The jasmonate-inducible transcription factor MYC2 activates the expression of three MYC2-TARGETED BHLH (MTB) proteins, which function as competitive repressors of MYC2-mediated transcription of jasmonate-responsive genes in tomato.
Abstract
In tomato (Solanum lycopersicum), as in other plants, the immunity hormone jasmonate (JA) triggers genome-wide transcriptional changes in response to pathogen and insect attack. These changes are largely regulated by the basic helix-loop-helix (bHLH) transcription factor MYC2. The function of MYC2 depends on its physical interaction with the MED25 subunit of the Mediator transcriptional coactivator complex. Although much has been learned about the MYC2-dependent transcriptional activation of JA-responsive genes, relatively less studied is the termination of JA-mediated transcriptional responses and the underlying mechanisms. Here, we report an unexpected function of MYC2 in regulating the termination of JA signaling through activating a small group of JA-inducible bHLH proteins, termed MYC2-TARGETED BHLH1 (MTB1), MTB2, and MTB3. MTB proteins negatively regulate JA-mediated transcriptional responses via their antagonistic effects on the functionality of the MYC2-MED25 transcriptional activation complex. MTB proteins impair the formation of the MYC2-MED25 complex and compete with MYC2 to bind to its target gene promoters. Therefore, MYC2 and MTB proteins form an autoregulatory negative feedback circuit to terminate JA signaling in a highly organized manner. We provide examples demonstrating that gene editing tools such as CRISPR/Cas9 open up new avenues to exploit MTB genes for crop protection.
INTRODUCTION
As a major immunity hormone, jasmonate (JA) promotes plant defense in response to mechanical wounding, insect attack, and pathogen infection. In addition, JA acts as a growth hormone to repress vegetative growth and promote reproductive development (Browse, 2009; Wasternack and Hause, 2013; Chini et al., 2016; Goossens et al., 2016; Zhai et al., 2017b; Howe et al., 2018). Underling these juxtaposing physiological functions, JA orchestrates a genome-wide transcriptional program controlling resource allocation between growth- and defense-related processes, thus optimizing plant fitness according to the rapidly changing and often hostile environment (Major et al., 2017; Guo et al., 2018a, 2018b).
Decades of studies in the model systems of Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum) have elucidated a core JA signaling pathway consisting of multiple interconnected protein-protein interaction modules that govern the transcriptional state of hormone-responsive genes. Among the most studied JA-inducible transcription factors is the basic helix-loop-helix (bHLH) protein MYC2, which acts as a master regulator of diverse aspects of JA responses both in Arabidopsis (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007; Kazan and Manners, 2013; Zhai et al., 2013, 2017b) and in tomato (Yan et al., 2013; Du et al., 2017; Zhai et al., 2017b). In the resting (i.e., “repressed”) stage of JA signaling, a group of JASMONATE-ZIM DOMAIN (JAZ) proteins physically recruit the general corepressor TOPLESS to form a repression complex, thereby preventing MYC2 and its close homologs MYC3, MYC4, and MYC5 from activating the expression of JA-responsive genes (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Pauwels et al., 2010; Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011; Qi et al., 2015). In the presence of the bioactive JA ligand jasmonoyl-isoleucine (JA-Ile), JAZ proteins form a JA-Ile-dependent coreceptor complex with CORONATINE-INSENSITIVE1 (COI1), the F-box subunit of the SCFCOI1 ubiquitin ligase (Xie et al., 1998; Devoto et al., 2002; Xu et al., 2002; Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Fonseca et al., 2009; Sheard et al., 2010). The SCFCOI1-dependent degradation of JAZ repressors leads to the “derepression” of MYC2 (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Fonseca et al., 2009; Sheard et al., 2010). In turn, free MYC2 forms a transcriptional activation complex with the MED25 subunit of the plant Mediator transcriptional coactivator complex to activate the expression of JA-responsive genes (Çevik et al., 2012; Chen et al., 2012; An et al., 2017; Zhai et al., 2017a).
We recently showed that, in addition to interacting with MYC2 for the assembly of the preinitiation complex, the multitalented Mediator subunit MED25 physically interacts with and coordinates the actions of multiple regulators during different stages of JA signaling. For example, MED25 physically brings the hormone receptor COI1 to promoters of MYC2 target genes during the resting stage and facilitates COI1-dependent degradation of JAZ repressors in the presence of JA-Ile (An et al., 2017). Furthermore, upon hormone elicitation, MED25 physically recruits the epigenetic regulator HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY1, which selectively regulates hormone-induced acetylation of Lys-9 of histone H3 in MYC2 target promoters (An et al., 2017). All of these mechanistically related functions of MED25 favor the hormone-induced activation of MYC2 function. Together, these findings indicate a central role of the MYC2-MED25 complex in the activation of JA-responsive genes.
Although JA-mediated defense responses facilitate the adaptation of plants to a wide range of biotic and abiotic stresses, these responses can be detrimental if they continue excessively and are not terminated in a timely manner. Emerging evidence suggests that plants have evolved diverse and sophisticated mechanisms that ensure the proper termination of JA-mediated defense responses. For example, an evolutionarily conserved metabolic network converts the bioactive JA-Ile into an inactive or less active form (Miersch et al., 2008; Kitaoka et al., 2011; Koo et al., 2011; VanDoorn et al., 2011; Heitz et al., 2012), thus providing an efficient route to switch off the JA signaling (Koo and Howe, 2012). Most JAZ genes contain a highly conserved Jas intron, whose alternative splicing generates a repertoire of JAZ splice variants lacking the intact C-terminal Jas motif (Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2010; Moreno et al., 2013). These dominant JAZ splicing variants are unable to recruit SCFCOI1 but still retain the ability to repress MYC2 and their interacting transcription factors, thus contributing to the desensitization of JA signaling (Zhang et al., 2017a). In addition, the Arabidopsis JAZ8 and JAZ13 are noncanonical JAZ repressors that are less sensitive to hormone-induced degradation and can recruit the corepressor TOPLESS in a NOVEL INTERACTOR OF JAZ-independent manner (Shyu et al., 2012; Thireault et al., 2015). Furthermore, the bHLH subclade IIId of MYC-related DNA binding proteins, termed JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3, play a negative role in JA-mediated defense responses via competing with the DNA binding capacity of MYC2 (Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Song et al., 2013; Fonseca et al., 2014). Collectively, these observations support that the appropriate termination of JA-mediated defense response is an integral part of JA signaling.
Here, we report the mechanistic action of a small group of JA-inducible bHLH proteins, termed MYC2-TARGETED BHLH1 (MTB1), MTB2, and MTB3, in terminating JA signaling. Interestingly, these MTB proteins function as negative regulators of JA signaling and are direct transcriptional targets of the master activator MYC2. We demonstrate that MTB proteins impair the formation of the MYC2-MED25 transcriptional activation complex and compete with MYC2 to bind to its target gene promoters, thus deactivating MYC2-dependent gene transcription. Therefore, MYC2 and MTB proteins form a negative feedback circuit to terminate JA signaling.
RESULTS
Tomato MED25 Acts as a Coactivator of MYC2
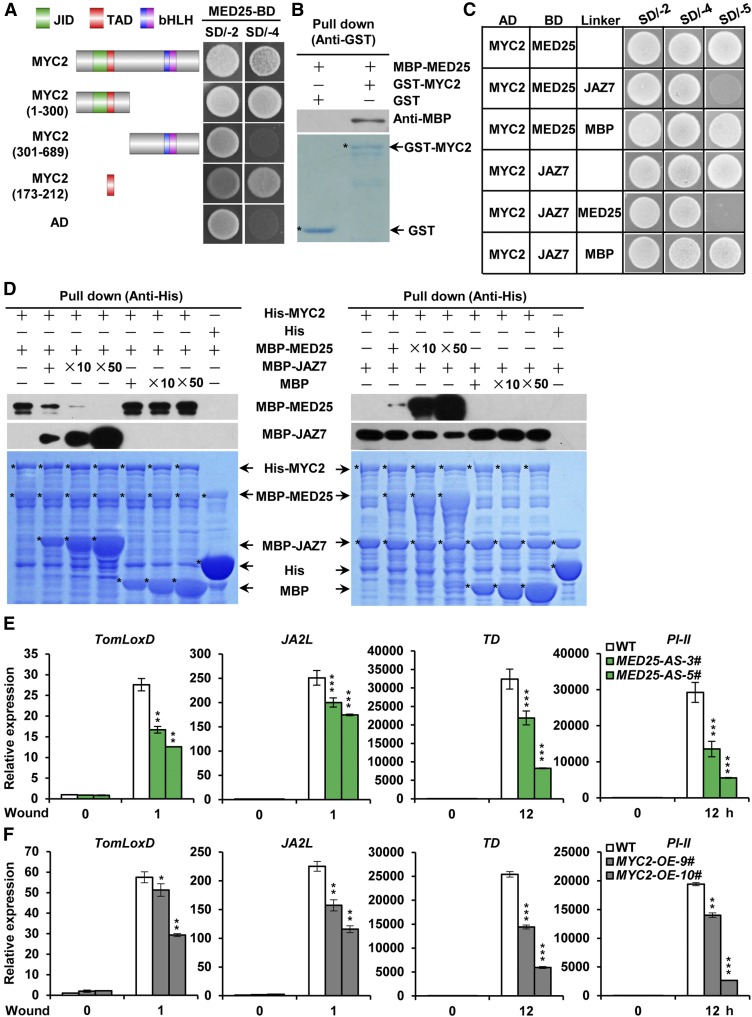
High similarity between tomato and Arabidopsis MED25 proteins (Supplemental Figure 1A) prompted us to test whether tomato MED25 acts as a coactivator of MYC2, which controls a transcriptional regulatory cascade involved in JA-mediated plant immunity (Du et al., 2017). In yeast two-hybrid (Y2H) assays, we found that MED25 interacts with the transcriptional activation domain (TAD) of MYC2 (Figure 1A). To confirm the physical interaction between MED25 and MYC2, we performed in vitro pull-down experiments using a purified maltose binding protein (MBP)-tagged MED25 fragment from amino acid 243 to 806 (MBP-MED25) and glutathione S-transferase (GST)-tagged MYC2 (GST-MYC2). As shown in Figure 1B, GST-MYC2 pulled down MBP-MED25, indicating that MED25 interacts with MYC2 in vitro.
Figure 1.
MED25 and JAZ7 Compete to Interact with MYC2.
(A) Y2H assays examining interactions between the GAL4 DNA activation domain (AD) fusions of MYC2 or MYC2 variants and GAL4 DNA binding domain (BD) fusion of MED25. The transformed yeast cells were plated on SD/-4. The schematic diagram shows the MYC2 domain constructs. The conserved JAZ-interacting domain (JID), TAD, basic (b) domain, and helix-loop-helix (HLH) domain are indicated with green, red, blue, and pink boxes, respectively.
(B) In vitro pull-down assays to verify the interaction between MED25 and MYC2. Purified MBP-MED25 was incubated with GST or GST-MYC2 for the GST pull-down assay and detected by immunoblotting using anti-MBP antibody. The positions of purified GST and GST-MYC2 separated by SDS-PAGE are marked with asterisks on the Coomassie Brilliant Blue (CBB)-stained gel.
(C) Y3H assays showing that JAZ7 and MED25 compete with each other to interact with MYC2. Yeast cells cotransformed with AD-MYC2 and pBridge-MED25-JAZ7/MBP (top three rows) or with AD-MYC2 and pBridge-JAZ7-MED25/MBP (bottom three rows) were grown on SD/-4 medium to assess MYC2-MED25 interaction or MYC2-JAZ7 interaction, respectively. The cotransformed yeast cells were grown on SD/-5 medium to induce the expression of JAZ7/MBP (top three rows) or MED25/MBP (bottom three rows).
(D) In vitro pull-down assays testing for JAZ7 and MED25 competition to interact with MYC2. Fixed amounts of His-MYC2 and MBP-MED25 fusion proteins were incubated with an increasing amount of MBP-JAZ7 fusion protein and MBP protein (left), and fixed amounts of His-MYC2 and MBP-JAZ7 fusion proteins were incubated with an increasing amount of MBP-MED25 fusion protein and MBP protein (right). Protein samples were immunoprecipitated with anti-His antibody and immunoblotted with anti-MBP antibody. CBB staining shows the amount of recombinant proteins loaded on the gel. Asterisks indicate the specific bands of recombinant proteins.
(E) and (F) RT-qPCR assays characterizing wound-induced expression of TomLoxD, JA2L, TD, and PI-II in wild-type and MED25-AS plants (E) and in wild-type and MYC2-OE plants (F). Eighteen-day-old seedlings of the indicated tomato genotypes with two fully expanded leaves were mechanically wounded using a hemostat on both leaves for the indicated times before extracting total RNAs for RT-qPCR assays. Data represent means ± sd (n = 3). Asterisks indicate significant differences from the wild type according to Student’s t test at *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (Supplemental Data Set 2).
Since the TAD of MYC2 is also required for the binding of JAZ repressors (Du et al., 2017), we investigated the possibility that MED25 competes with JAZ repressors to bind to MYC2. Yeast three-hybrid (Y3H) assays revealed MED25-MYC2 interaction on the synthetic defined (SD) medium lacking Ade, His, Trp, and Leu (SD/-4); however, the induction of JAZ7 expression on SD medium lacking Ade, His, Trp, Leu, and Met (SD/-5) led to a dramatic reduction in MED25-MYC2 interaction (Figure 1C), suggesting that JAZ7 interferes with MED25-MYC2 interaction. On the other hand, JAZ7-MYC2 interaction was detected on SD/-4 medium, and this interaction was dramatically reduced by inducing the expression of MED25 on SD/-5 medium (Figure 1C), suggesting that MED25 also impairs JAZ7-MYC2 interaction. In parallel Y3H experiments, we showed that induction of MBP expression did not affect the MED25-MYC2 interaction and the JAZ7-MYC2 interaction (Figure 1C). Together, these results support that MED25 and JAZ7 compete with each other to bind to MYC2.
