PAPST2 imports the stress signaling molecule PAP into the chloroplast and mitochondria, providing a missing link in our understanding of the organelle-to-nucleus PAP retrograde signaling pathway.
Abstract
The compartmentalization of PAPS (the sulfate donor 3′-phosphoadenosine 5′-phosphosulfate) synthesis (mainly in plastids), PAPS consumption (in the cytosol), and PAP (the stress signaling molecule 3′-phosphoadenosine 5′-phosphate) degradation (in plastids and mitochondria) requires organellar transport systems for both PAPS and PAP. The plastidial transporter PAPST1 (PAPS TRANSPORTER1) delivers newly synthesized PAPS from the stroma to the cytosol. We investigated the activity of PAPST2, the closest homolog of PAPST1, which unlike PAPST1 is targeted to both the plastids and mitochondria. Biochemical characterization in Arabidopsis thaliana revealed that PAPST2 mediates the antiport of PAP, PAPS, ATP, and ADP. Strongly increased cellular PAP levels negatively affect plant growth, as observed in the fry1 papst2 mutant, which lacks the PAP-catabolizing enzyme SALT TOLERANCE 1 and PAPST2. PAP levels were specifically elevated in the cytosol of papst2 and fiery1 papst2, but not in papst1 or fry1 papst1. PAPST1 failed to complement the papst2 mutant phenotype in mitochondria, because it likely removes PAPS from the cell, as demonstrated by the increased expression of phytosulfokine genes. Overexpression of SAL1 in mitochondria rescued the phenotype of fry1 but not fry1 papst2. Therefore, PAPST2 represents an important organellar importer of PAP, providing a piece of the puzzle in our understanding of the organelle-to-nucleus PAP retrograde signaling pathway.
INTRODUCTION
Sulfur is a constituent of the amino acids Cys and Met, proteins and cofactors, and multiple sulfated metabolites and compounds of unknown function (Schwacke et al., 2007). Unlike animals, which use proteins as their main source of sulfur, plants generally acquire inorganic sulfate from the soil (Takahashi et al., 2011). In the cytosol and plastids, sulfate is activated by adenylation with ATP to form adenosine 5′-phosphosulfate (APS). In the stroma, APS enters the sulfate reduction pathway, forming sulfite and sulfide and finally Cys, or it becomes phosphorylated to 3′-phosphoadenosine 5′-phosphosulfate (PAPS) by APS kinases (APK)1, 2, and 4 (Mugford et al., 2009; Koprivova and Kopriva, 2016). APK-induced activation of PAPS may also take place in the cytosol (via APK3). Interestingly, analyses of mutant plants lacking the different APK isoforms suggested that plastidial APK1 and APK2 produce the majority of cellular PAPS, whereas cytosolic APS phosphorylation appears to be of minor physiological importance (Mugford et al., 2009, 2010).
PAPS serves as a cofactor in post-translational protein modification and in the generation of various sulfated molecules, such as secondary metabolites and hormone-like peptides (Koprivova and Kopriva, 2016). The fact that these PAPS-consuming sulfotransferase reactions take place outside the chloroplast, whereas the majority of cellular PAPS is produced in the stroma, is indicative of the existence of a plastidial PAPS export system. A member of the mitochondrial carrier family (MCF), termed PAPS TRANSPORTER1 (PAPST1), was recently identified and shown to mediate the provision of newly synthesized PAPS from the chloroplast to the cytosol (Gigolashvili et al., 2012). Therefore, it is not surprising that Arabidopsis thaliana papst1 loss-of-function mutants resemble plants impaired in PAPS synthesis (double apk1 apk2 mutants) in several aspects. These plants exhibit a stunted growth phenotype, contain reduced levels of sulfotransferase products such as glucosinolates, and show increased feeding of excess APS into the reductive sulfate assimilation pathway (Gigolashvili et al., 2012). However, the phenotypic symptoms are generally of lower intensity compared with those of apk1 apk2 mutants (Mugford et al., 2009 2010). Therefore, the absence of PAPST1 reduces but does not totally abolish plastidial PAPS provision, which suggests that chloroplasts have additional PAPS export capacity.
Sulfate transfer from PAPS to acceptor molecules results in the release of 3′-phosphoadenosine 5′-phosphate (PAP). Efficient hydrolysis of PAP is apparently mandatory to drive the energetically unfavorable process of PAPS biosynthesis and to counteract the accumulation of PAP, which may competitively inhibit sulfotransferase reactions (Rens-Domiano and Roth, 1987). PAP was originally considered to be only a waste product of PAPS-dependent sulfotransferase reactions. However, recent studies have revealed that PAP acts as a signaling molecule (Estavillo et al., 2011; Chan et al., 2016). PAP accumulates during periods of drought and stress from salt and light; it inhibits 5′- to 3′-exoribonuclease activity, and it induces stress-responsive gene expression in the nucleus (Estavillo et al., 2011). Consequently, tight control of cellular PAP levels is mandatory to guarantee normal plant development and appropriate stress responses.
The enzyme SAL1 (salt tolerance in yeast), which regulates the cellular PAP concentration via its hydrolysis into AMP and phosphate, is located in chloroplasts and mitochondria (Estavillo et al., 2011). SAL1 not only plays a central role in PAP detoxification, but also it apparently represents the key player in PAP-dependent organelle-to-nucleus retrograde signaling. Arabidopsis mutants impaired in SAL1 activity (the different alleles are named sal1 [salt tolerance 1], fry1 [fiery 1], high expression of osmotically responsive genes 2, altered APX2 expression 8, rotunda 1, or fatty acid oxygenation upregulated 8, according to the diverse genetic screens that led to their isolation) show enhanced PAP levels and morphologically resemble 5′- to 3′-exoribonuclease loss-of-function mutants (Emanuelsson et al., 1999; Hirsch et al., 2011). These mutants are markedly affected in growth and development and exhibit increased tolerance toward various abiotic stressors (Quintero et al., 1996; Bodén and Hawkins, 2005; Höglund et al., 2006; Wilson et al., 2009a; Robles et al., 2010; Rodríguez et al., 2010). Some of these features (shorter petioles, wrinkled leaves, and anthocyanin accumulation) have previously been associated with enhanced jasmonate pathway activity (Bonaventure et al., 2007; Yan et al., 2007; Zhang and Turner, 2008). SAL1 also plays a role in integrating other hormonal signaling pathways, such as the regulation of stomatal closure and seed germination in Arabidopsis (Pornsiriwong et al., 2017). The phenotype of SAL1-loss-of-function mutants could be compensated for by a reduction of chloroplastidic but not cytosolic PAPS biosynthesis (Rodríguez et al., 2010; Lee et al., 2012). This observation implies that defects in the major PAPS production pathway decelerate extra-plastidial sulfotransferase reactions due to substrate limitation. The correspondingly restricted release of PAP apparently alleviates the adverse effects of defective PAP degradation. Moreover, targeting of SAL1 to either the chloroplast or cytosol significantly reduced PAP accumulation (Kim and von Arnim, 2009) and the expression of a high-light–responsive gene in loss-of-function mutants (Estavillo et al., 2011), demonstrating that cytosolic PAP levels help control nuclear gene expression.
Although the functional relevance of SAL1 in stress signaling and other fundamental physiological and developmental processes is widely accepted (Estavillo et al., 2011; Chan et al., 2016; Phua et al., 2018), our understanding of its mechanistic role in retrograde signaling is still limited. We have only a preliminary understanding of how information about organelle status becomes translated into the appropriate PAP concentration. Studies have revealed that oxidative stress in Arabidopsis chloroplasts is sensed and transduced by a series of interconnected redox-mediated structural and biochemical modifications of SAL1 (Chan et al., 2016). These alterations result in PAP accumulation and the associated cellular responses.
In this context, an important question arises: how does the signaling molecule PAP move between the different compartments? Or, more precisely, which components contribute to its membrane passage in chloroplasts and mitochondria? PAP uptake into chloroplasts and mitochondria is clearly required to shuttle PAP into degradation and thus to reduce cellular PAP levels. Inhibition of SAL1 activity or down-regulation of organellar PAP uptake may result in an increase in cytosolic PAP levels. More needs to be known about PAP transport in plants and the subcellular distribution of this potential signaling molecule.
The discrete cytosolic localization of PAP formation (by sulfotransferases) and organellar degradation (by SAL1) necessitates PAP uptake into mitochondria and plastids. PAPST1 was proposed to be involved in plastidial PAP import (Gigolashvili et al., 2012). However, two observations suggest that PAPST1 is not the only transporter that channels PAP into degradation. First, PAPST1 is restricted to plastids, whereas SAL1 is also located in mitochondria. Second, the phenotype of papst1 mutants (Gigolashvili et al., 2012) differs from that of SAL1 loss-of-function mutants in many aspects (Emanuelsson et al., 2000; Xiong et al., 2004; Kim and von Arnim, 2009; Wilson et al., 2009a; Rodríguez et al., 2010), indicating that PAP transport and degradation are still functional in papst1.
Here, we focus on the biochemical and physiological characterization of Arabidopsis PAPST2, which is 78% identical in amino acid sequence to PAPST1. PAPST2, like PAPST1, belongs to the MCF and shares important structural similarities with human and yeast MCF members, which transport adenine nucleotides and possibly also CoA and PAP across the mitochondrial membrane (Palmieri et al., 2011). Therefore, PAPST2 represents an excellent candidate PAPS and PAP transporter. Prediction of its subcellular targeting has been ambiguous (Schwacke et al., 2003, 2007), and proteome studies identified PAPST2 in the chloroplast envelope fraction (Ferro et al., 2003; Kleffmann et al., 2004) as well as in mitochondrial membrane preparations (Millar and Heazlewood, 2003). Here, with the help of the reporter protein green fluorescent protein (GFP), we verified the dual localization of PAPST2 in chloroplasts and mitochondria. Moreover, heterologous expression in Escherichia coli and uptake studies with the recombinant protein allowed us to identify its PAP, PAPS, ATP, and ADP transport capacity and mode of transport. Detailed analysis of selected mutant plants provided deeper insights into the physiological role of PAPST2 in the regulation of PAP concentrations in the cell. Although PAPST2, in contrast to PAPST1, appears to play a minor role in secondary sulfur metabolism, it clearly represents a key component of the PAP-degradation and PAP-signaling pathway in the plant cell.
RESULTS
PAPST2 Transports a Broad Spectrum of Adenylated Compounds
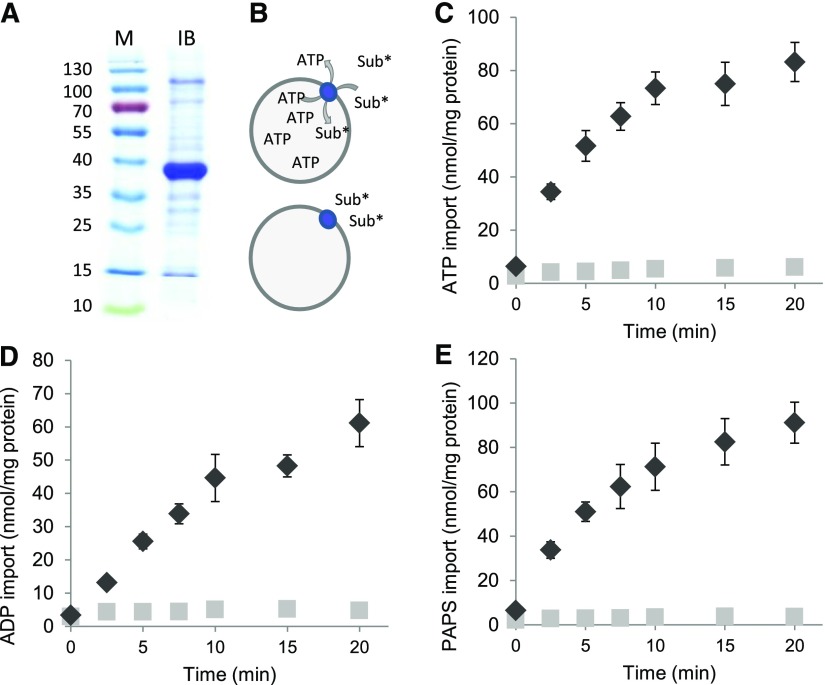
To biochemically characterize PAPST2, we heterologously expressed this transporter in E. coli cells. High amounts of recombinant PAPST2 were deposited in the form of inclusion bodies (Figure 1A). These protein aggregates were purified, solubilized, and refolded during reconstitution into artificial lipid vesicles. Transport measurements with radioactively labeled ATP revealed that the recombinant transporter was active in the liposomal environment. PAPST2 catalyzed considerable uptake of [α32P]ATP, [α32P]ADP, and [35S]PAPS into proteoliposomes loaded with ATP, whereas no (or only marginal) accumulation of the label was detectable when vesicles contained only the buffer medium (Figures 1B to 1E). This observation not only indicates that PAPST2 accepts ATP, ADP, and PAPS as substrates but also that it acts in antiport mode.
