The APETALA2-like transcription factor SUPERNUMERARY BRACT confers seed shattering by positively regulating qSH1 and SH5, while altering seed size by modulating longitudinal cell elongation in rice glumes.
Abstract
The elimination of seed shattering was a crucial event during crop domestication. Improving and fine-tuning the regulation of this process will further enhance grain yield by avoiding seed losses during crop production. In this work, we identified the loss-of-shattering mutant suppression of shattering1 (ssh1) through a screen of mutagenized wild rice (Oryza rufipogon) introgression lines with naturally high shattering. Using the MutMap approach and transformation experiments, we isolated a genetic factor for seed shattering, SSH1, which is an allele of SUPERNUMERARY BRACT (SNB), a gene encoding a plant-specific APETALA2-like transcription factor. A C-to-A point mutation in the ninth intron of SNB altered the splicing of its messenger RNA, causing the reduced shattering of the ssh1 mutant by altering the development of the abscission layer and vascular bundle at the junction between the seed and the pedicel. Our data suggest that SNB positively regulates the expression of two rice REPLUMLESS orthologs, qSH1 and SH5. In addition, the ssh1 mutant had larger seeds and a higher grain weight, resulting from its increased elongation of the glume longitudinal cells. The further identification of favorable SNB alleles will be valuable for improving rice seed shattering and grain yield using molecular breeding strategies.
INTRODUCTION
Seed shattering is determined by the development of the abscission zone (AZ), which consists of several layers of isodiametrically flattened and thin-walled cells at the junction of the seed and the mother tissue (Jin, 1986; McKim et al., 2008). In the past, several important genes regulating the development of the AZ and the corresponding dispersal of plant organs have been identified in various species, shedding light on the regulatory mechanisms underlying these processes (Dong and Wang, 2015; Pourkheirandish et al., 2015; Ballester and Ferrándiz, 2017). The natural shattering of mature seeds has great ecological importance for offspring dispersal, whereas in domesticated crops, mutations that reduce shattering and cause the seeds to be retained longer on the parent plant were selected and fixed to facilitate efficient harvesting and avoid yield loss. The reduction of seed shattering was one of most important advances in the history of crop domestication (Fuller, 2007; Onishi et al., 2007).
Previous studies have identified several genetic factors controlling the key transition from shattering to nonshattering during crop domestication. Rice (Oryza sativa) was one of the first crops to be domesticated. In this species, mutations in two genes, SH4 (the quantitative trait locus seed shattering on chromosome4)/SHA1 (Shattering 1, an allele of SH4) and qSH1 (the quantitative trait locus seed shattering on chromosome1), were mainly responsible for the transition away from seed shattering during domestication (Konishi et al., 2006; Li et al., 2006; Lin et al., 2007). A single amino acid change in the MYB3 DNA binding domain of SH4 resulted in decreased seed shattering due to the incomplete formation of the AZ in cultivated rice (Li et al., 2006). A single nucleotide polymorphism (SNP) in the 5′ regulatory region of qSH1, a rice ortholog of the BEL1-type homeobox gene REPLUMLESS (RPL) in Arabidopsis (Arabidopsis thaliana), caused the absence of the abscission layer in temperate japonica varieties (Konishi et al., 2006). The genetic control of seed shattering domestication has also been revealed for other species, including wheat (Triticum aestivum), barley (Hordeum vulgare), sorghum (Sorghum bicolor), and soybean (Glycine max; Faris and Gill, 2002; Faris et al., 2003; Simons et al., 2006; Zhang et al., 2011, 2013; Lin et al., 2012; Dong et al., 2014; Funatsuki et al., 2014; Katkout et al., 2015; Pourkheirandish et al., 2015; Debernardi et al., 2017). These findings provide important insights into the molecular mechanisms and evolutionary trajectories underlying seed shattering domestication.
Notably, some genes affecting seed shattering have a pleiotropic effect on spikelet development; for example, in African cultivated rice (Oryza glaberrima), the selection of an independent sh4 mutant resulted in the convergent evolution of the nonshattering trait observed in Asian cultivated rice (O. sativa); however, the sh4 mutant allele in African rice resulted in smaller seeds than its progenitor Oryza barthii, owing to a reduced elongation of the glume longitudinal cells (Wu et al., 2017). The mutation of SHATTERING ABORTION1 (SHAT1) caused both the loss of shattering and spikelet developmental defects in rice (Zhou et al., 2012). In addition, the mutation of a wheat domestication gene, Q, encoding an APETALA2 (AP2) transcription factor, affects a broad range of phenotypic characters, including subcompact inflorescences, glume shape, and free-threshing grains (Faris and Gill, 2002; Faris et al., 2003; Simons et al., 2006; Debernardi et al., 2017). The further identification of other nonshattering mutants would therefore facilitate the elucidation of the molecular basis underlying seed shattering while also providing a novel favorable genetic resource for high-yielding crop breeding programs.
In the present study, we identified a genetic suppressor of seed shattering, SUPPRESSION OF SHATTERING1 (SSH1), in a screen of mutagenized wild rice (Oryza rufipogon) introgression lines with naturally high levels of shattering, using a combination of MutMap analysis and transformation experiments. SSH1, which is an allele of SUPERNUMERARY BRACT (SNB) encoding a transcription factor with two plant-specific AP2 domains, controls the development of the AZ and regulates seed shattering by promoting the expression of two rice RPL orthologs, qSH1 and SH5. In addition, the ssh1 mutant identified in this study has larger seeds and a higher grain yield, suggesting that this gene may be a useful target in rice breeding applications.
RESULTS
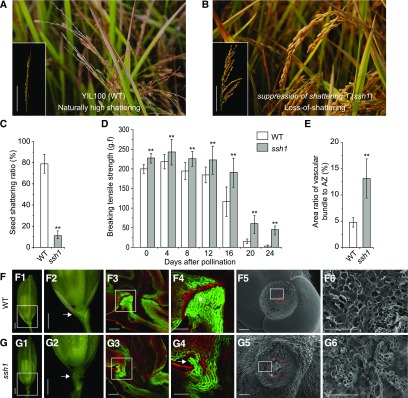
Identification of a Loss-of-Shattering Mutant from Mutagenized Wild Rice Introgression Lines with Naturally High Shattering
A wild rice introgression line, YIL100, referred to as the wild type below, was derived from the cross between the donor, O. rufipogon (accession YJCWR), and the recipient, an indica variety known as Teqing. The YIL100 introgression line carried SH4/SHA1 from the donor and the qSH1 locus from the recipient, resulting in a natural seed shattering phenotype (Figure 1A; Supplemental Figure 1). To study the regulatory mechanism underlying seed shattering in rice, we mutagenized the YIL100 line using EMS and screened the resulting M2 mutant generation, identifying the loss-of-shattering mutant ssh1 (Figure 1B). Phenotypic observation showed that the ssh1 mutant had less seed shattering at the harvest stage and a stronger pedicel breaking tensile strength (BTS) than the wild type (Figures 1C and 1D). Further observation of longitudinal sections of spikelets at the anthesis stage using confocal microscopy showed that the wild type had a complete AZ and a normal vascular bundle at the junction between the seed and pedicel, while the ssh1 mutant displayed a deficiency in AZ development and vascular bundle overgrowth (Figures 1F and 1G). The seed bases were observed using a scanning electron microscope (SEM), further revealing that the ssh1 mutation affected the development of both the abscission layer and the vascular bundle (Figures 1E to 1G). These results suggest that both the incomplete abscission layer and the larger vascular bundle in the ssh1 mutant might provide stronger support to the seed, decreasing shattering after seed ripening.
Figure 1.
Comparison of Seed Shattering and Floral AZ Morphology in the Wild Type and the ssh1 Mutant.
(A) and (B) Phenotypes of mature wild-type (YIL100) and ssh1 rice in the field. Panicles of the wild type and the ssh1 mutant are respectively illustrated in the lower left corners. Bars = 10 cm.
(C) Comparison of the seed shattering ratio of the mature wild type and the ssh1 mutant. Values are means ± sd (n = 10 main panicles).
(D) Comparison of the BTS in the wild type and the ssh1 mutant on the day after pollination and every 4 d thereafter during seed development. The g.f is the gravitational unit of force. Values are means ± sd (n = 50 grains).
(E) Comparison of the area ratio of the vascular bundle to AZ in the wild type and the ssh1 mutant. Values are means ± sd (n = 10 grains).
In (C) to (E), two-tailed Student’s t tests were used to compare the wild type and the ssh1 mutant (**, P < 0.01; Supplemental File 2).
(F) and (G) Characterization of floral AZ morphology in the wild type (F) and the ssh1 mutant (G). (F1) and (G1) show the spikelets. The white boxes indicate the junction between the seed and the pedicel and are enlarged in (F2) and (G2), where the arrows indicate the position of the AZ. (F3) and (G3) show the fluorescence images of longitudinal sections across the flower and pedicel junction. (F4) and (G4) are enlargements of the white boxes in (F3) and (G3), respectively. Arrows point to the AZ in the wild type or the corresponding region in the ssh1 mutant. (F5) and (G5) show SEM photographs of the seed bases where the pedicels attach. The red circles indicate vascular bundles. (F6) and (G6) are magnifications of the white boxes in (F5) and (G5), respectively. The surface of the seed base in the wild type is smooth, whereas the surfaces in the ssh1 mutant are broken and rough. Bars = 1 mm in panels (1) and (2), 100 μm in panels (3) and (5), and 50 μm in panels (4) and (6).
