In many bacteria, two or more homologous chemosensory pathways control several cellular functions, such as motility and gene regulation, in response to changes in the cell’s microenvironment. Cross talk between signal transduction systems is poorly understood; while generally it is considered to be undesired, in some instances it might be beneficial for coregulation of complex behaviors. We demonstrate that several receptors from the pathway controlling motility can physically interact with downstream components of the pathway controlling biofilm formation. We further show that a kinase from the pathway controlling motility can also phosphorylate a response regulator from the pathway controlling biofilm formation. We propose that cross talk between two chemosensory pathways might be involved in coordination of two types of cell behavior—chemotaxis and biofilm formation.
KEYWORDS: Comamonas, biofilms, chemoreceptors, chemotaxis, phosphotransfer, signal transduction
ABSTRACT
Complex chemosensory systems control multiple biological functions in bacteria, such as chemotaxis, gene regulation, and cell cycle progression. Many species contain more than one chemosensory system per genome, but little is known about their potential interplay. In this study, we reveal cross talk between two chemosensory pathways that modulate chemotaxis and biofilm formation in Comamonas testosteroni. We demonstrate that some chemoreceptors that govern chemotaxis also contribute to biofilm formation and these chemoreceptors can physically interact with components of both pathways. Finally, we show that the chemotaxis histidine kinase CheA can phosphorylate not only its cognate response regulator CheY2 but also one of the response regulators from the pathway mediating biofilm formation, FlmD. The phosphoryl group transfer from CheA to CheY2 is much faster than that from CheA to FlmD, which is consistent with chemotaxis being a fast response and biofilm formation being a much slower developmental process. We propose that cross talk between chemosensory pathways may play a role in coordination of complex behaviors in bacteria.
INTRODUCTION
Chemotaxis and biofilm formation are survival strategies that allow microorganisms to successfully find and dwell in environments. Chemotaxis is a process of active swimming toward attractants or away from repellents, which allows flagellated bacteria to monitor changes in the environment. Chemotaxis is best understood in model organisms Escherichia coli and Salmonella enterica (1). A chemotactic response is initiated by chemoreceptors, also called methyl-accepting chemotaxis proteins (MCPs) (2, 3) that detect various environmental signals through their sensory domains (4). Chemoreceptor signaling domains modulate the activity of the chemotaxis histidine kinase CheA. Following autophosphorylation, CheA transfers the phosphoryl group to the response regulator CheY. Phosphorylated CheY-P interacts with the flagellar switch protein FliM, causing a change in the direction of flagellar rotation (2, 5). Within the same chemotaxis pathway, the phosphatase CheZ (6), the methyltransferase CheR (7), and the methylesterase CheB (8) contribute to signal termination and adaptation.
In contrast to E. coli, which has only five chemoreceptors, many bacteria have a larger number of chemoreceptors; on average, fourteen chemoreceptor genes per bacterial genome were reported (9). In addition to chemotaxis, chemoreceptors and associated chemosensory pathways are implicated in regulation of twitching motility (10, 11), cell differentiation (12, 13) and aggregation (14), and biofilm formation (15–17). In Pseudomonas putida, the polyamine chemoreceptor McpU and the L-amino acids chemoreceptor McpA mediate chemotaxis and also contribute to biofilm formation (16).
Biofilm formation, a process of cell attachment and growing in aggregates on surfaces, is a regulated process that has been extensively investigated in model organisms, such as Pseudomonas species (18–22). In Pseudomonas aeruginosa, cyclic diguanosine-5′-monophosphate (c-di-GMP)-mediated signaling is the key regulatory circuit in biofilm formation. As a second messenger, c-di-GMP regulates biofilm formation by promoting the production of exopolysaccharides (23) and/or repressing synthesis of bacterial flagella (24). The Wsp chemosensory pathway in P. aeruginosa recognizes signals via its dedicated chemoreceptor WspA and contributes to biofilm formation via its response regulator WspR, which has diguanylate cyclase activity (15, 25). Together with other diguanylate cyclases and phosphodiesterases that modulate c-di-GMP levels as well as quorum sensing and small RNA signaling pathways, this chemosensory system contributes to a complex network that regulates biofilm formation (reviewed by Fazli et al. [20]).
Comamonas testosteroni CNB-1 belongs to a class of betaproteobacteria; it was isolated from a wastewater treatment bioreactor and grows on organic acids and aromatic compounds (26, 27). C. testosteroni is studied primarily as a promising organism for bioremediation of organics-contaminated environments: it forms organic-pollutant-degrading biofilms in natural ecosystems and water treatment systems (28). The process and mechanisms of biofilm formation in C. testosteroni are not well understood. C. testosteroni CNB-1 genome contains one chemotaxis (che) gene cluster and one chemotaxis-like (flm) gene cluster and nineteen chemoreceptor genes (29). In this study, we show that the flm cluster is involved in modulating biofilm formation in C. testosteroni and identify the FlmD protein as a response regulator for this behavior. We demonstrate that seven chemoreceptors contribute to biofilm formation, including those that are known to mediate chemotaxis. We also demonstrate that the CheA kinase can phosphorylate the FlmD response regulator, albeit at a much lower rate than its cognate response regulator CheY2. Therefore, we propose that chemotaxis and biofilm formation could be coregulated by the interplay between Che and Flm chemosensory pathways in C. testosteroni. Cross talk between chemosensory pathways in many other bacteria might play a similar role in coregulation of these and other types of cell behavior.
