PLPBP mutations have recently been associated with B6-responsive epilepsy, however the disease mechanism is not fully understood. Johnstone et al. report evidence for a genotype-phenotype correlation using 12 previously unidentified patients, expand the clinical spectrum, and model the disease using zebrafish, yeast, and cells to help elucidate the underlying pathophysiology.
Keywords: PLPBP, PROSC, epilepsy, pyridoxine, vitamin B6-responsive epilepsy
Abstract
Biallelic pathogenic variants in PLPBP (formerly called PROSC) have recently been shown to cause a novel form of vitamin B6-dependent epilepsy, the pathophysiological basis of which is poorly understood. When left untreated, the disease can progress to status epilepticus and death in infancy. Here we present 12 previously undescribed patients and six novel pathogenic variants in PLPBP. Suspected clinical diagnoses prior to identification of PLPBP variants included mitochondrial encephalopathy (two patients), folinic acid-responsive epilepsy (one patient) and a movement disorder compatible with AADC deficiency (one patient). The encoded protein, PLPHP is believed to be crucial for B6 homeostasis. We modelled the pathogenicity of the variants and developed a clinical severity scoring system. The most severe phenotypes were associated with variants leading to loss of function of PLPBP or significantly affecting protein stability/PLP-binding. To explore the pathophysiology of this disease further, we developed the first zebrafish model of PLPHP deficiency using CRISPR/Cas9. Our model recapitulates the disease, with plpbp−/− larvae showing behavioural, biochemical, and electrophysiological signs of seizure activity by 10 days post-fertilization and early death by 16 days post-fertilization. Treatment with pyridoxine significantly improved the epileptic phenotype and extended lifespan in plpbp−/− animals. Larvae had disruptions in amino acid metabolism as well as GABA and catecholamine biosynthesis, indicating impairment of PLP-dependent enzymatic activities. Using mass spectrometry, we observed significant B6 vitamer level changes in plpbp−/− zebrafish, patient fibroblasts and PLPHP-deficient HEK293 cells. Additional studies in human cells and yeast provide the first empirical evidence that PLPHP is localized in mitochondria and may play a role in mitochondrial metabolism. These models provide new insights into disease mechanisms and can serve as a platform for drug discovery.
Introduction
The vitamin B6-responsive disorders (B6RDs) are a clinically and genetically heterogeneous group of rare, autosomal recessive conditions (Clayton, 2006) with the hallmark feature of seizures uniquely responsive to treatment by the B6 vitamers pyridoxine and/or pyridoxal-5′-phosphate (PLP) (Baumgartner-Sigl et al., 2007; Basura et al., 2009). PLP is a cofactor for over 160 distinct catalytic functions (Percudani and Peracchi, 2009), including enzymes involved in glucose, lipid and amino acid metabolism (John, 1995; Percudani and Peracchi, 2003; Eliot and Kirsch, 2004), and for the synthesis of neurotransmitters, making it an essential vitamer for normal brain function (Surtees et al., 2006).
The B6RDs are characterized by recurrent seizures in the prenatal, neonatal, or postnatal period and are resistant to anti-epileptic medication (Walker et al., 2000; Mills et al., 2005, 2006; Baumgartner-Sigl et al., 2007). Intellectual disability, behavioural abnormalities, and psychiatric disturbances, as well as abnormalities in brain structure and myelination are frequently observed (Stockler et al., 2011). If untreated, B6RDs may lead to status epilepticus and death (Gospe, 2017). B6RDs have been attributed to a number of genetic variants disrupting B6 metabolism, including those leading to the accumulation of toxic metabolites that inactivate PLP [ALDH7A1 (MIM#266100), ALDH4A1, (MIM#239510)], those interfering with the interconversion of B6 vitamers [PNPO (MIM#610090), TNSALP (MIM#171760)] (Hamosh et al., 2005; Mills et al., 2005, 2006; Clayton, 2006; Stockler et al., 2011), and those impairing PLP homeostasis [(PLPBP (encoding PLP homeostasis protein, PLPHP), MIM#604436, previously named PROSC] (Darin et al., 2016; Plecko et al., 2017).
In bacteria (YggS) and yeast (YBL036C), the structures of PLPHP orthologous proteins show PLP covalently bound to a lysine residue, phosphate-binding motifs, and a typical triosephosphate isomerase (TIM)-barrel domain (Eswaramoorthy et al., 2003; Ito et al., 2013). Purified human PLPHP is also bound to PLP in the native state, but little is known about the molecular function of this protein (Tremino et al., 2018). Studies in YggS-deficient Escherichia coli revealed growth impairment and disrupted amino and keto acid homeostasis (Ito et al., 2013; Prunetti et al., 2016). In cyanobacteria, it has been suggested that the C-terminal helix may play a role in PLP exchange with apoenzymes (Tremino et al., 2017). B6 vitamer levels were significantly altered in human PLPHP loss-of-function patient samples, and it has been hypothesized this protein has a key role in B6 homeostasis (Darin et al., 2016; Prunetti et al., 2016), possibly acting as a PLP carrier that prevents PLP from reacting with other molecules, supplying it to dependent enzymes and/or protecting PLP from phosphatases.
PLPHP deficiency in humans is manifested by early-onset intractable seizures responsive to pyridoxine and/or PLP, developmental delay, and structural brain abnormalities, most notably simplified gyral pattern and cyst-like structures adjacent to the anterior horns (Darin et al., 2016). We undertook a comprehensive genetic and biochemical study of PLPHP deficiency in a cohort of 12 previously undescribed patients, highlighting a unique movement disorder phenotype (without epilepsy) as well as fatal mitochondrial encephalopathy phenotype, both of which, to our knowledge, have not been described previously. To characterize the pathophysiology of this neurometabolic disease, we generated knockout models in zebrafish (Danio rerio), yeast (Saccharomyces cerevisiae) and HEK293 cells, providing insights into the biochemical consequences of PLPHP deficiency.
Materials and methods
Patients
This study was approved by the clinical research ethics board of BC Children’s and Women’s Hospital, University of British Columbia (H12–00067), the Children’s Hospital of Eastern Ontario Research Ethics Board, and local institutional review boards at the University of Colorado. Many of the patients were recruited through international collaboration as part of an ongoing TIDEX neurometabolic gene discovery project (Tarailo-Graovac et al., 2016). After obtaining signed informed parental consent, referring clinicians provided detailed reports of clinical, MRI and EEG features of study patients.
Whole-exome sequencing, Sanger sequencing and in silico analysis
Detailed descriptions of whole-exome sequencing, bioinformatic analyses, Sanger sequencing and in silico analysis strategies are provided in the Supplementary material. All exomes were aligned to the human reference genome, February 2009 assembly (GRCh37/hg19).
Structural model of human PLPHP
The 3D model of PLPHP protein (NP_009129.1) was obtained by homology modelling using MODELLER (Webb and Sali, 2014) and the yeast orthologue [YBL036C, PDB 1CT5, (Eswaramoorthy et al., 2003), 41% identical, 57% similar] as template. The DOPE (discrete optimized protein energy) score was used to select the best model for subsequent refinement using Coot (v0.8.6.1; Emsley et al., 2010). Prosa-Web (Wiederstein and Sippl, 2007) and Coot’s Ramachandran plot analysis module were used to validate model quality. PyMOL (Schrodinger, 2015) was used for structural superimposition of the human PLPHP model with yeast 1CT5, and the coordinates of the PLP co-crystalized with the yeast orthologue were transferred to the PLPHP model, with PLP covalently bound to p.Lys47. Images were prepared using PyMOL. Arpeggio was used to calculate contacts (Jubb et al., 2017). DUET (Pires et al., 2014) was used to calculate stability changes.
Clinical severity score
We assessed the clinical severity of patients within this study and previous studies (Darin et al., 2016; Plecko et al., 2017) based on published data. We adapted a scoring system of patients with B6RD due to pathogenic variants in ALDH7A1 (Al Teneiji et al., 2017). The following criteria were used: (i) global and/or intellectual delay: 0, normal; 1, mild; 2, moderate; 3, severe; (ii) age of onset of seizures and/or movement disorder: 0, absent; 1, >1 month; 2, ≥7 days; 3, <7 days; and (iii) therapeutic response: 0, full cessation of seizures and normalization of EEG (if available) on <200 mg B6 (pyridoxine and/or PLP) total daily; 1, no clinical seizures or abnormal movements on ≥200 mg B6 total daily, with or without electrographic normalization OR clinical response to <200 mg B6 total daily dose with persistently abnormal EEG; 2, no seizures with B6 (any dose) AND other antiepileptic drug medication, with or without EEG normalization; 3, breakthrough seizures and/or persistent movement disorder, no responsiveness. We calculated the sum for each clinical feature (i–iii), and classified each patient as mild (1–3), moderate (4–6) or severe (7–9) (Al Teneiji et al., 2017).
Isolation of pure mitochondrial fractions and western blotting
Pure mitochondrial fractions were isolated from HeLa cells having hamenagglutinin (HA)-tagged mitochondria using an immunoprecipitation protocol as outlined previously (Chen et al., 2017). Whole cell and pure mitochondrial fractions were run on SDS-PAGE, and western blots were blocked in Tris-buffered saline-Tween (TBS-T) 5% milk and probed with the following primary antibodies: rabbit anti-PROSC (Proteintech 25154–1-AP; 1:1000), rabbit anti-SHMT2 (Sigma HPA020549; 1:1000), rabbit anti-VDAC (Cell Signaling 4661S; 1:1000), mouse anti-LAMP2 (Abcam ab25631; 1:1000), mouse anti-GAPDH (Santa Cruz sc-47724; 1:2000), and rabbit anti-GOLGIN-97 (Cell Signaling 13132; 1:1000). All antibodies were prepared fresh in TBS-T 5% bovine serum albumin. Horseradish peroxidase-conjugated goat anti-mouse (cat. no. sc-2055) and anti-rabbit (cat. no. sc-2054) secondary antibodies obtained from Santa Cruz Biotechnology were used at 1:3000.
Yeast strains and culture conditions
S. cerevisiae BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) was used as the wild-type strain along with derivative strains: fox1::KAN, carrying a deletion of peroxisomal acyl-CoA oxidase and ybl036C::KAN mutant (Euroscaft). Yeast strains and transformants containing the expression plasmids pPROSC1a and pPROSC2a (human PLPBP) were selected and grown in minimal medium containing 6.7 g/l yeast nitrogen base without amino acids (YNB-WO), supplemented with 5 g/l glucose and amino acids (20 mg/l), and growth was measured. For the induction of peroxisome and mitochondrial proliferation, cells were shifted to ethanol (YPE) 20 g/l, glycerol (YPG) 20 g/l, or oleate (YPO) medium containing 5 g/l potassium phosphate buffer, pH 6.0, 3 g/l yeast extract, 5 g/l peptone. YPO media were supplemented with 1.2 g/l oleate, and 2 g/l Tween-80. Prior to shifting to these media, the cells were grown in minimal medium with 5 g/l glucose for at least 24 h.
