Phosphatidic acid in the plasma membrane interacts with PINOID and regulates auxin redistribution during the plant’s response to salt stress.
Abstract
Remodeling of auxin distribution during the integration of plant growth responses with the environment requires the precise control of auxin influx and efflux transporters. The plasma membrane-localized PIN-FORMED (PIN) proteins facilitate auxin efflux from cells, and their activity is regulated by reversible phosphorylation. How PIN modulates plant cellular responses to external stresses and whether its activity is coordinated by phospholipids remain unclear. Here, we reveal that, in Arabidopsis (Arabidopsis thaliana), the phosphatidic acid (PA)-regulated PINOID (PID) kinase is a crucial modulator of PIN2 activity and auxin redistribution in response to salt stress. Under salt stress, loss of phospholipase D function impaired auxin redistribution and resulted in markedly reduced primary root growth; these effects were reversed by exogenous PA. The phospholipase D-derived PA interacted with PID and increased PID-dependent phosphorylation of PIN2, which activated auxin efflux and altered auxin accumulation, promoting root growth when exposed to salt stress. Ablation of the PA binding motif not only diminished PID accumulation at the plasma membrane but also abolished PA-promoted PID phosphorylation of PIN2 and its function in coping with salt stress; however, this ablation did not affect inflorescence and cotyledon development or PIN2-dependent gravitropic and halotropic responses. Our data indicate a role for PA in coupling extracellular salt signaling to PID-directed PIN2 phosphorylation and polar auxin transport, highlighting the importance of lipid-protein interactions in the spatiotemporal regulation of auxin signaling.
INTRODUCTION
Salinity impairs plant metabolic processes and decreases photosynthetic efficiency, thus limiting productivity (Zhu, 2001; Roy et al., 2014). To withstand salt stress, plants constantly exclude Na+ or Cl– and regulate cellular responses to acclimate to and/or recover from ionic and osmotic damage (Munns and Tester, 2008; Deinlein et al., 2014). The physiological and molecular mechanisms of tolerance to salt stress, including the stress sensing and signaling components, Na+ transporters, and detoxification system, have been investigated extensively at the cell, organ, and whole-plant levels (Xiong et al., 2002; Golldack et al., 2014; Zhu, 2016; Choudhury et al., 2017). Plants minimize the harmful effects of Na+ via sequestering Na+ into the vacuoles by the tonoplast-localized Na+/H+ exchanger1, exporting intracellular Na+ from the cytosol via the Na+/H+ antiporter regulated by the salt overly sensitive (SOS) pathway and removing Na+ from the xylem via members of the high-affinity K+ transporter family (Zhu, 2003; Ji et al., 2013; Deinlein et al., 2014).
The hormone auxin coordinates many of the key processes in plant development and adaptive growth (Adamowski and Friml, 2015; Strader and Zhao, 2016). Auxin is associated with specific biosynthetic, homeostasis, transport, and signal transduction pathways (Feraru and Friml, 2008). Both the plasma membrane-localized and intracellular auxin transporters regulate auxin homeostasis and dynamic rearrangement. Among these transporters, the PIN-FORMED (PIN) proteins are key regulators of auxin efflux from cells, and their asymmetric intracellular localization confers the directionality of intercellular auxin flow (Müller et al., 1998; Feraru and Friml, 2008; Feraru et al., 2011; Rakusová et al., 2015). The transport activity of PINs is regulated by PP2A phosphatase and the Arabidopsis (Arabidopsis thaliana) AGCVIII kinase family, including D6 PROTEIN KINASE (D6PK) and PINOID/WAVY ROOT GROWTH (PID/WAG) kinases, through reversible phosphorylation (Michniewicz et al., 2007; Huang et al., 2010; Galvan-Ampudia et al., 2013; Haga et al., 2014; Zourelidou et al., 2014).
Lipids are the building blocks of biological membranes. The plasma membrane heterogeneity generated by the accumulation of specific lipids and proteins is essential for polar PIN localization (Men et al., 2008; Stanislas et al., 2015; Naramoto, 2017). For instance, the sterol biosynthesis mutant cyclopropylsterol isomerase1-1 is defective in polar PIN2 localization, and sterol composition reportedly affects the postcytokinetic acquisition of PIN2 polarity by endocytosis (Men et al., 2008). Phosphoinositide (PI) asymmetry in the plasma membrane is required for the establishment and maintenance of the polar growth of cells that exhibit tip growth. Before root hair formation, D6PK is translocated to the polar and membrane domains, which are enriched in phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] and sterol, unraveling lipid-dependent D6PK localization during planar polarity formation (Stanislas et al., 2015). The polybasic Lys-rich motif of D6PK mediates its binding to polyacidic phospholipids, which is required for PIN3 phosphorylation and auxin transport-dependent tropic growth (Barbosa et al., 2016). In addition, mutation of the Arabidopsis PIs phosphatidylinositol 4-phosphate (PtdIns4P)-kinase PIP5K1 or PIP5K2 leads to apolar PIN distribution and instructive auxin gradients (Tejos et al., 2014).
Phosphatidic acid (PA), a minor membrane phospholipid, is an essential mediator of plant cell growth and development and responds to various environmental stresses such as cold, salinity, drought, and pathogens (Li et al., 2004; Hong et al., 2008; Yu et al., 2010; Zhang et al., 2012b). Phospholipase D (PLD) and phospholipase C/diacylglycerol kinase play a major role in the cellular generation of PA (Wang, 2005). Arabidopsis PLDα1-derived PA binds to and activates MITOGEN-ACTIVATED PROTEIN KINASE6 (MPK6) and enhances the phosphorylation level of the Na+/H+ antiporter SOS1, conferring salinity tolerance to plants (Yu et al., 2010). PLDζ2 and PA regulate PIN2 cycling and auxin transport, which mediate root gravitropism (Li and Xue, 2007). Salt-induced stimulation of PLD activity increases the clathrin-mediated endocytosis of PIN2 at the side of the root facing the higher salt concentration, suggesting that PA controls the polar distribution of PIN and auxin polarity during halotropism in plants (Galvan-Ampudia et al., 2013). Moreover, both PA and PtdIns4P interact with PID and D6PK, mediating their functions at the plasma membrane (Zegzouti et al., 2006; Barbosa et al., 2016; Simon et al., 2016).
Although crosstalk between lipid signaling and auxin via the regulation of auxin transport or auxin homeostasis has been reported (Li and Xue, 2007; Galvan-Ampudia et al., 2013), little is known about the upstream signaling pathways that control PIN activity during auxin transport. Here, we report in Arabidopsis the functional regulation of PID by membrane PA, which modulates PIN2 activity and polar auxin transport. PLD-derived PA binds to PID and enhances its phosphorylation of PIN2, hence promoting auxin polar efflux and redistribution to increase plant root growth.
RESULTS
Decreased PA Content and Increased Sensitivity to NaCl in PLD Knock Out Arabidopsis
In Arabidopsis, PLDα and PLDδ are the most abundant PLDs, and their activities increase upon exposure to salt stress (Bargmann et al., 2009; Yu et al., 2010; Zhang et al., 2012b). The absence of PLDs in pldα1, pldδ-1, and pldδ-2 single mutants and in pldα1 pldδ-1 and pldα1 pldδ-2 double mutants was verified by immunoblotting using anti-PLDα1 and anti-PLDδ antibodies (Figure 1A; Guo et al., 2012; Zhang et al., 2012b, 2017). Deletion of PLDα1 or PLDδ decreased total PLD (PLDα1 and PLDδ) activity, and PLD activity was decreased markedly in the pldα1 pldδ-1 and pldα1 pldδ-2 double mutants (Figure 1B). Consistent with previous results (Bargmann et al., 2009; Yu et al., 2010), treatment with 125 mM sodium chloride (NaCl) led to a significant increase of PLD activity in the wild-type seedlings within 3 min and a maximum peak of activity at 5 min followed by a decrease after 30 min of NaCl treatment (Figure 1C).
Figure 1.
PLD Production and Enzymatic Activity, and the PA Level in Arabidopsis.
(A) Immunoblot analysis of PLDα1 and PLDδ proteins in the wild type, pldα1, pldδ (pldδ-1 and pldδ-2) single mutants, and pldα1 pldδ (pldα1 pldδ-1 and pldα1 pldδ-2) double mutants. Total proteins were extracted from 6-d-old seedlings, and Arabidopsis PLDα1 and PLDδ proteins were detected by immunoblotting using PLDα1 and PLDδ antibodies. The Rubisco large subunit was used as a loading control.
(B) Total PLD (PLDα1 and PLDδ) activity in the wild type and pld mutants. Data are means ± sd of five independent experiments. Asterisks indicate significant differences from the wild type (*, P < 0.05 and **, P < 0.01) as determined by one-way ANOVA.
(C) Effects of NaCl on PLD activity. The total PLD (PLDα and PLDδ) activity of wild-type seedlings treated with 125 mM NaCl for the indicated periods was measured.
(D) Total amounts of PA in seedlings and the effects of NaCl. Six-day-old plate-grown Arabidopsis seedlings were treated with 125 mM NaCl for the indicated periods. Data are means ± sd of five independent experiments. Different letters indicate statistically significant differences as determined by two-way ANOVA (corrected with Tukey’s multiple comparisons test) for P < 0.05. DW, dry weight.
(E) Changes in PA species due to NaCl treatment in the wild type and the pldα1 pldδ-1 double mutant. Asterisks indicate significant differences from the control without NaCl treatment (P < 0.05) as determined by two-way ANOVA (corrected with Tukey’s multiple comparisons test).
Next, we analyzed the relative levels of PA molecular species in wild-type and pldα1 pldδ-1 double mutant seedlings using electrospray ionization tandem mass spectrometry (ESI-MS/MS). Under unstressed conditions, the PA level in pldα1 pldδ-1 double mutant seedlings was ∼12% of that of the wild type (Figure 1D). Treatment of wild-type seedlings with 125 mM NaCl resulted in a 50% increase in the PA level, but the PA level was not changed significantly in NaCl-treated pldα1 pldδ-1 seedlings (Figure 1D). The molecular species of PA in the wild-type seedlings with the greatest mass spectral signals were 34:2 (16:0-18:2) and 34:3 (16:0-18:3), followed by 36:5 (18:2-18:3), 36:4 (mainly 18:2-18:2), and 36:6 (18:3-18:3); the levels of these PA species increased significantly after 10 min of NaCl treatment (Figure 1E). The levels of all the major PA species were substantially lower in pldα1 pldδ-1 seedlings than in wild-type seedlings, irrespective of salt treatment (Figure 1E).
