteria were used: (i) layer V mPFC pyramidal neurons with comparable membrane capacitances were selected; (ii) stimulating electrode was positioned at the same location (layer VI, ∼100 µm horizontally) from the neuron under recording, and the electrode tip was cleaned after every recording to allow precise stimulation capacity; and (iii) recordings from wild-type versus FAD mice were interleaved throughout the course of all experiments. For paired-pulse ratios, AMPAR-EPSC was evoked by double pulses with various intervals. For voltage-dependent calcium current recordings, a modified artificial CSF (3 mM CaCl2, 20 mM CsCl, 0.5 µM tetrodotoxin) was used.
Hippocampal slices (400 µm) were collected and incubated in a modified artificial CSF with lower (1.2 mM) Mg2+ and higher (2 mM) Ca2+. Field excitatory postsynaptic potential (fEPSP) was recorded with a metallic electrode (FHC) from the stratum radiatum layer of CA1 area, and the Schaffer collateral afferents were stimulated with 100 μs test pulses via a bipolar cluster electrode (FHC). To evaluate basal synaptic transmission, we applied different stimulation strengths (10–150 μA) and plotted fEPSP slopes versus the stimulation intensities.
Lentivirus generation and delivery
The short hairpin (sh)RNA oligonucleotide targeting mouse Ehmt1 sequence (5′-TTATTCATCATCATCATA-GCG-3′, Open Biosystem) or Ehmt2 (5-′AAG AGCTATGAACTCTCTCGG-3′, Open Biosystem) was inserted to the lentiviral vector pLKO.3G (Addgene), which contains an eGFP marker. For the production of lentiviral particles, a mixture containing the pLKO.3G shRNA plasmid (against Ehmt1/2), psPAX2 packaging plasmid and pMD2.G envelope plasmid (Addgene) was transfected to HEK-293FT cells using Lipofectamine® 2000. The transfection reagent was removed 12–15 h later, and cells were incubated in fresh Dulbecco’s modified Eagle medium (DMEM) (containing 10% foetal bovine serum + penicillin/streptomycin) for 24 h. The media harvested from the cells, which contained lentiviral particles, was concentrated by centrifugation (2000g, 20 min) with Amicon Ultra Centrifugal Filter (Ultracel-100K, Millipore). In vivo delivery of the viral suspension (2 µl) was achieved by stereotaxic injection bilaterally into PFC (AP: 2.0 mm, ML: ±0.25 mm, DV: 2.0 mm) with a Hamilton syringe (needle gauge 31) as described previously (Yuen et al., 2012; Wei et al., 2016). After 10–14 days of viral expression, animals were subjected to behavioural, biochemical or electrophysiological studies. To test the specific knockdown effect, slice punches containing the viral infected regions were immunoblotted with anti-EHMT1 (Millipore, 1:500, 09878) or anti-EHMT2 (Abcam, 1:1000, ab180815).
Immunohistochemistry
Mice were anaesthetized and transcardially perfused with PBS, followed by 4% paraformaldehyde (PFA) prior to brain removal. Brains were stored overnight in 30% sucrose-containing PBS buffer at 4°C, then were cut into slices. Slices were transferred to PBS solution, and incubated in 0.3% H2O2 for 30 min. After three washes in PBS, slices were blocked for 30 min in PBS containing 10% normal goat serum.
For H3K9me2 and CaMKII staining, after washing, slices (30 µm) were incubated with the primary antibodies (anti-H3K9me2: Abcam, ab1220, 1:500; anti-CaMKII: Abcam, ab134041, 1:500) for 24 h at 4°C. After washing three times in PBS, slices were incubated with two secondary antibodies (Alexa Fluor® 488: Invitrogen, A-21202, 1:300; Alexa Fluor® 568: Invitrogen A10042, 1:300) for 1 h at room temperature, followed by three washes with phosphate-buffered saline (PBS). Slices were mounted on slides with VECTASHIELD mounting media (Vector Laboratories). Images were acquired using a 40× objective on a Zeiss LSM 510 Confocal Microscope. All specimens were imaged under identical conditions and analysed with identical parameters.
For the staining of amyloid-β plaques, after washing, slices (100 µm) were incubated with the purified antibody against β-Amyloid (1:1000, Biolegend, 803001) overnight at 4°C. After washing 3 times in PBS, slices were incubated with the secondary antibody (1:200, Vector Lab, BA-9200) for 40 min at 37°C. Following washing, slices were incubated with ABC mix (ABC Elite kit, Vector Lab) for 30 min at 37°C. Colour development was performed with DAB solution for ∼1 min, and the reaction was stopped with PBS. After dehydration, slices were mounted on slides with VECTASHIELD mounting media (Vector Lab). Images were acquired using a 20× objective on a Nikon microscope.
Chromatin immunoprecipitation-sequencing
Chromatin immunoprecipitation was performed with PFC samples from two wild-type mice, two FAD mice and two BIX01294-treated FAD mice. After cluster generation (cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS, Illumina), the library preparations were sequenced on an Illumina Hiseq platform and 50 bp single-end reads or 150 bp paired-end reads were generated. Raw data (raw reads) of fastq format were first processed through in-house Perl scripts. In this step, clean data (clean reads) were obtained by removing reads as follows: (i) remove reads with mean quality value <20; (ii) remove reads that contain <15% N bases; (iii) remove reads that align with the adapter sequence; and (iv) drop reads below 18 bases long after trimming. The remaining reads that passed all the filtering steps were counted as clean reads and all subsequent analyses were based on the clean reads. Then, the clean reads were aligned to the mouse genome (GRCm38/mm10) using BWA mem v 0.7.12.
After mapping reads to reference genome, we used the MACS2 version 2.1.0 (model-based analysis of ChIP-seq) peak finding algorithm to identify regions of immunoprecipitation enrichment over background. The position of peak summit around transcript start sites of genes was used to predict interaction sites of H3K9me2 and genes. Fold enrichment of H3K9me2 value was RPM (reads per 1 M bp) of ChIP normalized to RPM of Input. Genes with altered H3K9me2 enrichment at 5000 bp upstream and downstream of transcription start sites were determined by the value of Log2 fold-change of ChIP signal over corresponding input that was more than +0.2 (increase) or less than −0.2 (decrease), with the default FDR (false discovery rate) of 0.05. Enrichment analyses of genes were undertaken using gene sets derived from the Biological Process Ontology from Bioconductor.
Behavioural tests
Behavioural assessments were carried out by investigators blindly (with no prior knowledge of genotypes and treatments). In some experiments, heat maps illustrating the time of the test animal spent at different locations of the arena were generated by a computer running the ANY-maze behaviour tracking software (Stoelting, Wood Dale, IL).
Novel object recognition task was conducted with minor modifications (Bevins and Besheer, 2006). The first day of each experiment consisted of two to three habituation trials (3 min each, 5 min apart), during which mice explored the arena (44.5 cm in diameter) alone (no objects). Twenty-four hours later, the experimental trials began, which consisted of a familiarization phase (phase 1) and a test phase separated by a delay period. During the familiarization phase, mice were placed in the arena containing two copies of an object (A) and allowed to explore freely (3 min per trial). After a short (5 min) delay period, a test trial (3 min) was conducted. Mice were returned to the arena containing one of the original objects (‘familiar-A’) and a new, different object (‘novel-B’). All objects were made of plastic toys (height: ∼5 cm) with similar textures, colours, and sizes, but distinctive shapes. The objects were positioned in two adjacent corners (10 cm from the walls) counterbalanced. The arena and objects were cleaned between each trial with 70% alcohol to mask any olfactory cues. The room was illuminated by indirect white light. Exploration was defined by sniffing or touching the object with the nose. Sitting on the object was not considered exploration. Total exploration time of the familiar and novel objects was recorded and used to calculate a discrimination index [time spent with novel object (B) − time spent with familiar object (A)] / [total time exploring both objects (B + A)] for test sessions. This index was used to measure recognition memory.
T-maze delayed alternation task was used to test working memory as we previously described (Yuen et al., 2011, 2009). Mice were subjected to restricted diet and maintained at ∼85% of their original weight for 1–2 weeks. They were habituated to a T-maze until they voluntarily ate a sucrose pellet placed at the end of each arm. T-maze training began only after habituation had finished and when mice showed no visual signs of distress of being in the maze. On the first trial, animals were rewarded for entering either arm. Thereafter, for a total of 11 trials per session, animals were rewarded only if they entered the arm opposite to the one that was previously chosen. Between trials the choice point was wiped with alcohol to remove olfactory cues. In the initial one to two training sessions, the delay between trials started at 5 s, and was subsequently raised in 5-s intervals. In the later training sessions, the delay was fixed at 30 s, and animals were examined daily until establishing baseline performance of 60–70% correct for two consecutive days. The first trial was never included in assessing performance.
The Barnes maze test was used to examine spatial memory with modifications (Pompl et al., 1999). In brief, the animal was placed on a round platform with eight equally-spaced holes at the edge, one of which was attached with an escape box. A weak aversive stimulation, light, was applied to increase the motivation to escape from the circular platform. During the two learning phases (information acquisition), the animal was allowed to explore the platform until finding the correct hole using distal visual cues and entering the escape box. The interval between each learning phase was 5 min. After two learning sessions, the animal was placed in its home cage to rest for 15 min. In the memory phase (information retention and retrieval), the escape box was removed, and the animal was put back on the platform to explore for 5 min. The time spent on the correct hole (T1) and the other seven incorrect holes (T2) were counted. Spatial memory index was calculated by T1/T2.
Statistical analysis
Data analyses were performed with Clampfit (Molecular Devices, Sunnyvale, CA), Kaleidagraph (Synergy Software, Reading, PA) and GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA). All data are expressed as the mean ± standard error of the mean (SEM). For statistical significance, experiments with two groups were analysed using two-tailed Student’s t-tests (unpaired, unless otherwise stated). Experiments with more than two groups were subjected to one-way ANOVA, two-way ANOVA or two-way repeated measure ANOVA (rmANOVA), followed by post hoc Bonferroni tests for multiple comparisons.
Data availability
Genomic data will be deposited in a public repository. The access number and the dataset will be available for access.