RESULTS
Two chemosensory pathways modulate the chemotactic response and biofilm formation in C. testosteroni.
Analysis of the C. testosteroni CNB-2 (identical to CNB-1 except for the loss of the pCNB plasmid) complete genome using the MiST.2 database (30) revealed two genetic clusters (Fig. 1; see also Fig. S1 in the supplemental material) encoding chemosensory pathways. On the basis of the results obtained in this study, we termed them che and flm clusters. The che cluster contained a complete set of genes coding for chemotaxis proteins, including the flagellar motor switch components (FliG, FliM, and FliN), the histidine kinase CheA, two response regulators CheY1 and CheY2, the phosphatase CheZ, the adaptor protein CheW, the methyltransferase CheR, the methylesterase CheB, and the deamidase CheD. Matching CheA, CheW, CheB, and CheR sequences to hidden Markov models designed for specific chemotaxis pathways revealed that the pathway encoded by the che cluster belongs to the evolutionary class F7 (31). The best-studied chemotaxis pathway in the model organism E. coli belongs to this class. Consequently, this pathway in C. testosteroni was also predicted to mediate chemotaxis. We have previously deleted the cheA gene (32), and in-frame deletions of cheW and cheY genes were made in this study. All the mutants were characterized for chemotaxis, and results are shown in Fig. S1B. As expected, CheA and CheW were essential for chemotaxis in strain CNB-1. Two response regulators, CheY1 and CheY2, are encoded in the che gene cluster. Deletion of cheY2 resulted in a complete loss of the chemotactic response, while the deletion of cheY1 only partly reduced chemotaxis (Fig. S1B). We also found that CheA and CheY2 from strain CNB-1 were able to restore chemotactic response in the corresponding E. coli mutants (Fig. S1C).
FIG 1.
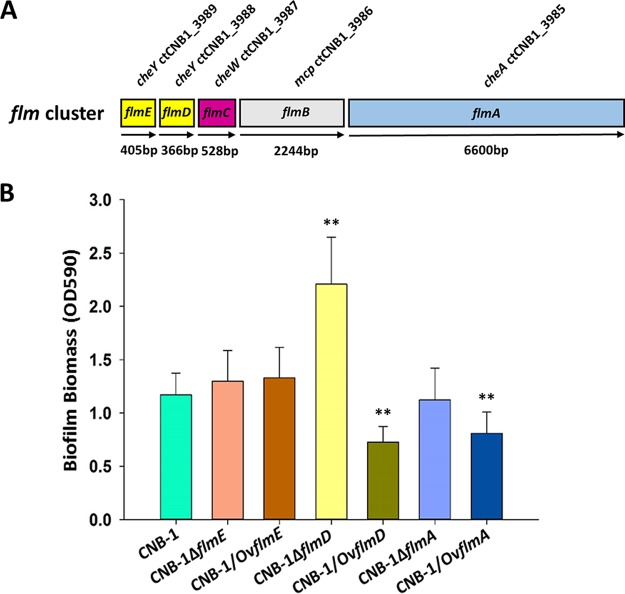
Flm pathway regulates biofilm formation. (A) Diagram of the flm genetic cluster. (B) Biofilm formation by flm gene deletion and overexpression mutants measured by a crystal violet assay. Data are the mean values plus standard deviations from triplicates. Values that are significantly different by Student’s t test are indicated by asterisks as follows: **, P < 0.01.
Che pathway regulates chemotaxis. (A) Diagram of the che genetic cluster. (B and C) Functional characterization of the che cluster in C. testosteroni (B) and E. coli (C) by swimming plate. Shown are the means and standard deviations from three independent experiments conducted in triplicate. Download FIG S1, TIF file, 0.4 MB (418.6KB, tif) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The flm gene cluster had not been studied previously. It contains genes encoding a CheA-like histidine kinase (termed FlmA), a CheW-like adaptor protein (termed FlmC), a chemoreceptor (ctCNB1_3986, termed FlmB), and two CheY-like response regulators that contain the conserved aspartyl residue, which serves as the phosphor-acceptor site (termed FlmD and FlmE). Matching FlmA (CheA-like) and FlmC (CheW-like) sequences to hidden Markov models designed for specific chemotaxis pathways revealed that the pathway encoded by the flm cluster belongs to the evolutionary class Tfp (31), named after type IV pilus-mediated motility. The Tfp pathway in P. aeruginosa (also known as Chp/Pil) regulates twitching motility (11) and causes alterations in the cAMP levels (33). Orthologous relationships between Chp/Pil and Flm were established by showing that ChpA-FlmA, PilJ-FlmB, PilI-FlmC, PilH-FlmD, and PilG-FlmE are mutual best BLAST hits when searched against the respective genomes (see Table S1 in the supplemental material). Although Flm is orthologous to Chp/Pil, it lacks MCP-modifying enzymes CheB and CheR (searches with Tfp-specific CheB and CheR sequences failed to identify any homologs in the C. testosteroni genome), and FlmB, a chemoreceptor associated with this pathway, lacks methylation sites (Fig. S3 and Table S2) that are conserved in its P. aeruginosa ortholog PilJ (34).