Generation of mutant zebrafish lines
Zebrafish were maintained following standard protocols (Westerfield, 1993), and experiments were in accordance with the animal care guidelines of the Canadian Council on Animal Care, the University of Ottawa animal care committee (protocol BL-2678), and the ARRIVE guidelines (Kilkenny et al., 2012). Handling, treatments, husbandry and nursery were performed as outlined previously (Pena et al., 2017). CRISPR/Cas9 was used to induce targeted indel mutations in the plpbp gene of zebrafish embryos as previously described (Hwang et al., 2013), using ZiFiT targeter (Sander et al., 2010) to select CRISPR targets and design oligonucleotides (5′-TAGGTGGAGCGGGTGAATCAAG-3′ and 5′-AAACCTTGATTCACCCGCTCCA-3′) in the first exon. The target was chosen as having the fewest predicted off-targets (minimum three mismatches with any predicted off-target sequence). Generation of the sgRNA and CRISPR/Cas9 injection, as well as screening for mutants by PCR/HMA-PAGE (heteroduplex melting assay polyacrylamide gel electrophoresis), were performed as previously described (Zhu et al., 2014; Pena et al., 2017). Genotyping PCR was performed as described in Kosuta et al. (2018) and in the Supplementary material. F0 larvae were raised to adulthood and backcrossed with wild-type to generate heterozygous F1 fish. These were again backcrossed with wild-type to minimize off-targets. Experimental compound heterozygous animals were obtained by crossing F2 heterozygotes.
Behavioural phenotyping
Sixteen 11 days post-fertilization (dpf) larvae per group were dispensed (one per well) in 48-well flat-bottomed culture dishes (Corning) containing 500 µl of system water. Behaviour was monitored as previously described (Pena et al., 2017) using a ZebraBox system (ViewPoint Behavior Technology). Videos were also analysed blindly by two observers to classify seizure scores using the S0-S3 system (Baraban et al., 2005).
Electrophysiology and c-fos expression
Electrophysiological local field potential recordings of activity in the optic tectum of five 11 dpf larvae per group selected randomly were obtained as previously described (Pena et al., 2017). Since c-fos expression can be used as a biomarker for increased neuronal activity and is known to increase with seizure activity (Baraban et al., 2005), we measured c-fos mRNA expression in pools of five 11 dpf larvae (mutants and wild-type) as well as in wild-type larvae treated with 15 mM pentylenetetrazol (PTZ) as a positive control. RNA was extracted, reverse transcribed and quantified by qPCR as previously described (Pena et al., 2017). Primers used were: cfos-F 5′-AACTGTCACGGCGATCTCTT-3′ and cfos-R 5′-TCTTCTGGAGAAAGCTGTTC-3′ with β-actin as internal control: actin-F 5′-CATCCATCGTTCACAGGAAGTG-3’ and actin-R 5′-TGGTCGTTCGTTTGAATCTCAT-3′.
Metabolite extraction and mass spectrometry
For analysis of B6 vitamers, three pools of six 10 dpf larvae (plpbp−/−, wild-type) were analysed as previously described (Pena et al., 2017). Measurement of amino acid panels was performed using three pools of five larvae per group (plpbp−/−, wild-type, heterozygotes) following established protocols (van der Ham et al., 2012; Pena et al., 2017), with the modification that 10 dpf larvae were fasted for 24 h prior to collection with metabolite extraction at 11 dpf. Neurotransmitter analytes (five pools of plpbp−/− and four pools of wild-type; six larvae per pool) were measured following established methods (van Vliet et al., 2015).
Statistical analysis
All statistical analyses and graphing were performed using GraphPad Prism. Where appropriate, one-way ANOVA with Tukey’s test, or Krustal-Wallis with Dunn’s post hoc test was performed. Student’s t-test was used for pairwise comparisons.
Data availability
Sequences, plasmids, cell and zebrafish lines are available upon request. All data necessary for confirming the conclusions are represented fully within the article or its online Supplementary material.
Results
Phenotypic spectrum of patient cohort with biallelic pathogenic PLPBP variants
The 12 previously unreported patients described here presented with encephalopathic phenotypes comprising neonatal-onset of refractory epilepsy (or a movement disorder in one case), with or without additional clinical features (Table 1 and Supplementary material). This cohort comprised six male and six female patients from seven different ethnic backgrounds. For Patients 1 and 6, the pregnancy history was notable for excessive foetal movements, possibly indicating seizures in utero. Three patients experienced respiratory insufficiency after birth, including Patient 3 who had progressive respiratory failure.
Table 1.
Clinical features of PLPHP-deficient patients
| Patient’s ID Sex, current age | Patient 1 Male, 3 11/12 y | Patient 2 Male, 14 y | Patient 3 Female, 5 2/12 y | Patient 4 Female, died at 2 w | Patient 5 Female, died at 8 w | Patient 6 Male, 4 3/12 y | Patient 7 Male, 23 mo | Patient 8 Male, 8 1/12 y | Patient 9 Male, 14 mo | Patient 10 Female, 10 6/12 y (sister of Patient 11) | Patient 11 Female, 6 10/12 y (sister of Patient 10) | Patient 12 Female, 5 mo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ancestry (domicile) | Arab (Oman) | Arab (Oman) | African/Creole (Curacao) | Dutch (Netherlands) | Cree First Nation (Canada) | Arab (United Arab Emirates) | Hispanic (Guatemala) | Arab (Oman) | Arab (Oman) | Kurdish (USA) | Kurdish (USA) | African American (USA) |
| Consanguinity (degree) | + (first cousin) | + (first cousin) | + (degree NA) | – | + (second cousin) | + (second cousin) | – | + (first cousin) | + (first cousin) | + (first cousins) | + (first cousin) | 1st degree relatives |
| PLPBP cDNA change (NM–007198) | 347C>T (homozygous) 823C>G (homozygous VUS) | 122G>A (homozygous) | 199G>A (homozygous) | 320–2A>G; 671G>C | 370–373del (homozygous) | 347C>T (homozygous) | 280A>T (homozygous) | 122G>A (homozygous) | 122G>A (homozygous) | 199G>A (homozygous) | 199G>A (homozygous) | 370–373del (homozygous) |
| Amino acid change | p.Thr116Ile; p.His275Asp (VUS) | p.Arg41Gln | p.Glu67Lys | Splicing; p.Gly224Ala | p.Asp124Lysfs*2 | p.Thr116Ile | p.Ile94Phe | p.Arg41Gln | p.Arg41Gln | p.Glu67Lys | p.Glu67Lys | p.Asp124Lysfs*2 |
| Pregnancy/delivery complications | Abnormal foetal movements | – | C-section due to foetal distress | DCDA-gemelli pregnancy | C-section due to foetal distress | Abnormal foetal movements | – | – | – | C-section due to foetal decelerations and meconium stained amniotic fluid | – | – |
| Birth HC percentile | 66th centile | 10th centile | <2nd centile | 1st centile | 82nd centile | NA | 12.5th centile | 50th centile | 50th centile | NA | 2nd centile | 22nd centile |
| Lactic acidosis | – | – | + | + | + | NA | + | – a | NA | – b | NA | + |
| Seizure onset | Day 5 | Day 7 | Day 2 | Day 1 | Day 1 | Day 4 | 2 mo | 1st w | Day 5 | Day 1 | Day 1 | Day 1 |
| Seizure typee: myoclonic | – | – | – | – | + | – | – f | + | – | + | + | |
| Seizure type: tonic | – | – | + | + | + | – | – | – | + | – | – | + |
| Seizure type: clonic | – | – | – | – | – | + | – | – | – | – | + | – |
| Seizure type: tonic-clonic | + | + | + | – | – | – | – | – | – | – | – | – |
| Seizure type: infantile spasms | – | – | – | + | – | + | – | – | – | + | + | – |
| Initial EEG pattern (at age) | Burst suppression (1 w) | NA | Discontinuous with tendency to burst suppression (5 days) | Discontinuous (Day 1) | Burst suppression (age NA) | Multifocal epileptiform activity (4 mo) | Continuously disorganized background with bursts of higher-amplitude activity (2 mo) | Burst suppression (3 w) | Burst suppression (10 days) | Discontinuous (2 days) | Discontinuous with multifocal sharps (age NA) | Burst suppression (2 days) |
| Response to initial AED treatmentc | Partial response | No response | NA | Partial response | Partial response | Partial response | Partial response | No response | Not tried | Partial response | No response | Partial response |
| Initial B6 treatment (age/responsec) | PN (5 w) PLP (2 y, 6 mo/partial response) | PN (<1 mo/ (seizure free) | PLP (5 days) PN (3 y, 10 mo/good response) | Not tried | Not tried | PN (6 mo/seizure freed) | PN and PLP (2.5 mo/seizure free) | PN (25 days/seizure free) | PN (2nd week/seizure free) | PN (2nd week/ seizure freed) | PN (age NA/ good response) | PN (age NA/no response) PLP (1 mo/seizure free) |
| B6 withdrawal (vitamer/response) | – | – | + (PLP/ seizure relapsed) | Not applicable | Not applicable | + (PN/ seizure relapsed) | – | – | – | + (PN/ seizure relapsed) | + (PN/ increased seizures) | – |
| B6 vitamer switch (type/response) | + (PN→PLP/ no improvement) | – | + (PLP→PN/ no improvement) | Not applicable | Not applicable | – | – | – | – | – | – | + (PN→PLP/ complete response) |
| Current treatment (dose) | PLP (58 mg/kg/day) Folinic acid (2 mg/kg/day) | PN (5 mg/kg/day) | PN (9 mg/kg/day) Midazolam (used during seizures only) | Not applicable | Not applicable | PN (12.8 mg/kg/day) Oxcarbazepine (53.8 mg/kg/day) | PN (23 mg/kg/day) PLP (30 mg/kg/day) | PN (6 mg/kg/day) | PN (8.5 mg/kg/day) | PN (4.7 mg/kg/day) Lamotrigine (3.5 mg/kg/day) Clobazam (0.75 mg/kg/day) | PN (7.8 mg/kg/day) Lamotrigine (4.5 mg/kg/day) (1.25 mg/kg/day) | PLP (40 mg/kg/day) Phenobarbital (9 mg/kg/day) |
| Breakthrough seizures with fever | + | + | + (seizure relapse on viral infections or sleep deprivation) | Not applicable | Not applicable | + | – | + | – | + | + | – |
| Motor neurological exam | Unremarkable | Unremarkable | Hypertonia, stereotypies | NA | NA | Mild axial hypotonia, stereotypies | Unremarkable | Unremarkable | Hyperreflexia of all limbs | Hypotonia, mild dysmetria, wide based gait | Hypotonia, mild dysmetria, wide based and ataxic gait | Mild hypotonia |
| Developmental delay | +, with ASD | – | + | Not applicable | Not applicable | +, with ASD | – | – | – | + | + | – |
| Speech delay | + | – | + | Not applicable | Not applicable | + | – | – | NA | + | + | Not applicable |
| School performance or IQ | NA | Average school performance | NA | Not applicable | Not applicable | DQ = 70, 2nd percentile (Bayley-III Cognitive Composite score) | NA | Excellent school performance | NA | NA | NA | Not applicable |
| Minor dysmorphic features | – | – | + g | – | – | – | + h | – | – | + i | – | – |
| Neuro-imaging (age)j | MRI (6 w): mild WM changes MRI (9 mo): mild hydrocephalus. MRI (3.5 y): normal | Not performed | MRI (Day 10): WM changes, large paraventricular (pseudo)-cysts, thin posterior CC, PLIC is not myelinated. | MRI (Day 1): WM changes, large paraventricular (pseudo)-cysts, thin posterior CC, PLIC is not myelinated. | MRI (Day 6): cystic leukencephalopathy | MRI (8 mo): normal. | MRI (2 mo): normal | MRI (4 wks): normal | MRI (10 mo): normal | MRI (2 days): underdeveloped frontal gyri. Subsequent MRI (age NA): thin posterior CC. | Initial MRI (age NA): normal. Subsequent MRI (age NA): slight asymmetry in height of the hippocampi, WM changes. | 2 MRI’s (2 days and 3 w): WM changes, mild dilatation of the lateral and third ventricles, PLIC is not myelinated. |
aElevated lactate but normal pH; bFirst measured after B6 treatment; cTreatment response is graded as follows: no response, partial (= mild or only short-term reduction), good (= marked long-term reduction), seizure free; dWhen combined with AEDs; eRefers to initial seizures; fThis patient has a movement disorder and lacks true epileptic seizures; gStrabismus, slight upslant of eyes and a slightly prominent forehead; hBilateral syndactyly of the third and fourth fingers; iJoint laxity; jShowing only main findings here, detailed MRI features are described in Supplementary Table 1.