Results
FAD mice exhibit significant elevation of repressive histone methylation H3K9me2 at glutamate receptor subunits
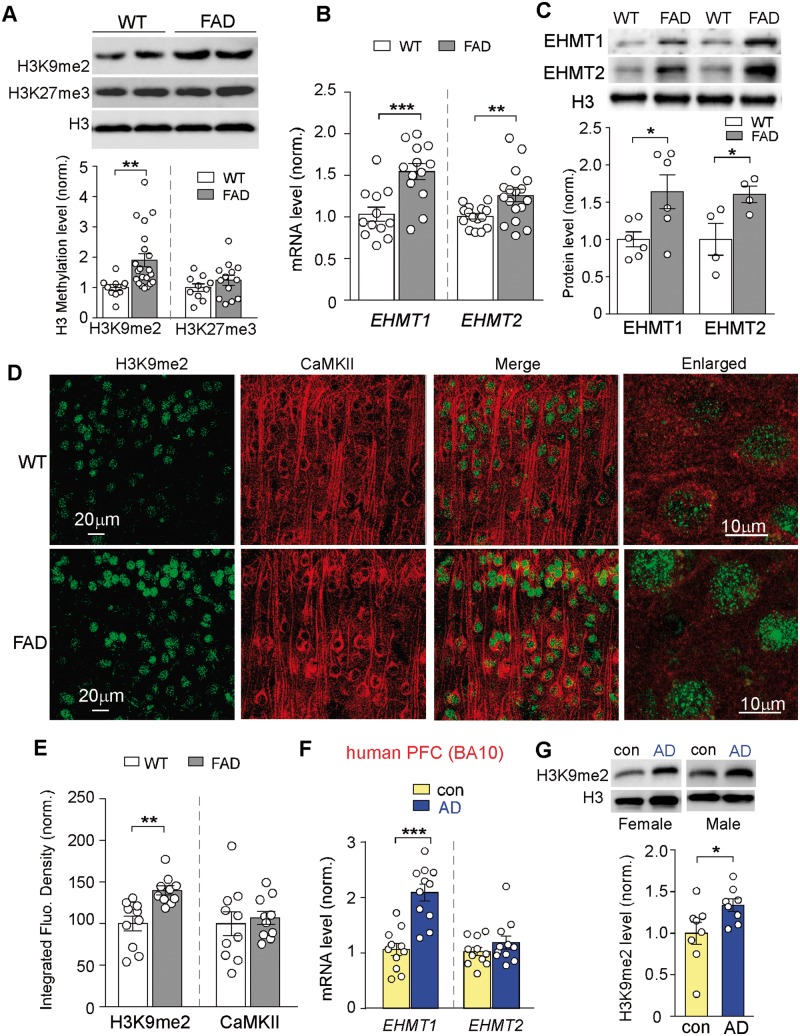
To reveal the role of histone methylation in Alzheimer’s disease pathophysiology, we examined global levels of histone methylation markers linked to transcriptional repression (H3K9me2, H3K27me3) in PFC slices of FAD mice (5–6 months old). Western blot assays of PFC lysates indicated that the level of H3K9me2 was significantly increased in aged FAD mice (Fig. 1A and Supplementary Fig. 1A; wild-type: 1.0 ± 0.09, n = 10, FAD: 1.91 ± 0.21, n = 21, P < 0.01, t-test), while levels of H3K27me3 were largely unaltered (wild-type: 1.0 ± 0.12, n = 10, FAD: 1.24 ± 0.16, n = 13, P > 0.05, t-test).
Figure 1.
Aged FAD mice exhibited the elevated histone methylation H3K9me2 and the upregulation of EHMT1 and EHMT2 in PFC, and human patients with Alzheimer’s disease also had the increased H3K9me2 in PFC. (A) Immunoblots and quantitative analysis of H3K9me2 and H3K27me3 levels in PFC lysates from wild-type versus 5xFAD mice (5–6 months old). **P < 0.01, t-test. (B) Quantitative real-time RT-PCR data on the mRNA level of Ehmt1 and Ehmt2 in PFC from wild-type versus FAD mice. **P < 0.01, ***P < 0.001, t-test. (C) Immunoblots and quantification analysis of protein levels of EHMT1 and EHMT2 in the nuclear fraction of PFC neurons from wild-type versus FAD mice. Nuclear protein levels were normalized to H3. *P < 0.05, t-test. (D) Confocal images of prefrontal cortical neurons immunostained with H3K9me2 and CaMKII in wild-type or 5xFAD mice. (E) Quantification of the intensity of H3K9me2 and CaMKII signals in PFC neurons from wild-type or 5xFAD mice. **P < 0.01, t-test. (F) Quantitative real-time RT-PCR data on the mRNA level of EHMT1 and EHMT2 in post-mortem prefrontal cortical tissue (Brodmann area 10) from human patients with Alzheimer’s disease versus age- and sex-matched control subjects. ***P < 0.001, t-test. (G) Immunoblots and quantitative analysis of H3K9me2 in the nuclear fraction of post-mortem PFC tissue from human patients with Alzheimer’s disease versus control subjects. *P < 0.05, t-test. Each set of the experiments was replicated between three and five times. Full immunoblots are in Supplementary Fig. 9.
To reveal the potential basis for the observed increase of H3K9me2 in aged FAD mice, we examined the expression of EHMT2 (G9a) and EHMT1 (GLP), the two histone methyltransferases that specifically catalyse H3K9 dimethylation. Quantitative PCR analyses indicated that the mRNA levels of Ehmt1 and Ehmt2 were significantly higher in PFC lysates from FAD mice (Fig. 1B; Ehmt1, wild-type: 1.0 ± 0.09, n = 12, FAD: 1.55 ± 0.10, n = 13, P < 0.001, t-test; Ehmt2, wild-type: 1.0 ± 0.03, n = 15; FAD: 1.26 ± 0.08, n = 17, P < 0.01, t-test). Protein levels of EHMT1 and EHMT2 in the nucleus fraction of PFC tissue samples were also significantly increased in FAD mice (Fig. 1C and Supplementary Fig. 1B; EHMT1, wild-type: 1.0 ± 0.10, n = 6, FAD: 1.64 ± 0.23, n = 6, P < 0.05, t-test; EHMT2, wild-type: 1.0 ± 0.21, n = 4, FAD: 1.61 ± 0.11, n = 4, P < 0.05, t-test). These data suggest that upregulation of EHMT1 and EHMT2 is responsible for the abnormally high H3K9 dimethylation in the Alzheimer’s disease mouse model.
Since western blotting lacks the cellular resolution, we further performed immunohistochemical staining of H3K9me2 and CaMKII (a marker of glutamatergic neurons) in PFC slices. As shown in Fig. 1D and E, the intensity of H3K9me2 signal in the nucleus of PFC pyramidal neurons was significantly increased in FAD mice (wild-type: 100 ± 8.9, n = 10 slices, FAD: 139.9 ± 5.4, n = 10 slices, P < 0.01, t-test), confirming that H3K9me2 abnormality occurs in PFC neurons.
To find out whether the levels of EHMT1/2 and H3K9me2 are also changed in patients with Alzheimer’s disease, we examined post-mortem human tissue from Brodmann area 10, which is the anterior-most portion of the prefrontal cortex in the human brain. Patients with Alzheimer’s disease (65–88 years old) were compared with age- and sex-matched control subjects (68–84 years old, Supplementary Table 1). As shown in Fig. 1F and Supplementary Fig. 1C, the mRNA level of EHMT1 in PFC of patients with Alzheimer’s disease was significantly higher, compared to normal controls (wild-type: 1.0 ± 0.11, n = 11, Alzheimer’s disease: 2.09 ± 0.15, n = 11, P < 0.001, t-test), while the mRNA level of EHMT2 was largely unchanged (wild-type: 1.0 ± 0.07, n = 11, Alzheimer’s disease: 1.19 ± 0.12, n = 11, P > 0.05, t-test). The level of H3K9me2 in the nuclear fraction of PFC lysates from patients with Alzheimer’s disease was also significantly elevated (Fig. 1G; control: 1.0 ± 0.13, n = 8, Alzheimer’s disease: 1.34 ± 0.08, n = 8, P < 0.05, t-test), consistent with results from Alzheimer’s disease mouse models.
To find out whether the alteration of histone methylation is an age-dependent phenomenon, we compared young FAD mice (3 months old) with age-matched wild-type mice. As shown in Supplementary Fig. 2A and B, the level of global H3K9me2, as well as mRNA levels of Ehmt1 and Ehmt2, were not elevated in young FAD mice, suggesting age dependence of this epigenetic change. Moreover, the levels of H3K9me2 in striatal neurons of aged FAD mice (5–6 months old) were largely unchanged compared to wild-type mice (Supplementary Fig. 2C and D), suggesting region specificity of this epigenetic change.
Next, we examined the level of H3K9me2 in another Alzheimer’s disease mouse model. Other than amyloid deposits, a hallmark of Alzheimer’s disease is the formation of neurofibrillary tangles formed by hyperphosphorylated microtubule-associated protein tau (Giannakopoulos et al., 2003; Forman et al., 2004). P301S tau gene mutation causes early-onset frontotemporal dementia (Sperfeld et al., 1999; Morris et al., 2001), thus, P301S Tau transgenic mice were developed to study the early progression of tauopathies. Synaptic dysfunction and cognitive deficits are manifested in P301S Tau transgenic mice at 5–6 months before neuron loss and neurofibrillary tangle formation (Yoshiyama et al., 2007; Takeuchi et al., 2011), so we used P301S Tau transgenic mice (6 months old) as another Alzheimer’s disease model. Immunohistochemical experiments indicated that the intensity of H3K9me2 signal in the nucleus of PFC pyramidal neurons was significantly higher in Tau transgenic mice, compared to wild-type mice (Supplementary Fig. 3A and B, P < 0.001, t-test). Western blot assays also demonstrated that the levels of H3K9me2 were significantly increased in PFC lysates of Tau transgenic mice (Supplementary Fig. 3C and D, P < 0.01, t-test). In contrast, immunohistochemical staining and immunoblotting did not find significant alterations of H3K9me2 in the hippocampus of Tau transgenic mice (Supplementary Fig. 4A–D). These results have confirmed the elevation of H3K9me2 in PFC of two different Alzheimer’s disease models.
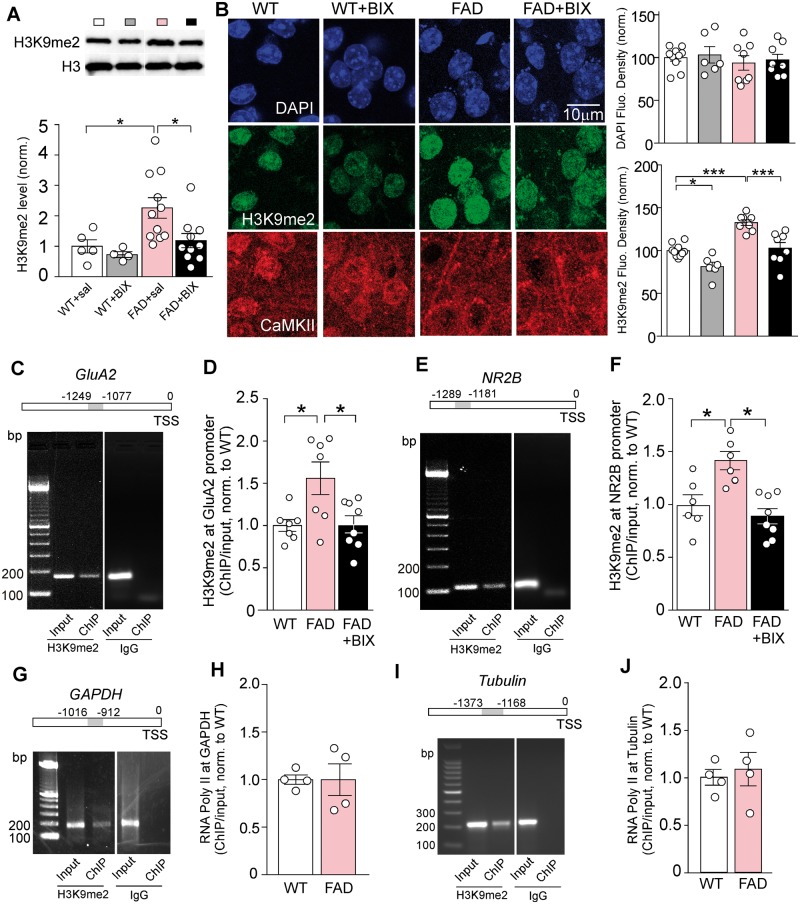
Next, we examined whether the aberrant histone methylation in aged FAD mice could be normalized by specific inhibition of EHMT1 and EHMT2. The selective EHMT1/2 inhibitor BIX01294 (Kubicek et al., 2007; Malmquist et al., 2012) was systemically administered [1 mg/kg, subcutaneously (s.c.) once daily for 3 days]. As shown in Fig. 2A, the elevated H3K9me2 level in PFC of FAD mice was significantly brought down by BIX01294 treatment [wild-type: 1.0 ± 0.21, n = 5, wild-type + BIX: 0.73 ± 0.10, n = 4, FAD + saline: 2.26 ± 0.32, n = 11, FAD + BIX: 1.19 ± 0.23, n = 10, F(1,26) (genotype) = 7.0, F(1,26) (treatment) = 4.0, P < 0.05, two-way ANOVA]. Immunohistochemical staining also indicated the significant reduction of H3K9me2 signal in the nucleus of PFC pyramidal neurons by BIX01294 treatment in FAD mice (Fig. 2B, wild-type: n = 10, wild-type + BIX: n = 6, FAD: n = 8, FAD + BIX: n = 8, F(1,28) (genotype) = 36.4, F(1,28) (treatment) = 29.1, P < 0.001, two-way ANOVA).