Orthologous relationships between components of the Flm and Chp/Pil pathways. Results of mutual best BLAST hits against corresponding genomes are shown along with their E values, percentage of identity, and length coverage. Download Table S1, PDF file, 0.01 MB (11KB, pdf) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Chemosensory apparatus of C. testosteroni CNB-2. Chemotaxis class definition for chemoreceptors is according to Alexander and Zhulin (35), where the number of helical heptads in signaling domain defines the class. Chemotaxis class definition for other proteins is according to Wuichet and Zhulin (31). Conserved methylation sites (M1 through M4) and a CheB/CheR-binding pentapeptide are deduced from the multiple sequence alignment shown in Fig. S3. Download Table S2, PDF file, 0.04 MB (38.3KB, pdf) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
While the Chp pathway in P. aeruginosa regulates twitching motility, we did not detect twitching motility in C. testosteroni strain CNB-1 under any condition tested. The Flm pathway had no effect on chemotaxis (Fig. S1D). The wild-type strain CNB-1 cells form a pellicle biofilm at the boundary of medium and air when grown in broth, and we tested whether the Flm pathway is involved in biofilm formation. We found that deletion of flmD resulted in a significant increase in biofilm formation (Fig. 1B), which suggested that FlmD functioned as a negative response regulator. We also observed that overexpression of the kinase FlmA and the response regulator FlmD resulted in a significant reduction of biofilm formation (Fig. 1B). Neither deletion nor overexpression of flmE (coding for another response regulator) had a significant effect on biofilm formation (Fig. 1B).
Multiple chemoreceptors modulate biofilm formation in C. testosteroni.
The early draft of C. testosteroni genome listed 20 chemoreceptor genes (reflected in the name of the chemoreceptor-null mutant CNB-1Δ20). The latest, high-quality whole genome contains 19 chemoreceptors that have diverse domain architectures (Fig. S2). Using previously described hidden Markov models (35), we assigned FlmB (MCP3986) to the 40H class (contains 40 helical heptads in the cytoplasmic signaling domain) and all other chemoreceptors (except for MCP0846, which did not match confidently to any model) to the 36H class (contains 36 helical heptads in the cytoplasmic signaling domain) (Table S2 and Fig. S3). While FlmB (MCP3986) is an ortholog of PilJ and it is predicted to interact with the Flm pathway, the 36H class chemoreceptors are known and predicted to interact with the F7 chemotaxis class (31, 34), i.e., the Che pathway in C. testosteroni. We analyzed the biofilm formation abilities of C. testosteroni CNB-1 mutants deficient in chemoreceptor genes. Deletions of individual chemoreceptor genes did not result in significant changes in biofilm formation (Fig. S4): however, the chemoreceptor-null mutant CNB-1Δ20 was severely affected in biofilm formation (Fig. 2), while its growth rate was not affected (Fig. S5). We then complemented the CNB-1Δ20 mutant with each of the 19 chemoreceptor genes, and the biofilm formation was assessed by using crystal violet staining. Unexpectedly, not only FlmB (MCP3986) but also six other chemoreceptors, namely, MCP0838, MCP0955, MCP2201, MCP2983, MCP3064, and MCP4715, restored biofilm formation to at least 80% of the wild-type CNB-1 (Fig. 2A). MCP2201 and MCP2983 were previously identified as chemoreceptors for chemotaxis (32, 36). Using confocal laser scanning microscopy, we showed that not only the adhesion ability but also the pellicle formation was restored by MCP2201 and MCP2983 (Fig. 2B and C). We tested further whether the ligands that are recognized by these chemoreceptors and trigger chemotactic responses would also affect biofilm formation. The addition of cis-aconitate, which is the sole ligand for MCP2983 (36), resulted in a significant increase in biofilm formation. Similarly, 2-ketoglutarate, cis-aconitate, fumarate, and oxaloacetate that are known ligands for MCP2201 (32) significantly promoted biofilm formation (Fig. 2D). These effects were seen only in the presence of the corresponding chemoreceptors (Fig. 2D), and the presence of a ligand did not significantly alter cell growth (Fig. S6).
FIG 2.
Chemoreceptors are involved in biofilm formation. (A) Biofilm formation by the CNB-1Δ20 mutant complemented with individual chemoreceptor genes measured by a crystal violet assay. (B) Average biofilm thickness by strain CNB-1, CNB-1Δ20, and chemoreceptor-complemented strains, calculated from confocal images. (C) Representative images of biofilm by confocal laser scanning microscopy (front view, x-axis profile, and y-axis profile). (D) Biofilm formation in the presence and absence of MCP2983 and MCP2201 ligands (final concentration, 2 mM). The values are means plus standard deviations from three independent experiments conducted in triplicate. Values that are significantly different are indicated by asterisks as follows: *, P < 0.05 by Student’s t test or rank sum test; **, P < 0.01 by Student’s t test or rank sum test.
Domain architectures of C. testosteroni chemoreceptors revealed by using CDvist. Transmembrane regions and Pfam domains are shown. Download FIG S2, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Multiple sequence alignment of signaling domains from C. testosteroni chemoreceptors. Predicted methylation sites are shown in red, and adjucent consensus sequences are shown in gray; predicted C-terminal pentapeptides are highlighted in yellow. Download FIG S3, PDF file, 0.06 MB (61.9KB, pdf) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Single chemoreceptor deletion does not affect the biofilm formation. Biofilm formation by each single chemoreceptor mutant measured by a crystal violet assay. Download FIG S4, PDF file, 0.5 MB (543.7KB, pdf) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth rates of the receptor-free mutant (Δ20) and the wild type (CNB-1) on different carbon sources. Gen, gentisate; 4HB, 4-hydroxybenzoate; 3HB, 3-hydroxybenzoate; PCA, protocatechuate. Download FIG S5, TIF file, 2.0 MB (2MB, tif) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of chemoreceptor mutants in the presence and absence of their ligand cis-aconitate. CNB-1, wild type; Δ20/mcp2201, Δ20 mutant complemented with the corresponding chemoreceptor gene; Δ20/mcp2983, Δ20 mutant complemented with the corresponding chemoreceptor gene; Δ20, mutant lacking all chemoreceptor genes. Growth conditions are described in Materials and Methods. Download FIG S6, PDF file, 1.2 MB (1.2MB, pdf) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Physical interactions between the Che and Flm proteins.