AEDs = anti-epileptic drugs; ASD = autism spectrum disorder; CC = corpus callous; C-section = Caesarean section; DCDA = dichorionic diamniotic twin pregnancy; DQ = developmental quotient; HC = head circumference; mo = month(s); NA = not available; PLIC = posterior limb of the internal capsule; PN = pyridoxine; PLP = pyridoxal 5’-phosphate; VUS = variant of uncertain significance; w = weeks; WM = white matter; y = years.
Epileptic seizures started within the first week of life in all affected infants except Patient 7, who instead presented with a movement disorder (opisthotonos, oculogyric crises) at 2 months of age. Patients manifested multiple seizure types, and initial EEG showed various patterns of abnormal electrographic activity with burst suppression being common (6/11 reported). Seizures were refractory to anti-epileptic drug treatment in all patients (Table 1 and Supplementary material). All patients who received vitamin B6 (10/12) showed responsiveness and improvement of seizures or abnormal movements upon its institution. Vitamin B6 therapy was first trialled as pyridoxine in eight patients, PLP in one patient and a combination of both vitamers in another patient (Table 1). The incomplete response to pyridoxine or PLP in Patient 1 prompted the clinicians to add folinic acid to his treatment, which produced a marked reduction in seizure frequency (only two brief episodes in a 3-month period). In Patient 3, PLP was initially started but failed to exert sufficient seizure control, and adjuvant AED treatment was necessary. A similar picture was seen for Patients 6, 11 and 12, who required treatment with pyridoxine and adjuvant AED.
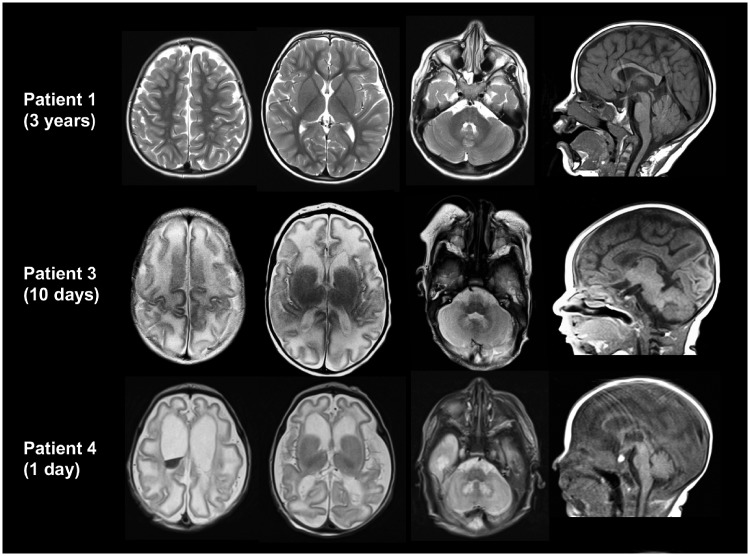
Patients 1 (Fig. 1) and 6–9 had normal brain MRI studies (with the exception of mild T2-hyperintense white matter signal in the neonatal period for Patient 1) (Supplementary Table 1). The remainder (6/11 patients for whom brain imaging was done) had structural brain abnormalities (Fig. 1 and Supplementary Table 1). Four patients (Patients 3–5 and 12) had simplified gyral pattern, suggesting prenatal onset of the disease and possible effect of PLPHP-deficiency on neuronal migration. In addition, these patients displayed large cysts adjacent to the anterior horns. In two patients, a lactate doublet was present in single voxel magnetic resonance spectroscopy of the basal ganglia.
Figure 1.
Axial T2 (first three columns) and sagittal (last column) T1-weighted images of Patients 1, 3 and 4. At age 3 years, the MRI of Patient 1 is normal. Patients 3 and 4 show a simplified gyral pattern, cyst-like structures connected to the anterior horns and a T2-hyperintense signal of the hilus of the dentate nuclei. White matter signal is T2 hyperintense and appears swollen. These abnormalities are more severe in Patient 4 (who additionally has intraventricular blood) than in Patient 3. The corpus callosum lacks the most posterior part.
Clinical presentations deviating from previous descriptions of this disease were also reported. Patient 7 showed a prominent movement disorder and biochemical picture resembling aromatic l-amino acid decarboxylase (AADC) deficiency (MIM#608643) (Supplementary material). This patient had no pathogenic variant in DDC on exome sequencing. Patients 4 and 5 presented with signs and symptoms suggestive of severe mitochondrial disease with fatal epileptic encephalopathy, lactic acidosis and brain white matter lesions. Both patients deteriorated rapidly and died at 2 and 8 weeks of age, respectively, due to uncontrolled seizures and respiratory failure. In neither case was the presentation deemed typical of pyridoxine-dependent epilepsy, nor was a trial of B6 vitamers administered (Supplementary material).
Genotypic spectrum, variant effect prediction and clinical severity
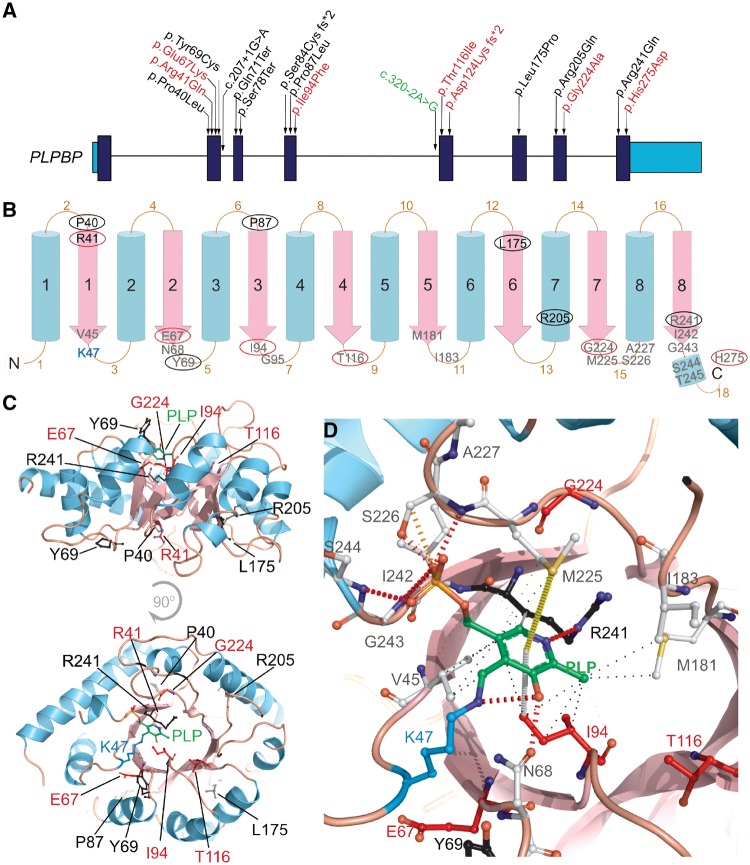
Eight variants in PLPBP were identified in our patient cohort, mostly novel missense variants (Fig. 2 and Supplementary Table 2). The exceptions are a novel homozygous frameshift deletion (c.370_373del) leading to a premature stop codon in two patients (Patients 5 and 12) and the splice site variant (c.320–2A>G) previously reported by Darin et al. (2016) (Supplementary Table 2) in another patient. To investigate potential genotype–phenotype correlations, we developed a clinical severity score to classify patients into three categories: mild, moderate and severe (Table 2). This score reflects the broad spectrum of clinical severity observed, ranging from B6-responsive epilepsy with normal developmental outcome, to perinatal lethality with lactic acidosis and structural brain malformations (e.g. Patients 4 and 5). All truncating variants leading to complete loss-of-function of PLPHP (c.207+1G>A, c.320–2A>G; p.Ser78Ter, p.Gln71Ter and p.Asp124Lysfs*2) are associated with the most severe forms of the disease (Table 2). In our cohort, this is evidenced in Patients 5 (deceased) and 12, both affected by biallelic exon 5 frameshift variants (p.Asp124Lysfs*2) leading to absence of protein expression in patient fibroblasts (Supplementary Fig. 1).
Figure 2.
Pathogenic variants reported so far and their genetic location, predicted secondary structure and 3D structure in the PLPHP protein in the context of PLP-binding. (A) Human PLPBP gene structure, protein coding exons shown in dark blue and 5′ and 3′ UTR shown in light blue. Position of the variants reported previously by Darin et al. (2016) and Plecko et al. (2017) are shown in black, seven novel variants identified by this study are shown in red and a splicing variant reported previously but also observed in our cohort is shown in green. (B) 2D graphical representation of the PLPHP protein based on secondary structure prediction and the tridimensional model (shown in D). Blue cylinders represent the outer α-helices and pink arrows represent the inner β-strands that comprise the (β/α)8-TIM barrel structure. Residues observed mutated in PLPHP-deficiency are shown in circles, black for variants reported previously or red for novel variants reported here. Residues located within 6Å of the modelled PLP position are shown in grey. (C) 3D structure of the human PLPHP model showing the PLP molecule in green, the lysine 47 residue in blue and the positions of the residues found mutated in PLPHP deficiency in black or red according to A. (D) Predicted PLP-binding pocket showing the key lysine 47 (K47) as a PLP-Lys adduct (blue and green), PLP-pocket residues (<6Å radius) and the residues found mutated in PLPHP deficiency in black or red according to A. Non-covalent contacts as calculated by Arpeggio are shown; black dashed lines indicate hydrophobic interactions, orange and red dashed lines represent weak and strong hydrogen bonds, grey dashed line represents carbon-pi interaction and a yellow dashed line indicates a methyl-sulphur-pi interaction. Note that the variant p.His275Asp was co-inherited homozygously with p.Thr116Ile in Patient 1, we report this as a variant of unknown significance.