Salt stress inhibits primary root elongation (Galvan-Ampudia and Testerink, 2011). To evaluate the contribution of PLD on root growth under NaCl treatment, 6-d-old seedlings germinated in half-strength Murashige and Skoog (MS) medium were transferred to fresh MS medium with or without NaCl. Under NaCl-free conditions, no overt differences in primary root growth were observed among the tested genotypes (Figures 2A and 2B), whereas the primary root growth of all the genotypes was inhibited to various degrees under salt stress. Among them, the primary roots of pldα1, pldδ-1, and pldδ-2 single mutants were ∼25% shorter than those of wild-type plants, and suppression of primary root growth was of greater magnitude in the pldα1 pldδ double mutants under salt stress (∼33% of the wild-type level; Figures 2A and 2B), which implies an association between PLD activity and primary root growth in response to salt. Moreover, exogenous application of PA restored primary root growth in the pldα1 pldδ double mutants to the wild-type level in the presence of NaCl (Figures 2A and 2C). Salt stress inhibits primary root elongation by suppressing root meristem activity (West et al., 2004; Liu et al., 2015). Therefore, we compared root meristem length and cell number of primary roots in the two genotypes. Both meristem length and cell number in wild-type roots were reduced by NaCl stress, and greater reductions were observed in pldα1 pldδ double mutants (Figures 2D to 2F). Using propidium iodide to stain dead and dying cells (Duan et al., 2010), we found more stained cells in the roots of pldα1 pldδ mutants than in those of the wild type, suggesting that the root tip of pldα1 pldδ mutants is damaged after salt stress, while the application of PA alleviated the damaging effects of salt treatment (Supplemental Figure 1).
Figure 2.
Arabidopsis pld Mutants Have Reduced Salt Tolerance.
(A) Phenotypes of the wild type and pld mutants under NaCl treatment. Six-day-old seedlings were transferred to half-strength MS medium supplemented with 125 mM NaCl. For PA treatment, the seedlings were grown in half-strength MS medium containing 25 μM PA for 6 d before NaCl treatment. Photographs were obtained 7 d after the transfer to half-strength MS medium containing 125 mM NaCl. The dashed black lines indicate the root length when transferred to salt treatment.
(B) and (C) Increased primary root length in the wild type and pld mutants after the treatments indicated in (A). Data are means ± sd of four independent experiments (n = 55–60 roots). Asterisks indicate significant differences from the wild type under the same conditions (*, P < 0.05 and **, P < 0.01) as determined by two-way ANOVA (corrected with Tukey’s multiple comparisons test).
(D) Root tip morphology of the wild type and pldα1 pldδ mutants after the treatments indicated in (A). Black arrowheads indicate quiescent center cells and red arrowheads indicate cortex transition zones. Insets show magnified images of the transition zones. Bars = 25 µm.
(E) and (F) Root meristem length (E) and root meristem cell number (F) of wild-type and pldα1 pldδ seedlings treated with or without 125 mM NaCl (n > 35 roots). Different letters indicate statistically significant differences as determined by two-way ANOVA (corrected with Tukey’s multiple comparisons test) for P < 0.05.
PLDζ2 has been found to participate in gravitropic, hydrotropic, and halotropic responses via auxin signaling (Li and Xue, 2007; Taniguchi et al., 2010; Galvan-Ampudia et al., 2013). By contrast, the pldα1 pldδ double mutants were not affected in gravitropic or halotropic responses in roots (Supplemental Figure 2). Instead, the pldα1 pldδ double mutants showed salt sensitivity in root elongation, which was not found in the pldζ2 mutant (Supplemental Figure 3). Taken together, our results suggest the specificity of PLDα1 and PLDδ in the positive regulation of root growth under salt stress.
PA Binds to PID
PA reportedly influences several kinases, such as MPK6 and 3-phosphoinositide-dependent protein kinase1 (PDK1), during the responses of plants to developmental or environmental cues (Anthony et al., 2004; Yu et al., 2010). PID and PP2A phosphatase have antagonistic effects on PIN phosphorylation to direct PIN polarity and intercellular auxin fluxes (Michniewicz et al., 2007). However, recent studies reveal that PIN phosphorylation is not linked to polar distribution but rather to PIN activity (Weller et al., 2017). PA interacts with the PP2AA1 subunit of PP2A and modulates the PP2A-mediated dephosphorylation of PIN1 and auxin distribution (Gao et al., 2013).
To examine whether PA binds to PID and regulates its activity, a 6× His tag was fused to the N terminus of the PID, PID2, WAG1, and WAG2 AGCVIII kinases. The recombinant proteins were produced and purified in Escherichia coli (Figure 3A) and subsequently subjected to a filter binding assay using a nitrocellulose filter spotted with natural PA from soybean (Glycine max; Bögre et al., 2003; Haga et al., 2014). PID exhibited stronger PA binding than PID2, WAG1, or WAG2 (Figure 3B), while D330 and D100 from Arabidopsis respiratory burst oxidase homolog D (RbohD) were used as positive and negative controls, respectively (Zhang et al., 2009); this result was confirmed by an ELISA (Figures 3C and 3D). PID weakly bound to phosphatidylglycerol, phosphatidylserine, and PtdIns4P but not to diacylglycerol, phosphatidylinositol, phosphatidylethanolamine, phosphatidylcholine (PC), phosphatidylinositol 3-phosphate, phosphatidylinositol 5-phosphate, phosphatidylinositol-3,4-bisphosphate, phosphatidylinositol-3,5-bisphosphate, or PtdIns(4,5)P2 (Figures 3E and 3F). In addition, the binding of PID to PA was dose dependent, and PID bound specifically to PA with dioleoyl, dilinoleoyl, palmitoyl-oleoyl, palmitoyl-linoleoyl, stearoyl-oleoyl, stearoyl-linoleoyl, or natural PA fatty acyl chains but not to PA dipalmitoyl or distearoyl fatty acyl chains (Figures 3G and 3H).
Figure 3.
PA Binds to PID.
(A) Immunoblot of His-tagged PID, PID2, WAG1, and WAG2 produced in E. coli Rosetta (DE3).
(B) Binding of PA to PID, PID2, WAG1, and WAG2. The D330 and D100 Arabidopsis RbohD fragments were used as positive and negative controls, respectively.
(C) and (D) ELISAs of the binding of PID, PID2, WAG1, and WAG2 to PA (C) and PC (D). Data are means ± sd of three replicates.
(E) and (F) Binding of PID to the indicated lipids (10 μg) on nitrocellulose filters. DAG, diacylglycerol; PS, phosphatidylserine; PI, phosphatidylinositol; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PA, natural PA from soybean; PtdIns3P, phosphatidylinositol 3-phosphate; PtdIns4P, phosphatidylinositol 4-phosphate; PtdIns5P, phosphatidylinositol 5-phosphate; PtdIns(3,4)P2, phosphatidylinositol-3,4-bisphosphate; PtdIns(3,5)P2, phosphatidylinositol-3,5-bisphosphate; PtdIns(4,5)P2, phosphatidylinositol-4,5-bisphosphate. PID binding to lipids was assayed by immunoblot analysis using an anti-His antibody. The protein from the pET28a vector was used as a control.
(G) Binding of PID to the following PA species: dipalmitoyl PA (di16:0), distearoyl PA (di18:0), dioleoyl PA (di18:1), dilinoleoyl PA (di18:2), palmitoyl-oleoyl PA (16:0-18:1), palmitoyl-linoleoyl PA (16:0-18:2), stearoyl-oleoyl PA (18:0-18:1), stearoyl-linoleoyl PA (18:0-18:2), and natural PA from soybean (Soy).
(H) Dose-dependent binding of PID to natural PA.
The PA-PID Interaction Enhances PID-Mediated Phosphorylation of PIN2
PID is a potent regulator of PIN phosphorylation, controlling the auxin efflux activity of PIN (Zourelidou et al., 2014). Accordingly, to investigate whether PA promotes the PID phosphorylation of PIN2, in vitro kinase assays were performed on recombinant GST-tagged PID using the PIN2 cytoplasmic loop (PIN2CL) fragment as a substrate. Treatment with natural PA led to a significant increase in the activity of PID kinase in a dose-dependent manner (Figure 4A). Furthermore, 16:0-18:2 PA increased PID phosphorylation of PIN2, while dipalmitoyl and distearoyl PA, PC, and PtdIns4P had no effect (Figures 4B to 4D), consistent with their reduction or loss of PID binding activity (Figures 3E and 3F). Moreover, a lower level of PIN2 phosphorylation was detected in PID2, WAG1, and WAG2, with or without PA application, compared with that in PID (Supplemental Figure 4). To determine whether PLD/PA enhances PID kinase activity in planta, we transformed the PID-GFP (green florescent protein) construct into wild-type and pldα1 pldδ-1 plants under the control of the Super promoter (a hybrid promoter incorporating the Agrobacterium tumefaciens octopine synthase activator sequences and the mannopine synthase activator elements fused to the mannopine synthase promoter; Ni et al., 1995) and performed a kinase assay to assess PID kinase activity. Treatment with 125 mM NaCl resulted in a rapid activation of PID in wild-type seedlings, whereas this activation was impaired significantly in pldα1 pldδ-1 (Figure 4E). However, the PID content remained unchanged in both genotypes during salt stress (Figure 4E, bottom). No phosphorylation activity or PID-GFP band was detected in the control line from nontransgenic wild-type plants with or without salt treatment (Supplemental Figure 5). Additionally, PID exhibited higher expression than PID2, WAG1, and WAG2 in Arabidopsis seedlings (Supplemental Figure 6). Therefore, the NaCl-induced promotion of PID activity is due mainly to kinase activation rather than protein synthesis and is dependent on PLD/PA.
Figure 4.
PA Enhances PID-Mediated Phosphorylation of PIN2.
(A) PID-dependent phosphorylation of the PIN2CL fragment. GST-tagged PID and PIN2CL were produced in E. coli and purified. Natural PA from soybean was added to purified GST-PID and GST-PIN2CL in the presence of [32P]ATP for 30 min. Phosphorylated GST-PIN2CL was detected by autoradiography (top panel). Relative band intensities, normalized relative to the intensity with the PID and PIN2 reaction without PA treatment, are indicated by numbers in boxes below the bands. Immunoblots of GST-PID and GST-PIN2CL were used as protein-loading controls (bottom panel).
(B) Effects of various PA species on PID-mediated phosphorylation of PIN2CL in vitro.
(C) PtdIns4P did not affect PID phosphorylation of PIN2CL in vitro.
(D) PC did not affect PID phosphorylation of PIN2CL in vitro.
(E) PID kinase activity in wild-type and pldα1 pldδ-1 double mutant plants after treatment with 125 mM NaCl for the indicated periods. PID-GFP immunoprecipitated from wild-type or pldα1 pldδ-1 seedlings was incubated with GST-PIN2CL in the presence of [32P]ATP and subjected to autoradiography (top panel). Relative band intensities, normalized relative to the corresponding intensity with the wild type at the 0-min time point, are indicated by numbers in boxes below the bands. The amounts of PID-GFP protein loaded are shown in the bottom panel.
PLD/PA Modulates the Abundance of PID and PIN2 at the Plasma Membrane under Salt Stress
Both PID and PIN2 are expressed predominantly at the epidermal and cortical cells in roots, and PID and its homologs phosphorylate PIN2 at the plasma membrane (Feraru and Friml, 2008; Kleine-Vehn et al., 2009; Dhonukshe et al., 2010). Thus, we investigated the abundance of PID and PIN2 at the plasma membrane. PID was polarly localized to the plasma membrane of the epidermis and cortex in both the wild type and the pldα1 pldδ-1 double mutant (Figure 5A). Treatment with NaCl led to a decrease in PID fluorescence at the plasma membrane in a time-dependent manner (Supplemental Figure 7), with a 67% decrease in the PID level at the epidermal layer in the pldα1 pldδ-1 double mutant and a 37% decrease in the wild type after 6 h of salt treatment (Figures 5A and 5B). Application of PA alleviated the decrease in PID fluorescence intensity in the wild type and the pldα1 pldδ-1 double mutant to an equal extent (Figures 5A and 5B).
