The High Expression Of Osmotically Responsive Gene 15–Evening Complex–histone deacetylase 9 histone-modifying complex regulates photoperiodic flowering via transcriptional repression of GIGANTEA.
Abstract
In plants, seasonal inputs such as photoperiod and temperature modulate the plant’s internal genetic program to regulate the timing of the developmental transition from vegetative to reproductive growth. This regulation of the floral transition involves chromatin remodeling, including covalent modification of histones. Here, we report that HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 15 (HOS15), a WD40 repeat protein, associates with a histone deacetylase complex to repress transcription of the GIGANTEA (GI)-mediated photoperiodic flowering pathway in Arabidopsis (Arabidopsis thaliana). Loss of function of HOS15 confers early flowering under long-day conditions because elevated GI expression. LUX ARRHYTHMO (LUX), a DNA binding transcription factor and component of the Evening Complex (EC), is important for the binding of HOS15 to the GI promoter. In wild type, HOS15 associates with the EC components LUX, EARLY FLOWERING 3 (ELF3), and ELF4 and the histone deacetylase HDA9 at the GI promoter, resulting in histone deacetylation and reduced GI expression. In the hos15-2 mutant, the levels of histone acetylation are elevated at the GI promoter, resulting in increased GI expression. Our data suggest that the HOS15–EC–HDA9 histone-modifying complex regulates photoperiodic flowering via the transcriptional repression of GI.
INTRODUCTION
Flowering time, a crucial trait for crop productivity (Jung and Müller, 2009), is regulated by environmental conditions such as daylength (photoperiod) and temperature (cold), as well as internal cues such as gibberellic acid levels and microRNAs. In Arabidopsis (Arabidopsis thaliana), flowering is promoted in long days (LD; Amasino, 2010; Song et al., 2015). The mechanism regulating the photoperiodic control of flowering involves the floral activator, CONSTANS (CO) and its target, the floral integrator, FLOWERING LOCUS T (FT; Samach et al., 2000). FT is transcribed in the leaf phloem. FT protein then moves to the shoot apical meristem, where it interacts with the bZIP transcription factor FD, to induce the expression of floral meristem identity genes such as APETALA1, FRUITFULL, and CAULIFLOWER (CAL; Corbesier et al., 2007; Jaeger and Wigge, 2007).
CO expression and stability are controlled by light and the circadian clock (Amasino, 2010; Song et al., 2015). GIGANTEA (GI) is a key clock component that is central to the photoperiodic (LD-induced) expression of CO. In LD, GI interacts with FLAVIN BINDING, KELCH REPEAT, F-BOX1 (FKF1). The light-dependent formation of the GI–FKF1 complex promotes the degradation of CYCLING DOF FACTOR1, relieving the repression of CO transcription (Imaizumi et al., 2005; Sawa et al., 2007). The stability of GI protein is regulated via its association with the clock-associated proteins EARLY FLOWERING 3 (ELF3) and CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1); the association of GI with ELF3 and COP1 leads to GI degradation in the dark and shapes the circadian oscillation pattern of its abundance (Yu et al., 2008).
ELF3, like GI, links the circadian clock with photoperiodic flowering (Hicks et al., 2001). ELF3 is part of the Evening Complex (EC), which comprises ELF3, ELF4, and LUX ARRHYTHMO (LUX, also known as PHYTOCLOCK1 [PCL1]), and functions as an important transcriptional repressor in the circadian clock (Nusinow et al., 2011; Herrero et al., 2012). The loss of ELF3 results in arrhythmicity under constant light, although rhythmicity is maintained under constant darkness, albeit with long period (Hicks et al., 2001). The elf3, elf4, and lux mutants share similar phenotypes, such as arrhythmicity, abnormal hypocotyl growth in diurnal cycles, and early flowering (Song et al., 2015). The expression of GI, CO, and FT is higher in the elf3-1 mutant than in wild type, suggesting that ELF3 negatively regulates the transcription of these genes (Kim et al., 2005). The elf3-1 co-1 double mutants flower much earlier than the co-1 single mutants under LD, although FT expression is still low in these plants, suggesting that ELF3 suppresses flowering (partially) in a CO-independent manner in addition to its role as an adaptor protein for COP1 and GI (Liu et al., 2008; Yu et al., 2008).
Epigenetic factors including DNA methylation, histone modifications, and microRNAs are involved in regulating flowering in Arabidopsis, especially in plants exposed to abiotic stress conditions such as cold, drought, and high salinity (Yaish et al., 2011). Histone proteins are subject to covalent modifications such as acetylation, methylation, phosphorylation, and ubiquitination (Yaish et al., 2011). For example, SHRINK 1 KINASE BINDING PROTEIN 1/ARGININE METHYLTRANSFERASE 5 suppresses the transcription of FLOWERING LOCUS C (FLC) and several stress-responsive genes via its association with chromatin and increases the levels of histone H4 ARG3 symmetric demethylation; thus, the skb1 mutant exhibits hypersensitivity to salt and flowers late (Wang et al., 2007; Deng et al., 2010; Zhang et al., 2011).
We previously identified HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE15 (HOS15) in a forward screen for stress response or signaling genes in Arabidopsis (Zhu et al., 2008). The hos15 mutant shows increased expression of stress-induced genes such as RESPONSIVE TO DESICCATION 29A (RD29A), COLD-REGULATED 15A, and ALCOHOL DEHYDROGENASE 1 and is hypersensitive to freezing (Zhu et al., 2008). HOS15 contains a WD-40 repeat and shares high similarity with human TRANSDUCIN (BETA)-LIKE 1X-LINKED (TBL1), a component of the chromatin repressor complex of histone deacetylase (Zhu et al., 2008). HOS15 interacts with histone H4, and acetylated H4 protein is more abundant in the hos15 mutant than wild type, suggesting that HOS15 mediates histone deacetylation, which activates or represses genes involved in cold stress tolerance (Zhu et al., 2008; Park et al., 2018a). In the current study, we show that HOS15 forms a complex with EC components and the histone deacetylase HDA9. This complex associates with the GI promoter through the direct binding of LUX to DNA and represses GI transcription, resulting in late flowering through HDA9-mediated chromatin remodeling.
RESULTS
HOS15 Is Involved in GI-Mediated Photoperiodic Flowering
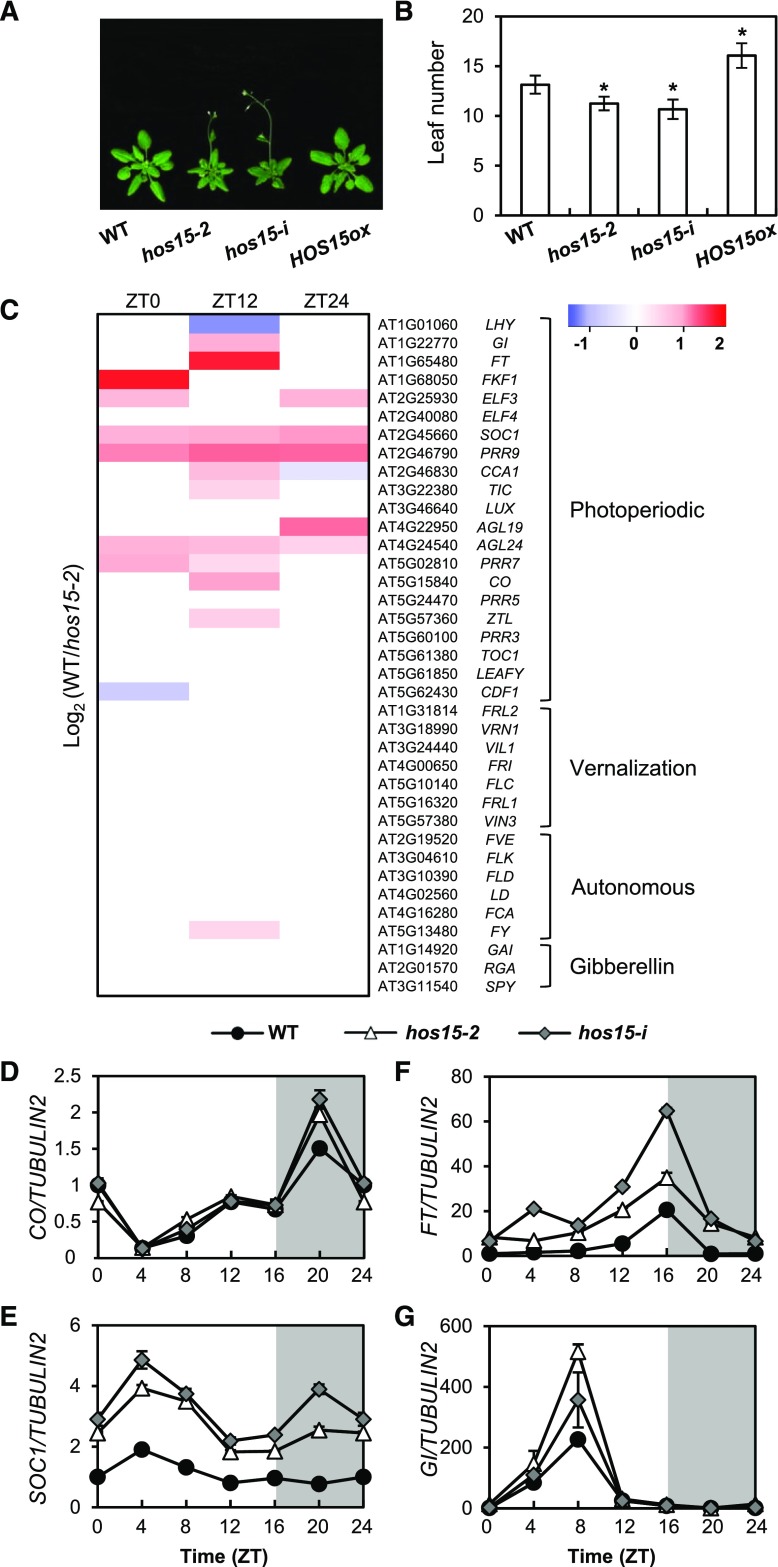
HOS15 is implicated in the regulation of flowering time because hos15-2 (GABI_785B10) mutant and hos15-i (micro RNA interference line) plants, which exhibit reduced HOS15 expression compared with wild type (Supplemental Figure 1), flower early in LD (Figures 1A and 1B). By contrast, the flowering time of HOS15 overexpressing plants (HOS15ox) is delayed relative to that of wild type (Figures 1A and 1B).