Table 2.
Clinical severity scores based on system adapted from Al Teneiji et al. (2017)
| Patient ID | Variant type | Amino acid change | First seizure/movement episode score | GDD/ID score | B6 response score | Severity score sum | Protein effect |
|---|---|---|---|---|---|---|---|
| Patients reported in this study | |||||||
| 1 | Homozygous missense | p.Thr116Ile; | 3 | 2 | 2 | Severe (7) | Predicted LOF - variant likely impacts PLP binding |
| Homozygous missense | p.His275Asp | Variant of unknown significance: variant likely impacts PLP binding | |||||
| 2 | Homozygous missense | p.Arg41Gln | 2 | 0 | 0 | Mild (2) | Predicted to still bind PLP, but stability is reduced |
| 3 | Homozygous missense | p.Glu67Lys | 3 | 3 | 2 | Severe (8) | Predicted LOF - variant likely impacts PLP binding |
| 4 | Compound heterozygous nonsense and missense | c.320–2A>G; | 3 | NAa | NAa | Deceased: Severe (9) | LOF - Truncated proteine |
| p.Gly224Ala | Predicted LOF - Variant likely disrupts loop 15 structure and orientation of several PLP binding residues | ||||||
| 5 | Homozygous nonsense | p.Asp124Lys fs*2 | 3 | NAb | NAb | Deceased: Severe (9) | LOF - Truncated protein (band absent as in Supplementary Fig 1) |
| 6 | Homozygous missense | p.Thr116Ile | 3 | 2 | 3 | Severe (8) | Predicted LOF - variant likely impacts PLP binding |
| 7 | Homozygous missense | p.Ile94Phe | 1 | 1 | 1 | Mild (3) | Predicted LOF? Variant likely impacts PLP binding, but it is possible Phe could still establish aromatic/hydrophobic contacts with PLP; |
| 8 | Homozygous missense | p.Arg41Gln | 3 | 0 | 0 | Mild (3) | Predicted to still bind PLP, but stability is reduced |
| 9 | Homozygous missense | p.Arg41Gln | 3 | 0 | 0 | Mild (3) | Predicted to still bind PLP, but stability is reduced |
| 10 | Homozygous missense | p.Glu67Lys | 3 | 2 | 2 | Severe (7) | Predicted LOF - variant likely impacts PLP binding |
| 11 | Homozygous missense | p.Glu67Lys | 3 | 2 | 2 | Severe (7) | Predicted LOF - variant likely impacts PLP binding |
| 12 | Homozygous deletion | p.Asp124Lys fs*2 | 3 | NAd | 2 | NAd | LOF - Truncated protein (band absent as in Supplementary Fig 1) |
| Patients reported by Darin et al. (2016) | |||||||
| 1 | Homozygous nonsense | p.Ser78Ter | 3 | NAc | NAc | Deceased: Severe (9) | LOF - Truncated proteine |
| 2 | Homozygous nonsense | p.Ser78Ter | 3 | 2 | 3 | Severe (8) | LOF - Truncated proteine |
| 3 | Homozygous nonsense | p.Ser78Ter | 3 | 3 | 3 | Severe (9) | LOF - Truncated proteine |
| 4 | Homozygous missense | p.Leu175Pro | 3 | 3 | 2 | Severe (8) | LOF - Misfolded proteine,g |
| 5 | Compound heterozygous missense and missense | c.207+1G>A; | 3 | 3 | 2 | Severe (8) | LOF - Truncated proteine; absent band in western blote |
| c.320–2A>G; | |||||||
| 6 | Homozygous nonsense | p.Gln71Ter | 3 | 2 | 3 | Severe (8) | LOF - Truncated proteine |
| 7 | Compound heterozygous missense | p.Pro87Leu; | 1 | 1 | 1 | Mild (3) | Lower solubility and some precipitated; folded forms still binds to PLPg |
| p.Arg241Gln | LOF - variant abolishes PLP bindingg, drastic reduction in stability (Tm shift −14°C)g | ||||||
| Patients reported by Plecko et al. (2017) | |||||||
| 1 | Compound heterozygous missense and missense | p.Pro40Leu; | 2 | 0 | 1 | Mild (3) | Reduced stability (Tm shift −6°C); Still binds to PLPg |
| p.Arg241Gln | LOF - variant abolishes PLP binding, drastic reduction in stability (Tm shift −14°C)g | ||||||
| 2 | Compound heterozygous truncating and missense | p.Ser84Cysfs*21; | 2 | 1 | 1 | Moderate (4) | LOF - Truncated proteinf |
| p.Arg205Gln | Reduced stability (Tm shift −7°C); Still binds to PLPg | ||||||
| 3 | Homozygous missense | p.Pro87Leu | 3 | 3 | 1 | Severe (7) | Lower solubility and some precipitated; Folded forms still binds to PLPg |
| 4 | Homozygous missense | p.Tyr69Cys | 2 | 0 | 2 | Moderate (4) | Cys forms disulfide bridges, which creates an artificial dimer that hides PLP. Decreased PLP binding in 30%g |
Variants are organized by whether seen homozygously versus compound heterozygous, then based on variant type (missense, truncating, splicing). Note that truncating variants are associated with the most severe phenotypes. aNA, bNA , cNA: full clinical scores could not be calculated due to early death of these patients but assumed severe based on lethality. dNA full clinical score could not yet be calculated due to early age of patient, so GDD/ID cannot yet be assessed. eVariant reported by Darin et al. (2016).f Variant reported by Plecko et al. (2017). gVariant experimentally studied by Tremino et al. (2018). LOF = loss-of-function.
To study the pathogenic effect of the missense variants in our cohort on PLPHP function (here based on PLP-binding, folding or stability), we modelled the 3D structure of the human PLPHP protein (Fig. 2B–D). The model indicates that PLPHP folds in a typical (β/α)8-TIM barrel structure, with PLP covalently bound to Lys47 (Schiff base). As with several TIM barrel fold members, a structurally conserved ‘phosphate binding motif’ exists; this is formed by the end of β-strands and loops, especially at the C-terminal (Nagano et al., 2002). Bound PLP interacts with R241, M225, S226, I242, G243, S244, V45, N68, I94 and M181 (Fig. 2C and D). Combining the novel and previously described variants (Darin et al., 2016; Plecko et al., 2017), 12 missense PLPHP variants have been reported so far in B6RD patients (Fig. 2A), seven in homozygosity (Table 2). Patients 1, 3, 6, 10 and 11 from our cohort were classified as severe, with either p.Glu67Lys or p.Thr116Ile homozygous variants identified. Both substitutions were computationally predicted as damaging (Table 2 and Supplementary Table 2). Residues 67 and 116 are conserved (Supplementary Fig. 5) and adjacently located to the predicted PLP-binding site (Fig. 2B–D); variants to Lys and Ile, respectively, likely lead to disruption of PLP-binding properties (Supplementary Table 2). Patients 4 and 2 were also classified as severe; Patient 4 is compound heterozygous for the splice variant leading to loss-of-function (c.320–2A>G) and the substitution of the highly conserved Gly224 (Supplementary Fig. 5) to Ala (Supplementary Table 2). The p.Gly224Ala variant likely impacts loop 15 structure and orientation of key PLP-binding residues, especially due to alanine’s reduced degree of freedom (ϕ and ψ angles). Patient 1 is uniquely homozygous for two missense variants; p.Thr116Ile (discussed above) and p.His275Asp (an American College of Medical Genetics and Genomics variant of uncertain significance). The importance of the C-terminal residues for ligand binding, stability and activity of proteins that fold as a TIM barrel is well known (Wierenga, 2001; Dias-Lopes et al., 2013); therefore, a drastic chemical change like replacing a positively-charged amino acid by a negatively-charged amino acid at the C-terminus in the p.His275Asp variant may negatively impact these functions.
Of the four mild cases reported here, three patients (Patients 2, 8 and 9) are homozygous for p.Arg41Gln. Normal intellectual development, average-excellent school performance, seizures that are well controlled with relatively low doses of pyridoxine, and normal brain structure on MRI were reported in each of these patients. Arg41 is not an invariant residue (Supplementary Fig. 5) and is located in the distal face of the TIM-Barrel structure, not directly involved in PLP-binding. The p.Pro40Leu variant (adjacent residue) seen in a previously reported mild case (Plecko et al., 2017) still binds PLP but has reduced stability (Tremino et al., 2018); it is possible that p.Arg41Gln has similar impact.
Patient 7 was also classified as mild and is homozygous for a p.Ile94Phe variant. This substitution is predicted to be damaging, destabilizing and likely inducing misplacement of PLP due to the large size of Phe compared to Ile (Table 2 and Supplementary Table 2). Although Phe has not been observed at this position among known orthologues (Supplementary Fig. 5), the milder phenotype in our patient with a p.Ile94Phe variant suggests that a hydrophobic/aromatic residue can be accommodated within the PLP-binding site.
Biochemical and vitamer profiles of patients with PLPHP deficiency
Biochemical investigations performed in patients prior to B6 treatment uncovered several abnormal profiles. The most consistently observed alterations were hyperlactatemia (six patients) and hyperglycinaemia (three patients). Urine organic acids investigation in Patient 7 revealed the presence of vanillactic acid, vanillpyruvic acid, and n-acetyl-vanilalanine, similar to what is seen in AADC deficiency. Minor elevations of urine lactic, malic, 2-ketoglutaric, and N-acetylaspartic acids were also observed. Pre-treatment CSF metabolomics analysis showed elevated 3-methoxytyrosine (Z-score = 4.2) with normal 3-methoxytyramine levels, and mild elevations of: palmitoyl-GPA 16:0 (Z-score = 3.7), α-ketoglutarate (Z-score = 3.2), adenosine (Z-score = 2.6), 2-aminooctanoate (Z-score = 2.6) and tryptophan (Z-score = 2.5).