Figure 1.
Purified, Reconstituted PAPST2 Mediates Time-Dependent ATP, ADP, and PAPS Uptake.
(A) SDS-PAGE of the purified inclusion body proteins (5 µg) used for reconstitution. Recombinant PAPST2 has a calculated molecular weight of ∼40 kD. Approximate molecular weights (in kDa) are shown on the left. IB, purified inclusion body proteins; M, pre-stained molecular weight marker (Thermo Fisher).
(B) Illustration displaying the transport of radiolabeled substrate (Sub*) into ATP-loaded liposomes with reconstituted PAPST2 (blue circle). No transport occurs into nonloaded vesicles. Time-dependent import of 50 µM [α32P]ATP (C), [α32P]ADP (D), [35S]PAPS (E) by the purified and reconstituted PAPST2 protein into liposomes loaded with 5 mM ATP (black diamonds) or buffer alone (light gray squares) nonloaded, negative control. Data are the mean of three independent experiments; standard errors are given.
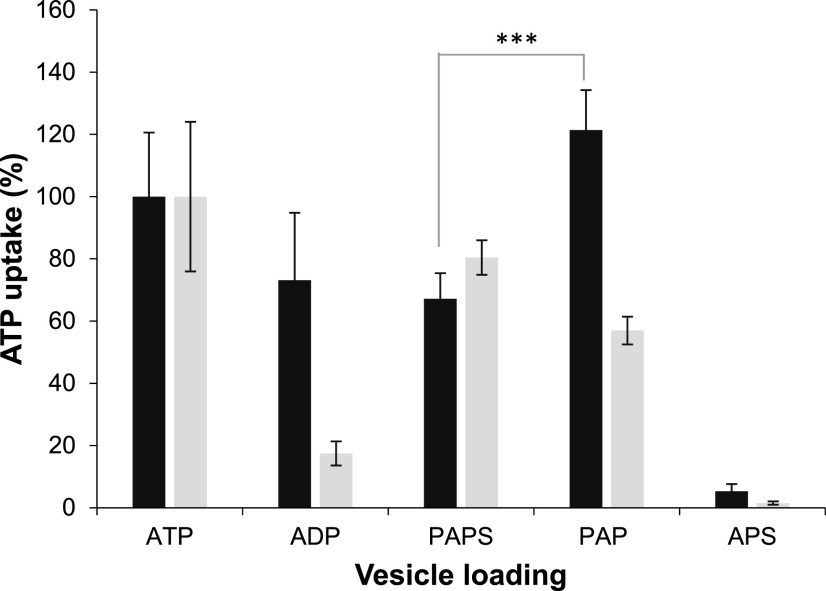
To gain deeper insights into the substrate spectrum of PAPST2 and particularly to test whether it might transport PAP, we conducted [α32P]ATP import studies in the presence of moderate (10-fold) excess of different nonlabeled adenine and guanine nucleotides, adenine nucleotide derivatives, precursors, and cofactors. Besides the three proven substrates ATP, ADP, and PAPS, only PAP and dATP substantially reduced the uptake of labeled substrate into proteoliposomes. The other tested molecules had no (or much weaker) effects on [α32P]ATP uptake (Table 1) and thus were not considered to be possible substrates. The high impact of PAP on [α32P]ATP import (Table 1), however, suggests that this molecule either acts as a potent inhibitor of PAPST2 transport or as a substrate that competes successfully with ATP at the binding center or during translocation. Uptake studies with radiolabeled PAP were not feasible because it is not commercially available. Therefore, we analyzed whether PAP, when present in the liposomal lumen, may induce (as a counter-exchange substrate) the uptake of [α32P]ATP. In fact, PAP was highly efficient in driving [α32P]ATP uptake (Figure 2). Therefore, we can conclude that PAP represents a substrate of PAPST2. Moreover, the rate of ATP/PAPS hetero-exchange was similar to that of ATP/ATP homo-exchange and slightly higher than the rates of ATP/PAPS and ATP/ADP hetero-exchange. This observation suggests that PAPST2 efficiently transports all four substrates and that it slightly prefers PAP and ATP over ADP and PAP (Figure 2). The significant difference between ATP/PAP and ATP/PAPS transport implies that it prefers PAP over PAPS transport. In this context, it is important to mention that recombinant PAPST1 preferentially transports [α32P]ATP during PAPS/ATP hetero-exchange, followed by PAP—whereas ADP is a rather inferior substrate (Figure 2). In liposomes containing reconstituted PAPST2 or PAPST1, APS, despite its structural similarity to the identified substrates, neither competed with [α32P]ATP for import (Table 1) nor induced any significant exchange when present in the liposomal lumen (Figure 2).
Table 1. Analysis of the Competing Effects of Potential Substrates on PAPST2-Mediated Import.
| Effector | Import of [α32P]-ATP (%) | SE (±) |
|---|---|---|
| None | 100 | - |
| ATP | 9.4 | 1.1 |
| ADP | 22.8 | 1.8 |
| PAP | 17.9 | 3.5 |
| PAPS | 38.4 | 7.4 |
| dATP | 17.8 | 3.3 |
| AMP | 73.9 | 11.6 |
| Adenine | 76.2 | 9.6 |
| Adenosine | 85.6 | 12.9 |
| ADP-Glc | 84.9 | 7.4 |
| 2,3 cyclo-AMP | 86.4 | 9.9 |
| 3,5 cyclo-AMP | 95.4 | 12.3 |
| GTP | 79.9 | 10.6 |
| GDP | 96.3 | 12.5 |
| GMP | 82.6 | 12.4 |
| dGTP | 75.2 | 5.4 |
| CoA | 56.1 | 7.1 |
| FAD | 72.6 | 10.6 |
| NAD | 88.6 | 14.9 |
| NADP | 85.4 | 12.4 |
| APS | 87.4 | 12.9 |
Uptake of [α32P]-ATP by PAPST2 into liposomes loaded with 20 mM ATP was measured at a substrate concentration of 50 µM. Unlabeled effectors were applied in 10-fold excess, and transport was allowed for 10 min. Rates of nucleotide uptake represent net values (minus control: liposomes containing solely buffer) and are given as percentage compared with nonaffected transport (none; set to 100%). Data are the mean of at least three independent experiments; standard errors (SE) are given.
Figure 2.
PAPST2 and PAPST1 Mediate Uptake of [α32P]ATP into Differently Loaded Liposomes.
Import of 50 µM [α32P]-ATP into PAPST2 (dark gray bars) or PAPST1 (light gray bars; data from Gigolashvili et al., 2012) liposomes loaded with 5 mM of the indicated substrates or into liposomes containing APS. Uptake was stopped at 2.5 min. The data represent net values (minus control values, nonloaded proteoliposomes). Homo-exchange (ATP/ATP transport) was set to 100% and transport into the remaining liposomes was calculated accordingly. Data are the mean of three independent experiments; standard errors are displayed. Asterisks indicate significant difference (Student´s two-tailed t test, P < 0.01).
Transport studies with rising concentrations of labeled substrates revealed relatively high affinity of PAPST2 for ATP (∼86 µM), whereas ADP and PAPS apparently represent low affinity substrates (∼255 µM and ∼334 µM, respectively; Supplemental Table 1). ATP, however, is transported with lower maximal velocity (∼1100 nmol mg protein−1 h−1) than ADP (∼2600 nmol mg protein−1 h−1) or PAPS (∼5000 nmol mg protein−1 h−1). Analyses of binding affinity implied that even moderate PAP concentrations (Ki ∼176 µM) can compete with the high-affinity import substrate ATP. These data suggest that PAPST2 may preferentially mediate the antiport of PAP and ATP. However, PAPS and ADP might compete with these two substrates, depending on their actual concentrations.
PAPST2 Targets GFP to Chloroplasts and Mitochondria
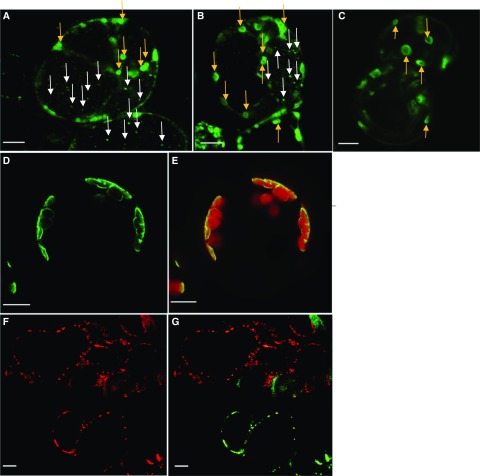
Plant MCF proteins often possess an N-terminal extension that supports proper subcellular targeting. The amino acid sequence of PAPST2 is also N-terminally extended, and bioinformatic predictions, as analyzed using seven different bioinformatic tools, pointed to the localization of PAPST2 in chloroplasts and mitochondria (Supplemental Table 2; Schwacke et al., 2003, 2007). Proteomic analyses have also identified PAPST2 in the chloroplast envelope fraction (Ferro et al., 2003; Kleffmann et al., 2004), as well as in the inner mitochondrial membrane (Millar and Heazlewood, 2003; Supplemental Table 2). We used C-terminal GFP fusion constructs to investigate the subcellular localization of PAPST2. The full-length coding sequence of PAPST2 driven by the Cauliflower mosaic virus (CaMV) 35S promoter was expressed in Arabidopsis root suspension cell cultures (Figures 3A and 3B) and in mesophyll protoplasts (Figures 3D and 3E). While in Arabidopsis mesophyll protoplasts, GFP fluorescence was detectable in chloroplasts only (Figures 3D and 3E); in cultured root cells, it appeared that the fusion protein was targeted to both plastids and mitochondria (Figures 3A and 3B). The plastid localization pattern of PAPST2 in cultured cells highly resembles that of triosephosphate/phosphate translocator the positive control for plastid localization (Figure 3C; Fischer et al., 1994). The mitochondrial localization of PAPST2-GFP (Supplemental Table 2) is further supported by the observation that pPAPST2-PAPST2-GFP (PAPST2 expression driven by its own promoter) co-localized with the mitochondrial marker protein Heat Shock Protein 90-mCHERRY/RFP (mCHERRY-tag of Red Fluorescent Protein) in Arabidopsis suspension cells derived from mesophyll but grown in darkness (Figures 3F and 3G). Thus, PAPST2-GFP appeared to mark plastids and mitochondria, with plastids being better visualized in photosynthetically active mesophyll cells (Figures 3D and 3E) and with mitochondria being preferentially labeled in root suspension cells (Figures 3A and 3B) and cells grown in darkness (Figures 3F and 3G). These findings imply that PAPST2 is differentially targeted and thus may fulfill different functions in photosynthetic and nonphotosynthetic tissues. In the chloroplasts of mesophyll cells, PAPST2 might exchange PAP with stromal PAPS; whereas in mitochondria and heterotrophic plastids of root cells, PAPST2 might instead use organellar ATP or ADP as an export substrate to drive PAP uptake. Alternatively, it is also plausible that we failed to detect the dual localization of PAPST2-GFP in photosynthetic tissue due to the known limitations of expression and detection systems in general (Yin et al., 2007). Taken together, bioinformatic predictions, proteome data, and GFP-targeting studies suggest that PAPST2 is present in the plastid envelope as well as the mitochondrial membrane.
Figure 3.
Subcellular Localization of PAPST2 Observed by Confocal Fluorescence Microscopy.
(A) and (B) Transient expression of the PAPST2-GFP fusion protein under the control of the 35S CaMV promoter in Arabidopsis suspension cells derived from roots (Berger et al., 2007). (A) and (B) show two independent experiments performed using the same cell culture. The green fluorescence labels plastids (green dots— indicated with yellow arrows) and mitochondria (tiny dots—indicated with white arrows).
(C) Localization of triosephosphate/phosphate translocator -GFP—a positive control for plastids (Gigolashvili et al., 2009, 2012).
(D) and (E) PAPST2-GFP under the control of the 35S CaMV promoter in Arabidopsis mesophyll protoplasts; the green fluorescence surrounds the red autofluorescence of the chloroplasts.