Figure 1.
HOS15 Regulates Photoperiodic Flowering Time.
(A) and (B) Flowering in hos15 mutants and HOS15 overexpressing plants (HOS15-ox) in LD. The numbers of rosette and cauline leaves were counted at bolting as an indicator of flowering time (B). Data are presented as mean ± sd of duplicate experiments, each including at least 15 plants. Asterisks indicate significantly different values from the wild type (*0.01< p-value ≤0.05; Student’s t test).
(C) RNA-seq analysis of 10-d-old hos15-2 plants. Samples were harvested at Zeitgeber Time 0 (ZT0), ZT12, and ZT24. The transcript levels of genes involved in the photoperiodic, vernalization, autonomous, and hormonal flowering pathways are shown.
(D) to (G) Transcript levels of photoperiodic flowering pathway genes in 10-d-old hos15 mutants. Transcript levels of CO (D), SOC1 (E), FT (F), and GI (G) were determined by RT-qPCR and normalized to TUBULIN2. Bars = means ± sd from three biological replicates, each with three technical replicates.
Flowering is regulated by multiple pathways, including the vernalization, autonomous, and photoperiodic pathways. To determine the pathway(s) in which HOS15 functions, we performed high-throughput RNA-seq analysis (Figure 1C; Supplemental Data Set 1). The transcript levels of floral pathway integrator genes such as SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), FT, AGAMOUS-LIKE 19 (AGL19), and AGL24, and several meristem-identity genes such as APETALA1 and CAL were higher in the hos15-2 mutant than in wild type, as were the transcript levels of photoperiodic-dependent flowering genes such as GI, FKF1, and CO (Figure 1C; Supplemental Figures 2A and 2B; Supplemental Data Set 1). By contrast, the expression of vernalization genes including FLC, VERNALIZATION INSENSITIVE3, VERNALIZATION INSENSITIVE3 LIKE1, and REDUCED VERNALIZATION RESPONSE, as well as genes in the autonomous floral promotion pathway such as FLOWERING LOCUS D, MULTICOPY SUPPRESSOR OF IRA1 4 (MSI 4), FLOWERING TIME CONTROL PROTEIN FY, FLOWERING TIME CONTROL PROTEIN FCA, and FLOWERING TIME CONTROL PROTEIN FPA were unchanged in hos15-2 relative to wild type (Figure 1C; Supplemental Data Set 1). To validate the RNA-seq data, we performed reverse transcription quantitative PCR (RT-qPCR) to measure mRNA levels in wild type and hos15 plants under LD conditions (Figures 1D to 1G). Consistent with the RNA-seq data, the photoperiodic genes GI, CO, and FT and the floral integrator gene SOC1 were more highly expressed in hos15-2 than in wild type (Figures 1D to 1G). The expression levels of the genes encoding ELF3 (which acts upstream of GI), ELF4 (which interacts with ELF3), and PSEUDO RESPONSE REGULATORs (PRRs) such as PRR7 and PRR9 (which stabilize CO protein and promote flowering) were higher in hos15-2 mutants (Supplemental Figure 2; Supplemental Data Set 1; Hayama et al., 2017). These results suggest that HOS15 is involved in GI-mediated photoperiodic flowering.
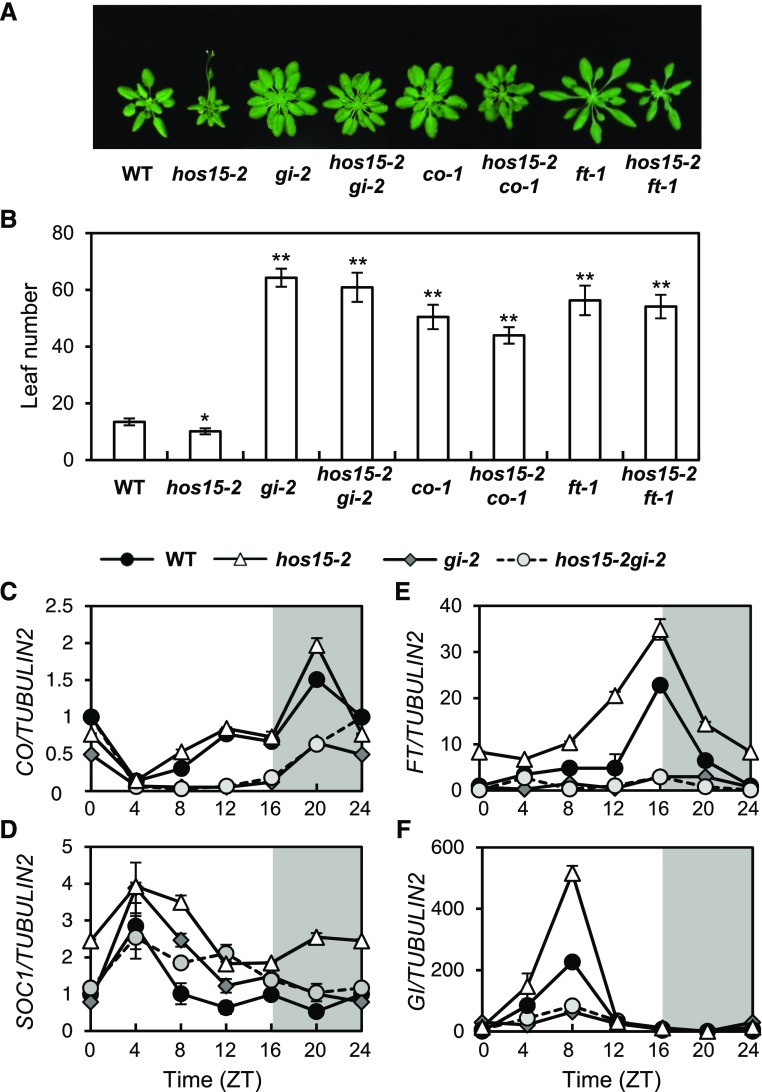
To investigate the genetic relationship between HOS15 and genes in the photoperiodic pathway, we crossed hos15-2 with gi-2, co-1, and ft-1. All double mutants, including hos15-2 gi-2, hos15-2 co-1, and hos15-2 ft-1, were late flowering in LD, like gi-2, co-1, and ft-1 single mutants (Figures 2A and 2B), suggesting that HOS15 functions upstream of GI, CO, and FT. The expression patterns of CO, FT, and SOC1 in hos15-2 gi-2 were very similar to their patterns in gi-2 (Figures 2C to 2F), suggesting that GI is epistatic to HOS15 in regulating the expression of FT, CO, and SOC1. However, the early flowering of the hos15-2 mutant under non-inductive short-day (SD) conditions is independent of GI (Supplemental Figure 3). First, the hos15-2 gi-2 double mutant flowers early under non-inductive SD conditions (Supplemental Figures 3G and 3H). Second, GI transcript levels were unchanged in hos15-2, although FT and SOC1 were upregulated (Supplemental Figures 3C to 3F). Therefore, in addition to photoperiodic flowering, HOS15 might be involved in other flowering pathways. In this study, we focused on the function of HOS15 in GI-mediated photoperiodic flowering in LD.
Figure 2.
HOS15 Is Involved in the GI-Mediated Photoperiodic Flowering Pathway.
(A) and (B) Flowering of hos15-2 and other mutants in LD. The numbers of rosette and cauline leaves were counted at bolting as an indicator of flowering time (B). Data are presented as means ± sd of three independent experiments, each including at least 10 plants. Asterisks indicate significantly different values from the wild type (*0.01 < p-value ≤ 0.05; **p-value < 0.01; Student’s t test).
(C) to (F) Transcript abundance of photoperiodic flowering genes in hos15-2 and other mutants. CO (C), SOC1 (D), FT (E), and GI (F) transcript abundances were determined by RT-qPCR and normalized to TUBULIN2. Bars = means ± sd from three biological replicates, each with three technical replicates.