Figure 2.
The repressive histone methylation H3K9me2 at Gria2/GluA2 and Grin2b/NR2B gene promoters was significantly increased in aged FAD mice, which was reversed by the treatment with an EHMT1/2 inhibitor. (A) Immunoblots and quantitative analysis of the H3K9me2 level in PFC lysates from wild-type and FAD mice treated with the EHMT1/2 inhibitor BIX01294 (1 mg/kg, s.c. 3×) or saline control. *P < 0.05, two-way ANOVA. Full blots are in Supplementary Fig. 9. (B) Confocal images and quantification of the fluorescent signal intensity of prefrontal cortical neurons immunostained with DAPI, H3K9me2, and CaMKII in wild-type or 5xFAD mice without or with BIX01294 treatment. *P < 0.05, ***P < 0.001, ANOVA. (C and E) PCR images showing the input (total DNA) and ChIP (H3K9me2-occupied DNA) signals at the upstream transcription start sites region of Gria2/GluA2 (C) and Grin2b/NR2B (E). Mouse IgG was used as the negative control. Top: Diagram showing the location of primers for detecting H3K9me2 occupancy. TSS = transcriptional start site. (D and F) ChIP-qPCR data comparing the enrichment of H3K9me2 at Gria2/GluA2 (D) and Grin2b/NR2B (F) in PFC lysates from wild-type versus FAD mice (5–6 months old) without or with the treatment of BIX01294 (1 mg/kg, s.c. 3×). *P < 0.05, ANOVA. (G and I) PCR images showing the input (total DNA) and ChIP (RNA polymerase II-occupied DNA) signals at the upstream transcription start site region of Gapdh (G) and tubulin (I). Top: Diagram showing the location of primers for detecting RNA polymerase II occupancy. Mouse IgG was used as the negative control. (H and J) ChIP-qPCR data comparing the RNA polymerase II occupancy at Gapdh (H) and tubulin (J) in PFC lysates from wild-type versus FAD mice. Each set of the experiments was replicated between three and four times.
The elevated global histone methylation in aged FAD mice prompted us to examine H3K9me2 at promoter regions of target proteins that are critically involved in cognitive processes. Given the key role of glutamate receptor-mediated synaptic transmission in PFC-dependent functions, such as working memory (Goldman-Rakic, 1995; Popoli et al., 2011; Yuen et al., 2012), we speculated that AMPA and NMDA receptors were potentially altered in aged FAD mice due to aberrant epigenetic regulation. To test this, we designed primers against the proximal and immediate upstream transcription start sites regions of genes, and measured the occupancy of H3K9 dimethylation at these regions using ChIP assays. We found that H3K9me2 was detectable at the immediate ∼1200 bp upstream region of Gria2/GluA2 and Grin2b/NR2B genes (Fig. 2C and E), and IgG controls gave no ChIP signals. The enrichment of H3K9me2 at Gria2/GluA2 and Grin2b/NR2B was significantly increased in PFC of FAD mice (5–6 months old), which was reversed by BIX01294 (1 mg/kg, s.c., 3×) treatment [Fig. 2D and F; Gria2/GluA2: wild-type: 1.0 ± 0.07, n = 7, FAD: 1.56 ± 0.19, n = 7, FAD + BIX: 1.02 ± 0.10, n = 8, F(2,19) = 5.96, P < 0.01, one-way ANOVA; Grin2b/NR2B: wild-type: 1.0 ± 0.10, n = 6, FAD: 1.41 ± 0.09, n = 6, FAD + BIX: 0.89 ± 0.07, n = 8, F(2,17) = 10.2, P < 0.01, one-way ANOVA]. No significant differences between wild-type and FAD mice were observed in ChIP assays of RNA polymerase II occupancy in the upstream transcription start sites region of Gapdh (Fig. 2G and H; n = 4 wild-type, n = 4 FAD, P > 0.05, t-test) or tubulin (Fig. 2I and J; n = 4 wild-type, n = 4 FAD, P > 0.05, t-test), ruling out the possibility of global transcriptional suppression. In addition, the enrichment of H3K9me2 was undetectable at synaptophysin (Supplementary Fig. 5A and B), and was unaltered at Map2 in FAD mice (Supplementary Fig. 5C and D). These data indicate that the repressive histone methylation H3K9me2 at AMPA receptor Gria2/GluA2 and NMDAR receptor Grin2b/NR2B subunits is specifically elevated in the PFC of aged FAD mice.
FAD mice show loss of glutamate receptor transcription and expression that is rescued by EHMT1/2 inhibition
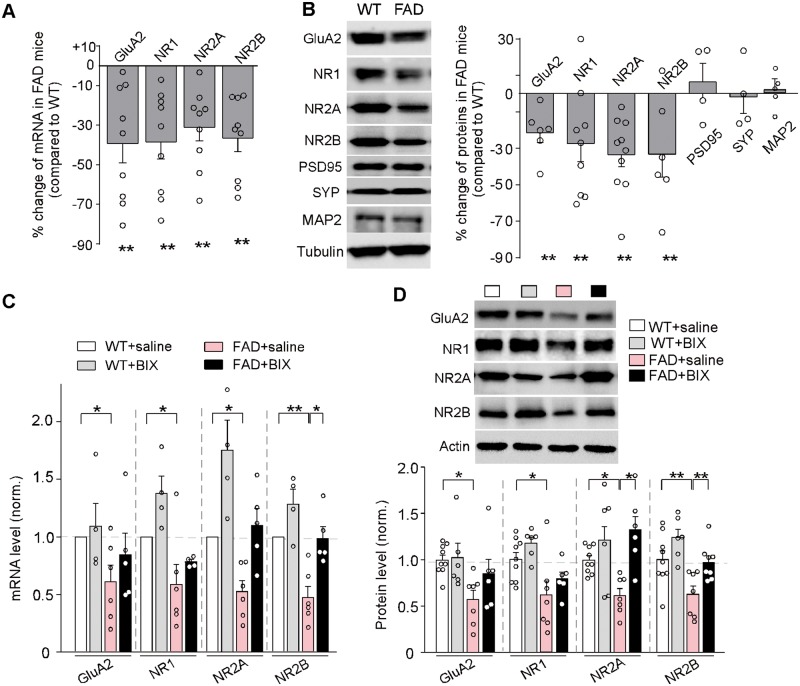
The hyperactive repressive histone methylation in FAD mice could lead to the transcriptional downregulation of glutamate receptor subunits. To test this, we performed quantitative PCR analyses to examine the mRNA levels of glutamate receptor subunits in the PFC. As shown in Fig. 3A, compared to wild-type mice, FAD mice (5–6 months old) exhibited significantly reduced mRNA levels of AMPA receptor and NMDA receptor subunits (Gria2/GluA2, 39% ± 9.8% decrease, NR1, 38 ± 8.7% decrease, Grin2a/NR2A: 31% ± 6.9% decrease, Grin2b/NR2B: 37% ± 6.9% decrease, n = 9 wild-type, n = 9 FAD, P < 0.01, t-test).
Figure 3.
Aged FAD mice exhibited the significant loss of glutamate receptor transcription and expression, which was rescued by the treatment with EHMT1/2 inhibitor. (A) Quantitative real-time RT-PCR data on the mRNA levels of Gria2/GluA2, Grin1/NR1, Grin2a/NR2A, and Grin2b/NR2B in PFC from wild-type versus FAD mice (5–6 months old). **P < 0.01, t-test, compared to wild-type. (B) Immunoblots and quantification analysis of the protein level of glutamate receptor subunits (GluA2, NR1, 2A, 2B), synaptic and dendritic markers (PSD-95, synaptophysin, MAP2), and cytoskeletons (tubulin, actin) in PFC from wild-type versus FAD mice. Protein levels were normalized to actin. **P < 0.01, t-test, compared to wild-type. (C) Quantitative real-time RT-PCR data on the mRNA level of Gria2/GluA2, Grin1/NR1, Grin2a/NR2A, and Grin2b/NR2B in PFC from wild-type and FAD mice treated with saline or BIX01294 (1 mg/kg, s.c. 3×). *P < 0.05, **P < 0.01, ANOVA. (D) Immunoblots and quantification analysis of the protein level of glutamate receptor subunits (GluA2, NR1,2A,2B) in PFC from wild-type and FAD mice treated with saline or BIX01294. *P < 0.05, **P < 0.01, ANOVA. Each set of the experiments was replicated between four and five times. Full immunoblots are in Supplementary Fig. 9.
Quantitative immunoblotting experiments were also performed to detect the total level of AMPAR and NMDAR subunits in PFC slices from FAD mice (5–6 months old). As shown in Fig. 3B, FAD mice exhibited a significant decrease in the amount of total GluA2 subunit (21.4% ± 5.5% decrease, n = 6 wild-type, n = 6 FAD, P < 0.01, t-test), as well as the level of total NR1, NR2A and NR2B subunits (NR1: 27.4% ± 9.0% decrease, P < 0.01, NR2A: 32.2% ± 6.1% decrease, P < 0.01; NR2B: 33.2% ± 12.7% decrease, P < 0.01, n = 6–10 pairs, t-test). No significant changes were found on the synaptic markers PSD-95 and synaptophysin, nor the dendritic marker MAP2, in FAD mice (n = 4-5 pairs, P > 0.05, t-test). These data indicate that the transcription and expression of glutamate receptors are significantly and specifically diminished in PFC of aged FAD mice.
To examine whether histone methyltransferase inhibitors could restore the mRNA and protein levels of AMPAR and NMDAR subunits in FAD mice, we injected the selective EHMT1/2 inhibitor BIX01294 (1 mg/kg, s.c., 3×) into FAD mice, and subsequently conducted quantitative PCR and immunoblotting analyses. As shown in Fig. 3C, in BIX01294-treated FAD mice, the mRNA levels of Gria2/GluA2, NR1, Grin2a/NR2A and Grin2b/NR2B in PFC were restored to that close to wild-type mice (Gria2/GluA2, 84% ± 19% of wild-type; NR1, 78% ± 2% of wild-type; Grin2a/NR2A, 110% ± 15% of wild-type; Grin2b/NR2B, 99% ± 10% of wild-type, n = 4–6 each group, two-way ANOVA). Western blots also showed a similar recovery of glutamate receptor proteins in PFC of BIX01294-treated FAD mice (Fig. 3D; GluA2, 85% ± 16% of wild-type; NR1, 79% ± 7% of wild-type; NR2A, 132% ± 14% of wild-type; NR2B, 97% ± 7% of wild-type, n = 6–9 each group, two-way ANOVA). These results indicate that EHMT1/2 inhibition is capable of restoring the transcription and expression of glutamate receptors in PFC of aged FAD mice.
