Current influenza vaccines are primarily strain specific, requiring annual updates, and offer minimal protection against drifted seasonal or pandemic strains. The highly conserved stem region of hemagglutinin (HA) of group 2 influenza A virus subtypes is a promising target for vaccine elicitation of broad cross-group protection against divergent strains. We used structure-guided protein engineering employing multiple protein stabilization methods simultaneously to develop group 2 HA stem-based candidate influenza A virus immunogens displayed as trimers on self-assembling nanoparticles. Characterization of antigenicity, thermostability, and particle formation confirmed structural integrity. Group 2 HA stem antigen designs were identified that, when displayed on ferritin nanoparticles, activated B cells expressing inferred unmutated common ancestor (UCA) versions of human antibody lineages associated with cross-group-reactive, broadly neutralizing antibodies (bNAbs). Immunization of mice led to protection against a lethal homosubtypic influenza virus challenge. These candidate vaccines are now being manufactured for clinical evaluation.
KEYWORDS: hemagglutinin, influenza, influenza vaccines, nanoparticle, protein engineering, vaccine design, ferritin
ABSTRACT
Influenza vaccines targeting the highly conserved stem of the hemagglutinin (HA) surface glycoprotein have the potential to protect against pandemic and drifted seasonal influenza viruses not covered by current vaccines. While HA stem-based immunogens derived from group 1 influenza A viruses have been shown to induce intragroup heterosubtypic protection, HA stem-specific antibody lineages originating from group 2 may be more likely to possess broad cross-group reactivity. We report the structure-guided development of mammalian-cell-expressed candidate vaccine immunogens based on influenza A virus group 2 H3 and H7 HA stem trimers displayed on self-assembling ferritin nanoparticles using an iterative, multipronged approach involving helix stabilization, loop optimization, disulfide bond addition, and side-chain repacking. These immunogens were thermostable, formed uniform and symmetric nanoparticles, were recognized by cross-group-reactive broadly neutralizing antibodies (bNAbs) with nanomolar affinity, and elicited protective, homosubtypic antibodies in mice. Importantly, several immunogens were able to activate B cells expressing inferred unmutated common ancestor (UCA) versions of cross-group-reactive human bNAbs from two multidonor classes, suggesting they could initiate elicitation of these bNAbs in humans.
INTRODUCTION
Influenza continues to be a significant global health burden, typically resulting in about 500,000 deaths worldwide annually (1), even though the technology for conventional egg-grown, whole inactivated influenza virus vaccines was developed more than 70 years ago. Constant antigenic drift of the influenza virus hemagglutinin (HA) coupled with immunodominant strain-specific antibody responses directed to the variable HA head domain results in conventional vaccine effectiveness ranging from 10 to 60% (2) and the need for seasonal updates of virus strains included in licensed vaccines. Furthermore, current vaccine approaches provide minimal protection against pandemic influenza virus strains (3, 4). Improved influenza vaccines would not be produced in eggs, would be designed to induce broad immunity against future drifted and pandemic strains without seasonal reformulation, and would elicit durable immune responses, avoiding the need for annual vaccination (4). One approach for achieving broadly cross-reactive influenza virus-specific antibodies is to target highly conserved neutralization-sensitive epitopes in the stem region of the influenza A virus HA surface glycoprotein (5). Monoclonal antibodies (MAb) have been identified that bind conserved HA stem epitopes and possess broad neutralizing activity across diverse HA subtypes within an influenza A virus group, as well as some that have cross-group neutralizing activity (6–16). We and others have recently developed influenza A virus group 1 HA stem trimer antigens (17, 18). One candidate displaying H1-based HA stem trimers on self-assembling ferritin nanoparticles protected mice and ferrets from a lethal heterosubtypic H5N1 influenza challenge (17). However, these group 1 HA stem immunogens did not provide cross-group protection against group 2 viruses, such as H3N2 and H7N9. The need for a better group 2 vaccine is particularly acute, as vaccine effectiveness against H3N2 over the past decade has averaged only 33% (19), recent H3N2 strains have exhibited increased virulence (20), and H7 viruses represent one of the greatest pandemic threats from nonseasonal strains. H7N9 immunization was recently shown to induce multiple HA stem-directed antibody lineages in humans that recognized both group 1 and group 2 HA molecules, whereas H5N1 immunization induced primarily HV1-69 antibody lineages that cross-reacted primarily within group 1 influenza A virus subtypes (13). This suggests that group 2 HA stem immunogens may be more effective at inducing cross-group responses than group 1 immunogens in humans with preexisting influenza immunity or at priming multidonor antibody lineages with the capacity for broad neutralizing activity in influenza-naive infants. In our studies, we used iterative structure-based design to develop headless group 2 HA stabilized stem trimers displayed on ferritin nanoparticles based on hemagglutinin sequences from H3N2 (H3ssF) and H7N9 (H7ssF) viruses as candidate vaccines. Notably, structural subtleties ultimately required the use of a set of design strategies to stabilize group 2 HA stem immunogens that was completely different than the simpler hydrophobic repacking used to stabilize group 1 immunogens. Ultimately, H3ssF and H7ssF immunogen designs were identified that could activate B cells expressing inferred unmutated common ancestor (UCA) of HA stem-directed human broadly neutralizing antibodies (bNAbs), suggesting the potential for inducing cross-reactive antibody lineages with the capacity to develop broad influenza virus neutralizing activity in humans.
RESULTS
Design of group 2 stabilized HA stem nanoparticle immunogens.
The initial design of group 2 stabilized HA stem nanoparticles incorporated some features of group 1 H1-stabilized HA stem nanoparticle immunogens (17) (referred to as H1ssF herein). This comprised replacing the HA head region (HA1 residues 43 to 313, H3 numbering [21]) with a GSG loop, replacing the HA2 interhelical region (residues 60 to 92) with a 6-residue glycine-rich loop, including two repacking substitutions in the HA2 hydrophobic core (replacing K with M at position 103 [K103M] and E with L at position 51 [E51L]), and connecting the C-terminal HA2 residue 174 to the N-terminal residue 5 of bacterial ferritin with a short SGG linker (Fig. 1A; Fig. S1 and S2 in the supplemental material). In the group 1 context, this strategy was sufficient for displaying antigenically correct H1, H2, and H5 HA stem trimers on ferritin nanoparticles (17). However, multiple additional steps were needed to achieve the expression of group 2 H3 and H7 HA stem nanoparticles in mammalian HEK293 cells (Fig. 1). Although there is structural similarity between H1 and H3 HA stem regions (e.g., root mean square deviation [RMSD] of 1.0 Å for 134 stem residues between A/California/04/2009 [H1N1; PDB identification code [ID] 3UBQ] and A/Victoria/361/2011 [H3N2; PDB ID 4O5N]) and sequence identity of approximately 55%, phylogenetic analysis indicates groups 1 and 2 comprise two distinct branches with consistent and predictable amino acid differences even for the stem region of HA (Fig. 1B). We hypothesized that further structural stabilization was required for expression, and therefore, an iterative structure-based design effort was employed to stabilize the HA stem region of H3ssF immunogens, using the 1.9-Å-resolution crystal structure of HA for A/Finland/486/2004 (H3N2; PDB ID 2YP2) (22) as a template. To screen for expression and correct antigenicity, designs were expressed in HEK293T cells using a high-throughput 96-well format (23). Supernatants containing HA stem trimers genetically fused to ferritin nanoparticles were assessed directly for antigenic recognition either by an immunoprecipitation assay using the human cross-group HA stem-specific bNAb FI6v3 or by an enzyme-linked immunosorbent assay (ELISA) using the bNAbs CR9114, FI6v3, and CT149. CT149 was specifically included to assess trimer integrity because its epitope incorporates two adjacent protomers of the HA stem and, thus, may be sensitive to quaternary structure (15).
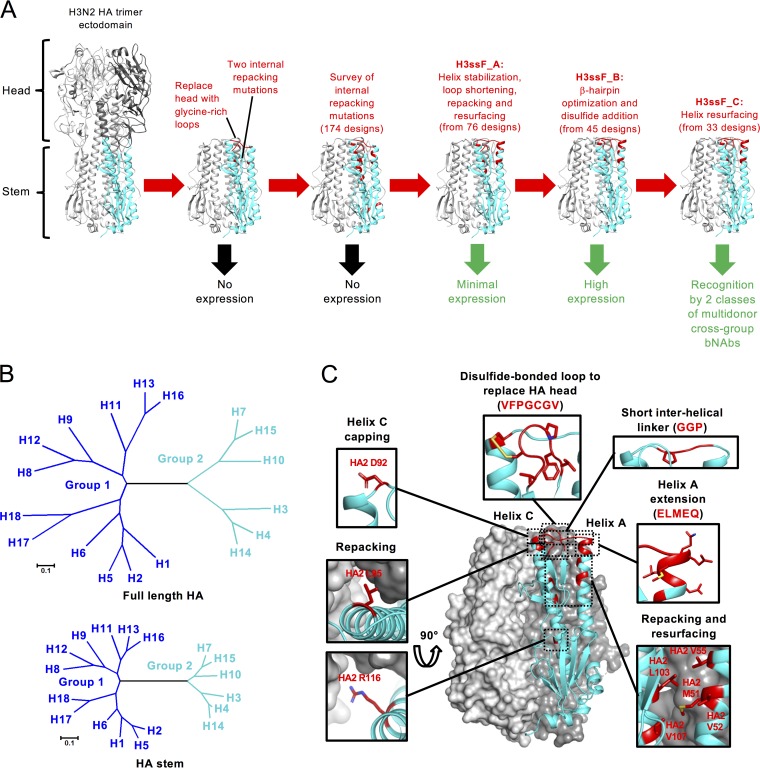
FIG 1.
Structure-based design of group 2 HA stem nanoparticles. (A) Ribbon models depicting the HA stem nanoparticle design pathway for stabilizing the HA stem starting from an H3N2 HA trimer ectodomain. Two of the three protomers are gray, and one protomer is cyan with the design modifications in red. Positive or negative outcomes are indicated by green or black arrows, respectively, below each step. bNAb, broadly neutralizing antibody. (B) Maximum-likelihood phylogenetic trees for protein sequences from representative full-length HAs (top) and HA stems (bottom) of the 18 different subtypes. Group 1 subtypes are dark blue, and group 2 subtypes are cyan. (C) A molecular model of the HA stem of an H3ssF_C design is shown colored as described for panel A. Two of the three protomers are depicted as surface representations, and one protomer is displayed as a ribbon diagram. Insets show close-ups of the modified regions with stick representations for side chains of modified residues. Surface representations are removed from some insets for clarity. PDB entry 2YP2 was used as an H3N2 model template.
Design of group 1 HA stem nanoparticle immunogens. Ribbon diagrams depict the design of HA stem-ferritin nanoparticles by (1) removing the head region of HA and replacing with glycine-rich linkers, (2) making two internal packing mutations, and (3) genetically fusing to a ferritin nanoparticle by a 3-residue SGG linker. Structures are shown by ribbon diagrams. One monomer of each HA and HA stem trimer is colored blue, and mutated regions or residues are colored red. The nanoparticle core is brown. Download FIG S1, PDF file, 2.3 MB (2.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Sequence alignments of select H3ssF (A) and H7ssF (B) variants with the full-length HA ectodomains for A/Finland/486/2004 (H3N2) (H3_FI04) and A/Shanghai/2/2013 (H7_SH13), respectively. Mutations are in red. Residues identical to the full-length sequence are represented by dots, and deletions represented by dashes. H3N2 numbering is shown above each alignment. For clarity, the HA head regions and the ferritin sequence are not shown. In each construct, the C-terminal HA2 residue 174 is connected to ferritin residue Asp5 by a short SGG linker (not shown). Download FIG S2, PDF file, 0.04 MB (44.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Since internal hydrophobic repacking mutations were critical for the stabilization of the group 1 HA stem immunogens (17) and the burying of just a single -CH2- group in a protein interior can add approximately 1.1 kcal/mol of free energy to protein stability (24, 25), we explored additional repacking mutations selected computationally using Rosetta (26). In combination with some of the repacking mutations, we also explored limited residue swapping between H1N1 and H3N2 subtypes (Table S1). Surprisingly, none of these 174 mutants could be expressed and recognized by the bNAb CR9114 or FI6v3 (Table S1), indicating that novel approaches for structural stabilization would be required.