Cloning and Characterization of SSH1
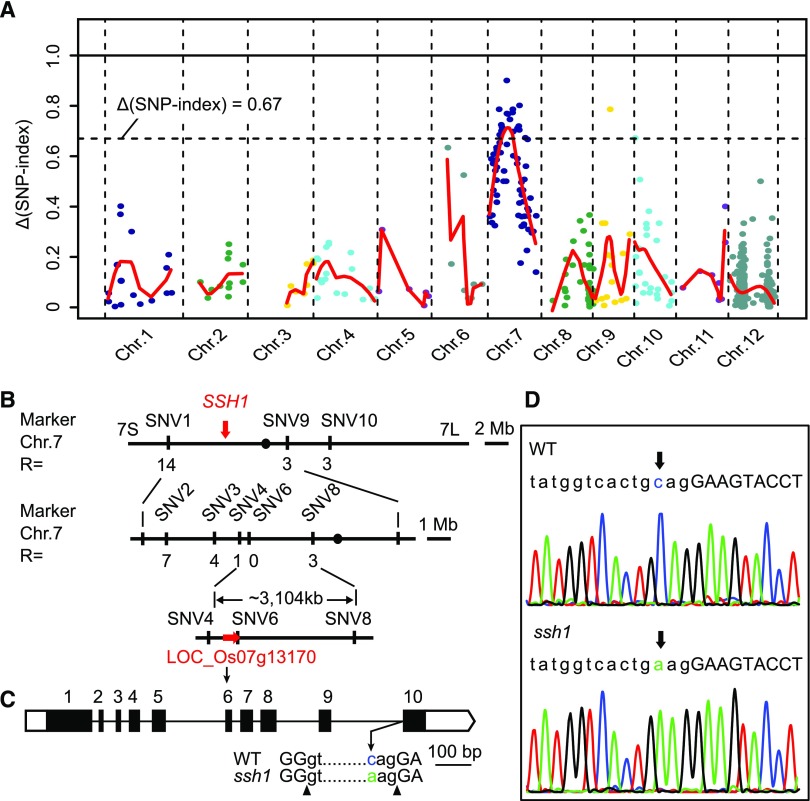
To evaluate the genetic characteristics of the phenotypes affected in the ssh1 mutant, we developed an F2 population derived from a cross between the wild type and the ssh1 mutant. Phenotypic observation of the F2 population (205 wild-type plants and 72 mutant plants; χ2 = 0.10 < χ20.05,1 = 3.84) revealed that nonshattering in ssh1 was controlled by a single recessive gene. We further bulked DNA from 20 recessive homozygous nonshattering plants and 20 dominant plants with natural shattering and then sequenced the pooled DNA on an Illumina HiSeq2500 platform. Using the MutMap approach (Abe et al., 2012; Takagi et al., 2013; Lu et al., 2014), a single peak was detected on chromosome 7, in which the Δ(SNP-index) was more than 0.67, indicating that the SNPs are associated with SSH1 (Figure 2A; Supplemental Table 1). Using a total of 72 recessive homozygous plants from the same F2 population, SSH1 was further mapped to an ∼3104-kb interval between SNP markers SNV4 and SNV8 and was found to cosegregate with SNV6 (Figure 2B).
Figure 2.
Mapping of SSH1.
(A) Δ(SNP-index) plot of the whole genome generated using MutMap. The red curves represent Δ(SNP-index) plot regression lines.
(B) The mapping location of the SSH1 gene was narrowed down to an ∼3104-kb interval between SNP markers SNV4 and SNV8 using 72 recessive F2 individuals. It was found to cosegregate with the SNP SNV6, which is located in the ninth intron of LOC_Os07g13170. The vertical red arrow indicates the location of SSH1. The horizontal red arrow represents the gene LOC_Os07g13170.
(C) The exon-intron structure of LOC_Os07g13170 and the location of the SNP SNV6. Black boxes represent exons, white boxes represent UTRs, and thin black lines indicate introns. Uppercase letters and lowercase letters below the gene structure represent exons and introns, respectively. Blue and green lowercase letters represent the C in the wild type and the A in the ssh1 mutant, respectively. Black triangles indicate the 5′ and 3′ splicing sites of the ninth intron of LOC_Os07g13170.
(D) The C-to-A point mutation (SNV6) in LOC_Os07g13170, detected using Sanger sequencing. Lowercase letters and uppercase letters above the DNA sequencing peak chromatograms represent the 9th intron and the 10th exon of LOC_Os07g13170, respectively.
Integrating data from whole-genome sequencing and SNP validation using Sanger sequencing, we identified three SNPs between the wild type and ssh1 mutant within the 3104-kb SSH1 fine-mapped region (Supplemental Table 1). We further compared the expression levels of four genes associated with these three SNPs in the wild type and ssh1 mutant using RNA-seq. The expression of LOC_Os07g13170 was significantly downregulated in ssh1 compared with the wild type (fold change ≥ 2, false discovery rate [FDR] < 0.001), while the expression levels of the three genes associated with the other two SNPs were not significantly different between the ssh1 mutant and the wild type (Supplemental Table 1). Based on the annotation of the rice reference genome (http://rice.plantbiology.msu.edu), we found that the SNP SNV6, a single nucleotide transversion from cytosine (C) to adenine (A) in the ssh1 mutant, was located in the ninth intron (+3473 bp) of LOC_Os07g13170 (Figures 2C and 2D). Therefore, we speculated that the SNP SNV6 might alter the expression profile of LOC_Os07g13170. Additionally, LOC_Os07g13170 is the rice heterochronic gene SNB, which encodes an AP2 domain-containing protein regulating the transition from the spikelet meristem to the floral meristem (Lee et al., 2007; Zhu et al., 2009; Lee and An, 2012). Taken together, these results suggest that LOC_Os07g13170 is a strong candidate gene for SSH1.
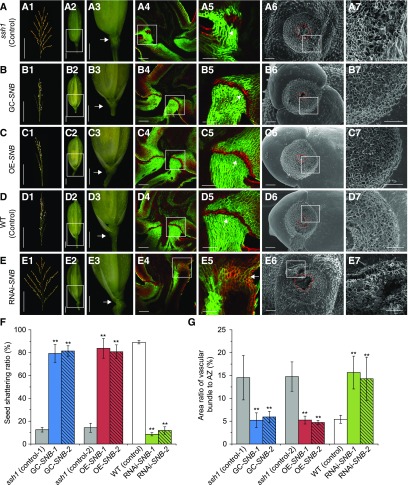
To test whether the phenotype of the ssh1 mutant was caused by this SNP in the SNB gene, we complemented the mutant with a 7206-bp wild-type genomic fragment containing the entire coding region of SNB and the 2309-bp 5′-flanking and 1209-bp 3′-flanking regions. A total of 11 independent positive transgenic complementation lines (GC-SNB) were generated, all of which had natural shattering and normal development of both the abscission layer and the vascular bundle at the junction between the seed and the pedicel, indicating complementation of the ssh1 phenotype (Figures 3A, 3B, 3F, and 3G). We also developed both overexpression and RNA interference (RNAi) constructs based on the SNB (LOC_Os07g13170) complementary DNA (cDNA) sequence from the wild type and introduced these into the ssh1 mutant and the wild-type plants, respectively. We found that all 17 independent overexpression transgenic plants (OE-SNB) showed an integral abscission layer, smaller vascular bundles at the seed base, and increased shattering compared with the ssh1 mutant (Figures 3C, 3F, and 3G). By contrast, all eight independent RNAi transgenic plants (RNAi-SNB) with significantly downregulated SNB transcripts (Supplemental Figure 2) showed a deficiency in AZ development, larger vascular bundles, and decreased shattering compared with the wild type, displaying similar phenotypes to the ssh1 mutants (Figures 3D to 3G). The results of these transformation experiments demonstrate that the C-to-A point mutation in the intron of SNB resulted in defective abscission layer development and vascular bundle overgrowth, leading to a decrease of seed shattering in rice.
Figure 3.
Characterization of Seed Shattering in the GC-SNB, OE-SNB, and RNAi-SNB Transgenic Plants.
(A) to (E) The morphological analyses of the ssh1 mutant (control; [A]), the GC-SNB transgenic plants (B), the OE-SNB transgenic plants (C), the wild type (control; [D]), and the RNAi-SNB transgenic plants (E). (A1), (B1), (C1), (D1), and (E1) show the seed shattering phenotypes on the main panicle. (A2), (B2), (C2), (D2), and (E2) show the spikelets. The white boxes indicating the junction between the seed and the pedicel are magnified in (A3), (B3), (C3), (D3), and (E3), respectively, in which arrows indicate the position of the AZ. (A4), (B4), (C4), (D4), and (E4) show fluorescence images of longitudinal sections across the flower and pedicel junction. The white boxes are magnified in (A5), (B5), (C5), (D5), and (E5), respectively, and the arrows point to the AZ or corresponding regions. (A6), (B6), (C6), (D6), and (E6) are SEM photographs of the seed base. The red circles indicate the vascular bundles. (A7), (B7), (C7), (D7), and (E7) are magnifications of the white boxes in (A6), (B6), (C6), (D6), and (E6), respectively. Bars = 50 μm in panels (5) and (7), 100 μm in panels (4) and (6), 1 mm in panels (2) and (3), and 10 cm in panel (1).
(F) Comparison of seed shattering ratios. Values are means ± sd (n = 10 main panicles).
(G) Comparison of the area ratios of vascular bundle to the AZ of the seed bases. Values are means ± sd (n = 10 grains).
In (F) and (G), two-tailed Student’s t tests were used to compare the transgenic lines and the corresponding controls (**, P < 0.01; Supplemental File 2).
SNB/SSH1 Encodes a Transcription Factor with Two AP2 Domains
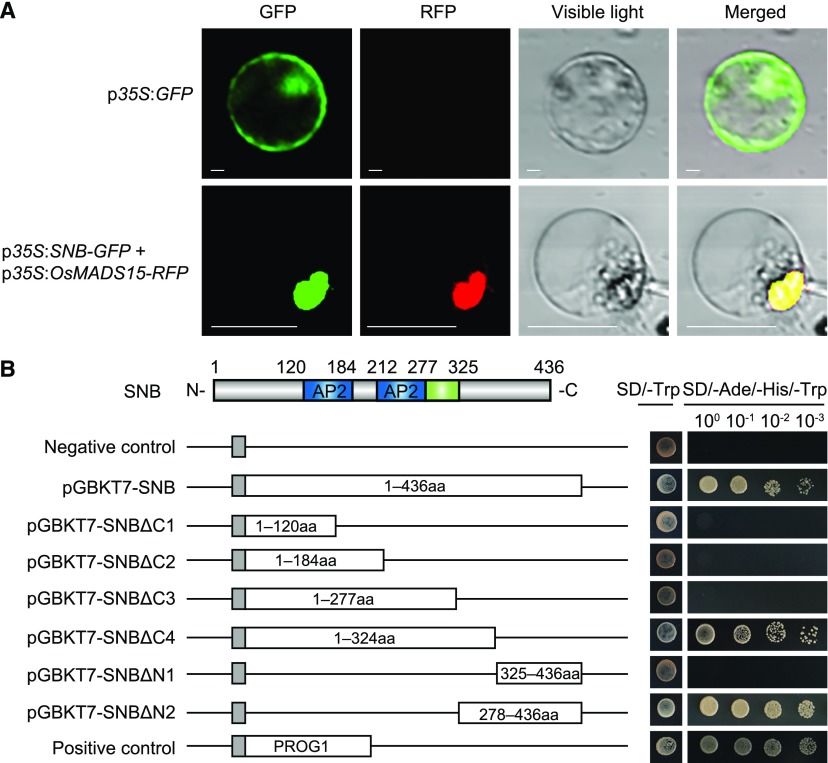
The full-length cDNA of SNB/SSH1 is 1909 bp and is divided into 10 exons interspersed with 9 introns (Supplemental Figure 3). The open reading frame of SNB is 1311 bp and encodes a protein of 436 amino acid residues, forming two plant-specific AP2 domains between residues 121 and 184 and residues 213 and 277, respectively (Supplemental Figure 4). Transient expression experiments indicated that the SNB-green fluorescent protein (GFP) fusion protein was specifically localized to the nucleus in rice protoplasts (Figure 4A). A transcriptional activation assay showed that the SNB and DNA binding domain (BD) fusion protein in yeast activated the expression of the reporter genes, implying that SNB has strong transcriptional activity and is a transcription factor. A truncation analysis revealed that residues 278 to 324 of SNB are required for its transcriptional activity (Figure 4B). A phylogenetic analysis showed that SNB is a rice ortholog of SID1 (SISTER OF INDETERMINATE SPIKELET1) in maize (Zea mays; Chuck et al., 2008) and has high amino acid similarity to rice OsIDS1 (Lee et al., 2007), maize IDS1 (INDETERMINATE SPIKELET1; Chuck et al., 1998), and wheat Q (Supplemental Figure 4; Supplemental File 1; Faris et al., 2003), indicating that the AP2 transcription factors have conserved functions in regulating the development of the AZ and inflorescences in cereal crops.