The observation that MCP2201 and MCP2983 that are known to mediate chemotaxis also affected biofilm formation suggested there could be potential cross talk between Che and Flm pathways. We used bacterial two-hybrid systems (BACTH) to identify possible protein-protein interactions between the components of the two pathways (Fig. 3). As expected, interactions between 36H class chemoreceptors (MCP2201, MCP2901, MCP2983, and MCP4715) and the histidine kinase CheA and the adaptor protein CheW were observed. Unexpectedly, we also observed interactions of these chemoreceptors with the kinase FlmA and the adaptor FlmC (Fig. 3), which suggests that physical interactions of chemoreceptors from the Che pathway with a kinase and an adaptor from the Flm pathway might be one of the potential mechanisms for observed cross talk.
FIG 3.
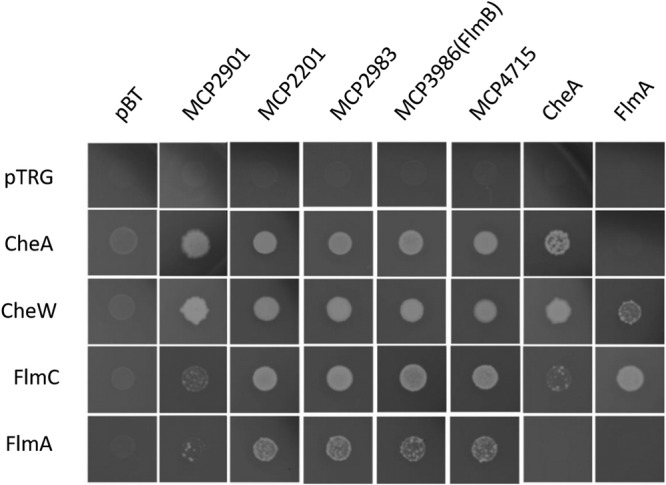
Chemoreceptors interact with other components of Che and Flm pathways. The growth of bacterial two-hybrid system cotransformants is shown on selective screening medium plates. Better growth represents a stronger interaction.
Phosphotransfer between CheA and response regulators of the Che and Flm pathways.
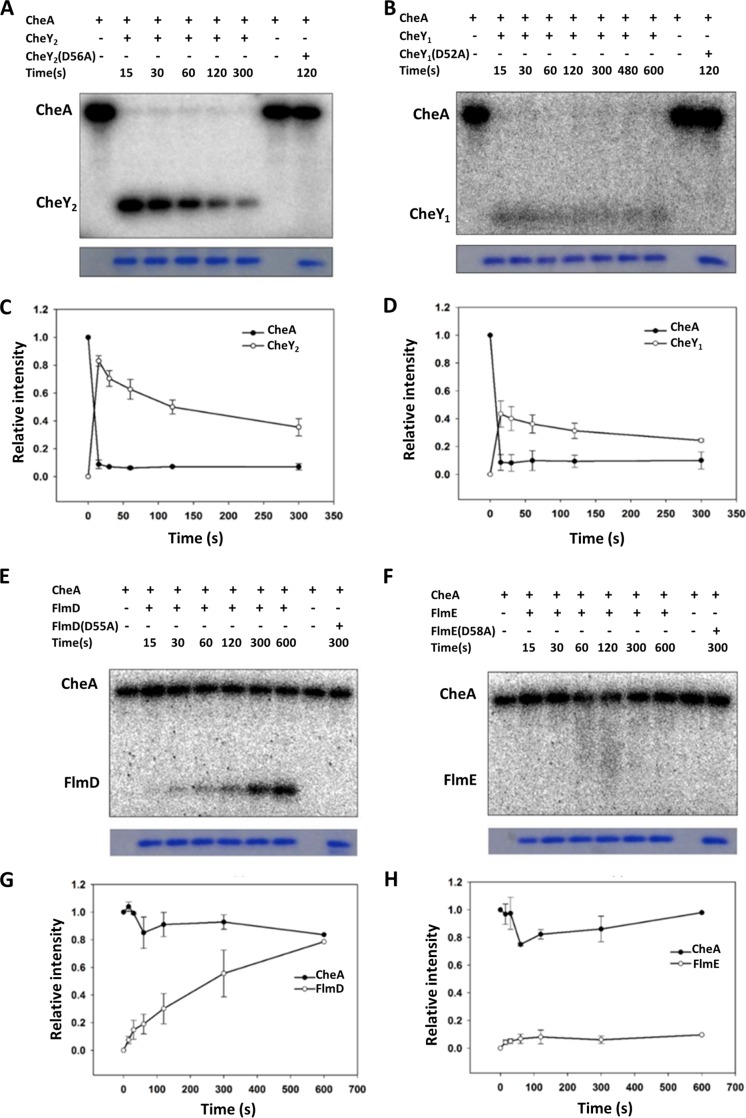
The phosphotransfer from the histidine kinase CheA to the response regulator CheY is the first step in signal transduction during chemotaxis. In order to measure phosphotransfer between kinases and response regulators of the two pathways, we purified recombinant CheA, CheY1, and CheY2, as well as FlmD and FlmE. Efforts to purify the histidine kinase FlmA were unsuccessful. As expected, we recorded a strong and clear phosphotransfer from CheA to one of the chemotaxis response regulators, CheY2 (Fig. 4A). We also observed a phosphotransfer from CheA to CheY1, but with much faster CheY autodephosphorylation rate (Fig. 4C and D), which implied that CheY1 might play a role as a phosphate sink, as previously suggested for other chemotaxis pathways with two CheY response regulators (37, 38). Unexpectedly, we observed that the kinase CheA phosphorylated FlmD (but not another response regulator, FlmE) (Fig. 4E and F). Compared to CheY2, the phosphotransfer from CheA to FlmD occurred at a significantly lower rate. FlmD received an equivalent level of phosphorylation from CheA at 600 s, while CheY2 achieved it at 15 s (Fig. 4C and G). CheY2 was quickly phosphorylated by CheA, and the phosphorylated CheY2 decayed after 15 s (Fig. 4A). FlmD was continuously phosphorylated even after 600 s of incubation.