Summary of group 2 nanoparticle designs. Download Table S1, XLSX file, 0.04 MB (44.3KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Upon further investigation of multiple protein stabilization strategies involving another 76 designs (Table S1), a lead design was successfully expressed (H3ssF_A) that was recognized by bNAb HA stem antibody FI6v3 by immunoprecipitation and self-assembled into nanoparticles (Table 1; Table S1, Fig. S2 and S3). H3ssF_A included the initial H1ssF-based mutations in addition to (i) helix stabilization, (ii) loop optimization, and (iii) point mutations of select H3 residues to improve internal packing (Fig. 1A and C; Fig. S2A). Each of the successful alterations was focused on the membrane distal end of the two central helices (A and C) of the HA2 fold. The outer helix A in H3N2 HA is approximately 5 residues shorter at its C terminus than in H1 HA, and it is known that helix length is related to stability (27). Therefore, helix A was extended at its C terminus by 5 residues (ALMAQ) with high helix-forming propensities (28). Likewise, the N terminus of the adjacent helix C was stabilized by adding an N-terminal Asp (HA2 92) as a helix-capping residue (29, 30), and two other mutations (HA2 S93A and N95L) were made at the N terminus of helix C to increase its helix-forming propensity and improve interprotomer packing (Fig. 1C). Biophysical studies have shown that excess loop length can destabilize protein folds by up to several kcal/mol (31), and in the initial H1ssF and H3ssF designs, helices A and C were connected by a 6-residue linker even though they are separated by only 12 Å. Therefore, the glycine-rich linker was shortened to a 4-residue GGPD linker to more closely match the interhelical distance. Finally, 4 residues (HA2 52, 55, 107, and 116) on helices A and C were mutated to improve side-chain packing (Fig. 1C; Fig. S2A). Three of these (HA2 52, 55, and 116) comprised mutations to H1 residues.
TABLE 1.
Physical characteristics of the H3ssF and H7ssF nanoparticles
| Subtype | Design | Design characteristic |
Relative yieldf |
Tm (°C) | EM characterizationg
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| HA1 loopa |
Helix A extensionb |
Interhelix loopc |
Helix C mutationd |
Predicted disulfide(s)e |
Ferritin core size (Å) |
HA stem size (Å) |
||||
| H3 | H3ssF_A | --GSG-- | ALMAQ | GGP | DAYL | NA | − | NDh | ND | ND |
| H3 | H3ssF_B | VFPGCGV | ALMAQ | GGP | DCYL | DS1 | +++ | 66.2 | 124.0 ± 7.0 | 82.0 ± 9.0 |
| H7 | H7ssF_B | +++ | 58.6 | 126.9 ± 6.8 | 71.6 ± 6.5 | |||||
| H3 | H3ssF_C | VFPGCGV | ELMEQ | GGP | DCYL | DS1 | ++ | 66.6 | 123.2 ± 3.4 | 65.0 ± 5.2 |
| H7 | H7ssF_C | +++ | 61.6 | 124.4 ± 4.4 | 66.9 ± 6.4 | |||||
| H3 | H3ssF_D | VFPGC-V | ALMAQ | GGP | DCYL | DS1 | ++ | 66.3 | 125.3 ± 5.1 | 67.7 ± 12.4 |
| H7 | H7ssF_D | +++ | 61.2 | 139.7 ± 4.8 | 64.8 ± 3.4 | |||||
| H3 | H3ssF_E | VFP-CGV | ALMAQ | GGP | DCYL | DS1 | ++ | 63.0 | 130.1 ± 5.3 | 66.9 ± 7.2 |
| H7 | H7ssF_E | + | 56.7 | 124.7 ± 6.5 | 68.1 ± 2.7 | |||||
| H3 | H3ssF_F | VFPGCGV | ALMEE | GGP | DCYL | DS1 | + | 65.6 | 127.1 ± 6.0 | 67.0 ± 4.1 |
| H7 | H7ssF_F | +++ | 59.9 | 130.2 ± 6.3 | 65.1 ± 5.5 | |||||
| H3 | H3ssF_G | CFNGIC- | ALMAQ | GGP | DAYL | DS2 | +++ | 59.9 | 126.3 ± 4.0 | 68.5 ± 3.3 |
| H3 | H3ssF_H | VFPGCGV | ELMEQ | GGP | DCYL | DS1, DS3 | ++ | 56.6 | 126.9 ± 3.5 | 67.8 ± 4.5 |
| H7 | H7ssF_I | VFPNCGV | ALMAQ | GGP | DCYL | DS1 | +++ | 58.7 | 124.3 ± 4.1 | 68.4 ± 6.6 |
| H7 | H7ssF_J | VFPGCGV | ALMAQ | GPP | DCYL | DS1 | +++ | 57.0 | 131.4 ± 5.1 | 62.6 ± 7.6 |
Between HA1 residues 42 and 314 (H3 numbering).
Directly following HA1 56.
Directly following the helix A extension.
Region between the interhelix loop and HA2 96.
DS1, HA1 C145-HA2 C93; DS2, HA1 C141-C146; DS3, HA1 C23-C322; NA, not applicable.
Based on gel electrophoresis analysis of unpurified supernatants following transient transfection.
EM, electron microscopy.
ND, not determined.
Characterization of H3ssF_A. (A) Superose 6 gel filtration chromatograms for H3ssF_A (magenta) and H3ssF_B (red) nanoparticles. (B) SDS-PAGE analysis of the results from an FI6v3 immunoprecipitation of the supernatants from H3ssF_A expressed in HEK293 cells. Molecular weight standards (MW) are designated kDa. The band indicative of H3ssF_A is boxed in magenta. H3ssF_null is a previous design iteration that did not express. H1ssF is shown as a positive control. (C) Negative-stain electron microscopy 2-D class averages of gel filtration-purified H3ssF_A. The white bar represents a 10-nm size marker. Download FIG S3, PDF file, 0.2 MB (250KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To further improve expression and stability, another 45 designs were explored, focusing on the short β-hairpin connected to a 3-residue loop which replaced the HA head in HA1 (Fig. 1). Since the stability of short, two-stranded β-sheets has been observed to increase with length (32), we replaced the GSG loop with longer loops of 6 or 7 residues designed to double the hairpin length. Residues with high strand-forming propensities (28), such as Val, Ile, and Phe, were used for strand extension, and dipeptides with propensities to form β-turns (33, 34), such as PG and NG, were used to connect them. Disulfides were also designed into the hairpin loops for added stability (35). From an initial survey of loop extensions, H3ssF_B, designed with a 7-residue hairpin loop extension disulfide bonded to the N terminus of helix C, resulted in expression levels of more than 5 mg/liter and recognition by CT149 (Fig. 1, Table 1; Table S1).
Further loop optimization of the H3ssF_B design included removing a glycine from either side of the cysteine in the β-hairpin loop (H3ssF_D and H3ssF_E) and incorporating a different β-hairpin sequence with an internal disulfide bond engineered to link both strands together (H3ssF_G). Additional H3ssF_B variants incorporated hydrophilic, helix-preferring Glu into the relatively hydrophobic helix A extension motif, for improved solubility (H3ssF_C, H3ssF_F, and H3ssF_H), and an engineered intraprotomer disulfide between HA1 residues 23 and 322 (H3ssF_H). All six variants resulted in nanoparticles recognized by CT149 in ELISAs but did not improve overall expression beyond that of H3ssF_B. Based on the successful expression and antigenic recognition of multiple H3ssF variants, we transferred mutations from constructs H3ssF_B, H3ssF_C, H3ssF_D, H3ssF_E, and H3ssF_F into analogous H7ssF nanoparticles using sequences from the human H7N9 strain (A/Shanghai/2/2013) and also created two additional loop variants (H7ssF_I and H7ssF_J) (Table 1; Fig. S2B). All seven of these H7ssF designs expressed milligram-per-liter quantities of nanoparticles recognized by CT149 (Table S1).
Physical characterization of H3ssF and H7ssF nanoparticles.
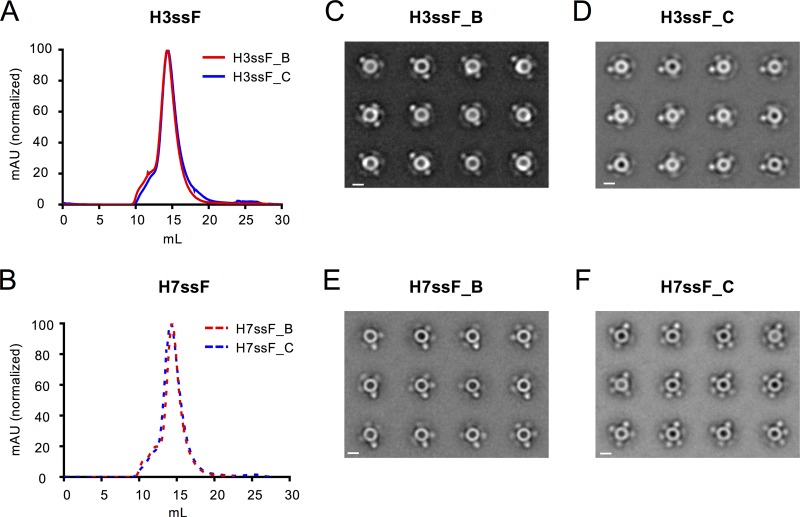
Fourteen nanoparticles were selected for further characterization. These were purified by lectin chromatography followed by size exclusion chromatography, which resulted in distinct peaks consistent with approximately 1.2-MDa particles (Fig. 2A and B; Fig. S4). Negative-stain electron microscopy of purified nanoparticles followed by reference-free 2-dimensional (2-D) classification and averaging revealed spherical particles with five or six visible, regularly spaced, protruding spikes (Fig. 2C to F; Fig. S4). All nanoparticles contained ferritin cores consistent with the expected 12- to 13-nm diameter and HA stems with the expected length of approximately 7 nm (Table 1).
FIG 2.
Physical characterization of group 2 HA stem nanoparticles. Gel filtration chromatograms for lectin-purified H3ssF (A) and H7ssF (B) nanoparticles reveal single peaks. (C to F) Negative-stain electron microscopy 2-D class averages for H3ssF (C, D) and H7ssF (E, F) B and C variants demonstrate the formation of particles with visible arrangements of HA stem trimers projecting from hollow spheres. White bars represent 10-nm size markers.
Physical characterization of H3ssF (A to E) and H7ssF (F to J) nanoparticles. Superose 6 gel filtration chromatograms (left panels) for lectin-purified nanoparticles reveal single peaks. Negative-stain electron microscopy 2-D class averages (right panels) demonstrate the formation of particles with visible arrangements of HA stem trimers projecting from hollow spheres. White bars represent 10-nm size markers for right panels. Download FIG S4, PDF file, 1.0 MB (1,023.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Thermostability of H3ssF and H7ssF nanoparticles.