Figure 4.
Subcellular Localization and Transcription Activity of SNB.
(A) Subcellular localization of the SNB-GFP fusion protein in a rice protoplast. The OsMADS15-RFP fusion protein was used as a nuclear localization marker, and the GFP protein alone was used as the control. Bars = 20 μm.
(B) Transcription activity assay of full-length or truncated SNB in yeast. pGBKT7-SNB, pGBKT7-SNBΔC (1–4), and pGBKT7-SNBΔN (1 and 2) had the GAL4 BD (gray) in the pGBKT7 vector fused with sequences encoding full-length, N-terminal, or C-terminal SNB, respectively. pGBKT7 was used as the negative control, and the transcription factor PROSTRATE GROWTH1 was fused with the GAL4 BD as the positive control. The numbers in the boxes indicate the SNB amino acid residues used for construction.
Posttranscriptional Regulation of SNB
The mutant gene ssh1 identified in this study is a novel allele of SNB. Compared with the knockout snb mutant (Lee et al., 2007), ssh1 had a weaker pleiotropic phenotype for both inflorescence and spikelet development, including forming fewer rudimentary glumes (2.9% in ssh1, 31.3% in snb), aberrant lemma/palea-like structures (8.7% in ssh1, 35.6% in snb), and ectopic lodicules (2.3% in ssh1, 14.3% in snb; Table 1; Supplemental Figure 5). We therefore speculated that the C-to-A point mutation (SNV6) in the ninth intron of SNB in the ssh1 mutant might alter its transcription level or messenger RNA (mRNA) splicing rather than cause a complete loss of function, which is consistent with the weaker phenotype of ssh1 than the loss-of-function snb mutant.
Table 1. Comparison of Floral Organ Numbers in the Wild Type and the ssh1 Mutant.
| No. of Organs | Glume | Palea and Lemma | Lodicule | Stamen | Carpel | Stigma | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild Type | ssh1 | Wild Type | ssh1 | Wild Type | ssh1 | Wild Type | ssh1 | Wild Type | ssh1 | Wild Type | ssh1 | |
| 0 | – | – | – | – | – | 12 | – | 13 | – | 13 | – | 13 |
| 1 | – | – | – | 6 | – | – | – | 1 | 516 | 495 | – | – |
| 2 | – | – | 516 | 471 | 516 | 492 | – | 5 | – | 8 | 516 | 494 |
| 3 | – | – | – | 30 | – | 6 | – | 3 | – | – | – | 1 |
| 4 | 516 | 502 | – | 8 | – | 3 | – | 6 | – | – | – | 7 |
| 5 | – | 11 | – | – | – | 1 | – | 6 | – | – | – | – |
| 6 | – | 1 | – | 1 | – | – | 516 | 480 | – | – | – | 1 |
| 7 | – | 1 | – | – | – | 1 | – | – | – | – | – | – |
| 8 | – | 1 | – | – | – | 1 | – | 2 | – | – | – | – |
A total of 516 spikelets each were investigated in the wild type and the ssh1 mutant. The dashes indicate no spikelets showing the corresponding phenotype.
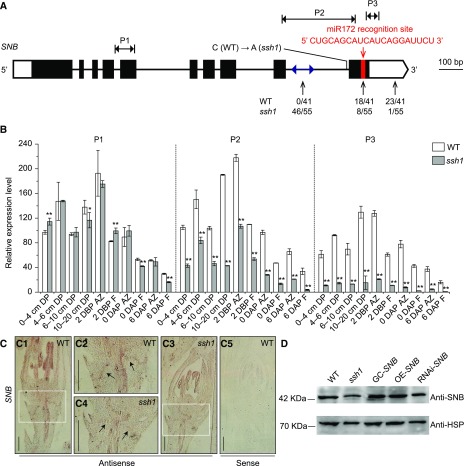
Like other AP2-like genes, previous studies revealed that the transcript levels of SNB are negatively regulated by a microRNA; microRNA172 (miR172) recognizes the SNB mRNA sequence at 1196 to 1216 bp (Lee et al., 2007; Zhu et al., 2009; Lee and An, 2012; Wang et al., 2015). To distinguish the effects of the SNP SNV6 and miR172 on SNB expression, we performed an reverse transcription quantitative PCR (RT-qPCR analysis to detect the expression levels of SNB using three primer sets (P1, P2, and P3; locations shown in Figure 5A). RT-qPCR analysis using primer set P1 (spanning the fourth intron) showed that the expression of SNB was similar between the ssh1 mutant and the wild-type plants (Figure 5B). This indicates that the SNP in the ssh1 mutant did not affect the transcription of this gene. In addition, among the organs investigated, SNB expression was highest in the AZ (the junctions between the seed and the pedicel) at 2 d before pollination, which is consistent with its function in regulating AZ development.
Figure 5.
The Point Mutation SNV6 Alters the mRNA Splicing of SNB.
(A) The posttranscriptional regulation of SNB caused by the SNP SNV6 and miR172. The 3′ end of SNB/ssh1 was determined by cloning and sequencing the amplicons generated using RACE. The blue double-sided arrow in the 9th intron represents the randomly distributed region of the 3′ ends of SNB transcripts detected in the ssh1 mutant. The red bar in the 10th exon indicates the miR172 target site. The black vertical arrows indicate the inferred 3′ ends of the SNB/ssh1 mRNAs detected in the wild type and the ssh1 mutant. The numbers below each arrow indicate the proportion of clones that possess these sites. The regions were targeted using RT-qPCR with three primer sets, P1, P2, and P3, indicated by the black double-sided arrows.
(B) Comparison of SNB expression in various tissues of the wild type and the ssh1 mutant using RT-qPCR with three primer sets, P1, P2, and P3 (n = 3 pooled tissues, five plants per pool). DP, developing panicles; F, florets; DBP, day before pollination; DAP, day after pollination. The rice housekeeping gene UBIQUITIN was used as an internal control to normalize gene expression data. Values are means ± sd. Two-tailed Student’s t tests were used to compare the wild type and the ssh1 mutant (*, P < 0.05 and **, P < 0.01; Supplemental File 2).
(C) mRNA in situ hybridization of SNB at spikelet developmental stage Sp8. (C1) and (C3) show that stronger SNB expression is maintained in the spikelet of the wild type than the ssh1 mutant. (C2) and (C4) are magnifications of the white boxes in (C1) and (C3), respectively, in which arrows indicate the abscission layer. (C5) shows a negative control using the sense probe. Bars = 200 μm.
(D) Immunoblot analysis of SNB in the wild type, the ssh1 mutant, GC-SNB, OE-SNB, and RNAi-SNB. The rice housekeeping protein HSP served as the loading control.
RT-qPCR analysis using primer set P2 (spanning the 9th intron) indicated significantly lower levels of SNB transcripts in the ssh1 mutant than in the wild type, indicating that the SNP SNV6 might alter mRNA splicing (Figure 5B). Additionally, the expression levels of SNB detected using primer set P3 (across the target site of miR172) were lower than those detected using P2 in both the ssh1 mutant and the wild type, implying that miR172 negatively regulates the expression of SNB by facilitating mRNA cleavage (Figure 5B). A further rapid amplification of cDNA ends (RACE) experiment was performed to investigate the 3′ ends of the SNB transcripts in both the ssh1 mutant and the wild type. Sequencing the clones of the RACE amplicons revealed that, in the wild type, 56.1% (23/41) and 43.9% (18/41) of the 3′ ends of the SNB transcripts were located in the 3′-untranslated region (UTR) and the miR172 target site, respectively (Figure 5A; Supplemental Figure 6), while in the ssh1 mutant, only 1.8% (1/55) and 14.5% (8/55) were located at the 3′-UTR and the miR172 target site, respectively, with the remaining (83.6%) distributed randomly throughout the 9th intron. These results indicate that the SNP SNV6, which is close to the 3′ acceptor site between the 9th intron and the 10th exon, caused aberrant splicing of the 9th intron in the ssh1 mutant (Figure 5A; Supplemental Figure 6).
An in situ hybridization analysis using an antisense probe designed to target the 3′-subterminal region of SNB was used to compare the transcript levels of SNB in the ssh1 mutant and the wild-type plants. We found that SNB was expressed in both the anthers and the AZ in the ssh1 mutant, but to a lesser extent than in the wild type (Figure 5C). Additionally, immunoblot analysis showed that the ssh1 mutant produced less SNB protein than the wild type, consistent with the decrease in the abundance of full-length SNB transcript (Figure 5D). Taken together, the SNP SNV6 reduces the number of entire SNB transcripts in ssh1 by altering its mRNA splicing, leading to a decrease in seed shattering.