Figure 5.
PID Abundance Is Decreased in the pldα1 pldδ-1 Double Mutant under Salt Stress.
(A) Confocal micrographs of ProPID:PID-GFP in 6-d-old seedlings of the wild type and the pldα1 pldδ-1 double mutant. Six-day-old seedlings were transferred to half-strength MS medium containing 125 mM NaCl; photographs were obtained 6 h after the transfer. For PA treatment, the seedlings were grown in half-strength MS medium containing 25 µM 16:0-18:2 PA before NaCl and 0.1% 1-But treatment. White arrowheads indicate the PID protein at the plasma membrane. Co, cortex; Ep, epidermis. Bar = 5 µm.
(B) Quantification of PID-GFP fluorescence intensity at the plasma membrane according to the treatments in (A) (n > 45 cells from 10 roots). Data are means ± sd of three independent experiments. Different letters indicate statistically significant differences as determined by two-way ANOVA (corrected with Tukey’s multiple comparisons test) for P < 0.05.
Next, we used 1-butanol (1-But), which reduces the cellular PA level by promoting the production of phosphatidylalcohol instead of PA from PLD (Munnik et al., 1995), to evaluate the effects of PA on the abundance of PID. In the presence of NaCl, 0.1% 1-But decreased the PID fluorescence intensity at the plasma membrane of wild-type epidermal cells to a level similar to that in the pldα1 pldδ-1 double mutant (Figures 5A and 5B). Moreover, application of PA alleviated the salt- and 1-But-induced reduction in PID fluorescence intensity in both genotypes (Figures 5A and 5B). By contrast, as a negative control, 2-butanol (2-But), which is an isomer of 1-But, did not affect the PID fluorescence signal in either genotype (Supplemental Figure 8).
PA depletion alters PIN1 localization and auxin distribution by regulating the PP2AA subunit (Gao et al., 2013). Thus, we analyzed PIN2 distribution in the roots by evaluating native promoter-driven PIN2-GFP fluorescence (Xu and Scheres, 2005). The polar localization of PIN2 in the epidermis and cortex exhibited a similar pattern in the wild type and the pldα1 pldδ-1 mutant, irrespective of NaCl stress (Figures 6A and 6D). However, a stronger reduction in PIN2-GFP signal was observed in the plasma membrane in pldα1 pldδ-1 than in the wild type at 6 h after NaCl application (Figures 6A and 6C). Consistent with the PID-GFP results, PA also alleviated the NaCl-induced reduction in PIN2 fluorescence in both genotypes, while 1-But treatment enhanced the NaCl-induced reduction of PIN2 fluorescence in the wild type (Figures 6B and 6C). As a negative control, 2-But did not affect the PIN2-GFP fluorescence signal in either genotype (Supplemental Figure 9). Additionally, PA had no effect on the polar-to-lateral ratio of PIN2 in the epidermis (Figure 6D). These results indicate that PLD-derived PA is essential for maintaining PID and PIN2 at the plasma membrane during salt stress.
Figure 6.
Effects of Salt Stress on the Abundance of PIN2-GFP in the Wild Type and the pldα1 pldδ-1 Double Mutant.
(A) and (B) Representative confocal micrographs of PIN2-GFP in the roots of seedlings of the wild type and the pldα1 pldδ-1 mutant treated with 125 mM NaCl and 1-But in the absence (A) or presence (B) of PA. For PA treatment, the seedlings were grown on half-strength MS medium containing 25 µM 16:0-18:2 PA for 6 d before treatment with NaCl and 0.1% 1-But. White and red arrowheads indicate PIN polarity. Co, cortex; Ep, epidermis. Bars = 5 µm.
(C) Quantification of PIN2-GFP fluorescence intensity at the plasma membrane (n > 50 cells from 12 roots). Data are means ± sd of three independent experiments. Different letters indicate statistically significant differences as determined by two-way ANOVA (corrected with Tukey’s multiple comparisons test) for P < 0.05.
(D) Polar-to-lateral PIN2-GFP intensity ratios in the epidermis of the wild type and the pldα1 pldδ-1 mutant (n > 50 cells from at least 10 roots).
PA Promotes PID Activity in a Manner Dependent on Lys-119 to Lys-121
To map the hypothetical PA binding motif in PID, the PA binding activity of three truncated fragments of PID (named a, b, and c) tagged with His was evaluated (Figures 7A and 7B, top). A filter binding assay showed that the PA binding moiety was present in fragment c (amino acids 1–228; Figure 7B, bottom). Because the classic PA binding sites comprise basic residues adjacent to hydrophobic residues (Kooijman et al., 2007; Zhang et al., 2012b), we mutated Arg and Lys residues into Gly in full-length PID (Figure 7C). Compared with the binding activity of the wild type, only the M2 mutation (residues 119KKK121 to 119GGG121) resulted in a loss of the PA binding ability of PID (Figure 7D); the other mutations did not reduce, or only slightly reduced, the binding activity (Figure 7D). This was confirmed by ELISA (Figure 7E), in which PC (as a control) did not bind to wild-type or mutant PID (Figure 7F). Moreover, PID is recruited to the plasma membrane by the electrostatic field generated by PtdIns4P (Simon et al., 2016), and the M2 mutation did not affect PID binding to PtdIns4P (Supplemental Figure 10). The three Lys residues also were found in PID2, while two Lys residues were present in WAG1 and WAG2, which showed weak binding to PA, suggesting that other residues also may be involved in the PA-PID interaction (Figure 7C). In vitro kinase assays revealed that the mutant PID (M2) lost PA-promoted kinase activity (Figure 7G). Moreover, the NaCl-promoted PID kinase activity in Arabidopsis was suppressed substantially in the mutant transgenic PID line after 5 min of NaCl treatment, although a slight increase of PID activity was found after 10 min of NaCl treatment, which might occur in a PLD/PA-independent manner (Figure 7H). These data suggest that the 119KKK121 residues are essential for PID binding to PA, which is required for the PA- and salt-induced activity.
Figure 7.
Identification of the PA Binding Region and Amino Acid Residues in PID.
(A) Schematic of serial deletions of PIDs. FL, full-length PID (amino acids 1–438); a, amino acids 280 to 438; b, amino acids 1 to 279; c, amino acids 1 to 228. ABD, ATP binding domain; PBD, PtdIns4P binding domain; PIF, PDK1-interacting fragment.
(B) Immunoblot of His-PID fragments (top panel) and their binding to 16:0-18:2 PA on a filter (bottom panel).
(C) Schematic representation of mutant PID. Arrows indicate site-directed amino acid mutations.
(D) Immunoblot of wild-type and mutant PID expressed in E. coli (top panel) and binding of the mutant PID proteins to 16:0-18:2 PA on a filter (bottom panel).
(E) and (F) ELISAs of the binding of wild-type and mutated PID proteins to 16:0-18:2 PA (E) and PC (F). Data are means ± sd of three replicates.
(G) Blocking of PA-PID binding impaired the PA-enhanced phosphorylation of PIN2 by PID in vitro. The kinase activity of wild-type and mutant PID against PIN2CL as a substrate was assayed in the presence or absence of 5 µM 16:0-18:2 PA (top panel). Relative band intensities, normalized relative to the corresponding intensity with wild-type PID and PIN2CL, are indicated by numbers in boxes below the bands. Immunoblots of GST-PID and GST-PIN2CL were used as protein-loading controls (bottom panel).
(H) PID kinase activity in plants after treatment with 125 mM NaCl for the indicated periods. The wild-type or mutant PID-GFP immunoprecipitated from seedlings was incubated with GST-PIN2CL in the presence of [32P]ATP and subjected to autoradiography (top panel). Relative band intensities, normalized relative to the corresponding intensity with wild-type PID at the 0-min time point, are indicated by numbers in boxes below the bands. The amounts of PID-GFP protein loaded are shown in the bottom panel.
Mutation of the PA Binding Domain Disrupted the Functions of PID in Salt Responses
To analyze the roles of the PA-PID interaction in plants, we isolated the PID insertional mutants pid-1 (SALK_082564) and pid-2 (SALK_049736) and confirmed the reduction of PID expression in these mutants by RT-PCR (Figures 8A and 8B). Phenotypic analysis showed that, compared with the wild type, primary root elongation in the pid-1 and pid-2 mutants was hypersensitive to salt and did not respond to PA treatment (Figures 8C and 8D), and similar salt phenotypes also were observed in the pin2 mutants (Supplemental Figure 11), whereas the wag roots generally were similar to the wild type under salt stress (Supplemental Figure 12). Consequently, we introduced GFP-tagged wild-type PID (ProPID:PID-GFP) and non-PA binding mutant PID (ProPID:mPID-GFP, 119KKK121 to 119GGG121) into the pid-1 mutant to complement its salt hypersensitivity. More than 25 transgenic lines for each construct were obtained, and 2 per construct (ProPID:PID-GFP; pid-1 and ProPID:mPID-GFP; pid-1) were used for further analysis based on our RT-PCR results that the expression of both constructs rescued PID expression (Supplemental Figure 13). The salt hypersensitivity of primary root elongation in the pid-1 mutant was completely restored by wild-type PID but not by mutant PID, irrespective of the presence or absence of exogenous PA (Figures 8C and 8D). Furthermore, the expression of wild-type PID, rather than mutant PID, in the pid-1 mutant restored the salt stress-induced reduction in PID abundance at the plasma membrane, irrespective of PA treatment (Figures 8E and 8F). Notably, blocking of PA binding to PID did not reduce the membrane affinity of PID under control conditions (Figure 8E). As expected, increased reductions in meristem length and cell number and more PI-stained cells were observed in the pin2-1 and pid-1 mutants compared with those in the wild type, and the wild-type PID rather than the mutant PID rescued the pid-1 phenotypes under salt stress (Supplemental Figures 14 and 15). These results are mechanistically consistent with our conclusion that Lys-119 to Lys-121 are necessary and sufficient for the PA-mediated function of PID during salt stress in plants.
Figure 8.
Disruption of PA Binding Impairs the PA-Enhanced Functions of PID in Plant Salt Tolerance.
(A) Arabidopsis PID contains two exons and one intron. Arrows indicate the sites of T-DNA insertion into PID in the pid-1 (SALK_082564) and pid-2 (SALK_049736) mutants. UTR, untranslated region.
(B) RT-PCR analysis of full-length PID expression in the wild type and the pid-1 and pid-2 mutants. 18S rRNA was used as an internal control.
(C) Phenotypes of the wild type, pid mutants, and pid-complemented lines after NaCl treatment. Wild-type (ProPID:PID-GFP) and mutant (ProPID:mPID-GFP) PID-complemented vectors were introduced into the pid-1 mutant. Six-day-old seedlings were transferred to half-strength MS medium containing 125 mM NaCl; photographs were obtained 7 d after the transfer. The dashed red lines indicate the root length when transferred to salt treatment, and the dashed black lines indicate the distance on the plates.