We next investigated immunogen thermostability using differential scanning calorimetry (DSC). DSC measurements revealed thermal melting temperatures (Tm) for the earliest transitions ranging from 56.6 to 66.6°C for H3ssF variants and 56.7 to 61.6°C for H7ssF variants (Table 1; Fig. S5A and B). Three other, higher temperature transitions at approximately 78, 96, and 106°C were consistently observed for all the immunogens tested. In contrast, the ferritin nanoparticle alone had a primary transition at 105.7°C and a minor one at 84°C. Since the earliest transitions were the most variable, we hypothesized that these were related to the HA stem structures.
Differential scanning calorimetry (DSC) plots for group 2 HA stem immunogens. (A) H3ssF. (B) H7ssF. (C) Ferritin alone. Plots of CP (heat capacity at constant pressure) versus temperature depict melting transition for each protein. The CP values on the y axis are shown with an arbitrary scale. Download FIG S5, PDF file, 0.9 MB (950.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The three H3ssF designs with the highest Tms, ranging from 66.2 to 66.6°C, had either the 7-residue β-hairpin extension disulfide bonded to helix C (H3ssF_B and H3ssF_C) or the same loop with one glycine removed just after the cysteine (H3ssF_D). Glycine removal just prior to the cysteine (H3ssF_E) was less well tolerated thermodynamically. Curiously, H3ssF_H with the same hairpin loop as H3ssF_B and H3ssF_C but designed to contain an additional disulfide bond to stabilize the HA1 region of the stem had the lowest Tm, 56.6°C, indicating that this variant had less than optimal stability. Design H3ssF_G, engineered to have an internal disulfide bond in the β-hairpin extension, had a Tm 6°C lower than those of H3ssF_B and H3ssF_C. Incorporation of Glu into the two Ala positions in the 5-residue helix A extension had little effect on stability (H3ssF_C), but Glu incorporation to replace the last two residues decreased the Tm by 1°C (H3ssF_F). For the five immunogen designs (B to F) made in both H3 and H7 formats, the Tm values were on average 5.9°C higher for the H3ssF than for the H7ssF designs, and similar trends were observed between designs. Finally, two additional H7ssF variants not represented in H3ssF, with an Asn incorporated into the HA1 β-hairpin extension (H7ssF_H) and a Pro incorporated into the HA2 GGP loop (H7ssF_I), did not significantly alter the Tm. While improved thermostability will facilitate vaccine transport and storage, increasing rigidity and stability of antigens can sometimes be counterproductive for immunogenicity (36). Therefore, multiple designs with various levels of DSC-determined stability were advanced for further evaluation.
Antigenicity of H3ssF and H7ssF nanoparticles.
Biolayer interferometry (BLI) was used to measure the binding kinetics and affinities of nanoparticles for antigen-binding fragments (Fabs) of bNAbs MEDI8852, CT149, and CR8020 (Table 2; Fig. S6 and S7). To avoid avidity effects, the nanoparticles were immobilized to anti-Fc-decorated sensors using the bNAb CR9114 IgG and monovalent Fabs were used as the analyte. CR9114 Fab was observed to recognize H3ssF and H7ssF immunogens with affinities of 94 and 48 nM, respectively (Table 2; Fig. S6E). However, given the bivalent nature of the CR9114 IgG and the 24 epitope sites on each nanoparticle, the avidity of this complex was more than enough to stably immobilize nanoparticles to the sensor for measurements of high-affinity bNAb Fabs.
TABLE 2.
Antigenicity of the H3ssF and H7ssF nanoparticles for Fab forms of bNAbs as determined by BLIa
| Design | CT149 |
MEDI8852 |
CR8020 |
CR9114 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
KD
(nM) |
Kon
(M−1s−1) |
Koff
(s−1) |
KD (nM) |
Kon
(M−1s−1) |
Koff
(s−1) |
KD (nM) |
Kon
(M−1s−1) |
Koff
(s−1) |
KD (nM) |
Kon
(M−1s−1) |
Koff
(s−1) |
|
| H3ssF_A | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| H3ssF_B | NB | NB | NB | 0.7 | 2.9 × 105 | 2.2 × 10−4 | 20.9 | 2.8 × 105 | 5.8 × 10−3 | ND | ND | ND |
| H7ssF_B | 7.0 | 4.2 × 105 | 3.0 × 10−3 | 1.0 | 2.6 × 105 | 2.6 × 10−4 | NB | NB | NB | ND | ND | ND |
| H3ssF_C | 21.3 | 8.1 × 105 | 1.7 × 10−2 | 0.8 | 3.0 × 105 | 2.4 × 10−4 | 23.3 | 2.4 × 105 | 5.5 × 10−3 | 93.6 | 7.0 × 104 | 6.5 × 10−3 |
| H7ssF_C | 0.9 | 6.0 × 105 | 5.3 × 10−4 | 1.0 | 2.7 × 105 | 2.6 × 10−4 | NB | NB | NB | 47.9 | 1.0 × 105 | 4.8 × 10−3 |
| H3ssF_D | NB | NB | NB | 1.1 | 2.9 × 105 | 3.2 × 10−4 | 21.7 | 2.8 × 105 | 6.0 × 10−3 | ND | ND | ND |
| H7ssF_D | 9.0 | 3.7 × 105 | 3.3 × 10−3 | 0.9 | 2.7 × 105 | 2.3 × 10−4 | NB | NB | NB | ND | ND | ND |
| H3ssF_E | NB | NB | NB | 1.2 | 3.2 × 105 | 3.7 × 10−4 | 18.7 | 3.0 × 105 | 5.7 × 10−3 | ND | ND | ND |
| H7ssF_E | 8.2 | 3.7 × 105 | 3.0 × 10−3 | 0.8 | 2.9 × 105 | 2.4 × 10−4 | NB | NB | NB | ND | ND | ND |
| H3ssF_F | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| H7ssF_F | 2.7 | 6.7 × 105 | 1.8 × 10−3 | 0.9 | 2.9 × 105 | 2.4 × 10−4 | NB | NB | NB | ND | ND | ND |
| H3ssF_G | NB | NB | NB | 0.9 | 3.0 × 105 | 2.6 × 10−4 | 17.9 | 2.7 × 105 | 4.8 × 10−3 | ND | ND | ND |
| H3ssF_H | 22.2 | 8.2 × 105 | 1.8 × 10−2 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| H7ssF_I | 7.2 | 4.1 × 105 | 2.9 × 10−3 | 1.0 | 2.6 × 105 | 2.7 × 10−4 | NB | NB | NB | ND | ND | ND |
| H7ssF_J | 9.8 | 3.5 × 105 | 3.4 × 10−3 | 1.9 | 2.5 × 105 | 4.9 × 10−4 | NB | NB | NB | ND | ND | ND |
| H3 HA | 42.2 | 7.5 × 104 | 3.2 × 10−3 | 1.2 | 1.3 × 105 | 1.5 × 10−4 | ND | ND | ND | ND | ND | ND |
| H7 HA | 3.7 | 3.6 × 105 | 3.1 × 10−3 | 1.2 | 1.1 × 105 | 1.3 × 10−4 | ND | ND | ND | ND | ND | ND |
KD, equilibrium dissociation constant; Kon, association rate constant; Koff, dissociation rate constant; ND, not determined; NB, no detectable binding.
BLI binding curves for MEDI8852 (A and B) and CT149 (C and D) Fab recognition of H3ssF and H7ssF immunogens, respectively. Binding constants and kinetic parameters for each plot are shown in Table 2. (E) Binding of H3ssF_C and H7ssF_C to CR9114 Fab. Nanoparticles were immobilized to the sensor tip by binding to CR9114 IgG coupled by human anti-Fc antibody and HA trimers were immobilized on HIS1K sensors through C-terminal His tags. Data curves are in red; fitting for a 1 to 1 interaction are in black. Download FIG S6, PDF file, 0.8 MB (802.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Antigenic recognition of group 2 HA stem nanoparticles. (A) ELISA binding of H3ssF and H7ssF designs B and C by six broadly neutralizing HA stem antibodies. (B) Relative recognition of H3ssF (top) and H7ssF (bottom) by IgG and Fab forms of CT149 as measured by ELISA. In both (A) and (B), the nanoparticles were immobilized on the plate. (C) BLI binding curves for CR8020 Fab recognition of H3ssF (left) and H7ssF (right) immunogens. Binding constants and kinetic parameters for each plot are shown in Table 2. (D) Relative antigenic recognition of H3ssF and H7ssF by mature and inferred UCAforms of HA stem human antibodies as measured by BLI. CR9114 and the HA head antibody CH65 served as positive and negative controls, respectively. Antibody recognition was determined by Octet and binding relative to CR9114 was plotted on the y axis as a percentage. In (C) and (D), all nanoparticles were immobilized to the sensor tip by binding to CR9114 IgG coupled by human anti-Fc antibody. Download FIG S7, PDF file, 0.4 MB (391.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The first two antibodies (MEDI8852 and CT149) recognize similar HA stem epitopes centered on the HA2 helix A and fusion peptide loop supersite and represent two multidonor classes of cross-group bNAbs (HV6-1+HD3-3 class and HV1-18 QXXV class, respectively) recently identified in healthy human adults vaccinated with H5N1 (12, 14, 15). CR8020 is a group 2-specific bNAb that recognizes a less conserved, more membrane-proximal HA stem epitope (11). The HV6-1+HD3-3 class MAb MEDI8852 recognized all H3ssF and H7ssF nanoparticles, as well as full-length H3 and H7 HA trimers, with similar nanomolar affinities and kinetics (Table 2; Fig. S6A and B). The HV1-18 QXXV class MAb CT149 also recognized all H7ssF nanoparticles with approximately 1 to 10 nanomolar affinity. Of these, H7ssF_C was recognized with approximately 10-fold-greater affinity (0.9 nM) and a 10-fold-slower off rate (5.3 × 10−4 s−1) than the other H7ssF nanoparticles. However, among the H3ssF nanoparticles, only H3ssF_C and H3ssF_H were recognized by CT149, with affinities of 21.3 to 22.2 nM; the other four H3ssF immunogens showed no measurable affinity to CT149 (Table 2; Fig. S6). This was surprising, since each of these designs was initially chosen based on strong binding to CT149 in ELISAs. Furthermore, ELISAs performed on purified H3 and H7 immunogens for designs B and C also indicated a high affinity to each of the group 2-reactive HA stem antibodies tested (Fig. S7A). This apparent disparity between BLI and ELISA was potentially due to utilization of bivalent IgGs in ELISAs as opposed to monovalent Fabs in BLI assays, which provides an avidity advantage for apparent binding affinity of up to 2 orders of magnitude (37–39). Consistent with this idea, we observed in ELISAs that the Fab form of CT149 did not bind to H3ssF_B or H3ssF_C and bound only modestly to H7ssF_B and H7ssF_C (Fig. S7B). Moreover, ELISAs used higher densities of immobilized antigen than BLI, and there are inherent differences in the methodologies of the two assays. Full-length H3 and H7 HA trimers were observed to bind CT149 with affinities and kinetics consistent with those of the C and H versions of the H3ssF and H7ssF nanoparticles (Fig. S6C to D). Interestingly, the nanoparticles with greatest affinity for CT149, H3ssF_C, H3ssF_H, and H7ssF_C were each designed to have two outwardly facing Glu residues at the C-terminal end of helix A, whereas the other designs had Ala at these positions.