FIG 4.
The phosphoryl group transfers from CheA to CheY1, CheY2, and FlmD. (A to H) Representative phosphotransfer images (A, B, E, and F) and time courses of the phosphotransfer from CheA-P to CheY1, CheY2, FlmD, and FlmE (C, D, G, and H). The data are presented as the mean values of three independent experiments. Error bars represent the standard deviations (SD).
The conserved aspartate residues serving as phosphorylation sites (39) were unchanged in all three response regulators: D52 in CheY1, D56 in CheY2, and D55 in FlmD. When the phosphorylation site was mutated from aspartate to alanine, the phosphotransfer from CheA to each mutant response regulator was no longer observed (Fig. 4).
FlmD modulates biofilm formation in the presence and absence of FlmA.
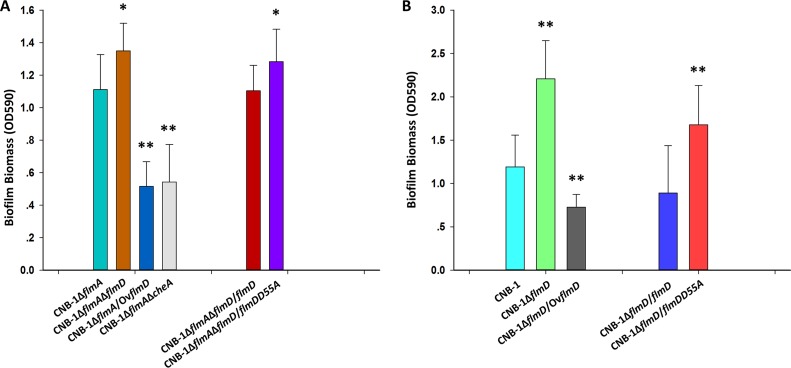
On the basis of the observation that kinase CheA can phosphorylate the response regulator FlmD (potential signal cross talk between Che and Flm pathways), we carried out experiments using mutants to demonstrate in vivo that such interplay might affect biofilm formation. To exclude the influence from the kinase FlmA (whose cognate target is FlmD/FlmE) on the potential phosphotransfer from CheA to the response regulator FlmD, we used the flmA knockout mutant. As shown in Fig. 5A, deletion of flmD resulted in upregulation of biofilm formation, whereas overexpression of FlmD resulted in significant reduction of biofilm formation, and deletion of cheA also caused a significant decrease in biofilm formation. These results demonstrated that the response regulator FlmD and the kinase CheA modulate biofilm formation in the absence of the FlmA kinase. This was further confirmed by the fact that the D55A FlmD mutant deficient in the phosphor-acceptor site showed enhanced biofilm formation compared to the FlmA mutant and the wild type (Fig. 5A and B). On the basis of these results, we conclude that a phosphorylated FlmD negatively regulates biofilm formation. This further supported by experiments showing that in the presence of the kinase FlmA, the phosphorylated response regulator FlmD (overexpressed as wild-type FlmD) also negatively regulates biofilm formation (Fig. 5B).
FIG 5.
FlmD regulates biofilm formation in vivo in the presence and absence of FlmA. (A) Functional characterization of FlmD mutants in the absence of FlmA (A) and in the presence of FlmA (B) measured by crystal violet assay. Shown are the means and standard deviations from three independent experiments conducted in triplicate. *, P < 0.05 by Student’s t test or rank sum test; **, P < 0.01 by Student’s t test or rank sum test.
DISCUSSION
Chemotaxis and biofilm formation are processes that are important for different lifestyles in bacteria. Chemotaxis is a rapid response to fluctuating conditions in the microenvironment, such as gradients of nutrients. Biofilm formation is a response to persistent changes, such as transition from a liquid environment to a surface. Actively moving chemotactic cells live in a planktonic state, whereas cells in biofilms live in a sessile state. Switching from one lifestyle to another requires coordinated regulation, and cross talk between regulatory systems might be one type of such coordination. We found that the same signals—organic acids—serve as chemoattractants and stimulate biofilm formation, which might seem counterintuitive, because typically, these are inversely regulated processes. One possible explanation is that chemotaxis allows C. testosteroni cells to detect low concentrations of organic acids and by moving along their gradients to find higher concentrations that sustain metabolism and proliferation and trigger biofilm formation. In such a scenario, biofilm helps bacteria to establish themselves and to remain in a favorable microenvironment.