HOS15 Regulates the H3 Acetylation Status of the GI Promoter through an Interaction with LUX
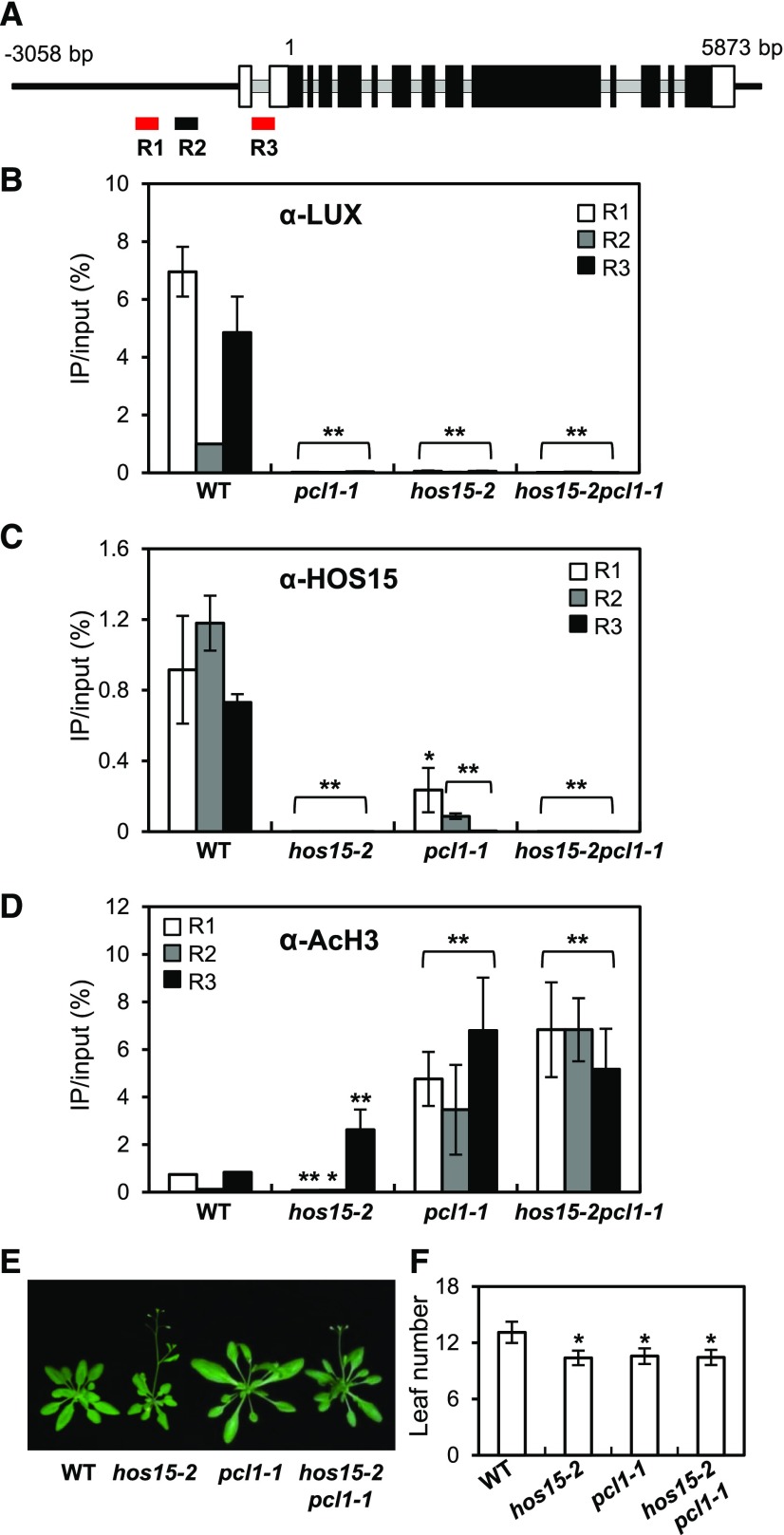
HOS15 physically interacts with H4 and associates with histone deacetylase complexes (HDAC) to repress the expression of cold-induced downstream genes (Zhu et al., 2008; Park et al., 2018a). This finding, together with the increased expression of GI in hos15-2 mutants (Figure 1G), prompted us to investigate whether HOS15 represses GI transcription by recruiting HDACs to the GI promoter. Because GI transcription is regulated by the EC through its binding to a LUX binding site (LBS, GATWCG, where W is A or T) in the GI promoter (Berns et al., 2014; Mizuno et al., 2014), we designed two amplicons containing putative LBSs (R1 and R3) and another amplicon without a putative LBS (R2; Figure 3A). Mizuno et al. (2014) showed that ELF3 and LUX associate with the GI-c region of GI, i.e., R3.
Figure 3.
HOS15 Represses GI Expression through De-Acetylating the Histones at the GI Promoter.
(A) Schematic representation of the GI locus and the locations of the three amplicons used for ChIP analysis. R1 and R3 (red) include LUX binding sites. Introns are indicated by gray lines, and exons are indicated by boxes; black boxes indicate the translated coding sequences, and white boxes indicate the untranslated regions.
(B) to (D) ChIP analysis of 2-week-old wild type, hos15-2, pcl1-1, and hos15-2 pcl1-1 plants. Samples were harvested at ZT8. Genomic DNA was immunoprecipitated by α-LUX (B), α-HOS15 (C), or α-AcH3 (D) antibodies. Results were normalized to the input DNA (n = 3), and values represent means ± sd from three biological replicates, each with three technical replicates. Asterisks indicate significantly different values from the wild type (*0.01 < p-value ≤ 0.05; **p-value < 0.01; Student’s t test).
(E) and (F) pcl1-1 and hos15-2 pcl1-1 plants growing under LD were photographed (E), and flowering time was quantified by counting rosette and cauline leaves at bolting (F). Data are presented as means ± sd from two biological replicates, each including at least 15 plants. Asterisks indicate significantly different values from the wild type (*0.01 < p-value ≤ 0.05; **p-value < 0.01; Student’s t test).
We first investigated whether LUX indeed binds to or associates with R3 in the GI promoter region in vivo by performing chromatin immunoprecipitation (ChIP) using 2-week-old wild type and pcl1-1 (a loss of function mutation in LUX, which is also known as PCL1) plants probed with α-LUX antibody, followed by quantitative PCR (qPCR). Indeed, LUX associated with R1 and R3 in the GI promoter, and these fragments contain the LBSs (Figure 3B). We then performed ChIP analysis using wild type and hos15-2 plants, finding that HOS15 associated with the R1 and R3 regions in the GI promoter, as well as the non-LBS R2 region (Figure 3C). The interaction was not detected in the hos15-2 mutant, indicating that it is specific (Figure 3C; Supplemental Figure 4A). Interestingly, HOS15 and LUX each appear to be required for the other to bind to the GI promoter, because the association of LUX and HOS15 with all regions of the GI locus was lost in the hos15-2 and pcl1-1 mutants (Figures 3B and 3C).
We also performed ChIP assays with α-AcH3 antibody to examine whether H3 acetylation (AcH3) in the GI promoter is affected in hos15-2 and pcl1-1 mutants. The ChIP results showed that the levels of AcH3 of the R3 region were higher in hos15-2 and pcl1-1 mutants than in wild type. Also, the H3 levels on the GI promoter region in hos15-2 and pcl1-1 mutants were similar to the wild type levels (Figure 3D; Supplemental Figures 4B and 5A). These results suggest that HOS15 is involved in H3 deacetylation of the R3 site in the GI promoter, perhaps leading to the downregulation of GI transcription.
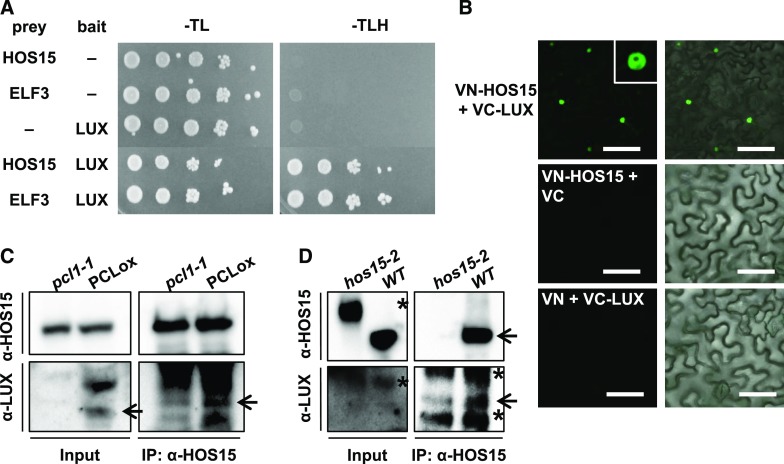
Because HOS15 associated with the same LBS regions on the GI promoter as LUX, we investigated whether HOS15 can bind to LUX. To test the HOS15–LUX interaction, we used a yeast two-hybrid assay, which showed that HOS15 does bind to LUX (Figure 4A). In addition, we performed bimolecular fluorescence complementation (BiFC) assays and in vivo co-immunoprecipitation (Co-IP) to confirm the interaction of HOS15 with LUX in planta (Figures 4B to 4D). For BiFC, we transiently expressed HOS15 fused to the N-terminal fragment of the eYFP fluorescent protein (VN-HOS15) and LUX fused to the N-terminal fragment of the eYFP fluorescent protein (VC-LUX) in Nicotiana benthamiana epidermal cells. The fluorescence was detected in the nucleus, indicating that HOS15 associates with LUX in the nucleus (Figure 4B). The interaction of HOS15 with LUX was verified in Arabidopsis by Co-IP with PCL-overexpressing transgenic plants and hos15-2 mutants, and α-HOS15 and α-LUX antibodies (Figures 4C and 4D; Supplemental Figure 6A). These results suggest that HOS15 interacts with LUX and contributes not only to the ability of HOS15 to associate with the GI promoter, but also to histone deacetylase activity at the GI promoter. Furthermore, like hos15-2, the pcl1-1 and hos15-2 pcl1-1 mutants exhibited early flowering (Figures 3E and 3F), likely because the high levels of acetylated H3 on the GI promoter and the consequent strong expression of GI (Figure 3D).
Figure 4.
HOS15 Associates with LUX.
(A) HOS15 interacts with LUX in a yeast two-hybrid assay. Shown are the results of a yeast two-hybrid assay using HOS15 or ELF3 as prey and LUX as bait. Empty vector was used as a negative control. Growth on SD-TLH medium indicates a positive interaction. ELF3, which interacts with LUX, was used as a positive control.
(B) BiFC analysis of proteins transiently expressed in Nicotiana benthamiana leaves. VN and VC indicate the N and C termini, respectively, of Venus (eYFP). Fluorescence (left panels) was detected by confocal microscopy. Right panels are overlay of fluorescence and differential interference contest images. Bars = 100 μm.