SNB Positively Regulates the Expression of Two Rice RPL Orthologs, qSH1 and SH5
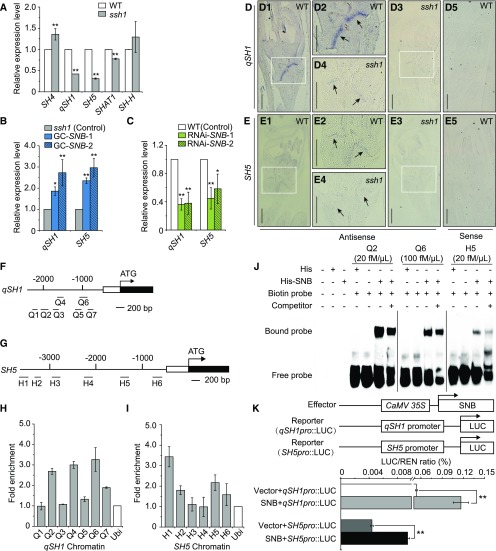
To elucidate the regulatory pathway involving SNB, we compared the expression of the five known rice genes related to seed shattering in the ssh1 mutant (Supplemental Figure 7). Compared with the wild type, the two rice orthologs of RPL, qSH1 and SH5, were dramatically downregulated in the young panicles (0–4 cm, the stage of abscission layer formation) of the ssh1 mutant (Figure 6A). We further investigated the expression levels of qSH1 and SH5 in the GC-SNB and the RNAi-SNB lines. Consistent with the changes in qSH1 and SH5 expression between ssh1 and the wild type, both qSH1 and SH5 were significantly upregulated and downregulated in the GC-SNB and RNAi-SNB lines, respectively, compared with their controls (Figures 6B and 6C). In addition, an mRNA in situ hybridization demonstrated that the transcripts of both qSH1 and SH5 were reduced in the AZ of the ssh1 mutant compared with the wild type (Figures 6D and 6E). These results indicated that SNB positively regulates the expression of qSH1 and SH5.
Figure 6.
SNB Positively Regulates the Expression of qSH1 and SH5.
(A) Comparison of the expression levels of five rice seed shattering genes in 0- to 4-cm young panicles of the wild type and the ssh1 mutant. The rice housekeeping gene UBIQUITIN was used as an internal control to normalize gene expression data. Values are means ± sd (n = 3 pooled tissues, five plants per pool). Two-tailed Student’s t tests were used to compare the wild type and the ssh1 mutant (**, P < 0.01).
(B) Comparison of expression levels of qSH1 and SH5 in the young panicles (0–4 cm) of the GC-SNB lines and the ssh1 mutant (control).
(C) Comparison of the expression levels of qSH1 and SH5 in the young panicles (0–4 cm) of the RNAi-SNB plants and the wild type (control).
In (B) and (C), the rice housekeeping gene UBIQUITIN was used as an internal control to normalize gene expression data. Values are means ± sd (n = 3 pooled tissues, five plants per pool). Two-tailed Student’s t tests were used to compare the transgenic lines and the corresponding controls (*, P < 0.05 and **, P < 0.01; Supplemental File 2).
(D) and (E) Stronger mRNA hybridization signals of qSH1 (D) and SH5 (E) are detected in the AZ of the wild type than the ssh1 mutant. (D2), (D4), (E2), and (E4) are magnifications of the white boxes in (D1), (D3), (E1), and (E3), respectively, and the arrows indicate the abscission layer. (D5) and (E5) show the hybridization signal using the sense probes as a negative control. Bars = 200 μm.
(F) to (I) ChIP-qPCR assays of qSH1 and SH5 using ChIP-DNA complexes isolated from 0- to 4-cm young panicles of the SNB-GFP transgenic plants. (F) and (G) show the genomic structures of qSH1 and SH5, respectively. The numbers (Q1 to Q7 and H1 to H6) indicate the tested regions. (H) and (I) show the enrichment of SNB on the qSH1 and SH5 chromatin, indicated as the fold change in the immunoprecipitation sample over the control containing no antibodies. Values are means ± sd (n = 3 pooled tissues, 10 plants per pool).
(J) EMSA revealed that the His-SNB recombinant protein was able to bind to both the Q2 and Q6 fragments of the qSH1 promoter and the H5 fragment of the SH5 promoter.
(K) Dual luciferase reporter assays in rice protoplasts showed that the SNB protein promoted the expression of the LUC gene through binding to the qSH1 and SH5 promoters. Vector represents the pGreenII 62-SK empty vector, and REN represents the Renilla luciferase gene. Values are means ± sd (n = 3 biological replicates). Difference significance analysis was conducted with two-tailed Student’s t tests (**, P < 0.01; Supplemental File 2).
To examine whether SNB affects the development of the abscission layer by directly regulating qSH1 and SH5 in vivo, we first generated a construct containing an SNB-GFP fusion gene under the control of the Cauliflower mosaic virus (CaMV) 35S promoter and introduced it into the ssh1 mutant. The transgenic plants constitutively expressing the SNB-GFP fusion gene (termed SNB-GFP) had wild-type levels of seed shattering, implying that the SNB-GFP fusion protein had a similar function to SNB (Supplemental Figure 8). Furthermore, we performed a chromatin immunoprecipitation qPCR (ChIP-qPCR) assay using 0- to 4-cm young panicles from the SNB-GFP transgenic plants. Our data showed that the DNA fragments were more than 3-fold enriched at the promoter (Q2, Q4, and Q6) of the qSH1 gene and the promoter (H1) of the SH5 gene (Figures 6F to 6I). We subsequently performed an electrophoretic mobility shift assay (EMSA) using the His-SNB recombinant protein to determine whether the promoters of qSH1 and SH5 were bound by SNB in vitro. The probes were synthesized based on the nucleotide sequences of the DNA fragments (Q1 through Q7 and H1 through H6) and labeled with biotin at the 3ʹ end, while the unlabeled probes were used as competitors. The EMSA revealed that the His-SNB recombinant protein was able to bind to both the Q2 and Q6 fragments of the qSH1 promoter and the H5 fragment of the SH5 promoter (Figure 6J; Supplemental Figure 9). Additionally, dual luciferase reporter assays in rice protoplasts showed that SNB can promote the expression of the LUC gene through binding to both the qSH1 and SH5 promoters (Figure 6K). These results suggest that SNB controls the development of the AZ by directly regulating the expression of two rice RPL orthologs, qSH1 and SH5.
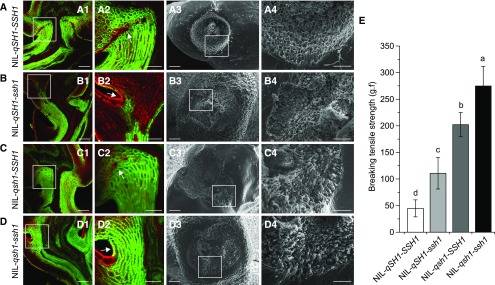
To investigate the AZ developmental effects of the regulatory pathway involving SNB/SSH1 and qSH1, we further developed four near-isogenic lines (NILs) in the genetic background of the indica variety Teqing with the inactive SH4 gene: NIL-qSH1-SSH1, NIL-qSH1-ssh1, NIL-qsh1-SSH1, and NIL-qsh1-ssh1 (Supplemental Figure 10). Longitudinal sections of the spikelets at anthesis were observed using confocal microscopy, which revealed that, in NIL-qSH1-SSH1, the AZ cells were formed in the basal area near the sterile lemmas, but the abscission layer was discontinuous (Figure 7A). In NIL-qSH1-ssh1, the formation of the AZ cells was repressed on one side, indicating defective AZ development (Figure 7B). The AZ cells of NIL-qsh1-SSH1 were visible but their development was incomplete (Figure 7C). In NIL-qsh1-ssh1, the abscission layer was completely absent, the surfaces of the seed base were broken and rough, and the pedicel BTS was the strongest among the four NILs (Figures 7D and 7E), indicating that the mutation of SNB in NIL-qsh1-ssh1 might cause a lower level of qSH1 expression than was present in NIL-qsh1-SSH1, completely suppressing the development of the AZ.
Figure 7.
Characterization of Floral AZ Morphology in the Four NILs of qSH1 and SNB.
(A) to (D) Morphological analyses of NIL-qSH1-SSH1 (A), NIL-qSH1-ssh1 (B), NIL-qsh1-SSH1 (C), and NIL-qsh1-ssh1 (D). (A1), (B1), (C1), and (D1) show fluorescence images of longitudinal sections across the flower and pedicel junction. (A2), (B2), (C2), and (D2) are magnifications of the white boxes in (A1), (B1), (C1), and (D1), respectively, and the arrows point to the AZ or the corresponding region. (A3), (B3), (C3), and (D3) show SEM images of the seed base. The white boxes in these images are magnified in (A4), (B4), (C4), and (D4), respectively. Bars = 50 μm in panels (2) and (4) and 100 μm in panels (1) and (3).
(E) Comparison of BTS in the four NILs at 35 d after pollination. Values are means ± sd (n = 50 grains). The g.f is the gravitational unit of force. Different letters denote significant differences (P < 0.01) determined using Tukey’s honestly significant difference analysis (Supplemental File 2).
SNB Affects Lignin Deposition in the AZ
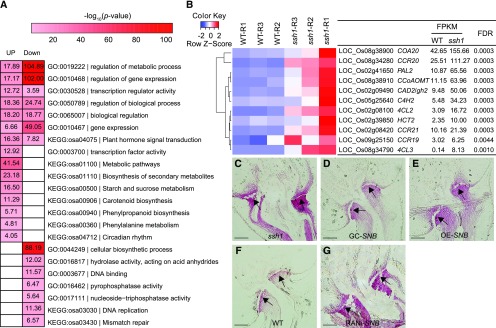
To analyze the molecular functions of SNB, we performed RNA-seq experiments using 0- to 4-cm young panicles from the wild type and the ssh1 mutant. We identified a total of 2402 differentially expressed genes (DEGs), including 1548 upregulated and 854 downregulated DEGs (fold change ≥ 2, FDR < 0.001; Supplemental Data Set 1). Further Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses revealed that these DEGs were enriched in multiple biological processes, including the regulation of metabolic processes, regulation of gene expression, transcription regulator activity, plant hormone signal transduction, and phenylpropanoid biosynthesis (Figure 8A), suggesting that SNB is involved in a complex network regulating rice inflorescence development.
Figure 8.
SNB Negatively Regulates Lignin Biosynthesis and Metabolism.
(A) GO and KEGG analysis of 1403 upregulated and 999 downregulated genes in the young panicles (0–4 cm) of the wild type and the ssh1 mutant.
(B) Hierarchical clustering of 11 DEGs involved in lignin biosynthesis. The average fragments per kilobase of exon per million mapped reads (FPKM) value of all three biological replicates is shown.
(C) to (G) Comparison of lignin deposition in the pedicel junction at spikelet developmental stage Sp8, revealed using phloroglucinol staining. The arrows indicate the AZ. Bars = 100 μm.
Previous studies have shown that lignin content is an important determinant of seed shattering in rice and fruit dehiscence in Arabidopsis (Mele et al., 2003; Yoon et al., 2014, 2017; Lee et al., 2018). Notably, SH5 was previously found to negatively regulate the expression of CINNAMYL ALCOHOL DEHYDROGENASE, an important gene for lignin biosynthesis, by directly binding to its promoter, thus affecting seed shattering in rice (Yoon et al., 2014). Eleven genes involved in lignin biosynthesis, including CINNAMYL ALCOHOL DEHYDROGENASE, were upregulated in the young panicles of the ssh1 mutant (Figure 8B).