The group 2-specific HA stem bNAb CR8020 recognized all of the H3ssF nanoparticles tested with affinities of 17.9 to 23.3 nM but did not recognize any of the H7ssF nanoparticles (Table 2; Fig. S7C). This is consistent with the previously reported lack of CR8020 binding to full-length A/Shanghai/2/2013 (H7N9) HA, the same strain used in the H7ssF designs (40). The CR8020 epitope is not completely conserved even among group 2 HAs, as there are six epitope mutations between the A/Hong Kong/1/1968 (H3N2) HA and the A/Shanghai/2/2013 (H7N9) HA.
Newly recombined, naive B cell receptors (BCRs) need to first recognize an immunogen to initiate affinity maturation and B cell proliferation (41). Therefore, we also used BLI to assess the relative affinities of three sets of mature stem bNAbs and their inferred UCAs to HA stem nanoparticle immunogens (Fig. S7D). The UCAs were each inferred phylogenetically from multiple clonal family members and include unmutated CDRH3 loops (Fig. S8). Two of these (54-1G07 and 09-1B12) were from the human multidonor HV6-1+HD3-3 class and HV1-18 QXXV class, respectively (13). The third antibody, 04-1D02, was from an HV3-53+HD3-3 lineage. Measurable H3ssF and H7ssF immunogen recognition by inferred UCAs for each of the three antibodies was not considerably different from the binding of mature forms of the antibodies or the CR9114 positive control (Fig. S7D). This is interesting, considering that there are 2 to 5 unmutated CDRH3 residues in these UCAs (Fig. S8) and crystal structures of MAb complexes with HA show considerable CDRH3 binding for members of the HV1-18 QXXV and HV6-1 HD3-3 multidonor antibody classes (12, 14, 15).
Sequence alignments of mature and inferred UCA for mAbs 09-1B12, 16.a.26, 54-1G07, 04-1D02 and 01.a.44. Dots indicate identical residues. CDR loops are boxed according to Kabat numbering and convention. Shading indicates the antibody classes labeled in the upper left of each box. Download FIG S8, PDF file, 0.04 MB (44.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Immunogenicity of H3ssF and H7ssF nanoparticles.
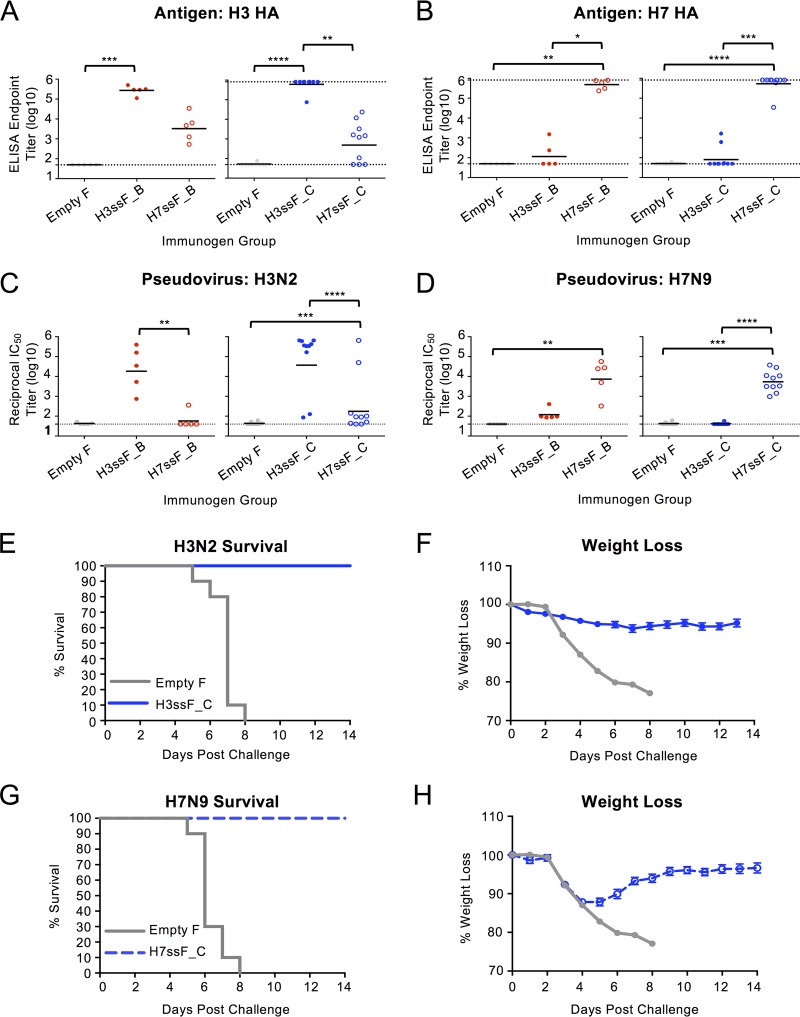
To evaluate the immunogenicity of the group 2 HA stem nanoparticles, we immunized mice three times intramuscularly at 4-week intervals with the B and C designs of H3ssF and H7ssF. Both H3ssF and H7ssF elicited robust homosubtypic antibody responses against H3 and H7, respectively (Fig. 3A and B). Similarly, strong homotypic H3ssF and H7ssF neutralizing antibody responses were elicited against H3N2 A/Wisconsin/67/2005 and H7N9 A/Anhui/1/2013 pseudoviruses, respectively (Fig. 3C and D). For both H3 and H7, B and C designs elicited similar levels of antibody responses (Fig. 3A to D). Following homosubtypic challenges, H3ssF-vaccinated mice were fully protected against H3N2 A/Philippines/2/1982 influenza virus (Fig. 3E, H3ssF_C); likewise, H7ssF vaccination fully protected mice from H7N9 A/Anhui/1/2013 (Fig. 3G, H7ssF_C). Overall, these animal studies demonstrated robust immunogenicity and protective efficacy of our group 2 HA stem nanoparticles against homotypic viruses, with limited cross-recognition of heterosubtypic strains by elicited sera. Although the stabilized group 2 HA stem antigens are immunogenic and protective, the murine species tested does not have the appropriate B cell repertoire to evaluate the potential for eliciting bNAbs with specific characteristics similar to those of known classes of human MAbs with cross-subtype binding (12, 42, 43).
FIG 3.
Immunogenicity of group 2 HA stem nanoparticles in mice. BALB/c mice were immunized with H3ssF or H7ssF. B and C design groups were tested in separate experiments. (A to D) Serum was assessed for binding antibodies to H3 A/Hong Kong/1/1968 HA (A) and H7 A/Anhui/1/2013 HA (B) and neutralizing antibodies against H3N2 A/Wisconsin/67/2005 (C) or H7N9 A/Anhui/1/2013 (D) pseudovirus. (E to H) H3ssF_C- and H7ssF_C-immunized mice were challenged with lethal doses of H3N2 A/Philippines/2/1982 or H7N9 A/Anhui/1/2013, respectively. Survival (E, G) and weight loss (F, H) were recorded postchallenge. Dotted lines in panels A to D represent assay limits of detection. One-way ANOVA with Kruskal-Wallis posttest was used to compare mean ELISA and IC50 titers between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
B cell activation by H3ssF and H7ssF nanoparticles.
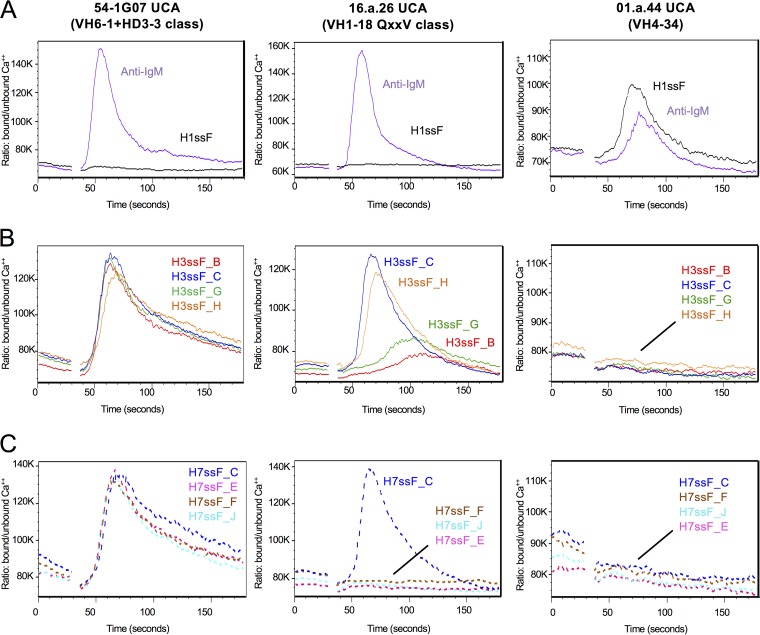
Recent studies have indicated that characterization of in vitro engagement of V gene-reverted BCRs by antigen can be used to assess and assign rank order to vaccine immunogens (43). Therefore, to better understand how the immunogens may be able to engage elements of the human antibody repertoire, Ca++ flux assays were used to measure activation of Ramos B cells expressing IgM versions of the inferred UCAs of the human antibodies 16.a.26 and 54-1G07, representing two multidonor classes of cross-group human bNAbs (HV1-18 QXXV class and HV6-1+HD3-3 class, respectively) (12, 13, 43). All eight H3ssF and H7ssF designs tested strongly activated the 54-1G07 UCA BCRs, with magnitudes comparable to that of the anti-IgM control, whereas a group 1 H1ssF negative control resulted in no activation (Fig. 4, left column). In contrast, only two H3ssF immunogens (H3ssF_C and H3ssF_H) and one H7ssF immunogen (H7ssF_C) were observed to elicit a strong activation of the 16.a.26 UCA BCR (Fig. 4, middle column). No activation of the HV4-34 group 1-specific 01.a.44 UCA BCR was observed (Fig. 4, right column) (12). Notably, these results were consistent with the BLI binding data. In both sets of experiments, the representatives of the multidonor HV6-1+HD3-3 class bNAbs engaged all of the H3ssF and H7ssF immunogens tested and the representatives of the multidonor HV1-18 QXXV class bNAbs engaged only the C and H design immunogens that contain added Glu in the helix A C-terminal extension. Our results suggest that these C and H designs could potentially activate nascent B cells expressing unmutated versions of multidonor HV6-1+HD3-3 or HV1-18 QXXV class bNAbs to initiate B cell proliferation and affinity maturation, thereby eliciting cross-group-reactive bNAb responses.
FIG 4.
Group 2 HA stem immunogens activate B cells expressing the inferred UCA versions of multidonor cross-group-reactive human BCRs. The results from Ca++ flux assays indicate B cell activation by the IgM positive control and a group 1 H1ssF (A), H3ssF variants (B), and H7ssF variants (C). BCR activation is displayed for the group 2-specific 54-1G07 UCA, group 2-specific 16.a.26 UCA activation, and group 1-specific 01.a.44 control BCRs. Ca++ flux was measured by flow cytometry as the ratio (y axis) of the Ca++-bound/-unbound states of the Ca++-sensitive dye fura red over time in seconds (x axis). Each experiment was repeated three times, and representative curves from one of the repeats are shown for each cell line.
DISCUSSION
To address the need for influenza virus immunogens that can elicit broad heterosubtypic responses in humans, particularly against group 2 influenza A virus, we used an iterative, structure-based design to develop headless group 2 HA stem-only nanoparticle immunogens from both H3 and H7 influenza virus subtype sequences. Stabilizing the group 2 HA stem was unexpectedly difficult compared to doing so for group 1 HA stem antigens. In addition to modifications used to achieve antigenically correct and immunogenic H1ssF candidate vaccines (17), multiple combined protein engineering strategies, including helix stabilization, loop optimization, and side-chain repacking, were required to achieve the first lead design candidate. Without all three stabilization strategies, the HA stem trimers were not stable enough to be expressed and displayed on self-assembling ferritin nanoparticles. This suggests that screening multiple stabilization methods concurrently may be necessary to design vaccine antigens from metastable proteins or proteins that have had significant portions removed.