EHMT1/2 inhibition or knockdown restores glutamate receptor function in FAD mice
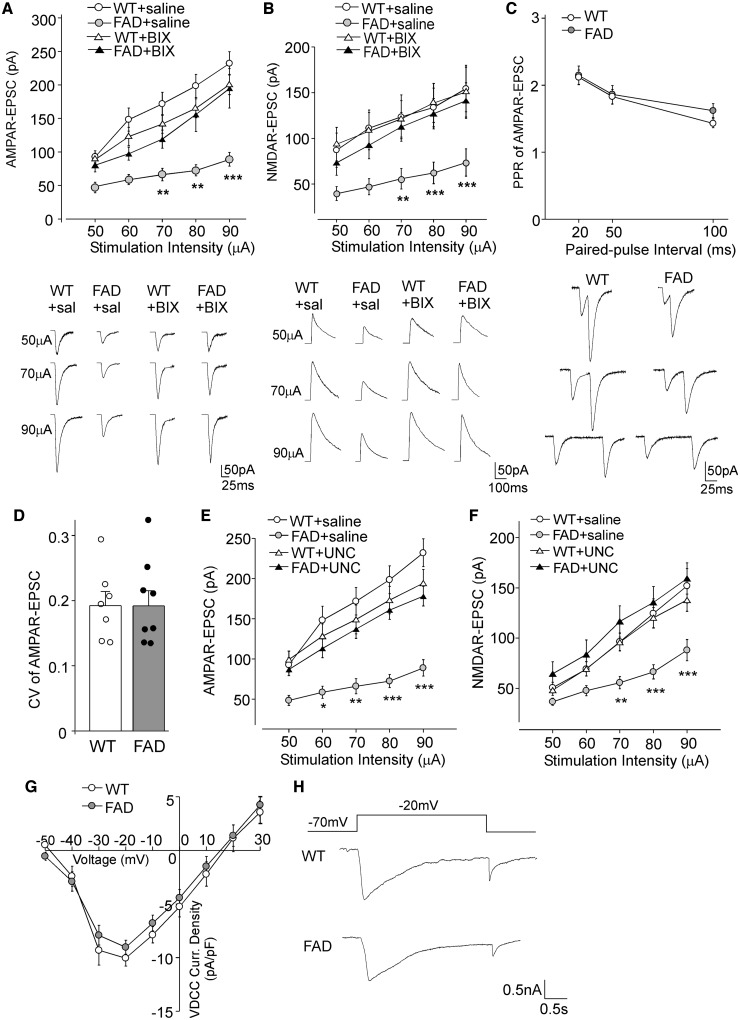
The loss of glutamate receptor transcription and expression in PFC of aged FAD mice could lead to diminished glutamatergic transmission. To test this, whole-cell patch-clamp recordings were performed to measure AMPAR- and NMDAR-mediated EPSC in PFC slices from age-matched wild-type versus FAD mice (5–6 months old). As shown in Fig. 4A and B, AMPAR-EPSC and NMDAR-EPSC induced by a series of stimulus intensities were markedly reduced in PFC pyramidal neurons from FAD mice, compared to wild-type mice (AMPA: 48–63% decrease, NMDA: 52–58% decrease, n = 10–14 per group, P < 0.001, two-way ANOVA). FAD mice injected with BIX01294 (1 mg/kg, s.c., 3×) exhibited a substantial increase of the input/output curves of AMPAR-EPSC and NMDAR-EPSC, compared to saline-injected FAD mice [AMPA: 65–118% increase, n = 10–13 per group, F(3,44) (group) = 10.5, P < 0.001, two-way rmANOVA; NMDA: 85–104% increase, n = 11-12 per group, F(3,39) (group) = 13.8, P < 0.001, two-way rmANOVA], bringing glutamatergic responses to the level of wild-type mice. The rescuing effect of BIX01294 disappeared at 7 days post-injection (AMPA: n = 11, NMDA: n = 11, P > 0.05, two-way ANOVA). Paired-pulse ratio (PPR) of AMPAR-EPSC and coefficient of variation (CV) of AMPAR-EPSC, two readouts for presynaptic changes, were not significantly altered in FAD mice [Fig. 4C and D; PPR: n = 12 per group, F(1,66) (group) = 0.46, P > 0.05, two-way ANOVA; CV: n = 7–8 per group, P > 0.05, t-test], pointing to a postsynaptic locus for the altered glutamatergic signalling.
Figure 4.
AMPAR- and NMDAR-mediated synaptic transmission was significantly diminished in FAD mice, which was restored by EHMT1/2 inhibitors. (A and B) Input-output curves (mean ± SEM) of AMPAR-EPSC (A) and NMDAR-EPSC (B) in response to a series of stimulation intensities in PFC pyramidal neurons from wild-type and FAD mice (5–6 months old) treated with BIX01294 (1 mg/kg, s.c. 3×) or saline control. **P < 0.01, ***P < 0.001, two-way rmANOVA. Bottom: Representative EPSC traces at different stimuli. (C) Paired-pulse ratio (PPR) of AMPAR-EPSC in PFC pyramidal neurons of wild-type (WT) versus FAD mice. Bottom: Representative traces of AMPAR-EPSC evoked by paired pulses at different intervals. (D) Bar graphs showing coefficient of variations (CV) of AMPAR-EPSC in PFC pyramidal neurons of wild-type versus FAD mice. (E and F) Input-output curves (mean ± SEM) of AMPAR-EPSC (E) and NMDAR-EPSC (F) in response to a series of stimulation intensities in PFC pyramidal neurons from wild-type and FAD mice (5–6 months old) treated with the EHMT1/2 inhibitor UNC0642 (1 mg/kg, i.p. 3×) or saline control. *P < 0.05, **P < 0.01, ***P < 0.001, two-way rmANOVA. (G) I-V curves of calcium currents through voltage-dependent calcium channels (VDCCs) in PFC pyramidal neurons from wild-type versus FAD mice. (H) Representative VDCC current traces. Each set of the experiments was replicated between four and six times.
To confirm the rescuing effect of histone methyltransferase inhibition, we applied another potent and selective EHMT1/2 inhibitor, UNC0642. UNC0642 exhibits >2000-fold to >20 000-fold selectivity for EHMT1/2 over other methyltransferases and displays modest brain penetration in vivo (Liu et al., 2013). As shown in Fig. 4E and F, after a brief administration of UNC0642 [1 mg/kg, intraperitoneally (i.p.) once daily for 3 days], FAD mice had a significant recovery of AMPAR- and NMDAR-mediated synaptic responses [AMPA: n = 11–13 per group, F(3,45) (group) = 12.5, P < 0.001, two-way rmANOVA; NMDA: n = 12–18 per group, F(3,56) (group) = 6.7, P < 0.001, two-way rmANOVA].
No significant change in voltage-dependent calcium channels was observed in PFC pyramidal neurons of FAD mice [Fig. 4G and H; n = 12 wild-type; n = 10 FAD, F(1,20) (group) = 0.32, P > 0.05, two-way rmANOVA], suggesting that the neuronal health state of FAD mice is generally intact, while AMPA and NMDA receptors show specific vulnerability.
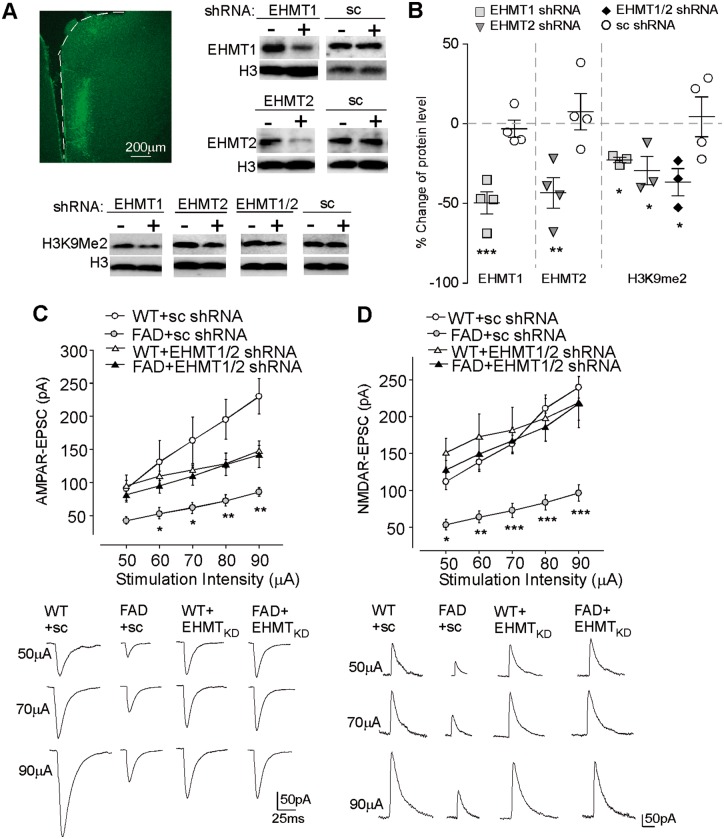
These pharmacological results have implicated EHMT1/2 in Alzheimer’s disease. Given the limited selectivity of pharmacological agents in inhibiting histone methyltransferases, we performed RNA interference-mediated knockdown of EHMT1 and EHMT2 to examine the rescue of synaptic functions in FAD mice. Lentiviral vectors containing shRNAs against EHMT1 and EHMT2 were generated and stereotaxically injected into PFC of FAD mice. As shown in Fig. 5A and B, Ehmt1 or Ehmt2 shRNA, but not the scrambled shRNA, caused a significant in vivo suppression of the expression of these histone methyltransferases (Ehmt1: 49.6% ± 7.0% knockdown, n = 4, P < 0.001, t-test, Ehmt2: 43.4% ± 9.5% knockdown, n = 4, P < 0.01, t-test). Moreover, the level of H3K9me2 was significantly inhibited by Ehmt1 and Ehmt2 shRNAs (22.7–36.7% reduction, n = 3, P < 0.05, t-test). FAD mice injected with Ehmt1 plus Ehmt2 shRNA lentiviruses exhibited the significant recovery of AMPAR-EPSC and NMDAR-EPSC, compared to FAD mice injected with the scrambled shRNA lentivirus [Fig. 5C and D; AMPA: n = 13–16 per group, F(3,55) (group) = 10.0, P < 0.001, NMDA: n = 7–10 per group, F(3,31) (group) = 10.1, P < 0.001, two-way rmANOVA]. Taken together, these data suggest that EHMT1/2 inhibition or knockdown is capable of rescuing glutamatergic synaptic function in FAD mice.
Figure 5.