To determine whether SNB suppresses lignin deposition in the AZ, we detected the AZ lignin contents of the wild type, the ssh1 mutant, and the transgenic plants at spikelet developmental stage Sp8 using phloroglucinol staining. Lignin deposition was lower in the wild-type, GC-SNB, and OE-SNB plants than in the ssh1 mutant and RNAi-SNB plants (Figures 8C to 8G). Thus, SNB positively regulates the expression of SH5 and qSH1, suppressing lignin deposition in the AZ and thereby modulating seed shattering in rice.
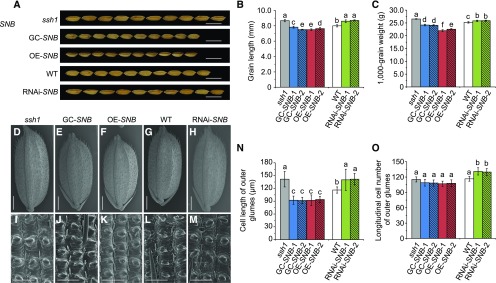
The ssh1 Mutant Has Larger Seeds and a Higher Grain Yield Than the Wild Type
In addition to the decreased shattering observed in the ssh1 mutant, these plants were also found to have increased seed lengths (+7.0%) and 1000-grain weights (+6.1%) compared with the wild-type plants (Figures 9A to 9C). To determine whether the mutated SNB gene positively modulated seed length, we investigated the seed size and grain weights of the transgenic plants. We found that, compared with the controls, the GC-SNB and OE-SNB lines had shorter seed lengths and lower 1000-grain weights, while downregulating the expression of SNB (RNAi-SNB) resulted in longer seeds and dramatically increased 1000-grain weights (Figures 9A to 9C). Further histological examination using SEM revealed that both the mutation and the downregulation of SNB significantly increased the longitudinal lengths of outer epidermis cells in the lemma. By contrast, the overexpression of SNB decreased the cell lengths of these tissues (Figures 9D to 9N). The numbers of outer epidermis cells in the lemma showed no significant increase in the ssh1 mutant relative to the wild type (Figure 9O), indicating that SNB regulates the seed size mainly by modulating the longitudinal cell lengths. In addition, an evaluation of other yield-related traits showed that, although the ssh1 mutant had fewer primary branches and a lower seed set ratio, ssh1 had an increased number of secondary branches, resulting in a significant increase in overall grain yield (Supplemental Figure 11).
Figure 9.
The ssh1 Mutation Positively Regulates Seed Size and Grain Weight.
(A) Comparison of seed size between the wild type, the ssh1 mutant, and the GC-SNB, OE-SNB, RNAi-SNB transgenic plants. Bars = 1 cm.
(B) Grain length of the wild type, the ssh1 mutant, and the GC-SNB, OE-SNB, and RNAi-SNB transgenic plants. Values are means ± sd (n = 30 grains).
(C) The 1000-grain weight of the wild type, the ssh1 mutant, and the GC-SNB, OE-SNB, and RNAi-SNB transgenic plants. Values are means ± sd (n = 10 plants).
(D) to (H) SEM images of grains from the wild type, the ssh1 mutant, and the GC-SNB, OE-SNB, and RNAi-SNB transgenic plants. Bars = 1 mm.
(I) to (M) SEM images of the lemma outer epidermis cells from the wild type, the ssh1 mutant, and the GC-SNB, OE-SNB, and RNAi-SNB transgenic plants. Bars = 100 μm.
(N) and (O) Comparison of the longitudinal cell lengths and cell numbers of the outer glumes in the wild type, the ssh1 mutant, and the GC-SNB, OE-SNB, and RNAi-SNB transgenic plants. Values are means ± sd (n = 10 grains).
In (B), (C), (N), and (O), different letters denote significant differences (P < 0.01) determined using Tukey’s honestly significant difference analysis (Supplemental File 2).
To investigate whether the introgression of the EMS-generated ssh1 allele into the current cultivars would have a positive effect on grain weight, we introduced the ssh1 allele into a nonshattering indica variety, 93-11, by backcrossing it with the ssh1 mutant. We then selected 10 plants homozygous for the SSH1 allele and 10 plants homozygous for the ssh1 allele from the BC1F2 population and measured their seed lengths and weights. The 93-11ssh1 plants had increased seed lengths (+9.5%) and 1000-grain weights (+7.7%) compared with the 93-11SSH1 plants (Supplemental Figure 12), suggesting the ssh1 allele as a possible target for efforts to enhance grain yields.
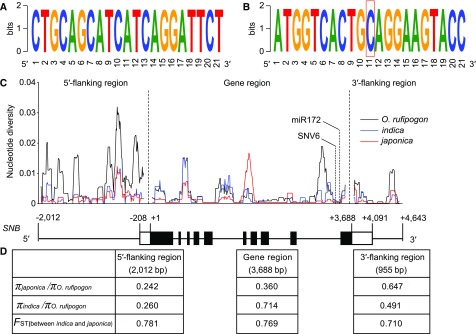
Nucleotide Diversity and Selection Signature in SNB
To investigate the nucleotide diversity of SNB/SSH1 in wild and cultivated rice, we sequenced a 6655-bp genomic fragment covering the entire SNB gene (3688 bp), a 2012-bp 5′-flanking region, and a 955-bp 3′-flanking region, from 46 accessions of O. rufipogon (the wild ancestor of Asian cultivated rice) and 86 Asian rice cultivars (48 indica and 38 japonica cultivars collected from 16 countries). Nucleotide alignment showed that the wild and cultivated rice had identical sequences at both the miR172 target site and the SNV6 SNP identified in this study (Figures 10A and 10B). We also recovered the reads that mapped to the miR172 target site and the 21 bp flanking the SNP SNV6 from publicly available genome resequencing data of 446 accessions of the wild rice species O. rufipogon and 1083 cultivated indica and japonica varieties (Huang et al., 2012). The sequence alignment of these reads showed that both the cultivated and wild rice genomes had highly conserved sequences at these two sites (Supplemental Figures 13A and 13B), which was consistent with the results generated using Sanger sequencing. However, the nucleotide diversity at the 5′-flanking region of SNB was strongly reduced in cultivated rice compared with wild rice (Figure 10C). The percentage of nucleotide diversity at the 5′-flanking regions between japonica and O. rufipogon and between indica and O. rufipogon was 24.2 and 26.0%, respectively (Figure 10D), much lower than the average levels observed using random gene fragments across the rice genome (42% in japonica/O. rufipogon and 48% in indica/O. rufipogon, respectively; Caicedo et al., 2007), suggesting that the 5′-flanking region of SNB was subjected to selection during rice domestication. In addition, the fixation index (FST), which was used to evaluate the genetic divergence between populations, was more than 0.7 at the coding, 5′-flanking, and 3′-flanking regions of SNB between japonica and indica (Figure 10D). We also analyzed the FST on chromosome 7 using the SNP data between geographically diverse cultivated rice (Huang et al., 2012). The FST level in SNB/SSH1 was higher between the indica and japonica subspecies than it was between the temperate japonica and tropical japonica subpopulations (Supplemental Figures 13C and 13D); therefore, we propose that differences in SNB/SSH1 might exist between indica and japonica subspecies.
Figure 10.
Nucleotide Diversity and indica-japonica Differentiation Analysis of SNB using 46 Accessions of O. rufipogon, 48 indica Varieties, and 38 japonica Varieties.
(A) Conservation analysis showing that both wild and cultivated rice lines have identical sequences at the miR172 target site.
(B) Conservation analysis of the 10-bp upstream and downstream sequences surrounding the SNP SNV6, showing that both wild and cultivated rice have a conserved C at the SNV6 site. The SNP SNV6 is boxed in red.
(C) Sliding-window analysis of nucleotide polymorphism (π) in SNB. The values were calculated for each sliding window of 100 bp with an increment of 10 bp. Black boxes represent exons, white boxes represent UTRs, and thin black lines indicate flanking regions and introns.
(D) The values of πjaponica/πO. rufipogon, πindica/πO. rufipogon, and FST (between indica and japonica) were each calculated for the 5′-flanking region, gene region, and 3′-flanking region of SNB.
DISCUSSION
The AP2/Ethylene Responsive Element Binding Factor gene family contains plant-specific transcription factors with one or two conserved AP2 domain(s), which are divided into two major lineages: euAP2 and AINTEGUMENTA (Riechmann and Meyerowitz, 1998; Kim et al., 2006; Zumajo-Cardona and Pabón-Mora, 2016). The AP2-like genes execute a series of floral development functions, including specifying the identity of the floral organs and modulating the expression of genes related to flower development (Kim et al., 2006; Ripoll et al., 2011; Samad et al., 2017). The AP2-like genes also play important roles in fruit dehiscence in Arabidopsis (Ripoll et al., 2011) and seed shattering in wheat and rice (Simons et al., 2006; Zhou et al., 2012; Debernardi et al., 2017). In the present study, we found that SNB, a transcription factor belonging to the euAP2 group and closely related to maize SID1, regulates seed shattering by conferring abscission layer identity, suppressing lignin deposition in AZ, and repressing vascular bundle overgrowth in rice. In addition, the ssh1 mutant identified in this study represents a new mutant allele of the rice heterochronic gene SNB, which was previously reported to regulate the transition from the spikelet meristem to the floral meristem (Lee et al., 2007; Zhu et al., 2009; Lee and An, 2012). These findings support the suggestion that the AP2 transcription factors have at least partially conserved functions in regulating the development of the AZ and inflorescences in dicots and monocots.
Previous studies have shown that the Arabidopsis AP2 gene (At4g36920) negatively regulates the homeobox gene RPL, which controls the development of the replum, a structure associated with pod dehiscence and seed dispersal (Ripoll et al., 2011). In rice, two RPL orthologs, qSH1 and SH5, play an important role in the development of the AZ. The downregulation or dysfunction of qSH1 and SH5 resulted in an aberrant abscission layer and decreased seed shattering (Konishi et al., 2006; Yoon et al., 2014). SHAT1, another AP2-like transcription factor, is known to regulate the formation of the abscission layer. The expression of SHAT1 in the AZ was positively regulated by SH4, while qSH1 functions downstream of SHAT1 and SH4 to maintain their expression in the AZ, thus promoting AZ differentiation (Zhou et al., 2012). Despite this, our comparison of SNB expression in NIL-SH4-SSH1 and NIL-sh4-SSH1 did not indicate that SH4 regulates the expression of SNB (Supplemental Figure 14). Consistent with the fact that qSH1 functions downstream of SHAT1, we found that SNB positively regulates qSH1 and SH5 expression by directly binding to their promoters, thus controlling seed shattering. The NIL-qsh1-ssh1 plants, harboring both the inactive variation in the cis-element of qSH1 and the ssh1 mutation, had a complete loss of the abscission layer (Figure 7D). Based on these results, we speculated that the AP2 genes in Arabidopsis and rice might exhibit different regulatory patterns when modulating the expression of the RPL genes to promote organ abscission, because of the structural differences between the replum in Arabidopsis and the pedicel in rice.