(F) and (G) Co-localization of mitochondrial HSP90 protein fused to mCHERRY with the PAPST2-GFP fusion protein driven by the PAPST2 promoter in Arabidopsis dark-grown suspension cells derived from mesophyll cells. (F) Red fluorescence of the mitochondrial HSP90-mCHERRY. (G) Co-expression of PAPST2-GFP driven by the PAPST2 promoter (green) with the mitochondrial HSP90-mCHERRY (red). Cells co-expressing both constructs are shown in yellow. Red, green, and yellow dots label mitochondria.
(A) to (G) Bars = 10 µm.
Absence of PAPST2 Affects Plant Growth
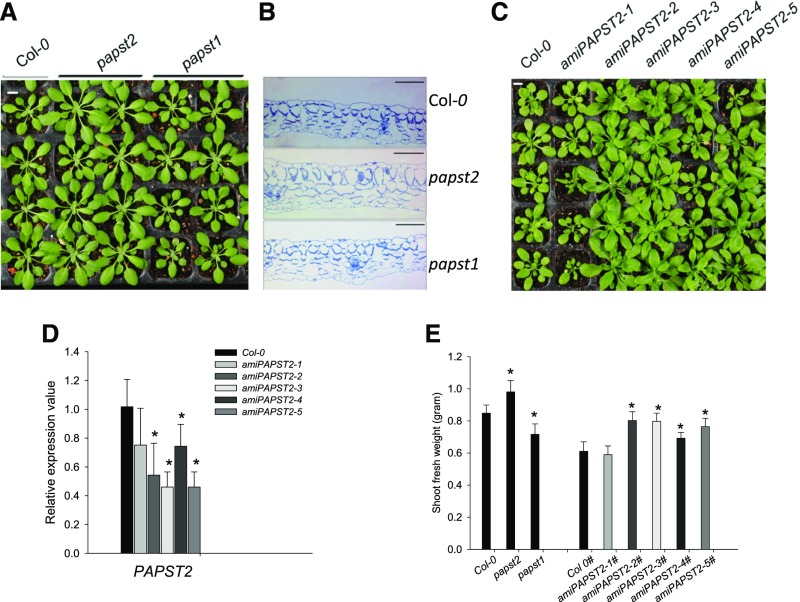
We analyzed three putative homozygous mutant lines (SALK_098190, SALK_012760, and SALK_009348) with a T-DNA insertion in the PAPST2 (At3g51870) gene as described in Methods. Only one out of three lines (SALK_009348), hereafter referred to as papst2, was used for most of the experiments presented in this work (Supplemental Figure 1A). The papst2 plants grew larger than Col-0 and papst1 plants, as shown by the analysis of plant fresh weight (Figures 4A and 4E) and dry weight (Supplemental Figure 1B). This phenotype was observed under different growth conditions, especially under short day cycles (8 h light/16 h dark). To further substantiate this observation, we made anatomical cross-sections of papst2 rosette leaves and compared the cell sizes of papst2 with the wild type and papst1 mutant plants. In agreement with the increased leaf size, the sizes of both palisade and spongy cells were also increased in papst2 mutant plants (Figure 4B), especially spongy parenchyma cells, which appeared to double in size in papst2 compared with the wild type.
Figure 4.
Phenotypes of the papst2 T-DNA Insertion Mutant and amiRNA Lines.
(A) The papst2 T-DNA insertion line shows larger rosettes than the wild type (Col-0) and papst1 mutant. Bar = 1 cm.
(B) Anatomical cross-sections of rosette leaves of papst2 compared with the wild type (Col-0) and papst1 showing increased cell size in the papst2 mutant. Bar = 100 µm.
(C) Phenotype of amiPAPST2 plants. The larger growth phenotype depends on the reduction in PAPST2 transcript level. The amiRNA lines 1 and 4 resemble the wild type. Plants with highest reduction in PAPST2 transcript levels (lines 2, 3, and 5) exhibit larger rosettes than the wild type. Bar = 1 cm.
(D) Determination of the PAPST2 transcript level in 5-week-old amiRNA lines by quantitative RT-PCR. Relative gene expression values are normalized to the wild type (set to 1). Data show means ±sd, (n = 3). Asterisks indicate significant differences compared with the wild-type Col-0 (Student´s t test, P < 0.05). Both papst2 and amiPAPST2 plants were grown in soil under short day conditions for approximately four weeks.
(E) Shoot fresh weight of papst2 knockout and amiPAPST2 plants. Plants were grown for 5 weeks in soil under a short day light cycle in a controlled environmental chamber. Data show means ±sd (n = 9). The amiPAPST2 plants were cultivated in different growth chambers (marked with #) from papst2 and had their own Col-0 control. Asterisks indicate significant differences compared with the respective wild types (Student´s t test, P < 0.05).
To obtain additional plant lines with reduced PAPST2 transcript levels, we made use of artificial microRNA (amiRNA) technology. We generated five amiRNA lines showing different degrees of reduction in PAPST2 levels (Figure 4C). Interestingly, all amiRNA lines (with 30% to 50% PAPST2 messenger RNA levels relative to the wild type) also grew better than the wild type, as shown by the analysis of plant fresh and dry weights (Figures 4C to 4E; Supplemental Figure 1B). Only one artificial microPAPST1-1 line out of five did not show improved plant growth (Figures 4C to 4E). This line (amiPAPST1-1) had only marginally (statistically not significant) reduced levels of PAPST2 expression (Figures 4D to 4E). Notably, the expression of the homologous PAPST1 gene was not affected by the presence of the amiPAPST2 construct (Supplemental Figure 1C). Thus, both knock-out and amiRNA PAPST2 lines showed improved growth compared with both the wild-type and papst1. It can be speculated that this phenotype is caused by moderately increased cytosolic PAP levels and the modulation of organelle-to-nuclear signaling.
PAPST1 Cannot Complement the papst2 Mutant Phenotype
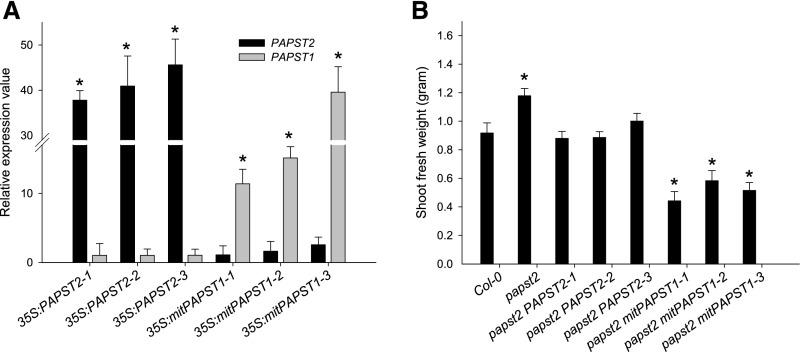
To address the question of whether the papst2 phenotype could be reversed by overexpression of PAPST2 or PAPST1, we performed complementation experiments by overexpressing these genes (Figure 5; Supplemental Figure 2). Previous studies (Mugford et al., 2009, 2010) showed that reducing PAPS levels in chloroplasts can have dramatic effects on plant growth or can be lethal. To avoid the manipulation of PAPS transport in chloroplasts by PAPST1, we used a mitochondrial targeting peptide to localize PAPST1 to mitochondria. PAPST1 localization to mitochondria was expected to affect PAP transport in these organelles. As shown in Figure 5A, high expression levels of PAPST2 and mitochondrialPAPST1 (mitPAPST1), respectively, were detected in the papst2 background. While PAPST2 overexpression converted the papst2 mutant phenotype to the wild-type growth (Figure 5B; Supplemental Figure 2), the overexpression of mitPAPST1 in papst2 led to strong growth retardation, bleaching of leaf blades (Figure 5B; Supplemental Figure 2), and even plant death. This phenotype is likely caused by the depletion of PAPS in the cytosol for the following reasons: (1) Transport assays demonstrated that PAPST1 prefers PAPS to ATP or PAP (Figures 1 and 2; Supplemental Table 1) and therefore, PAPS can be imported into the mitochondria. (2) PAPS in the mitochondria will be degraded by SAL1, as SAL1 is known to equally use both PAPS and PAP as substrates (Gläser et al., 1993; Murguia et al., 1995; Quintero et al., 1996). (3) As a result of PAPS degradation in the mitochondria, phytosulfokines 2 and 4 (PSK2 and PSK4) were significantly upregulated in mitPAPST1 lines (Supplemental Figure 3). PSK2 and PSK4 encode small secreted peptides that require sulfation via PAPS. In addition to PSKs, PSY and RGFs also encode small secreted peptides with hormone-like activity. These sulfated molecules regulate plant growth, development, and differentiation in a complex manner (Matsubayashi et al., 1999; Yang et al., 1999; Amano et al., 2007; Kutschmar et al., 2009; Matsuzaki et al., 2010; Matsubayashi, 2012). Together, these experiments demonstrate that PAPST1 and PAPST2 play distinct roles and that PAPST1 preferably serves as a PAPS transporter.
Figure 5.
Complementation of papst2 by PAPST1 and PAPST2.
(A) Expression of PAPST2 and PAPST1 in papst2 lines complemented by 35S:PAPST2 or 35S:mitPAPST1. PAPST2 and PAPST1 transcript levels in rosette leaves of 5-week-old mutant plants were determined by quantitative RT-PCR. Relative gene expression values are given compared with the wild type Col-0 = 1. Data show means ±sd (n = 3).
(B) Shoot fresh weight of the wild type, papst2, and complemented transgenic plants overexpressing PAPST2 and mitPAPST1. Plants were grown for 5 weeks in soil under a short day light period in a controlled environmental chamber. Data show means ±sd (n = 7).
Asterisks in (A) and (B) indicate significant differences compared with the wild-type Col-0 (Student´s t test, P < 0.05).
Glucosinolate Biosynthesis and Sulfate Reduction Pathway Are Only Marginally Affected in papst2 Plants
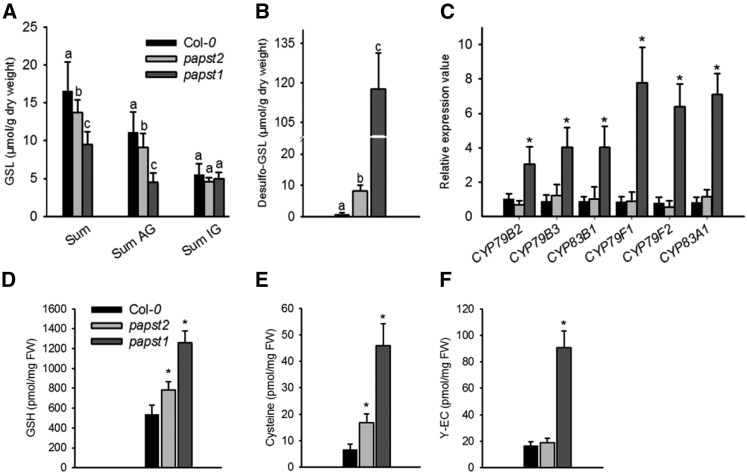
Glucosinolates represent a class of secondary metabolites that contribute to plant defense against pests and diseases. These sulfated organic compounds are derived from Glc and an amino acid and occur in almost all members of the order Brassicales, including Arabidopsis. Glucosinolate biosynthesis takes place in the cytosol, and the final step, which is catalyzed by sulfotransferases, requires PAPS as an activated sulfate donor (Mugford et al., 2009). PAPS generation and hence its cytosolic availability have a great impact on glucosinolate biosynthesis (Mugford et al., 2009). Moreover, several studies have revealed that impaired delivery of PAPS from the chloroplast to the cytosol decreases the production of sulfated glucosinolates and increases the accumulation of desulfo-precursors (Mugford et al., 2009, 2010; Gigolashvili et al., 2012).
To clarify whether PAPST2, like PAPST1, might be involved in plastidial PAPS export, we tested its impact on glucosinolate biosynthesis. Reduced PAPST2 expression (in the three strongest amiRNA lines) had no measurable effect on cellular glucosinolate levels (Supplemental Figure 4), and even a complete absence of the transcript (in the papst2 T-DNA insertion line) only led to a slight reduction in these levels (Figures 6A and 6B). Moreover, glucosinolate biosynthesis genes were upregulated only in papst1, but not in papst2 (Figure 6C).
Figure 6.
Glucosinolate Biosynthesis and Sulfate Assimilation Are Slightly Affected in papst2.