Ultimately, H3ssF and H7ssF immunogen designs were identified that provided uniform, thermostable nanoparticles recognized by HA stem-directed bNAbs with subnanomolar to nanomolar affinity. Additionally, H3ssF and H7ssF were immunogenic in mice, eliciting robust homotypic neutralizing antibody responses that fully protected mice against homosubtypic lethal challenges.
The unique antibody repertoire of humans makes it difficult to predict or model the human immune response of candidate influenza vaccines using currently available animal models like mice (42, 44) or ferrets, particularly given the specific classes of human V, D, or J gene combinations associated with broadly cross-reactive immune responses across multiple individuals (12). Therefore, instead of relying on a mouse model to assess the potential breadth of antibody response, we utilized in vitro Ca++ flux assays (43) to evaluate the ability of group 2 HA stem immunogens to activate B cells expressing IgM BCRs comprising inferred UCA forms of the human, cross-group-reactive, HA stem-directed bNAbs from two multidonor classes (12). Germline targeting with vaccines designed to initiate specific antibody lineages is a new concept being tested with HIV antigens and not yet proven as a way of achieving broad immunity in humans. Even when transgenic animals are used for preclinical evaluation, the artificial constraints imposed may lessen correlations with the human immune system. Furthermore, since this type of strategy is intended to enhance de novo antibody responses, it may be particularly difficult to evaluate in preimmune adults and may require clinical evaluation in naive children to demonstrate proof of concept.
Consistency with the BLI binding results informed the choice to focus on immunogens H3ssF_C, H3ssF_H, and H7ssF_C, each of which was designed to have two outwardly facing surface Glu’s (at positions i and i + 3) at the C terminus of helix A. The first Glu occurs at a position (HA2 57) that naturally has a Glu in most human H3 strains from before 2002 and the majority of human H7 and H10 strains, although it is not in a helical conformation in full-length HA. Modeling with CT149 suggests the two Glu residues might interact with light-chain CDRL1 and CDRL2 loops (Fig. S9).
Model of CT149 interaction with helix A of H3ssF_C. The left panel shows a ribbon diagram model of CT149 (green and brown) bound to H3ssF_C (blue and gray). Helix A from one protomer of the H3ssF_C model was superimposed onto HA helix A (chain B) of the CT149/H3N2 HA crystal structure (PDB entry 4UBD) using PyMOL. For clarity, the H3N2 HA is not shown and only one CT149 Fab (chains C and D) is depicted. The middle panel shows a close-up of the light chain interaction with HA helix A and the right panel shows the same close-up view rotated 90° along the x-axis. For clarity, the middle and right panels show only helix A from one chain of the H3ssF_C model. The two Glu side chains at the C-terminal end of H3ssF_C helix A are shown as stick models. Download FIG S9, PDF file, 0.7 MB (703.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The observation that H3ssF and H7ssF variants were able to activate inferred UCA versions of cross-reactive BCRs suggests that these immunogens could potentially induce broad cross-group multidonor antibodies in humans. Due to their broad heterosubtypic breadth (12, 14, 15), such antibodies could potentially protect against pandemic strains, in addition to drifted seasonal strains, and preclude the need for annual vaccine reformulations. Furthermore, group 2 HA immunogens based on conventional H7N9 monovalent split product formulations have been shown to induce more diverse lineages of cross-group-reactive HA stem antibodies than conventional group 1 H5N1 immunogens in humans (13). Immunological imprinting from earlier or first exposures has also been suggested to play a significant role in shaping the subsequent immune response to influenza virus (45, 46). Thus, in influenza virus-naive individuals, the order of antigen priming may influence the composition of the influenza virus-specific B cell repertoire. Current vaccine approaches combine multiple strains of influenza virus in a single formulation. While this may be an effective strategy in adults with preexisting immunity from prior infections and vaccinations, it is possible that using an influenza A virus group 2 HA stem-based vaccine as the primary antigen exposure in children could potentially imprint or prime for broader cross-group responses later in life. Moreover, since titers of anti-HA stem antibodies increase with age and are inversely correlated with influenza virus infection rates, any early robust induction of broad, cross-reactive HA stem antibodies has the potential to mitigate the relatively high incidence of severe influenza disease in children, in addition to augmenting protection from influenza later in life (47). For adults and the elderly who have likely already encountered influenza virus, the group 2 stem immunogens could selectively boost antibody responses with greater breadth by not engaging otherwise predominant group 1 HA stem-specific HV1-69 antibodies (48, 49). These group 2 immunogens complement our earlier development of group 1 HA stem immunogens, which were demonstrated to elicit within-group heterosubtypic protection in mice and ferrets (17). Group 1 HA stem immunogens could potentially be used as a boost for the group 2 HA stem immunogens to better focus the antibody response to the neutralization-sensitive epitope centered on helix A. Finally, group 1 and 2 HA stem immunogens could also be given together in a manner analogous to the current multivalent inactivated influenza vaccines (IIV).
Another approach for inducing an HA stem antibody response involves immunizing sequentially with heterologous chimeric HA immunogens comprising two or more HAs with the same stem but head regions from different subtypes. This strategy has elicited heterosubtypic group 1 and group 2 protection in animal models (50–53). However, the exotic HA heads may still be immunodominant and potentially interfere with or limit HA stem-specific responses, as it has been shown in H5 vaccine trials in humans that a transient vaccine-induced boost of the stem-directed antibody responses can be replaced by immunodominant head-directed responses over time (49, 54). From a practical clinical perspective, careful records would need to be maintained to ensure the correct order of administration of heterologous vaccine products.
Although bacterially produced HA stem-only immunogens from H1, H5, H3, and H7 subtypes have also been reported to protect mice from H3 or H7 influenza virus, complete protection has only been observed for some homotypic strains (55–57), indicating that the breadth of response is limited. The immunogens in our study more closely mimic the native H3N2 and H7N9 HA stems in that they include native N-linked glycans as well as the membrane-proximal region of the HA stem that are both missing in the bacterially expressed designs. Eukaryotic expression of HA stem immunogens may be of particular importance because each native HA stem trimer has nine glycans and glycans have been observed to guide antibody binding by either masking certain epitopes (58, 59) or participating as part of an epitope surface (60). Indeed, many cross-group-reactive HA stem-directed bNAbs are immediately proximal to the HA2 N38 glycan when recognizing group 2 HA (12). Interestingly, the HA stem-directed bNAbs most frequently elicited from group 1 influenza virus, which does not have N38 glycans, are of the HV1-69 class (48), which is generally incompatible with the N38 glycan, and therefore, they are not cross-group reactive. Thus, group 2 HA stem immunogens bearing native glycans may provide a selective advantage for eliciting broad influenza virus antibody responses. However, to date, no experimental data have shown glycosylated hemagglutinin immunogens to be more effective than nonglycosylated versions. A recently reported particulate vaccine comprising nonglycosylated HA stem cross-linked to desolvated tandem M2e segments has been shown to elicit broad cross-group protection in mice (61). However, these particles comprised proteins derived from multiple host species, strains, and subtypes of influenza virus with the goal of accumulating breadth, as opposed to the approach used herein of attempting to achieve breadth through targeting conserved antigenic supersites.
In summary, we have designed group 2 headless HA stem trimers with conformational and antigenic fidelity to native HA. These antigens were immunogenic in mice and provided protection against a lethal homosubtypic challenge. Since mice lack critical genetic elements found in human bNAbs targeting the HA stem, we used a B cell receptor activation assay to demonstrate the potential for eliciting broadly reactive human antibody responses. Based on activation of B cells expressing inferred UCA versions of human antibodies associated with broad cross-neutralizing activity, candidate vaccines with antigens displayed on self-assembling ferritin nanoparticles are now being manufactured for clinical evaluation. Additional animal studies will be performed in parallel, including testing in mice expressing specific human immunoglobulin genes and in nonhuman primates. These immunogens represent a step toward achieving the goal of a vaccine that can elicit protective immunity against drifted seasonal and pandemic strains of influenza A virus.
MATERIALS AND METHODS
Structure-based design.
The crystal structure of A/Finland/486/2004 (H3N2) HA (PDB ID 2YP2) was used as a template for subtype H3, and the crystal structure of A/Shanghai/2/2013 (H7N9) HA (PDB ID 4LN6) was used as a template for subtype H7. The complete sequences for A/Finland/486/2004 HA and A/Shanghai/2/2013 (H7N9) HA, including the signal sequence regions not included in the crystal structures, were obtained from the database records with GISAID accession number EPI397685 and GenBank accession number YP_009118475.1, respectively. The changes in free energy from point mutations were estimated using the program DDG_MONOMER (26) from the ROSETTA suite. Loops were modeled with LOOPY (62), superpositions were performed using UCSF Chimera (63) or LSQMAN (64), and energy minimization of final models was performed using RELAX (65) from the ROSETTA suite. The graphics programs PyMOL (66) and UCSF Chimera (63) were used for detailed visual inspection of structures and the generation of structural figures.
Phylogenetic tree generation for HA sequences.
Maximum-likelihood trees were created using the program MEGA7.0 (67) from 18 representative HA sequences (13) aligned with MUSCLE (68). The stem regions of HA sequences were defined as residues HA1 10 to 42 and 314 to 329 and HA2 1 to 59 and 93 to 174 (H3 numbering [21]).
Antigenic screening of nanoparticle immunogen designs.
Most designs were initially screened in a high-throughput ELISA format as described previously (23). However, the contributions of individual mutations within multiple-mutation designs were also assessed by expressing transmembrane versions of H3 HA stem containing the native C-terminal HA sequence starting with HA2 175 (SVELKSGYKDWILWISFAISCFLLCVALLGFIMWACQKGNIRCNICI) on the surface of HEK293F cells and measuring recognition of bNAbs FI6v3 and CR8020 using flow cytometry. Since HA stem was more readily expressed in this format, with less stabilization required, this strategy enabled a more sensitive evaluation of HA stem mutations than the nanoparticle format. Promising designs that exhibited greater FI6v3 and CR8020 binding were subsequently expressed in the nanoparticle format for final evaluation.
Expression and purification of immunogens and antibodies.
All H3ssF and H7ssF nanoparticles and FI6v3, CT149, CR8020, CR6261, MEDI8852, 315-53-1F12, 54-1G07, 04-1D02, and 09-1B12 MAbs were expressed in 293 Expi cells (Life Technologies) and purified by affinity chromatography (Galanthus nivalis lectin for the nanoparticles and protein A for MAbs), using previously described methods (13, 17). Assembly of nanoparticles was assessed by gel filtration with a Superose 6 increase 10/300 GL column (GE Healthcare). To make MEDI8852 and CR8020 Fabs, IgG was digested with endoproteinase Lys-C (New England Biolabs) overnight at room temperature (RT). To make CT149 Fab, CT149 IgG containing an HRV-3C cleavage site engineered in the heavy-chain hinge region was digested using HRV-3C enzyme overnight at RT. The reactions were assessed by SDS-PAGE and, upon completion, were quenched by the addition of protease inhibitor cocktail (MilliporeSigma), and Fc and Fab antibodies were separated by protein A affinity chromatography.
ELISA binding assays.