In addition to controlling the development of the flower and the dehiscence zone, Arabidopsis AP2 negatively regulates the size and number of embryonic cells, thereby affecting seed size (Jofuku et al., 2005; Ohto et al., 2005). Compared with the wild type, the Arabidopsis ap2 mutants have larger replum epidermal cells and form an enlarged replum (Ripoll et al., 2011). Consistent with this function in Arabidopsis, the mutation of SNB led to longer seed lengths and higher grain weights by increasing the longitudinal cell lengths of the glumes, suggesting this gene as a possible target for efforts to enhance grain yield. Similarly, the mutation of the SHAT1 gene, encoding an AP2 transcription factor regulating seed shattering, also increased seed lengths in rice (Zhou et al., 2012). Additionally, several genes controlling seed size, including GS2/OsGRF4 and GL4, also regulated seed shattering in rice (Sun et al., 2016; Wu et al., 2017). Notably, we found that the ssh1 mutation also affected plant height and panicle patterning in rice, consistent with a previous report (Wang et al., 2015). Plant hormones are known to play important roles in plant height, seed size, and the separation of the AZ (Wang and Li, 2008; Estornell et al., 2013; Zuo and Li, 2014), and our genome-wide expression profile analysis showed that SNB might be involved in plant hormone signal transduction. The future investigation of the involvement of SNB in plant hormone signal transduction could therefore further elucidate its roles in seed shattering and inflorescence development.
Favorable natural variants of AP2-like genes were selected during the domestication of wheat and barley. The wheat domestication gene Q encodes an AP2 transcription factor with a high amino acid similarity (64%) to SNB. The selection of two mutations, a G-to-A mutation in exon 8 and a C-to-T change within the miR172 target site in exon 10, resulted in the transition from the elongated spikes and hulled grain in wild wheats to the subcompact spikes and free-threshing grains in domesticated wheats (Debernardi et al., 2017). In addition, natural variations at the miR172 target site of barley AP2 were selected during its domestication and improvement, associated with the transition from noncleistogamy and subcompact spikes to cleistogamy and dense spikes (Nair et al., 2010; Houston et al., 2013). In the present study, we found that the level of sequence polymorphism at the 5′-flanking region of SNB was strongly reduced in both indica and japonica varieties relative to the wild progenitors, similar to domestication signatures for maize teosinte branched1 (Wang et al., 1999), teosinte glume architecture1 (Wang et al., 2005), and rice GRAIN INCOMPLETE FILLING1 (Wang et al., 2008). We therefore speculated that the natural variations in SNB, especially the cis-element variations at the 5′-flanking region, might be associated with the domestication and improvement of seed shattering and yield-related traits in rice, which is valuable information for the further exploration of the underlying evolutionary mechanisms of these traits in the process of rice domestication.
The ssh1 allele identified in this study has a positive effect on seed shattering and seed weight but a weak negative effect on primary branch number and seed set ratio. The application of SNB would therefore be challenging in crop breeding programs. To circumvent any undesirable pleiotropic effects and enhance crop production, one strategy would be to apply CRISPR/Cas9 genome editing to generate diverse alleles of the target genes and provide beneficial variations for crop breeding (Scheben and Edwards, 2018). Another approach would be to apply temporally and spatially specific promoters that optimize the expression of the target genes (Vanhaeren et al., 2016). Thus, identifying favorable alleles of SNB and optimizing SNB expression could facilitate the improvement of rice seed shattering and grain yields.
METHODS
Plant Materials
The wild rice (Oryza rufipogon) introgression line YIL100, which possesses a high-shattering trait, was derived from a cross between an indica variety (Teqing) and an O. rufipogon accession (YJCWR). The nonshattering ssh1 mutant was identified from the M2 plants of an EMS-mutagenized YIL100 population. The four NILs, NIL-qSH1-SSH1, NIL-qSH1-ssh1, NIL-qsh1-SSH1, and NIL-qsh1-ssh1, were developed in the genetic background of indica variety Teqing through the marker assistance selection of the progenies derived from crosses between the ssh1 mutant and a japonica variety (C418) with an inactive qSH1 allele. For the nucleotide polymorphism analysis, 46 accessions of O. rufipogon and 86 Asian rice (Oryza sativa) cultivars were used (Supplemental Data Set 2). All plants were grown in the field at Sanya (18°N, 109°E), China.
Phenotypic Evaluation
To evaluate seed shattering, the main panicles of 10 plants were bagged and isolated prior to heading and used to investigate the average ratio of shattered filled grains to the total filled grains on the main panicle after harvesting. For BTS, a total of 50 spikelets or grains from three panicles were measured with a digital force gauge (FGP-1; SHIMPO). A total of 10 wild-type and ssh1 plants were bagged before heading to investigate their yield-related traits. For each line, the seed lengths of 30 filled grains were measured with an electronic digital caliper. Two independent T2 lines (10 plants per line), GC-SNB and OE-SNB, and downregulation (RNAi-SNB) transgenic plants were used to investigate seed shattering and yield-related traits.
MutMap Analysis
The MutMap method was used to map SSH1, according to the description by Abe et al. (2012) with minor modifications. An F2 population containing 277 individuals was constructed by crossing the ssh1 mutant with the wild-type YIL100. Second, a recessive pool and a dominant pool of DNAs were constructed by mixing equal amounts of DNA from 20 nonshattering F2 individuals and 20 easily shattering F2 individuals, respectively. Finally, both the bulked DNA samples and the two parental DNA samples were subjected to whole-genome sequencing using an Illumina HiSeq2500 platform, which was performed by BerryGenomics. Approximately 5-Gb paired-end short reads were obtained from the ssh1 mutant and the wild-type plants, and ∼10 Gb of reads was generated from the two bulked DNA pools, respectively. These short reads were aligned to the Nipponbare reference sequence (http://rice.plantbiology.msu.edu/) to enable the identification of reliable SNPs. The true SNPs from the mutagenesis were further screened, and the SNP index and the Δ(SNP-index) were calculated based on the SNP information, following the method described by Lu et al. (2014).
Histological Analysis
To observe the AZ structure, spikelets at the anthesis stage were longitudinally sectioned by hand. The sections were covered with 0.01% (w/v) acridine orange for 10 min in the dark, rinsed three times in distilled water, and then observed with a 488-nm and a 543-nm laser line using an Olympus FV1000 laser scanning microscope, as described by Briggs and Morris (2008). To analyze lignin deposition, young panicles (0–4 cm) at spikelet developmental stage Sp8 were collected from wild-type, ssh1 mutant, and transgenic plants and then dehydrated, embedded, spliced, rehydrated, and stained with phloroglucinol in 20% (v/v) HCl, according to the methods described by Yoon et al. (2014).
Scanning Electron Microscopy
The bases of mature seeds and the glume surfaces were gold plated and observed using a Hitachi S-3400N SEM at 15 kV. The longitudinal cell numbers and cell lengths of the outer glumes, the vascular bundle area, and the AZ at the pedicle junctions were measured using ImageJ software.
Generation of Constructs and Transformation
A 7206-bp genomic fragment from the wild type, harboring the entire SNB gene with a 2309-bp 5′-flanking region and a 1209-bp 3′-flanking region, was amplified using the primers GC-SSH1-F and GC-SSH1-R (Supplemental Data Set 3) and inserted into the binary vector pCAMBIA1300 (http://www.cambia.org) between the KnpI and XbaI sites to form the genetic complementary construct pGC-SNB. The construct pUbi:SNB harbored the wild-type SNB open reading frame, which was amplified using the primers OE-SSH1-F and OE-SSH1-R (Supplemental Data Set 3) and cloned into the binary vector pCAMBIA1301 (http://www.cambia.org) between the BmaHI and SpeI sites under the control of the maize Ubiquitin promoter. The construct p35S:SNB-GFP contained the SNB coding sequence, except that the TGA terminator was fused with GFP at the C terminus, driven by the 35S promoter. An inverted repeat harboring a 323-bp fragment from the wild-type SNB cDNA was inserted into the vector pTCK303 (Wang et al., 2004) to develop the construct pRNAi-SNB. All plasmid constructs were introduced into the Agrobacterium tumefaciens strain EHA105. The constructs pGC-SNB, pUbi:SNB, and p35S:SNB-GFP were transferred into the ssh1 mutant, and the construct pRNAi-SNB was transferred into the wild-type plants. Primers used for all cloning are listed in Supplemental Data Set 3.
Subcellular Localization
To determine the subcellular localization of SNB, two plasmid constructs were generated: p35S:OsMADS15-RFP (Wang et al., 2010), a nuclear localization marker, and p35S:SNB-GFP. The two plasmid constructs were cotransformed into rice protoplasts as described by Bart et al. (2006). After a 16-h incubation at 28°C in the dark, GFP and RFP fluorescence were examined with 488- and 543-nm laser lines using an Olympus FV1000 laser scanning microscope.
Transcription Activity Assay
To generate the plasmid constructs for the transcription activity assay using the Matchmarker GAL4 Two-Hybrid System 3 (Clontech), full-length coding sequences and various truncations of SNB were amplified, using cDNA from the wild type as a template. The PCR products were cloned into EcoRI and PstI sites of pGBKT7 to fuse to the GAL4 BD, and the transcription factor PROSTRATE GROWTH1 (Tan et al., 2008) was fused with the GAL4 BD as the positive control. All plasmid constructs were transformed into the yeast strain AH109 to evaluate the transcription activity of SNB, following the manufacturer’s instructions.
RNA Extraction, RACE, and RT-qPCR
Total RNAs from various tissues were extracted using the Trizol reagent (Thermo Fisher Scientific) and purified using the RNeasy Mini Kit (Qiagen), following the manufacturer’s instructions. First-strand cDNA was synthesized using SuperScript reverse transcriptase (Thermo Fisher Scientific) with an oligo(dT)12–18 primer. The RACE was conducted with the 3′-Full RACE kit (TaKaRa), according to the instructions. RT-qPCR was performed using a CFX96 real-time system (Bio-Rad). The rice housekeeping gene UBIQUITIN (LOC_Os03g13170.1) was used as an internal control to normalize the gene expression data using the relative quantification method (2–ΔΔCT) described by Livak and Schmittgen (2001). Each set of experiments had three biological replicates containing a pool of cDNA from five plants.