To substantiate the above observations, we performed in vitro pull-down experiments using a constant protein concentration of His-MYC2 and increasing protein concentration of MBP-MED25 or MBP-JAZ7. Results showed that the ability of His-MYC2 to pull down MBP-MED25 decreased as the amount of MBP-JAZ7 increased (Figure 1D, left panel). Similarly, the ability of His-MYC2 to pull down MBP-JAZ7 decreased as the amount of MBP-MED25 increased (Figure 1D, right panel). As a negative control, we showed that the ability of His-MYC2 to pull down MBP-MED25 or MBP-JAZ7 was not obviously affected by increasing the amount of MBP (Figure 1D). These results corroborate that MED25 and JAZ7 compete with each other to bind to MYC2.
To explore the significance of MED25-MYC2 interaction in JA signaling, we generated plants expressing antisense (AS) MED25 (MED25-AS) in which the expression level of endogenous MED25 was substantially reduced compared with that in wild-type plants (Supplemental Figure 1B). The MED25-AS transgenic and wild-type plants were then subjected to a standard wounding assay. RT-qPCR analysis revealed that wound-induced expression of MYC2 direct target genes TOMATO LIPOXYGENASE D (TomLoxD) and JA2L (Yan et al., 2013; Du et al., 2014, 2017) was significantly reduced in MED25-AS plants compared with the wild type (Figure 1E). Similarly, wound-induced expression of the defense marker genes THREONINE DEAMINASE (TD; Chen et al., 2005) and PROTEINASE INHIBITOR II (PI-II; Ryan and Pearce, 1998; Ryan, 2000) was also significantly reduced in MED25-AS plants compared with the wild type (Figure 1E). These results indicate that MED25 plays a positive role in wound-induced activation of JA-responsive MYC2 target genes.
Overexpression of MYC2 Attenuates, Rather Than Enhances, JA-Mediated Defense Responses
The above results showed that MED25 competitively binds to MYC2, thereby activating JA-responsive gene expression upon wound-induced JA-Ile accumulation and subsequent JAZ degradation. Therefore, we hypothesized that the overexpression of MYC2 enhances JA-mediated defense responses. To test this hypothesis, we generated several transgenic lines overexpressing GFP-tagged MYC2; the expression of MYC2-GFP was elevated in MYC2-OE plants (Supplemental Figure 1C). Contrary to our hypothesis, wound-induced expression levels of TomLoxD, JA2L, TD, and PI-II were decreased in MYC2-OE plants compared with wild-type plants (Figure 1F), indicating that the overexpression of MYC2 weakens, rather than enhances, JA-mediated defense responses.
As a first step to understand why MYC2-OE plants are defective in wound-induced defense gene expression, we compared wound-induced JA accumulation between MYC2-OE plants and their wild-type counterparts. Wound-induced JA accumulation was significantly reduced in MYC2-OE plants compared with the wild type (Supplemental Figure 2A), suggesting that MYC2-OE plants are defective in wound-induced JA production. In addition, exogenous methyl jasmonate (MeJA)-induced expression of TomLoxD, JA2L, TD, and PI-II was also significantly decreased in MYC2-OE plants compared with the wild type (Supplemental Figure 2B), suggesting that MYC2-OE plants are also defective in JA signaling. Together, these observations led us to a hypothesis that, in addition to regulating the activation of JA signaling, MYC2 might also autoregulate the inactivation and/or termination of JA signaling via a hitherto unknown mechanism.
MTB1 to MTB3 Are Directly Targeted by MYC2
To understand how MYC2 regulates the termination of JA-mediated defense responses, we attempted to identify MYC2-targeted transcription factors (Du et al., 2017) that negatively regulate JA signaling. From our genome-wide binding profile of MYC2 (Du et al., 2017), we identified three MTB proteins, including MTB1 (bHLH113, Solyc01g096050), MTB2 (bHLH133, Solyc05g050560), and MTB3 (bHLH138, Solyc06g0839800). Phylogenetic analysis revealed that MTB1 to MTB3 proteins are homologs of the Arabidopsis JAM proteins (Supplemental Figure 3; Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Song et al., 2013; Fonseca et al., 2014; Goossens et al., 2017) and they share high sequence similarity with MYC2 (Supplemental Figure 3; Sun et al., 2015). In addition to tomato and Arabidopsis, MYC2- and MTB-like proteins are also present in other plant species, including Oryza sativa, Zea mays, Populus trichocarpa, Nicotiana tabacum, Catharanthus roseus, Musa acuminate, Malus domestica, and Hevea brasiliensis (Supplemental Figure 3A; Supplemental Data Set 1).
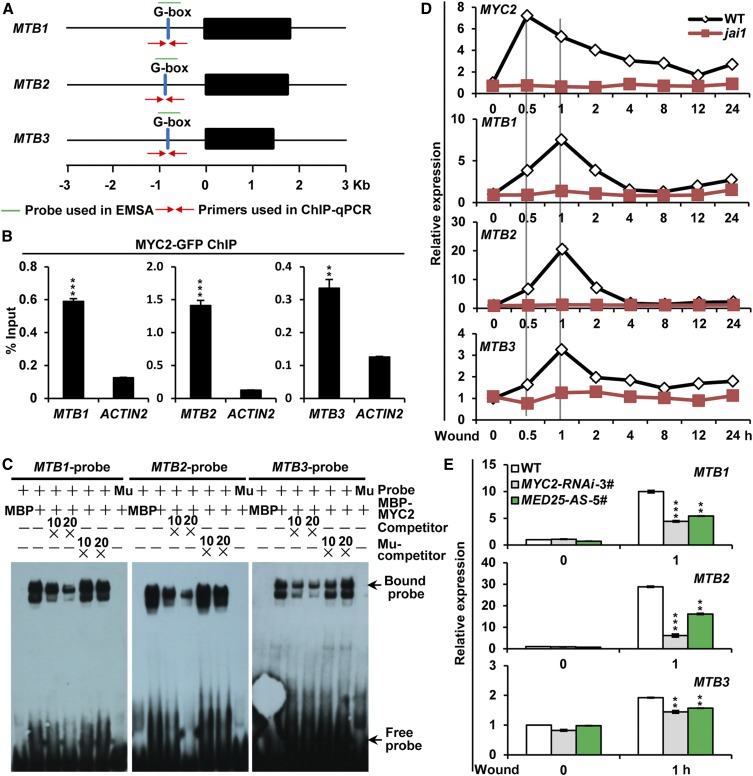
Consistent with the recent chromatin immunoprecipitation (ChIP)-sequencing data (Supplemental Figure 4; Du et al., 2017), our ChIP-qPCR assays using MYC2-GFP plants (Supplemental Figure 1C; Du et al., 2017) revealed an enrichment of MYC2-GFP recombinant protein on the G-box (CACATG) motifs in MTB gene promoters (Figures 2A and 2B). Electrophoretic mobility shift assays (EMSAs) showed that MBP-MYC2 recombinant protein bound a DNA probe containing the G-box motif but failed to bind a DNA probe in which the G-box motif was mutated (Figure 2C), indicating that MYC2 specifically binds the G-box motif of MTB promoters. Collectively, these results demonstrate that MTB1 to MTB3 are direct transcriptional targets of MYC2.
Figure 2.
MTB1 to MTB3 Are Direct Transcriptional Targets of MYC2.
(A) Schematic representations of MTB1, MTB2, and MTB3 genes showing the primers and probe used for ChIP-qPCR assay and EMSA.
(B) ChIP-qPCR analysis of MYC2 enrichment on the chromatin of MTB1, MTB2, and MTB3. Chromatin of MYC2-GFP-9# plants was immunoprecipitated using anti-GFP antibody, and the immunoprecipitated DNA was quantified by qPCR. The enrichment of target gene promoters is displayed as a percentage of input DNA. Data represent means ± sd (n = 3). ACTIN2 was used as a nonspecific target.
(C) EMSA showing that MBP-MYC2 directly binds to the promoters of MTB1, MTB2, and MTB3. The MBP protein was incubated with the labeled probe to serve as a negative control; mutated probes were used as a negative control. Ten- and 20-fold excesses of unlabeled or mutated probes were used for competition. Ten- and 20-fold excesses of MBP protein were used for competition. Mu, mutated probe in which the G-box motif 5ʹ-CACATG-3ʹ was replaced with 5ʹ-AAAAAA-3ʹ.
(D) and (E) RT-qPCR assays showing wound-induced expression of MYC2, MTB1, MTB2, and MTB3 in wild-type and jai1 plants (D) and MTB1, MTB2, and MTB3 in wild-type, MYC2-RNAi-3#, and MED25-AS-5# plants (E). Eighteen-day-old seedlings of the indicated tomato genotypes with two fully expanded leaves were mechanically wounded using a hemostat on both leaves for the indicated times before extracting total RNAs for RT-qPCR assays. Data represent means ± sd (n = 3).
In (B) and (E), asterisks indicate significant differences from the wild type according to Student’s t test at **, P < 0.01 and ***, P < 0.001 (Supplemental Data Set 2).
We then compared the temporal expression patterns of MTB1 to MTB3 and MYC2 in response to wounding. RT-qPCR assays revealed that, although wounding induced the expression of both MYC2 and MTB genes in wild-type plants, the timing of their induction was distinct; while the expression of MYC2 peaked at 0.5 h after wounding, the expression of MTB genes peaked at 1 h after wounding (Figure 2D). The wound-induced expression kinetics of MTB1 to MTB3 and MYC2 were overall conserved for their Arabidopsis homologs in response to MeJA (Hickman et al., 2017). Notably, wound-induced expression of MYC2 and MTB genes was largely abolished in the jai1 mutant, which harbors a null mutation of the JA-Ile receptor protein of tomato (Li et al., 2004), indicating that wound-induced expression of MTB genes depends on COI1. Additionally, wound-induced expression of all three MTB genes was significantly reduced in RNAi-mediated MYC2 knockdown plants (MYC2-RNAi-3#; Yan et al., 2013; Du et al., 2017) and MED25-AS-5# plants compared with the wild type (Figure 2E), indicating that wound-induced expression of MTB genes requires MYC2- and MED25-dependent JA signaling.
MTB1 to MTB3 Negatively Regulate Diverse Aspects of JA Responses
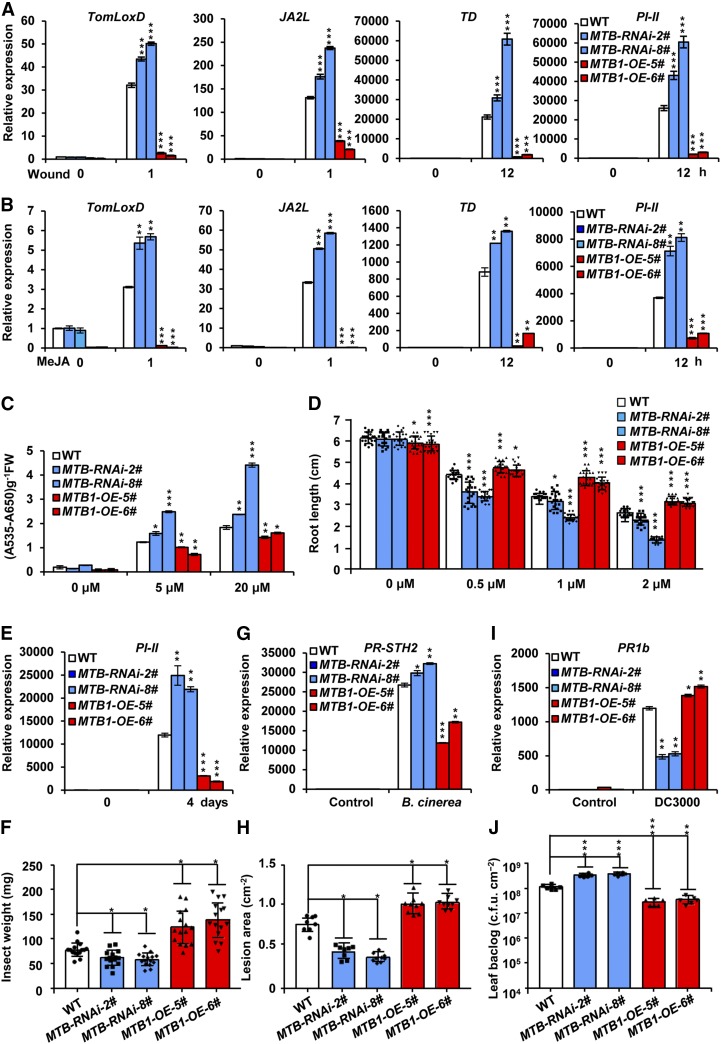
To determine the function of MTB genes in JA signaling, we generated MTB-RNAi transgenic tomato plants in which the expression of MTB1, MTB2, and MTB3 was downregulated (Supplemental Figure 5A). We also generated MTB1-OE plants overexpressing MTB1 cDNA fused with GFP under the control of the 35S promoter (Supplemental Figures 5B and 5C). Two lines of MTB-RNAi plants (MTB-RNAi-2# and MTB-RNAi-8#) and two lines of MTB1-OE plants (MTB1-OE-5# and MTB1-OE-6#) were selected for further analyses. In a standard wounding response assay of tomato seedlings, wound-induced expression levels of TomLoxD, JA2L, TD, and PI-II were increased in MTB-RNAi plants and decreased in MTB1-OE plants compared with wild-type plants (Figure 3A), indicating that MTB proteins negatively regulate the wound response of tomato plants.
Figure 3.
MTB1 to MTB3 Negatively Regulate Diverse Aspects of JA Responses.
(A) and (B) RT-qPCR assays of the expression of TomLoxD, JA2L, TD, and PI-II in wild-type, MTB-RNAi-2#, MTB-RNAi-8#, MTB1-OE-5#, and MTB1-OE-6# plants in response to mechanical wounding (A) and MeJA treatment (B). For (A), 18-d-old seedlings of the indicated tomato genotypes with two fully expanded leaves were mechanically wounded using a hemostat on both leaves for the indicated times before extracting total RNAs for RT-qPCR assays. For (B), 18-d-old seedlings of the indicated tomato genotypes with two fully expanded leaves were exposed to MeJA vapor for the indicated times before extracting total RNAs for RT-qPCR assays.