(A) Glucosinolate contents (µmol/mg dry weight) in papst2 plants relative to wild-type plants (Col-0) and the papst1 T-DNA insertion line.
(B) Desulfo-glucosinolates (µmol/mg dry weight) contents in papst2 compared with wild-type plants (Col-0) and the papst1 T-DNA insertion line.
AG = aliphatic glucosinolates; GSL = glucosinolates; IG = indolic glucosinolates.
The data are the sums of three independent biological replicates (independently grown plant trays), with GSLs isolated from six individual plants from each. Statistical analysis performed with ANOVA, Tukey’s test (Supplemental Data Set). Different letters indicate significant differences at P < 0.05.
(C) Glucosinolate biosynthesis gene expression is affected in papst1 but not in the papst2 T-DNA insertion line.
Relative gene expression values are given compared with the wild type Col-0 = 1. Data show means ±sd, (n = 3). Asterisks indicate significant differences compared with the wild-type Col-0 (Student´s t test, P < 0.05).
(D-F) Glutathione = GSH (D), Cys (E), and γ-glutamylcysteine or γ-EC (F) levels were determined in 4-week-olds as described in Methods and were compared with those of papst1. The data are the sums of two independent experiments, with five biological replicates. Asterisk indicates significant difference compared with the wild type (Col-0; Student’s t test, P < 0.05). FW = fresh weight.
However, reduced or absent PAPST2 expression led to ameasurable accumulation of desulfo-glucosinolates (Figure 6B; Supplemental Figure 4). Although papst2 exhibited a higher cellular desulfo-glucosinolate level than the three amiRNA lines, the concentration was still more than ten times lower than that of papst1 plants (Figure 6B; Supplemental Figure 4). The moderate increase in desulfo-precursor levels suggests that sulfotransferase activity is at least slightly affected in papst2. This may result from a reduced cofactor supply or the inhibition of sulfotransferase enzyme activity due to elevated PAP levels.
Interestingly, in the papst2 T-DNA insertion line, diverse sulfotransferase genes were strongly upregulated (Supplemental Figure 5), with transcript levels often exceeding those of the papst1 lines. Only AtST5a/SOT16, which is known to be involved in glucosinolate biosynthesis, showed higher expression levels in papst1 than in the papst2 mutants. This observation suggests that the increased abundance of sulfotransferase (due to enhanced gene expression), particularly in papst2 mutants, might help counteract the limited sulfotransferase enzyme activity in this line. This observation also supports the idea that PAPST1 rather than PAPST2 is associated with glucosinolate biosynthesis. In agreement with this notion, it was previously shown that the promoter of PAPST1, but not PAPST2, was activated “in trans” by myeloblastosis transcription factors when expressed in Arabidopsis cultured cells (Gigolashvili et al., 2012).
To address the possible role of PAPST2 in sulfur metabolism, we analyzed the reduction of sulfate. Impaired plastidial PAPS biosynthesis (via APKs) or transport (via PAPST1) stimulates the entry of APS into the reduction pathway (Mugford et al., 2009; Gigolashvili et al., 2012). Quantification of selected thiol-containing molecules demonstrated that papst2 contains higher Cys and glutathione levels than the wild type (Figures 6D to 6F). However, much higher values were measured in papst1, which also exhibited increased levels of the glutathione precursor γ-glutamylcysteine. These observations indicate that the absence of PAPST2 leads to the re-allocation of sulfate flux into the reduction pathway, but to a far lesser extent than the absence of PAPST1. Therefore, PAPST1 can be considered to be the main plastidial PAPS exporter, whereas PAPST2 is apparently of minor importance in this respect.
Loss of PAPST2 but Not of PAPST1 Increases PAP Levels in the fry1 Mutant
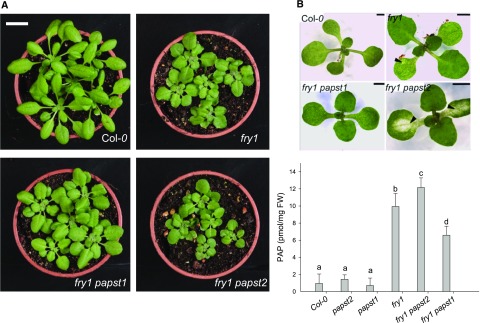
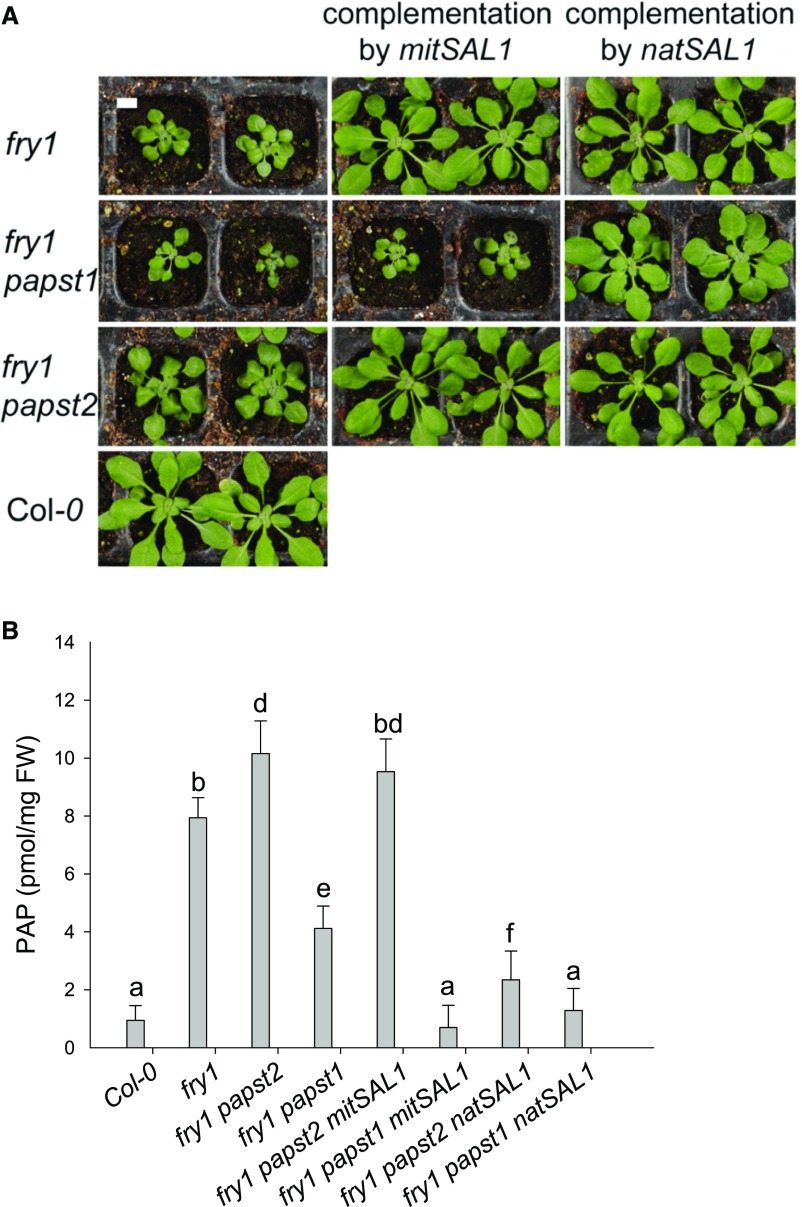
To further address the roles of PAPSTs in organellar PAP import, we generated double fry1 papst1 and fry1 papst2 mutants. Assuming that both transporters are capable of transporting PAP and that a defect in SAL1 may exacerbate the levels of PAP, we expected to detect an increase or decrease in the accumulation of PAP in the double mutants. Interestingly, in fry1 papst1 the strong morphological phenotype of fry1 was partly complemented (Figures 7A and 7B). This observation is in line with reduced PAP levels compared with fry1 (Figure 7C), which results from decreased PAPS supply in the absence of PAPST1. On the other hand, fry1 papst2 exhibited a stronger phenotype than fry1 (Figures 7A and 7B), which is in accordance with its increased PAP levels compared with fry1. PAP levels were significantly higher in fry1 papst2 seedlings than in all other mutants analyzed (Figure 7C). Interestingly, PAP levels were even slightly higher in fry1 papst2 than in fry1, supporting the notion that additional components beyond PAPST2 and SAL1 help regulate total PAP levels in the cell. These components include yet-unknown PAP transporters, PAP nucleotidases, and homologs of SAL1 or other PAP signaling and/or regulatory proteins and mechanisms. In addition, fry1 papst2 plants showed bleaching of cotyledons during the early stages of development (Figure 7B; Supplemental Table 3) and accumulated anthocyanins (Supplemental Table 3; Supplemental Figure 6), which could be related to the upregulation of stress-responsive genes. Bleaching of cotyledons was not observed in fry1 (Supplemental Table 3), and the chlorophyll content was not affected in adult fry1 papst2 plants compared with fry1 plants. Moreover and in contrast to fry1, the fry1 papst2 plants often failed to produce seeds, perhaps due to even stronger inhibition of sulfotransferase activity in fry1 papst2. The worsening of the fry1 phenotype in the fry1 papst2 mutant appears to be related to the reduced import of PAP into organelles. Altogether, these data indicate that cellular PAP levels are regulated by SAL1, PAPST2, and PAPST1 transporters. Due to their activities, plants with two different concentration ranges of PAP with different effects on plant growth can be generated. While a moderate increase in PAP levels (as in papst2 and the amiRNA lines) could stimulate plant growth, a strong increase in PAP levels would have the opposite effect (as observed in fry1 and fry1 papst2).
Figure 7.
The fry1 Phenotype Is Alleviated by the Absence of PAPST1 and Worsened by the Absence of PAPST2.
Phenotypes of fry1 papst1 and the fry1 papst2 double mutant lines compared with the wild-type plants and the fry1 mutant. Plants were grown in soil under short-day conditions.
(A) Adult plants (age: 5 weeks). Bar = 2 cm.
(B) Young seedlings (age: 1 week). Arrows in fry1 papst2 indicate bleaching along the veins of cotyledons. Note fry1 has only slightly more pale areas in cotyledons than the wild-type (Col-0). Bar = 2 mm.
(C) PAP levels in mutants with impaired PAPS and PAP transport. PAP levels in papst2, papst1, fry1 papst2, and fry1 papst1 mutants compared with those of the wild type and fry1. Results represent the means of two independent cultivations and eight biological replicates; and results were analyzed via ANOVA and Tukey’s test (Supplemental Data Set). Error bars indicate the ±sd of the mean. Different letters indicate significant differences at P < 0.05. Plants were grown in soil under short-day conditions for five weeks. FW = fresh weight.
Toward an Understanding of the Physiological Roles of PAPST2 and PAPST1 in Plastidial PAP Uptake
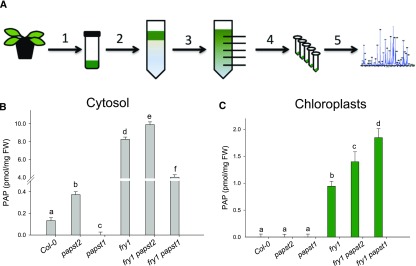
To dissect the contribution of PAPST2, PAPST1, and SAL1 to organellar PAP uptake and metabolism, we performed non-aqueous fractionation (NAF; Gerhardt and Heldt, 1984; Krueger et al., 2011) and analyzed PAP levels in the chloroplast and cytosol fractions of the respective mutant plants. While the total PAP content in crude extracts of papst1 and papst2 show no substantial differences compared with the wild type (Figure 7C), the picture was markedly different for cytosolic PAP levels: papst1 exhibited lower and papst2 higher cytosolic PAP levels than the wild type (Figure 8). The reduced cytosolic PAP levels of papst1 plants might result from reduced delivery of PAPS to the cytosol, and, as a consequence, reduced PAP biosynthesis in the cytosol. Moreover, in the absence of PAPST1 (i.e., in papst1), it can be assumed that PAPST2 stimulates PAP uptake into organelles (both chloroplasts and mitochondria). By contrast, the increased cytosolic PAP concentration in papst2 plants (Figure 8) is indicative of reduced PAPST2-mediated PAP uptake into the organelles.
Figure 8.
Concentrations of PAP in the Cytosol and Chloroplasts of papst1, papst2, fry1, fry1 papst1, and fry1 papst2 Mutants Compared with the Wild Type, as Detected by NAF.
Workflow for NAF of plant extracts.