ELISAs were done as previously described (17). Briefly, ELISA plates were coated with 100 ng/well H1ssF, H3ssF, or H7ssF antigens overnight at 4°C. The plates were then incubated with IgG or Fab forms of MAbs with dilution series starting at 10 μg/ml. For Fab, anti-CH1 MAb was then bound at 10 μg/ml. For testing of mouse sera, H3 HA and H7 HA antigens represented A/Hong Kong/1/1968 HA and A/Anhui/1/2013, respectively. Mouse serum serial dilutions started at 1:50. Appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies were used to detect primary antibody, followed by a colorimetric detection assay. Linear regression analysis of the absorbance values (optical density at 450 nm [OD450]) was completed using GraphPad Prism software. Serum endpoint titers were determined to be the serum dilution that resulted in a 4-fold increase in OD value above the background level. One-way analysis of variance (ANOVA) with Kruskal-Wallis posttest was used to compare mean endpoint titers between groups.
Immunoprecipitation assay.
Magnetic protein G Dynobeads (Thermo Fisher) were used to capture FI6v3 MAb according to the manufacturer’s protocol and allowed to incubate for 1 h at RT. FI6v3-bound Dynobeads were washed 2 times with 1× phosphate-buffered saline (PBS) and incubated with supernatants from H3ssF or H7ssF expressed in HEK293 cells or purified control protein for 1 h at RT. Beads were washed 2 times with 1× PBS and heated to 95°C for 10 min to detach bound protein. Proteins were then analyzed by SDS-PAGE.
Biolayer interferometry binding assays.
All biosensors were hydrated in PBS prior to use. CR9114 IgG (10 µg/ml diluted in 1% bovine serum albumin [BSA]–PBS) was immobilized on AHC biosensors through conjugated anti-human Fc antibody (fortéBio). After briefly dipping in assay buffer (1% BSA–PBS), the biosensors were dipped in various H3ssF and H7ssF constructs (10 µg/ml) to capture nanoparticles. The biosensors were then equilibrated in assay buffer for 1 min before being dipped in a 2-fold dilution series of CR8020, CT149, or MEDI8852 Fab for 5 min, followed by dipping in assay buffer to allow Fab to dissociate from nanoparticles for 5 min. All assay steps were performed at 30°C with agitation set at 1,000 rpm in the Octet HTX instrument (fortéBio). Correction to subtract a baseline drift was carried out by subtracting the measurements recorded for a sensor incubated with no Fab. Data analysis and curve fitting were carried out using Octet analysis software (version 9.0) as previously described (69, 70). Experimental data were fitted with the binding equations describing a 1:1 interaction. Global analyses of the complete data sets assuming binding was reversible (full dissociation) were carried out using nonlinear least-squares fitting, allowing a single set of binding parameters to be obtained simultaneously for all concentrations used in each experiment.
Electron microscopy analysis.
Samples were diluted to ∼0.02 mg/ml with buffer containing 10 mM HEPES, pH 7.0, and 150 mM NaCl and adsorbed to freshly glow-discharged carbon film grids for 15 s. The grids were washed with the same buffer and stained with 0.7% uranyl formate. Images were collected at a nominal magnification of 50,000 semiautomatically using SerialEM (71) on an FEI Tecnai T20 transmission electron microscope operated at 200 kV and equipped with a 2,048- by 2,048-pixel Eagle charge-coupled-device (CCD) camera. The pixel size was 0.44 nm. Particles were picked automatically using software developed in-house (Y.T., unpublished data). Particle alignment and reference-free 2-D classification were performed with SPIDER (72) and Relion 1.4 (73).
DSC.
Thermal melting points were determined using a MicroCal VP-capillary differential scanning calorimeter (DSC; Malvern Instruments). All protein samples were in the range of 0.2 to 0.5 mg/ml and were extensively dialyzed against PBS. Thermal denaturation was probed at a scan rate of 60°C/h from 30 to 120°C. Buffer correction, normalization, and baseline subtraction procedures were applied before the data were analyzed using the VP-capillary DSC automated data analysis software.
Mouse immunizations.
Animal experiments were carried out in compliance with all pertinent U.S. National Institutes of Health regulations and policies. The Animal Care and Use Committee of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, reviewed and approved all animal experiments. Female BALB/cJ mice aged 6 to 8 weeks (Jackson Laboratory) were immunized (n = 10) with 2 μg H3ssF or H7ssF and adjuvanted with the Sigma adjuvant system at 0, 4, and 8 weeks. Mice were inoculated with 100 μl intramuscularly, given as 50 μl into each hind leg. Two weeks after the final immunization, sera were collected for measurement of antibody responses. When appropriate, challenges were performed at 4 to 8 weeks postimmunization. Mice were inoculated intranasally with a 10× 50% lethal dose (LD50) of H3N2 A/Philippines/2/1982 or a 20× LD50 of H7N9 A/Anhui/1/2013. Weight loss was recorded daily for 14 days postchallenge.
Pseudovirus neutralization assays.
Lentivirus-based pseudoviruses displaying influenza HA and NA were produced as previously described (17). Mouse serum was treated with receptor-destroying enzyme (Denka Seiken). Serial dilutions of mouse sera (4-fold, starting point 1:40, 8 dilutions) were mixed with either H3N2 A/Wisconsin/67/2005 or H7N9 A/Anhui/1/2013 pseudovirus (whose titers to target 50,000 relative luciferase units [RLU] were previously determined) for 30 min at RT. The virus-pseudovirus mixture was then added to previously plated 293A cells (ATCC) in triplicate. After 2 h at 37°C and 5% CO2, fresh Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, and 1% penicillin–streptomycin was added. Cells were lysed at 72 h, and luciferase substrate (Promega) was added. Luciferase activity was measured as RLU at 570 nm on a SpectramaxL (Molecular Devices). Sigmoidal curves, taking averages of triplicates at each dilution, were generated from RLU readings; 50% neutralization (50% inhibitory concentration [IC50]) titers were calculated considering uninfected cells as 100% neutralization and cells transduced with only virus as 0% neutralization. One-way ANOVA with Kruskal-Wallis posttest was used to compare mean IC50 titers between groups.
Determination of inferred UCAs for HA stem antibodies.
Paired heavy- and light-chain DNA sequences for 58 members of the 54-1G07 clonal family (315-54-1G07) were isolated from single IgG+ B cells, along with 6 additional heavy-chain DNA sequences from cells for which light chains could not be amplified (13). A maximum-likelihood phylogenetic tree was constructed from concatenated heavy- and light-chain sequences and the most recent common ancestor (MRCA) of the lineage was inferred using DNAML in SONAR (74). This MRCA had 5 residual nucleotide mutations each in the heavy and light chain (resulting in 2 and 5 amino acid changes, respectively) compared to the sequences of the assigned germ line VH and VK genes. To construct the final UCA, these changes were reverted to the presumed germ line sequence. The same procedure was used for 57 paired heavy- and light-chain DNA sequences and 8 unpaired heavy-chain DNA sequences from the 09-1B12 clonal family. This MRCA had 1 residual nucleotide mutation (1 amino acid change) in the heavy chain and no residual mutations in the light chain. For the 04-1D02 clonal family, 29 paired heavy- and light-chain DNA sequences and 1 unpaired heavy-chain DNA sequence were used. The inferred MRCA contained 4 residual nucleotide mutations in the heavy chain and 6 nucleotide mutations in the light chain (4 and 5 amino acid changes, respectively). For this clonal family, only changes in VH genes were reverted; the lambda chain of 04-1D02-MRCA was used with the residual mutations included.
Ninety-three heavy-chain and 83 light-chain sequences were identified for the 16.a.26 clonal family and 51 heavy-chain and 49 light-chain sequences for the 01.a.44 clonal family from single sorted IgG+ B cells. For the heavy chain, a maximum-likelihood phylogenetic tree based upon the V gene sequence alone was constructed using Geneious (75), with each tree rooted to the respective germ line reference sequence as listed in the IMGT database. The VDJ junction of clonal members with the lowest mutation loads was inferred using IMGT V-quest, allowing identification of residues within the V, D, and J gene segments most likely subject to somatic mutation. To construct the heavy-chain UCA, these changes were reverted to the presumed germ line sequences. Due to the lack of precision in estimating N-nucleotide addition, mutations falling at V/D or D/J junctions were not reverted. For the light chains, an analogous process was used for V and J gene segments.
Generation of B cell lines.
B cell lines expressing inferred UCA versions of monoclonal antibodies were generated by transduction of surface-IgM-negative Ramos cells as previously described (43). Briefly, DNA encoding VDJ (heavy-chain) and VJ (light-chain) immunoglobulin regions was synthesized by GenScript and cloned, along with the consensus human IgM C region, into the pLVX-ZsGreen and the pLVX-mCherry expression vector (both from Clontech), respectively. Each of these vectors was then cotransfected with the lentivirus packaging plasmid psPAX2 and the vesicular stomatitis virus envelope protein G (VSV-G)-expressing plasmid pMD2.G (both from Addgene) into HEK293T cells for formation of lentiviral particles using Lipofectamine 2000 (Thermofisher). Supernatant was harvested 3 days after transfection and cleared by centrifugation before IgM-negative Ramos cells were cotransduced for both heavy and light chains. After 5 days, transduced Ramos cells were enriched by cell sorting for the expression of ZsGreen and mCherry using a FACSAria II interfaced to the FACSDiva software (BD Biosciences). This procedure was repeated until a pure double-positive population could be selected. Cell lines were further enriched for high IgM surface expression by using a fluorescently labeled anti-human IgM monoclonal antibody.
B cell activation assays.
The ability of the different particles to induce calcium flux upon BCR engagement was measured in vitro using inferred UCA monoclonal antibodies expressed as surface IgM in Ramos cells as previously described (76). Briefly, 1 million cells (per test) were stained with 0.35 μl fura red (Thermo Fisher Scientific) in 100 μl serum-free medium at ambient temperature in the dark for 30 min. After washing, cells were resuspended in 300 μl serum-free medium and heated to 37°C for 3 to 5 min in a heated water bath before being acquired on a FACSymphony interfaced to the FACSDiva software (BD Biosciences). Cells were first acquired for 30 s in the absence of antigen to record baseline intracellular calcium levels, and then tubes were removed (leaving the acquisition in progress) to add the antigen and quickly vortexed before being placed back on acquisition for a total time of 180 s. Nanoparticles were used at the final concentration of 50 nM. An anti-human unlabeled mouse F(ab′)2 anti-IgM (Southern Biotech) was used (1.5 μg/test) to determine the maximal induction of calcium flux measured by the change between the emission of fura red bound to calcium and that of unbound fura red (ratio) over time. The inferred UCA for 01.a.44 was chosen as a negative control since 01.a.44 is observed to bind only to group 1 HAs and not to group 2 HAs (not shown). Likewise, the inferred UCAs for 54-1D07 and 16.a.26 are observed to recognize group 2 HAs only (data not shown).
Accession number(s).
The sequences for the inferred UCA versions of the following HA stem antibodies have been deposited into GenBank under the accession numbers in parentheses: 54-1G07 (MH631450 and MH631451), 04-1D02 (MH997406 and MH997407), 09-1B12 (MH997404 and MH997405), 01.a.44 (MK291363 and MK291365), and 16.a.26 (MK291364 and MK291366).
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the Vaccine Research Center and the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Electron microscopy data collection and analyses were funded by federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract number HHSN261200800001E, and by Leidos Biomedical Research, Inc. (Y.T.). K.S.C.’s research fellowship was partially funded by the Undergraduate Scholarship Program, Office of Intramural Training and Education, Office of the Director, National Institutes of Health.
J.C.B., K.S.C., S.M.M., H.M.Y., M.K., L.W., J.R.M., and B.S.G. are named inventors on pending patent applications involving influenza vaccines.