(C) Anthocyanin contents of the 7-d-old seedlings in wild-type, MTB-RNAi-2#, MTB-RNAi-8#, MTB1-OE-5#, and MTB1-OE-6# plants grown on Murashige and Skoog (MS) medium containing indicated concentrations of JA. FW, fresh weight.
(D) Root growth inhibition assay of 7-d-old seedlings of wild-type, MTB-RNAi-2#, MTB-RNAi-8#, MTB1-OE-5#, and MTB1-OE-6# plants grown on MS medium supplied with indicated concentrations (µM) of JA. Data represent means ± sd (n = 20).
(E) Expression of PI-II in wild-type, MTB-RNAi-2#, MTB-RNAi-8#, MTB1-OE-5#, and MTB1-OE-6# plants subjected to H. armigera larvae. Plants were harvested at the indicated time points during the feeding trial for RNA extraction and RT-qPCR analysis.
(F) Average weight of larvae recovered at the end of day 4 of the feeding trial using whole plants of wild-type, MTB-RNAi-2#, MTB-RNAi-8#, MTB1-OE-5#, and MTB1-OE-6# genotypes. Data represent means ± sd (n = 15). Each symbol represents the weight of an individual larva.
(G) Expression of PR-STH2 in wild-type, MTB-RNAi-2#, MTB-RNAi-8#, MTB1-OE-5#, and MTB1-OE-6# plants treated with B. cinerea suspensions. Five-week-old plants were spotted with a 5-μL spore suspension (106 spores/mL). Leaves were harvested 24 h after inoculation for RNA extraction and RT-qPCR analysis.
(H) Growth of B. cinerea in wild-type, MTB-RNAi-2#, MTB-RNAi-8#, MTB1-OE-5#, and MTB1-OE-6# plants. Detached leaves from 5-week-old tomato plants were spotted with a 5-μL spore suspension (106 spores/mL). The lesion areas were analyzed at 3 d after inoculation. Data represent means ± sd (n = 9).
(I) Expression of PR1b in wild-type, MTB-RNAi-2#, MTB-RNAi-8#, MTB1-OE-5#, and MTB1-OE-6# plants infected with Pst DC3000. Five-week-old plants were vacuum infiltrated with Pst DC3000 (0.5 × 10−5 cfu/mL). Leaves were harvested 24 h after inoculation for RNA extraction and RT-qPCR analysis.
(J) Growth of Pst DC3000 in wild-type, MTB-RNAi-2#, MTB-RNAi-8#, MTB1-OE-5#, and MTB1-OE-6# plants. Data represent means ± sd (n = 6).
For (A), (B), (C), (E), (G), and (I), data represent means ± sd (n = 3). For all panels, asterisks indicate significant differences from the wild type according to Student’s t test at *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (Supplemental Data Set 2).
In the context that JA plays a critical role in regulating the plant wound response (Schilmiller and Howe, 2005; Sun et al., 2011; Campos et al., 2014; Zhai et al., 2017b; Howe et al., 2018), we reasoned that MTB proteins might attenuate the wound response by negatively regulating the JA signaling. Indeed, MeJA-induced expression levels of TomLoxD, JA2L, TD, and PI-II were increased in MTB-RNAi plants and decreased in MTB1-OE plants compared with wild-type plants (Figure 3B), confirming that MTB proteins negatively regulate MeJA-induced expression of these defense genes.
In line with previous observations (Li et al., 2004; Chen et al., 2011; Qi et al., 2015), we found that exogenous application of JA to wild-type seedlings led to anthocyanin accumulation (Figure 3C) and root growth inhibition (Figure 3D) in a dose-dependent manner. Notably, JA-induced anthocyanin accumulation was significantly increased in MTB-RNAi plants and significantly decreased in MTB1-OE plants compared with wild-type plants (Figure 3C). Similarly, JA-induced root growth inhibition was significantly enhanced in MTB-RNAi plants and significantly attenuated in MTB1-OE plants compared with wild-type plants (Figure 3D). Collectively, these results support that MTB proteins negatively regulate diverse aspects of JA responses.
To explore whether MTB proteins play a role in plant defense responses to herbivorous insects, we examined the expression pattern of MTB1 to MTB3 in response to insect attack. For these experiments, 18-d-old wild-type seedlings were prechallenged with newly hatched cotton bollworm (Helicoverpa armigera) larvae for 1 h; afterwards, seedlings were either continuously challenged for another 1 h or relieved of challenging by removing the insects from plants. RT-qPCR assays revealed that the expression of MTB1 to MTB3 was low at steady state and was induced to high levels at 1 h upon larvae challenge (Supplemental Figure 6A). Interestingly, the expression of the MTB genes was retained at similar levels when seedlings were continuously challenged by the larvae for another 1 h or not ( Supplemental Figure 6A).
Next, we examined the performance of MTB-RNAi and MTB1-OE plants to herbivorous insects. For these experiments, 5-week-old plants of each genotype and their wild-type counterparts were challenged with larvae. After the termination of the feeding trial, the expression of defense-related gene PI-II was measured in the remaining leaf tissues of damaged plants. Results showed that the expression of PI-II was significantly higher in herbivore-damaged MTB-RNAi leaves and markedly reduced in herbivore-damaged MTB1-OE leaves compared with herbivore-damaged wild-type leaves (Figure 3E). In turn, the average weight of larvae reared on MTB-RNAi plants was significantly lower and that of larvae reared on MTB1-OE plants was 2.0-fold greater than that of larvae reared on wild-type plants (Figure 3F). These results demonstrate that MTB proteins negatively regulate plant defense responses against chewing insects.
We have recently shown that MYC2-dependent JA-signaling plays a critical role in regulating plant resistance to the necrotrophic pathogen Botrytis cinerea (Du et al., 2017). We found that the expression of MTB1 to MTB3 was induced by B. cinerea infection (Supplemental Figure 6B). To test that MTB proteins may play a role in plant resistance to this pathogen, leaves of MTB-RNAi and MTB1-OE plants were inoculated with B. cinerea spore suspensions for 24 h and the expression of pathogen-related marker gene PR-STH2 (Marineau et al., 1987; Matton and Brisson, 1989; Despres et al., 1995; Du et al., 2017) was examined. The expression of PR-STH2 was significantly increased in B. cinerea-infected MTB-RNAi leaves and markedly reduced in B. cinerea-infected MTB1-OE leaves compared with that in the wild type (Figure 3G). Consistently, MTB-RNAi plants exhibited increased resistance against B. cinerea compared with wild-type plants, whereas MTB1-OE plants displayed more severe disease symptoms compared with wild-type plants, as revealed by the size of the necrotic lesions (Figure 3H). These results demonstrate that MTB proteins negatively regulate plant resistance against B. cinerea infection.
Previous studies revealed that JA induces plant susceptibility to the bacterium strain Pseudomonas syringae pv tomato (Pst) DC3000 by antagonizing salicylic acid (SA)-mediated plant defense responses (Kloek et al., 2001; Zhao et al., 2003; Brooks et al., 2005; Pieterse et al., 2012). The negative roles of MTB proteins in JA signaling prompted us to test whether MTB proteins are involved in the Pst DC3000-induced plant defense. RT-qPCR assays revealed that MTB genes were induced by Pst DC3000 infection (Supplemental Figure 6C). We examined the response of MTB-RNAi plants and MTB1-OE plants to infection by Pst DC3000. As shown in Figure 3I, Pst DC3000-induced expression of PR1b (Zhao et al., 2003) showed a dramatic reduction in MTB-RNAi plants and a slight yet significant increase in MTB1-OE plants compared with wild-type plants. Consistent with this pattern, MTB-RNAi plants were more susceptible than the wild type to Pst DC3000 infection, whereas MTB1-OE plants were more resistant than the wild type to Pst DC3000 infection, as revealed by pathogen growth (Figure 3J). These results suggest that MTB proteins play a positive role in plant resistance against Pst DC3000 infection.
Taken together, our results support that, in contrast to MYC2, which positively regulates JA signaling, MTB proteins play a negative role in regulating JA signaling.
MTB Proteins Interact with Most Tomato JAZ Proteins via a Conserved JID
The negative regulation of JA-mediated defense responses by MTB proteins prompted us to compare the structural domains of MTB proteins with that of MYC2, which positively regulates JA-mediated defense responses in tomato (Du et al., 2017). Sequence comparisons revealed that MTB proteins contained a conserved JID (Fernández-Calvo et al., 2011), and key amino acids required for interacting with JAZ proteins (Zhang et al., 2015) were largely conserved among MTB proteins, MYC2, and the Arabidopsis homologs AtMYC2 and AtMYC3 (Supplemental Figure 7A). Consistent with these data, Y2H assays revealed that MTB1 interacted with 10 of the 11 JAZ proteins encoded by the tomato genome, with JAZ9 being the only exception; this interaction between MTB1 and JAZ proteins occurred via the N-terminal fragment of MTB1 containing JID and TAD (Supplemental Figure 7B). These results are in line with the observations that the Arabidopsis JAM proteins are targets of JAZ repressors (Song et al., 2013; Fonseca et al., 2014). The interaction between MTB1 and JAZ proteins was further validated in pull-down experiments using transgenic plants expressing c-myc-tagged MTB1 (MTB1-myc) protein (Supplemental Figure 8) and purified epitope-tagged JAZ proteins. Ten MBP-JAZ fusions or GST-JAZ fusions, but not GST-JAZ9, pulled down MTB1-myc (Supplemental Figure 7C). Thus, the interaction of MTB proteins with most of the JAZ proteins in tomato suggests that, like MYC2 (Du et al., 2017), these bHLH proteins act at a high hierarchical position in the JA signaling pathway (i.e., immediately downstream of the COI1-JAZ coreceptor complex).
MTB Proteins Lack a Canonical MED25-Interacting Domain and Repress TomLoxD Transcription
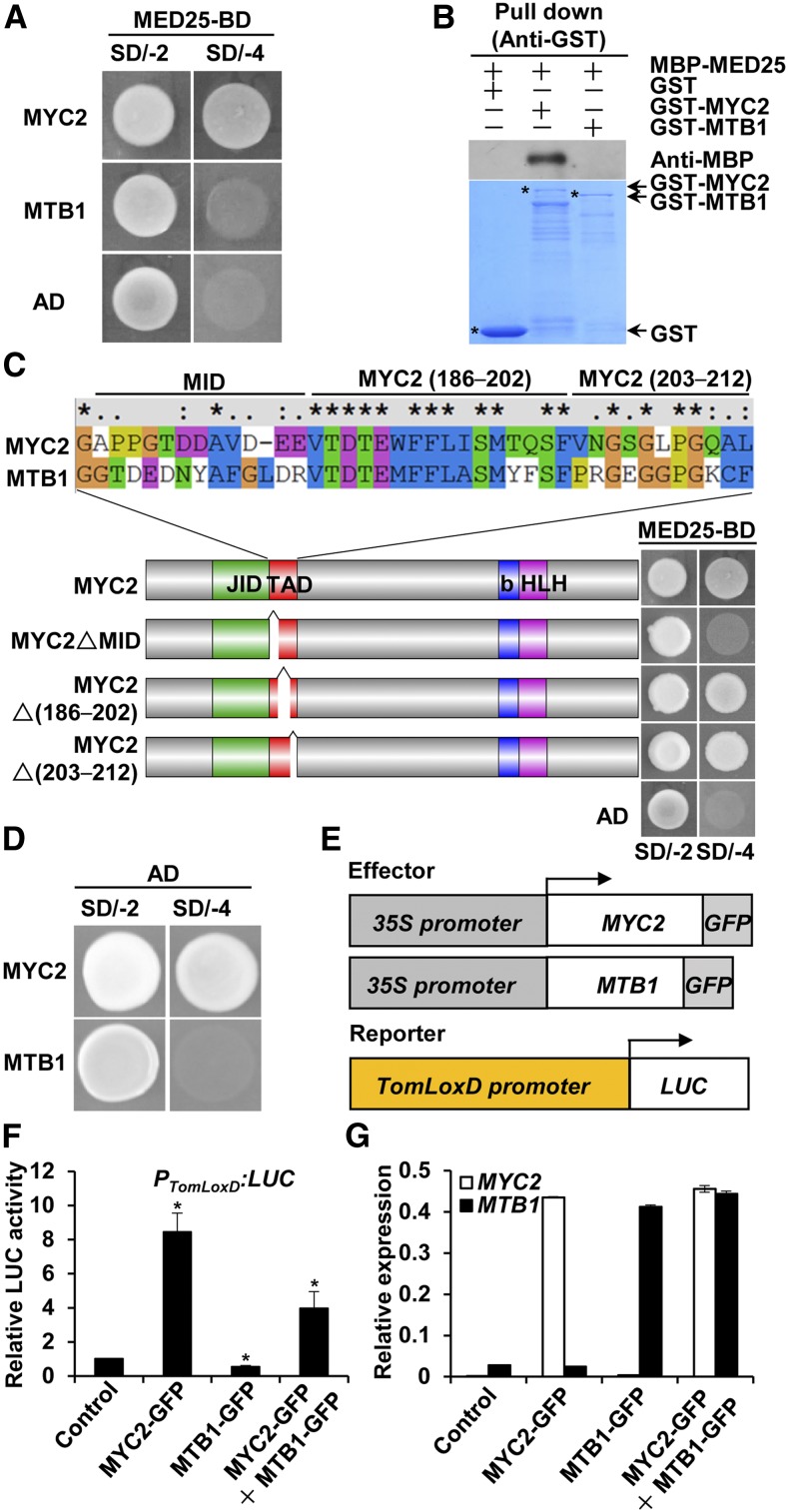
Amino acid sequence comparisons of MTB1 to MTB3, MYC2, and its Arabidopsis homologs AtMYC2 and AtMYC3 revealed sequence variation in the putative TAD of MTB proteins (Supplemental Figure 7A). Previously, TAD of MYC2 was shown to interact with MED25 in Arabidopsis (Chen et al., 2012; An et al., 2017). In Y2H assays, MYC2, but not MTB1, interacted with MED25 (Figures 1A and 4A). Consistent with these results, in an in vitro pull-down assay, recombinant GST-MYC2, but not GST-MTB1, pulled down MBP-MED25 (Figure 4B). Together, these observations indicated that the MED25-interacting domain (MID) of MYC2 must be localized within its TAD. Domain mapping with Y2H assays revealed that the deletion of a 12-amino acid fragment from the N terminus of MYC2 TAD abolished MED25-MYC2 interaction (Figure 4C), indicating that this 12-amino acid fragment represents the MID of MYC2 (Figure 4C). Notably, the MID-corresponding region in MTB1 (MTB1132–144) exhibits a high degree of sequence variation compared with the MID of MYC2 (Figure 4C), referred to here as altered MID (AMID). Notably, sequence alignment indicated that the MID is generally conserved in MYC2-like proteins and that the AMID is conserved in MTB-like proteins in different plant species (Supplemental Figure 3B).