(D) Increased primary root length of the wild type, pid mutants, and pid-complemented lines after the treatments indicated in (C). Data are means ± sd of five replicates. Asterisks indicate significant differences from the wild type under the same conditions (P < 0.05) as determined by two-way ANOVA (corrected with Tukey’s multiple comparisons test).
(E) Confocal images of wild-type and mutant PID-GFP fluorescence in the indicated backgrounds. Six-day-old seedlings grown in half-strength MS medium were transferred to medium containing 125 mM NaCl for 6 h. For PA treatment, the seedlings were grown in half-strength MS medium containing 25 µM 16:0-18:2 PA. White arrowheads indicate PID protein at the plasma membrane. Co, cortex; Ep, epidermis. Bar = 5 µm.
(F) Quantification of PID-GFP fluorescence intensity at the plasma membrane after the treatments indicated in (E). Data are means ± sd of three independent experiments (n > 50 cells from at least 10 roots). Different letters indicate statistically significant differences as determined by two-way ANOVA (corrected with Tukey’s multiple comparisons test) for P < 0.05.
PA Enhances the PID-Dependent Auxin Efflux Activity of PIN2
PIN1- and PIN3-mediated auxin transport requires PID kinase and D6PK (Zourelidou et al., 2014). To determine whether PA enhances the auxin-transport activity of PIN2 in a PID-dependent manner, a heterologous assay of auxin efflux in vivo using Xenopus laevis oocytes was performed as described previously (Zourelidou et al., 2014; Weller et al., 2017). Radiolabeled indole-3-acetic acid (IAA) was injected into oocytes at 3 d after cDNA injection, and the intracellular IAA content was determined at 15-min intervals. The IAA content of nontransformed oocytes decreased gradually over time (Figure 9A), which was considered background leakage of IAA. Compared with the background control, the expression of PIN2 alone in oocytes failed to activate auxin efflux, while a reduction in IAA level was detected in oocytes coexpressing PIN2 and wild-type PID (Figures 9A and 9C), which indicates that the auxin efflux activity of PIN2 requires PID phosphorylation. Interestingly, this PID-activated auxin efflux activity of PIN2 was increased further by PA but not by PC (Figures 9A and 9C). In addition, PA did not promote PIN2 activity in the absence of PID or in the presence of mutant PID (Figures 9A to 9C), suggesting that the Lys-119 to Lys-121 residues of PID are critical for PA interacting with PID to activate PIN2-mediated auxin efflux.
Figure 9.
PA Enhances the PID-Mediated Auxin Efflux Activity of PIN2 in X. laevis Oocytes.
(A) Auxin efflux in X. laevis oocytes expressing PIN2 and PID. Oocytes expressing no exogenous protein (indicated as H2O) were used as a background control. At 0, 15, and 30 min after radioactive IAA injection, the residual IAA in oocytes was measured. For lipid treatment, 16:0-18:2 PA or PC was injected into oocytes to an intracellular concentration of 10 μM. Data at each time point were normalized to the mean value at time 0 (1.0). Each data point is the mean and se of at least 10 individual oocytes.
(B) Disruption of PA binding impairs the PA-enhanced PID-induced auxin efflux activity of PIN2 in X. laevis oocytes. PA treatment was performed as described in (A). PID (mutant), mutant non-PA binding PID (M2, residues 119KKK121 to 119GGG121). Each data point is the mean and se of at least 10 individual oocytes.
(C) Auxin transport rates of oocytes with or without PA treatment. Linear regression was performed to calculate IAA transport rates (slope) (i.e., the change in relative IAA content over time). Data (slope) were normalized to the value of the H2O mock control (1.0). Values are means and se of three biological replicates. Different letters indicate statistically significant differences as determined by two-way ANOVA (corrected with Tukey’s multiple comparisons test) for P < 0.05.
The PA-PID Interaction Regulates Auxin Redistribution under Salt Stress
Changes in PIN activity affect auxin efflux and distribution, which influence primary root growth (Blilou et al., 2005; Grieneisen et al., 2007). We assessed auxin redistribution in the root meristem of the pldα1 pldδ-1 double mutant and the pid-1 mutant by introgressing a negative auxin response reporter, domain II (DII)-VENUS, which enables the detection of cellular changes in auxin levels following salt stress (Band et al., 2012; Brunoud et al., 2012). As a target for auxin interaction that triggers its degradation, DII-VENUS is degraded rapidly in response to increasing endogenous auxin levels (Brunoud et al., 2012). Confocal imaging revealed that, similar to the auxin changes in the pin2-1 mutant (Supplemental Figure 16), the DII-VENUS fluorescence intensity in epidermal cells was decreased markedly in the roots of pldα1 pldδ-1 and pid-1 mutants under control conditions (Figures 10A and 10B), which suggests that, in these tissues, defective polar auxin transport results in local auxin accumulation (Friml et al., 2004; Galvan-Ampudia et al., 2013). As expected from the decrease in cellular auxin levels as a consequence of enhanced auxin efflux, the DII-VENUS fluorescence intensity in the epidermis increased ∼1.3-fold in the root of the wild type after treatment with NaCl for 6 h, whereas this increase was suppressed completely in the pldα1 pldδ-1 double mutant and the pid-1 mutant (Figures 10A and 10B). By contrast, PA application promoted the NaCl-induced reduction in auxin level in the wild type and the pldα1 pldδ-1 double mutant but not in the pid-1 mutant (Figures 10A and 10B), in agreement with their primary root phenotypes in the presence of PA (Figures 2A and 8C). Similar NaCl- and PA-induced changes in DII-VENUS signal also were observed in the cortex of the wild type, pldα1 pldδ-1, and pid-1, except for their comparable auxin levels under unstressed conditions (Figures 10A and 10C). The DII-VENUS fluorescence in the pid-1 mutant could be complemented by ProPID:PID-GFP but not by mutant PID, irrespective of PA application. Thus, the regulation of auxin redistribution in the root response to salt could be mediated by the PA-PID module.
Figure 10.
Salt Stress Induces the Redistribution of Auxin.
(A) Confocal DII-VENUS images of the root tip of the wild type, pldα1 pldδ-1, pid-1, and pid-1 mutants complemented with wild-type or non-PA binding mutant PID. Seedlings in the indicated backgrounds grown in half-strength MS medium were transferred to medium containing 125 mM NaCl with or without 25 μM 16:0-18:2 PA for 6 h. Bar = 20 µm.
(B) and (C) Relative DII-VENUS fluorescence intensity at the epidermis (B) and cortex (C) cell layers, as indicated in (A). Data are means ± sd of three replicates (n > 60 cells from 15 roots). Different letters indicate statistically significant differences as determined by two-way ANOVA (corrected with Tukey’s multiple comparisons test) for P < 0.01.
The PA-PID Interaction Is Not Involved in Auxin-Dependent Organ Formation or in Gravitropic or Halotropic Root Growth
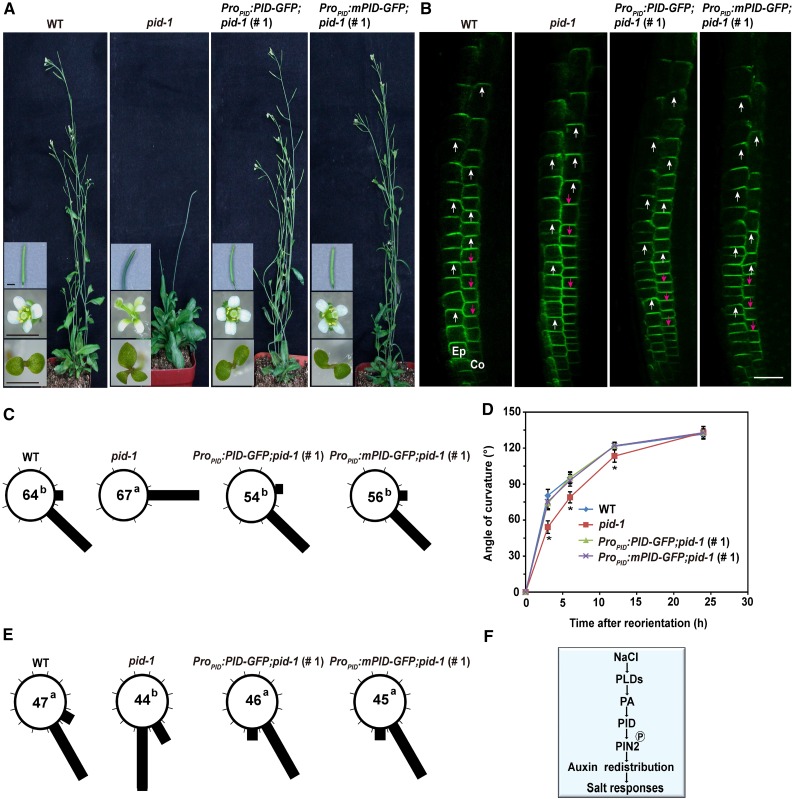
We next investigated whether PA bound to PID affects other PID-involved biological processes. To address this, we examined the developmental phenotypes of the pid-1 mutant and its complementation lines. Consistent with previous studies (Bennett et al., 1995; Christensen et al., 2000; Dhonukshe et al., 2010), the pid-1 mutant developed pin-like inflorescences, aberrant flowers, and abnormal cotyledons, with three cotyledons being the most common phenotype (40% penetrance; Figure 11A). Both the wild-type and mutant transgene were able to complement the PID-dependent developmental defects of the pid-1 mutant. Confocal observation revealed that PIN2 in wild-type cells displayed apical (shootward) localization in the upper cortex and basal (rootward) localization in the lower cortex (Figure 11B). Of the pid-1 mutant seedlings, 23% exhibited an apical-to-basal shift in PIN2 localization in the upper cortex, while the expression of wild-type and mutant PID was sufficient to complement the phenotypes of PIN2 localization (Figure 11B). Moreover, the pid-1 mutant exhibited reduced gravitropic and halotropic responses, and no obvious differences were found between the wild type and the pid-complemented lines (Figures 11C to 11E).
Figure 11.
Phenotypes of pid-1 and pid-Complemented Plants.
(A) Phenotypes of 8-week-old adult plants and cotyledons (bottom insets), inflorescences (middle insets), and floral organs (top insets) of the pid-1 and pid-complemented plants. Bars = 0.25 mm.
(B) PIN2-GFP localization in the epidermis (Ep) and cortex (Co). White and red arrowheads indicate PIN polarity. Bar = 40 µm.
(C) Root gravitropic response histogram in angle categories after 3 h, with number of roots measured. Six-day-old seedlings were transferred to medium with 135° reorientation for gravitropic assay, and root curvatures were measured and assigned to one of the eight 45° sectors. Different letters indicate significant differences (P < 0.05) as determined by one-way ANOVA.
(D) Gravitropism analysis of pid-1 and pid-complemented roots. Asterisks indicate significant differences from the wild type at the same time point (P < 0.05) as determined by two-way ANOVA (corrected with Tukey’s multiple comparisons test).
(E) Halotropic responses of the pid-1 and pid-complemented roots. Quantification is shown for the percentage of curved roots by angle categories after 48 h, with number of roots measured. The curvature of each root was assigned to 1 of the 12 30° sectors. Different letters indicate significant differences (P < 0.05) as determined by one-way ANOVA.