B6 vitamer analysis in plasma from Patient 4 (on no B6 treatment) revealed low levels of PLP (1.1 nM, reference >20.5 nM) and elevation of 4-pyridoxic acid (PA) (130 nM, reference <84 nM). In a plasma sample from Patient 3 collected during treatment with vitamin B6, accumulations of PLP (685 nM), 4-pyridoxic acid (365 nM), and pyridoxal (276 nM) were observed (Supplementary Table 3). Analysis of B6 vitamers from Patient 5 primary skin fibroblast lysates revealed significant decreases in PLP (P < 0.0001), pyridoxamine 5′-phosphate (PMP) (P = 0.007) and pyridoxine (P = 0.0018), along with accumulation of pyridoxine 5′-phosphate (PNP) (P < 0.0001, ANOVA) in the patient cells compared to the controls, whereas pyridoxal, pyridoxamine (PM) and 4-pyridoxic acid showed no difference (Supplementary Fig. 2). Similarly, in PLPHP-deficient HEK293 cells, PLP was markedly decreased (P < 0.0001) and PNP was greatly increased (P < 0.0001) (Supplementary Fig. 3).
PLPHP mitochondrial localization and effects on energy metabolism
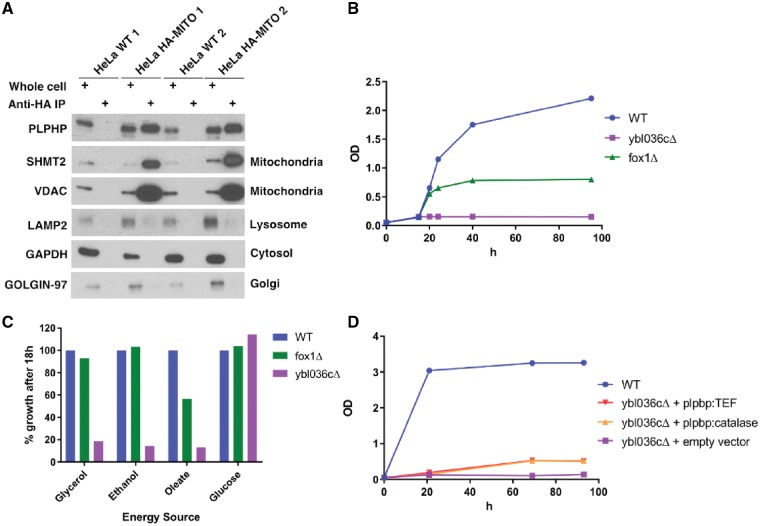
To provide further insights on PLPHP function, we investigated its subcellular localization in human cells. Some evidence suggests that PLPHP resides primarily in the cytoplasm (Ikegawa et al., 1999; Uhlen et al., 2015) (Human Protein Atlas available from www.proteinatlas.org). The MitoCarta 2.0 database, however, suggests a mitochondrial localization for human and mouse PLPHP (Pagliarini et al., 2008; Calvo et al., 2016). Furthermore, MitoMiner 4.0 rates the protein as ‘known mitochondrial’ (Integrated Mitochondrial Protein Index score 0.991), based on mass-spectrometry evidence (Smith and Robinson, 2016). To test if PLPHP does indeed localize to the mitochondria, we purified mitochondrial fractions using a recently developed method for immunoprecipitation of HA-tagged mitochondria in HeLa cells (Chen et al., 2017). The pure mitochondrial fractions were enriched for PLPHP, further evidencing the mitochondrial localization of this protein (Fig. 3A). Cytosolic and mitochondrial localization were also evidenced by immunofluorescence assays (Supplementary Fig. 3B and C).
Figure 3.
Evidence of mitochondrial enrichment of PLPHP in HeLa cells and growth defects in yeast null for the PLPHP ortholog in several energy sources requiring active mitochondrial metabolism. (A) Western blot of wild-type HeLa cells and HeLa cells with HA-tagged mitochondria (HeLa HA-MITO) that were immunoprecipitated for mitochondrial purification, showing PLPHP enrichment in the mitochondrial fraction, other antibodies show minimal contamination from the cytosol or other organelles. (B) Growth curves of wild-type yeast cells and mutant strains on rich oleate medium. The strains shown are: wild-type (WT, BY4742, blue), fox1Δ (green) and ybl036cΔ (purple). (C) Growth of wild-type and mutant cells after 18 h on 20 g/l glucose and non-fermentable carbon sources: rich oleate, 2% ethanol and 2% glycerol medium. Values are expressed as % growth relative to wild-type. The strains shown are: wild-type (BY4742) (blue), fox1Δ (green) and ybl036cΔ (purple). (D) Growth of wild-type cells and mutant cells on 2% ethanol medium. The strains shown are: wild-type (BY4742) (blue), ybl036cΔ + pPROSC1a (human PLPHP under catalase promoter) (orange) or pPROSC2a (human PLPHP under Tef promoter) (red) and ybl036cΔ + empty vector (purple).
Furthermore, we observed that the skin fibroblast cell line obtained from Patient 5 displays reduced growth in the presence of galactose as carbon source in the culture medium while normal growth was observed in the presence of glucose (Supplementary Fig. 3C and D). Patient 5 fibroblasts also showed an elevated lactate-to-pyruvate ratio (41.65 ± 7.13 standard deviations; reference 9.57–26.49), which is consistent with NADH accumulation. Activities of mitochondrial pyruvate dehydrogenase and respiratory complexes II-IV were normal, as were mitochondrial morphology and inner membrane potential (data not shown). Extracellular flux testing showed an apparent reduction of carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP)-stimulated spare respiratory capacity. These data may indicate that a direct role in electron transport is unlikely. However, considering that primary skin fibroblasts do not always replicate the disease phenotype in mitochondrial disorders (Soiferma and Saada, 2015), we decided to test other cell models.
Yeast is a well-established model to study mitochondrial function and disease (Lasserre et al., 2015). In yeast cells, ATP is produced through two mechanisms. In the presence of glucose, ATP is primarily generated via glycolysis, while gluconeogenesis and mitochondrial respiration are repressed. In the absence of fermentable carbon sources, the cell resorts to oxidative phosphorylation (OXPHOS) for the production of ATP. As a result, mutations affecting OXPHOS components are not lethal and the levels of expression of these components can be manipulated simply by changes in culture conditions (Barrientos, 2003).
To determine if PLPHP could play a role in energy metabolism, we studied the function of the PLPHP orthologue of S. cerevisiae: YBL036C. Growth of ybl036Δ yeast cells was completely normal on glucose medium but markedly reduced under conditions in which either glycerol, oleate, or ethanol was used as a carbon source (Fig. 3B–D). Since oxidation of the latter three substrates (but not glucose) is fully dependent on the proper functioning of the mitochondrial citric acid cycle and oxidative phosphorylation system, these findings suggest that YBL036C affects mitochondrial metabolism. Introduction of human PLPHP in ybl036cΔ yeast partially rescued the growth phenotype, which is consistent with a conserved function (Fig. 3D). Because PLP is a cofactor for key mitochondrial metabolism enzymes (Percudani and Peracchi, 2003) including aspartate aminotransferase (AST) in the malate-aspartate shuttle and serine hydroxymethyltransferase (SHMT2) involved in one-carbon metabolism, the metabolic pleiotropy of PLPHP deficiency is expected, although the mechanisms through which PLPBP variants produce mitochondrial dysfunction remain to be elucidated in detail.
Loss of Plphp in zebrafish leads to spontaneous seizures and early death
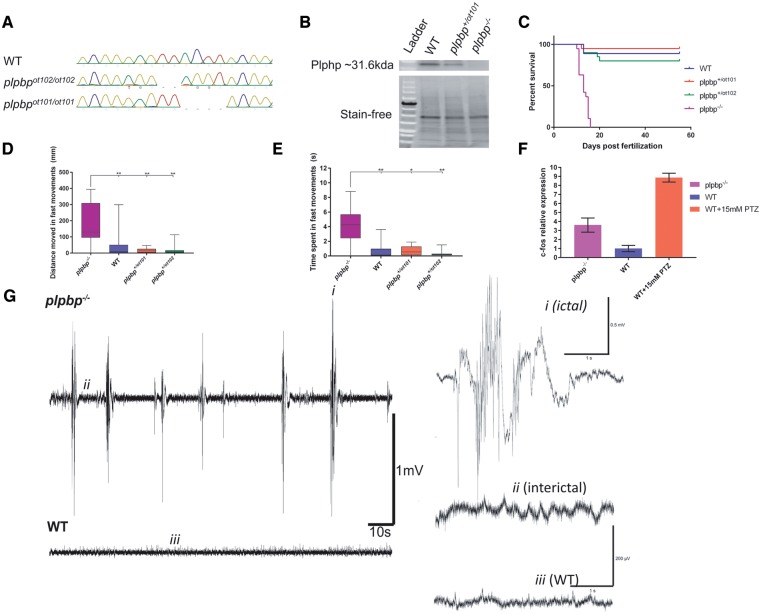
We developed zebrafish lines carrying two different plpbp mutant alleles: a 4-bp deletion (chr23:34037190–chr23:34037193) (NM_001126409; p.Asp23Lysfs*138) (plpbpot101) and the mutation CGGGTGAATCAA >CGGTGG–TGGA (chr23:34037185–34037192) (plpbpot102), the latter resulting in a 2-bp frameshift, in the transcript (NM_001126409; p.Asp23Trpfs*56) (Fig. 4A). We crossed the F2s from each heterozygous line (plpbp+/ot101 × plpbp+/ot102) to generate compound heterozygous plpbpot101/ot102 (henceforth referred to as plpbp−/−). F3 homozygous mutants and/or compound-heterozygous plpbp−/− displayed loss-of-function of Plphp as evidenced by western blot analysis (Fig. 4B). There were no phenotypic differences between homozygous and compound heterozygous mutants (Supplementary Fig. 8), and the latter was used for experiments due to the relative ease of genotyping (Supplementary material). In the F3 generation, there were no obvious morphological or behavioural differences between genotypes until ∼9 dpf. As early as 10 dpf, plpbp−/− larvae showed spontaneous seizure-like behaviour, and all mutants died by 16 dpf (Fig. 4C).
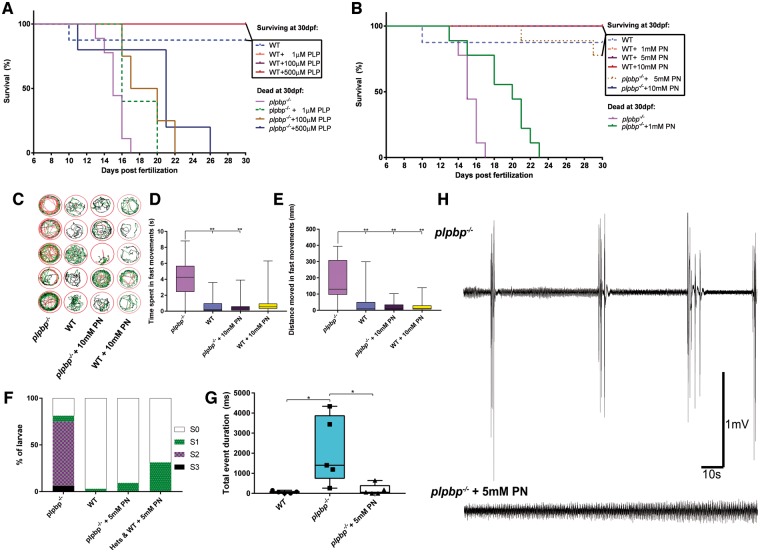
Figure 4.