Figure 4.
MTB Proteins Lack a Canonical MID and Act as Transcriptional Repressors.
(A) Y2H assays of the interaction of MED25 with MYC2 and MTB1. Full-length coding sequence (CDS) of MED25 was fused with the BD in pGBKT7, and full-length CDS of MYC2 or MTB1 was fused with the AD in pGADT7. Transformed yeast cells were grown on SD/-4 to determine protein-protein interactions.
(B) In vitro pull-down assays of the interaction of MED25 with MYC2 and MTB1. Purified MBP-MED25 was incubated with GST, GST-MYC2, or GST-MTB1 for the GST pull-down assay and detected by immunoblotting using anti-MBP antibody. The positions of various purified proteins separated by SDS-PAGE are marked with asterisks on the CBB-stained gel.
(C) Y2H assays of MYC2 interaction with MED25 through MID. According to the sequence alignment of the TAD of MYC2 and MTB1, MYC2 TAD was divided into three fragments labeled as MID, MYC2 (186–202), and MYC2 (203–212). Full-length MYC2 CDS and MYC2 variants lacking the MID, MYC2 (186–202), and MYC2 (203–212) fragments were fused with the AD in pGADT7, and full-length MED25 CDS was fused with the BD in pGBKT7. Transformed yeast cells were grown on SD/-4 medium to determine protein-protein interactions. Cotransformation of the AD vector with MED25 was used as a negative control.
(D) Yeast assays used to detect the transcriptional activity of MYC2 and MTB1. Yeast cells cotransformed with BD-MYC2 (or BD-MTB1) and AD vector were grown on SD/-4 medium.
(E) to (G) Transient expression assays to detect the effect of MTB1 on the transcriptional activity of MYC2.
(E) Schematic representations of effector constructs and LUC reporter construct used in transient expression assays.
(F) Quantification of luminescence intensity of LUC in N. benthamiana leaves 72 h after coinfiltration with PTomLoxD:LUC reporter construct and effector constructs or infiltration of only the PTomLoxD:LUC reporter construct (control).
(G) RT-qPCR analysis of MYC2 and MTB1 expression in N. benthamiana leaves infiltrated with the indicated constructs.
For (F) and (G), data represent means ± sd (n = 5). Statistically significant differences between control and test samples were determined using Student’s t test and are indicated using asterisks (*, P < 0.05; Supplemental Data Set 2).
Notably, deletion of MID abolished the interaction of MYC2 with all of the JAZ proteins in Y2H assays (Supplemental Figure 9A). Protein gel analysis indicated that MYC2△MID and all JAZ fusion proteins were expressed in yeast cells (Supplemental Figure 9B), indicating that the absence of interaction between MYC2△MID and JAZ proteins cannot be due to a lack of protein expression. These results support that the MID is also involved in MYC2-JAZ interaction. In parallel Y2H experiments, we found that deletion of the AMID also abolished the interaction of MTB1 with JAZ proteins (Supplemental Figures 9C and 9E), and protein gel analysis indicated that MTB1, MTB1△AMID, and all JAZ fusion proteins were expressed in yeast cells (Supplemental Figures 9D and 9F), indicating that the absence of interaction between MTB1△AMID and JAZ proteins cannot be due to a lack of protein expression. These results support that the AMID is involved in MTB1-JAZ interaction.
Because MTB proteins lacked a canonical MID and negatively regulated JA signaling, we hypothesized that these bHLH proteins function as transcriptional repressors of JA-responsive genes. To test this hypothesis, we used a yeast assay (Zhai et al., 2013) to examine the transcriptional activation capability of MTB1 and MYC2. Results showed that MYC2 functioned as a strong transcriptional activator, whereas MTB1 did not (Figure 4D).
Next, we examined the effect of MTB1 and MYC2 on transient expression of the firefly luciferase (LUC) gene driven by the JA-responsive TomLoxD promoter in tobacco (Nicotiana benthamiana) leaves (Yan et al., 2013). We previously demonstrated that the JA biosynthetic gene TomLoxD is a direct target of MYC2 (Yan et al., 2013; Du et al., 2017). In line with our previous observation (Yan et al., 2013), when the PTomLoxD:LUC reporter was coexpressed with MYC2-GFP in N. benthamiana leaves, LUC activity was greatly increased compared with that in the empty vector control (Figures 4E to 4G). By contrast, coexpression of MTB1-GFP and PTomLoxD:LUC reporter construct resulted in reduced LUC activity (Figures 4E to 4G). When MTB1-GFP and MYC2-GFP were simultaneously coexpressed with the PTomLoxD:LUC reporter, MYC2-mediated enhancement of LUC activity was significantly inhibited (Figures 4E to 4G). Together, these data substantiate that, unlike the transcriptional activator MYC2, MTB1 likely functions as a transcriptional repressor of JA-responsive genes and counteracts the function of MYC2 in regulating these genes.
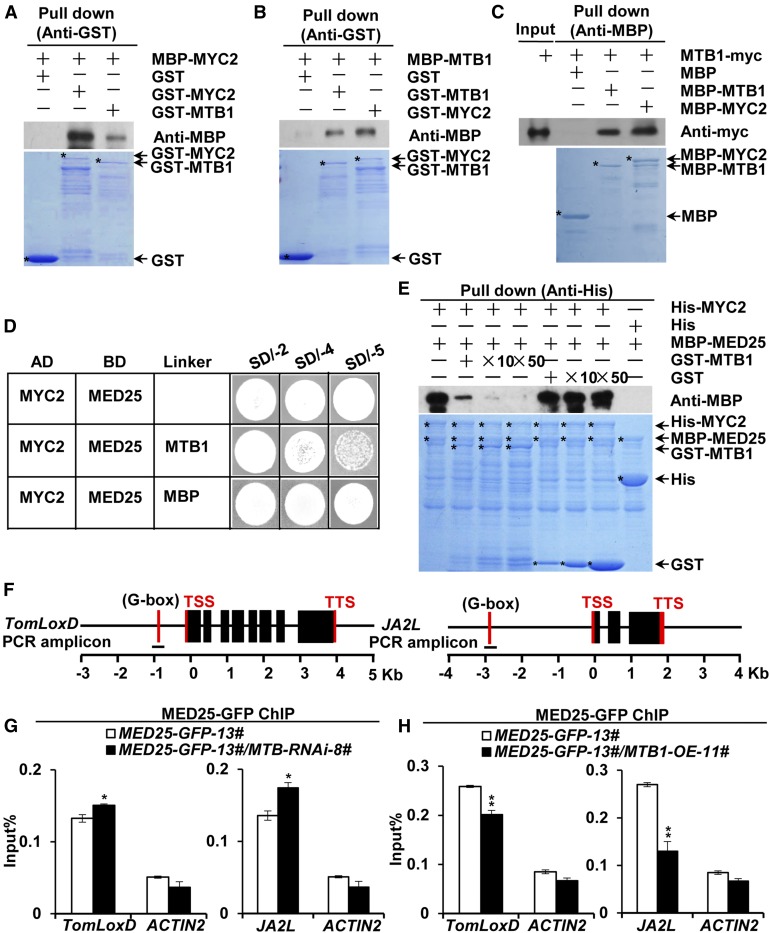
MTB1 Physically Interacts with MYC2 and Interferes with the MED25-MYC2 Interaction
Compared with MYC2 and other bHLH proteins, MTB proteins contain a conserved HLH domain (Supplemental Figure 7A), which is believed to form homo- and heterodimers (Carretero-Paulet et al., 2010; Fernández-Calvo et al., 2011; Goossens et al., 2016). This was evident in our in vitro pull-down assays, in which both GST-MYC2 and GST-MTB1 pulled down recombinant MBP-MYC2 (Figure 5A) as well as recombinant MBP-MTB1 (Figure 5B). These results suggest that, like MYC2, the bHLH protein MTB1 forms homodimers with itself and heterodimers with MYC2 in vitro. Moreover, MTB1-myc fusion proteins from protein extracts of MTB1-myc-11# plants (Supplemental Figure 8) were pulled down by both MBP-MTB1 and MBP-MYC2 (Figure 5C), corroborating that MTB1 forms homodimers with itself and heterodimers with MYC2.
Figure 5.
MTB1 Interacts with MYC2 and Disrupts MED25-MYC2 Interaction.
(A) In vitro pull-down assays of MYC2 formation of homo- and heterodimers. Purified MBP-MYC2 was incubated with GST, GST-MYC2, or GST-MTB1 for the GST pull-down assay and detected by immunoblotting using anti-MBP antibody.
(B) and (C) Pull-down assays of MTB1 formation of homo- and heterodimers. Purified MBP-MTB1 was incubated with GST, GST-MTB1, or GST-MYC2 for the GST pull-down assay and detected by immunoblotting using anti-MBP antibody (B), and protein extracts prepared from MTB1-myc seedlings were incubated with MBP, MBP-MTB1, or MBP-MYC2 for the MBP pull-down assay and detected by immunoblotting using anti-myc antibody (C).
For (A) to (C), positions of various purified proteins separated by SDS-PAGE are indicated with asterisks on CBB-stained gels.
(D) Y3H assays showing that MTB1 interferes with MYC2-MED25 interaction. Yeast cells cotransformed with pGADT7-MYC2 and pBridge-MED25-MTB1/MBP were plated on SD/-4 medium to assess the MYC2-MED25 interaction and on SD/-5 medium to induce MTB1/MBP.
(E) In vitro pull-down assays of MTB1 interference with MED25-MYC2 interaction. Fixed amounts of His-MYC2 and MBP-MED25 fusion proteins were incubated with an increasing amount of GST-MTB1 fusion protein or GST protein. Protein samples were immunoprecipitated with anti-His antibody and immunoblotted with anti-MBP antibody.
(F) Schematic diagrams of PCR amplicons of TomLoxD and JA2L used for ChIP-qPCR. Positions of the transcription start site (TSS) and transcription termination site (TTS) are indicated.
(G) and (H) ChIP-qPCR assays of MTB-RNAi (G) and MTB1-OE (H) impairment of the enrichment of MED25 on the G-boxes of TomLoxD and JA2L promoters upon wounding. MED25-GFP-13# and MED25-GFP-13#/MTB-RNAi-8# plants (G) and MED25-GFP-13# and MED25-GFP-13#/MTB1-OE-11# plants (H) were treated with mechanical wounding for 1 h before cross-linking, and chromatin of each sample was immunoprecipitated using anti-GFP antibody. Immunoprecipitated DNAs were quantified by qPCR. The enrichment of target gene promoters is displayed as a percentage of input DNA. ACTIN2 was used as a nonspecific control. Data represent means ± sd (n = 3). Statistically significant differences between MED25-GFP-13# and other genotypes were determined using Student’s t test and are indicated using asterisks (*, P < 0.05 and **, P < 0.01; Supplemental Data Set 2).
The heterodimerization of MTB1 with MYC2 raised the possibility that MTB1 disrupts the interaction of MYC2 with its coactivator MED25, which plays a critical role in MYC2-regulated transcriptional activation of JA-responsive genes (Chen et al., 2012; An et al., 2017). Y3H assays detected MED25-MYC2 interaction on SD/-4 medium (Figure 5D). However, the induction of MTB1 expression on SD/-5 medium significantly impaired the MED25-MYC2 interaction (Figure 5D); by contrast, the induction of MBP expression on SD/-5 medium showed negligible effects on the MED25-MYC2 interaction (Figure 5D). These results suggest that MTB1 competitively inhibits MED25-MYC2 interaction in yeast cells. These observations were further confirmed by pull-down experiments in which both His-MYC2 and MBP-MED25 fusion proteins were maintained at a constant amount in each sample and the concentration of GST-MTB1 was increased in a gradient. In this assay, the ability of His-MYC2 to pull down MBP-MED25 decreased as the amount of GST-MTB1 increased (Figure 5E); as a control, we showed that the ability of His-MYC2 to pull down MBP-MED25 was not affected as the amount of GST increased (Figure 5E). These results corroborate our hypothesis that MTB1 interferes with MED25-MYC2 interaction.
The interruption of MED25-MYC2 interaction due to MTB1 further suggested the possibility that MTB1 affects the enrichment of MED25 on MYC2 target gene promoters. To test this possibility, we generated several transgenic lines expressing MED25-GFP (Supplemental Figure 10) and subsequently transferred the MED25-GFP transgene into MTB-RNAi and MTB1-OE genetic backgrounds by crossing MED25-GFP-13# plants with both MTB-RNAi-8# and MTB1-OE-11# plants. ChIP-qPCR analysis revealed the enrichment of MED25-GFP on the G-box of TomLoxD and JA2L promoters upon wounding (Figures 5F to 5H). Notably, the enrichment of MED25-GFP on these promoters was significantly increased in MTB-RNAi-8# plants and significantly reduced in MTB1-OE-11# plants compared with wild-type plants (Figures 5F to 5H). These results indicate that MTB1 impairs MYC2-dependent recruitment of MED25 to MYC2 target promoters.