(F) Schematic illustration of the interaction between PA and PID in the regulation of auxin signaling under salt stress.
DISCUSSION
Auxin is involved in regulating the growth and adaptive responses of plants to environmental stresses such as cold (Shibasaki et al., 2009; Du et al., 2012; Rahman, 2013), salinity (Sun et al., 2008; Galvan-Ampudia et al., 2013; Liu et al., 2015), drought (Leymarie et al., 1996; Zhang et al., 2012a; Remy et al., 2013), and metal ions (Kopittke et al., 2015; Li et al., 2015a, 2015b). Plant root systems show high developmental plasticity and exhibit an adaptive response to salt stress, which involves actively prioritizing growth away from salinity by modulating gravity signaling and maintaining continuous root elongation (Sun et al., 2008; Galvan-Ampudia et al., 2013). In this study, we revealed that PLDα1/PLDδ are activated by salt stress and that their product, PA, binds specifically to PID, enhancing PIN2 transport activity to maintain the auxin distribution required for root growth under salt stress (Figure 11F).
Roles of PLD/PA and Auxin Signaling in Plant Response to Salt Stress
The 12 Arabidopsis PLD members have distinct enzymatic properties and are involved in diverse plant processes (Hong et al., 2016). Among them, PLDα and PLDδ are the most abundant in planta. Mutation of either results in hypersensitivity to salt stress, and double mutation of both leads to additive sensitivity to salt (Figure 1; Bargmann et al., 2009; Yu et al., 2010; Zhang et al., 2012b). The mechanisms underlying this process include the regulation of enzyme activities, reorganization of the cytoskeleton, accumulation of osmoprotectants, and transport of transcription factors into nuclei, where they affect gene expression (Yu et al., 2010; Ghars et al., 2012; McLoughlin et al., 2012; Zhang et al., 2012b; Yao et al., 2013). PA accumulates soon after exposure to external stresses (Figure 1; Hong et al., 2008; Bargmann et al., 2009; Yu et al., 2010; Zheng et al., 2011) and may dramatically increase membrane electrostatics, reinforcing PA-directed protein interaction (Wallis and Browse, 2002). PLDα1-derived PA interacts with MPK6 kinase to enhance its phosphorylation of SOS1 (Yu et al., 2010), which is an active antiporter pumping toxic Na+ out of cells (Zhu, 2003). In the salt response, another potential PA target kinase is a member(s) of the sucrose nonfermenting1-related protein kinase2 (SnRK2) family, SnRK2.4 (and/or SnRK2.10) (McLoughlin et al., 2012). Upon exposure of roots to salt, the recruitment of endogenous SnRK2.4/2.10 to membranes was observed, supporting their in vitro binding affinity for PA. However, no in vivo phosphorylation target has been identified for SnRK2.4/2.10 (McLoughlin et al., 2012).
Previous studies have demonstrated the role of salt stress in root growth, lateral root development, root hair formation, and root gravitropism. Many, if not all, of these processes are regulated by auxin signaling (West et al., 2004; Sun et al., 2008; Zhao et al., 2011; McLoughlin et al., 2013; Liu et al., 2015). Salt stress induces the redistribution of auxin and suppresses root meristem activity (Zhao et al., 2011; Liu et al., 2015). The pin2 mutation resulted in inefficient basal transport and increased auxin concentrations in the roots (Ottenschläger et al., 2003; Supplemental Figure 16), as observed in the pldα1 pldδ-1 and pid-1 mutants (Figure 10), and exogenous PA alleviated auxin accumulation in pldα1 pldδ-1, but not in pid-1 or its mutant complementation lines, relative to that of the control or salt treatment (Figures 8 to 10). The genetic evidence suggests that both lipid PA and auxin signaling are involved in the response to salt stress in Arabidopsis.
When wild-type plants were challenged with salt stress, a transient PA increase was observed (Figure 1). The increased PA could bind to PID and activate its activity, because salt-induced PID activation was impaired in the pldα1 pldδ-1 double mutant, in which no salt-induced PA increase was found (Figure 4). The activation of PID leads to the phosphorylation of PIN2, thus affecting auxin transport and redistribution during root development (Michniewicz et al., 2007). During the salt stress response, such an auxin redistribution was maintained for at least 6 h, and the PA-PID interaction regulated this process (Figure 10). The changes of auxin level in cells could be sensed by direct binding to its receptor protein, followed by promoting the degradation of auxin/IAA proteins, releasing auxin response factors, and activating the expression of auxin-responsive genes (Reed, 2001). Using genetic pins mutants, Liu et al. (2015) found that salt stress inhibited root meristem development by stabilizing IAA17. Although salt-induced PA increase as well as PID activation are transient, their mediated auxin level changes and auxin signaling have long-term effects on meristem development and root growth. The detailed components in these processes should be investigated in future work.
Galvan-Ampudia et al. (2013) found that another PLD, PLDζ2, also is involved in the salt stress response (halotropism), but in other mechanisms. Knockout of PLDζ2 results in reduced PIN2 internalization upon salt treatment (Galvan-Ampudia et al., 2013) and a lower halotropic response to NaCl (150 mM; Supplemental Figure 2), suggesting that PLDζ2 is involved in these processes. These results also are consistent with the finding that PA plays a regulatory role in clathrin-mediated endocytosis (McLoughlin et al., 2013). PLDζ2 also is required for the trafficking of PIN2-containing vesicles and functions in auxin transport and distribution, and hence auxin responses, such as gravitropism (Li and Xue, 2007).
By contrast, pldα1 pldδ double mutants exhibited wild-type gravitropic or halotropic phenotypes (Supplemental Figure 2), suggesting that the two genes probably are not involved directly in gravitropism or halotropism. On the other hand, the sensitivity of pldζ2 seedlings to salt stress did not change according to root growth (Zhu, 2001), as compared with the wild type (Supplemental Figure 3), although they showed suppressed gravitropism induced by auxin (Supplemental Figure 2; Li and Xue, 2007). These results imply that, although both PLDζ2 and PLDα1/PLDδ participate in auxin-mediated processes, they show distinctly different physiological roles via unique pathways. Based on our and previous work (Li and Xue, 2007; McLoughlin et al., 2013; Liu et al., 2015), we propose that different PLDs may interact with auxin in different pathways during the active salt-responsive developmental plasticity of roots: i.e., PLDα1/PLDδ mediate PIN2 activation and auxin efflux for growth maintenance, while PLDζ2 mediates PIN2 internalization, asymmetric auxin signaling, and organ expansion for halotropism and/or gravitropism. It is noteworthy that PLDα1/PLDδ and PLDζ2 belong to two distinctive subfamilies, C2-PLD and PX/PH-PLD, respectively (Wang, 2005), and their different domain structures might affect their detailed functions in auxin signaling.
PID-Dependent PIN Phosphorylation by PA Is Essential for Auxin Transport and Root Growth under Salt Stress
The Arabidopsis PIN family consists of eight members, which are polarly localized in various cell types (Feraru and Friml, 2008). PIN2 is phosphorylated by AGCVIII protein kinase family PID and D6PK and by MPKs (Naramoto, 2017). Notably, PID protein levels are higher in root epidermal and cortical cells, where PIN2, but not other PINs, localizes to the poles (Feraru and Friml, 2008; Figures 5 and 6), making it possible for PID and PIN2 to interact physically. PID phosphorylated PIN2 in vitro (Figure 4), resulting in enhanced auxin efflux activity of PIN2 in X. laevis oocytes (Figure 9). Importantly, the PID activity on PIN2 was activated by PA, and this activation is dependent on PA binding (Figure 7). Salt-induced PID activation was impaired in the pldα1 pldδ double mutants and the pid mutant harboring PID that cannot bind PA (Figures 4 and 7), suggesting that PA binding and activation of PID exists in Arabidopsis cells responding to salt stress.
However, there is a basic PID localization at the plasma membrane under normal conditions (Figure 5), even after mutation of PA binding amino acids in PID (Figure 8E). Furthermore, the mutation of PA binding amino acids in PID did not affect PID-dependent inflorescence and cotyledon development or PIN2-dependent gravitropic and halotropic responses (Figure 11). These phenomena suggest that other factors mediate the association of PID with the plasma membrane, especially for its normal functions. Indeed, PID was bound by other anionic lipids, such as PtdIns4P (Simon et al., 2016; Figure 3F). Nine polybasic amino acids (residues 248 to 270) in PID are essential for its direction to the plasma membrane (Simon et al., 2016), which differ from PA binding sites (residues Lys-119 to Lys-21; Figure 7). In contrast to PA binding, however, PtdIns4P binding did not affect PID activities (Figure 4C). In addition, compared with other AGCVIII kinases, PA binding with PID was strongest (Figure 3), and alignments of the amino acid sequences showed that the three Lys residues were found in PID and PID2, while only two were found in WAG1 and WAG2, consistent with their PA binding activity, suggesting that other residues also may contribute to PA binding except for the KKK sites (Figure 7). Although PA also enhanced the WAG2 phosphorylation of PIN2 in vitro (Supplemental Figure 4), the wag mutants exhibited similar salt responses to the wild type in terms of root elongation (Supplemental Figure 12), suggesting that the PA-WAG interaction is not involved directly in the root’s response to salt stress (Santner and Watson, 2006; Willige et al., 2012). Hence, we propose that salt-induced PA that binds to and stimulates PID activity and PIN2 phosphorylation is required specifically for the auxin-mediated maintenance of root growth under salt stress.
Polar PIN distribution is important for developmental processes (Adamowski and Friml, 2015). Bilayer-forming lipids create the structural matrix for proteins in a membrane (Wallis and Browse, 2002; Okazaki and Saito, 2014), and the membrane surface charge is carried by anionic phospholipids (Zhao et al., 2007; Bigay and Antonny, 2012; Simon et al., 2016). PtdIns4P and PtdIns(4,5)P2 as well as PIP5K, which mediates their interconversion, are enriched specifically at the apical and basal polar plasma membrane domains, determining the asymmetric distribution of PIN proteins (Ischebeck et al., 2013; Tejos et al., 2014). In the pip5k1 pip5k2 double mutants, the polar localization of PIN1 and PIN2 is disrupted and embryonic and postembryonic patterning are severely compromised (Tejos et al., 2014). Such local regulation of PIs may influence the formation of clathrin-coated vesicles, thus being decisive for PIN endocytosis and its polarity (Ischebeck et al., 2013). In this work, we determined that PA influences the abundance of PIN2 in the plasma membrane under salt stress (Figure 6). PIN2 localization in pldα1 pldδ differed from that in the wild type by consistently displaying weaker membrane association under salt stress, and the altered PIN2 localization could be recovered by the application of PA (Figure 6). These observations suggest that knocking out both PLDα1 and PLDδ altered the membrane association of PIN2, which might result from perturbed endocytic trafficking regulated by PA, similar to the PIN2 endocytosis regulated by PIs (Ischebeck et al., 2013). In yeast and animals, PA has been found to be an essential mediator of membrane trafficking and clathrin-mediated endocytosis (Freyberg et al., 2003; Jenkins and Frohman, 2005). In a quantitative proteomics screen for PA targets, several clathrin machinery proteins were identified (McLoughlin et al., 2013), suggesting that PA-mediated trafficking and endocytosis reconfigured by salt stress also may exist in plant cells. Finally, the PID activated by PA in turn phosphorylates PIN proteins, and this phosphorylation probably is involved in salt-dependent protein ubiquitylation and degradation (Baral et al., 2015; Barbosa et al., 2018), associated with the requirement of PID-specific protein interactors rather than the polarity control of PINs (Figure 6D; Weller et al., 2017).