(A) PAP concentrations in the cytosol (B) and chloroplasts (C) of papst1, papst2, fry1, fry1 papst1, and fry1 papst2 calculated based on the relative subcellular distribution of PAP and the respective absolute values. Data show means from three pools of plants (6 g fresh weight each) grown in two independent experiments. Error bars indicate the SD of the mean. Different letters indicate significant differences among means based on t tests at P < 0.05. FW = fresh weight; PAP, 3′-phosphoadenosine 5′-phosphate.
In line with the expected role of SAL1 in PAP degradation, the fry1 mutant had elevated PAP levels in the chloroplasts and cytosol (Figure 8). However, when PAPST1 was also absent (i.e., in fry1 papst1), the cytosolic PAP content was significantly reduced (Figure 8B), which is in agreement with the phenotypic complementation of this mutant (Figure 7). This decrease in PAP levels can be explained by both the effective removal of PAP from the cytosol by PAPST2 and by a decrease in PAPS export from chloroplasts—and thus a decrease in PAP formation in the absence of PAPST1, as reported previously (Gigolashvili et al., 2012). Furthermore, the reduced cytosolic PAP levels of fry1 papst1 versus fry1 were accompanied by significant increases in PAP levels in chloroplasts, likely due to increased PAPST2 activity (Figure 8C).
As shown in Figure 7, the fry1 phenotype was exacerbated by the absence of PAPST2 (i.e., in fry1 papst2). This phenotypic worsening appears to be related to increased accumulation of PAP, especially in the cytosol (Figure 8). The cytosolic PAP levels were significantly higher in these plants than in fry1, pointing to the inability of PAPST2 to deliver PAP from the cytosol to organelles. On the contrary, it appears that PAPST1 is able to import PAP into chloroplasts in the absence of PAPST2, as observed in fry1 papst2 (Figure 8). However, PAP levels were significantly increased in chloroplasts of fry1 papst2 versus fry1, because PAP could not be metabolized in chloroplasts in the absence of SAL1.
Altogether, these observations corroborate the notion that PAPST2 plays a greater role than PAPST1 in removing PAP from the cytosol. In the absence of PAPST2, the PAP accumulation-related phenotype of the fry1 mutant was further exacerbated. Furthermore, the data suggest that the worsening of the fry1 phenotype in fry1 papst2 is likely also caused by increased PAP accumulation in the cytosol. Altogether, these experiments showed that PAPST2 plays a pivotal role in the removal of PAP from the cytosol and that PAPST1 plays a dominant role in facilitating PAPS export from plastids.
SAL1 Expressed in Mitochondria Fails to Complement the fry1 Phenotype of the fry1 papst2 Mutant
To clarify the physiological relevance of mitochondrial PAP uptake and degradation, we analyzed whether the growth defects of fry1 papst1 and fry1 papst2 could be complemented by dually targeted SAL1 or by SAL1 targeted exclusively to mitochondria.
In fry1 papst2, the expression of the dually targeted SAL1 led to normal plant growth and substantially reduced the cellular PAP concentration to approximately the wild-type levels (Figures 9A and 9B). Although PAP could not be imported into the mitochondria for degradation, it was sufficiently detoxified by plastidial SAL1 following import via PAPST1 (Figures 9 and 10). By contrast, when SAL1 was only present in the mitochondria of fry1 papst2 plants, PAP levels remained high, and the corresponding mutant plants were still small. Therefore, mitochondrial PAP degradation is apparently impossible in the absence of PAPST2. We conclude that PAPST2 is the only transporter that efficiently supplies the mitochondrial SAL1 enzyme with PAP (Figure 10).
Figure 9.
FRY1 Exclusively Targeted to Mitochondria Cannot Complement the fry1 papst2 Mutant Phenotype.
The fry1 papst2 and fry1 papst1 mutants express either mitochondrial (mitSAL1) or dually targeted FRY1 (natSAL1). Plants were grown in soil under short-day conditions for four weeks before analysis.
(A). Plant phenotypes. Bar = 1 cm.
(B). PAP concentrations. The data are the sums of two independent experiments, with five biological replicates each. They were analyzed via ANOVA and Tukey’s test (Supplemental Data Set). Error bars indicate the sd of the mean. Different letters indicate significant differences at P < 0.05. FW = fresh weight.
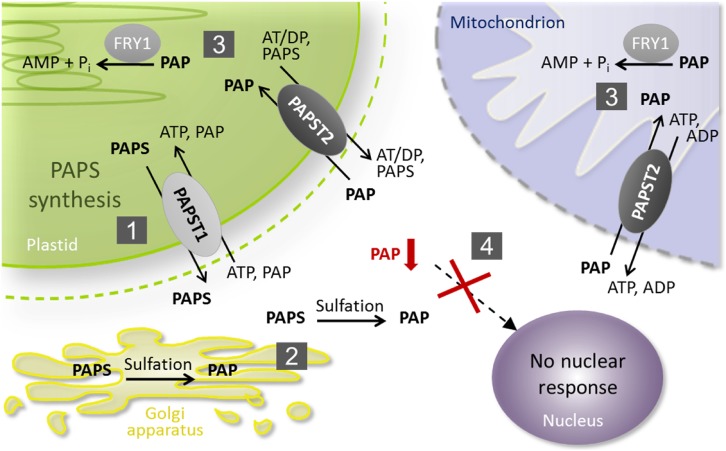
Figure 10.
Schematic Illustration of PAP Metabolism and Transport.
(1) PAPST1 is the major transporter that delivers newly synthesized PAPS from the plastid to the cytosol.
(2) In the cytosol and the Golgi apparatus, PAPS consumption via sulfotransferases results in PAP release.
(3) PAPST2 is the major transporter that imports cytosolic PAP into mitochondria and plastids, where it becomes degraded via FRY1.
(4) The concerted interaction between PAP production, transport, and degradation prevents cytosolic PAP accumulation (marked in red), the associated nuclear responses, and phenotypic symptoms.
Interestingly, the mutant lines fry1 (positive control) and fry1 papst1 showed no obvious growth impairment and had wild-type appearances when expressing either native or mitochondrial SAL1. Moreover, the expression of both native and mitochondrial SAL1 reduced PAP accumulation in fry1 papst1 to the wild-type levels. These observations suggest that SAL1 in mitochondria is sufficient to detoxify the PAP produced under normal conditions and to prevent dwarfism. These findings confirm the idea that PAPST2 is a key component in the PAP degradation pathway, particularly mitochondrial PAP uptake (Figure 10).
DISCUSSION
PAPST1 and its closest homolog PAPST2 are members of the MCF and form a distinct subgroup among the clade of adenine nucleotide transporters. These two transporters are related to MCF proteins in plants whose functions have not been characterized and to putative CoA and PAP transporters in animals and yeast (Kaundal et al., 2010; Palmieri et al., 2011). Initially, PAPST1 was thought to act as an ATP/ADP exchanger in the thylakoid membrane (Thuswaldner et al., 2007). However, more recently, PAPST1 was shown to also accept PAPS and PAP as substrates and to be present in the inner plastid envelope (Figure 10; Gigolashvili et al., 2012). Analyses of papst1 loss-of-function mutants revealed that PAPST1 fulfills an important function in the provision of newly synthesized PAPS from the plastid to the cytosol. Interestingly, the absence of PAPST1 lowers but apparently does not totally block plastidial PAPS export, which argues for the existence of additional PAPS transport activity in the inner plastid envelope. Due to its close relationship with PAPST1, we hypothesize that PAPST2 fulfills this function. In fact, the dual localization of PAPST2 also includes the chloroplast (Figure 3).
Biochemical characterization revealed that PAPST2 shows approximately nine times lower affinity for PAPS (∼340 µM; Supplemental Table 1) than PAPST1 (∼40 µM; Gigolashvili et al., 2012). Transport studies also revealed relatively high affinity of PAPST2 for ATP. However, ATP was transported with lower maximal velocity than ADP or PAPS. Furthermore, analysis of binding affinities indicated that PAP can successfully compete with the high-affinity import substrate ATP. Altogether, these data suggest that PAPST2 may preferentially mediate PAP/ATP antiport, but PAPS and ADP might compete with these two substrates, depending on their actual concentrations in the respective plant compartments. Altogether, the biochemical data reveal that the differences in the uptake and kinetics of potential substrates are not very striking in vitro. That is why we performed careful comparative analysis of papst2 and papst1 Arabidopsis mutants and measured the levels of potential substrates in different plant compartments for our study.
While the absence of PAPST1 significantly reduced the biosynthesis of secondary sulfated compounds and stimulated the allocation of sulfur into primary reductive sulfate assimilation, the absence of PAPST2 had a comparatively low impact on sulfur metabolism (Figure 6). These observations indicate that PAPST1 (in the papst2 mutant) alone is able to deliver sufficient PAPS to the cytosol and that this protein represents the main plastidial PAPS exporter. By contrast, PAPST2 cannot compensate for the absence of PAPS export activity in the papst1 mutant and thus might fulfill an accessory function in this context (Figure 6). This assumption is further supported by the strong upregulation of glucosinolate biosynthesis gene expression in papst1 mutants, which was not detectable in papst2 plants (Figure 6C). Interestingly, targeting PAPST1 to the mitochondria failed to complement the papst2 phenotype but caused the worsening of plant growth and strong bleaching of leaves (Figure 5; Supplemental Figure 2). We believe that the high affinity of PAPST1 for PAPS causes PAPS sequestration in the mitochondria, leading to depletion of its cytosolic pool and the bleaching phenotype. Accordingly, the levels of sulfated hormone-like peptides also appeared to be reduced in the papst2 mutants, as PSK precursor genes were strongly upregulated in these plants (Supplemental Figure 3).
The moderate decline in PAPS levels was previously shown to cause the reduced growth of papst1 (Gigolashvili et al., 2012) and apk1 apk2 (Mugford et al., 2009). Further declines in cytosolic PAPS levels are thought to cause growth retardation in multiple apk mutants (Mugford et al., 2010), as well as growth decline and bleaching in mitPAPST1 papst2 plants. The inability of PAPST1 to rescue the papst2 mutant phenotype provided additional evidence for the distinct roles of PAPST1 and PAPST2. This failure demonstrated that PAPST1 preferentially acts as a PAPS transporter while PAPST2 functions as a PAP importer.
Indeed, PAPST1 is less important for PAP import than PAPST2, as neither papst1 nor fry1 papst1 accumulated higher PAP levels than the respective control plants (Col-0 and fry1) (Figures 7 and 8). On the other hand, both papst2 and fry1 papst2 accumulated higher PAP levels, demonstrating the major role of PAPST2 in PAP transport. The finding that PAP levels in crude extracts of fry1 papst2 were even slightly higher than in fry1 alone appeared to be counterintuitive. On one hand, this finding further substantiates the important role of PAPST2 in PAP import, but on the other hand it also indicates that more components beyond the enzymatic activities of FRY1/SAL1 might also contribute to PAP status in the cell. These components could be less well-studied PAP catabolizing enzymes and SAL1/FRY1 homologs and/or yet unknown Golgi-localized PAP transporters or sulfotransferases. The Golgi-localized sulfotransferase TPST (Komori et al., 2009) is a central player in the sulfation of hormone-like peptides; however, as the phenotype of this mutant is not as dramatic as that of the APK mutants (Mugford et al., 2010), additional proteins with sulfotransferase activity are expected to be found in the cell. Furthermore, the modulation of PAP by regulatory or signaling proteins via redox-mediated structural and biochemical modifications of SAL1/FRY1 (Chan et al., 2016) is an emerging topic of study in plant science. All of these components could also contribute to PAP accumulation in the cell; we hope the ways in which this is achieved will attract more attention in the future.
In chloroplasts, both PAPST1 and PAPST2 are potentially capable of importing PAP in exchange for PAPS. The moderate affinity of PAPST2 for PAPS, however, indicates that ATP and ADP also represent suitable export substrates that may be preferred, particularly under conditions of reduced PAPS availability in the chloroplast. The latter appears to be the case in fry1, which accumulates not only high PAP levels, but also high ATP levels in chloroplasts (Figure 8; Supplemental Figure 7). Increased PAP levels in chloroplasts restrict further import of PAP against the concentration gradient and stimulate ATP import in exchange for PAPS, allowing PAPS to leave the chloroplasts.
The situation is different in mitochondria. Because there are no indications that substantial amounts of PAPS exist in the mitochondrial matrix, PAP uptake into this organelle is most likely exclusively driven by adenine nucleotides. Moreover, the higher apparent affinity of PAPST2 for ATP than for PAP may suppress the export of previously imported PAP, particularly in energized mitochondria. The low affinity of PAPST2 for PAPS also reduces the chances for competition of cytosolic PAPS with PAP during import. Therefore, PAPST2 appears to be well suited to channel cytosolic PAP into degradation (Figure 10).