MTB1 Antagonizes MYC2 for DNA Binding
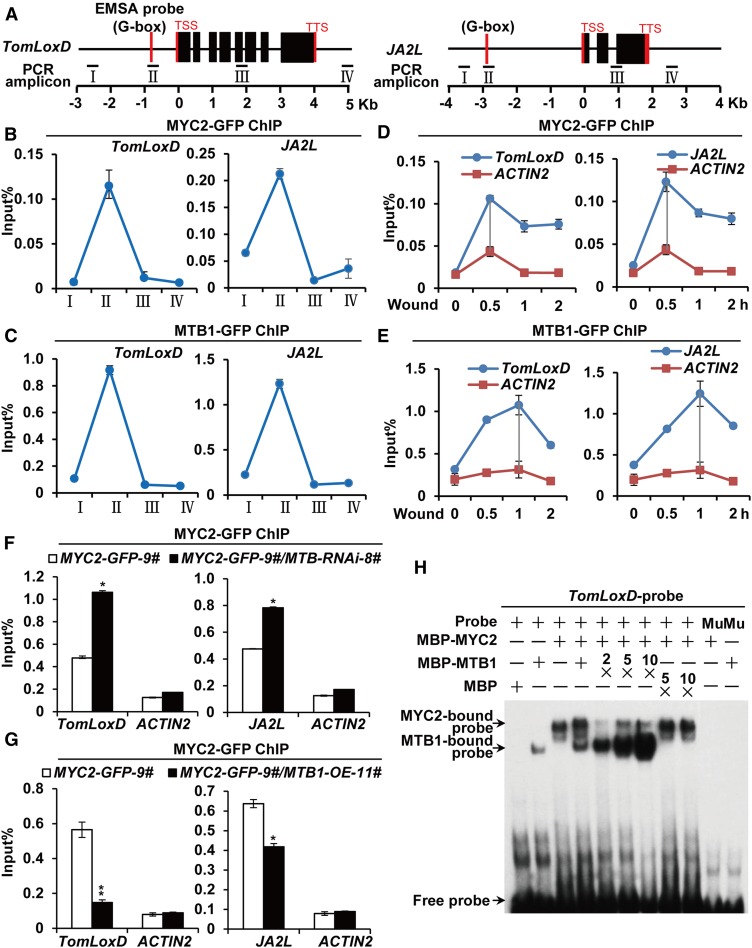
Sequence alignment revealed that the basic domain is highly conserved between MTB proteins and MYC2 as well as other bHLH proteins (Supplemental Figure 7A). Given that the basic domain is involved in DNA binding (Carretero-Paulet et al., 2010; Fernández-Calvo et al., 2011; Goossens et al., 2016), we hypothesized that MTB proteins bind to the same promoter region as MYC2. ChIP-qPCR assays revealed that, similar to MYC2-GFP, MTB1-GFP was preferentially enriched on the G-box motifs of TomLoxD and JA2L promoters instead of on the upstream promoter regions and coding sequences of these genes (Figures 6A to 6C). We then compared the wound-induced binding kinetics of MTB1-GFP and MYC2-GFP with the G-box regions of TomLoxD and JA2L gene promoters. As shown in Figures 6D and 6E, the enrichment of MYC2-GFP on the G-box motifs of TomLoxD and JA2L reached a peak at 0.5 h after wounding, whereas that of MTB1-GFP on the same promoter regions reached a peak at 1 h after wounding, indicating that the enrichment of MTB proteins at its target sites occurs later than that of MYC2.
Figure 6.
MTB1 Antagonizes MYC2 for DNA Binding.
(A) Schematic representations of TomLoxD and JA2L showing the amplicons and probe used for ChIP-qPCR assay and EMSA. Positions of the transcription start site (TSS) and transcription termination site (TTS) are indicated.
(B) and (C) ChIP-qPCR analysis of the enrichment of MYC2 (B) and MTB1 (C) on the chromatin of TomLoxD and JA2L in MYC2-GFP-9# and MTB1-GFP-5# plants, respectively.
(D) and (E) ChIP-qPCR analysis of the enrichment of MYC2 (D) and MTB1 (E) on the G-box regions of TomLoxD and JA2L in MYC2-GFP-9# and MTB1-GFP-5# plants, respectively, upon wounding. Plants were treated with mechanical wounding for the indicated times before cross-linking.
(F) and (G) ChIP-qPCR assays of the impaired enrichment of MYC2 on the G-boxes of TomLoxD and JA2L due to MTB-RNAi in MYC2-GFP-9# and MYC2-GFP-9#/MTB-RNAi-8# plants (F) and MTB1-OE in MYC2-GFP-9# and MYC2-GFP-9#/MTB1-OE-11# plants (G) upon wounding.
Plants were treated with mechanical wounding for 1 h before cross-linking. For (B) to (G), chromatin of each sample was immunoprecipitated using anti-GFP antibody and quantified by qPCR. The enrichment of target gene promoters is displayed as a percentage of input DNA. Data represent means ± sd (n = 3). For (D) to (G), ACTIN2 was used as a nonspecific control. For (F) and (G), asterisks indicate significant differences detected using Student’s t test (*, P < 0.05 and **, P < 0.01; Supplemental Data Set 2) when compared with MYC2-GFP.
(H) EMSA showing that MBP-MTB1 interferes with MBP-MYC2 to bind to DNA probes from the TomLoxD promoter in vitro. Biotin-labeled probes were incubated with a fixed amount of MBP-MYC2 and an increasing amount of MBP-MTB1 or MBP, and the free and bound DNAs were separated on an acrylamide gel. The MBP protein was incubated with the labeled probe to serve as a negative control; mutated probes were used as a negative control. Mu, mutated probe in which the G-box motif 5ʹ-ACCATGTG-3ʹ was deleted.
To test whether MTB proteins antagonize the binding of MYC2 to its target promoters, we transferred the MYC2-GFP transgene into the backgrounds of MTB-RNAi-8# and MTB1-OE-11# through crossing. The results of ChIP-qPCR assays revealed that, upon wounding, the enrichment of MYC2-GFP on the G-box of TomLoxD and JA2L was significantly increased in MTB-RNAi-8# plants compared with wild-type plants (Figure 6F) but significantly decreased in MTB1-OE-11# plants compared with wild-type plants (Figure 6G). We then performed EMSAs to examine the effect of MTB1 on the binding of MYC2 to a DNA probe containing the G-box motif of the TomLoxD promoter (Yan et al., 2013; Du et al., 2017). In these experiments, fixed amounts of MBP-MYC2 recombinant protein and the DNA probe were added to an increasing amount of MBP-MTB1 recombinant protein. As shown in Figure 6H, the amount of MBP-MYC2-bound DNA probe decreased as the amount of MBP-MTB1 fusion protein increased, revealing a competition effect of MBP-MTB1 on the DNA binding ability of MBP-MYC2. As a control, we showed that MBP protein did not influence the DNA binding ability of MBP-MYC2 (Figure 6H). Taken together, these data demonstrate that MTB proteins antagonize MYC2 by binding to its target gene promoters.
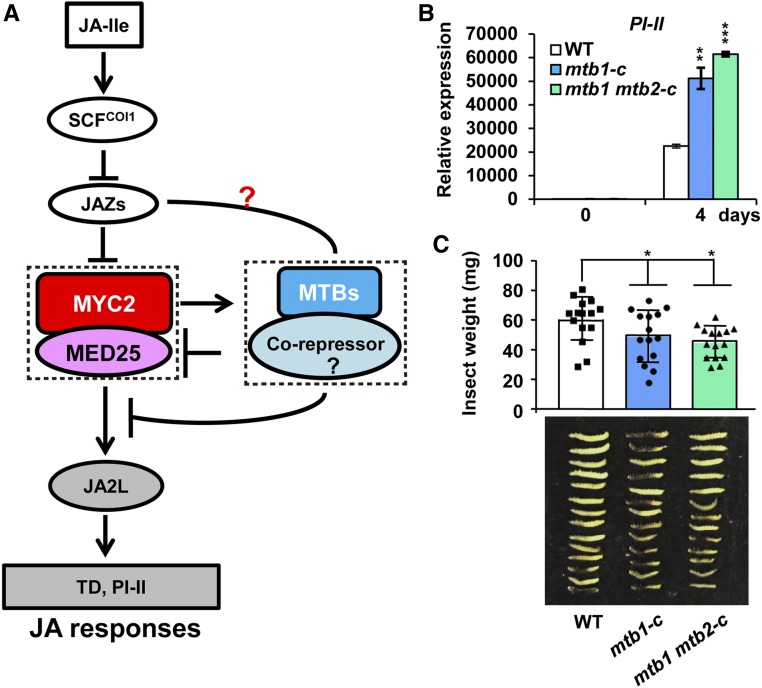
MTB Genes Represent a Potential Crop Protection Tool
Our results showed that MYC2 and its direct transcriptional targets, MTB proteins, form a feedback loop to terminate the JA responses (Figure 7A). Considering that MTB proteins negatively regulate JA signaling, we reasoned that the manipulation of MTB genes via gene editing would provide an effective tool for crop protection against insect attack in an environmentally friendly manner. We used the CRISPR/Cas9 gene editing system (Deng et al., 2018) to generate mtb1-c single mutant and mtb1 mtb2-c double mutant plants (Supplemental Figures 11A to 11D). Sequence analyses indicated that mtb1-c carries a 5-bp deletion in the MTB1 open reading frame (ORF), which leads to frame shift and the generation of a premature stop codon TAA (Supplemental Figures 11A and 11B). In the mtb1 mtb2-c double mutant, the MTB1 ORF contains the same mutation as that in mtb1-c (Supplemental Figures 11A and 11B), and the MTB2 ORF contains a T insertion at nucleotide 490, which also leads to frame shift and the generation of a premature stop codon TAA (Supplemental Figures 11C and 11D). RT-qPCR assays indicated that the MTB1 and/or MTB2 transcript levels do not show obvious change in mtb1-c or mtb1 mtb2-c compared with the wild type (Supplemental Figure 11E). However, protein gel analysis using anti-MTB1 antibodies failed to detect MTB1 protein accumulation in the mtb1-c single mutant and the mtb1 mtb2-c double mutant (Supplemental Figure 11F).
Figure 7.
MTB Genes Represent a Tool for Crop Protection.
(A) Proposed mechanism by which MYC2 and MTB proteins form a feedback loop to terminate JA signaling. MYC2 and MED25 activate the expression of MTB genes; in turn, MTB proteins interfere with the MED25-MYC2 interaction and DNA binding activity of MYC2 to terminate JA-triggered defense responses.
(B) Expression of PI-II in wild-type, mtb1-c, and mtb1 mtb2-c plants exposed to H. armigera larvae. Plants were harvested at the indicated time points during the feeding trial for RNA extraction and RT-qPCR analysis. Data represent means ± sd (n = 3). Asterisks indicate significant differences from the wild type according to Student’s t test at **, P < 0.01; and ***, P < 0.001
(C) Average weight (top) and images (bottom) of larvae recovered at the end of day 4 of the feeding trial using whole plants of the wild type, mtb1-c, and mtb1 mtb2-c. Data represent means ± sd (n = 15). Each symbol denotes the weight of an individual larva. Asterisks indicate significant differences from the wild type according to Student’s t test at *, P < 0.05 (Supplemental Data Set 2).
Five-week-old mtb1-c, mtb1 mtb2-c, and wild-type plants were challenged with newly hatched H. armigera larvae. After terminating the feeding trial, the expression of defense-related gene PI-II was measured in the remaining leaf tissues of damaged plants. The expression of PI-II was significantly higher in herbivore-damaged mtb1-c and mtb1 mtb2-c leaves compared with herbivore-damaged wild-type leaves (Figure 7B). Consistent with this, the average weight of larvae reared on mtb1-c and mtb1 mtb2-c plants was significantly lower than that of larvae reared on wild-type plants (Figure 7C), indicating that these mutants are more resistant to herbivore attack than wild-type plants. Apart from resistance to insect attack, mtb1-c and mtb1 mtb2-c plants did not exhibit visible differences from the wild-type plants in terms of overall plant growth, fertility, and fruit set (Supplemental Figure 12). These results suggest that MTB genes have a great potential for application in crop protection.
DISCUSSION
Although it is well recognized that the JA signaling pathway is subject to multiple layers of regulation to accurately control the amplitude, duration, and timing of defense- and growth-related responses, an understanding of the underlying mechanism(s) has been lacking. Here, we describe a highly organized feedback circuit that controls the accurate termination of JA signaling in tomato. This study not only provides mechanistic insights into the broader role of JA in controlling physiological tradeoffs but also promises to open new avenues for the application of these basic insights in the field of crop protection.
MYC2 and MTB Proteins Form an Autoregulatory Feedback Loop to Terminate JA Signaling
We provide evidence that JA-induced expression of the three MTB genes (MTB1 to MTB3) depends on the MYC2-MED25 transcriptional activation complex. First, wound-induced expression of these MTB genes was delayed compared with that of MYC2. Second, wound-induced expression of MTB genes was impaired in MYC2- or MED25-knockdown plants. Third, results of ChIP-qPCR analysis and EMSA indicated that MYC2 directly bound the MTB promoters. These results demonstrated that the wound-induced expression of MTB genes was directly controlled by the MYC2-MED25 transcriptional activation complex. Considering that the MYC2-MED25 complex plays a central role in the initiation and amplification of JA-mediated transcriptional responses (Chen et al., 2012; An et al., 2017; Du et al., 2017), our results suggest that the activation of MTB genes is a default mechanism that inactivates the MYC2-MED25 transcriptional activation complex. In other words, the formation of the MYC2-MTB feedback loop is already preprogrammed during the induction phase of JA signaling. Thus, in addition to controlling the initiation and amplification of JA-mediated transcriptional responses, the MYC2-MED25 transcriptional activation complex also executes intrinsic termination of JA-mediated defense responses. Therefore, MYC2 and MTB proteins form an autoregulatory feedback loop, which temporally and spatially terminates JA signaling.
Considering that MTB1 to MTB3 operate as a termination step of the JA signaling, it is reasonable to speculate that their induction may only be seen when the invading insects are removed from the plant. To test this, we designed experiments to examine whether the induction of MTB1 to MTB3 is absent when plants are continuously challenged with insects. Contrary to this speculation, our insect feeding experiments indicated that the MTB1 to MTB3 induction is still present in conditions of continuous insect feeding (Supplemental Figure 6A). These, together with the fact that the induction of MTB1 to MTB3 depends on the MYC2-MED25 transcriptional activation complex, reinforce our scenario that the operation of these termination regulators is already preprogrammed during the induction phase of JA signaling.