Based on the above-described previous reports together with our results, we propose that the lipid PA regulation of PIN2 transporters via PID-mediated phosphorylation is an essential mechanism in the adaptive responses of plants to external stresses. Our work also provides insight into auxin signaling pathways, in which PIN abundance and activity could be regulated via PA metabolism.
METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) seeds used in this study were of the Col-0 ecotype. The pid-1 (SALK_082564), pid-2 (SALK_049736), pin2-1 (SALK_042899), pin2-2 (CS8058), and wag1 (SALK_002056) mutants were obtained from the ABRC at Ohio State University. The homozygous lines were confirmed by PCR. The homozygous pld mutants (pldα1, pldδ-1, pldδ-2, and pldζ2) and wag2 (SALK_070240) were described previously (Li et al., 2006; Zhang et al., 2012b, 2017; Miao et al., 2018). The pid mutant is sterile and therefore must be propagated as a heterozygote. Approximately 40% of homozygous pid mutants develop three symmetrically arranged cotyledons, and selecting three cotyledons serves as an effective screen for homozygous pid seedlings during their further phenotypical analysis, and the heterozygous lines were used for complementation experiments. The pldα1 pldδ-1 and pldα1 pldδ-2 double mutants were generated by crossing and verified by immunoblotting using anti-PLDα1 and anti-PLDδ antibodies (Zhang et al., 2012b, 2017), respectively. The Arabidopsis lines ProPIN2:PIN2-GFP and DII-VENUS have been described previously (Blilou et al., 2005; Xu and Scheres, 2005; Brunoud et al., 2012). The primers used to check T-DNA insertions were designed using Salk T-DNA verification primer design software (http://signal.salk.edu/tdnaprimers.2.html) (Supplemental Table 1).
Growth Conditions and Salt Treatment
Arabidopsis seeds were surface sterilized and germinated on half-strength MS medium [1% (w/v) sucrose and 1% (w/v) agar] at 4°C for 2 d. The seedlings were grown vertically in a chamber with an illumination intensity of 160 μmol m–2 s–1 using a 30-W fluorescent lamp and a day/night regime of 14 h/10 h (23/21°C). Seedlings were transferred to soil after 10 to 14 d for use in experiments. For salt treatment, 6-d-old seedlings were transferred to half-strength MS medium with or without 125 mM NaCl containing 25 μM natural PA from soybean (Glycine max; Avanti Polar Lipids).
RNA Extraction, RT-PCR, and Vector Construction
Total RNA was isolated from leaves or seedlings of 6-d-old Arabidopsis plants using RNAiso Plus reagent (TaKaRa) according to the manufacturer’s instructions. For reverse transcription, first-strand cDNA was synthesized from 1 μg of total RNA using a PrimeScript RT reagent kit with gDNA eraser (TaKaRa). PCR amplifications were performed according to standard protocols in thermocyclers using SYBR Premix ExTaq II (TaKaRa). To determine the transcript levels of PID and PIN2, comparative RT-PCR was performed using the primers P1/P2 and P3/P4, respectively. 18S rRNA was used as an internal control and was amplified using the primers 18S-S/18S-X (Supplemental Table 1).
For pid-1 mutant complementation, the fragment 1830 bp upstream of the initiation codon (ATG) and the entire sequence of PID containing exons and introns were amplified and ligated into the pSuper1300GFP vector using the ProPID:PID-1/ProPID:PID-2 primers. To generate site-directed mutants for complementation, the QuickChange site-directed mutagenesis kit (Stratagene) was used. To construct wild-type and mutant ProSuper:PID-GFP, the PCR fragments of the coding DNA sequence (CDS) of wild-type and M2 mutant PIDs were amplified and ligated into the pSuper1300GFP vector using the primers ProSuper:PID-1/ProSuper:PID-2 (Supplemental Table 1). To complement the pin2-1 mutant, the fragment 1396 bp upstream of the initiation codon (ATG) and the CDS of wild-type or M4 mutant PIN2 were amplified and ligated into the pSuper1300GFP vector using the primer pairs PIN2-promoter-1/PIN2-promoter-2 and PIN2-CDS-1/PIN2-CDS-2, respectively. All of the constructed vectors were transformed into Agrobacterium tumefaciens strain C58C1 (GV3101) by electroporation. Arabidopsis plants were transformed using the floral dip method as described previously (Clough and Bent, 1998). PID (AT2G34650), PID2 (AT2G26700), WAG1 (AT1G53700), WAG2 (AT3G14370), and PIN2CL (AT5G57090) were cloned from Arabidopsis cDNA by PCR using the primers in Supplemental Table 1 and ligated into the pET28a (+) or pGEX-4T vector. Truncated PID proteins (amino acids 1–228, 1–279, and 280–438) were cloned using the primer pairs PID-1-228-1/PID-1-228-2, PID-1-279-1/PID-1-279-2, and PID-280-438-1/PID-280-438-2, respectively (Supplemental Table 1). The full-length and truncated genes were ligated into the pET28a (+) vector. Site-directed mutagenesis of PID was performed using the QuickChange site-directed mutagenesis kit (Stratagene). Using His-PID as a template, the primers used for the M1, M2, M3, M4, and M5 mutations were PID-M1-1/PID-M1-2, PID-M2-1/PID-M2-2, PID-M3-1/PID-M3-2, PID-M4-1/PID-M4-2, and PID-M5-1/PID-M5-2, respectively (Supplemental Table 1).
Analysis of Gravitropism and Halotropism
For the root gravitropism assay, 6-d-old seedlings were transferred to half-strength MS medium, and the plates were reoriented by 135°. Photographs of roots were taken at the indicated time points after reorientation. The angles of root tip curvature were measured using ImageJ software (National Institutes of Health). The root halotropism assay was done as described previously with minor modifications (Galvan-Ampudia et al., 2013). Briefly, NaCl gradients were generated by cutting and removing the lower portion of solidified medium on a plate and replacing it with half-strength MS medium supplemented with 150 mM NaCl [1% (w/v) sucrose and 1% (w/v) agar]. Six-day-old seedlings were transferred to the gradient medium with root tips ∼15 mm from the plate border. Root images were taken after vertical growth for 48 h and were analyzed and assembled using ImageJ.
Recombinant Protein Production and Purification and Protein-Lipid Binding Analysis
PID, PID2, WAG1, WAG2, RbohD330, and RbohD100 were amplified and ligated into the pET28a (+) vector, expressed in Escherichia coli strain BL21 (DE3; TaKaRa), and purified using Ni-affinity agarose (Qiagen; Zhang et al., 2009). The protein from empty pET28a vectors was used as a control. Protein binding to lipids was assayed using nitrocellulose filters and ELISA, as described previously (Zhang et al., 2012b).
Confocal Microscopy, Measurement of Root Meristem Size, and Propidium Iodide Staining
ProPID:PID-GFP, ProPIN2:PIN2-GFP, and DII-VENUS fluorescence signals in the root tips of 6-d-old seedlings were visualized using the LSM 780 Exciter confocal laser scanning microscope (Zeiss) with the following parameters: GFP (excitation, 488 nm; inspection, 505–555 nm) and VENUS (excitation, 514 nm; inspection, 525–555 nm). All scans were conducted at a 1024- × 1024-pixel resolution with repeated scanning of two lines. Images were analyzed and quantified using Zen software, blue edition.
The root meristem zone was defined according to published methods (Dello Ioio et al., 2007). Digital images of wild-type and pld mutant seedlings were captured with a Zeiss differential interference contrast microscope with a clearing solution (50 g of chloral hydrate, 15 mL of water, and 10 mL of glycerol). At least 35 seedlings were analyzed per treatment and genotype.
For the detection of cell death, propidium iodide was dissolved in distilled water (1 μg/mL), and the roots were incubated with the solution for 2 min and then washed with distilled water before fluorescence observation. Propidium iodide staining was quantified using the propidium iodide fluorescence:area ratio in the meristem.
PID Phosphorylation Assays
GST-tagged proteins (PID and PIN2) were expressed and purified using glutathione resin as described previously (Zhao and Wang, 2004). GST-PID (0.2 μg) and GST-PIN2 (0.5 μg) were added to 30 μL of kinase reaction buffer (25 mM Tris-HCl, 5 mM MgCl2, and 0.2 mM EDTA, pH 7.5) containing ATP solution (100 μM MgCl2 and 1 μCi of [32P]γ-ATP). Reactions were incubated at 30°C for 30 min and stopped by adding 10 μL of protein-loading buffer (310 mM Tris-HCl, 10% [v/v] SDS, 50% [v/v] glycerol, 750 mM β-mercaptoethanol, and 0.125% [v/v] bromophenol blue, pH 6.8). The reaction sample was separated by SDS-PAGE on 10% SDS gels, and the gels were washed three times in washing buffer (5% trichloroacetic acid and 1% Na2H2P2O7) for 30 min. The gels were dried for 24 h and subjected to autoradiography. For kinase assays, total proteins were extracted from 6-d-old seedlings. The extracts (100 μg) were incubated with 15 μL of GFP-Trap (ChromoTek) and rotated at 4°C for 5 h. The GFP-Trap was collected by centrifugation at 4000g and 4°C for 1 min and washed three times with immunoprecipitation buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.5% [v/v] Nonidet P-40, 2 mM DTT, phosphatase inhibitor cocktail, and 0.5 mM PMSF, pH 8.0) and then twice with kinase buffer. Reaction mixtures (final volume, 30 μL) containing 0.5 μg of GST-PIN2 substrate and 1 μCi of [32P]γ-ATP were incubated for 30 min at 30°C, the reaction was stopped by adding 10 μL of protein-loading buffer, and the samples were subjected to autoradiography.
Oocyte Auxin Transport Assay
The coding sequences of PIN2, PID, and mutant PID were cloned into the pGEMHE vector. Xenopus laevis oocytes were defolliculated manually and injected with the indicated cRNA mixtures. The complementary RNA (cRNA) was transcribed in vitro using the T7 RiboMAX Large-Scale RNA Production System (Promega), whose concentration was adjusted to 400 ng/μL for PIN2 and 200 ng/μL for wild-type and mutated PID. Oocytes were injected with 50 nL of a 1:1 (v/v) mixture of cRNAs for PIN2 and PID/RNase-free water. Following cRNA injection, oocytes were incubated in modified Barth’s saline (MBS) containing 88 mM NaCl, 1 mM KCl, 0.91 mM CaCl2, 0.33 mM Ca(NO3)2, 0.82 mM MgSO4, 2.4 mM NaHCO3, and 10 mM HEPES, pH 7.5, supplemented with gentamycin (100 mg/L) and tetracycline (25 mg/L). After incubation at 16°C for 3 d, the oocytes were used to examine auxin efflux activity as described by Zourelidou et al. (2014) with minor modifications. Briefly, at each time point, at least 10 oocytes were injected together with 50 nL of a 1:5 dilution (in MBS buffer) of [3H]IAA, 26 Ci/mmol and 1 mCi/mL (Perkin-Elmer), resulting in ∼1 μM intracellular [3H]IAA (estimated oocyte volume, 400 nL). The [3H]IAA-injected oocytes were placed immediately in chilled MBS buffer for 10 min. Next, the oocytes were washed twice and transferred to fresh MBS solution at room temperature to allow for auxin efflux. To stop auxin efflux at each time point (0, 15, and 30 min), oocytes were washed twice and lysed individually in 100 μL of 10% (w/v) SDS, and the residual amount of [3H]IAA in each oocyte was determined by liquid scintillation counting. For PA or PC treatment, the lipids were dried under a flow of N2 gas and sonicated in injection buffer (MBS). Next, lipid-containing injection buffer was added to [3H]IAA and injected immediately into oocytes to achieve an intracellular PA or PC concentration of 10 μM.