(C) and (D) HOS15 interacts with LUX. Total protein extracts from 10-d-old plants were immunoprecipitated with α-HOS15 antibodies and resolved by SDS-PAGE. Immunoblots were probed with α-HOS15, or with α-LUX antibodies to detect LUX. Wild type and hos15-2 plants used for (D) were cross-linked in 1% formaldehyde before sampling. *Nonspecific bands.
HOS15 Interacts with EC Components
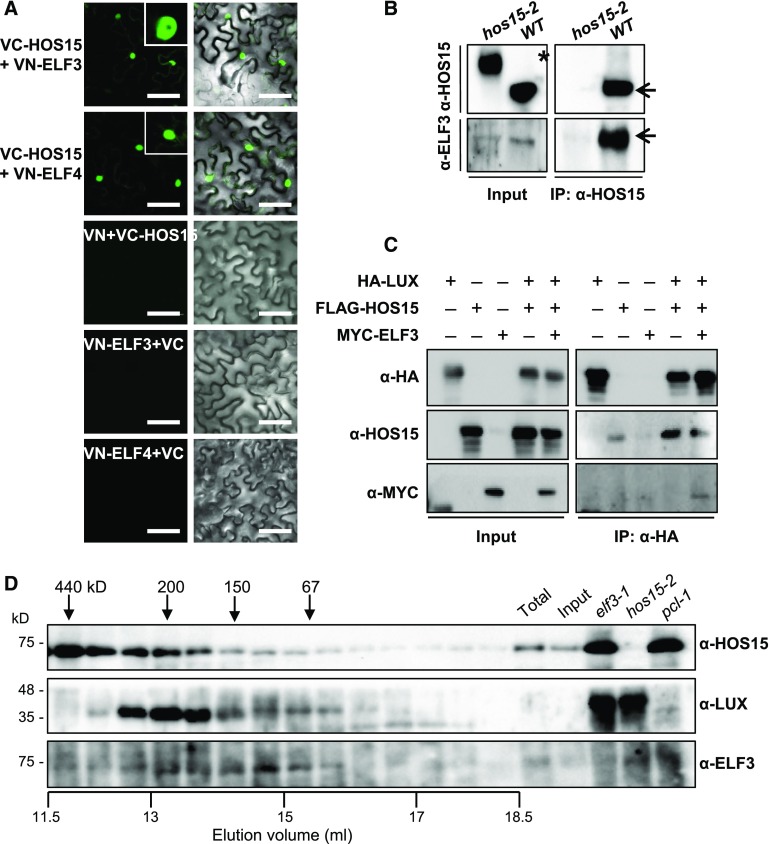
LUX forms a complex with ELF3 and ELF4; this EC functions as a nighttime repressor upstream of GI (Mizuno et al., 2014) and binds to the PRR9 promoter (Chow et al., 2012). The recruitment of ELF3 and ELF4 to chromatin is dependent on LUX and its homolog NOX, also known as BROTHER OF LUX ARRHYTHMO (Dai et al., 2011; Helfer et al., 2011; Chow et al., 2012; Herrero et al., 2012). Furthermore, the elf3 and pcl1-1 mutants exhibit early flowering (Fowler et al., 1999; Hazen et al., 2005; Yu et al., 2008). Therefore, we investigated whether HOS15 interacts with the other EC components ELF3, and ELF4. To this end, we used a BiFC assay in which we transiently expressed HOS15 fused to the C-terminal fragment of eYFP (VC-HOS15) and ELF3 fused to the N-terminal fragment of eYFP (VN-ELF3) in N. benthamiana epidermal cells. HOS15 binds not only to LUX (Figure 4), but also to ELF3 and ELF4 in the nucleus (Figure 5A). In vivo Co-IP assays confirmed the interactions of HOS15 with ELF3, using total protein extracts from N. benthamiana leaves transiently co-expressing HOS15 with ELF3 (Supplemental Figure 6B). The interactions were also confirmed in Arabidopsis by Co-IP with α-HOS15 antibodies (Figure 5B).
Figure 5.
HOS15 Associates with EC Components.
(A) BiFC analysis of proteins transiently expressed in N. benthamiana leaves. VN and VC indicate the N and C termini, respectively, of Venus (eYFP). Fluorescence (left panels) was detected by confocal microscopy. Right panels are overlay of fluorescence and differential interference contest images. Bars = 100 μm.
(B) HOS15 interacts with ELF3. Wild type (10 d old) and hos15-2 plants were cross-linked in 1% formaldehyde to preserve in vivo interaction after harvesting plant material. Total protein extracts were immunoprecipitated with α-HOS15 antibody and resolved by SDS-PAGE. Immunoblots were probed with α-HOS15, or with α-ELF3 to detect ELF3. *Nonspecific bands.
(C) HOS15 forms a complex with EC components. N. benthamiana plants were infiltrated with Agrobacterium harboring 35S:HA-LUX, 35S:FLAG-HOS15, and 35S:MYC-ELF3 for transient expression. The proteins were immunoprecipitated with α-HA antibody and resolved by SDS-PAGE. The immunoblots were probed with α-HA to detect LUX, α-HOS15, or α-MYC to detect ELF3.
(D) Gel-filtration analysis of HOS15, LUX, and ELF3. Total proteins extracted from 2-week-old wild type plants were fractionated by size-exclusion chromatography using a Superdex 200 10/300GL column equilibrated with elution buffer [50 mM Tris-Cl (pH 7.5), 100 mM NaCl, 0.02% sodium azide]. The samples were separated by SDS-PAGE and subjected to immunoblotting analysis using α-HOS15, α-LUX, or α-ELF3. Aliquots of total protein extracts from wild type, input after ammonium sulfate precipitation of wild type total extracts, and total protein extracts individually isolated from elf3-1, hos15-2, or pcl1-1 mutants were used as input controls. Molecular mass markers (ferritin, 440 kD; β-amylase, 200 kD; alcohol dehydrogenase, 150 kD; BSA, 67 kD) were independently eluted using the same equilibrated column. Total, total protein extract; Input, input after ammonium sulfate precipitation.
Furthermore, we conducted Co-IP assays using transient co-expression of HA-LUX, FLAG-HOS15, and MYC-ELF3 in N. benthamiana leaves with α-HA. These Co-IPs showed that HOS15 forms a complex with LUX and ELF3 (Figure 5C). Finally, a gel filtration assay demonstrated that HOS15, LUX, and ELF3 co-eluted in complexes with molecular masses of ∼300–150 kD (Figure 5D). Although we did not directly test for the presence of the third component of the EC, ELF4, we think that the co-eluted complex likely comprised HOS15:LUX:ELF3:ELF4 in a 1:1:1:1 ratio, because the sum of the molecular masses of these heterogeneous protein complexes is ∼195 kD, which corresponds to the 150–300 kD fractions in which these proteins co-eluted. Together, these results suggest that HOS15, in association with the EC, represses the expression of GI.
HDA9 Is Involved in HOS15–GI-Mediated Photoperiodic Flowering
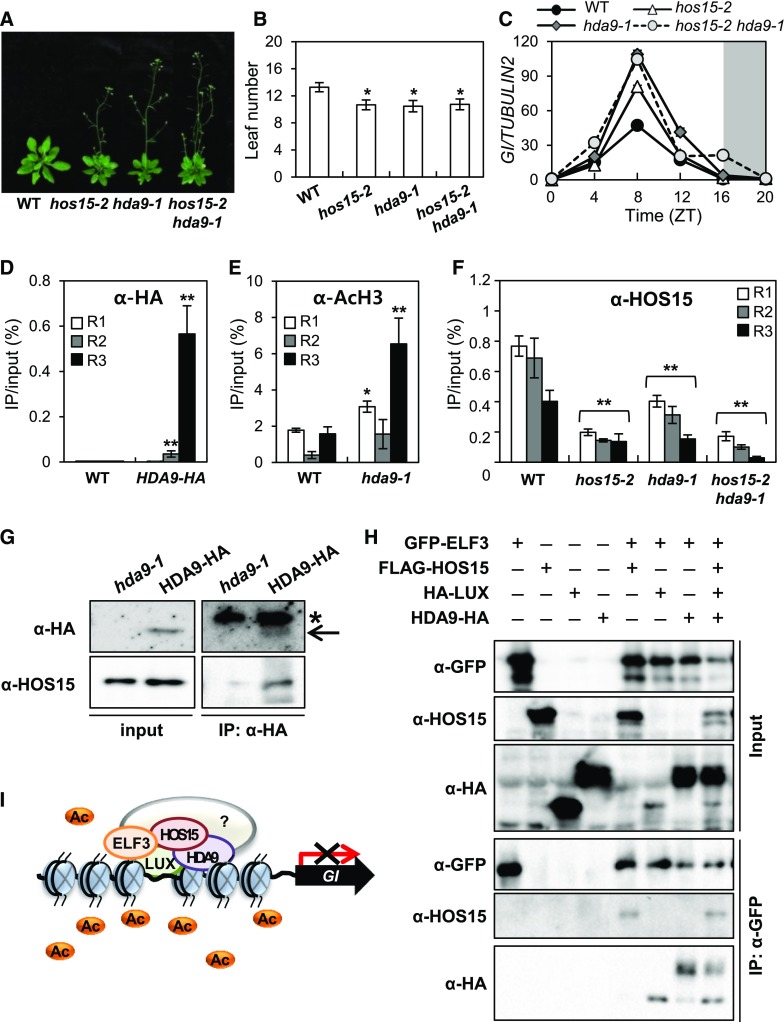
Histone acetylation is regulated by histone acetyltransferases and HDACs, which control transcription in various systems. Our data suggest that HOS15 functions as a transcriptional co-repressor in conjunction with an HDAC (Figure 3; Supplemental Figure 7). Arabidopsis HDACs related to members of the yeast REDUCED POTASSIUM DEPENDENCY 3 (RPD3) family include HDA6, HDA7, HDA9, and HDA19; among these, hda6, hda9, and hda19 mutants exhibit altered flowering time, with HDA9 negatively regulating this trait (Kim et al., 2013; Kang et al., 2015). We confirmed that hda9-1 flowers earlier than wild type under LD (Figures 6A and 6B; Supplemental Figure 8).