Knockdown of EHMT1/2 attenuated the synaptic deficits in FAD mice. (A and B) Immunoblots and quantitative analysis of EHMT1, EHMT2, and H3K9me2 in PFC slices from mice with the stereotaxic injection of Ehmt1 shRNA, Ehmt2 shRNA or a scrambled shRNA lentivirus. *P < 0.05, **P < 0.01, ***P < 0.001, paired t-test, compared with PFC from the ipsilateral non-injection site (−). (A) Top left: Image shows the infected brain region (medial PFC). Full blots are in Supplementary Fig. 9. (C and D) Input-output curves (mean ± SEM) of AMPAR-EPSC (C) and NMDAR-EPSC (D) in PFC pyramidal neurons from wild-type and FAD mice injected with EHMT1 plus EHMT2 shRNA lentiviruses or a scrambled shRNA lentivirus. *P < 0.05, **P < 0.01, ***P < 0.001, two-way rmANOVA. Bottom: Representative EPSC traces at different stimuli. Each set of the experiments was replicated between three and four times.
In addition to the glutamate system, we examined the effect of EHMT1/2 inhibition on the pathological phenotypes that define Alzheimer’s disease, such as extracellular deposition of amyloid-β and intracellular accumulation of tau protein, in aged FAD mice (5–6 months old). As shown in Supplementary Fig. 6A and B, amyloid-β plaques were prominent in PFC of FAD mice, and BIX01294 (1 mg/kg, s.c., 3×) treatment failed to significantly reduce the density of amyloid-β plaques (FAD: 37.3 ± 1.4/cm2, n = 12 slices/4 mice; FAD + BIX: 34.0 ± 1.4/cm2, n = 12 slices/4 mice, P > 0.05, t-test). The level of hyper-phosphorylated tau was significantly elevated in PFC of aged FAD mice, but was not reversed by BIX01294 treatment (Supplementary Figs 6C and 3D; wild-type: 1.0 ± 0.15, n = 4, FAD: 2.90 ± 0.39, n = 6, FAD + BIX: 2.47 ± 0.40, n = 6, P < 0.05, one-way ANOVA). The level of total tau was unchanged. These results suggest that the rescuing effect of BIX01294 on synaptic function is independent of the typical Alzheimer’s disease pathology.
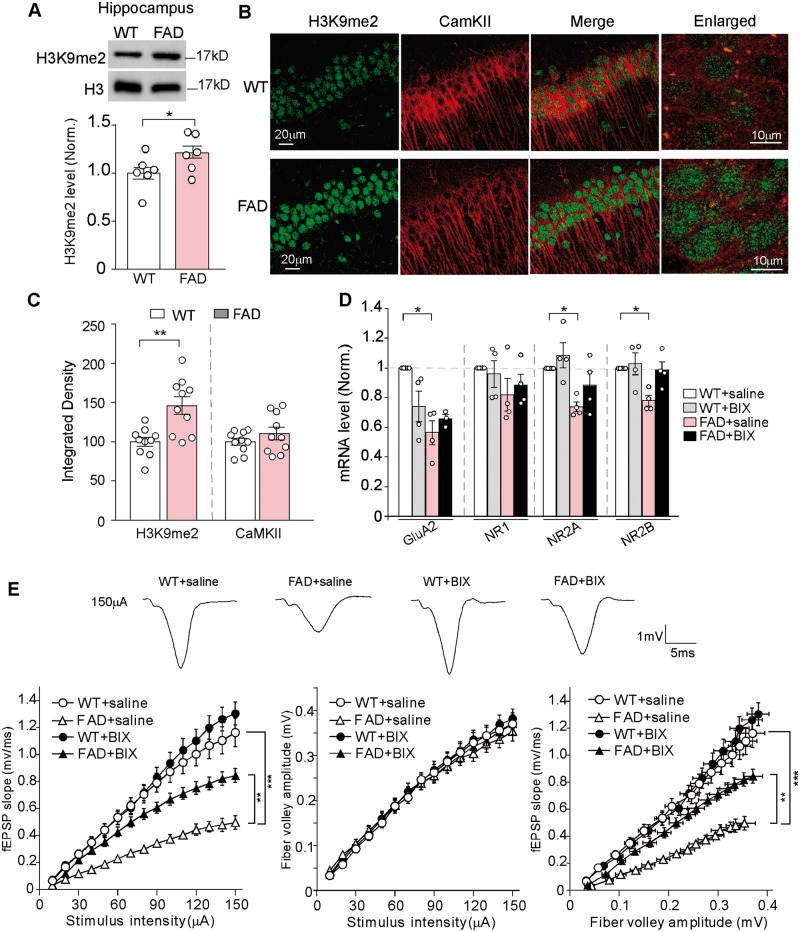
Besides PFC, another brain region that is significantly impaired in Alzheimer’s disease is the hippocampus. Therefore, we examined the changes in H3K9me2 and glutamate receptor subunits in the hippocampus of FAD mice (6 months old) without or with drug treatment. As shown in Fig. 6A, the level of hippocampal H3K9me2 was modestly but significantly increased (wild-type: 1.0 ± 0.06, n = 6, FAD: 1.22 ± 0.06, n = 6, P < 0.05, t-test) in FAD mice. Immunohistochemical staining of H3K9me2 and CaMKII (a marker of glutamatergic neurons) in hippocampal slices also indicated that the intensity of H3K9me2 signal in the nucleus of CA1 pyramidal neurons was significantly increased in FAD mice (Fig. 6B and C; wild-type: 100 ± 5.7, n = 10 slices, FAD: 146.1 ± 11.2, n = 10 slices, P < 0.01, t-test).
Figure 6.
Hippocampus of FAD mice showed the elevated H3K9me2 and the diminished glutamate receptor transcription and excitatory synaptic transmission, which was ameliorated by BIX01294 treatment. (A) Immunoblots and quantitative analysis of H3K9me2 levels in hippocampal lysates from wild-type (WT) versus 5xFAD mice (5–6 months old). *P < 0.05, t-test. Full blots are in Supplementary Fig. 9. (B) Confocal images of hippocampal CA1 neurons immunostained with H3K9me2 and CaMKII in wild-type or 5xFAD mice. (C) Quantification of the intensity of H3K9me2 and CaMKII signals in hippocampal neurons from wild-type or 5xFAD mice. **P < 0.01, t-test. (D) Quantitative real-time RT-PCR data on the mRNA level of Gria2/GluA2, Grin1/NR1, Grin2a/NR2A, and Grin2b/NR2B in hippocampus from wild-type and FAD mice treated with saline or BIX01294 (1 mg/kg, s.c. 3×). *P < 0.05, ANOVA, compared with wild-type + saline. (E) Input/output (I/O) curves relating the slope of fEPSP to the amplitude of presynaptic fibre volley at various stimulus intensities in the CA1 region of wild-type versus 5xFAD mice (6 months old) treated with saline or BIX01294. **P < 0.01, ***P < 0.001, ANOVA. Top: Representative fEPSP traces. Each set of the experiments was replicated three times.
Quantitative PCR experiments indicated that the mRNA levels of Gria2/GluA2, Grin2a/NR2A and Grin2b/NR2B were significantly diminished in the hippocampus of FAD mice, and were partially or fully restored by BIX01294 (1 mg/kg, s.c., 3×) treatment (Fig. 6D; FAD + saline, Gria2/GluA2: 43.7% ± 8.1% decrease, Grin2a/NR2A: 26.2% ± 3.3% decrease, Grin2b/NR2B: 21.9% ± 3.6% decrease; FAD + BIX, Gria2/GluA2: 65.6% ± 3.0% of wild-type, Grin2a/NR2A: 88.6% ± 9.0% of wild-type, Grin2b/NR2B: 98.7% ± 5.6% of wild-type, n = 4 each group, two-way ANOVA). Electrophysiological data also demonstrated that glutamatergic synaptic transmission was markedly diminished in the hippocampus of FAD mice (6 months old), as indicated by the reduced fEPSPs at Schaffer collateral-CA1 synapses in response to different stimulations, and BIX01294 treatment significantly elevated the synaptic strength [Fig. 6E, n = 18–20 each group, F(3,70) (group) = 19.8, P < 0.001, two-way rmANOVA]. Taken together, these data suggest that the hippocampus of FAD mice exhibit similar alteration of histone methylation and glutamate receptors as the PFC, which can be normalized by EHMT1/2 inhibition.
FAD mice exhibit genome-wide increase of H3K9me2 enrichment that is reversed by EHMT1/2 inhibition
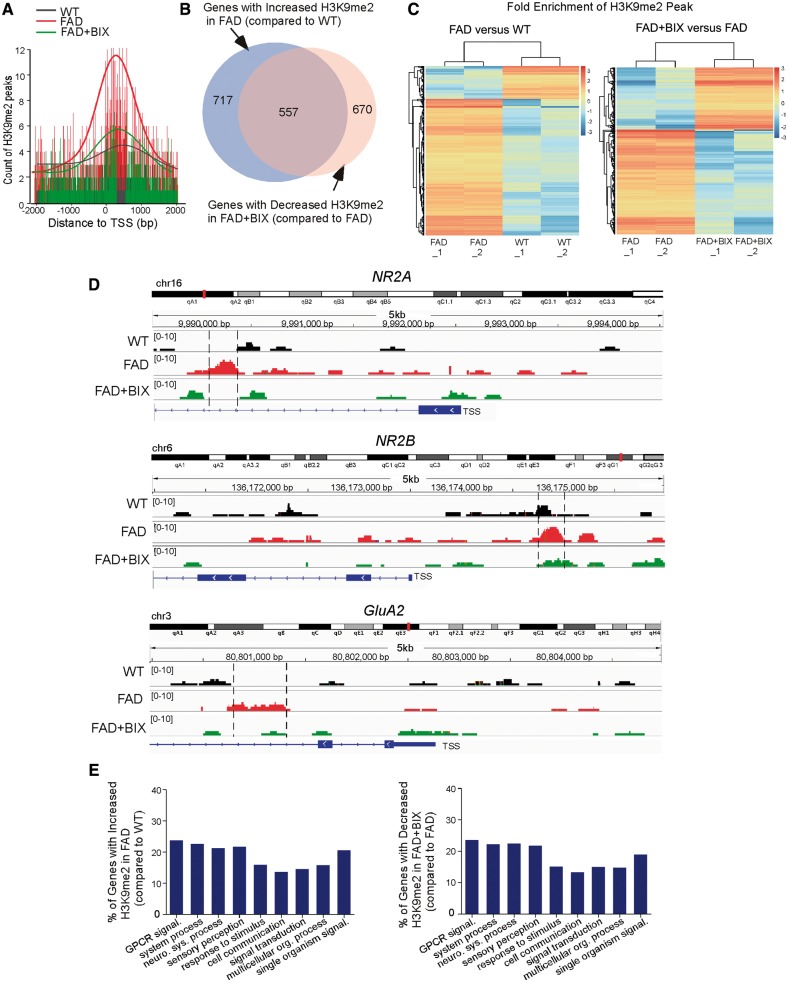
Finally, to determine whether the genome-wide aberration of histone methylation occurs in FAD mice and whether it can be normalized by EHMT1/2 inhibition, we performed ChIP-seq experiments to examine H3K9me2 occupancy using PFC samples from wild-type and FAD mice without or with BIX01294 treatment. Compared to wild-type mice, the DNA fragments occupied by H3K9me2 around the transcription start site across the genome were markedly increased in FAD mice, which was substantially brought down by BIX01294 treatment (Fig. 7A). About 717 genes showed the increased H3K9me2 enrichment in FAD mice, and BIX01294 treatment reduced H3K9me2 enrichment in the majority (77.7%) of these genes (557; Fig. 7B and Supplementary Table 2). From the hierarchical clustering analysis of H3K9me2 peaks (Fig. 7C), it was also evident that the majority of H3K9me2 peaks had increased enrichment in FAD mice (compared to wild-type mice), while many of the H3K9me2 peaks had decreased enrichment in BIX01294-treated FAD mice (compared to FAD mice). These data indicate that FAD mice exhibit a genome-wide increase of H3K9me2 occupancy around transcription start sites regions of multiple genes, which is largely reversed by BIX01294 treatment.