Footnotes
Citation Corbett KS, Moin SM, Yassine HM, Cagigi A, Kanekiyo M, Boyoglu-Barnum S, Myers SI, Tsybovsky Y, Wheatley AK, Schramm CA, Gillespie RA, Shi W, Wang L, Zhang Y, Andrews SF, Joyce MG, Crank MC, Douek DC, McDermott AB, Mascola JR, Graham BS, Boyington JC. 2019. Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor B cell receptors of broadly neutralizing antibody lineages. mBio 10:e02810-18. https://doi.org/10.1128/mBio.02810-18.
REFERENCES
- 1.Fischer WA II, Gong M, Bhagwanjee S, Sevransky J. 2014. Global burden of influenza as a cause of cardiopulmonary morbidity and mortality. Glob Heart 9:325–336. doi: 10.1016/j.gheart.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2018. Seasonal influenza vaccine effectiveness, 2005–2018. CDC, Atlanta, GA. https://www.cdc.gov/flu/professionals/vaccination/effectiveness-studies.htm. Accessed 15 May 3018.
- 3.Dormitzer PR, Galli G, Castellino F, Golding H, Khurana S, Del Giudice G, Rappuoli R. 2011. Influenza vaccine immunology. Immunol Rev 239:167–177. doi: 10.1111/j.1600-065X.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- 4.Erbelding EJ, Post D, Stemmy E, Roberts PC, Augustine AD, Ferguson S, Paules CI, Graham BS, Fauci AS. 2018. A universal influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J Infect Dis 218:347–354. doi: 10.1093/infdis/jiy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews SF, Graham BS, Mascola JR, McDermott AB. 2017. Is it possible to develop a “universal” influenza virus vaccine? Immunogenetic considerations underlying B-cell biology in the development of a pan-subtype influenza A vaccine targeting the hemagglutinin stem. Cold Spring Harb Perspect Biol 2:a029413. doi: 10.1101/cshperspect.a029413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67:2552–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 10.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, Druz A, Georgiev IS, Gillespie RA, Kanekiyo M, Kong WP, Leung K, Narpala SN, Prabhakaran MS, Yang ES, Zhang BS, Zhang Y, Asokan M, Boyington JC, Bylund T, Darko S, Lees CR, Ransier A, Shen CH, Wang LS, Whittle JR, Wu XL, Yassine HM, Santos C, Matsuoka Y, Tsybovsky Y, Baxa U, Mullikin JC, Subbarao K, Douek DC, Graham BS, Koup RA, Ledgerwood JE, Roederer M, Shapiro L, Kwong PD, Mascola JR, McDermott AB. 2016. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell 166:609–623. doi: 10.1016/j.cell.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrews SF, Joyce MG, Chambers MJ, Gillespie RA, Kanekiyo M, Leung K, Yang ES, Tsybovsky Y, Wheatley AK, Crank MC, Boyington JC, Prabhakaran MS, Narpala SR, Chen X, Bailer RT, Chen G, Coates E, Kwong PD, Koup RA, Mascola JR, Graham BS, Ledgerwood JE, McDermott AB. 2017. Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci Immunol 2:eaan2676. doi: 10.1126/sciimmunol.aan2676. [DOI] [PubMed] [Google Scholar]
- 14.Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, Vorlaender MK, Bianchi S, Guarino B, De Marco A, Vanzetta F, Agatic G, Foglierini M, Pinna D, Fernandez-Rodriguez B, Fruehwirth A, Silacci C, Ogrodowicz RW, Martin SR, Sallusto F, Suzich JA, Lanzavecchia A, Zhu Q, Gamblin SJ, Skehel JJ. 2016. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell 166:596–608. doi: 10.1016/j.cell.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Cho M, Shore D, Song M, Choi J, Jiang T, Deng YQ, Bourgeois M, Almli L, Yang H, Chen LM, Shi Y, Qi J, Li A, Yi KS, Chang M, Bae JS, Lee H, Shin J, Stevens J, Hong S, Qin CF, Gao GF, Chang SJ, Donis RO. 2015. A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus. Nat Commun 6:7708. doi: 10.1038/ncomms8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura G, Chai N, Park S, Chiang N, Lin Z, Chiu H, Fong R, Yan D, Kim J, Zhang J, Lee WP, Estevez A, Coons M, Xu M, Lupardus P, Balazs M, Swem LR. 2013. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe 14:93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, Lingwood D, Moin SM, Andersen H, Okuno Y, Rao SS, Harris AK, Kwong PD, Mascola JR, Nabel GJ, Graham BS. 2015. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 18.Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RMB, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding Z, Apetri A, Kukrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu W, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, Rado Evi K. 2015. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 19.Belongia EA, Simpson MD, King JP, Sundaram ME, Kelley NS, Osterholm MT, McLean HQ. 2016. Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis 16:942–951. doi: 10.1016/S1473-3099(16)00129-8. [DOI] [PubMed] [Google Scholar]
- 20.Garten R, Blanton L, Elal AIA, Alabi N, Barnes J, Biggerstaff M, Brammer L, Budd AP, Burns E, Cummings CN, Davis T, Garg S, Gubareva L, Jang Y, Kniss K, Kramer N, Lindstrom S, Mustaquim D, O’Halloran A, Sessions W, Taylor C, Xu X, Dugan VG, Fry AM, Wentworth DE, Katz J, Jernigan D. 2018. Update: influenza activity in the United States during the 2017-18 season and composition of the 2018-19 influenza vaccine. MMWR Morb Mortal Wkly Rep 67:634–642. doi: 10.15585/mmwr.mm6722a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson IA, Skehel JJ, Wiley DC. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366–373. [DOI] [PubMed] [Google Scholar]
- 22.Lin YP, Xiong X, Wharton SA, Martin SR, Coombs PJ, Vachieri SG, Christodoulou E, Walker PA, Liu J, Skehel JJ, Gamblin SJ, Hay AJ, Daniels RS, McCauley JW. 2012. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci U S A 109:21474–21479. doi: 10.1073/pnas.1218841110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang GY, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko SY, Todd JP, Rao S, Graham BS, Kwong PD. 2013. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kellis JT Jr, Nyberg K, Sali D, Fersht AR. 1988. Contribution of hydrophobic interactions to protein stability. Nature 333:784–786. doi: 10.1038/333784a0. [DOI] [PubMed] [Google Scholar]
- 25.Pace CN, Fu H, Fryar KL, Landua J, Trevino SR, Shirley BA, Hendricks MM, Iimura S, Gajiwala K, Scholtz JM, Grimsley GR. 2011. Contribution of hydrophobic interactions to protein stability. J Mol Biol 408:514–528. doi: 10.1016/j.jmb.2011.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellogg EH, Leaver-Fay A, Baker D. 2011. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins 79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scholtz JM, Baldwin RL. 1992. The mechanism of alpha-helix formation by peptides. Annu Rev Biophys Biomol Struct 21:95–118. doi: 10.1146/annurev.bb.21.060192.000523. [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara K, Toda H, Ikeguchi M. 2012. Dependence of alpha-helical and beta-sheet amino acid propensities on the overall protein fold type. BMC Struct Biol 12:18. doi: 10.1186/1472-6807-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aurora R, Rose GD. 1998. Helix capping. Protein Sci 7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serrano L, Fersht AR. 1989. Capping and alpha-helix stability. Nature 342:296–299. doi: 10.1038/342296a0. [DOI] [PubMed] [Google Scholar]
- 31.Nagi AD, Regan L. 1997. An inverse correlation between loop length and stability in a four-helix-bundle protein. Fold Des 2:67–75. doi: 10.1016/S1359-0278(97)00007-2. [DOI] [PubMed] [Google Scholar]
- 32.Stanger HE, Syud FA, Espinosa JF, Giriat I, Muir T, Gellman SH. 2001. Length-dependent stability and strand length limits in antiparallel beta-sheet secondary structure. Proc Natl Acad Sci U S A 98:12015–12020. doi: 10.1073/pnas.211536998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hutchinson EG, Thornton JM. 1994. A revised set of potentials for beta-turn formation in proteins. Protein Sci 3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madan B, Seo SY, Lee SG. 2014. Structural and sequence features of two residue turns in beta-hairpins. Proteins 82:1721–1733. doi: 10.1002/prot.24526. [DOI] [PubMed] [Google Scholar]
- 35.Trivedi MV, Laurence JS, Siahaan TJ. 2009. The role of thiols and disulfides on protein stability. Curr Protein Pept Sci 10:614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westhof E, Altschuh D, Moras D, Bloomer AC, Mondragon A, Klug A, Van Regenmortel MH. 1984. Correlation between segmental mobility and the location of antigenic determinants in proteins. Nature 311:123–126. [DOI] [PubMed] [Google Scholar]
- 37.Borrebaeck CAK, Malmborg AC, Furebring C, Michaelsson A, Ward S, Danielsson L, Ohlin M. 1992. Kinetic-analysis of recombinant antibody-antigen interactions: relation between structural domains and antigen-binding. Biotechnology (N Y) 10:697–698. doi: 10.1038/nbt0692-697. [DOI] [PubMed] [Google Scholar]
- 38.Roggenbuck D, Konig H, Niemann B, Schoenherr G, Jahn S, Porstmann T. 1994. Real-time biospecific interaction analysis of a natural human polyreactive monoclonal Igm antibody and its Fab and Scfv fragments with several antigens. Scand J Immunol 40:64–70. [DOI] [PubMed] [Google Scholar]
- 39.Crespillo S, Casares S, Mateo PL, Conejero-Lara F. 2014. Thermodynamic analysis of the binding of 2F5 (Fab and immunoglobulin G forms) to its gp41 epitope reveals a strong influence of the immunoglobulin Fc region on affinity. J Biol Chem 289:594–599. doi: 10.1074/jbc.C113.524439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tharakaraman K, Subramanian V, Cain D, Sasisekharan V, Sasisekharan R. 2014. Broadly neutralizing influenza hemagglutinin stem-specific antibody CR8020 targets residues that are prone to escape due to host selection pressure. Cell Host Microbe 15:644–651. doi: 10.1016/j.chom.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, Schief WR, Nemazee D, Crotty S. 2018. Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity 48:133. doi: 10.1016/j.immuni.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroeder HW., Jr. 2006. Similarity and divergence in the development and expression of the mouse and human antibody repertoires. Dev Comp Immunol 30:119–135. doi: 10.1016/j.dci.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Weaver GC, Villar RF, Kanekiyo M, Nabel GJ, Mascola JR, Lingwood D. 2016. In vitro reconstitution of B cell receptor-antigen interactions to evaluate potential vaccine candidates. Nat Protoc 11:193–213. doi: 10.1038/nprot.2016.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnaout R, Lee W, Cahill P, Honan T, Sparrow T, Weiand M, Nusbaum C, Rajewsky K, Koralov SB. 2011. High-resolution description of antibody heavy-chain repertoires in humans. PLoS One 6:e22365. doi: 10.1371/journal.pone.0022365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gostic KM, Ambrose M, Worobey M, Lloyd-Smith JO. 2016. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354:722–726. doi: 10.1126/science.aag1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cobey S, Hensley SE. 2017. Immune history and influenza virus susceptibility. Curr Opin Virol 22:105–111. doi: 10.1016/j.coviro.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nachbagauer R, Choi A, Izikson R, Cox MM, Palese P, Krammer F. 2016. Age dependence and isotype specificity of influenza virus hemagglutinin stalk-reactive antibodies in humans. mBio 7:e01996-15. doi: 10.1128/mBio.01996-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whittle JRR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, Ledgerwood JE, Wei CJ, McDermott AB, Graham BS, Koup RA, Nabel GJ. 2014. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol 88:4047–4057. doi: 10.1128/JVI.03422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wheatley AK, Whittle JR, Lingwood D, Kanekiyo M, Yassine HM, Ma SS, Narpala SR, Prabhakaran MS, Matus-Nicodemos RA, Bailer RT, Nabel GJ, Graham BS, Ledgerwood JE, Koup RA, McDermott AB. 2015. H5N1 vaccine-elicited memory B cells are genetically constrained by the IGHV locus in the recognition of a neutralizing epitope in the hemagglutinin stem. J Immunol 195:602–610. doi: 10.4049/jimmunol.1402835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krammer F, Pica N, Hai R, Margine I, Palese P. 2013. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 87:6542–6550. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Margine I, Krammer F, Hai R, Heaton NS, Tan GS, Andrews SA, Runstadler JA, Wilson PC, Albrecht RA, Garcia-Sastre A, Palese P. 2013. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J Virol 87:10435–10446. doi: 10.1128/JVI.01715-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krammer F, Margine I, Hai R, Flood A, Hirsh A, Tsvetnitsky V, Chen D, Palese P. 2014. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J Virol 88:2340–2343. doi: 10.1128/JVI.03183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nachbagauer R, Liu WC, Choi A, Wohlbold TJ, Atlas T, Rajendran M, Solorzano A, Berlanda-Scorza F, Garcia-Sastre A, Palese P, Albrecht RA, Krammer F. 2017. A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. NPJ Vaccines 2:26. doi: 10.1038/s41541-017-0026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellebedy AH, Krammer F, Li GM, Miller MS, Chiu C, Wrammert J, Chang CY, Davis CW, McCausland M, Elbein R, Edupuganti S, Spearman P, Andrews SF, Wilson PC, Garcia-Sastre A, Mulligan MJ, Mehta AK, Palese P, Ahmed R. 2014. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci U S A 111:13133–13138. doi: 10.1073/pnas.1414070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mallajosyula VV, Citron M, Ferrara F, Temperton NJ, Liang X, Flynn JA, Varadarajan R. 2015. Hemagglutinin sequence conservation guided stem immunogen design from influenza A H3 subtype. Front Immunol 6:329. doi: 10.3389/fimmu.2015.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valkenburg SA, Mallajosyula VVA, Li OTW, Chin AWH, Carnell G, Temperton N, Varadarajan R, Poon LLM. 2016. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci Rep 6:22666. doi: 10.1038/srep22666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sutton TC, Chakraborty S, Mallajosyula VVA, Lamirande EW, Ganti K, Bock KW, Moore IN, Varadarajan R, Subbarao K. 2017. Protective efficacy of influenza group 2 hemagglutinin stem-fragment immunogen vaccines. NPJ Vaccines 2:35. doi: 10.1038/s41541-017-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skehel JJ, Stevens DJ, Daniels RS, Douglas AR, Knossow M, Wilson IA, Wiley DC. 1984. A carbohydrate side-chain on hemagglutinins of Hong-Kong influenza-viruses inhibits recognition by a monoclonal-antibody. Proc Natl Acad Sci U S A 81:1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tate MD, Job ER, Deng YM, Gunalan V, Maurer-Stroh S, Reading PC. 2014. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 6:1294–1316. doi: 10.3390/v6031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O’Dell S, Patel N, Shahzad-Ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang G-Y, Diwanji D, Georgiev I, Do Kwon Y, Lee D, Louder MK, Moquin S, Schmidt SD, Yang Z-Y, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang L-X, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng L, Mohan T, Chang TZ, Gonzalez GX, Wang Y, Kwon YM, Kang SM, Compans RW, Champion JA, Wang BZ. 2018. Double-layered protein nanoparticles induce broad protection against divergent influenza A viruses. Nat Commun 9:359. doi: 10.1038/s41467-017-02725-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiang Z, Soto CS, Honig B. 2002. Evaluating conformational free energies: the colony energy and its application to the problem of loop prediction. Proc Natl Acad Sci U S A 99:7432–7437. doi: 10.1073/pnas.102179699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 64.Kleywegt GJ, Zou JY, Kjeldgaard M, Jones TA. 2001. Around O, p 353–367. In Rossmann MG, Arnold E (ed), International tables for crystallography, vol F. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 65.Nivon LG, Moretti R, Baker D. 2013. A pareto-optimal refinement method for protein design scaffolds. PLoS One 8:e59004. doi: 10.1371/journal.pone.0059004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schrodinger. 2018. Pymol by Schrodinger. https://pymol.org/2/. Accessed 18 May 2018.
- 67.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Corti D, Misasi J, Mulangu S, Stanley DA, Kanekiyo M, Wollen S, Ploquin A, Doria-Rose NA, Staupe RP, Bailey M, Shi W, Choe M, Marcus H, Thompson EA, Cagigi A, Silacci C, Fernandez-Rodriguez B, Perez L, Sallusto F, Vanzetta F, Agatic G, Cameroni E, Kisalu N, Gordon I, Ledgerwood JE, Mascola JR, Graham BS, Muyembe-Tamfun JJ, Trefry JC, Lanzavecchia A, Sullivan NJ. 2016. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- 70.Misasi J, Gilman MS, Kanekiyo M, Gui M, Cagigi A, Mulangu S, Corti D, Ledgerwood JE, Lanzavecchia A, Cunningham J, Muyembe-Tamfun JJ, Baxa U, Graham BS, Xiang Y, Sullivan NJ, McLellan JS. 2016. Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science 351:1343–1346. doi: 10.1126/science.aad6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 72.Shaikh TR, Gao HX, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. 2008. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheres SH. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schramm CA, Sheng Z, Zhang Z, Mascola JR, Kwong PD, Shapiro L. 2016. SONAR: a high-throughput pipeline for inferring antibody ontogenies from longitudinal sequencing of B cell transcripts. Front Immunol 7:372. doi: 10.3389/fimmu.2016.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Villar RF, Patel J, Weaver GC, Kanekiyo M, Wheatley AK, Yassine HM, Costello CE, Chandler KB, McTamney PM, Nabel GJ, McDermott AB, Mascola JR, Carr SA, Lingwood D. 2016. Reconstituted B cell receptor signaling reveals carbohydrate-dependent mode of activation. Sci Rep 6:36298. doi: 10.1038/srep36298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Design of group 1 HA stem nanoparticle immunogens. Ribbon diagrams depict the design of HA stem-ferritin nanoparticles by (1) removing the head region of HA and replacing with glycine-rich linkers, (2) making two internal packing mutations, and (3) genetically fusing to a ferritin nanoparticle by a 3-residue SGG linker. Structures are shown by ribbon diagrams. One monomer of each HA and HA stem trimer is colored blue, and mutated regions or residues are colored red. The nanoparticle core is brown. Download FIG S1, PDF file, 2.3 MB (2.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Sequence alignments of select H3ssF (A) and H7ssF (B) variants with the full-length HA ectodomains for A/Finland/486/2004 (H3N2) (H3_FI04) and A/Shanghai/2/2013 (H7_SH13), respectively. Mutations are in red. Residues identical to the full-length sequence are represented by dots, and deletions represented by dashes. H3N2 numbering is shown above each alignment. For clarity, the HA head regions and the ferritin sequence are not shown. In each construct, the C-terminal HA2 residue 174 is connected to ferritin residue Asp5 by a short SGG linker (not shown). Download FIG S2, PDF file, 0.04 MB (44.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Summary of group 2 nanoparticle designs. Download Table S1, XLSX file, 0.04 MB (44.3KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Characterization of H3ssF_A. (A) Superose 6 gel filtration chromatograms for H3ssF_A (magenta) and H3ssF_B (red) nanoparticles. (B) SDS-PAGE analysis of the results from an FI6v3 immunoprecipitation of the supernatants from H3ssF_A expressed in HEK293 cells. Molecular weight standards (MW) are designated kDa. The band indicative of H3ssF_A is boxed in magenta. H3ssF_null is a previous design iteration that did not express. H1ssF is shown as a positive control. (C) Negative-stain electron microscopy 2-D class averages of gel filtration-purified H3ssF_A. The white bar represents a 10-nm size marker. Download FIG S3, PDF file, 0.2 MB (250KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Physical characterization of H3ssF (A to E) and H7ssF (F to J) nanoparticles. Superose 6 gel filtration chromatograms (left panels) for lectin-purified nanoparticles reveal single peaks. Negative-stain electron microscopy 2-D class averages (right panels) demonstrate the formation of particles with visible arrangements of HA stem trimers projecting from hollow spheres. White bars represent 10-nm size markers for right panels. Download FIG S4, PDF file, 1.0 MB (1,023.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Differential scanning calorimetry (DSC) plots for group 2 HA stem immunogens. (A) H3ssF. (B) H7ssF. (C) Ferritin alone. Plots of CP (heat capacity at constant pressure) versus temperature depict melting transition for each protein. The CP values on the y axis are shown with an arbitrary scale. Download FIG S5, PDF file, 0.9 MB (950.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
BLI binding curves for MEDI8852 (A and B) and CT149 (C and D) Fab recognition of H3ssF and H7ssF immunogens, respectively. Binding constants and kinetic parameters for each plot are shown in Table 2. (E) Binding of H3ssF_C and H7ssF_C to CR9114 Fab. Nanoparticles were immobilized to the sensor tip by binding to CR9114 IgG coupled by human anti-Fc antibody and HA trimers were immobilized on HIS1K sensors through C-terminal His tags. Data curves are in red; fitting for a 1 to 1 interaction are in black. Download FIG S6, PDF file, 0.8 MB (802.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Antigenic recognition of group 2 HA stem nanoparticles. (A) ELISA binding of H3ssF and H7ssF designs B and C by six broadly neutralizing HA stem antibodies. (B) Relative recognition of H3ssF (top) and H7ssF (bottom) by IgG and Fab forms of CT149 as measured by ELISA. In both (A) and (B), the nanoparticles were immobilized on the plate. (C) BLI binding curves for CR8020 Fab recognition of H3ssF (left) and H7ssF (right) immunogens. Binding constants and kinetic parameters for each plot are shown in Table 2. (D) Relative antigenic recognition of H3ssF and H7ssF by mature and inferred UCAforms of HA stem human antibodies as measured by BLI. CR9114 and the HA head antibody CH65 served as positive and negative controls, respectively. Antibody recognition was determined by Octet and binding relative to CR9114 was plotted on the y axis as a percentage. In (C) and (D), all nanoparticles were immobilized to the sensor tip by binding to CR9114 IgG coupled by human anti-Fc antibody. Download FIG S7, PDF file, 0.4 MB (391.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Sequence alignments of mature and inferred UCA for mAbs 09-1B12, 16.a.26, 54-1G07, 04-1D02 and 01.a.44. Dots indicate identical residues. CDR loops are boxed according to Kabat numbering and convention. Shading indicates the antibody classes labeled in the upper left of each box. Download FIG S8, PDF file, 0.04 MB (44.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Model of CT149 interaction with helix A of H3ssF_C. The left panel shows a ribbon diagram model of CT149 (green and brown) bound to H3ssF_C (blue and gray). Helix A from one protomer of the H3ssF_C model was superimposed onto HA helix A (chain B) of the CT149/H3N2 HA crystal structure (PDB entry 4UBD) using PyMOL. For clarity, the H3N2 HA is not shown and only one CT149 Fab (chains C and D) is depicted. The middle panel shows a close-up of the light chain interaction with HA helix A and the right panel shows the same close-up view rotated 90° along the x-axis. For clarity, the middle and right panels show only helix A from one chain of the H3ssF_C model. The two Glu side chains at the C-terminal end of H3ssF_C helix A are shown as stick models. Download FIG S9, PDF file, 0.7 MB (703.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.