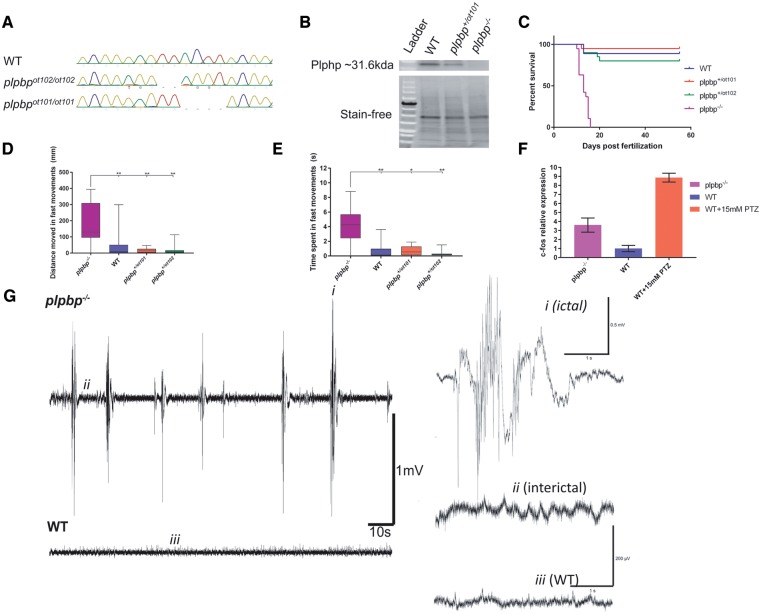
Figure 4.
Development of plpbp−/− zebrafish model by CRISPR/Cas9 and epileptic phenotypic analysis. (A) Chromatograms of zebrafish larvae showing wild-type and the genotypes for homozygous mutants plpbpot101/ot101 and plpbpot102/ot102. Compound heterozygous mutant larvae (plpbpot101/ot102) (not shown) were used for most experiments with the same phenotype as the homozygotes. (B) Cropped western blot (for clarity) showing that no Plphp protein was detected in mutant larvae. Total protein (stain free blot) is shown underneath for standardization. Full blot available in the Supplementary material. (C) Survival curves showing reduced survival of mutant larvae compared to wild-type and the two heterozygous parental types (n = 20 larvae per group). (D and E) Mutant larvae moved a greater total distance during fast speed (>20 mm/s) movements and spent more time in fast movements, respectively (n = 16 larvae per group). (F) Relative mRNA expression showing increased expression of c-fos in mutant larvae compared to wild-type larvae, pentylenetetrazol (PTZ) treatment was used as a positive control. (G) Example electrophysiology recordings of mutant (top) and wild-type (bottom) larvae showing increased number of ictal-like events. Insets are magnified examples (4 s) of ictal-like, interictal and wild-type recordings. Significance: **P < 0.01, *P < 0.05.