Figure 7.
FAD mice exhibited genome-wide increase of H3K9me2 enrichment at genes involved in neural signalling, which was reversed by EHMT1/2 inhibition. (A) Profile plot of H3K9me2-enriched DNA fragment reads around 2000 bp of transcription start site (TSS) across the genome in PFC samples from wild-type mice and FAD mice without or with BIX01294 treatment (1 mg/kg, s.c. 3×). The trend lines are the average of two samples in each group. The lower bars represent the counts of H3K9me2 at the location of the highest part of the peak. (B) Venn diagram illustrating the overlap of genes with increased H3K9me2 enrichment in FAD mice (compared to wild-type) and decreased H3K9me2 enrichment in BIX01294-treated FAD mice (compared to FAD). (C) Hierarchical clustering map showing the fold enrichment of H3K9me2 peaks in FAD versus wild-type mice, and FAD + BIX versus FAD mice. (D) ChIP-seq data showing the enrichment of H3K9me2 around transcription start sites regions of Grin2a/NR2A, Grin2b/NR2B and Gria2/GluA2 genes in PFC from wild-type, FAD and BIX01294-treated FAD mice. The putative H3K9me2 binding sites are highlighted with dotted lines. (E) Enrichment analysis using gene sets derived from the Biological Process Ontology for the genes that had increased H3K9me2 enrichment in FAD mice (compared to wild-type) and decreased H3K9me2 enrichment in BIX01294-treated FAD mice (compared to FAD). ChIPseq experiments were performed with two biological replicates for each group.
Interestingly, the NMDAR subunits Grin2a/NR2A and Grin2b/NR2B were among this set of genes (Supplementary Table 1), consistent with our molecular and biochemical results. We further examined ChIP-seq data for Grin2a/NR2A and Grin2b/NR2B to identify the correlation between H3K9me2 binding and gene expression. As shown in Fig. 7D, H3K9me2 binding peaks around the transcription start sites region of Grin2a/NR2A and Grin2b/NR2B were increased in FAD mice, comparing to wild-type, which was reversed in BIX01294-treated FAD mice. Given the role of H3K9me2 in gene repression, it is possible that changes in H3K9me2 binding at these loci contribute to the decreased Grin2a/NR2A and Grin2b/NR2B expression in FAD and the normalization of their expression by BIX01294 treatment.
Gene Ontology Biological Process analyses (Fig. 7E) revealed that the genes with increased H3K9me2 in FAD mice were enriched in neuronal signalling, including cell communications, signal transduction, neurological system processes, G-protein coupled receptor (GPCR) signalling, and sensory perception. Interestingly, the genes with H3K9me2 occupancy reversed by BIX01294 treatment also fell into the same categories. Molecular Function analyses confirmed the enrichment of receptors among these genes. Cellular Compartment analyses indicated that most of these genes were located at membranes. In addition to genes encoding glutamate receptors, we have found a few other genes implicated in learning, memory, synaptic function, cognition or neurodegeneration that show the normalized H3K9me2 enrichment by BIX01294 treatment (Supplementary Table 3). The recovery of these genes may collectively contribute to the therapeutic effects of BIX01294 on synaptic function in FAD mice.
EHMT1/2 inhibition rescues memory deficits in FAD mice
Since glutamatergic transmission in PFC and hippocampus mediates cognitive processes, we next performed behavioural experiments to examine the therapeutic potential of EHMT inhibitors on memory tasks in FAD mice. Novel object recognition memory, T-maze alternation task for working memory, and Barnes maze for spatial memory were measured before and after drug treatment.
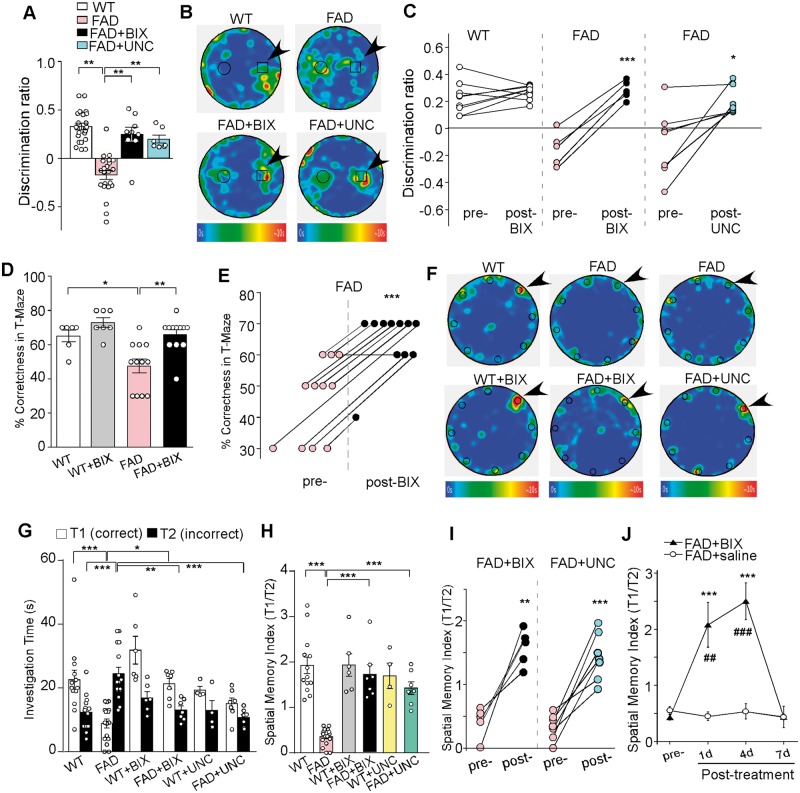
As shown in Fig. 8A, FAD mice exhibited novel object recognition memory impairment, as reflected by the significantly decreased discrimination ratio (DR) of the novel object over the familiar object (wild-type: 0.33 ± 0.04, n = 22; FAD: −0.17 ± 0.05, n = 22, P < 0.001, one-way ANOVA). A short treatment of FAD mice with BIX01294 (1 mg/kg, s.c. 3×) or UNC0642 (1 mg/kg, i.p. 3×) rescued the novel object recognition deficits [FAD + BIX: 0.25 ± 0.07, n = 9, FAD + UNC: 0.20 ± 0.04, n = 7, F(3,56) = 28.8, P < 0.001, one-way ANOVA]. The time of representative animals in each group spent on exploring novel versus familiar objects is illustrated in the heat maps (Fig. 8B). The improvement of novel object recognition memory by BIX01294 or UNC0642 was consistently observed in almost every FAD mouse examined (Fig. 8C, BIX: n = 5, P < 0.001, UNC: n = 7, P < 0.05, paired t-test). In the T-maze alternation task, BIX01294 treatment ameliorated working memory deficits in FAD mice, as indicated by a significant increase in the rate of correct choices [Fig. 8D, wild-type: 65% ± 4%, n = 6; wild-type + BIX: 72.9% ± 3%, n = 7; FAD: 47% ± 4%, n = 12; FAD + BIX: 65.8% ± 3%, n = 12, F(1,33) (treatment) = 11.9, P < 0.01, two-way ANOVA]. Working memory improvement was consistently observed in almost all BIX01294-treated FAD mice (Fig. 8E; n = 11, P < 0.001, paired t-test). BIX01294 treatment of FAD mice did not alter locomotion or anxiety-like behaviours in the open-field tests [Supplementary Fig. 7, n = 8 wild-type; n = 7 FAD; n = 5 FAD + BIX, F(2,17) (group) = 0.4, P > 0.05, one-way ANOVA].
Figure 8.
FAD mice exhibited deficits in novel object recognition memory, working memory and spatial memory, which was rescued by EHMT1/2 inhibitors. (A) Bar graphs (mean ± SEM) showing the discrimination ratio of novel object recognition (NOR) tests in wild-type (WT) versus FAD mice (5–6 months old) without or with the treatment of BIX01294 (1 mg/kg, s.c. 3×) or UNC0642 (1 mg/kg, i.p. 3×). **P < 0.01, one-way ANOVA. (B) Representative heat maps illustrating the time spent in different locations of the arena for novel object recognition tests of all groups (blue: 0 s; red: ∼10 s). Locations of novel (denoted by an arrow) and familiar objects are labelled with the circles or squares. (C) Scatter plots showing the discrimination ratio of novel object recognition tests in each of the examined wild-type or FAD mice before and after the treatment with BIX01294 or UNC0642. *P < 0.05, ***P < 0.001, paired t-test. (D) Bar graphs (mean ± SEM) showing the percentage correctness in T-maze working memory (WM) tests in wild-type or FAD mice with or without BIX01294 treatment. *P < 0.05, **P < 0.01, two-way ANOVA. (E) Scatter plots showing the percentage correctness in T-maze tests in each of the examined FAD mice before and after BIX01294 treatment. ***P < 0.001, paired t-test. (F) Representative heat maps illustrating the time spent in different locations of the arena for Barnes maze tests during the memory phase (escape box removed) in wild-type versus FAD mice without or with the treatment of BIX01294 (1 mg/kg, s.c. 3×) or UNC0642 (1 mg/kg, i.p. 3×) (blue: 0 s; red: ∼10 s). Locations of the correct (indicated by an arrow) and seven incorrect holes are labelled with circles. (G) Bar graphs (mean ± SEM) showing the time spent on exploring the correct hole (T1) versus the seven incorrect holes (T2) in the memory phase of Barnes maze tests of all groups. *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA. (H) Bar graphs (mean ± SEM) showing the spatial memory index (T1/T2) of Barnes maze tests in wild-type versus FAD mice without or with the treatment of BIX01294 or UNC0642. ***P < 0.001, two-way ANOVA. (I) Scatter plots showing the spatial memory index in Barnes maze tests in each of the examined FAD mice before and after the treatment with BIX01294 or UNC0642. **P < 0.01, ***P < 0.001, paired t-test. (J) Plots (mean ± SEM) of spatial memory index in FAD mice treated with BIX01294 (1 mg/kg, s.c. 3×) or saline at different time points. **P < 0.01, ***P < 0.001, saline versus BIX01294; ##P < 0.01, ###P < 0.001, pre- versus post-injection, two-way ANOVA. Each set of the experiments was replicated between four and five times.