Elevated PAP levels are known to inhibit sulfotransferase activity. The increased expression of sulfotransferases might help counteract their inhibition by PAP. Therefore, the expression patterns of sulfotransferase genes (Supplemental Figure 5) detected in the study are in line with the finding that papst2 exhibited higher cytosolic PAP levels than papst1 (Figure 8).
Cellular PAP levels can be regulated by SAL1, PAPST2, and PAPST1 transporters. It appears that depending on the mode of regulation, two different concentration ranges of PAP activate different sets of target genes and affect plants differently. While a moderate increase in cytosolic PAP levels (physiological range) activates the expression of genes related to growth promotion, higher PAP levels induce the expression of stress-responsive genes and inhibit growth. In this study, we were confronted with both scenarios: a moderate increase in cytosolic PAP levels in papst2 or amiRNA lines, as well as strongly increased cytosolic PAP levels in fry1 and fry1 papst2 mutants (Figure 8). The upregulation of stress-responsive genes in response to high PAP concentrations impairs plant growth, as previously documented for sal1 (Estavillo et al., 2011). Notably, neither moderately increased PAP accumulation in the cytosol (and correspondingly improved plant growth) nor strong PAP accumulation and growth inhibition were observed in papst1.
The analysis of double mutants lacking SAL1 and either PAPST1 (fry1 papst1) or PAPST2 (fry1 papst2) further strengthened the assumption that the two transporters fulfill distinct and only partially overlapping physiological functions. The absence of PAPST1 attenuated the growth phenotypes of fry1, including PAP accumulation (Supplemental Figure 8). This observation is fully in line with the proposed dominant role of PAPST1 in plastidial PAPS export. In the fry1 papst1 mutant, decreased PAPS export is thought to substantially decrease cytosolic PAP concentrations (Figure 8; Supplemental Figure 9). The reduced availability of PAPS slows down sulfotransferase reactions and thus decreases the generation of PAP. Therefore, the reduced PAP formation can partially compensate for the defect in PAP degradation in the fry1 papst1 mutant.
Various data indicate that PAPST1 and PAPST2 are expressed in many plant organs and cell types (Supplemental Figure 10; Figures 3a and 3b in Winter et al., 2007; Gigolashvili et al., 2012). The highest activities of the corresponding promoters were detected in shoot and root meristems as well as vascular tissue (Supplemental Figure 11). Interestingly, the bundle sheaths represent the main sites of sulfur and glucosinolate metabolism in Arabidopsis (Aubry et al., 2014), and they also show considerable SAL1 promoter activity (Robles et al., 2010). Therefore, the expression patterns of PAPST1 and PAPST2 nicely overlap with the sites of PAPS consumption, PAP generation, and PAP degradation. The transcription factors MYB51 and MYB28, which represent the main regulators of glucosinolate biosynthesis, activate the promoter of PAPST1 but not PAPST2 (Gigolashvili et al., 2012). These observations suggest that PAPST1 rather than PAPST2 provides the cofactors for the corresponding sulfotransferase reactions during glucosinolate biosynthesis. Interestingly, transcriptomic studies have revealed elevated transcription of SAL1 in young seedlings as well as during inflorescence development (Zimmermann et al., 2005). In particular, at these stages, the fry1 papst2 mutant exhibited even more severe morphological symptoms than fry1 (Figure 7B; Supplemental Table 3; Supplemental Figure 6).
To obtain unequivocal proof for the role of PAPST2 in mitochondria, we complemented the fry1 papst2 mutant with SAL1 that was specifically targeted to mitochondria. As shown in Figure 9, mitSAL1 was unable to rescue the fry1 papst2 mutant. On the other hand, in plants containing dually targeted SAL1, a phenotype similar to that of the wild type was obtained, including substantially reduced cellular PAP levels (Figure 9B).
Thus, the cellular PAP concentration is not only tightly controlled by SAL1, but it also appears to be fine-tuned by PAPST2. Changes in cytosolic PAP levels can influence many biological processes, including sulfation reactions, RNA quality control, plant growth and development, drought and stress tolerance, and even jasmonate signaling (Wilson et al., 2009b; Rodríguez et al., 2010; Estavillo et al., 2011). Therefore, plastidial and mitochondrial PAP transport systems serve as important links not only in the PAP degradation pathway but also in PAP-dependent signaling (Figure 10). Here, we showed that PAPST2 represents an essential component in the regulation of cellular PAP concentration. PAPST2 functions as a PAP shuttle that mediates the import as well as export of PAP in exchange for ATP, ADP, or PAPS. The direction of transport is most likely determined by the cytosolic and organellar concentrations of the individual substrates. Because the velocity of transport is influenced by the abundance of the corresponding transporters, it is tempting to speculate that changes in PAPST1 and PAPST2 expression may regulate organellar entry and efflux of PAP in accordance with cellular demands.
In summary, PAPST2 represents an important component in the regulation of cellular PAP levels and is therefore an important component of cellular and organellar PAP signaling.
METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis thaliana ecotype Col-0, the corresponding T-DNA insertion lines, and transgenic plants were sown on soil or culture plates containing 1/2 Murashige and Skoog medium. The seeds were stratified for two to three days in the dark, and the plants were cultivated under short day (8 h light and 16 h dark) or long day (16 h light and 8 h dark) conditions with an average photon flux density of 100 to 150 µmol m–2 s–1. White light was provided by Fluora L58W/77 fluorescent tubes. The temperature was kept at 22°C during the light period and 18°C during the dark period. The relative humidity was ∼40%.
Identification of Homozygous T-DNA Insertion Mutants and Generation of fry1 papst1 and fry1 papst2 Double Mutants
Arabidopsis mutant plants carrying a T-DNA insertion in PAPST2 (At3g51870, SALK_098190, SALK_012760, and SALK_009348) or FRY1/SAL1 (At5g63980, SALK_020882, and SALK_005741) were obtained from the European Arabidopsis Stock Centre (NASC). For simplicity, we refer to only to one line (SALK_020882 line), called fry1. Homozygous mutant lines were identified using primers proposed by the SALK Institute (http://signal.salk.edu/cgi-bin/tdnaexpress) and gene-specific primers (Supplemental Table 4). The insertion position of the T-DNA in the target gene was confirmed by sequencing. The loss of PAPST2 transcripts could be confirmed by RT-PCR in SALK_009348 and SALK_020882 (Supplemental Table 5), whereas insertions in SALK_098190 and SALK_012760 (in 5′ untranslated region [UTR]) did not reduce gene expression (Supplemental Figure 1). The fry1 mutant isolated in this work is morphologically identical to the previously described fry1 alleles: altered APX2 expression 8 (Emanuelsson et al., 2000; Wilson et al., 2009a), high expression of osmotically responsive genes 2 (Xiong et al., 2004), fatty acid oxygenation upregulated 8 (Rodríguez et al., 2010), and fiery1 (Kim and von Arnim, 2009). The double mutants fry1 papst1 and fry1 papst2 were generated by crossing of the homozygous fry1 T-DNA insertion line (SALK_020882) with the papst1 (PAPST1 At5g01500) T-DNA insertion line (Gigolashvili et al., 2012) and the papst2 line, respectively. Segregating populations from all crosses were PCR genotyped for the parental loci. Harvested seeds from each cross were planted and screened at one segregating locus for the insertions in SAL1, PAPST1, and PAPST2 by PCR. Plants found to be homozygous at one locus were then screened with a marker for the second segregating locus.
Generation of PAPST2 amiRNA Lines
The artificial microRNA fragment 5′-UUUAAACUAGCACUCGAGCAC-3′ against PAPST2 was designed as recommended by Web MicroRNA designer (http://wmd.weigelworld.org) and inserted into the Gateway pDONR207 vector. The miR319a precursor was amplified from the RS300 plasmid kindly provided by D. Weigel (Max Planck Institute for Developmental Biology). Prior to recombination into the Gateway destination pGWB2 vector, correctness of the amiRNA insertion in pDONR207 was verified by sequencing. Transgenic Arabidopsis plants were generated by Agrobacterium-mediated transformation (Clough and Bent, 1998) with the destination vector, and positive transformants were selected with kanamycin (50 mg per mL).
RNA Extraction and qRT-PCR
To measure transcript levels in the wild type and the different mutant plants, total RNA was isolated from leaves using TRIzol reagent (Thermo Fisher). The cDNA was reversely transcribed using SuperScript II Reverse Transcriptase (Thermo Fisher) according to the manufacturer's instructions. The qRT-PCR was performed with a SYBR-Green Master kit (Thermo Fisher) and a GeneAmp 7300 sequence detection system (Thermo Fisher) as described previously (Gigolashvili et al., 2009). Expression levels were quantified with the comparative cycle threshold method and normalized to the wild-type expression level (set to 1). Means were calculated from at least two or three independent biological replicates (independently grown plant trays), and the different cDNAs synthesis reactions were performed from the RNA isolated from three different plants grown in each biological replicate (technical replicates). Figure legends indicate details specific for each experiment.
GFP-Based Localization Studies
The PAPST2 coding sequence and the PAPST2 full-length genomic fragment were amplified without the stop codon from cDNA and genomic DNA, respectively. The amplicons were inserted into the entry vector pDONOR207 (Thermo Fisher) via attP1 and attP2 sites and verified by sequencing. The obtained entry clone was recombined with the binary vectors pGWB5 or pGWB4 (Nakagawa et al., 2007) to allow expression of the GFP fusion construct under the control of the 35S cauliflower mosaic virus promoter (cDNA amplicon) or the PAPST2 promoter (genomic amplicon). To obtain a mitochondrial marker protein, the coding sequence of the mitochondrial chaperone HEAT SHOCK PROTEIN90 (HSP90.6, At3g07770) (Krishna and Gloor, 2001; Sangster and Queitsch, 2005; Prassinos et al., 2008) amplified from Arabidopsis cDNA was fused with mCHERRY in the pAubergine vector (M. Jakoby, GenBank ID: FR695418). Agrobacterium-mediated transformation of Arabidopsis suspension cell culture and transfection of Arabidopsis mesophyll protoplasts were performed as described previously (Gigolashvili et al., 2012) and details for the suspension cells are described in the following. The subcellular distribution of the marker proteins was analyzed by fluorescence microscopy (Eclipse E800; Nikon) with a filter required for the analysis of Green Fluorescent and Red Fluorescent proteins using confocal laser scanning microscopy (Zeiss). Results were documented with Discus and Zeiss software.
Arabidopsis Suspension Cell Culture: Cultivation and Transformation
To localize fluorescent proteins, Arabidopsis suspension cell cultures were used, allowing high-efficiency transfection (Koroleva et al., 2005; Berger et al., 2007). Arabidopsis cells derived from root or mesophyll cells were grown in the dark at 22°C with gentle shaking at 160 rpm. The cells were inoculated weekly at a 1:5 dilution into fresh medium (4.3 g/L MS basal salts (Duchefa); 4 mL/L Gamborg’s vitamin solution (Sigma); 1 mg/L 2,4-dichlorophenoxyacetic acid (2,4-D); 30 g/L sucrose, pH 5.8). The agrobacteria strains used were the hypervirulent strain LBA4404.pBBR1MCS virGN54D (van der Fits et al., 2000) for the effector and reporter vectors and the antisilencing strain 19K (Voinnet et al., 1999).
Freshly diluted, cultured Arabidopsis cells were distributed into multi-well culture plates, insuring homogenous conditions for all parallel experiments. Agrobacterium cells containing the construct of interest were grown overnight in liquid culture, washed once, and resuspended in plant cell culture medium. The highest efficient transfection rate was achieved using 4 mL setups in 6-well plates, and microscopic analysis of cells was performed after 3 to 4 days of co-culture.