PLD Immunoblotting and Activity Assay
Total proteins from 6-d-old Arabidopsis seedlings were isolated and immunoblotted using an anti-PLDα1 or anti-PLDδ antibody (Zhang et al., 2012b, 2017) and incubated with a secondary antibody conjugated to alkaline phosphatase (Sigma-Aldrich; catalog No. SAB3700957). The proteins were visualized by staining for alkaline phosphatase activity (Zhang et al., 2012b). PLDα1 activity was determined according to a previous method using 1,2-dipalmitoyl-3-phosphatidyl-[methyl-3H]choline (Zhang et al., 2004). PLDδ activity was measured using 1,2-dipalmitoyl-3-phosphatidyl-[methyl-3H]choline mixed with phosphatidylethanolamine, PtdIns(4,5)P2, and oleic acid, as described previously (Zhang et al., 2017).
Lipid Preparation and ESI-MS/MS Analysis
For lipid preparation, PA or other lipids purchased from Avanti Polar Lipids in chloroform were dried in a stream of N2 and emulsified by sonication in distilled water on ice, followed by filter sterilization. The lipids were added to half-strength MS medium or to the kinase reaction buffer at the indicated concentrations.
For lipid profiling, 7-d-old seedlings grown in half-strength MS medium were transferred to liquid half-strength MS medium with or without 125 mM NaCl for the indicated periods. Lipid extraction, ESI-MS/MS analysis, and quantification were performed as described previously (Devaiah et al., 2006; Zhang et al., 2009). Briefly, Arabidopsis seedlings were collected and immersed immediately in 3 mL of isopropanol containing 0.01% butylated hydroxytoluene at 75°C to inhibit lipolytic activity. The seedlings were extracted five times with chloroform:methanol (2:1, v/v), and the remaining plant tissues were dried at 110°C and weighed. Lipid samples were analyzed using an electrospray ionization triple quadrupole mass spectrometer (API 4000; Applied Biosystems). Five independent pools of each treatment per phenotype were analyzed.
Statistical Analysis
PLD activity, plant growth, and confocal data were analyzed by one-way or two-way ANOVA corrected with Tukey’s multiple comparisons test at a significance level of P > 0.05. ANOVA tables are provided in Supplemental File 1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PLDα1, At3g15730; PLDδ, At4g35790; PID, At2g34650; PID2, At2g26700; WAG1, At1g53700; WAG2, At3g14370; and PIN2, At5g57090.
Supplemental Data
Supplemental Figure 1. Root phenotypes of the wild type and pldα1 pldδ mutants after salt stress.
Supplemental Figure 2. Gravitropic and halotropic responses of the wild type and pld mutants.
Supplemental Figure 3. Effect of salt on the phenotype of the pldζ2 mutant.
Supplemental Figure 4. Effect of PA on PID, PID2, WAG1, and WAG2 kinase activity.
Supplemental Figure 5. No PID kinase activity was detected from the wild type.
Supplemental Figure 6. Expression of PID, PID2, WAG1, and WAG2 in Arabidopsis.
Supplemental Figure 7. Effect of salt on the abundance of PID-GFP fluorescence in roots.
Supplemental Figure 8. Effect of 2-but on the abundance of PID-GFP in the wild type and the pldα1 pldδ-1 double mutant.
Supplemental Figure 9. The abundance of PIN2-GFP in wild type and pldα1 pldδ-1 double mutant seedlings unaffected by 2-but.
Supplemental Figure 10. Mutation of the PA binding domain in PID did not affect its binding to PtdIns4P.
Supplemental Figure 11. Reduced salt tolerance in Arabidopsis pin2 mutants.
Supplemental Figure 12. Phenotypes of wag mutants under salt stress.
Supplemental Figure 13. PID expression in the wild type and pid-complemented lines.
Supplemental Figure 14. Root phenotypes of pin2, pid, and the pid-complemented lines after salt stress.
Supplemental Figure 15. PI-stained meristem cells in pin2, pid, and the pid-complemented lines after salt stress.
Supplemental Figure 16. Auxin redistribution in the pin2-1 mutant.
Supplemental Table 1. PCR primers used in the article.
Supplemental File 1. ANOVA tables.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Xuemin Wang (Donald Danforth Plant Science Center) for kindly providing pldζ2 seeds. We also thank Chengbin Xiang (University of Science and Technology of China) for kindly providing the wag2 mutant. This work was supported by grants from the National Natural Science Foundation of China (31670263) and the Fundamental Research Funds for the Central Universities (KYZ201858 and KYTZ201402) to Q.Z., the National Natural Science Foundation of Jiangsu Province (BK20160720) and the National Natural Science Foundation of China (31700237) to Y.Q., and the National Natural Science Foundation of China (31770294) to W.Z.
AUTHOR CONTRIBUTIONS
P.W., W.Z., and Q.Z. designed the research and wrote the article. P.W., J.G., L.S., W.J., Y.Q., W.L., R.B., and W.Z. performed specific experiments and analyzed the data. Q.Z., W.Z., W.X., and L.S. revised and edited the article.
References
- Adamowski M., Friml J. (2015). PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 27: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony R.G., Henriques R., Helfer A., Mészáros T., Rios G., Testerink C., Munnik T., Deák M., Koncz C., Bögre L. (2004). A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 23: 572–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band L.R., et al. (2012). Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl. Acad. Sci. USA 109: 4668–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral A., Irani N.G., Fujimoto M., Nakano A., Mayor S., Mathew M.K. (2015). Salt-induced remodeling of spatially restricted clathrin-independent endocytic pathways in Arabidopsis root. Plant Cell 27: 1297–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa I.C., Shikata H., Zourelidou M., Heilmann M., Heilmann I., Schwechheimer C. (2016). Phospholipid composition and a polybasic motif determine D6 PROTEIN KINASE polar association with the plasma membrane and tropic responses. Development 143: 4687–4700. [DOI] [PubMed] [Google Scholar]
- Barbosa I.C.R., Hammes U.Z., Schwechheimer C. (2018). Activation and polarity control of PIN-FORMED auxin transporters by phosphorylation. Trends Plant Sci. 23: 523–538. [DOI] [PubMed] [Google Scholar]
- Bargmann B.O., Laxalt A.M., ter Riet B., van Schooten B., Merquiol E., Testerink C., Haring M.A., Bartels D., Munnik T. (2009). Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 50: 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett S.R.M., Alvarez J., Bossinger G., Smyth D.R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8: 505–520. [Google Scholar]
- Bigay J., Antonny B. (2012). Curvature, lipid packing, and electrostatics of membrane organelles: Defining cellular territories in determining specificity. Dev. Cell 23: 886–895. [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Bögre L., Okrész L., Henriques R., Anthony R.G. (2003). Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 8: 424–431. [DOI] [PubMed] [Google Scholar]
- Brunoud G., Wells D.M., Oliva M., Larrieu A., Mirabet V., Burrow A.H., Beeckman T., Kepinski S., Traas J., Bennett M.J., Vernoux T. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106. [DOI] [PubMed] [Google Scholar]
- Choudhury F.K., Rivero R.M., Blumwald E., Mittler R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90: 856–867. [DOI] [PubMed] [Google Scholar]
- Christensen S.K., Dagenais N., Chory J., Weigel D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100: 469–478. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Deinlein U., Stephan A.B., Horie T., Luo W., Xu G., Schroeder J.I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19: 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682. [DOI] [PubMed] [Google Scholar]
- Devaiah S.P., Roth M.R., Baughman E., Li M., Tamura P., Jeannotte R., Welti R., Wang X. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dα1 knockout mutant. Phytochemistry 67: 1907–1924. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Huang F., Galvan-Ampudia C.S., Mähönen A.P., Kleine-Vehn J., Xu J., Quint A., Prasad K., Friml J., Scheres B., Offringa R. (2010). Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255. [DOI] [PubMed] [Google Scholar]
- Du H., Wu N., Fu J., Wang S., Li X., Xiao J., Xiong L. (2012). A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 63: 6467–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y., Zhang W., Li B., Wang Y., Li K., Sodmergen, Han C., Zhang Y., Li X. (2010). An endoplasmic reticulum response pathway mediates programmed cell death of root tip induced by water stress in Arabidopsis. New Phytol. 186: 681–695. [DOI] [PubMed] [Google Scholar]
- Feraru E., Friml J. (2008). PIN polar targeting. Plant Physiol. 147: 1553–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E., Feraru M.I., Kleine-Vehn J., Martinière A., Mouille G., Vanneste S., Vernhettes S., Runions J., Friml J. (2011). PIN polarity maintenance by the cell wall in Arabidopsis. Curr. Biol. 21: 338–343. [DOI] [PubMed] [Google Scholar]
- Freyberg Z., Siddhanta A., Shields D. (2003). “Slip, sliding away”: Phospholipase D and the Golgi apparatus. Trends Cell Biol. 13: 540–546. [DOI] [PubMed] [Google Scholar]
- Friml J., Yang X., Michniewicz M., Weijers D., Quint A., Tietz O., Benjamins R., Ouwerkerk P.B., Ljung K., Sandberg G., Hooykaas P.J., Palme K., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865. [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia C.S., Testerink C. (2011). Salt stress signals shape the plant root. Curr. Opin. Plant Biol. 14: 296–302. [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia C.S., Julkowska M.M., Darwish E., Gandullo J., Korver R.A., Brunoud G., Haring M.A., Munnik T., Vernoux T., Testerink C. (2013). Halotropism is a response of plant roots to avoid a saline environment. Curr. Biol. 23: 2044–2050. [DOI] [PubMed] [Google Scholar]
- Gao H.B., Chu Y.J., Xue H.W. (2013). Phosphatidic acid (PA) binds PP2AA1 to regulate PP2A activity and PIN1 polar localization. Mol. Plant 6: 1692–1702. [DOI] [PubMed] [Google Scholar]
- Ghars M.A., Richard L., Lefebvre-De Vos D., Leprince A.S., Parre E., Bordenave M., Abdelly C., Savouré A. (2012). Phospholipases C and D modulate proline accumulation in Thellungiella halophila/salsuginea differently according to the severity of salt or hyperosmotic stress. Plant Cell Physiol. 53: 183–192. [DOI] [PubMed] [Google Scholar]
- Golldack D., Li C., Mohan H., Probst N. (2014). Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 5: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen V.A., Xu J., Marée A.F., Hogeweg P., Scheres B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013. [DOI] [PubMed] [Google Scholar]
- Guo L., Devaiah S.P., Narasimhan R., Pan X., Zhang Y., Zhang W., Wang X. (2012). Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 24: 2200–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K., Hayashi K., Sakai T. (2014). PINOID AGC kinases are necessary for phytochrome-mediated enhancement of hypocotyl phototropism in Arabidopsis. Plant Physiol. 166: 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Pan X., Welti R., Wang X. (2008). Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20: 803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Zhao J., Guo L., Kim S.C., Deng X., Wang G., Zhang G., Li M., Wang X. (2016). Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 62: 55–74. [DOI] [PubMed] [Google Scholar]