We then used a more challenging memory task, the Barnes maze, to examine the spatial memory in FAD mice before and after treatment with EHMT1/2 inhibitors. The Barnes maze tests the animal’s spatial memory by recalling the location of one correct hole (where an escape box was attached before) from seven other incorrect holes on a round platform. The spatial memory index is calculated by the time spent on exploring the correct hole divided by the time spent on exploring all the other incorrect holes. As shown in Fig. 8F and G, wild-type mice (6 months old) spent significantly more time exploring the correct hole than the incorrect holes during the memory test phase, while 5xFAD mice (6 months old) failed to remember the correct location, as indicated by the significantly less time on exploring the correct hole and more time on exploring the incorrect holes. Remarkably, after treatment with the specific EHMT1/2 inhibitor BIX01294 (1 mg/kg, s.c. 3×) or UNC0642 (1 mg/kg, i.p. 3×), the spatial memory was significantly improved [Fig. 8H; spatial memory index, wild-type: 1.94 ± 0.18, n = 13, FAD: 0.35 ± 0.05, n = 16; wild-type + BIX: 1.93 ± 0.25, n = 6, FAD + BIX: 1.71 ± 0.21, n = 7; wild-type + UNC: 1.69 ± 0.28, n = 4, FAD + UNC: 1.43 ± 0.14, n = 7, F(1,47) (genotype) = 21.2, F(2,47) (treatment) = 9.5, P < 0.001, two-way ANOVA]. The EHMT1/2 inhibitor-induced improvement of spatial memory was consistent in individual FAD mice (Fig. 8I; BIX, n = 5, P < 0.01, paired t-test; UNC, n = 7, P < 0.001, paired t-test). The rescuing effect persisted for ∼4 days and vanished at 7 days post-injection [Fig. 8J; FAD + saline: n = 7; FAD + BIX: n = 8, F(1,52) (treatment) = 20.5, F(3,52) (time) = 8.6, P < 0.001, two-way ANOVA]. These data suggest that the selective inhibition of EHMT1/2 is able to restore a variety of cognitive behaviours in the mouse model of Alzheimer’s disease.
Discussion
Despite the unclear aetiology of Alzheimer’s disease, it is thought that a combination of genetic risk factors (e.g. APP and presenilin mutations) and environmental factors (e.g. age), which lead to deregulation of vulnerability genes, may underlie Alzheimer’s disease pathogenesis, and epigenetic mechanisms might be central to mediate gene-environment interactions relevant to disease progression (Mitchell et al., 2014; Lardenoije et al., 2015). The present study has found that the repressive histone methylation H3K9me2 (linked to gene silencing) is significantly elevated in PFC and hippocampus of late-stage FAD mice, which correlates with the abnormal epigenetic regulation of glutamate receptor transcription. Consequently, synaptic transmission is significantly impaired, leading to a variety of Alzheimer’s disease-like cognitive deficits. More importantly, we have found that inhibiting the euchromatic histone methyltransferases, EHMT1 and EHMT2, which catalyse H3K9me2 to repress transcription, leads to the recovery of glutamate receptors and synaptic function and the amelioration of cognitive impairment in aged FAD mice (Supplementary Fig. 8).
Alterations of gene expression resulting from aberrant epigenetic mechanisms, such as DNA methylation and histone modifications, are thought to play an important role in the onset and progression of neurodegeneration (Coppedè, 2014; Fischer, 2014). Dysregulation of microRNAs, which interfere with the translation of target mRNAs, is also linked to core pathophysiological processes underlying Alzheimer’s disease (Millan, 2017). Both transcriptionally activating and transcriptionally repressive global histone changes in different regions of post-mortem Alzheimer’s disease brains have been observed (Narayan and Dragunow, 2017). The role of histone acetylation in ageing and neurodegeneration is well documented (Chuang et al., 2009; Fischer et al., 2010; Gräff and Tsai, 2013). For example, it has been found that memory disturbances in the ageing mouse brain are associated with the deregulation of histone acetylation during learning and the failure to initiate a hippocampal gene expression program associated with memory consolidation (Peleg et al., 2010). Histone deacetylase (HDAC) inhibitors, which act to restore histone acetylation, can improve the cognitive performance in an Alzheimer’s disease model (Gräff et al., 2012) and older mice (Peleg et al., 2010). Histone methylation marks and enzymes are implicated in senescence and cognitive functioning (Greer and Shi, 2012); however, no links with Alzheimer’s disease have been firmly identified yet (Fischer, 2014; Lardenoije et al., 2015).
Clinical study has reported changes in H2B K108 and H4 R55 methylation in frontal cortex from human donors with Alzheimer’s disease (Anderson and Turko, 2015), supporting our idea that targeting histone methylation is a new avenue to treat Alzheimer’s disease-related abnormalities. Among the most extensively studied site-specific methylation of histones, tri- and dimethylated forms of histone H3 lysines K4, K9 and K27 are particularly interesting for post-mortem research, because of their link to the activity state of chromatin and their stable preservation during tissue autolysis (Akbarian and Huang, 2009). In this study, we have found that H3K9me2, a repressive chromatin mark (Barski et al., 2007), is specifically increased in the PFC of 5xFAD mice, tau transgenic mice, and Alzheimer’s disease human post-mortem tissues. Furthermore, our large-scale analyses have revealed a genome-wide increase of H3K9me2 enrichment around transcription start sites regions of multiple genes in PFC of FAD mice, which is reversed by EHMT1/2 inhibition. Consistent with our findings, H3K9me2 in the occipital cortex was found to be significantly elevated in post-mortem Alzheimer’s disease, compared to non-demented and age-matched control subjects (Lithner et al., 2013).
On the other hand, loss of heterochromatin and increased gene expression has also been reported in Alzheimer’s disease conditions (Frost et al., 2014). Multiple reasons may account for the discrepancies. First, different animals and cell types were mainly examined, with our study focusing on PFC of mice expressing mutant APP/PS1 and Frost et al.rsquo;s study focusing on Drosophila with mutant tau and motor neurons in mice with mutant tau. Second, different preparation methods of human samples were used, with our study using nuclear fraction of prefrontal cortex and Frost et al.rsquo;s study using the whole lysates of sorted hippocampal neurons. Third, different human samples in terms of age and sex were used in the two studies. The heterogeneity of human genomes may contribute to different results. Besides the presence of some clinical features, no information is available on genetic background of the patients (familial Alzheimer’s disease and mutants or sporadic Alzheimer’s disease) nor the presence of specific pathological hallmarks, so it is difficult to directly compare different studies. Another study using middle-aged controls to compare with Alzheimer’s disease patients also found the reduction of H3K9me2 in CA1 (Hernández-Ortega et al., 2016).
One question remaining to be answered is how H3K9me2 is elevated in aged 5xFAD mice. Recently it has been found that chronic activity deprivation increases the amount of neuronal H3K9me2 (Benevento et al., 2016). Excessive amyloid-β depresses excitatory synaptic transmission (Kamenetz et al., 2003), resulting in chronic activity deprivation. Consequently, the level of repressive H3K9me2 is elevated, leading to the loss of glutamate receptor transcription. This vicious cycle causes more synaptic depression, which contributes to cognitive impairment.
Given the epigenetic changes in Alzheimer’s disease models or patients with Alzheimer’s disease, is the aberrant histone methylation a causal factor for the synaptic and cognitive deficits in Alzheimer’s disease? Here we show that the elevated global H3K9me2 is correlated with the significantly increased enrichment of H3K9me2 at transcription start sites regions of glutamate receptors, suggesting that the loss of glutamate receptor transcription in Alzheimer’s disease could be caused by the aberrant histone methylation. In agreement with this, reducing H3K9me2 with EHMT1/2 inhibitors leads to the restoration of glutamate receptors. The importance of epigenetic mechanisms in regulating NMDAR expression is further supported by the previous finding that developmental changes in histone methylation at Grin2b/NR2B promoter mediate the decline of NR2B expression at mature synapses (Rodenas-Ruano et al., 2012). In support of our discovery on the loss of synaptic proteins due to the increased repressive H3K9me2 in Alzheimer’s disease, a recent genetic study of ageing human brains has found that molecular modules for neuronal or synaptic function are negatively correlated with Alzheimer’s disease pathology and cognitive decline, and the module that is most strongly associated with cognitive decline is enriched with genes involved in chromatin modification (Mostafavi et al., 2018).
Other than glutamate receptor genes, additional genes implicated in Alzheimer’s disease have also shown the rescued H3K9me2 enrichment by BIX01294 treatment (Supplementary Tables 2 and 3). One example is SHANK2, which encodes a key scaffolding protein and signalling adaptor enriched at glutamatergic synapses (Sheng and Kim, 2000). Genetics studies have found that patients with SHANK2 deficiency have intellectual disability at infancy and tend to have Alzheimer’s disease at the elderly stage (Guilmatre et al., 2014). The NMDAR hypofunction induced by loss of SHANK2 (Won et al., 2012) may contribute to the cognitive impairment. Given the complexity of Alzheimer’s disease pathophysiology, it is likely that BIX01294 treatment leads to the restoration of synaptic function in FAD mice by normalizing multiple targets.
Drosophila EHMT/G9a has been identified as a key regulator of cognition that orchestrates the epigenetic control of learning and memory genes (Kramer et al., 2011). Human studies have revealed a causal role of EHMT1 in intellectual disability (Kleefstra et al., 2006, 2012), and mice with EHMT1 haploinsufficiency exhibit learning deficits and synaptic dysfunction (Balemans et al., 2013). Thus, we speculate that targeting EHMT1/2-regulated chromatin-modifications might be able to restore transcriptional homeostasis and intellectual function in neurodegenerative disorders. Indeed, we have found that treatment of FAD mice with EHMT1/2 inhibitors leads to the remarkable restoration of cognitive behaviours, including novel object recognition memory, working memory, and spatial memory. No reduction of the amyloid load in aged FAD mice was observed following treatment with an EHMT1/2 inhibitor, suggesting that this epigenetic perturbation is working directly on the synaptic proteins, such as AMPA- and NMDA-type glutamate receptors, which control synaptic transmission and cognitive function. This explains the rapid onset and robust potency of these epigenetic drugs for the therapeutic intervention of Alzheimer’s disease. Our results have provided the first experimental evidence that restoring the homeostasis of histone methylation could be a promising therapeutic strategy for Alzheimer’s disease-related neurodegenerative disorders. Considering the multi-factorial feature of Alzheimer’s disease, manipulation of an individual or couple of misregulated genes will unlikely ameliorate the physiological and behavioural deficits. The epigenetic-based treatment strategy offers the advantage of having broad, multi-functional actions and being able to normalize a network of genes important for neuronal survival and synaptic function.
Supplementary Material
Acknowledgement
We would like to thank Xiaoqing Chen for her excellent technical support.
Glossary
Abbreviations
- ChIP
chromatin immunoprecipitation
- EPSC
excitatory postsynaptic current
- FAD
familial Alzheimer’s disease
- PFC
prefrontal cortex
Funding
This work was supported by the VA Merit Award (I01BX001633) to Z.Y., the National Institutes of Health grant (R01-AG056060) to Z.Y., the National Key Research and Development Program of China (2016YFC1306300) to X.W., NSFC grants (81571038, 81771145) to Y.Z., and NSFC grants (81171197, 81220108010) to G.J.C. None of the authors have the financial interest related to this work.
Competing interests
The authors report no competing interests.