Figure 6.
HOS15 Regulates Histone Modification with HDA9 in the GI-Mediated Photoperiodic Flowering Pathway.
(A) and (B) The hda9-1 mutant exhibits early flowering in LD. The numbers of rosette and cauline leaves were counted at bolting as an indicator of flowering time (B). Data are presented as means ± sd (n > 15). Asterisks indicate significantly different values from the wild type (*0.01 < p-value ≤ 0.05; Student’s t test).
(C) GI transcript levels increase in hos15, hda9, and hos15 hda9 mutants. Transcript levels of GI were determined in 10-d-old plants grown under LD by RT-qPCR and normalized to TUBULIN2. Bars = means ± sd from two biological replicates, each with three technical replicates.
(D) to (F) ChIP analysis of 2-week-old plants. Samples were harvested at ZT8 (D) and (E) or ZT12 (F). Genomic DNA was immunoprecipitated by α-HA (D), α- AcH3 (E), or α-HOS15 (F) antibodies, respectively. Specific regions of the GI promoter were amplified by qPCR. Results were normalized to the input DNA (n = 3), and values represent means ± sd from three biological replicates, each with three technical replicates. Asterisks indicate significantly different values from the wild type (*0.01 < p-value ≤ 0.05; **p-value < 0.01; Student’s t test).
(G) HOS15 interacts with HDA9. The total proteins extracted from proHDA9:HDA9-HA transgenic plants were immunoprecipitated with α-HA antibody and resolved by SDS-PAGE. The immunoblots were probed with α-HOS15 and α-HA, respectively. The input protein was extracted in urea/SDS buffer. *Nonspecific bands.
(H) HOS15 and HDA9 form a complex with the EC. N. benthamiana plants were infiltrated with Agrobacterium harboring 35S:GFP-ELF3, 35S:FLAG-HOS15, 35S:HA-LUX, and 35S:HDA9-HA for transient expression. The proteins were immunoprecipitated with α-GFP antibody and resolved by SDS-PAGE. The immunoblots were probed with α-HA to detect LUX (bottom band) and HDA9 (top band), α-HOS15, and α-GFP to detect ELF3.
(I) Model of the role of HOS15 in the photoperiodic flowering pathway. A histone deacetylase complex composed of HOS15, HDA9, and EC components binds to the GI promoter through LUX, and the level of acetylated H3 at the GI locus is reduced, thereby repressing its transcription.
The early flowering phenotype of the hos15-2 hda9-1 double mutant was similar to that of the hos15-2 single mutants (Figures 6A and 6B), and the early flowering is associated with elevated GI expression (Figure 6C). Furthermore, ChIP analysis showed that HDA9 associates with the R3 region of the GI promoter, and the level of AcH3 on the GI promoter was increased in hda9-1 mutant, especially at the R3 region (Figures 6D and 6E). Finally, Co-IP analysis with proHDA9:HDA9-HA/hda9-1 transgenic plants revealed that HDA9 indeed interacts with HOS15 in planta (Figure 6G; Park et al., 2018b). In addition, we found that the binding of HOS15 to the GI promoter was reduced in hda9-1 and hos15-2 hda9-1 plants by ChIP analysis (Figure 6F). These results suggest that HOS15 and HDA9 regulate flowering via a single pathway.
Next, we investigated whether the HOS15–HDA9 complex is associated with the EC. We performed Co-IP analysis of N. benthamiana epidermal cells coexpressing GFP-ELF3, FLAG-HOS15, HA-LUX, and HDA9-HA (Figure 6H). The HOS15–HDA9 complex was detected in the same complex as ELF3 and LUX, suggesting that the HDAC, HOS15, and HDA9 function together with the EC to regulate flowering. Collectively, our findings suggest that an HDAC composed of HOS15, HDA9, and the EC binds to the GI promoter through LUX and regulates the acetylation status of histones at the GI locus, resulting in compacted chromatin structure and repressed GI transcription, thereby delaying flowering in Arabidopsis (Figure 6I).
DISCUSSION
HOS15 Negatively Regulates GI Transcription under LD through the EC
Plants modulate the regulatory network controlling the time of flowering in response to seasonal cues such as daylength and ambient temperatures, and environmental and internal cues such as sugars, hormones, plant age, and stress (Bouché et al., 2016). Four distinct pathways, i.e., the autonomous, gibberellin, photoperiod, and vernalization pathways, regulate flowering through common floral integrators such as FT, SOC1, and LEAFY (Amasino, 2010; Song et al., 2015). The photoperiod pathway integrates the circadian clock and light signals as inputs, with seasonal changes causing fluctuations in daylength and temperature (Song et al., 2015). In polar and temperate regions, temperatures are lower and daylength is shorter in winter than in summer. Arabidopsis is an LD plant that flowers late under winter-like conditions (Song et al., 2015), suggesting that it senses these environmental cues as signals in parallel. Plants also interpret environmental changes such as abiotic stresses as inputs in the regulation of flowering. Developmental process leading to the floral transition and stress responses are extensively regulated by epigenetic processes such as RNA metabolism, DNA modifications, and histone modifications (Yaish et al., 2011; Nguyen et al., 2016).
We previously isolated several Arabidopsis mutants with altered expression of RD29A:LUC, a stress marker that is induced by low temperatures, abscisic acid (ABA), or osmotic treatment. We found that hos15 plants with high levels of RD29A:LUC expression were sensitive to freezing stress and showed upregulated expression of stress-responsive genes, especially the cold-responsive gene COLD-REGULATED 15A (Zhu et al., 2008; Park et al., 2018a). Here, we found that the hos15-2 mutant and the RNA interference line hos15-i showed early flowering in LD because upregulated expression of GI and FT, which function in photoperiodic flowering (Figure 1). Genetic analysis of HOS15 and photoperiodic flowering genes (GI, CO, and FT) revealed that HOS15 functions upstream of GI (Figure 2).
GI is downregulated by the EC at night via interaction of the EC with LUX binding sequences (LBS) in the GI promoter (Mizuno et al., 2014). We found that HOS15 associates with the GI promoter, through the interaction of HOS15 with the EC, which is composed of ELF3 (a negative regulator of GI; Kim et al., 2005), ELF4, and LUX (Figures 3 to 5). The association of HOS15 with the GI promoter was reduced in the pcl1-1 mutant, which lacks LUX (Figure 3C), suggesting that a repressor complex composed of HOS15 and the EC suppresses GI transcription via the binding of LUX to the GI promoter.
A study examining the freezing sensitivity of hos15 mutants suggested that HOS15 functions as a repressor of cold tolerance genes via histone deacetylation (Zhu et al., 2008). Similarly, in hos15-2, pcl1-1, and hos15-2 pcl1-1 mutants, the GI promoter contained higher levels of acetylated histone H3 than in wild type (Figure 3D; Supplemental Figure 4B). In addition, in early morning (ZT0) when the GI transcript levels are low in wild type and hos15-2 mutants, the association of HOS15 to the GI promoter is low with low abundance of acetylated H3 on the same region, although HOS15 targets strongly to the GI promoter and acetylated H3 increases at ZT8 (Figure 1; Supplemental Figure 4). These results suggest that the complex formed of HOS15 and the EC represses GI transcription via histone deacetylation in the afternoon when the EC starts to form (Figure 6I).
LUX/PCL1, an evening-phased circadian clock component, participates in a feedback loop with CCA1 and LHY (Hazen et al., 2005; Onai and Ishiura, 2005), which directly bind to the evening element motif within the LUX promoter to inhibit its expression (Hazen et al., 2005). In turn, LUX promotes the expression of CCA1 and LHY via an unknown mechanism (Hazen et al., 2005). In addition, ELF3 and ELF4 form a feedback loop with CCA1 and LHY, which inhibit ELF3 and ELF4 expression through direct binding to the evening element motifs within their promoters (Hicks et al., 2001; Huang and Nusinow, 2016). Like LUX, ELF3 and ELF4 promote the expression of CCA1 and LHY via an indirect mechanism (Hicks et al., 2001; Huang and Nusinow, 2016). LUX selectively binds to the LBS of its own promoter and inhibits its own expression (Dixon et al., 2011; Helfer et al., 2011; Chow et al., 2012). The EC represses PHYTOCHROME INTERACTING FACTOR4 (PIF4) and PIF5 transcription via the direct binding of LUX to their promoters around dusk, thereby limiting their expression until dawn. Consequently, hypocotyl growth is inhibited in the early evening and resumes later in the night as this repression is relieved (Nusinow et al., 2011). The EC also associates with the promoters of PRR7 and PRR9, which encode transcriptional repressors of the circadian clock, through direct binding of LUX to the LBS of these loci (Helfer et al., 2011; Chow et al., 2012; Huang and Nusinow, 2016). Consistent with the EC–HOS15 complex functioning as a negative regulator, expression of PRR7, PRR9, ELF3, and ELF4 is increased in hos15-2 mutants (Supplemental Figure 2; Supplemental Data Set 2).