Cross talk between chemotaxis and other signaling pathways, such as pili-mediated surface motility or virulence induction signaling system, has been proposed (40, 41) but not demonstrated. In this study, we showed that two chemosensory pathways in C. testosteroni modulate chemotaxis and biofilm formation. Comparative genomic analysis revealed that the che pathway in C. testosteroni belongs to the most abundant type of the chemotaxis signal transduction class, F7, which controls flagellar motility in a closely related model organism E. coli. Seventeen chemoreceptors from the C. testosteroni genome were predicted computationally to feed into the Che pathway, and three of them were previously shown to govern chemotaxis (32, 36, 42). By showing that the chemotaxis response was lost in cheA (32), cheW, and cheY (this study) mutants, we firmly established the role for this signal transduction pathway.
Computational analysis showed that the second chemosensory pathway in C. testosteroni, which we termed Flm, belongs to the evolutionary class Tfp (31), and it is orthologous to the Chp/Pil pathway, which modulates twitching motility and virulence in P. aeruginosa (11, 33, 43). In comparison with Chp/Pil, the Flm pathway lacks three components: the additional adaptor protein ChpC, the methyltransferase CheR, and the methylesterase CheB. Furthermore, FlmB, the only chemoreceptor predicted to feed into the Flm pathway, lacks methylation sites, which is consistent with the loss of methylation/demethylation enzymes. Therefore, Flm function was expected to be somewhat different from that of the Chp/Pil pathway. We have found that the Flm pathway modulates biofilm formation in C. testosteroni and that the response regulator FlmD, which is the preferred target of FlmA kinase phosphorylation, serves as a negative response regulator. FlmD is orthologous to the response regulator PilH of the Chp/Pil pathway, which is also the preferred target of ChpA phosphorylation (44), but its function is not well understood. PilG, another response regulator of the Chp/Pil pathway, is required for Tfp function as a motility organelle and mechanosensor in P. aeruginosa (33, 45). We did not detect twitching motility in C. testosteroni under any condition tested, but we showed that FlmE, the PilG ortholog, has no role in biofilm formation. We searched for CyaA or CyaB homologs in the C. testosteroni genome that would potentially suggest that the Flm pathway might regulate cAMP levels, as does the orthologous pathway in P. aeruginosa (33); however, these searches failed to identify any proteins orthologous to CyaA and CyaB.
A chemosensory pathway modulating biofilm formation (namely, Wsp) was previously identified in P. aeruginosa (15, 25) and P. putida (16). The response regulator WspR contains a c-di-GMP cyclase domain (GGDEF) as the pathway output. As a result of chemosensory signal transduction, increased levels of c-di-GMP enhance biofilm formation (15, 25). The Wsp pathway belongs to a different evolutionary class—ACF (named ACF for alternative cellular functions) (31), and the Flm response regulators do not contain GGDEF domains. Furthermore, they are both comprised of a single response regulator receiver domain, similar to the chemotaxis response regulator CheY. Such single domain response regulators are ubiquitous, and they might have multiple, spatially separated targets (46). Biofilm formation is a very complex process (47), and the target for the response regulator FlmD remains to be identified.
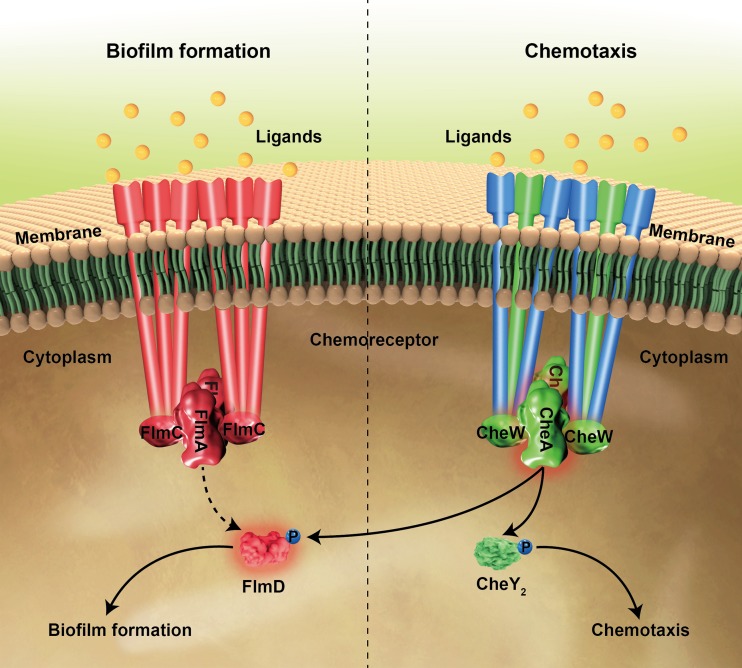
We documented cross talk between the two chemosensory systems in C. testosteroni at two potential sites (Fig. 6): (i) chemoreceptor interaction with a nonpartner pathway and (ii) phosphotransfer from a kinase to a nonpartner response regulator. Results obtained in bacterial two-hybrid screens raise the possibility that chemoreceptors from the Che pathway may interact with the adaptor protein FlmC and the histidine kinase FlmA. These interactions might be insignificant in vivo, because chemoreceptors from Che and Flm pathways belong to different length classes. Chemoreceptors of different length classes in orthologous systems in P. aeruginosa were found to possess pathway specificity determinants (34) that likely target them to “preferred” partners in spatially separated signaling arrays.
FIG 6.