Generation of Constructs and Complementation of papst2 by PAPST2 and PAPST1
To complement the papst2 plants, binary vectors containing 35S:PAPST2 and 35S:mitPAPST1 were generated. To obtain plants with PAPST1 restricted only to mitochondria, the PAPST1 coding sequence without the predicted organellar (http://www.cbs.dtu.dk/services/TargetP/) targeting sequence was amplified from Arabidopsis cDNA. The truncated PAPST1 missing 60 amino acids was then fused with the 65 amino acid sequence of the mitochondrial transit peptide of HSP90.6 (Krishna and Gloor, 2001; Prassinos et al., 2008) by fusion PCR. The PAPST2 coding sequence, which was also used for subcellular localization together with mitPAPST1, was recombined from the entry vector pDONOR207 into pGWB2. Both PAPST2 and mitPAPST1 were used to transform Agrobacterium strain GV3101. Transgenic Arabidopsis plants were generated by Agrobacterium-mediated transformation, and positive transformants were selected with kanamycin. Approximately 77 independent transgenic lines per line were isolated for 35S:PAPST2 and 35S:mitPAPST1. All 35S:mitPAPST1 lines showed growth retardation and bleaching of leaves but to varying degrees, depending on the amount of PAPST1 transgene expression. All 35S:PAPST2 plants had a wild-type-like appearance after complementation.
Generation of mitSAL1 and natSAL1 Constructs for Complementation of fry1 papst2 and fry1 papst1
To obtain plants with SAL1 restricted only to mitochondria, a truncated SAL1 sequence (without the predicted organellar targeting sequence http://www.cbs.dtu.dk/services/TargetP/ ; Kim and von Arnim, 2009) was fused with the 65 amino acid sequence of the mitochondrial transit peptide of HSP90.6 (Krishna and Gloor, 2001; Sangster and Queitsch, 2005; Prassinos et al., 2008). Both the complete SAL1 coding sequence (natSAL1) and the truncated version with mitochondrial targeting information (mitochondrialSAL1) were cloned into entry vector pDONOR207 followed by sequencing. The entry clones were recombined with the pGWB5 vector for GFP localization studies or with the pAMPAT vector to generate transgenic plants expressing natSAL1 and mitSAL1 under the control of the CaMV 35S promoter. The correct targeting of SAL1-GFP (to chloroplasts and mitochondria or solely to mitochondria) in cultured Arabidopsis cells was confirmed prior to the generation of the transgenic natSAL1 and mitSAL1 expressing lines. The homozygous 35S:mitSAL1 or 35S:natSAL1 lines were crossed with the fry1 papst2 and fry1 papst1 mutants, and segregation populations were checked for homozygosity of the parental loci.
Generation of ProPAPST2-uidA Plants
Two different fusion constructs were generated to investigate PAPST2 promoter activity, with the help of the reporter protein β‑glucuronidase (GUS). The first construct contained the putative 946 bp promoter region (−946 + 327 bp), whereas the second was longer and included the promoter and all seven introns (−946 + 1773 bp). The two gene fragments were amplified by PCR and inserted into pDONR207. After confirmation, the promoter constructs were recombined into the GUS-encoding binary vector pGWB3i. Plants stably transformed with the promoter-reporter sequences were used for the GUS activity assays. Histochemical detection of GUS enzyme activity was performed as described previously (Gigolashvili et al., 2009).
Extraction and Measurement of Selected Sulfated Compounds
Glucosinolates and their desulfo-precursors were extracted from lyophilized plant material using 80% methanol (v/v) supplemented with the internal standard 4-hydroxybenzyl-glucosinolate. Glucosinolates and desulfo-precursors were purified as described previously and quantified based on UV light absorption at 229 nm relative to the internal standard (Gigolashvili et al., 2012). Thiols were extracted from Arabidopsis leaves according to Wirtz and Hell (2003))), derivatized with monobromobimane (Calbiochem, EMD Chemicals), separated by HPLC, and detected with a fluorescent detector (Dionex).
Quantification of PAP and Other Adenylates in Plant Extracts
Frozen plant material was homogenized in 500 μL of hot (80°C) extraction/derivatisation buffer (62 mM citric acid, 76 mM K2HPO4, pH 4) and incubated for 5 min at 80°C. The samples were cooled on ice for 15 min and centrifuged at 14,000 rpm for 10 min at 4°C. The supernatants were transferred to new Eppendorf tubes and used for derivatization or stored at –20°C.
Chromatographic determination of adenylates was performed according to Haink and Deussen (2003))). For derivatization, the plant extracts and standards for ATP, ADP, AMP, and PAP (20 µM, 10 µM, and 5 µM) were mixed with 500 μL extraction/derivatization buffer and chloracetaldehyde and incubated for 40 min at 60°C. The derivatization was stopped by cooling the samples on ice for 15 min. Subsequently, the samples were centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant was diluted 1:1 with HPLC-grade water. Separation and analysis of the nucleotide derivatives was performed using a reversed-phase HyperClone C18 base-deactivated silica column HPLC system with a tetrabutylammonium bisulfate-buffer (TBAS) and acetonitrile as an elution buffer. The elution was performed by applying a gradient (A: TBAS; B: water:acetonitrile mixture (40:60) from apolar A to polar B solvent). The DionexUltiMate HPLC system was used with a 3 micron C18 column (Phenomenex) and a Dionex RF 2000 fluorescence detector. TBAS buffer (5.7 mM TBAS and 30.5 mM KH2PO4 pH to 5.8) was used as solution A, and 60% acetonitrile was used as solution B. The gradient for PAP separation was optimized as follows: column equilibration for 0.5 min with 100% of buffer A, followed by 0 to 15% B (2.5 min), 15 to 42% B (7.5 min), 42 to 44% B (9 min), 44 to 55% B (13 min), 55 to 80% B (15 min), and 80 to 90% B (15.5 min), followed by a cleaning cycle using 90 to 0% B for 4 min. The chromatograms were recorded and processed with Chromeleon software (Thermo Fisher).
Non-Aqueous Fractionation
To investigate the subcellular compartmentalization of plant adenylates, cellular compartments were separated using NAF (Gerhardt and Heldt, 1984; Krueger et al., 2011). NAF was performed on three independent replicates of pooled, freeze-dried rosette leaves of 5-week-old plants, as described by Krueger et al. (2011) but with the following modifications. To determine the subcellular metabolite levels, cellular compartments were separated by density gradient centrifugation under non-aqueous conditions. Compartment enrichment and compartmental separation between fractions were calculated based on the activity of compartment-specific markers. The best fit computational algorithm was applied to define the subcellular compartments. Uridinediphosphate-glucose-pyrophosphorylase was used as a marker for cytosol; glyceraldehyde-3-phosphate dehydrogenase or chlorophyll concentrations were used as markers for chloroplasts. Unfortunately, the use of potential mitochondrial marker enzymes did not allow reliable detection of mitochondrial activities in six of the fractions tested. We therefore presented the data for PAP accumulation in chloroplasts and the cytosol. Adenosine nucleotide levels were measured in six fractions by HPLC, and the amounts of nucleotides in the fractions were calculated based on absolute concentrations determined in the crude extracts.
Statistical Analysis
Comparison of means was performed to determine statistical significance using a two sample Student’s t test or an ANOVA (Tukey’s test; Supplemental Data Set). Constant variance and normal distribution of data were verified prior to statistical analysis. Differences between two unpaired means were identified with unequal variances assumed.
Heterologous Expression of PAPST2 in E. coli and Enrichment of Inclusion Bodies
Heterologous PAPST2 synthesis was conducted using the isopropyl β-D-thiogalactopyranoside-inducible T7 RNA polymerase pET-vector/RosettaTM2 expression system (Merck Biosciences, Novagen). For this, the coding sequence of PAPST2 (without the targeting sequence) was amplified from Arabidopsis cDNA with primers allowing in-frame-insertion (via the XhoI and BamHI restriction sites) with the His-tag sequence of the pET16b vector. Correctness of the expression construct was verified by sequencing. RosettaTM2 expression cells containing the expression construct were cultivated in 50 mL standard Luria-Bertani medium at 37°C under vigorous shaking. Heterologous protein synthesis was induced by the addition of β-D-thiogalactopyranoside (1 mM) during exponential cell growth (at an OD600 of 0.5). Three hours after induction, cells were harvested by centrifugation (3000 g, 5 min, 4°C). Cell disruption and purification of inclusion bodies were conducted exactly as described for PAPST1 (Gigolashvili et al., 2012). Purity of the inclusion proteins was documented by SDS-PAGE.
Solubilization and Reconstitution of PAPST2 into Liposomes
PAPST2 inclusion bodies were solubilized by resuspension in 10 mM Tris, pH 7.0, 0.1 mM EDTA, 1.67% n-lauroylsarcosine, 1 mM DTT, and 0.05% (w/v) polyethylene glycol 4000, followed by incubation for 15 min on ice. After threefold dilution with 10 mM Tris, pH 7.0, the solubilized proteins were separated from nonsolubilized aggregates by centrifugation (12,000g, 4 min, 4°C). Then 100 µg of the solubilized proteins were mixed with a buffer medium (20 mM HEPES, pH 7.0, and 1 mM phenylmethylsulfonyl fluoride). To obtain vesicles with internal counter-exchange substrates, the indicated concentrations of adenylated compounds were added to the protein mixture. Lipid/detergent micelles (100 mM PIPES, pH 7.0, 25 mg phosphatidylcholine, 16 µg cardiolipine, 28 µg C10E5) and Amberlite XAD-2 beads were prepared and step-wise added to the solubilized inclusion proteins as described by Heimpel et al. (2001). The samples were incubated overnight at 4°C to enable detergent removal and the formation of proteoliposomes.
Import Studies with Radiolabeled Substrates
External counter-exchange nucleotides and non-incorporated carrier proteins were removed from the proteoliposomes by desalting (NAP-5 columns; GE Healthcare). The proteoliposomes were eluted with an import buffer (50 mM NaCl and 10 mM PIPES, pH 7.0). For transport measurements, 50 μL of the eluate was mixed with 50 μL of import buffer supplemented with the indicated concentrations of [35S]PAPS, [α32P]ATP, or [α32P]ADP and incubated at 30°C. Transport was stopped at a given time point by removing external substrate medium. Then [32P]-labeled substrates were removed by ion-exchange chromatography and [35S]PAPS by vacuum filtration as described previously (Gigolashvili et al., 2012; Lorenz et al., 2015). Radioactivity in the samples was quantified by scintillation counting (Canberra-Packard).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under accession numbers PAPST2 (At3g51870); PAPST1 (At5g01500); SAL1/FRY1 (At5g63980); CYP79B2 (At4g39950); CYP79B3 (At2g22330); CYP83B1 (At4g31500); CYP79F1 (At1g16410); CYP79F2 (At1g16400); and CYP83A1 (At4g13770).
Supplemental Data
Supplemental Figure 1. Analysis of papst2 knock-out and amiRNA lines.
Supplemental Figure 2. papst2 can be rescued by 35S:PAPST2 but not by 35S:mitPAPST1.
Supplemental Figure 3. Effects of overexpression of PAPST2 and mitPAPST1 on the expression of PSK precursor genes.
Supplemental Figure 4. Glucosinolate and desulfo-glucosinolate levels in amiRNA lines with substantially reduced PAPST2 transcript levels.
Supplemental Figure 5. Sulfotransferase (SOT) genes are strongly upregulated in papst2.
Supplemental Figure 6. Anthocyanin accumulation is induced in adult fry1 papst2 plants.
Supplemental Figure 7. Subcellular distribution of adenylates is changed in papst1, papst2, fry1, fry1 papst1, and fry1 papst2 mutants.
Supplemental Figure 8. Schematic illustration of impaired PAP catabolism in the fry1 mutant.
Supplemental Figure 9. Schematic illustration of impaired PAPS transport in the fry1 papst1 mutant.
Supplemental Figure 10. Schematic illustration of impaired PAP transport in the fry1 papst2 mutant.
Supplemental Figure 11. Determination of GUS activity in tissues of ProPAPST2:uidA plants (promoter length, 946 + 327 bp).
Supplemental Table 1. Kinetic parameters of PAPST2 vs. PAPST1.
Supplemental Table 2. Bioinformatic predictions and experimental evidence indicating both the chloroplastidic and mitochondrial localizations of PAPST2.
Supplemental Table 3. Number of seedlings and adult plants showing bleaching of cotyledons in fry1 papst2 compared with fry1 and fry1 papst1.
Supplemental Table 4. Sequences of primers used for genotyping and cloning.
Supplemental Table 5. Sequences of primers used for qRT-PCR analysis.
Supplemental Data Set. ANOVA and Tukey’s test tables.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by Deutsche Forschungsgemeinshaft grant (GI-824/2-1) and theDeutsche Forschungsgemeinshaft Heisenberg-Fellowship (GI-824/3-1) given to T. Gigolashvili. We greatly acknowledge Stephan Krueger for teaching us the non-aqueous-fractionation method and Richard Jakoby for carefully proofreading the article. We very much appreciate financial support from the Cluster of Excellence on Plant Sciences (EXC 1028) and the Deutsche Forschungsgemeinshaft grant (FL-126/24-1). Research in S.K.´s lab is supported by Deutsche Forschungsgemeinschaft (EXC 1028).