We compared the expression profile of hos15-2 with the putative target list of EC components, which were selected by ChIP seq and belong to the top 5% differentially expressed genes in elf3-1 and lux-4 mutants even though the experiments used different sampling times, developmental stages, or growth conditions (Supplemental Data Set 2; Ezer et al., 2017). The upregulated genes in lux-4 and hos15-2 mutants include PRR7, PRR9, GI, CIPK20 (CBL [CALCINEURIN B]-INTERACTING PROTEIN KINASE20), BBX28, and CXIP1 (CAX [CATION / H+ EXCHANGERS] INTERACTING PROTEIN1). The genes that overlapped with the up-regulated genes in elf3-1 and hos15-2 mutants are PRR7, PRR9, and RTF21 (ROTUNDIFOLIA-LIKE21). The genes shared by elf4 and hos15-2 mutants are PRR7, PRR9, GI, LHB1B1, CIPK20, BBX28, and CXIP1. Interestingly the target genes of LUX in ambient cold (17°C for 2 h, shifted from 22°C to 17°C at ZT8 and harvested at ZT10) include PRR7, PRR9, GI, CIPK20, CXIP1, FT, LIPID TRANSFER PROTEIN3 (LTP3), and LTP4. LTP3 and LTP4 are also upregulated in hda9-1 mutants (Supplemental Data Set 3, third sheet: hda9-1 → hos15-2). We checked the expression of CIPK20 and CXIP1 in hos15-2 mutants, because these two genes are putative targets of the EC and HOS15 (Supplemental Figure 2G and 2H). Indeed, the transcripts of CIPK20 and CXIP1 are higher in hos15-2 and hda9-1 mutants, but how HOS15 regulates the expression of CIPK20 and CXIP1 remains to be tested.
The Role of HOS15–HDA9-Mediated Chromatin Remodeling beyond the Transcriptional Control of Flowering Time
HOS15 is thought to function as a transcriptional repressor in association with an HDAC (Lee et al., 2008; Zhu et al., 2008). HOS15 has high sequence similarity to the human protein TBL1, a component of SILENCING MEDIATOR OF RETINOID AND THYROID RECEPTOR/NUCLEAR RECEPTOR CO-REPRESSOR (N-CoR) repressor complexes, which function in chromatin modification by associating with HDACs (Yoon et al., 2003; Zhu et al., 2008). Both hda6 (axe1-5) and hda19-1 are hypersensitive to salt and ABA and exhibit reduced expression of stress-responsive genes (Chen et al., 2010; Chen and Wu, 2010), which is opposite to the salt-resistance and elevated stress-responsive gene expression seen in hos15 mutants (Zhu et al., 2008). HDA9 functions in various biological processes including flowering (Kim et al., 2013; Kang et al., 2015), seed germination (van Zanten et al., 2014), salt and drought stress responses (Zheng et al., 2016), and leaf senescence (Chen et al., 2016).
HDA9 targets the promoters of transcribed genes and represses their expression through H3-deacetylation-mediated chromatin remodeling in cooperation with POWERDRESS (PWR), which possesses a pair of SWI3/DAD2/N-CoR/TFIII-B domains highly similar to that of the HDAC subunits SILENCING MEDIATOR OF RETINOID AND THYROID RECEPTOR and N-CoR (52% and 63%, respectively; Yumul et al., 2013; Chen et al., 2016; Kim et al., 2016). The pwr and hda9-1 mutants have small leaves, early flowering, and bulging siliques, as well as high levels of acetylated H3 (Kim et al., 2013, 2016; Kang et al., 2015; Suzuki et al., 2018). In addition, hda9-1 and hos15-2 hda9-1 mutants flower as early as hos15-2, whereas hda6 (axe1-5) and hda19-3 plants flower later than wild type (Figures 6A and 6B; Supplemental Figure 8; Tian and Chen, 2001; Tian et al., 2003; Yu et al., 2011). We found that HOS15 interacts with HDA9 together with ELF3 and LUX and that the association of HOS15 with the GI promoter is dramatically reduced in hos15-2 hda9-1 plants (Figure 6).
The SWI3/DAD2/N-CoR/TFIII-B domains of PWR interact with wide range of modified H3s but not with H4 or H2A/B (Kim et al., 2016). The levels of acetylated H3, but not of acetylated H4, are higher in pwr-2 and hda9-1 mutants than in wild type (Kim et al., 2013, 2016). Not only does PWR interact with HDA9 physically, but it also shares many targets with HDA9 (Kim et al., 2016; Suzuki et al., 2018), including AGL19, whose expression is elevated in hda9-1 and pwr-2 mutants, resulting in early flowering (Kim et al., 2013; Kang et al., 2015). Similarly, the floral promoter genes SOC1, FT, AGL24, and AGL19 are highly expressed in hos15-2 mutants (Figure 1C; Supplemental Figure 2A and 2B; Supplemental Data Set 1, second sheet: flowering-Countif), consistent with the set of upregulated genes in pwr-2, suggesting that HOS15 belongs to an HDAC that contains PWR (Supplemental Data Set 3, first sheet: pwr-2 upregulated → hos15-2; Yumul et al., 2013).
The pattern of differentially expressed genes in hda9-1 does not match that of hos15-2, except for a few genes such as lipid transfer transposable element genes (Supplemental Data Set 3, third sheet: hda9-1 → hos15-2), and AGL19 (Supplemental Figure 2A). There are several possible reasons for this. RPD3-type Class I histone deacetylases appear to have functional redundancy as transcriptional repressors and, thus, the loss of HDA9 itself would not be sufficient to release the transcriptional repression of their targets (Chen et al., 2016). Another possible reason for this difference is that gene expression is induced or repressed according to developmental stage and plant tissue, as well as internal and/or external inputs or signals. Indeed, ∼30% of the transcriptome is regulated by environmental conditions (Kreps et al., 2002). Also, gene expression is dependent on the time of day, i.e., the diurnal cycle, with ∼30% of transcripts under clock control (Covington et al., 2008). The expression of stress-responsive genes such as salt-, drought-, and osmotic stress-regulated genes is under circadian control (Harmer et al., 2000; Kreps et al., 2002; Covington et al., 2008). As a subject for future studies, integrated analysis of RNA seq and whole-genome ChIP seq will be required to compare the global binding of HOS15, HDA9, and EC components and to understand the genes and pathways regulated by HOS15-HDA9-EC. Moreover, careful experimental design that considers multiple factors such as plant stage, tissue, and sampling time will be essential for understanding the biological function of each component of this complex, together and separately.
HOS1, a RING finger E3 ligase (Lee et al., 2001), helps regulate flowering time via chromatin remodeling (Jung et al., 2013), like HOS15. HOS1 degrades ICE1, a positive regulator of the cold stress response (Dong et al., 2006), and CO, a floral promoter (Jung et al., 2012; Lazaro et al., 2012), as well as a chromatin remodeling factor that regulates the status of FLC chromatin under short-term cold stress in cooperation with FVE/MSI4, a component of the Arabidopsis autonomous floral promotion pathway (Jung et al., 2013; Jung and Park, 2013). Cold induces the physical interaction of HOS1 with FVE (a negative regulator of FLC) on the FLC promoter. HOS1 also inhibits the association of HDA6 (a HOS15-interacting partner) with FVE and the FLC locus, thereby maintaining high levels of acetylation and the subsequent expression of FLC (Jung et al., 2013).
In addition, FVE, which contains WD40 motifs like those of HOS15 (van Nocker and Ludwig, 2003; Lee et al., 2008), associates with the CUL4–DAMAGED DNA BINDING PROTEIN 1 (DDB1) complex (Pazhouhandeh et al., 2011), suggesting that HOS1 in the FVE–CUL4–DDB1 E3 ligase complex functions as both an E3 ligase and a transcriptional repressor via its effect on chromatin remodeling. FVE/MSI4 also contains WD40 motifs. In conjunction with the HOS1–FVE1–HDA6 complex, HDA9, an interactor of HOS15, appears to interact with DDB1A and DDB1B, which serve as substrate adaptor interacting subunits for a CUL4-based E3 ubiquitin ligase within the ubiquitin proteasome pathway (Geisler-Lee et al., 2007), suggesting that HOS15–HDA9 might be involved with a CUL4-based E3 ligase.
HDA9 helps control flowering time, especially under non-inductive SD conditions (Supplemental Figure 2K and 2L), through chromatin remodeling of AGL19 independently of CO, SOC1, and FLC. Indeed, hda9 mutants flower early because high expression levels of AGL19 and its downstream gene FT, with high levels of AcH3 on AGL19 chromatin (Kim et al., 2013; Kang et al., 2015). The early flowering of the hos15-2 mutant under non-inductive SD conditions is independent of GI (Supplemental Figures 2A to H). Therefore, HOS15 is thought to function with diverse floral regulators; other putative HOS15 interactors include histones (H2B, HTA12, HTA9, H3.1, and H3.3), other HDACs (HDA6, HDA18, and HAC11), and chromatin-remodeling factors (CHR5 and ASHH3), as well as E3 ubiquitin ligase SCF complex components such as ASK2, as revealed using a predicted interactome database (https://bar.utoronto.ca/interactions; Geisler-Lee et al., 2007).
In this study, we demonstrated that the EC–HOS15–HDA9 complex mediates histone deacetylation of the rhythmically expressed gene GI, which helps to maintain the circadian clock and regulates floral integrator genes involved in photoperiodic flowering. This histone deacetylation dampens the expression of GI, especially in the late afternoon under LDs in Arabidopsis. Our results suggest that HOS15 forms a complex with HDA9, an HDAC, and the EC in the late afternoon. This complex associates with the GI promoter through the direct binding of LUX, thereby repressing GI expression (Figure 6I).
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col-0, hos15-2 (GABI_785B10), gi-2 (Fowler et al., 1999), pcl1-1 (Onai and Ishiura, 2005), hda6-1/axe1-5 (Chen et al., 2010), hda9-1 (Kang et al., 2015), and hda19-1 (Chen and Wu, 2010) mutants, and proHDA9:HDA9-HA/hda9-1 (Kang et al., 2015) and PCL-overexpressing transgenic plants (Onai and Ishiura, 2005) transgenic plants were used in this study. The hos15-2 gi-2, hos15-2 pcl1-1, and hos15-2 hda9-1 double mutants were generated by crossing (Supplemental Table 1). Agrobacterium tumefaciens GV3101 carrying 35S:FLAG-HOS15 in the pEarleyGate202 vector was used to transform the hos15-2 mutant by the floral dip method to generate HOS15ox. The plants were incubated in a growth room (LD cycles, 100 μmol photons m−2 s−1 of cool white fluorescent light) at 23°C.