It is generally believed that, compared with insect attack or mechanical wounding, pathogen infection could be more continuous and durable. Further support to our hypothesis that the operation of MTB1 to MTB3 is a preprogrammed process came from our physiological assays with different pathogens. Our results indicated that MTB1 to MTB3 were induced by B. cinerea infection and they played a negative role in plant resistance to this necrotrophic pathogen, which is controlled by JA-inducible defenses.
In addition, MTB1 to MTB3 were also induced by Pst DC3000 infection and they played a positive role in plant resistance to this hemibiotrophic pathogen, which is controlled by SA-inducible defenses. In the context that Pst DC3000 infection leads to increased accumulation of endogenous JA (Spoel et al., 2003; Van der Does et al., 2013) and that this pathogen activates the JA signaling by producing the JA-Ile mimic coronatine (reviewed in Pieterse et al., 2012; Zhang et al., 2017b), it is easy to understand that this hemibiotrophic invader also induces MTB1 to MTB3 expression. Considering that the antagonism between SA and JA plays a central role in the modulation of the plant immune signaling network (Kloek et al., 2001; Zhao et al., 2003; Brooks et al., 2005; Pieterse et al., 2012; Zhang et al., 2017b), it is reasonable to speculate that MTB1 to MTB3 may play a role in SA-mediated suppression of JA signaling. In this context, our finding that MTB1 to MTB3 are direct transcriptional targets of MYC2 is consistent with the observation that the SA-mediated inhibition of the JA pathway is executed downstream of the SCFCOI1-JAZ coreceptor complex through targeting JA-responsive transcription factors (Van der Does et al., 2013).
Notably, the wound-triggered induction of MTB genes was delayed relative to that of MYC2, indicating that the negative feedback regulation by MTB proteins is temporally delayed. Characteristic delays are also observed in other negative feedback loops, including the de novo synthesis of stabilized JAZ repressors (Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2010; Moreno et al., 2013) and induction of JA-Ile catabolic pathways (Miersch et al., 2008; Kitaoka et al., 2011; Koo et al., 2011; VanDoorn et al., 2011; Heitz et al., 2012). We speculate that the delayed termination of JA signaling ensures the complete execution of an already-initiated JA response. In future studies, it is critical to determine whether the MYC2-MTB circuit operates synergistically with or independent of the induction of stabilized JAZ repressors or JA-Ile catabolic pathways to deactivate JA-mediated defense responses.
MTB Proteins Impinge Their Effect on the MYC2-MED25 Transcriptional Activation Complex
Like the master transcriptional activator MYC2, MTB1 physically interacted with most of the JAZ proteins in tomato via the conserved JID, implying that MTB proteins act at a high hierarchical level (i.e., immediately downstream of the hormone coreceptor complex) to deactivate JA signaling. Our results revealed two mechanisms by which MTB proteins imposed their negative effect on the MYC2-MED25 transcriptional activation complex. First, MTB1 physically interacted with MYC2 and competitively inhibited the interaction of MYC2 with its coactivator MED25; because the MYC2-MED25 complex is essential for MYC2-dependent preinitiation complex formation, the interference of MTB1 determines the transcriptional output of the MYC2-MED25 transcriptional activation complex (Chen et al., 2012; An et al., 2017; Du et al., 2017). In this regard, the action mechanism of MTB proteins is different from the Arabidopsis JAM proteins, which did not show physical interaction with AtMYC2 in Y2H and coimmunoprecipitation assays (Song et al., 2013; Fonseca et al., 2014). Second, MTB1 directly bound the G-box motifs of MYC2 target genes via its conserved basic domain and competitively inhibited the binding of MYC2 to its target gene promoters.
Arabidopsis MYC2 forms a homotetramer, and the tetramerization of AtMYC2 enhances its DNA binding and transcriptional activation capacity (Lian et al., 2017). Taking these data into consideration, it is reasonable to speculate that MTB repressors might also form homotetramers, which would further inhibit both the DNA binding and transcriptional activation capacity of MYC2. Consistent with this speculation, our sequence analysis revealed that several key residues of the AtMYC2 tetramer interface (Lian et al., 2017) were conserved in SlMYC2 as well as all three SlMTB proteins (Supplemental Figure 5A). Further structural investigations should reveal key insights into the mechanisms by which MTB transcriptional repressors counteract the function of MYC transcriptional activators in regulating JA-responsive gene transcription.
Importantly, our results revealed that the MID couples the MYC2 interactions with both its coactivator MED25 and its repressor JAZ proteins. This finding, which is consistent with our recent observation that MED25 and JAZ1 form a ternary complex with MYC2 and thereby fine-tune the transcriptional output of the JA signaling in Arabidopsis (An et al., 2017), supports a scenario that the MID of MYC2 is evolved to keep the activation and repression of this master transcriptional regulator in check. Consistent with their negative role in regulating JA signaling, MTB1 to MTB3 do not harbor a canonical MID, but rather harbor an AMID, and therefore fail to physically interact with the MED25 coactivator. Intriguingly, we found that, as the MID plays a critical role for the MYC2-JAZ interaction, the AMID also plays a critical role for the MTB1-JAZ interaction. We predict that these findings will stimulate future research to elucidate the functional relevance of “repressor-repressor interactions” in JA signaling or other signal transduction pathways. In the context that both MTB genes and JAZ genes are induced by wounding and JA, it is possible that JAZ proteins might “help” MTB proteins to repress gene expression and to terminate JA signaling. At this stage, however, we could not rule out an opposite possibility that MTB-JAZ interaction might result in a positive effect on JA signaling (i.e., repressing a repressor may lead to a positive effect). Anyway, we believe that our findings provide a starting point to address these possibilities. For example, we could manipulate the AMID and test the resulting effects on the function of MTB1. Another interesting direction for future exploration is to identify the cofactors (i.e., corepressors) that are involved in the action of MTB proteins and to test whether the AMID plays an important role for the interaction of MTB proteins with these cofactors.
METHODS
Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum) cv M82 was used as the wild type for all plant materials except for jai1, mtb1-c, and mtb1 mtb2-c. For jai1, cv Castlemart was used as the wild type, and for mtb1-c and mtb1 mtb2-c, cv Ailsa Craig was used as the wild type. The following tomato genotypes were used in this study: MED25-AS, MED25-GFP, MYC2-OE (MYC2-GFP; Du et al., 2017), MYC2-RNAi (Du et al., 2017), MTB1-OE (MTB1-GFP or MTB1-myc), MTB-RNAi, mtb1-c, and mtb1mtb2-c. The MYC2-GFP and MED25-GFP transgenes were introduced into the MTB-RNAi and MTB1-OE backgrounds via crossing. Homozygous plants were selected by genotyping. The original jai1 mutant, which is in the genetic background of cv Micro-Tom (Li et al., 2004), was backcrossed into the cv Castlemart background through at least five generations. Homozygous jai1 plants were identified as described previously (Li et al., 2004). Tomato seeds were placed on moistened filter paper for 48 h for germination. Tomato seedlings were transferred to growth chambers and maintained under a long-day photoperiod (16 h of light/8 h of dark) with a white light intensity of 200 mmol photons m−2 s−1 at 25°C during the subjective day and at 18°C during the subjective night. Nicotiana benthamiana plants were grown at 28°C under a long-day photoperiod.
Plasmid Construction and Plant Transformation
To generate the MED25-AS construct, the coding sequence (CDS) of MED25 was PCR amplified, cloned into pENTR in the reverse orientation using a pENTR Directional TOPO Cloning Kit (Invitrogen), and recombined with the binary vector PGWB2 (35S promoter). To generate the 35Spro:MED25-GFP construct, MED25 CDS was cloned into pENTR and recombined with the binary vector PGWB5 (35S promoter, c-GFP). To generate the 35Spro:MTB1-GFP construct, MTB1 CDS was PCR amplified, cloned into pENTR, and recombined with the binary vector PGWB5 (35S promoter, c-GFP). To generate the 35Spro:MTB1-myc construct, pENTR-MTB1 was recombined with the binary vector PGWB17 (35S promoter, c-4myc). To generate the MTB-RNAi construct, fragments of the ORFs of MTB1 (1401–1600 bp), MTB2 (81–280 bp), and MTB3 (601–800 bp) were PCR amplified and fused to generate a 658-bp PCR product containing the attB1 and attB2 sites. The fused PCR product was cloned into pHellsgate2 (Invitrogen). All DNA constructs were generated following standard molecular biology protocols and using Gateway (Invitrogen) technology (Nakagawa et al., 2007). Primers used for plasmid construction are listed in Supplemental Table. All constructs were introduced into tomato cv M82 via Agrobacterium (Agrobacterium tumefaciens)-mediated transformation (Du et al., 2014). Transformants were selected based on their resistance to hygromycin B or kanamycin. Homozygous T2 or T3 transgenic plants were used for phenotypic and molecular characterization.
Generation of mtb1-c and mtb1 mtb2-c Using CRISPR/Cas9 Technology
A 19-bp fragment of the MTB1 CDS (240–258 bp) was used as the targeting sequence for genome editing of MTB1. To synthesize two target guide RNA (gRNA) sequences, PCR was performed using forward and reverse primers containing the gRNAs (Supplemental Table) and pHSE401 vector as the template. The tomatoU6-26-MTB1-gRNA cassette and the CRISPR/Cas9 binary vector pCBC-DT1T2_tomatoU6 were digested with BsaI, and the cassette was cloned into the binary vector to generate pCBC-DT1T2_tomatoU6-MTB1. To generate CRISPR/Cas9 construct carrying two gRNAs targeting MTB1 and MTB2, 19-bp fragments of each CDS (MTB1, 240–258 bp; MTB2, 473–491 bp) were used as target gRNAs. The pCBC-DT1T2_tomatoU6-MTB1MTB2 construct was generated in the same way as the pCBC-DT1T2_tomatoU6-MTB1 construct. The final binary vectors were introduced into tomato cv Ailsa Craig via Agrobacterium-mediated transformation (Du et al., 2014). CRISPR/Cas9-induced mutations were genotyped by PCR amplification and DNA sequencing. Primers used for plasmid construction are listed in Supplemental Table. Cas9-free T2 plants carrying mutations were identified for further experiments.
Y2H Assays
Y2H assays were performed using the MATCHMAKER GAL4 Two-Hybrid System (Clontech). To verify the interaction of MED25 with MYC2 and MTB1, MYC2 CDS, MTB1 CDS, and MYC2 derivatives were fused to GAL4 AD in pGADT7, and MED25 CDS was fused to GAL4 BD in pGBKT7. To verify the interaction of JAZ proteins with MYC2 and MTB1, MYC2 CDS, MTB1 CDS, MYC2 derivatives, and MTB1 derivatives were fused to GAL4 AD in pGADT7, and CDSs of JAZ genes were fused to GAL4 BD in pGBKT7. Primers used for plasmid construction are listed in Supplemental Table. Constructs used to test protein-protein interactions were cotransformed into yeast (Saccharomyces cerevisiae) strain AH109. Cotransformation of empty pGBKT7 and pGADT7 vectors was used as a negative control. The presence of transgenes in yeast cells was confirmed by growing these cells on plates containing solid SD medium lacking Leu and Trp (SD/-2). To assess protein-protein interactions, transformed yeast cells were suspended in liquid SD/-2 medium to an OD600 of 1.0. Samples (5 µL) of suspended yeast cells were spread on plates containing SD/-4. To detect protein-protein interactions, plates were examined after 3 d of incubation at 30°C.
Y3H Assays
Y3H assays were performed based on the MATCHMAKER GAL4 Two-Hybrid System (Clontech). To construct pBridge-MED25-JAZ7, MED25 CDS was cloned into the multiple cloning site (MCS) I of pBridge vector (Clontech) fused to the GAL4 BD domain, and JAZ7 CDS was cloned into MCS II of the pBridge vector and expressed as the “bridge” protein only in the absence of Met. Constructs used for testing protein-protein interactions were cotransformed into yeast strain AH109. The presence of transgenes was confirmed by growing the yeast cells on SD/-2 medium. Transformed yeast cells were spread on plates containing SD/-4 medium to assess the MYC2-MED25 interaction without the expression of JAZ7 and on plates containing SD/-5 medium to induce JAZ7 expression. Interactions were observed after 3 d of incubation at 30°C. To construct pBridge-JAZ7-MED25, JAZ7 and MED25 CDSs were cloned into MCS I and MCS II, respectively, of pBridge vector, and MED25 was expressed as the bridge protein to test its effect on MYC2-JAZ7 interaction. To construct pBridge-MED25-MTB1, MED25 and MTB1 CDSs were cloned into MCS I and MCS II, respectively, of pBridge vector, and MTB1 was expressed as the bridge protein to test its effect on the MYC2-MED25 interaction. To construct pBridge-MED25-MBP, MED25 and MBP CDSs were cloned into MCS I and MCS II, respectively, of pBridge vector, and MBP was expressed as the bridge protein to test its effect on the MYC2-MED25 interaction. To construct pBridge-JAZ7-MBP, JAZ7 and MBP CDSs were cloned into MCS I and MCS II, respectively, of pBridge vector, and MBP was expressed as the bridge protein to test its effect on the MYC2-JAZ7 interaction. The experimental procedures were the same as those described above.
Transactivation Activity Assay in Yeast
Full-length CDSs of MYC2 and MTB1 were fused to the GAL4 BD in pGBKT7. The resulting constructs were then cotransformed with pGADT7 into the yeast strain AH109. The MATCHMAKER GAL4-based Two-Hybrid System 3 (Clontech) was used for the transactivation activity assay. Transformed yeast cells were suspended in liquid SD/-2 medium to an OD600 = 0.6, and 5 μL of each dilution was spread onto plates containing SD/-4 medium.
Pull-Down Assays
To produce MBP-JAZ and GST-JAZ fusion proteins, full-length JAZ CDSs were PCR amplified and cloned into pMAL-c2X and pGEX-4T-3, respectively. The recombinant vectors were transformed into Escherichia coli BL21 (DE3) cells. The MBP-JAZ and GST-JAZ fusion proteins were expressed by adding 0.5 mM IPTG and then purified using the amylose resin (NEB) or GST Bind Resin (Millipore), respectively. A similar approach was used to produce MBP-MTB1, GST-MTB1, MBP-MYC2, GST-MYC2, and MBP-MED25243–806 (MBP-MED25) fusion proteins. To produce His-MYC2 fusion protein, full-length CDS of MYC2 was PCR amplified and cloned into pCold TF DNA (Takara). Primers used for plasmid construction are listed in Supplemental Table.