- Huang F., Zago M.K., Abas L., van Marion A., Galván-Ampudia C.S., Offringa R. (2010). Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T., et al. (2013). Phosphatidylinositol 4,5-bisphosphate influences PIN polarization by controlling clathrin-mediated membrane trafficking in Arabidopsis. Plant Cell 25: 4894–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G.M., Frohman M.A. (2005). Phospholipase D: A lipid centric review. Cell. Mol. Life Sci. 62: 2305–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Pardo J.M., Batelli G., Van Oosten M.J., Bressan R.A., Li X. (2013). The Salt Overly Sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 6: 275–286. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Huang F., Naramoto S., Zhang J., Michniewicz M., Offringa R., Friml J. (2009). PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman E.E., Tieleman D.P., Testerink C., Munnik T., Rijkers D.T., Burger K.N., de Kruijff B. (2007). An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J. Biol. Chem. 282: 11356–11364. [DOI] [PubMed] [Google Scholar]
- Kopittke P.M., et al. (2015). Identification of the primary lesion of toxic aluminum in plant roots. Plant Physiol. 167: 1402–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leymarie J., Damerval C., Marcotte L., Combes V., Vartanian N. (1996). Two-dimensional protein patterns of Arabidopsis wild-type and auxin insensitive mutants, axr1, axr2, reveal interactions between drought and hormonal responses. Plant Cell Physiol. 37: 966–975. [DOI] [PubMed] [Google Scholar]
- Li G., Xue H.W. (2007). Arabidopsis PLDζ2 regulates vesicle trafficking and is required for auxin response. Plant Cell 19: 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Song H., Li B., Kronzucker H.J., Shi W. (2015a). Auxin Resistant1 and PIN-FORMED2 protect lateral root formation in Arabidopsis under iron stress. Plant Physiol. 169: 2608–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Xu H.H., Liu W.C., Zhang X.W., Lu Y.T. (2015b). Ethylene inhibits root elongation during alkaline stress through AUXIN1 and associated changes in auxin accumulation. Plant Physiol. 168: 1777–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Welti R., Wang X. (2006). Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation: Roles of phospholipases D ζ1 and D ζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 142: 750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li M., Zhang W., Welti R., Wang X. (2004). The plasma membrane-bound phospholipase Dδ enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 22: 427–433. [DOI] [PubMed] [Google Scholar]
- Liu W., Li R.J., Han T.T., Cai W., Fu Z.W., Lu Y.T. (2015). Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 168: 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin F., Galvan-Ampudia C.S., Julkowska M.M., Caarls L., van der Does D., Laurière C., Munnik T., Haring M.A., Testerink C. (2012). The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 72: 436–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin F., Arisz S.A., Dekker H.L., Kramer G., de Koster C.G., Haring M.A., Munnik T., Testerink C. (2013). Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem. J. 450: 573–581. [DOI] [PubMed] [Google Scholar]
- Men S., Boutté Y., Ikeda Y., Li X., Palme K., Stierhof Y.-D., Hartmann M.-A., Moritz T., Grebe M. (2008). Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat. Cell Biol. 10: 237–244. [DOI] [PubMed] [Google Scholar]
- Miao Z.Q., Zhao P.X., Mao J.L., Yu L.H., Yuan Y., Tang H., Xiang C. (2018). Arabidopsis HB52 mediates the crosstalk between ethylene and auxin signaling pathways by regulating PIN2, WAG1, and WAG2 during primary root elongation. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056. [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T., Arisz S.A., De Vrije T., Musgrave A. (1995). G protein activation stimulates phospholipase D signaling in plants. Plant Cell 7: 2197–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59: 651–681. [DOI] [PubMed] [Google Scholar]
- Naramoto S. (2017). Polar transport in plants mediated by membrane transporters: Focus on mechanisms of polar auxin transport. Curr. Opin. Plant Biol. 40: 8–14. [DOI] [PubMed] [Google Scholar]
- Ni M., Cui D., Einstein J., Narasimhulu S., Vergara C.E., Gelvin S.B. (1995). Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J. 7: 661–676. [Google Scholar]
- Okazaki Y., Saito K. (2014). Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J. 79: 584–596. [DOI] [PubMed] [Google Scholar]
- Ottenschläger I., Wolff P., Wolverton C., Bhalerao R.P., Sandberg G., Ishikawa H., Evans M., Palme K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100: 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A. (2013). Auxin: A regulator of cold stress response. Physiol. Plant. 147: 28–35. [DOI] [PubMed] [Google Scholar]
- Rakusová H., Fendrych M., Friml J. (2015). Intracellular trafficking and PIN-mediated cell polarity during tropic responses in plants. Curr. Opin. Plant Biol. 23: 116–123. [DOI] [PubMed] [Google Scholar]
- Reed J.W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6: 420–425. [DOI] [PubMed] [Google Scholar]
- Remy E., Cabrito T.R., Baster P., Batista R.A., Teixeira M.C., Friml J., Sá-Correia I., Duque P. (2013). A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 25: 901–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.J., Negrão S., Tester M. (2014). Salt resistant crop plants. Curr. Opin. Biotechnol. 26: 115–124. [DOI] [PubMed] [Google Scholar]
- Santner A.A., Watson J.C. (2006). The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. Plant J. 45: 752–764. [DOI] [PubMed] [Google Scholar]
- Shibasaki K., Uemura M., Tsurumi S., Rahman A. (2009). Auxin response in Arabidopsis under cold stress: Underlying molecular mechanisms. Plant Cell 21: 3823–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M.L.A., Platre M.P., Marquès-Bueno M.M., Armengot L., Stanislas T., Bayle V., Caillaud M.C., Jaillais Y. (2016). A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat. Plants 2: 16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislas T., Hüser A., Barbosa I.C., Kiefer C.S., Brackmann K., Pietra S., Gustavsson A., Zourelidou M., Schwechheimer C., Grebe M. (2015). Arabidopsis D6PK is a lipid domain-dependent mediator of root epidermal planar polarity. Nat. Plants 1: 15162. [DOI] [PubMed] [Google Scholar]
- Strader L.C., Zhao Y. (2016). Auxin perception and downstream events. Curr. Opin. Plant Biol. 33: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Zhang W., Hu H., Li B., Wang Y., Zhao Y., Li K., Liu M., Li X. (2008). Salt modulates gravity signaling pathway to regulate growth direction of primary roots in Arabidopsis. Plant Physiol. 146: 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y.Y., Taniguchi M., Tsuge T., Oka A., Aoyama T. (2010). Involvement of Arabidopsis thaliana phospholipase Dζ2 in root hydrotropism through the suppression of root gravitropism. Planta 231: 491–497. [DOI] [PubMed] [Google Scholar]
- Tejos R., Sauer M., Vanneste S., Palacios-Gomez M., Li H., Heilmann M., van Wijk R., Vermeer J.E., Heilmann I., Munnik T., Friml J. (2014). Bipolar plasma membrane distribution of phosphoinositides and their requirement for auxin-mediated cell polarity and patterning in Arabidopsis. Plant Cell 26: 2114–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J.G., Browse J. (2002). Mutants of Arabidopsis reveal many roles for membrane lipids. Prog. Lipid Res. 41: 254–278. [DOI] [PubMed] [Google Scholar]
- Wang X. (2005). Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 139: 566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller B., Zourelidou M., Frank L., Barbosa I.C., Fastner A., Richter S., Jürgens G., Hammes U.Z., Schwechheimer C. (2017). Dynamic PIN-FORMED auxin efflux carrier phosphorylation at the plasma membrane controls auxin efflux-dependent growth. Proc. Natl. Acad. Sci. USA 114: E887–E896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West G., Inzé D., Beemster G.T. (2004). Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 135: 1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige B.C., Ogiso-Tanaka E., Zourelidou M., Schwechheimer C. (2012). WAG2 represses apical hook opening downstream from gibberellin and PHYTOCHROME INTERACTING FACTOR 5. Development 139: 4020–4028. [DOI] [PubMed] [Google Scholar]
- Xiong L., Schumaker K.S., Zhu J.K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 (suppl.): S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Wang G., Guo L., Wang X. (2013). Phosphatidic acid interacts with a MYB transcription factor and regulates its nuclear localization and function in Arabidopsis. Plant Cell 25: 5030–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Nie J., Cao C., Jin Y., Yan M., Wang F., Liu J., Xiao Y., Liang Y., Zhang W. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188: 762–773. [DOI] [PubMed] [Google Scholar]
- Zegzouti H., Anthony R.G., Jahchan N., Bögre L., Christensen S.K. (2006). Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 6404–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Li J., Zhang W., Yan S., Wang R., Zhao J., Li Y., Qi Z., Sun Z., Zhu Z. (2012a). The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J. 72: 805–816. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Lin F., Mao T., Nie J., Yan M., Yuan M., Zhang W. (2012b). Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24: 4555–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Song P., Qu Y., Wang P., Jia Q., Guo L., Zhang C., Mao T., Yuan M., Wang X., Zhang W. (2017). Phospholipase Dδ negatively regulates plant thermotolerance by destabilizing cortical microtubules in Arabidopsis. Plant Cell Environ. 40: 2220–2235. [DOI] [PubMed] [Google Scholar]
- Zhang W., Qin C., Zhao J., Wang X. (2004). Phospholipase D α 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 101: 9508–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhu H., Zhang Q., Li M., Yan M., Wang R., Wang L., Welti R., Zhang W., Wang X. (2009). Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21: 2357–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wang X. (2004). Arabidopsis phospholipase Dα1 interacts with the heterotrimeric G-protein α-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J. Biol. Chem. 279: 1794–1800. [DOI] [PubMed] [Google Scholar]
- Zhao C., Du G., Skowronek K., Frohman M.A., Bar-Sagi D. (2007). Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol. 9: 706–712. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wang T., Zhang W., Li X. (2011). SOS3 mediates lateral root development under low salt stress through regulation of auxin redistribution and maxima in Arabidopsis. New Phytol. 189: 1122–1134. [DOI] [PubMed] [Google Scholar]
- Zheng G., Tian B., Zhang F., Tao F., Li W. (2011). Plant adaptation to frequent alterations between high and low temperatures: Remodelling of membrane lipids and maintenance of unsaturation levels. Plant Cell Environ. 34: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2001). Plant salt tolerance. Trends Plant Sci. 6: 66–71. [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6: 441–445. [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2016). Abiotic stress signaling and responses in plants. Cell 167: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M., et al. (2014). Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3: e02860. [DOI] [PMC free article] [PubMed] [Google Scholar]