RNA-Seq Analysis
Total RNA was isolated from the young panicles (0–4 cm) of the wild type and the ssh1 mutant, with three biological replicates each containing five plants. Paired-end libraries were constructed and sequenced using an Illumina HiSeq2500 at BerryGenomics. The raw reads were mapped to the reference genome (Os-Nipponbare-Refrence-IRGSP-1.0, MSU7) using TopHat2 with the default parameters (Kim et al., 2013). Cuffdiff was used to calculate the fragments per kilobase of exon per million mapped reads of each gene and identify the DEGs (fold change ≥ 2, FDR < 0.001) between the ssh1 mutant and the wild type (Trapnell et al., 2010). The functional category analysis of the DEGs was performed using agriGO and KEGG (Yi et al., 2013; Tian et al., 2017).
Immunoblot Analysis
Leaves of rice seedlings were ground into powder in liquid nitrogen and then suspended in protein extraction buffer (62.5 mM Tris-HCl [pH 7.4], 10% [v/v] glycerol, 2% [w/v] SDS, 2 mM EDTA [pH 7.4], 1 mM PMSF, and 5% [v/v] β-mercaptoethanol). The samples were boiled, and the supernatants were resolved on a 12% (w/v) SDS-PAGE gel. The separated proteins were transferred to a nitrocellulose membrane (GE Healthcare) and detected by immunoblotting with the antibodies Anti-SNB (CUSABIO, DK70) and Anti-HSP (Beijing Protein Innovation, AbM51099-31-PU), respectively.
mRNA in Situ Hybridization
Young panicles (0–4 cm) were collected from the wild type and the ssh1 mutant, then fixed in 3.7% (v/v) Formalin-Acetic Acid-Alcohol solution, dehydrated, embedded in paraffin (Sigma-Aldrich), and sliced into 8-μm sections using a microtome (Leica RM2145). Three ∼300-bp fragments of SNB, qSH1, and SH5 cDNA were amplified and used as templates to generate sense and antisense digoxigenin-labeled RNA probes, which were prepared using a DIG RNA labeling kit (Roche). The mRNA hybridization and the immunological detection of the hybridized probes were performed as described previously (Zhang et al., 2007), with minor modifications.
ChIP-qPCR Analysis
Young panicles (0–4 cm) of the SNB-GFP transgenic lines were collected and fixed in 1% (v/v) formaldehyde under vacuum. Chromatin was isolated from the samples using sucrose gradient centrifugation and sonicated to produce DNA fragments using a Qsonica Q700 (100% amplitude, 40 cycles of pulse-on 30 s and pulse-off 30 s). A 40-μL aliquot of the sonicated chromatin was reverse cross-linked and used as the total input DNA control. Immunoprecipitation was performed with anti-GFP (Abcam) or without any antibody as described by He et al. (2013). The amounts of immunoprecipitated genomic DNA were assayed using real-time qPCR, performed on a CFX96 real-time system (Bio-Rad) with three biological replicates (10 plants per replicate). The calculation of the relative fold enrichment was performed as described by Zhang et al. (2010). The corresponding samples without any antibodies were used as negative controls. Quantification involved the normalization of each immunoprecipitation (IP) or control (no antibodies) sample Ct to the input DNA sample Ct value to obtain a ΔCt (ΔCt IP or ΔCt control), and the relative enrichment of each fragment was calculated using 2–(ΔCt IP–ΔCt control). The relative enrichment of the unrelated DNA sequence from the rice UBIQUITIN gene (LOC_Os03g13170.1) was set to 1 and used as an internal control to normalize the relative fold enrichment of the investigated fragments.
EMSA
Full-length SNB cDNA was amplified using the primers His-SSH1-F and His-SSH1-R (Supplemental Data Set 3) and cloned into pET32a between the BmaHI and EcoRI sites. His and His-SNB recombinant proteins were expressed in the Escherichia coli Rosetta (DE3) strain and purified using Ni Sepharose Beads (GE Healthcare), following the manufacturer’s instructions. DNA gel shift assays were performed using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific). The biotin 3ʹ end-labeled DNA fragments listed in Supplemental Data Set 3 were synthesized and annealed to be used as DNA probes, while the corresponding unlabeled DNA probes were used as competitors. Each 20-μL binding reaction contained 2 μL of biotin-labeled dsDNAs, 3 μg of His-SNB protein, 2 μL of 10× binding buffer, and 1 μL of 50% (v/v) glycerol. The binding reactions were incubated for 30 min at room temperature and then resolved by electrophoresis on 6% (w/v) native polyacrylamide gels in 0.5× Tris-borate-EDTA buffer. The biotin-labeled probes were detected using chemiluminescence, according to the instructions provided by Thermo Fisher Scientific.
Dual Luciferase Reporter Assay
To construct the effector plasmid, the full coding sequence of SNB was inserted into the vector pGreenII 62-SK between the PstI and KpnI sites. For the reporter construct, a 2011-bp upstream fragment of qSH1 and a 1772-bp upstream fragment of SH5 were inserted into pGreenII 0800-LUC between the KpnI and PstI sites to drive the firefly luciferase (LUC) gene to get the qSH1pro:LUC and SH5pro:LUC plasmids, respectively. A CaMV 35S promoter-driven Renilla luciferase gene was used as an internal control. For each assay, 8 μg of effector plasmid DNA and 8 μg of reporter plasmid DNA were cotransformed into rice protoplasts with the polyethylene glycol-mediated method (Bart et al., 2006). After incubating for 16 h at 28°C in the dark, the relative luciferase activities were measured by the Dual-Luciferase Reporter Assay System (Promega). Three biological replicates were performed for each assay.
Phylogenetic Analysis
The SNB protein sequence was used to perform a BLASTP search for homologs in other plant species. An amino acid multiple sequence alignment (Supplemental File 1) was conducted using ClustalX (version 2.1; Larkin et al., 2007). The phylogenetic tree of the AP2 subgroup genes was constructed by MEGA6 using the neighbor-joining method with a Jukes-Cantor model, pairwise deletion for missing data, and 1000 bootstrap repetitions (Tamura et al., 2013). The resulting tree was visualized and annotated using EvolView (Zhang et al., 2012).
Sequencing and Data Analysis
The fragments covering the coding region (3688 bp), the 5′-flanking region (2012 bp), and the 3′-flanking region (955 bp) of SNB were amplified using six PCR primer pairs and sequenced using the Sanger sequencing approach. The nucleic acid multiple sequence alignment was conducted using ClustalX (Larkin et al., 2007). Sequence conservation of the miR172 recognition site and the SNP SNV6 site identified in this study was assessed using WebLogo (Crooks et al., 2004). Both the average proportion of pairwise differences per base pair and the FST were calculated using DnaSP (version 5.1; Librado and Rozas, 2009) . In addition, a previously published data set of 1034 diverse rice accessions (550 indica, 409 temperate japonica, and 75 tropical japonica lines) was used to calculate the FST across chromosome 7 between different subspecies using VCFtools, with a 100-kb window size (Danecek et al., 2011; Huang et al., 2012).
Primers
The primers used in this study are listed in Supplemental Data Set 3.
Statistical Analysis
The two-tailed Student’s t tests used to compare data from two groups and the Tukey’s honestly significant difference analyses used to compare multiple groups were performed using SPSS version 17 (SPSS).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: SNB/SSH1, LOC_Os07g13170; SH4/SHA1, LOC_Os04g57530; qSH1, LOC_Os01g62920; SH5, LOC_Os05g38120; SHAT1, LOC_Os04g55560; SH-H, LOC_Os07g10690; and OsMADS15, LOC_Os07g01820. The RNA-seq data derived from the wild type and the ssh1 mutant have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus under accession number GSE116422.
Supplemental Data
Supplemental Figure 1. Genotype of the wild rice introgression line YIL100.
Supplemental Figure 2. Comparison of SNB expression in RNAi-SNB transgenic plants and the controls.
Supplemental Figure 3. Full-length cDNA of SNB and the deduced amino acid sequence.
Supplemental Figure 4. Phylogenetic tree of the AP2 subgroup genes from rice and other plant species.
Supplemental Figure 5. Floral morphology in the ssh1 mutant.
Supplemental Figure 6. Positions of the 3′ end of SNB/ssh1 in the wild type and the ssh1 mutant.
Supplemental Figure 7. Chromosome positions of seed shattering-related genes in rice.
Supplemental Figure 8. Overexpression of SNB-GFP fusion gene rescues the mutation phenotype of the ssh1 mutant.
Supplemental Figure 9. Screening of putatively bound sites in the promoters of qSH1 and SH5 using an EMSA.
Supplemental Figure 10. Graphical genotypes of four NILs in the genetic background of the indica variety teqing.
Supplemental Figure 11. Comparison of yield-related traits in the wild type and the ssh1 mutant.
Supplemental Figure 12. The ssh1 allele increases seed lengths and weights in the indica variety 93-11 background.
Supplemental Figure 13. Consensus sequences at the miR172 target site and the SNV6 site in SNB and the FST on chromosome 7 detected using publicly available genome data from wild and cultivated rice.
Supplemental Figure 14. Comparison of SSH1 expression in two NILs, NIL-SH4-SSH1 and NIL-sh4-SSH1.
Supplemental Table 1. SNP information in the SSH1 mapped region.
Supplemental Data Set 1. DEGs between the wild type and the ssh1 mutant detected using RNA-Seq.
Supplemental Data Set 2. Plant materials used in this study.
Supplemental Data Set 3. Primers used in this study.
Supplemental File 1. Text file of the alignment used for the phylogenetic analysis in Supplemental Figure 4.
Supplemental File 2. The results of statistical analyses.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank International Rice Research Institute, Chinese Rice Research Institute, and Institute of Crop Sciences of Chinese Academy of Agricultural Sciences for providing the wild and cultivated rice germplasm. This work was supported by National Natural Science Foundation of China (Grants 31771742 and 91435103), National Key Research and Development Program of China (Grant 2016YFD0100400), and Chinese Universities Scientific Fund (Grant 2017QC174).
AUTHOR CONTRIBUTIONS
L.T. conceived and designed the experiments; L.J. performed most of the experiments; X.M. analyzed the data of whole-genome resequencing; S.Z. and Y.T. identified the mutant; F.L., P.G., Y.F., Z.Z., H.C., and C.S. provided technical assistance and conducted the collection and maintenance of rice germplasm; L.T. and L.J. performed data analysis and wrote the article.