Model of signal transduction and cross talk between Che and Flm pathways. Proteins that are involved in chemotaxis only are shown in green, proteins that are involved in biofilm formation only are shown in red, and proteins that are involved in both chemotaxis and biofilm formation are shown in blue. Che and Flm pathways regulate chemotaxis and biofilm formation, respectively. Che pathway might also influence biofilm formation through the phosphotransfer from CheA to FlmD. The pathways that have not been genetically or biochemically confirmed are represented by dashed lines.
Phosphotransfer from the histidine kinase CheA to the response regulator FlmD is a more likely site for cross talk. Our results supporting FlmD phosphorylation by CheA (both in vitro and in vivo) were obtained in the absence of the histidine kinase FlmA. Similar cases of phosphotransfer from a histidine kinase to a noncognate response regulator in the absence of its own histidine kinase have been reported previously (48). However, a series of studies argue that such cross talk is physiologically irrelevant in wild-type cells in vivo (49–51). One of the key arguments is that in systems where cross talk was observed, the output of the system is blind to input stimulus (49, 52). In our case, the FlmD modulated output, i.e., biofilm formation, appears to be responsive to input stimulus: ligands specific to a chemoreceptor, which signals through CheA, modulated biofilm formation, and it was observed only in the presence of a corresponding chemoreceptor. Observations consistent with the proposition that components of chemosensory pathways controlling chemotaxis also modulate biofilm formation have been previously reported. The BdlA chemoreceptor in P. aeruginosa, which is predicted to feed into the chemotaxis pathway (34), is essential for biofilm dispersal (53, 54). Inactivation of the chemotaxis methyltransferase CheR (55) and the response regulator CheY (56) in P. aeruginosa also led to defects in biofilm formation, although the basis for this behavior is unknown. In a closely related bacterium Shewanella oneidensis, a chemosensory pathway was also implicated in biofilm formation (57), likely via the interaction of its response regulator CheY3 with the c-di-GMP-binding protein (58). The results described here provide potential mechanisms for these and other observations linking chemotaxis and biofilm formation and suggesting their coregulation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. C. testosteroni and its mutants were cultivated and maintained at 30°C in LB broth or on LB plates with 1.5% (wt/vol) agar; antibiotic (200 μg/ml kanamycin) was added when necessary. For E. coli, strains were grown at 37°C in LB, and kanamycin was used at 50 μg/ml when needed. Genetic disruption and complementation in C. testosteroni CNB-1 were conducted using pK18mobSacB and pBBR1MCS-2, respectively. The plasmids for overexpression were pBBR1MCS-2 derivative (pBBR1MCS2pfer) whose promoter was replaced with a strong promoter from C. testosteroni (Table 1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or description | Reference or source |
|---|---|---|
|
Comamonas testosteroni strains |
||
| CNB-1 | 26 | |
| CNB-1Δ20 | All putative chemoreceptor genes were disrupted in CNB-1 | 31 |
| CNB-1ΔcheY1 | CheY1(CtCNB1_0474) disrupted in CNB-1 | This work |
| CNB-1ΔcheY2 | CheY2(CtCNB1_0455) disrupted in CNB-1 | This work |
| CNB-1ΔcheA | CheA(CtCNB1_0475) disrupted in CNB-1 | 31 |
| CNB-1ΔcheW | CheW(CtCNB1_0476) disrupted in CNB-1 | This work |
| CNB-1ΔflmA | FlmA(CtCNB1_3985) disrupted in CNB-1 | This work |
| CNB-1ΔflmD | FlmD(CtCNB1_3988) disrupted in CNB-1 | This work |
| CNB-1ΔflmE | FlmE(CtCNB1_3989) disrupted in CNB-1 | This work |
| CNB-1ΔflmAΔflmD | FlmA FlmD double disruptions in CNB-1 | This work |
| CNB-1ΔflmAΔcheA | FlmA CheA double disruptions in CNB-1 | This work |
| Escherichia coli strains | ||
| DH5α | F− φ80d lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) supE44 λ−
thi-1 gyrA96 relA1 phoA; host for DNA manipulations |
TransGen |
| BL21(DE3) | F− ompT hsdS(rB− mB−) gal dcm (DE3) | Novagen |
| RP9535 | CheA disrupted in E. coli RP437 | Parkinson’s lab |
| RP5232 | CheY disrupted in E. coli RP437 | Parkinson’s lab |
| Plasmids | ||
| pBBR1MCS-2 | Kmr; lacPOZ′ broad-host vector with R-type conjugative origin | 66 |
| pBBR1MCS2-mcp0033 | Carries mcp0033 to generate complementation | This work |
| pBBR1MCS2-mcp0034 | Carries mcp0834 to generate complementation | This work |
| pBBR1MCS2-mcp0838 | Carries mcp0838 to generate complementation | This work |
| pBBR1MCS2-mcp0846 | Carries mcp0846 to generate complementation | This work |
| pBBR1MCS2-mcp0955 | Carries mcp0955 to generate complementation | This work |
| pBBR1MCS2-mcp1646 | Carries mcp1646 to generate complementation | This work |
| pBBR1MCS2-mcp1647 | Carries mcp1647 to generate complementation | This work |
| pBBR1MCS2-mcp2001 | Carries mcp2001 to generate complementation | This work |
| pBBR1MCS2-mcp2005 | Carries mcp2005 to generate complementation | This work |
| pBBR1MCS2-mcp2201 | Carries mcp2201 to generate complementation | 31 |
| pBBR1MCS2-mcp2342 | Carries mcp2342 to generate complementation | This work |