To detect MYC2-MED25 interaction using in vitro pull-down assays, GST-MYC2 and MBP-MED25 fusion proteins were affinity purified. For each reaction, 15 μL of agarose beads bound with 1 μg of GST-MYC2 was incubated with 1 μg of MBP-MED25 in 1 mL of reaction buffer (25 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM DTT, and Roche protease inhibitor cocktail) at 4°C for 2 h. Subsequently, beads were collected and washed three times with washing buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 1 mM DTT). After washing, samples were denatured using SDS loading buffer and separated using SDS-PAGE. The MBP-MED25 fusion protein was detected by immunoblotting with anti-MBP antibody (NEB). Purified GST was used as a negative control. Five-microliter aliquots of GST and GST-MYC2 fusion proteins were separated by SDS-PAGE, and the staining of polyacrylamide gels with CBB was used as a loading control. To detect MTB1-MED25, MTB1-MTB1, and MTB1-MYC2 interactions using in vitro pull-down experiments, sequential manipulations were similar to those described above.
To detect whether the effect of JAZ7 on MYC2-MED25 interaction was concentration-dependent, in vitro pull-down assays were performed by adding 1 μg each of purified His-MYC2 and MBP-MED25 proteins to increasing concentrations of MBP-JAZ7 fusion protein and MBP. The Ni-NTA His Bind Resin (Millipore) was used to pull down proteins. Sequential manipulation was performed as described above. The effect of MED25 on MYC2-JAZ7 interaction and the effect of MTB1 on MYC2-MED25 interaction were detected similarly.
To detect the MTB1-JAZ interaction, pull-down assays were conducted using protein extracts of 10-d-old MTB1-myc tomato seedlings. Seedlings were ground in liquid nitrogen and homogenized in extraction buffer containing 50 mM Tris-HCl (pH 7.4), 80 mM NaCl, 10% (v/v) glycerol, 0.1% (v/v) Tween 20, 1 mM DTT, 1 mM PMSF, 50 mM MG132 (Sigma-Aldrich), and protease inhibitor cocktail (Roche). After centrifugation at 16,000g and 4°C, the supernatant was collected. One milligram of total protein extract was incubated with resin-bound MBP-JAZ and GST-JAZ fusion proteins for 2 h at 4°C with rotation. Purified MBP and GST proteins were used as negative controls. After washing, samples were denatured using SDS loading buffer and separated by SDS-PAGE. The MTB1-myc protein was detected using anti-myc antibody (Abmart). Five-microliter aliquots of MBP-JAZ and GST-JAZ fusion proteins were separated by SDS-PAGE, and the staining of polyacrylamide gels with CBB was used as a loading control. To detect MTB1-MTB1 and MTB1-MYC2 interactions, pull-down assays were conducted by incubating 1 mg of total protein extract of 10-d-old MTB1-myc tomato seedlings with resin-bound MBP-MTB1 and MBP-MYC2 fusion proteins, respectively, for 2 h at 4°C with rotation. Purified MBP protein was used as a negative control. Sequential manipulation was similar to that described above.
Plant Treatment and Gene Expression Analysis
For wounding treatment, 18-d-old seedlings were wounded with a hemostat across the midrib of all leaflets of the lower and upper leaves. The same leaflets were wounded again, proximal to the petiole. Plants with wounded leaves were incubated under continuous light. Five whole leaves were harvested at each sampling time point and used for extracting total RNA. For MeJA treatment, 18-d-old seedlings were enclosed in a Lucite box (10 × 32 × 60 cm) containing 5 μL of MeJA applied to cotton wicks that were spaced evenly within the box. Plants exposed to MeJA vapor were harvested at each sampling time point and used for extracting total RNA (Li et al., 2004). RNA extraction and RT-qPCR analysis were performed as previously described (Du et al., 2014). Expression levels of target genes were normalized relative to that of the tomato ACTIN2 gene. Primers used to quantify gene expression levels are listed in Supplemental Table.
Anthocyanin Content Measurement
Anthocyanin content was measured as previously described (Deikman and Hammer, 1995). Seven-day-old tomato seedlings (50 mg) grown on 0.5× MS medium with 0, 5, or 20 μM JA were placed into 1 mL of extract buffer (propanol:HCl:H2O, 18:1:81, v/v/v). Then the sample was boiled for 3 min and incubated overnight at room temperature. Absorbance values (A535 and A650) of the extraction solution were measured using a spectrophotometer. The anthocyanin content is presented as (A535 − A650)/g fresh weight.
Root Length Measurement
Root lengths of 7-d-old tomato seedlings for each genotype grown on 0.5× MS medium with 0, 0.5, 1, or 2 μM JA were measured and presented.
Botrytis cinerea Inoculation Assays
B. cinerea isolate B05.10 was grown on 2× V8 agar (36% (v/v) V8 juice, 0.2% (w/v) CaCO3, and 2% (w/v) Bacto-agar) for 14 d at 20°C under a 12-h photoperiod prior to spore collection. Spore suspensions were prepared by harvesting the spores in 1% (w/v) Sabouraud Maltose Broth, filtering them through nylon mesh to remove hyphae, and adjusting the concentration to 106 spores/mL (Mengiste et al., 2003). B. cinerea inoculation of tomato plants was performed as previously described (El Oirdi et al., 2011; Yan et al., 2013; Zhai et al., 2013; Du et al., 2017; Lian et al., 2018), with minor modifications. For the pathogenicity test, detached leaves from 5-week-old tomato plants were placed in Petri dishes containing 0.8% (w/v) agar medium (agar dissolved in sterile water), with the petiole embedded in the medium. Each leaflet was spotted with a single 5-μL droplet of B. cinerea spore suspension at a concentration of 106 spores/mL. The trays were covered with lids and kept under the same conditions used for plant growth. Photographs were taken after 3 d, and the lesion sizes were recorded.
For RT-qPCR, inoculations were performed in planta: leaves of 5-week-old plants were spotted with a 5-μL B. cinerea spore suspension (106 spores/mL). The plants were then incubated in a growth chamber with high humidity. A similar experiment was performed using Sabouraud Maltose Broth-spotted plants as a control. Spotted leaves were harvested 24 h after inoculation for RT-qPCR. Error bars represent the sd of three biological replicates. Each biological replicate (sample) consisted of the pooled leaves of three spotted plants from one tray (different genotypes were grown together in a randomized design per tray). Biological replicates (trays) were grown at different locations in growth chambers and treated separately.
Pseudomonas syringae pv tomato DC3000 Infection Assays
Pst DC3000 (Melotto et al., 2006) was cultured at 28°C in King’s B medium (10 g/L tryptone, 5 g/L yeast extract, and 5 g/L NaCl) containing 50 mg/mL rifampicin until OD600 of 0.8 was reached. The bacteria were collected, centrifuged at 2500g for 10 min, washed twice, and resuspended in 10 mM MgCl2 solution containing 0.02% (v/v) Silwet L-77. For vacuum infiltration, 5-week-old plants were vacuum-infiltrated with Pst DC3000 at a concentration of 0.5 × 105 cfu/mL and then kept under high humidity until disease symptoms developed. Leaves infiltrated with 10 mM MgCl2 containing 0.02% (v/v) Silwet L-77 were used as controls. Spotted leaves were harvested 24 h after inoculation for RT-qPCR experiments. Error bars represent the sd of three biological replicates. Each biological replicate (sample) consisted of the pooled leaves of three spotted plants from one tray (different genotypes were grown together in a randomized design per tray). Biological replicates (trays) were grown at different locations in growth chambers and treated separately. Disease symptoms were observed at 3 d after inoculation. For quantification of Pst DC3000 growth, discs from the infected leaves were collected at 3 d after inoculation and ground in 10 mM MgCl2 with a Microfuge tube glass pestle. The samples were diluted 1:10 serially and spotted on solid King’s B medium. After growth at 28°C for 2 d, the cfu was counted.
JA Quantification
For JA content measurement, 18-d-old seedlings were wounded with a hemostat across the midrib of all leaflets of the lower and upper leaves. The same leaflets were wounded again, proximal to the petiole. Plants with wounded leaves were incubated under continuous light. Approximately 600 mg of leaf tissue (fresh weight) from five different plants was pooled for JA quantification as described previously (Fu et al., 2012). Leaf tissues were also harvested from unwounded plants as controls.
ChIP-qPCR Assay
Leaves of 18-d-old MTB1-GFP, MYC2-GFP, MYC2-GFP/MTB-RNAi, MYC2-GFP/MTB1-OE, MED25-GFP, MED25-GFP/MTB-RNAi, and MED25-GFP/MTB1-OE seedlings were wounded for the indicated times. Two grams of wounded or unwounded (control) leaves of each sample was harvested and cross-linked in 1% (v/v) formaldehyde at room temperature for 10 min, followed by neutralization with 0.125 M Gly. The chromatin-protein complex was isolated, resuspended in lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% (w/v) SDS, 1% (v/v) Triton X-100, 0.1% (w/v) sodium deoxycholate, 1 mM PMSF, and 1× Roche protease inhibitor mixture), and sheared by sonication to reduce the average DNA fragment size to ∼500 bp. Then, 50 μL of sheared chromatin was saved for use as input control. Anti-GFP antibody (Abcam) was incubated with Dynabeads Protein G (Invitrogen) at 4°C for at least 6 h and added to the remaining chromatin for overnight incubation at 4°C. The immunoprecipitated chromatin-protein complex was sequentially washed with low-salt buffer (20 mM Tris-HCl [pH 8.0], 2 mM EDTA, 150 mM NaCl, 0.5% (v/v) Triton X-100, and 0.2% (w/v) SDS), high-salt buffer (20 mM Tris-HCl [pH 8.0], 2 mM EDTA, 500 mM NaCl, 0.5% (v/v) Triton X-100, and 0.2% (w/v) SDS), LiCl buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.25 M LiCl, 0.5% (w/v) Nonidet P-40, and 0.5% (w/v) sodium deoxycholate), and TE buffer (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA). After washing, the immunoprecipitated chromatin was eluted with elution buffer (1% SDS and 0.1 M NaHCO3). Protein-DNA cross-links were reversed by incubating the immunoprecipitated complexes with 20 μL of 5 M NaCl at 65°C overnight. DNA was recovered using QIAquick PCR Purification Kit (Qiagen) and analyzed by qPCR. The enrichment of target gene promoters is shown as the percentage of the input DNA, which was calculated by determining the immunoprecipitation efficiency at TomLoxD and JA2L loci as the ratio of the amount of immunoprecipitated DNA to the normalized amount of starting material (percentage of input DNA). ACTIN2 was used as a nonspecific target gene. Primers for qPCR are listed in Supplemental Table.
EMSA
Full-length CDSs of MYC2 and MTB1 were PCR amplified and cloned into pMAL-c2X. The recombinant MBP fusion proteins were expressed in E. coli BL21 (DE3) cells and purified to homogeneity using an amylase resin column. Oligonucleotide probes were synthesized and labeled with biotin at the 5′ ends (Invitrogen). EMSAs were performed as previously described (Chen et al., 2011; Du et al., 2014). Briefly, biotin-labeled probes were incubated with MBP fusion proteins at room temperature for 20 min, and free and bound probes were separated via PAGE. Mutated MTB probes, in which the specific transcription factor binding motif 5′-CACATG-3′ was replaced by 5′-AAAAAA-3′, and mutated TomLoxD probes, in which the specific transcription factor binding motif 5′-ACCATGTG-3′ was deleted, were used as negative controls. Probes used for EMSA are listed in Supplemental Table.
Transient Expression Assays
Transient expression assays were performed in N. benthamiana leaves as previously described (Matsui et al., 2008; Shang et al., 2010; Chen et al., 2011). The TomLoxD promoter was PCR amplified, cloned into pENTR using a pENTR Directional TOPO Cloning Kit (Invitrogen), and then fused with the LUC reporter gene into the plant binary vector pGWB35 using Gateway cloning (Nakagawa et al., 2007) to generate the PTomLoxD:LUC reporter construct. MYC2-GFP (Du et al., 2014) and MTB1-GFP were used as effector constructs. Agrobacterium cells transformed with different constructs were incubated, harvested, and resuspended in infiltration buffer (10 mM MES, 0.2 mM acetosyringone, and 10 mM MgCl2) to a final concentration of OD600 = 0.5. Equal volumes of transformed Agrobacterium cells were mixed in different combinations and coinfiltrated into N. benthamiana leaves with a needleless syringe. Infiltrated plants were incubated at 28°C for 72 h before CCD imaging. A low-light cooled CCD imaging apparatus (NightOWL II LB983 with Indigo software) was used to capture images showing LUC expression and to quantify LUC luminescence intensity. Leaves were sprayed with 100 mM luciferin and incubated in the dark for 3 min before luminescence detection. Five independent determinations were performed.
Insect Feeding Trials
Cotton bollworm (Helicoverpa armigera) larvae were hatched at 26°C, as recommended by the supplier (Yanhui Lu, Institute of Plant Protection, Chinese Academy of Agricultural Sciences). Hatched larvae were reared on an artificial diet for 3 d followed by no diet for 2 d, before transfer to tomato plants. Fifteen third-instar H. armigera larvae were reared on five tomato plants per genotype in three independent replicates. The average weight of larvae at the beginning of the feeding trial was ∼5 mg. After the termination of the 4-d feeding trial, the weight gain of larvae was measured, and the expression of PI-II in the remaining leaf tissues was examined by RT-qPCR.
For monitoring insect attack-induced MTB expression, 18-d-old tomato seedlings that were exposed to third-instar H. armigera larvae were divided into three groups, and the expression of MTBs in the remaining leaf tissues was examined by RT-qPCR. In the first group, six attacked tomato plants were harvested after 1 h of feeding. In the second group, six attacked tomato plants were incubated with the insects for 1 h and the insects were removed for an additional 1 h and then leaves were harvested. In the third group, six attacked tomato plants were harvested after 2 h of continuous feeding.