Development of plpbp−/− zebrafish model by CRISPR/Cas9 and epileptic phenotypic analysis. (A) Chromatograms of zebrafish larvae showing wild-type and the genotypes for homozygous mutants plpbpot101/ot101 and plpbpot102/ot102. Compound heterozygous mutant larvae (plpbpot101/ot102) (not shown) were used for most experiments with the same phenotype as the homozygotes. (B) Cropped western blot (for clarity) showing that no Plphp protein was detected in mutant larvae. Total protein (stain free blot) is shown underneath for standardization. Full blot available in the Supplementary material. (C) Survival curves showing reduced survival of mutant larvae compared to wild-type and the two heterozygous parental types (n = 20 larvae per group). (D and E) Mutant larvae moved a greater total distance during fast speed (>20 mm/s) movements and spent more time in fast movements, respectively (n = 16 larvae per group). (F) Relative mRNA expression showing increased expression of c-fos in mutant larvae compared to wild-type larvae, pentylenetetrazol (PTZ) treatment was used as a positive control. (G) Example electrophysiology recordings of mutant (top) and wild-type (bottom) larvae showing increased number of ictal-like events. Insets are magnified examples (4 s) of ictal-like, interictal and wild-type recordings. Significance: **P < 0.01, *P < 0.05.
Epilepsy in zebrafish can be characterized by episodes of excessive locomotion, sustained rhythmic jerking (clonus), stiffening (tonus) and/or tonic-clonic seizures (Baraban et al., 2005, 2013; Hortopan et al., 2010; Teng et al., 2010). We measured the amount of high-speed movements as a correlate of hyperactivity and found that untreated plpbp−/− larvae spent significantly more time (P < 0.01) and moved a greater distance in high-speed movements (P < 0.01) than wild-type or heterozygous siblings (Fig. 4D and E). Eleven days post-fertilization plpbp−/− larvae displayed increased c-fos mRNA expression [a biomarker of neuronal activity (Baraban et al., 2005)] compared to wild-type larvae, but less than wild-type treated with 15 mM PTZ (Fig. 4F). Finally, tectal field recordings of agar-immobilized 11 dpf larvae showed that mutant larvae (n = 5) displayed spontaneous electrical discharges with high amplitude and duration, similar to ictal-like events previously reported in other zebrafish models, whereas wild-type siblings (n = 5) showed only normal activity (Figs 4G and 5G). We conclude that plpbp−/− larvae recapitulate a seizure phenotype.
Figure 5.
Vitamin B6-responsive epilepsy in plpbp−/− zebrafish larvae. Survival in mutants was moderately improved using PLP (A) but showed a better response that was clearly dose-dependent with pyridoxine (B). (C) Five-minute trace recordings of 11 dpf zebrafish larvae showing increased hyperactivity in the mutants which was alleviated with 10 mM pyridoxine treatment, as measured by (D) time spent in fast movements and (E) distance moved in fast movements. (F) Highest seizure-like behaviour category identified by blinded observers. Only untreated mutant larvae showed evidence of S2 or S3 seizure-like activity. (G) Electrographic activity in mutant larvae was normalized by treatment with 5 mM pyridoxine. (H) Example electrophysiology recordings of untreated and treated mutant larvae. Significance: **P < 0.01, *P < 0.05. PN = pyridoxine; WT = wild-type.
Vitamin B6 responsiveness and dependency in plpbp−/− larvae
We tested if seizures in plpbp-null zebrafish larvae show beneficial response to PLP and pyridoxine. Although we observed a PLP dose-dependent increase in the lifespan, all larvae died by 26 dpf, even at the highest dose (500 µM PLP) (Fig. 5A). Treatment with pyridoxine showed a more remarkable effect, with dose-dependent rescue of survival to nearly 100% until juvenile stages using 5 or 10 mM pyridoxine (Fig. 5B). Removal of pyridoxine daily treatments induced seizures and death within days, indicating B6-dependence, as previously reported for aldh7a1−/− larvae (Pena et al., 2017).
In agreement with the B6-dependency and rescue, pyridoxine treatment significantly reduced the number of hyperactive movements as measured by the time spent (P = 0.0028) and distance travelled in high-speed movements (P < 0.0001) (Fig. 5D and E). Additionally, by classifying larval movements as little movement (S0), increased spontaneous swim bursts (S1), whirlpool-like swimming (S2) or whole-body convulsions with loss of posture (S3) (Baraban et al., 2005) through blinded analysis, we observed that only untreated plpbp−/− larvae displayed S2 or S3 swimming behaviour (Fig. 5F). Similarly, treatment with 5 mM pyridoxine resulted in a 5-fold reduction of the number of high-amplitude spikes of electrographic activity in tectal field recordings (P = 0.0458) (Fig. 5G). We conclude that plpbp−/− larvae have B6-responsive and dependent epilepsy.
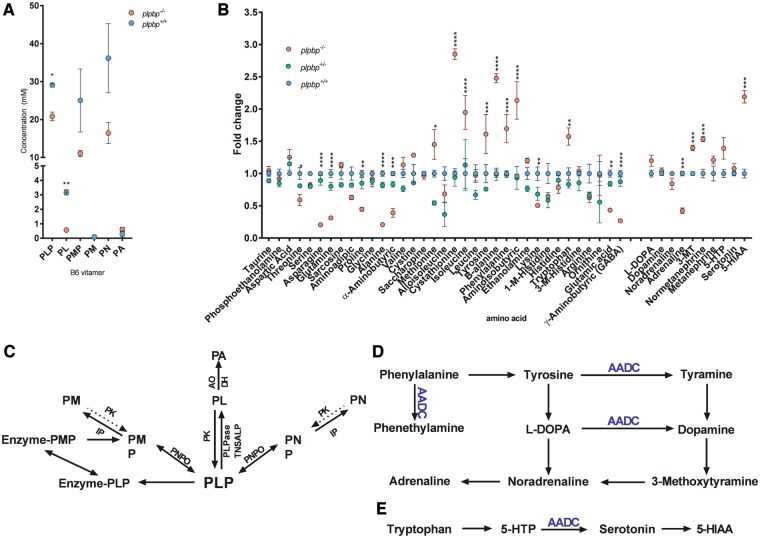
Biochemical abnormalities in plpbp−/− zebrafish
B6 vitamer levels were quantified in untreated 10 dpf larval lysates. The plpbp−/− larvae displayed significant reductions in systemic concentrations of PLP and pyridoxal (1.4- and 5.5-fold reductions, P = 0.0026 and P = 0.0003, respectively) compared to wild-type siblings, together with non-significant reductions in PMP and pyridoxine levels (Fig. 6A). PNP was not detectable in either group. As PLP was markedly low in plpbp−/− larvae, we hypothesized that neurotransmitter and amino acid metabolism would be greatly affected since most transamination/decarboxylation reactions require PLP. Neurotransmitters were also analysed in fasted 11 dpf larval lysates (Fig. 6B). We noted a significant decrease in levels of adrenaline (P < 0.001) as well as significant accumulations of 3-methoxytyramine (3-MT), normetanephrine and 5-hydroxyindoleacetic acid (5-HIAA) (P < 0.001).
Figure 6.
Targeted mass spectrometry studies of plpbp−/− zebrafish larvae indicates changes in B6 vitamer, amino acid, and neurotransmitter profiles. (A) B6 vitamer profile of mutant and wild-type 10 dpf larvae. (B) Amino acid and neurotransmitter profile of whole larval mutant, heterozygous and wild-type 11 dpf larvae after 24 h fasting. (C) Metabolic pathways for the synthesis and degradation of PLP. (D) Biosynthetic pathways of catecholamines and trace amines, highlighting (in blue) the role of AADC. (E) The serotonin biosynthesis pathway, highlighting the role of AADC. 3-MT = 3-methoxytyramine; 5-HIAA = 5-hydroxyindoleacetic acid; 5-HTTP = 5-hydroxytrytpophan; AADC = aromatic-l-amino acid decarboxylase; AO = aldehyde oxidase I; DH = β-NAD dehydrogenase; PA = 4-pyridoxic acid; PK = pyridoxal kinase; PL = pyridoxal; PM = pyridoxamine; PMP = pyridoxamine 5′-phosphate; PN = pyridoxine; PNPO = PNP oxidase. Significance: ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
Analysis of amino acid levels by liquid chromatography-mass spectrometry in fasted larvae revealed 17 analytes significantly different between homozygous mutants and the heterozygous/wild-type siblings (Fig. 6C). Nine analytes were found reduced in plpbp−/− larval extracts: threonine, asparagine, glutamate, glutamine, proline, alanine, α-aminobutyric acid, γ-aminobutyric acid (GABA), and lysine (Tukey’s post hoc comparison: P = 0.0315, P < 0.0001, P = 0.0015, P < 0.0001, P = 0.0020, P < 0.0001, P = 0.0006, P < 0.0001, and P = 0.0068, respectively). Eight compounds were significantly elevated in plpbp−/− larvae compared to wild-type: methionine, cystathionine, isoleucine, tyrosine, ß-alanine, phenylalanine, aminoisobutyric acid and tryptophan (P = 0.0147, P < 0.0001, P < 0.0001, P = 0.0005, P < 0.0001, P < 0.0001, P < 0.0001, and P = 0.0013, respectively). Low GABA levels were also observed in aldh7a1−/− zebrafish and could constitute part of the pathophysiologic mechanism for seizure occurrence. We conclude that Plphp deficiency leads to significant disruptions in amino acid and neurotransmitter metabolism and likely other metabolic pathways that are dependent on PLP in zebrafish.
Discussion
Here we report a cohort of 12 patients, six novel disease-causing variants in PLPBP, and experimental models to further elucidate the pathophysiology of this B6RD. Many of the clinical features of PLPHP deficiency in this new cohort of patients concur with those described by Darin et al. (2016) and Plecko et al. (2017), thus confirming the previously described phenotypic spectrum (Darin et al., 2016; Plecko et al., 2017). Additionally, our patients presented with novel features; one patient required folinic acid in addition to B6 for adequate seizure control, two patients suffered a lethal mitochondrial encephalopathy phenotype, while another patient presented with an AADC deficiency-phenocopy without clear epilepsy. Darin et al. (2016) described increased levels of AADC substrates in another PLPHP-deficient patient, and our zebrafish plpbp−/− model accumulated phenylalanine, tryptophan and tyrosine, in keeping with reduced AADC function. It is possible that reduction of AADC function may contribute to the clinical picture in PLPHP-deficient patients, given it is a PLP-dependent enzyme important in the biosynthesis of serotonin, dopamine, epinephrine and norepinephrine (Brun et al., 2010).