References
- Abe, A., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Ballester, P., Ferrándiz, C. (2017). Shattering fruits: Variations on a dehiscent theme. Curr. Opin. Plant Biol. 35: 68–75. [DOI] [PubMed] [Google Scholar]
- Bart, R., Chern, M., Park, C.J., Bartley, L., Ronald, P.C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, C.L., Morris, E.C. (2008). Seed-coat dormancy in Grevillea linearifolia: Little change in permeability to an apoplastic tracer after treatment with smoke and heat. Ann. Bot. 101: 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A.L., Williamson, S.H., Hernandez, R.D., Boyko, A., Fledel-Alon, A., York, T.L., Polato, N.R., Olsen, K.M., Nielsen, R., McCouch, S.R., Bustamante, C.D., Purugganan, M.D. (2007). Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 3: 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Meeley, R.B., Hake, S. (1998). The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 12: 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Meeley, R., Hake, S. (2008). Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135: 3013–3019. [DOI] [PubMed] [Google Scholar]
- Crooks, G.E., Hon, G., Chandonia, J.M., Brenner, S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek, P., et al. (2011). The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi, J.M., Lin, H., Chuck, G., Faris, J.D., Dubcovsky, J. (2017). microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144: 1966–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y., Wang, Y.Z. (2015). Seed shattering: From models to crops. Front. Plant Sci. 6: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y., Yang, X., Liu, J., Wang, B.H., Liu, B.L., Wang, Y.Z. (2014). Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat. Commun. 5: 3352. [DOI] [PubMed] [Google Scholar]
- Estornell, L.H., Agustí, J., Merelo, P., Talón, M., Tadeo, F.R. (2013). Elucidating mechanisms underlying organ abscission. Plant Sci. 199-200: 48–60. [DOI] [PubMed] [Google Scholar]
- Faris, J.D., Gill, B.S. (2002). Genomic targeting and high-resolution mapping of the domestication gene Q in wheat. Genome 45: 706–718. [DOI] [PubMed] [Google Scholar]
- Faris, J.D., Fellers, J.P., Brooks, S.A., Gill, B.S. (2003). A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics 164: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, D.Q. (2007). Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the Old World. Ann. Bot. 100: 903–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsuki, H., Suzuki, M., Hirose, A., Inaba, H., Yamada, T., Hajika, M., Komatsu, K., Katayama, T., Sayama, T., Ishimoto, M., Fujino, K. (2014). Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc. Natl. Acad. Sci. USA 111: 17797–17802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., Sidhu, G., Pawlowski, W.P. (2013). Chromatin immunoprecipitation for studying chromosomal localization of meiotic proteins in maize. Methods Mol. Biol. 990: 191–201. [DOI] [PubMed] [Google Scholar]
- Houston, K., et al. (2013). Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc. Natl. Acad. Sci. USA 110: 16675–16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, I.D. (1986). On the formation and development of abscission layer in rice plants, Oryza sativa. Jpn. J. Crop. Sci. 55: 451–457. [Google Scholar]
- Jofuku, K.D., Omidyar, P.K., Gee, Z., Okamuro, J.K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. USA 102: 3117–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katkout, M., Sakuma, S., Kawaura, K., Ogihara, Y. (2015). TaqSH1-D, wheat ortholog of rice seed shattering gene qSH1, maps to the interval of a rachis fragility QTL on chromosome 3DL of common wheat (Triticum aestivum). Genet. Resour. Crop Evol. 62: 979–984. [Google Scholar]
- Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., Salzberg, S.L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Soltis, P.S., Wall, K., Soltis, D.E. (2006). Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 23: 107–120. [DOI] [PubMed] [Google Scholar]
- Konishi, S., Izawa, T., Lin, S.Y., Ebana, K., Fukuta, Y., Sasaki, T., Yano, M. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396. [DOI] [PubMed] [Google Scholar]
- Larkin, M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee, Y., et al. (2018). A lignin molecular brace controls precision processing of cell walls critical for surface integrity in Arabidopsis. Cell 173: 1468–1480.e9. [DOI] [PubMed] [Google Scholar]
- Lee, D.Y., An, G. (2012). Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 69: 445–461. [DOI] [PubMed] [Google Scholar]
- Lee, D.Y., Lee, J., Moon, S., Park, S.Y., An, G. (2007). The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 49: 64–78. [DOI] [PubMed] [Google Scholar]
- Li, C., Zhou, A., Sang, T. (2006). Rice domestication by reducing shattering. Science 311: 1936–1939. [DOI] [PubMed] [Google Scholar]
- Librado, P., Rozas, J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lin, Z., et al. (2012). Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 44: 720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Z., Griffith, M.E., Li, X., Zhu, Z., Tan, L., Fu, Y., Zhang, W., Wang, X., Xie, D., Sun, C. (2007). Origin of seed shattering in rice (Oryza sativa L.). Planta 226: 11–20. [DOI] [PubMed] [Google Scholar]
- Livak, K.J., Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lu, H., Lin, T., Klein, J., Wang, S., Qi, J., Zhou, Q., Sun, J., Zhang, Z., Weng, Y., Huang, S. (2014). QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor. Appl. Genet. 127: 1491–1499. [DOI] [PubMed] [Google Scholar]
- McKim, S.M., Stenvik, G.E., Butenko, M.A., Kristiansen, W., Cho, S.K., Hepworth, S.R., Aalen, R.B., Haughn, G.W. (2008). The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135: 1537–1546. [DOI] [PubMed] [Google Scholar]
- Mele, G., Ori, N., Sato, Y., Hake, S. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17: 2088–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, S.K., et al. (2010). Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc. Natl. Acad. Sci. USA 107: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto, M.A., Fischer, R.L., Goldberg, R.B., Nakamura, K., Harada, J.J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 102: 3123–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi, K., Takagi, K., Kontani, M., Tanaka, T., Sano, Y. (2007). Different patterns of genealogical relationships found in the two major QTLs causing reduction of seed shattering during rice domestication. Genome 50: 757–766. [DOI] [PubMed] [Google Scholar]
- Pourkheirandish, M., et al. (2015). Evolution of the grain dispersal system in barley. Cell 162: 527–539. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379: 633–646. [DOI] [PubMed] [Google Scholar]
- Ripoll, J.J., Roeder, A.H., Ditta, G.S., Yanofsky, M.F. (2011). A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138: 5167–5176. [DOI] [PubMed] [Google Scholar]
- Samad, A.F.A., Sajad, M., Nazaruddin, N., Fauzi, I.A., Murad, A.M.A., Zainal, Z., Ismail, I. (2017). MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 8: 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheben, A., Edwards, D. (2018). Towards a more predictable plant breeding pipeline with CRISPR/Cas-induced allelic series to optimize quantitative and qualitative traits. Curr. Opin. Plant Biol. 45: 218–225. [DOI] [PubMed] [Google Scholar]
- Simons, K.J., Fellers, J.P., Trick, H.N., Zhang, Z., Tai, Y.S., Gill, B.S., Faris, J.D. (2006). Molecular characterization of the major wheat domestication gene Q. Genetics 172: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, P., Zhang, W., Wang, Y., He, Q., Shu, F., Liu, H., Wang, J., Wang, J., Yuan, L., Deng, H. (2016). OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 58: 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, H., et al. (2013). QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74: 174–183. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, L., Li, X., Liu, F., Sun, X., Li, C., Zhu, Z., Fu, Y., Cai, H., Wang, X., Xie, D., Sun, C. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40: 1360–1364. [DOI] [PubMed] [Google Scholar]
- Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., Xu, W., Su, Z. (2017). agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45: W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C., Williams, B.A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M.J., Salzberg, S.L., Wold, B.J., Pachter, L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaeren, H., Inzé, D., Gonzalez, N. (2016). Plant growth beyond limits. Trends Plant Sci. 21: 102–109. [DOI] [PubMed] [Google Scholar]
- Wang, E., et al. (2008). Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40: 1370–1374. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Li, J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59: 253–279. [DOI] [PubMed] [Google Scholar]
- Wang, H., Nussbaum-Wagler, T., Li, B., Zhao, Q., Vigouroux, Y., Faller, M., Bomblies, K., Lukens, L., Doebley, J.F. (2005). The origin of the naked grains of maize. Nature 436: 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K., Tang, D., Hong, L., Xu, W., Huang, J., Li, M., Gu, M., Xue, Y., Cheng, Z. (2010). DEP and AFO regulate reproductive habit in rice. PLoS Genet. 6: e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Sun, S., Jin, J., Fu, D., Yang, X., Weng, X., Xu, C., Li, X., Xiao, J., Zhang, Q. (2015). Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. USA 112: 15504–15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.L., Stec, A., Hey, J., Lukens, L., Doebley, J. (1999). The limits of selection during maize domestication. Nature 398: 236–239. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Chen, C., Xu, Y., Jiang, R., Han, Y., Xu, Z., Chong, K. (2004). A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 22: 409–417. [Google Scholar]
- Wu, W., et al. (2017). A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants 3: 17064. [DOI] [PubMed] [Google Scholar]
- Yi, X., Du, Z., Su, Z. (2013). PlantGSEA: A gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 41: W98–W103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, J., Cho, L.H., Kim, S.L., Choi, H., Koh, H.J., An, G. (2014). The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 79: 717–728. [DOI] [PubMed] [Google Scholar]
- Yoon, J., Cho, L.H., Antt, H.W., Koh, H.J., An, G. (2017). KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiol. 174: 312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Liang, W., Yang, X., Luo, X., Jiang, N., Ma, H., Zhang, D. (2010). Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22: 672–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Gao, S., Lercher, M.J., Hu, S., Chen, W.H. (2012). EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40: W569–W572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Liu, D., Wang, D., Zhang, R., Geng, S., Wu, L., Li, A., Mao, L. (2013). Over expression of the wheat BEL1-like gene TaqSH1 affects floral organ abscission in Arabidopsis thaliana. J. Plant Biol. 56: 98–105. [Google Scholar]
- Zhang, X., Madi, S., Borsuk, L., Nettleton, D., Elshire, R.J., Buckner, B., Janick-Buckner, D., Beck, J., Timmermans, M., Schnable, P.S., Scanlon, M.J. (2007). Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet. 3: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Belcram, H., Gornicki, P., Charles, M., Just, J., Huneau, C., Magdelenat, G., Couloux, A., Samain, S., Gill, B.S., Rasmussen, J.B., Barbe, V., et al. (2011). Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc. Natl. Acad. Sci. USA 108: 18737–18742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Lu, D., Li, C., Luo, J., Zhu, B.F., Zhu, J., Shangguan, Y., Wang, Z., Sang, T., Zhou, B., Han, B. (2012). Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1. Plant Cell 24: 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q.H., Upadhyaya, N.M., Gubler, F., Helliwell, C.A. (2009). Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol. 9: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumajo-Cardona, C., Pabón-Mora, N. (2016). Evolution of the APETALA2 gene lineage in seed plants. Mol. Biol. Evol. 33: 1818–1832. [DOI] [PubMed] [Google Scholar]
- Zuo, J., Li, J. (2014). Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48: 99–118. [DOI] [PubMed] [Google Scholar]