BiFC Assay
HOS15, ELF3, and ELF4 were fused in-frame to Venus aa 1–173, the N-terminal fragment of the eYFP fluorescent protein, in the pDEST-VYNE(R)GW vector, generating VN-HOS15, VN-ELF3, and VN-ELF4, respectively. LUX was fused in-frame to Venus aa 156–239, the C-terminal fragment of eYFP, in the pDEST-VYCE(R)GW vector (Supplemental Table 2; Gehl et al., 2009). For co-expression, Agrobacterium tumefaciens (GV 3101) cells harboring the construct for each protein were infiltrated into the leaves of 3-week-old Nicotiana benthamiana plants and grown for 2 d (Sparkes et al., 2006). Fluorescence was detected via confocal laser-scanning microscopy (Olympus FV1000, Olympus) with an excitation wavelength of 515 nm for YFP.
Co-IP Assay
Total proteins from N. benthamiana leaves expressing the indicated proteins (Supplemental Table 2) or 10-d-old Arabidopsis plants harvested at ZT8 were extracted in extraction buffer [100 mM Tris-HCl (pH 7.5), 150 mM sodium chloride (NaCl), 0.5% Nonidet P-40, 1 mM EDTA, 3 mM DTT, and protease inhibitors] and incubated with α-HA cross-linked to Protein G Agarose (Invitrogen), α-GFP cross-linked to protein A agarose (Invitrogen), or protein A agarose fused to α-HOS15 at 4°C for 2 h. For the Co-IP shown in Figures 4D and 5B, wild type and hos15-2 plants were cross-linked in 1% formaldehyde (10 min) to preserve in vivo interactions after harvesting. The cross-linking reaction was stopped by adding 0.125 M glycine and incubating for 5 min. After electrophoresis, the proteins were transferred onto a polyvinylidene fluoride membrane (Immobilon-P, Millipore). Immunoblotting was performed using α-HOS15 (Lee et al., 2012), α-ELF3 (Liu et al., 2001), α-LUX (Nusinow et al., 2011), α-HA (Roche, #11 867 423 001), α-GFP (Abcam, #ab6556), and α-FLAG (Sigma-Aldrich, # F1804) antibodies. The antigen protein was detected by chemiluminescence using ECL-detecting reagent (Thermo Fisher Scientific).
Gel Filtration Analysis
Gel filtration analysis was performed by size-exclusion chromatography using an ÄKTA fast performance liquid chromatography system with a Superdex 200 10/300GL column (GE Healthcare). Total proteins were extracted from wild type Arabidopsis using protein extraction buffer [100 mM Tris-Cl (pH 7.5), 150 mM NaCl, 0.5% Nonidet P-40, and 1 mM EDTA with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 5 μg/ml pepstatin, 5 μg/ml antipain, 5 μg/ml chymostatin, 2 mM sodium orthovanadate, 2 mM sodium floride, and 50 μM MG132)]. The proteins were precipitated using ammonium sulfate, resuspended in elution buffer [50 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 0.02% sodium azide] for gel filtration and dialyzed in a dialysis tube (7000 D cut-off, Thermo Fisher Scientific) overnight at 4°C. The proteins were eluted with elution buffer at a flow-rate of 0.5 mL/min. The eluted proteins were monitored at OD280. After gel filtration, each protein fraction (500 μl) was immediately precipitated with 10% (v/v) trichloroacetic acid and washed twice with 100% acetone. The precipitated protein pellets were dissolved in urea/SDS buffer and separated in an 8% SDS gel.
ChIP Assay
ChIP assays were performed as described (Saleh et al., 2008). The nuclei were isolated from 10-d-old plants (100 mg) harvested at ZT8 using CelLytic PN (Sigma-Aldrich), and chromatin was extracted from the nuclei by sonication using a Bioruptor (BMS; 10 × 30 s at low power). α-HOS15 (Lee et al., 2012), α-AcH3 (Abcam, #ab47915), α-H3 (Abcam, #ab1791), α-ELF3 (Liu et al., 2001), and α-LUX (Nusinow et al., 2011) antibodies were used for immunoprecipitation (Supplemental Figure 9). qPCR was used to determine the amounts of genomic DNA immunoprecipitated in the ChIP experiments. The primers used in the ChIP assay are listed in Supplemental Table 3.
Flowering Time Measurements
Flowering time analysis of the mutants was performed on plants grown under LD (16 h light/8 h dark) conditions at 22° to 23°C. Flowering time was measured by counting the total number of rosette and cauline leaves when the flower stalks were ∼3 cm to 5 cm tall, when the first floral buds were visible, and when the first flowers bloomed. Data from two independent experiments are presented as means ± sd, where n ≥ 15.
RNA-Seq Analysis
Wild type and hos15-2 (10-d-old) plants grown in LD conditions were harvested at ZT0, Z12, and Z24. Total RNA was extracted using an RNeasy Plant Mini kit (Qiagen). RNA-sequencing (RNA-seq) libraries were prepared using a TruSeq Stranded RNA Sample Prep kit (Illumina). Libraries with two biological replicates were bar-coded and sequenced in a single Illumina HiSeq2000 flow cell, generating > 597 million high-quality 100-nucleotide paired-ends RNA-seq reads per sample. RNA-seq reads from each sample were aligned to the A. thaliana reference genome using RNA-seq Unified Mapper(Grant et al., 2011). The number of reads uniquely aligned to each contig was counted for each sample. Genes with significantly different uniquely mapped RNA-seq reads between wild-type and mutant plants and at different ZT time points were identified as differentially expressed genes based on false discovery rate < 0.01 (Benjamini-Hochberg correction) using the DESeq pipeline (Anders and Huber, 2010).
RT-qPCR and qPCR Analysis
Total RNA was isolated from 10-d-old seedlings using an RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Total RNA (2 µg) was used for first-strand complementary DNA (cDNA) synthesis with a cDNA Synthesis kit (Invitrogen, Carlsbad, CA, USA). To measure levels of gene expression and quantify ChIP assay results, quantitative PCR was performed according to the manufacturer’s instructions. The QuantiMix SYBR kit (PKT, Philekorea Tech.) was used for RT-qPCR as follows: (1) 50°C for 10 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s for analysis of gene expression; (2) 50°C for 10 min, 95°C for 10 min, and 60 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s for ChIP assay. The values were automatically calculated using a CFX384 Real-time PCR Detection System and CFX Manager software (Bio-Rad, Hercules) following a standard protocol. The primers used for RT-qPCR analysis are listed in Supplemental Table 3.
Yeast Two-Hybrid Assay
Saccharomyces cerevisiae strain JD53 was used for the yeast two-hybrid assay. The constructs (Supplemental Table 2) were transformed into yeast using polyethylene glycol and heat shock following the manufacturer’s protocol (Clontech).
Generating α-ELF3 and α-LUX Antibodies
The α-ELF3 (Liu et al., 2001) and α-LUX antibodies (Nusinow et al., 2011) were generated as described in the references. The LUX cDNA was inserted into the modified gGEX4T-3 vector using the Gateway system to create an in-frame GST-fusion. gGEX4T-3:LUX was introduced into Escherichia coli strain BL21 (Merck). For protein expression, cells were induced for 5 h at 30°C with 0.4 mM IPTG. The induced cells were harvested, suspended in 1× GST Bind/Wash buffer [4.3 mM disodium phosphate, 1.47 mM monopotassium phosphate, 137 mM NaCl, 2.7 mM potassium chloride (pH 7.3)], incubated on ice for 20 min, and lysed by sonication. After centrifugation at 12,000 rpm and 4°C for 30 min, the supernatant was combined with 0.5 mL Glutathione Sepharose 4 Fast Flow (GE Healthcare) that had been equilibrated with 1× GST Bind/Wash buffer. The slurry was mixed gently by shaking at room temperature for 30 min. The resin was collected and washed 2 or 3 times with 10 mL of 1× GST Bind/Wash Buffer. GST-LUX was eluted in 1 mL of 1× GST elution buffer [50 mM Tris-HCl (pH 8.0) and 10 mM reduced glutathione].
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: GI: At1g22770, HOS15: At5g67320, ELF3: At2g25930, LUX: At3g46640, ELF4: At2g40080, HDA9: At3g44680.
Supplemental Data
Supplemental Figure 1. The levels of HOS15 transcript do not cycle under LD.
Supplemental Figure 2. Expression of AGL19, AGL24, ELF3, ELF4, PRR7, PRR9, CIPK20 and CXIP in hos15-2, hda9-1, hos15-2 hda9-1, and wild type.
Supplemental Figure 3. Flowering time of hos15-2 and other mutants in SD.
Supplemental Figure 4. The association of HOS15 and acetylated H3 level on the GI promoter R3 region at ZT0 and ZT8.
Supplemental Figure 5. Endogenous H3 levels on the GI locus are similar in plants used in this study.
Supplemental Figure 6. HOS15 interacts with LUX and ELF3.
Supplemental Figure 7. Histone acetylation is increased in hos15 mutants.
Supplemental Figure 8. hda9-1 exhibits early flowering in LD.
Supplemental Figure 9. Specificities of the α-ELF3 and α-LUX antibodies.
Supplemental Table 1. List of plants used in this study.
Supplemental Table 2. Constructs used in this study.
Supplemental Table 3. Primers used in this study.