The severe clinical presentation of Patients 4 and 5 with respiratory failure, chronic lactic acidosis, NADH accumulation, and periventricular cerebral cysts prompted us to investigate whether PLPHP could have a role in mitochondrial energy metabolism. We observed enrichment of PLPHP in pure mitochondrial fractions extracted from HA-tagged mitochondria in HeLa cells (Fig. 3). The mitochondrial enrichment was also evidenced by immunofluorescence studies (Supplementary Fig. 3). In silico prediction tools and previous high-throughput mass spectrometry experiments suggested intracellular localization of PLPHP for both the cytoplasm and mitochondria (Calvo et al., 2016; Smith and Robinson, 2016). Although we could not identify clear electron transport chain defects in in the primary skin fibroblast cell line obtained from Patient 5 by Seahorse assay, its reduced growth in galactose and our identified mitochondrial enrichment of PLPHP encouraged us to investigate other models.
S. cerevisiae is a well-established model to study mitochondrial defects (Lasserre et al., 2015), and we observed that energy metabolism is affected in yeast cells deficient for the PLPHP ortholog, YBL036C (ybl036cΔ cells) (Fig. 3B–D). It is not yet clear if this is due to a direct effect or to indirect changes in key energy metabolism substrates. Several PLP-dependent enzymes, such as SHMT2 (Giardina et al., 2015), AST and the glycine cleavage system (Kikuchi et al., 2008) have mitochondrial localization. It has also recently been shown that loss-of-function variants in KYNU, encoding a PLP-dependent enzyme, lead to deficiencies in the synthesis of NAD (Shi et al., 2017). The kynurenine pathway uses tryptophan as a precursor for NAD biosynthesis, and several PLP-dependent enzymes are involved (Rios-Avila et al., 2013). The multitude of enzymatic functions of PLP may explain the complex array of biochemical phenotypes associated with B6RDs, suggestive of a key role of PLPHP in PLP homeostasis.
By adapting a clinical severity score used for another B6RD (Al Teneiji et al., 2017), we observed that the patients with severe phenotypes (scores 7–9) and/or early mortality were usually associated with proven or predicted loss-of-function variants (Table 2). These included splicing defects, truncating variants, and missense variants predicted or experimentally proven (Tremino et al., 2018) to affect PLP binding negatively. A missense variant associated with a severe disease presentation (Darin et al., 2016), p.Leu175Pro, was experimentally proven to induce PLPHP loss-of-function due to protein misfolding (Tremino et al., 2018). In contrast, it seems that missense variants in residues not associated with the PLP-binding site are seen in patients with milder disease presentations (Table 2). When stability and folding are not drastically affected, it is possible that PLPHP is still able to bind PLP, as evidenced experimentally for p.Pro40Leu and p.Arg205Gln (Tremino et al., 2018). Residual PLP binding and PLPHP function may be associated with milder presentations of the disease. In silico molecular dynamics simulations or in vitro assessment of PLP binding, PLPHP folding and stability should be performed to further access these scenarios in the missense variants reported here. We acknowledge that the clinical data presented and used to assess clinical severity were collected retrospectively after patients were identified, which limited the level of detail available. Future prospective natural history studies would be valuable in further clarifying the phenotype.
In both lysates derived from patient fibroblasts and PLPHP-deficient HEK293 cells, decreases in intracellular PLP were observed. Intracellular PLP was found to accumulate as reported by Darin et al. (2016); further work may be necessary to resolve this discrepancy. A significant accumulation of PNP levels was found in PLPHP-deficient cells, but our methods were not sensitive enough for the detection of PNP in plasma, CSF or whole zebrafish larvae. PNP accumulation, therefore, may be of limited use as a biomarker of the disease, but it may help to unravel the functional role of PLPHP.
To enable analysis of the untreated biochemical status, improve our understanding of the pathophysiology of this disease, and establish a platform for potential drug discovery, we successfully developed a plpbp-null zebrafish model. The plpbp−/− larvae recapitulated the disease, and seizure activity was detected as early as 10 dpf, with 100% mortality by 16 dpf. Treatment with pyridoxine fully reversed these phenotypes, and treated plpbp−/− larvae often survived to adulthood, but PLP was not very effective, similar to aldh7a1−/− larvae (Pena et al., 2017). It is possible that low water solubility, instability, or light sensitivity of PLP play an important role in the ineffectiveness of PLP. Larvae showed significant changes in the levels of B6 vitamers, particularly PLP and pyridoxal, which lend further support to the hypothesis that PLPHP is important for PLP homeostasis (Darin et al., 2016; Prunetti et al., 2016). By quantifying systemic amino acid levels, our results indicate disruption of many key PLP-dependent enzymes. Furthermore, the reduction of GABA may provide a possible explanation for the increased neuronal activity of mutants, as has been previously reported in aldh7a1−/− zebrafish (Pena et al., 2017). Another mechanism to consider as part of disease pathophysiology is altered biosynthesis of catecholamines (especially adrenaline), likely due to reduced availability of PLP for AADC activity (Fig. 6D). This is further evidenced in the mutant animals by the accumulation of phenylalanine, tryptophan and tyrosine (precursors to monoamine neurotransmitter synthesis). PLPHP-deficiency patients with AADC deficiency-like symptoms may provide support to this observation. Given that systemic dopamine levels were unchanged, a reduction of metabolic flux towards AADC is likely taking place, rather than a complete inactivation of this enzyme; alternatively, small amounts of dopamine may be formed via tyramine hydroxylation by renal CYP2D6, as suggested by Wassenberg et al. (2010). Our results illustrate the dynamic and complex nature of PLP binding to dependent enzymes and its turnover in the context of PLPHP deficiency.
In conclusion, we presented detailed profiles of the clinical, genetic and biochemical alterations of PLPHP deficiency in a series of 12 new patients. Given the broad phenotypic spectrum of B6RDs, PLPHP deficiency should be considered in neonatal/infantile epilepsy and possibly also in patients who present with a movement disorder ‘only’ and/or a phenotype suggestive of mitochondrial epileptic encephalopathy. In the latter case, we note that patients with severe forms of this disease may show increased levels of glycine in combination with marked lactic acidosis, a finding not typical of similar presentations such as pyruvate dehydrogenase deficiency (Prasad et al., 2011). When PLPHP deficiency is suspected, B6 therapy should be initiated. A lack of response to pyridoxine may not rule out this condition, and PLP should be trialled as well. We recommend obtaining diagnostic samples prior to B6 treatment and screening for vitamer levels, with low PLP suggestive of this condition.
We report the first animal model organism for PLPHP deficiency, which replicated the human epileptic disorder. Research using the zebrafish plpbp−/− has added insight into which PLP-dependent pathways are mostly affected and increased our understanding of systemic B6 vitamer dysfunction. The pathophysiology of the seizure phenotype in zebrafish seems to be connected with impaired PLP-dependent neurotransmitter biosynthesis and homeostasis. This model may be used to investigate other disease mechanisms and to search for biomarkers that may facilitate diagnosis. Finally, our zebrafish model provides a stepping stone for preclinical treatment trials, which are necessary, given the poor developmental outcomes and incomplete seizure control seen in many patients with this form of B6-dependent epilepsy.
Supplementary Material
Acknowledgements
We gratefully acknowledge the patients and families living with pyridoxine-dependent epilepsy for participating in this study; they give our work meaning. We also thank the Canadian Rare Disease Models and Mechanism Network for their support, as well as the clinicians and laboratory specialists involved in the management of these families, as well as the following individuals for their contributions: Xiaohua Han for Sanger sequencing; Evelyn Lomba and Dora Pak for research management support; Michelle Higginson for DNA extraction, sample handling, and technical data; Lauren Muttumacoroe and Bryan Sayson for data management; Dr David M. Sabatini for kindly donating the HeLa HA-mito cells and Dr Grzegorz Sienski for the assistance with mitochondrial purification protocols; Alexanne Cuillerier for technical advice and support; Dr Wendy Mears for help with cell culture; Dr Wyatt Yue for critically reviewing the variant interpretation section; Vishal Saxena, Christine Archer and Bill Fletcher for the invaluable help to support the zebrafish protocols.
Glossary
Abbreviations
- AADC
aromatic l-amino acid decarboxylase
- B6RD
(vitamin) B6-responsive disorder
- dpf
days post-fertilization
- PLP
pyridoxal 5′-phosphate
- PNP
pyridoxine 5′-phosphate
- TIM
typical triosephosphate isomerase
Funding
Funding support was provided by the B.C. Children’s Hospital Foundation as ‘1st Collaborative Area of Innovation’ (www.tidebc.org); Genome British Columbia (grant number SOF-195); BC Clinical Genomics Network (Michael Smith Foundation for Health Research grant #00032); the Canadian Institutes of Health Research (CIHR) (grant #301221); the Rare Diseases Foundation; a catalyst grant from the Canadian Rare Diseases Models and Mechanism Network; the Care4Rare Canada Consortium (funded by Genome Canada, CIHR, Ontario Genomics, Ontario Research Fund, and the CHEO Foundation); National Ataxia Foundation; The Physicians’ Services Inc. (PSI) Foundation; and informatics infrastructure supported by Genome British Columbia and Genome Canada (ABC4DE Project). The Zebrabox was funded by a Natural Sciences and Engineering Research Council RTI grant. D.J. is supported by a Vanier Canada Graduate Scholarship. I.A.P. is supported by a CIHR postdoctoral fellowship award. K.M.B’s program is supported by a CIHR Foundation grant (# FDN-154279). H.H.A.S. is supported by doctoral scholarship from the Ministry of Higher Education, Oman, and Al Awael Overseas Company LLC, Oman. A.D. and R.K.O. were supported by TUBİTAK from Turkey (grant #111S217). C.D.M.vK. is a recipient of the Michael Smith Foundation for Health Foundation Research Scholar Award and a Metakids Foundation salary award.
Competing interests
The authors report no competing interests.