References
- Akbarian S, Huang HS. Epigenetic regulation in human brain-focus on histone lysine methylation. Biol Psychiatry 2009; 65: 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KW, Turko IV. Histone post-translational modifications in frontal cortex from human donors with Alzheimer’s disease. Clin Proteomics 2015; 12: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balemans MC, Kasri NN, Kopanitsa MV, Afinowi NO, Ramakers G, Peters TA, et al. Hippocampal dysfunction in the Euchromatin histone methyltransferase 1 heterozygous knockout mouse model for Kleefstra syndrome. Hum Mol Genet 2013; 22: 852–66. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129: 823–37. [DOI] [PubMed] [Google Scholar]
- Benevento M, Iacono G, Selten M, Ba W, Oudakker A, Frega M, et al. Histone methylation by the Kleefstra syndrome protein EHMT1 mediates homeostatic synaptic scaling. Neuron 2016; 91: 341–55. [DOI] [PubMed] [Google Scholar]
- Bevins RA, Besheer J. Object recognition in rats and mice: a one-trial non-matching-to-sample learning task to study ‘recognition memory’. Nat Protoc 2006; 1: 1306–11. [DOI] [PubMed] [Google Scholar]
- Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron 2008; 60: 961–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Zhang J, Wu Y, Wang D, Feng G, Tang YP, et al. Monoacylglycerol lipase is a therapeutic target for Alzheimer’s disease. Cell Rep 2012; 2: 1329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci 2009; 32: 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppedè F. The potential of epigenetic therapies in neurodegenerative diseases. Front Genet 2014; 5: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzin N, Baranger K, Cavalier M, Marchalant Y, Cohen-Solal C, Roman FS, et al. Area-specific alterations of synaptic plasticity in the 5XFAD mouse model of Alzheimer’s disease: dissociation between somatosensory cortex and hippocampus. PLoS One 2013; 8: e74667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wei J, Cheng J, Zhong P, Xiong Z, Liu A, et al. Partial amelioration of synaptic and cognitive deficits by inhibiting cofilin dephosphorylation in an animal model of Alzheimer’s disease. J Alzheimer’s Disease 2016; 53: 1419–32. [DOI] [PubMed] [Google Scholar]
- Duffney LJ, Zhong P, Matas E, Wei J, Cheng J, Dietz DM, et al. Autism-like deficits in Shank3-deficient mice are rescued by targeting actin regulators. Cell Rep 2015; 11: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A. Targeting histone-modifications in Alzheimer’s disease. What is the evidence that this is a promising therapeutic avenue? Neuropharmacology 2014; 80: 95–102. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci 2010; 31: 605–17. [DOI] [PubMed] [Google Scholar]
- Forman MS, Trojanowski JQ, Lee VM-Y. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med 2004; 10: 1055–1063. [DOI] [PubMed] [Google Scholar]
- Frost B, Hemberg M, Lewis J, Feany MB. Tau promotes neurodegeneration through global chromatin relaxation. Nat Neurosci 2014; 17: 357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido GE, Furuie SS, Buchpiguel CA, Bottino CM, Almeida OP, Cid CG, et al. Relation between medial temporal atrophy and functional brain activity during memory processing in Alzheimer’s disease: a combined MRI and SPECT study. J Neurol Neurosurg Psychiatry 2002; 73: 508–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakopoulos P, Herrmann FR, Bussière T, Bouras C, Kövari E, Perl DP, et al. Tangle and neuron numbers, but not amyloid load, predict cognitive status in Alzheimer’s disease. Neurology 2003; 60: 1495–1500. [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Cellular basis of working memory. Neuron 1995; 14: 477–85. [DOI] [PubMed] [Google Scholar]
- Gräff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 2012; 483: 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräff J, Tsai LH. The potential of HDAC inhibitors as cognitive enhancers. Annu Rev Pharmacol Toxicol 2013; 53: 311–30. [DOI] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13: 343–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Liu W, Yan Z. β-amyloid impairs AMPA receptor trafficking and function by reducing CaMKII synaptic distribution. J Biol Chem 2009; 284: 10639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilmatre A, Huguet G, Delorme R, Bourgeron T. The emerging role of SHANK genes in neuropsychiatric disorders. Dev Neurobiol 2014; 74: 113–22. [DOI] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. J. Neurosci 2010; 30: 3589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Agarwal S, Franklinm AV, Deramusm T, Wheelockm M, Davism RL, McMahonm LL, et al. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J Neurosci 2012; 32: 5440–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007; 39: 311–8. [DOI] [PubMed] [Google Scholar]
- Hernández-Ortega K, Garcia-Esparcia P, Gil L, Lucas JJ, Ferrer I. Altered machinery of protein synthesis in Alzheimer’s: from the nucleolus to the ribosome. brain pathology 2016; 26: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, Iwatsubo T, et al. APP processing and synaptic function. Neuron 2003; 37: 925–37. [DOI] [PubMed] [Google Scholar]
- Kerimoglu C, Agis-Balboa RC, Kranz A, Stilling R, Bahari-Javan S, Benito-Garagorri E, et al. Histonemethyltransferase mll2 (kmt2b) is required for memory formation in mice. J Neurosci 2013; 33: 3452–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura R, Ohno M. Impairments in remote memory stabilization precede hippocampal synaptic and cognitive failures in 5XFAD Alzheimer mouse model. Neurobiol Dis 2009; 33: 229–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet 2006; 79: 370–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Kramer JM, Neveling K, Willemsen MH, Koemans TS, Vissers LE, et al. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am J Hum Genet 2012; 91: 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, Kochinke K, Oortveld MA, Marks H, Kramer D, de Jong EK, et al. Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. Plos Biol 2011; 9: e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methytransferase. Mol Cell 2007; 25: 473–81. [DOI] [PubMed] [Google Scholar]
- Lardenoije R, Iatrou A, Kenis G, Kompotis K, Steinbusch HW, Mastroeni D, et al. The epigenetics of aging and neurodegeneration. Prog Neurobiol 2015; 131: 21–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 2007; 128: 707–19. [DOI] [PubMed] [Google Scholar]
- Lithner CU, Lacor PN, Zhao WQ, Mustafiz T, Klein WL, Sweatt JD, et al. Disruption of neocortical histone H3 homeostasis by soluble Aβ: implications for Alzheimer’s disease. Neurobiol Aging 2013; 34: 2081–90. [DOI] [PubMed] [Google Scholar]
- Liu F, Barsyte-Lovejoy D, Li F, Xiong Y, Korboukh V, Huang XP, et al. Discovery of an in vivo chemical probe of the lysine methyltransferases G9a and GLP. J Med Chem 2013; 56: 8931–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunnon K, Mill J. Epigenetic studies in Alzheimer’s disease: current findings, caveats, and considerations for future studies. Am J Med Genet B Neuropsychiatr Genet 2013; 162B: 789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet D, Rajah MN. Association between prefrontal activity and volume change in prefrontal and medial temporal lobes in aging and dementia: a review. Ageing Res Rev 2013; 12: 479–89. [DOI] [PubMed] [Google Scholar]
- Malmquist NA, Moss TA, Mecheri S, Scherf A, Fuchter MJ. Small-molecule histone methyltransferase inhibitors display rapid antimalarial activity against all blood stage forms in Plasmodium falciparum. Proc Natl Acad Sci USA 2012; 109: 16708–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol 2008; 86: 305–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan MJ. The epigenetic dimension of Alzheimer’s disease: causal, consequence, or curiosity? Dialogues Clin Neurosci 2014; 16: 373–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan MJ. Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer’s disease: an integrative review. Prog Neurobiol 2017; 156: 1–68. [DOI] [PubMed] [Google Scholar]
- Mitchell A, Roussos P, Peter C, Tsankova N, Akbarian S. The future of neuroepigenetics in the human brain. Prog Mol Biol Transl Sci 2014; 128: 199–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris HR, Khan MN, Janssen JC, Brown JM, Perez-Tur J, Baker M, et al. The genetic and pathological classification of familial frontotemporal dementia. Arch Neurol 2001; 58: 1813–6. [DOI] [PubMed] [Google Scholar]
- Mostafavi S, Gaiteri C, Sullivan SE, White CC, Tasaki S, Xu J, et al. A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat Neurosci 2018; 21: 811–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan P, Dragunow M. Alzheimer’s disease and histone code alterations. Adv Exp Med Biol 2017; 978: 321–36. [DOI] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 2010; 42: 790–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci 2006; 26: 10129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 2010; 328: 753–6. [DOI] [PubMed] [Google Scholar]
- Pompl PN, Mullan MJ, Bjugstad K, Arendash GW. Adaptation of the circular platform spatial memory task for mice: use in detecting cognitive impairment in the APP(SW) transgenic mouse model for Alzheimer’s disease. J Neurosci Methods 1999; 87: 87–95. [DOI] [PubMed] [Google Scholar]
- Popoli M, Yan Z, McEwen BS, Sanacora G. The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nat Rev Neurosci 2011; 13: 22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Ma K, Wang ZJ, Hu Z, Matas E, Wei J, et al. Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition. Nat Neurosci 2018; 21: 564–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rémy F, Mirrashed F, Campbell B, Richter W. Verbal episodic memory impairment in Alzheimer’s disease: a combined structural and functional MRI study. Neuroimage 2005; 25: 253–66. [DOI] [PubMed] [Google Scholar]
- Rodenas-Ruano A, Chávez AE, Cossio MJ, Castillo PE, Zukin RS. REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci 2012; 15: 1382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 2009; 64: 678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Giusti-Rodríguez P, Zhou Y, Rudenko A, Cho S, Ota KT, et al. Activity-dependent p25 generation regulates synaptic plasticity and Aβ-induced cognitive impairment. Cell 2014; 157: 486–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Kim E. The shank family of scaffold proteins. J Cell Sci 2000; 113: 1851–6. [DOI] [PubMed] [Google Scholar]
- Sperfeld AD, Collatz MB, Baier H, Palmbach M, Storch A, Schwarz J, et al. FTDP-17: an early-onset phenotype with parkinsonism and epileptic seizures caused by a novel mutation. Ann Neurol 1999; 46: 708–715. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Iba M, Inoue H, Higuchi M, Takao K, Tsukita K, et al. P301S mutant human tau transgenic mice manifest early symptoms of human tauopathies with dementia and altered sensorimotor gating. PLoS One 2011; 6(6): e21050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Xiong Z, Lee JB, Cheng J, Duffney LJ, Matas E, et al. Histone modification of Nedd4 ubiquitin ligase controls the loss of AMPA receptors and cognitive impairment induced by repeated stress. J Neurosci 2016; 36: 2119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemichael BT, Bohacek J, Gapp K, Mansuy IM. Epigenetics of memory and plasticity. Prog Mol Biol Transl Sci 2014; 122: 305–40. [DOI] [PubMed] [Google Scholar]
- Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature 2012; 486: 261–5. [DOI] [PubMed] [Google Scholar]
- Yoshiyama Y, Higuchi M, Zhang B, Huang SM, Iwata N, Saido TC, et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 2007; 53: 337–351. [DOI] [PubMed] [Google Scholar]
- Yuen EY, Liu W, Karatsoreos IN, Feng J, McEwen BS, Yan Z. Acute stress enhances glutamatergic transmission in prefrontal cortex and facilitates working memory. Proc Natl Acad Sci USA 2009; 106: 14075–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen EY, Liu W, Karatsoreos IN, Ren Y, Feng J, McEwen BS, et al. Mechanisms for acute stress-induced enhancement of glutamatergic transmission and working memory. Mol Psychiatry 2011; 16: 156–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen EY, Wei J, Liu W, Zhong P, Li X, Yan Z. Repeated stress causes cognitive impairment by suppressing glutamate receptor expression and function in prefrontal cortex. Neuron 2012; 73: 962–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Fu Y, Yasvoina M, Shao P, Hitt B, O’Connor T, et al. Beta-site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: implications for Alzheimer’s disease pathogenesis. J Neurosci 2007; 27: 3639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data will be deposited in a public repository. The access number and the dataset will be available for access.