Supplemental Data Set 1. RNA-seq analysis complementary to Figure 1C.
Supplemental Data Set 2. Comparison of the putative targets of the EC and list of up-regulated genes in hos15-2 and hda9-1.
Supplemental Data Set 3. Comparison of the pattern of differentially expressed genes in pwr-2 and hda9-2 to that of hos15-2.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Drs. Junghoon Park (Konkuk University, Korea) and Dong-Ha Oh (Department of Biological Sciences, Louisiana State University, LA) for help with mRNA-seq analysis. We thank Drs. Yoo-Sun Noh and Bosl Noh (Seoul National University, Korea) for the proHDA9:HDA9-HA/hda9-1 seeds and HDA9-HA plasmid. gGEX4T-3 was a kind gift from Dr. Hong Gil Nam (Postech, Korea). This work was supported by the Next Generation BioGreen21 Program, Rural Development Administration (RDA), Republic of Korea (SSAC, PJ01327301 to W.-Y.K. and PJ01327306 to C.R.M.); the National Research Foundation of Korea (NRF) funded by the Korean Government (MSIP 2016R1A2A1A05004931 to D.-J.Y) and Global Research Lab (2017K1A1A2013146 to D.-J.Y.).
AUTHOR CONTRIBUTIONS
H.J.P., D.B., D.-J.Y., and W.-Y.K. designed research; H.J.P., D.B., and X.L. performed research; H.J.P., J.-Y.C., S.Y.L., D.-J.Y., and W.-Y.K. analyzed data; S.-H.K. analyzed mRNA seq data; and H.J.P., D.B., J.-Y.C., C.R.M., and W.-Y.K. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Amasino R. (2010). Seasonal and developmental timing of flowering. Plant J. 61: 1001–1013. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106–R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns M.C., Nordström K., Cremer F., Tóth R., Hartke M., Simon S., Klasen J.R., Bürstel I., Coupland G. (2014). Evening expression of Arabidopsis GIGANTEA is controlled by combinatorial interactions among evolutionarily conserved regulatory motifs. Plant Cell 26: 3999–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché F., Lobet G., Tocquin P., Périlleux C. (2016). FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 44 (D1): D1167–D1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-T., Wu K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5: 1318–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-T., Luo M., Wang Y.-Y., Wu K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61: 3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Lu L., Mayer K.S., Scalf M., Qian S., Lomax A., Smith L.M., Zhong X. (2016). POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. eLife 5: e17214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow B.Y., Helfer A., Nusinow D.A., Kay S.A. (2012). ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal. Behav. 7: 170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S., Wei X., Pei L., Thompson R.L., Liu Y., Heard J.E., Ruff T.G., Beachy R.N. (2011). BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell 23: 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S., Cao X. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 107: 19114–19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.E., Knox K., Kozma-Bognar L., Southern M.M., Pokhilko A., Millar A.J. (2011). Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 21: 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.-H., Agarwal M., Zhang Y., Xie Q., Zhu J.-K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezer D., et al. (2017). The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 3: 17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S., Lee K., Onouchi H., Samach A., Richardson K., Morris B., Coupland G., Putterill J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18: 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C., Waadt R., Kudla J., Mendel R.-R., Hänsch R. (2009). New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant 2: 1051–1058. [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J., O’Toole N., Ammar R., Provart N.J., Millar A.H., Geisler M. (2007). A predicted interactome for Arabidopsis. Plant Physiol. 145: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant G.R., Farkas M.H., Pizarro A.D., Lahens N.F., Schug J., Brunk B.P., Stoeckert C.J., Hogenesch J.B., Pierce E.A. (2011). Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM). Bioinformatics 27: 2518–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.-S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113. [DOI] [PubMed] [Google Scholar]
- Hayama R., Sarid-Krebs L., Richter R., Fernández V., Jang S., Coupland G. (2017). PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 36: 904–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen S.P., Schultz T.F., Pruneda-Paz J.L., Borevitz J.O., Ecker J.R., Kay S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102: 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks K.A., Albertson T.M., Wagner D.R. (2001). EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Nusinow D.A. (2016). Into the evening: Complex interactions in the Arabidopsis circadian clock. Trends Genet. 32: 674–686. [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297. [DOI] [PubMed] [Google Scholar]
- Jaeger K.E., Wigge P.A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17: 1050–1054. [DOI] [PubMed] [Google Scholar]
- Jung C., Müller A.E. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci. 14: 563–573. [DOI] [PubMed] [Google Scholar]
- Jung J.-H., Park C.-M. (2013). HOS1-mediated activation of FLC via chromatin remodeling under cold stress. Plant Signal. Behav. 8: e27342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.-H., Seo P.J., Park C.-M. (2012). The E3 ubiquitin ligase HOS1 regulates Arabidopsis flowering by mediating CONSTANS degradation under cold stress. J. Biol. Chem. 287: 43277–43287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.-H., Park J.-H., Lee S., To T.K., Kim J.-M., Seki M., Park C.-M. (2013). The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell 25: 4378–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M.-J., Jin H.-S., Noh Y.-S., Noh B. (2015). Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in Arabidopsis. New Phytol. 206: 281–294. [DOI] [PubMed] [Google Scholar]
- Kim Y.J., et al. (2016). POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proc. Natl. Acad. Sci. USA 113: 14858–14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.-Y., Hicks K.A., Somers D.E. (2005). Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol. 139: 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Latrasse D., Servet C., Zhou D.-X. (2013). Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem. Biophys. Res. Commun. 432: 394–398. [DOI] [PubMed] [Google Scholar]
- Kreps J.A., Wu Y., Chang H.-S., Zhu T., Wang X., Harper J.F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130: 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A., Valverde F., Piñeiro M., Jarillo J.A. (2012). The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., et al. (2012). Monoclonal antibodies against recombinant AtHOS15. Hybridoma (Larchmt.) 31: 118–124. [DOI] [PubMed] [Google Scholar]
- Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., Zhu J.-K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo--cytoplasmic partitioning. Genes Dev. 15: 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Terzaghi W., Gusmaroli G., Charron J.-B.F., Yoon H.-J., Chen H., He Y.J., Xiong Y., Deng X.W. (2008). Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20: 152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.-J., Zhang Y.-C., Li Q.-H., Sang Y., Mao J., Lian H.-L., Wang L., Yang H.-Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.L., Covington M.F., Fankhauser C., Chory J., Wagner D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Nomoto Y., Oka H., Kitayama M., Takeuchi A., Tsubouchi M., Yamashino T. (2014). Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 55: 958–976. [DOI] [PubMed] [Google Scholar]
- Nguyen N.H., Jeong C.Y., Lee W.J., Lee H. (2016). Identification of a novel Arabidopsis mutant showing sensitivity to histone deacetylase inhibitors. Appl. Biol. Chem. 59: 855–860. [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai K., Ishiura M. (2005). PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10: 963–972. [DOI] [PubMed] [Google Scholar]
- Park J., et al. (2018a). Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. USA 115: E5400–E5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lim C.J., Khan I.U., Jan M., Khan H.A., Park H.J., Guo Y., Yun D.-J. (2018b). Identification and molecular characterization of HOS15-interacting proteins in Arabidopsis thaliana. J. Plant Biol. 61: 336–345. [Google Scholar]
- Pazhouhandeh M., Molinier J., Berr A., Genschik P. (2011). MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 3430–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sawa M., Nusinow D.A., Kay S.A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Shim J.S., Kinmonth-Schultz H.A., Imaizumi T. (2015). Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66: 441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Shinozuka N., Hirakata T., Nakata M.T., Demura T., Tsukaya H., Horiguchi G. (2018). OLIGOCELLULA1/HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES15 promotes cell proliferation with HISTONE DEACETYLASE9 and POWERDRESS during leaf development in Arabidopsis thaliana. Front. Plant Sci. 9: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Chen Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98: 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Wang J., Fong M.P., Chen M., Cao H., Gelvin S.B., Chen Z.J. (2003). Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics 165: 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S., Ludwig P. (2003). The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genomics 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M., Zöll C., Wang Z., Philipp C., Carles A., Li Y., Kornet N.G., Liu Y., Soppe W.J.J. (2014). HISTONE DEACETYLASE 9 represses seedling traits in Arabidopsis thaliana dry seeds. Plant J. 80: 475–488. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhang Y., Ma Q., Zhang Z., Xue Y., Bao S., Chong K. (2007). SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 26: 1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaish M.W., Colasanti J., Rothstein S.J. (2011). The role of epigenetic processes in controlling flowering time in plants exposed to stress. J. Exp. Bot. 62: 3727–3735. [DOI] [PubMed] [Google Scholar]
- Yoon H.-G., Chan D.W., Huang Z.-Q., Li J., Fondell J.D., Qin J., Wong J. (2003). Purification and functional characterization of the human N-CoR complex: The roles of HDAC3, TBL1 and TBLR1. EMBO J. 22: 1336–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.-W., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-W., Liu X., Luo M., Chen C., Lin X., Tian G., Lu Q., Cui Y., Wu K. (2011). HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 156: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumul R.E., Kim Y.J., Liu X., Wang R., Ding J., Xiao L., Chen X. (2013). POWERDRESS and diversified expression of the MIR172 gene family bolster the floral stem cell network. PLoS Genet. 9: e1003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., et al. (2011). Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23: 396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Ding Y., Sun X., Xie S., Wang D., Liu X., Su L., Wei W., Pan L., Zhou D.-X. (2016). Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 67: 1703–1713. [DOI] [PubMed] [Google Scholar]
- Zhu J., Jeong J.C., Zhu Y., Sokolchik I., Miyazaki S., Zhu J.-K., Hasegawa P.M., Bohnert H.J., Shi H., Yun D.-J., Bressan R.A. (2008). Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. USA 105: 4945–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]