References
- Al Teneiji A, Bruun TU, Cordeiro D, Patel J, Inbar-Feigenberg M, Weiss S, et al. Phenotype, biochemical features, genotype and treatment outcome of pyridoxine-dependent epilepsy. Metab Brain Dis 2017; 32: 443–51. [DOI] [PubMed] [Google Scholar]
- Baraban SC, Dinday MT, Hortopan GA. Drug screening in Scn1a zebrafish mutant identifies clemizole as a potential Dravet syndrome treatment. Nat Commun 2013; 4: 2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraban SC, Taylor MR, Castro PA, Baier H. Pentylenetetrazole induced changes in zebrafish behavior, neural activity and c-fos expression. Neuroscience 2005; 131: 759–68. [DOI] [PubMed] [Google Scholar]
- Barrientos A. Yeast models of human mitochondrial diseases. IUBMB Life 2003; 55: 83–95. [DOI] [PubMed] [Google Scholar]
- Basura GJ, Hagland SP, Wiltse AM, Gospe SM. Clinical features and the management of pyridoxine-dependent and pyridoxine-responsive seizures: review of 63 North American cases submitted to a patient registry. Eur J Pediatr 2009; 168: 697–704. [DOI] [PubMed] [Google Scholar]
- Baumgartner-Sigl S, Haberlandt E, Mumm S, Scholl-Bürgi S, Sergi C, Ryan L, et al. Pyridoxine-responsive seizures as the first symptom of infantile hypophosphatasia caused by two novel missense mutations (c.677T > C, p.M226T; c.1112C > T, p.T371I) of the tissue-nonspecific alkaline phosphatase gene. Bone 2007; 40: 1655–61. [DOI] [PubMed] [Google Scholar]
- Brun L, Ngu LH, Keng WT, Ch’ng GS, Choy YS, Hwu WL, et al. Clinical and biochemical features of aromatic L-amino acid decarboxylase deficiency. Neurology 2010; 75: 64–71. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 2016; 44: D1251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Freinkman E, Sabatini DM. Rapid immunopurification of mitochondria for metabolite profiling and absolute quantification of matrix metabolites. Nat Protoc 2017; 12: 2215–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton PT. B6-responsive disorders: a model of vitamin dependency. J Inherit Metab Dis 2006; 29: 317–26. [DOI] [PubMed] [Google Scholar]
- Darin N, Reid E, Prunetti L, Samuelsson L, Husain RA, Wilson M, et al. Mutations in PROSC disrupt cellular pyridoxal phosphate homeostasis and cause Vitamin-B6-dependent epilepsy. Am J Hum Genet 2016; 99: 1325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias-Lopes C, Neshich IA, Neshich G, Ortega JM, Granier C, Chavez-Olortegui C, et al. Identification of new sphingomyelinases D in pathogenic fungi and other pathogenic organisms. PLoS One 2013; 8: e79240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem 2004; 73: 383–415. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66 (Pt 4): 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy S, Gerchman S, Graziano V, Kycia H, Studier FW, Swaminathan S. Structure of a yeast hypothetical protein selected by a structural genomics approach. Acta Crystallogr D Biol Crystallogr 2003; 59 (Pt 1): 127–35. [DOI] [PubMed] [Google Scholar]
- Giardina G, Brunotti P, Fiascarelli A, Cicalini A, Costa MG, Buckle AM, et al. How pyridoxal 5’-phosphate differentially regulates human cytosolic and mitochondrial serine hydroxymethyltransferase oligomeric state. FEBS J 2015; 282: 1225–41. [DOI] [PubMed] [Google Scholar]
- Gospe SJ. Pyridoxine-Dependent Epilepsy. GeneReviews® [Internet] 2017 2017 Apr 13 [cited 2017; Available from https://www.ncbi.nlm.nih.gov/books/NBK1486/
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005; 33: D514–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortopan GA, Dinday MT, Baraban SC. Zebrafish as a model for studying genetic aspects of epilepsy. Dis Model Mech 2010; 3: 144–8. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013; 31: 227–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegawa S, Isomura M, Koshizuka Y, Nakamura Y. Cloning and characterization of human and mouse PROSC (proline synthetase co-transcribed) genes. J Hum Genet 1999; 44: 337–42. [DOI] [PubMed] [Google Scholar]
- Ito T, Iimori J, Takayama S, Moriyama A, Yamauchi A, Hemmi H, et al. Conserved pyridoxal protein that regulates Ile and Val metabolism. J Bacteriol 2013; 195: 5439–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John RA. Pyridoxal phosphate-dependent enzymes. Biochim Biophys Acta 1995; 1248(2): 81–96. [DOI] [PubMed] [Google Scholar]
- Jubb HC, Higueruelo AP, Ochoa-Montano B, Pitt WR, Ascher DB, Blundell TL. Arpeggio: a web server for calculating and visualising interatomic interactions in protein structures. J Mol Biol 2017; 429: 365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B Phys Biol Sci 2008; 84: 246–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. Osteoarthritis Cartilage 2012; 20: 256–60. [DOI] [PubMed] [Google Scholar]
- Kosuta C, Daniel K, Johnstone DL, Mongeon K, Ban K, LeBlanc S, et al. High-throughput DNA extraction and genotyping of 3dpf Zebrafish Larvae by Fin Clipping. J Vis Exp 2018(136). doi: 10.3791/58024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasserre JP, Dautant A, Aiyar RS, Kucharczyk R, Glatigny A, Tribouillard-Tanvier D, et al. Yeast as a system for modeling mitochondrial disease mechanisms and discovering therapies. Dis Model Mech 2015; 8: 509–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills PB, Struys E, Jakobs C, Plecko B, Baxter P, Baumgartner M, et al. Mutations in antiquitin in individuals with pyridoxine-dependent seizures. Nat Med 2006; 12: 307–9. [DOI] [PubMed] [Google Scholar]
- Mills PB, Surtees RA, Champion MP, Beesley CE, Dalton N, Scambler PJ, et al. Neonatal epileptic encephalopathy caused by mutations in the PNPO gene encoding pyridox(am)ine 5’-phosphate oxidase. Hum Mol Genet 2005; 14: 1077–86. [DOI] [PubMed] [Google Scholar]
- Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol 2002; 321: 741–65. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008; 134: 112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena IA, Roussel Y, Daniel K, Mongeon K, Johnstone D, Mendes HW, et al. Pyridoxine-dependent epilepsy in zebrafish caused by Aldh7a1 deficiency. Genetics 2017; 207: 1501–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep 2003; 4: 850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R, Peracchi A. The B6 database: a tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families. BMC Bioinformatics 2009; 10: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires DE, Ascher DB, Blundell TL. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res 2014; 42: W314–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plecko B, Zweier M, Begemann A, Mathis D, Schmitt B, Striano P, et al. Confirmation of mutations in PROSC as a novel cause of vitamin B 6 -dependent epilepsy. J Med Genet 2017; 54: 809–14. [DOI] [PubMed] [Google Scholar]
- Prasad C, Rupar T, Prasad AN. Pyruvate dehydrogenase deficiency and epilepsy. Brain Dev 2011; 33: 856–65. [DOI] [PubMed] [Google Scholar]
- Prunetti L, El Yacoubi B, Schiavon CR, Kirkpatrick E, Huang L, Bailly M, et al. Evidence that COG0325 proteins are involved in PLP homeostasis. Microbiology 2016; 162: 694–706. [DOI] [PubMed] [Google Scholar]
- Rios-Avila L, Nijhout HF, Reed MC, Sitren HS, Gregory JF, 3rd. A mathematical model of tryptophan metabolism via the kynurenine pathway provides insights into the effects of vitamin B-6 deficiency, tryptophan loading, and induction of tryptophan 2,3-dioxygenase on tryptophan metabolites. J Nutr 2013; 143: 1509–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res 2010; 38: W462–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger LLC. The PyMOL molecular graphics system, Version 1.8. 2015. https://pymol.org/ [Google Scholar]
- Shi H, Enriquez A, Rapadas M, Martin E, Wang R, Moreau J, et al. NAD deficiency, congenital malformations, and niacin supplementation. N Engl J Med 2017; 377: 544–52. [DOI] [PubMed] [Google Scholar]
- Smith AC, Robinson AJ. MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res 2016; 44: D1258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soiferman D, Saada A. The use of fibroblasts from patients with inherited mitochondrial disorders for pathomechanistic studies and evaluation of therapies. In: Gribkoff VK, Jonas EA, Hardwick JM, editors. The functions, disease-related dysfunctions, and therapeutic targeting of neuronal mitochondria. Hoboken, New Jersey: John Wiley & Sons, Inc; 2015. p. 378–98. doi: 10.1002/9781119017127.ch18. [Google Scholar]
- Stockler S, Plecko B, Gospe SM Jr., Coulter-Mackie M, Connolly M, van Karnebeek C, et al. Pyridoxine dependent epilepsy and antiquitin deficiency: clinical and molecular characteristics and recommendations for diagnosis, treatment and follow-up. Mol Genet Metab 2011; 104: 48–60. [DOI] [PubMed] [Google Scholar]
- Surtees R, Mills P, Clayton P. Inborn errors affecting vitamin B6 metabolism. Future Neurol 2006; 1: 615–20. [Google Scholar]
- Tarailo-Graovac M, Shyr C, Ross CJ, Horvath GA, Salvarinova R, Ye XC, et al. Exome sequencing and the management of neurometabolic disorders. N Engl J Med 2016; 374: 2246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Xie X, Walker S, Rempala G, Kozlowski DJ, Mumm JS, et al. Knockdown of zebrafish Lgi1a results in abnormal development, brain defects and a seizure-like behavioral phenotype. Hum Mol Genet 2010; 19: 4409–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremino L, Forcada-Nadal A, Contreras A, Rubio V. Studies on cyanobacterial protein PipY shed light on structure, potential functions, and vitamin B6 -dependent epilepsy. FEBS Lett 2017; 591: 3431–42. [DOI] [PubMed] [Google Scholar]
- Tremino L, Forcada-Nadal A, Rubio V. Insight into vitamin B6 -dependent epilepsy due to PLPBP (previously PROSC) missense mutations. Hum Mutat 2018; 39: 1002–13. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science 2015; 347: 1260419. [DOI] [PubMed] [Google Scholar]
- van der Ham M, Albersen M, de Koning TJ, Visser G, Middendorp A, Bosma M, et al. Quantification of vitamin B6 vitamers in human cerebrospinal fluid by ultra performance liquid chromatography-tandem mass spectrometry. Anal Chim Acta 2012; 712: 108–14. [DOI] [PubMed] [Google Scholar]
- van Vliet D, Bruinenberg VM, Mazzola PN, van Faassen MH, de Blaauw P, Kema IP, et al. Large neutral amino acid supplementation exerts its effect through three synergistic mechanisms: proof of principle in phenylketonuria mice. PLoS One 2015; 10: e0143833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker V, Mills GA, Peters SA, Merton WL. Fits, pyridoxine, and hyperprolinaemia type II. Arch Dis Child 2000; 82: 236–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenberg T, Willemsen MA, Geurtz PB, Lammens M, Verrijp K, Wilmer M, et al. Urinary dopamine in aromatic L-amino acid decarboxylase deficiency: the unsolved paradox. Mol Genet Metab 2010; 101: 349–56. [DOI] [PubMed] [Google Scholar]
- Webb B, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 2014; 47: 5.6.1–32. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio). Eugene, OR: M. Westerfield; 1993. [Google Scholar]
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 2007; 35: W407–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga RK. The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett 2001; 492: 193–8. [DOI] [PubMed] [Google Scholar]
- Zhu X, Xu Y, Yu S, Lu L, Ding M, Cheng J, et al. An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci Rep 2014; 4: 6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences, plasmids, cell and zebrafish lines are available upon request. All data necessary for confirming the conclusions are represented fully within the article or its online Supplementary material.