AUTHOR CONTRIBUTIONS
T.G., S.K., and U.I.F. conceived the experiments. N.A., M.L., H.F., A.K., E.H., N.S., I.H., and T.G. performed the experiments. N.A., M.L., H.F., I.H., U.I.F., S.K., and T.G. analyzed the data. T.G. and I.H. wrote the article with contributions from the other authors.
References
- Amano Y., Tsubouchi H., Shinohara H., Ogawa M., Matsubayashi Y. (2007). Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S., Smith-Unna R.D., Boursnell C.M., Kopriva S., Hibberd J.M. (2014). Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. Plant J. 78: 659–673. [DOI] [PubMed] [Google Scholar]
- Berger B., Stracke R., Yatusevich R., Weisshaar B., Flügge U.I., Gigolashvili T. (2007). A simplified method for the analysis of transcription factor-promoter interactions that allows high-throughput data generation. Plant J. 50: 911–916. [DOI] [PubMed] [Google Scholar]
- Bodén M., Hawkins J. (2005). Prediction of subcellular localization using sequence-biased recurrent networks. Bioinformatics 21: 2279–2286. [DOI] [PubMed] [Google Scholar]
- Bonaventure G., Gfeller A., Rodríguez V.M., Armand F., Farmer E.E. (2007). The fou2 gain-of-function allele and the wild-type allele of Two Pore Channel 1 contribute to different extents or by different mechanisms to defense gene expression in Arabidopsis. Plant Cell Physiol. 48: 1775–1789. [DOI] [PubMed] [Google Scholar]
- Chan K.X., Mabbitt P.D., Phua S.Y., Mueller J.W., Nisar N., Gigolashvili T., Stroeher E., Grassl J., Arlt W., Estavillo G.M., et al. (2016). Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase. Proceedings of the National Academy of Sciences, 113: E4567–E4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: Simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. The Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8: 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Estavillo G.M., et al. (2011). Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M., Salvi D., Brugière S., Miras S., Kowalski S., Louwagie M., Garin J., Joyard J., Rolland N. (2003). Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2: 325–345. [DOI] [PubMed] [Google Scholar]
- Fischer K., Arbinger B., Kammerer B., Busch C., Brink S., Wallmeier H., Sauer N., Eckerskorn C., Flügge U.I. (1994). Cloning and in vivo expression of functional triose phosphate/phosphate translocators from C3- and C4-plants: Evidence for the putative participation of specific amino acid residues in the recognition of phosphoenolpyruvate. Plant J. 5: 215–226. [DOI] [PubMed] [Google Scholar]
- Gerhardt R., Heldt H.W. (1984). Measurement of subcellular metabolite levels in leaves by fractionation of freeze-stopped material in nonaqueous media. Plant Physiol. 75: 542–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T., Yatusevich R., Rollwitz I., Humphry M., Gershenzon J., Flügge U.I. (2009). The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell 21: 1813–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T., Geier M., Ashykhmina N., Frerigmann H., Wulfert S., Krueger S., Mugford S.G., Kopriva S., Haferkamp I., Flügge U.-I. (2012). The Arabidopsis thylakoid ADP/ATP carrier TAAC has an additional role in supplying plastidic phosphoadenosine 5′-phosphosulfate to the cytosol. Plant Cell 24: 4187–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gläser H.U., Thomas D., Gaxiola R., Montrichard F., Surdin-Kerjan Y., Serrano R. (1993). Salt tolerance and methionine biosynthesis in Saccharomyces cerevisiae involve a putative phosphatase gene. EMBO J. 12: 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haink G., Deussen A. (2003). Liquid chromatography method for the analysis of adenosine compounds. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 784: 189–193. [DOI] [PubMed] [Google Scholar]
- Heimpel S., Basset G., Odoy S., Klingenberg M. (2001). Expression of the mitochondrial ADP/ATP carrier in Escherichia coli. Renaturation, reconstitution, and the effect of mutations on 10 positive residues. J. Biol. Chem. 276: 11499–11506. [DOI] [PubMed] [Google Scholar]
- Hirsch J., et al. (2011). A novel fry1 allele reveals the existence of a mutant phenotype unrelated to 5'->3' exoribonuclease (XRN) activities in Arabidopsis thaliana roots. PLoS One 6: e16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höglund A., Dönnes P., Blum T., Adolph H.-W., Kohlbacher O. (2006). MultiLoc: Prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics 22: 1158–1165. [DOI] [PubMed] [Google Scholar]
- Kaundal R., Saini R., Zhao P.X. (2010). Combining machine learning and homology-based approaches to accurately predict subcellular localization in Arabidopsis. Plant Physiol. 154: 36–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.H., von Arnim A.G. (2009). FIERY1 regulates light-mediated repression of cell elongation and flowering time via its 3′(2′),5′-bisphosphate nucleotidase activity. Plant J. 58: 208–219. [DOI] [PubMed] [Google Scholar]
- Kleffmann T., Russenberger D., von Zychlinski A., Christopher W., Sjölander K., Gruissem W., Baginsky S. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14: 354–362. [DOI] [PubMed] [Google Scholar]
- Komori R., Amano Y., Ogawa-Ohnishi M., Matsubayashi Y. (2009). Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 15067–15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A., Kopriva S. (2016). Sulfur metabolism and its manipulation in crops. J. Gen. Genom. 43: 623–629. [DOI] [PubMed] [Google Scholar]
- Koroleva O.A., Tomlinson M.L., Leader D., Shaw P., Doonan J.H. (2005). High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J. 41: 162–174. [DOI] [PubMed] [Google Scholar]
- Krishna P., Gloor G. (2001). The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6: 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger S., Giavalisco P., Krall L., Steinhauser M.-C., Büssis D., Usadel B., Flügge U.-I., Fernie A.R., Willmitzer L., Steinhauser D. (2011). A topological map of the compartmentalized Arabidopsis thaliana leaf metabolome. PLoS One 6: e17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschmar A., Rzewuski G., Stührwohldt N., Beemster G.T., Inzé D., Sauter M. (2009). PSK-α promotes root growth in Arabidopsis. New Phytol. 181: 820–831. [DOI] [PubMed] [Google Scholar]
- Lee B.-R., Huseby S., Koprivova A., Chételat A., Wirtz M., Mugford S.T., Navid E., Brearley C., Saha S., Mithen R., Hell R., Farmer E.E., et al. (2012). Effects of fou8/fry1 mutation on sulfur metabolism: is decreased internal sulfate the trigger of sulfate starvation response? PLoS One 7: e39425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y. (2012). MBSJ MCC Young Scientist Award 2010. Recent progress in research on small post-translationally modified peptide signals in plants. Genes Cells 17: 1–10. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Takagi L., Omura N., Morita A., Sakagami Y. (1999). The endogenous sulfated pentapeptide phytosulfokine-alpha stimulates tracheary element differentiation of isolated mesophyll cells of zinnia. Plant Physiol. 120: 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. (2010). Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067. [DOI] [PubMed] [Google Scholar]
- Millar A.H., Heazlewood J.L. (2003). Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol. 131: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford S.G., et al. (2009). Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21: 910–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford S.G., Matthewman C.A., Hill L., Kopriva S. (2010). Adenosine-5′-phosphosulfate kinase is essential for Arabidopsis viability. FEBS Lett. 584: 119–123. [DOI] [PubMed] [Google Scholar]
- Murguia J.R., Bellés J.M., Serrano R. (1995). A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267: 232–234. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Palmieri F., Pierri C.L., De Grassi A., Nunes-Nesi A., Fernie A.R. (2011). Evolution, structure and function of mitochondrial carriers: A review with new insights. Plant J. 66: 161–181. [DOI] [PubMed] [Google Scholar]
- Phua S.Y., Yan D., Chan K.X., Estavillo G.M., Nambara E., Pogson B.J. (2018). The Arabidopsis SAL1-PAP Pathway: A case study for integrating chloroplast retrograde, light and hormonal signaling in modulating plant growth and development? Front. Plant Sci. 9: 1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornsiriwong W., et al. (2017). A chloroplast retrograde signal, 3′-phosphoadenosine 5′-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. eLife 6: e23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prassinos C., Haralampidis K., Milioni D., Samakovli D., Krambis K., Hatzopoulos P. (2008). Complexity of Hsp90 in organelle targeting. Plant Mol. Biol. 67: 323–334. [DOI] [PubMed] [Google Scholar]
- Quintero F.J., Garciadeblás B., Rodríguez-Navarro A. (1996). The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 8: 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rens-Domiano S.S., Roth J.A. (1987). Inhibition of M and P phenol sulfotransferase by analogues of 3′-phosphoadenosine-5′-phosphosulfate. J. Neurochem. 48: 1411–1415. [DOI] [PubMed] [Google Scholar]
- Robles P., Fleury D., Candela H., Cnops G., Alonso-Peral M.M., Anami S., Falcone A., Caldana C., Willmitzer L., Ponce M.R., Van Lijsebettens M., Micol J.L. (2010). The RON1/FRY1/SAL1 gene is required for leaf morphogenesis and venation patterning in Arabidopsis. Plant Physiol. 152: 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez V.M., Chételat A., Majcherczyk P., Farmer E.E. (2010). Chloroplastic phosphoadenosine phosphosulfate metabolism regulates basal levels of the prohormone jasmonic acid in Arabidopsis leaves. Plant Physiol. 152: 1335–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster T.A., Queitsch C. (2005). The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr. Opin. Plant Biol. 8: 86–92. [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flügge U.I., Kunze R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R., Fischer K., Ketelsen B., Krupinska K., Krause K. (2007). Comparative survey of plastid and mitochondrial targeting properties of transcription factors in Arabidopsis and rice. Mol. Genet. Genomics 277: 631–646. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Kopriva S., Giordano M., Saito K., Hell R. (2011). Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annual Review of Plant Biology 62: 157–184. [DOI] [PubMed] [Google Scholar]
- Thuswaldner S., Lagerstedt J.O., Rojas-Stütz M., Bouhidel K., Der C., Leborgne-Castel N., Mishra A., Marty F., Schoefs B., Adamska I., Persson B.L., Spetea C. (2007). Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis. J. Biol. Chem. 282: 8848–8859. [DOI] [PubMed] [Google Scholar]
- van der Fits L., Deakin E.A., Hoge J.H.C., Memelink J. (2000). The ternary transformation system: Constitutive virG on a compatible plasmid dramatically increases Agrobacterium-mediated plant transformation. Plant Mol. Biol. 43: 495–502. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Pinto Y.M., Baulcombe D.C. (1999). Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96: 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson P.B., Estavillo G.M., Field K.J., Pornsiriwong W., Carroll A.J., Howell K.A., Woo N.S., Lake J.A., Smith S.M., Harvey Millar A., von Caemmerer S., Pogson B.J. (2009a). The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 58: 299–317. [DOI] [PubMed] [Google Scholar]
- Wilson P.B., Estavillo G.M., Field K.J., Pornsiriwong W., Carroll A.J., Howell K.A., Woo N.S., Lake J.A., Smith S.M., Harvey Millar A., von Caemmerer S., Pogson B.J. (2009b). The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 58: 299–317. [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M., Hell R. (2003). Comparative biochemical characterization of OAS-TL isoforms from Arabidopsis thaliana. In Sulfur Transport and Assimilation in Plants: Regulation, Interaction, Signaling, J.-C. Davidian, D. Grill, L.J. De Kok, I. Stulen, M.J. Hawkesford, E. Schnug and H. Rennenberg, eds (Leiden, The Netherlands: Backhuys; ), pp. 355–357. [Google Scholar]
- Xiong L., Lee H., Huang R., Zhu J.K. (2004). A single amino acid substitution in the Arabidopsis FIERY1/HOS2 protein confers cold signaling specificity and lithium tolerance. Plant J. 40: 536–545. [DOI] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Matsubayashi Y., Nakamura K., Sakagami Y. (1999). Oryza sativa PSK gene encodes a precursor of phytosulfokine-α, a sulfated peptide growth factor found in plants. Proc. Natl. Acad. Sci. USA 96: 13560–13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Li G., Ren X., Herrler G. (2007). Select what you need: A comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J. Biotechnol. 127: 335–347. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hennig L., Gruissem W. (2005). Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 10: 407–409. [DOI] [PubMed] [Google Scholar]