Antibody Generation
The region of MTB1 (amino acids 196–400, MTB1196–400) was PCR-amplified from wild-type cDNA using gene-specific primers (Supplemental Table). The resultant PCR product was cloned into vector pET28a (Novagen) to express the His-MTB1196–400 protein fusion in E. coli BL21 (DE3). The recombinant fusion protein was purified with Ni-NTA His•Bind Resin (Novagen) and used to raise polyclonal antibodies in mouse. Anti-MTB1 antibodies were used in protein gel blotting at a final dilution of 1:2000.
Accession Numbers
Sequence data from this article can be found in the Sol Genomics Network Initiative under the following accession numbers: MTB1 (Solyc01g096050), MTB2 (Solyc05g050560), MTB3 (Solyc06g083980), MYC2 (Solyc08g076930), MED25 (Solyc12g070100), JAZ1 (Solyc07g042170), JAZ2 (Solyc12g009220), JAZ3 (Solyc03g122190), JAZ4 (Solyc12g049400), JAZ5 (Solyc03g118540), JAZ6 (Solyc01g005440), JAZ7 (Solyc11g011030), JAZ8 (Solyc06g068930), JAZ9 (Solyc08g036640), JAZ10 (Soly08g036620), JAZ11 (Solyc08g036660), TomLoxD (Solyc03g122340), JA2L (Solyc07g063410), TD (Solyc09g008670), PI-II (NP_001234627.1), PR-STH2 (Solyc05g054380), PR1b (Solyc09g007010), and ACTIN2 (Solyc11g005330).
Supplemental Data
Supplemental Figure 1. Generation of MED25-AS and MYC2-OE (MYC2-GFP) plants.
Supplemental Figure 2. MYC2-OE plants show decreased JA accumulation in response to wound and reduced defense gene expression in response to MeJA treatment.
Supplemental Figure 3. Conservation of MYC2- and MTB-related bHLH proteins from tomato, Arabidopsis, and other plants.
Supplemental Figure 4. MTB1 to MTB3 are direct transcriptional targets of MYC2.
Supplemental Figure 5. Generation of MTB-RNAi and MTB1-OE (MTB1-GFP) plants.
Supplemental Figure 6. Expression of MTB1 to MTB3 in response to different stimuli.
Supplemental Figure 7. MTB proteins interact with most of the JAZ proteins in tomato.
Supplemental Figure 8. Generation of MTB1-OE (MTB1-myc) plants.
Supplemental Figure 9. MID of MYC2 and AMID of MTB1 are important for their interactions with JAZ proteins.
Supplemental Figure 10. Generation of MED25-GFP plants.
Supplemental Figure 11. Construction of mtb1-c and mtb1 mtb2-c plants using the CRISPR/Cas9 system.
Supplemental Figure 12. Growth and reproductive phenotypes of wild type, mtb1-c, and mtb1 mtb2-c plants.
Supplemental Table. List of the DNA primers used in this study.
Supplemental Table 2. Primers for RACE and real-time PCR.
Supplemental Data Set 1. Text file of the alignment used for the phylogenetic analysis in Supplemental Figure 3.
Supplemental Data Set 2. Statistical analysis.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Xia Cui for providing the pHSE401_tomatoU6 and the pCBC-DT1T2_tomatoU6 vectors. This work was supported by the National Key R&D Program of China (2017YFD0200400), the Ministry of Agriculture of China (2016ZX08009003-001), the National Natural Science Foundation of China (31730010 and 31672157), the National Basic Research Program of China (2015CB942900), and the Tai-Shan Scholar Program from the Shandong Provincial Government.
AUTHOR CONTRIBUTIONS
C.L., Q.Z., Q.W., and M.D. designed the research. YY.L., M.D., L.D., J.S., M.F., YH.L and Q.C. performed the research. YY.L., M.D., Q.Z., and C.L. analyzed data. Q.Z. and C.L. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- An C., Li L., Zhai Q., You Y., Deng L., Wu F., Chen R., Jiang H., Wang H., Chen Q., Li C. (2017). Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 114: E8930–E8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks D.M., Bender C.L., Kunkel B.N. (2005). The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 6: 629–639. [DOI] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205. [DOI] [PubMed] [Google Scholar]
- Campos M.L., Kang J.H., Howe G.A. (2014). Jasmonate-triggered plant immunity. J. Chem. Ecol. 40: 657–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L., Galstyan A., Roig-Villanova I., Martínez-García J.F., Bilbao-Castro J.R., Robertson D.L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153: 1398–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çevik V., Kidd B.N., Zhang P., Hill C., Kiddle S., Denby K.J., Holub E.B., Cahill D.M., Manners J.M., Schenk P.M., Beynon J., Kazan K. (2012). MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160: 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wilkerson C.G., Kuchar J.A., Phinney B.S., Howe G.A. (2005). Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc. Natl. Acad. Sci. USA 102: 19237–19242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Jiang H., Li L., Zhai Q., Qi L., Zhou W., Liu X., Li H., Zheng W., Sun J., Li C. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Sun L., Qi T., Zhang B., Peng W., Liu Y., Xie D. (2011). The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4: 279–288. [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Chini A., Gimenez-Ibanez S., Goossens A., Solano R. (2016). Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 33: 147–156. [DOI] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Cooke T.F., Depew C.L., Patel L.C., Ogawa N., Kobayashi Y., Howe G.A. (2010). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikman J., Hammer P.E. (1995). Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiol. 108: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Wang H., Sun C., Li Q., Jiang H., Du M., Li C.B., Li C. (2018). Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. J. Genet. Genomics 45: 51–54. [DOI] [PubMed] [Google Scholar]
- Despres C., Subramaniam R., Matton D.P., Brisson N. (1995). The activation of the potato PR-10a gene requires the phosphorylation of the nuclear factor PBF-1. Plant Cell 7: 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Nieto-Rostro M., Xie D., Ellis C., Harmston R., Patrick E., Davis J., Sherratt L., Coleman M., Turner J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32: 457–466. [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M., et al. (2014). Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 26: 3167–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M., et al. (2017). MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 29: 1883–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Oirdi M., El Rahman T.A., Rigano L., El Hadrami A., Rodriguez M.C., Daayf F., Vojnov A., Bouarab K. (2011). Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23: 2405–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Fonseca S., Fernández-Calvo P., Fernández G.M., Díez-Díaz M., Gimenez-Ibanez S., López-Vidriero I., Godoy M., Fernández-Barbero G., Van Leene J., De Jaeger G., Franco-Zorrilla J.M., Solano R. (2014). bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PLoS One 9: e86182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Chu J., Sun X., Wang J., Yan C. (2012). Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal. Sci. 28: 1081–1087. [DOI] [PubMed] [Google Scholar]
- Goossens J., Fernández-Calvo P., Schweizer F., Goossens A. (2016). Jasmonates: Signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 91: 673–689. [DOI] [PubMed] [Google Scholar]
- Goossens J., Mertens J., Goossens A. (2017). Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 68: 1333–1347. [DOI] [PubMed] [Google Scholar]
- Guo Q., Major I.T., Howe G.A. (2018a). Resolution of growth-defense conflict: Mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol. 44: 72–81. [DOI] [PubMed] [Google Scholar]
- Guo Q., Yoshida Y., Major I.T., Wang K., Sugimoto K., Kapali G., Havko N.E., Benning C., Howe G.A. (2018b). JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 115: E10768–E10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz T., Widemann E., Lugan R., Miesch L., Ullmann P., Désaubry L., Holder E., Grausem B., Kandel S., Miesch M., Werck-Reichhart D., Pinot F. (2012). Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 287: 6296–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman R., Van Verk M.C., Van Dijken A.J.H., Mendes M.P., Vroegop-Vos I.A., Caarls L., Steenbergen M., Van der Nagel I., Wesselink G.J., Jironkin A., Talbot A., Rhodes J., et al. (2017). Architecture and dynamics of the jasmonic acid gene regulatory network. Plant Cell 29: 2086–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Major I.T., Koo A.J. (2018). Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 69: 387–415. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2013). MYC2: The master in action. Mol. Plant 6: 686–703. [DOI] [PubMed] [Google Scholar]
- Kitaoka N., Matsubara T., Sato M., Takahashi K., Wakuta S., Kawaide H., Matsui H., Nabeta K., Matsuura H. (2011). Arabidopsis CYP94B3 encodes jasmonyl-L-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 52: 1757–1765. [DOI] [PubMed] [Google Scholar]
- Kloek A.P., Verbsky M.L., Sharma S.B., Schoelz J.E., Vogel J., Klessig D.F., Kunkel B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26: 509–522. [DOI] [PubMed] [Google Scholar]
- Koo A.J., Howe G.A. (2012). Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front. Plant Sci. 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo A.J., Cooke T.F., Howe G.A. (2011). Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Natl. Acad. Sci. USA 108: 9298–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J., Han H., Zhao J., Li C. (2018). In-vitro and in-planta Botrytis cinerea inoculation assays for tomato. Bio Protoc. 8: e2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian T.F., Xu Y.P., Li L.F., Su X.D. (2017). Crystal structure of tetrameric Arabidopsis MYC2 reveals the mechanism of enhanced interaction with DNA. Cell Rep. 19: 1334–1342. [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major I.T., Yoshida Y., Campos M.L., Kapali G., Xin X.F., Sugimoto K., de Oliveira Ferreira D., He S.Y., Howe G.A. (2017). Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 215: 1533–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marineau C., Matton D.P., Brisson N. (1987). Differential accumulation of potato tuber mRNAs during the hypersensitive response induced by arachidonic acid elicitor. Plant Mol. Biol. 9: 335–342. [DOI] [PubMed] [Google Scholar]
- Matsui K., Umemura Y., Ohme-Takagi M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55: 954–967. [DOI] [PubMed] [Google Scholar]
- Matton D.P., Brisson N. (1989). Cloning, expression, and sequence conservation of pathogenesis-related gene transcripts of potato. Mol. Plant Microbe Interact. 2: 325–331. [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980. [DOI] [PubMed] [Google Scholar]
- Mengiste T., Chen X., Salmeron J., Dietrich R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch O., Neumerkel J., Dippe M., Stenzel I., Wasternack C. (2008). Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 177: 114–127. [DOI] [PubMed] [Google Scholar]
- Moreno J.E., Shyu C., Campos M.L., Patel L.C., Chung H.S., Yao J., He S.Y., Howe G.A. (2013). Negative feedback control of jasmonate signaling by an alternative splice variant of JAZ10. Plant Physiol. 162: 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nakata M., Mitsuda N., Herde M., Koo A.J., Moreno J.E., Suzuki K., Howe G.A., Ohme-Takagi M. (2013). A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25: 1641–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C.M.J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C.M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Qi T., Wang J., Huang H., Liu B., Gao H., Liu Y., Song S., Xie D. (2015). Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27: 1634–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C.A. (2000). The systemin signaling pathway: Differential activation of plant defensive genes. Biochim. Biophys. Acta 1477: 112–121. [DOI] [PubMed] [Google Scholar]
- Ryan C.A., Pearce G. (1998). Systemin: a Polypeptide signal for plant defensive genes. Annu. Rev. Cell Dev. Biol. 14: 1–17. [DOI] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y., Jikumaru Y., Obayashi T., Saito H., Masuda S., Kamiya Y., Ohta H., Shirasu K. (2013). Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol. 163: 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Howe G.A. (2005). Systemic signaling in the wound response. Curr. Opin. Plant Biol. 8: 369–377. [DOI] [PubMed] [Google Scholar]
- Shang Y., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu C., Figueroa P., Depew C.L., Cooke T.F., Sheard L.B., Moreno J.E., Katsir L., Zheng N., Browse J., Howe G.A. (2012). JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24: 536–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Fan M., Zhang X., Gao H., Huang H., Wu D., Guo H., Xie D. (2013). The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 9: e1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Fan H.J., Ling H.Q. (2015). Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.Q., Jiang H.L., Li C.Y. (2011). Systemin/jasmonate-mediated systemic defense signaling in tomato. Mol. Plant 4: 607–615. [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Thireault C., Shyu C., Yoshida Y., St Aubin B., Campos M.L., Howe G.A. (2015). Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 82: 669–679. [DOI] [PubMed] [Google Scholar]
- Van der Does D., Leon-Reyes A., Koornneef A., Van Verk M.C., Rodenburg N., Pauwels L., Goossens A., Körbes A.P., Memelink J., Ritsema T., Van Wees S.C.M., Pieterse C.M.J. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDoorn A., Bonaventure G., Schmidt D.D., Baldwin I.T. (2011). Regulation of jasmonate metabolism and activation of systemic signaling in Solanum nigrum: COI1 and JAR4 play overlapping yet distinct roles. New Phytol. 190: 640–652. [DOI] [PubMed] [Google Scholar]
- Wasternack C., Hause B. (2013). Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Zhai Q., Wei J., Li S., Wang B., Huang T., Du M., Sun J., Kang L., Li C.B., Li C. (2013). Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 9: e1003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q., Yan L., Tan D., Chen R., Sun J., Gao L., Dong M.Q., Wang Y., Li C. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9: e1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q., Li L., An C., Li C. (2017a). Conserved function of mediator in regulating nuclear hormone receptor activation between plants and animals. Plant Signal. Behav. e1403709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q., Yan C., Li L., Xie D., Li C. (2017b). Jasmonates. In Hormone Metabolism and Signaling in Plants, Li J., Li C., and Smith S.M., eds, 7 (London: Elsevier; ), pp. 243–272. [Google Scholar]
- Zhang F., et al. (2015). Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525: 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Ke J., Zhang L., Chen R., Sugimoto K., Howe G.A., Xu H.E., Zhou M., He S.Y., Melcher K. (2017a). Structural insights into alternative splicing-mediated desensitization of jasmonate signaling. Proc. Natl. Acad. Sci. USA 114: 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang F., Melotto M., Yao J., He S.Y. (2017b). Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 68: 1371–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Thilmony R., Bender C.L., Schaller A., He S.Y., Howe G.A. (2003). Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 36: 485–499. [DOI] [PubMed] [Google Scholar]