| pBBR1MCS2-mcp2901 | Carries mcp2901 to generate complementation | 41 |
| pBBR1MCS2-mcp2923 | Carries mcp2923 to generate complementation | This work |
| pBBR1MCS2-mcp2983 | Carries mcp2983 to generate complementation | 35 |
| pBBR1MCS2-mcp3064 | Carries mcp3064 to generate complementation | This work |
| pBBR1MCS2-mcp3329 | Carries mcp3329 to generate complementation | This work |
| pBBR1MCS2-mcp3986 | Carries mcp3986 to generate complementation | This work |
| pBBR1MCS2-mcp4715 | Carries mcp4715 to generate complementation | This work |
| pBBR1MCS2-flmD | Carries flmD to generate complementation | This work |
| pBBR1MCS2-flmDD55A | A mutation from an aspartate to an alanine in 55th residue | This work |
| pBBR1MCS2-flmE | Carries flmE to generate complementation | This work |
| pBBR1MCS2-flmED58A | A mutation from an aspartate to an alanine in 58th residue | This work |
| pBBR1MCS2pfer | Adds a strong constitutive promoter in pBBR1MCS-2 | Our lab |
| pBBR1MCS2pfer-flmA | Carries flmA to overexpression | This work |
| pBBR1MCS2pfer-flmD | Carries flmD to overexpression | This work |
| pBBR1MCS2pfer-flmE | Carries flmE to overexpression | This work |
| pET28a-cheA | pET28a derivative for expression of CheA | 41 |
| pET28a-cheY1 | pET28a derivative for expression of CheY1 | This work |
| pET28a-cheY1(D52A) | pET28a derivative for expression of CheY1 with D52A mutation | This work |
| pET28a-cheY2 | pET28a derivative for expression of CheY2 | This work |
| pET28a-cheY2(D56A) | pET28a derivative for expression of CheY2 with D56A mutation | This work |
| pET28a-flmD | pET28a derivative for expression of FlmD | This work |
| pET28a-flmD(D52A) | pET28a derivative for expression of FlmD with D52A mutation | This work |
| pET28a-flmE | pET28a derivative for expression of FlmE | This work |
| pET28a-flmE(D58A) | pET28a derivative for expression of FlmE with D58A mutation | This work |
| pBT | Bacterial two-hybrid bait plasmid with λ repressor protein (λcI) | Stratagene |
| pBT-cheA | pBT derivative with λcl linked to the N-terminal region of CheA | This work |
| pBT-flmA | pBT derivative with λcl linked to the N-terminal region of FlmA | This work |
| pBT-mcp2201 | pBT derivative with λcl linked to the C-terminal region of MCP2201 | This work |
| pBT-mcp2901 | pBT derivative with λcl linked to the C-terminal region of MCP2901 | This work |
| pBT-mcp2983 | pBT derivative with λcl linked to the C-terminal region of MCP2983 | This work |
| pBT-mcp3986(flmB) | pBT derivative with λcl linked to the C-terminal region of MCP3986 | This work |
| pBT-mcp4715 | pBT derivative with λcl linked to the C-terminal region of MCP4715 | This work |
| pTRG | Bacterial two-hybrid bait plasmid with α-subunit of RNA polymerase (RNAp) | Stratagene |
| pTRG-cheA | pBT derivative with RNAp linked to the N-terminal region of CheA | This work |
| pTRG-flmA | pBT derivative with RNAp linked to the N-terminal region of FlmA | This work |
| pTRG-cheW | pBT derivative with RNAp linked to the N-terminal region of CheW | This work |
| pTRG-flmC | pBT derivative with RNAp linked to the N-terminal region of FlmC | This work |
Chemotaxis, twitching motility, and biofilm formation assays.
Chemotaxis assays were performed using semisoft agar plates with tryptone broth (TB) supplemented with 0.26% agar. Bacterial cells in logarithmic phase (OD600 of 0.4 to 0.7) from TB cultures were inoculated into the solidified agar and incubated at 30°C. Pictures were taken after 20 h of incubation. The twitching motility assay was performed as previously described (11). Briefly, colonies grown overnight on LB agar plates were picked using sterile toothpicks and stabbed into the bottom of petri dishes filled with medium and supplemented with 1% agar. Following incubation at 30°C in a humidified incubator for 24 h or 48 h, the agar and petri dish interface was inspected for a zone of motility. The biofilm formation assay was conducted by the method of O’Toole and Kolter (18) with slight modifications. Overnight cultures were diluted to an OD600 of 1.5, and 100 μl of the diluted sample was added to a 96-well PVC plate (Corning, MA, USA) as previously described (59). The plates were incubated at 30°C in a humidified incubator for 48 h. Planktonic cells were poured out carefully, and plates were washed with phosphate-buffered saline three times. One hundred twenty-five microliters of crystal violet (0.1%) was added to the wells and incubated for 30 min. After three washes, 150 μl of 30% acetic acid was added to dissolve the crystal violet, and the OD590 was measured on a multiwell plate reader. To determine whether the addition of ligands affects the growth of biofilm, bacteria were grown the same way as in the biofilm assay. Overnight cultures were diluted to an OD600 of 1.5, and the samples were divided into two equal parts. Ligands (final concentration, 2 mM) were added to one part, and the second part was left as a control. To determine growth, OD600 was measured in both samples. To compare growth of wild-type and mutant cells, 1% (vol/vol) bacteria from LB cultures were inoculated into minimal medium containing 2 mmol aromatic compounds as the sole carbon sources. A 200-ml mixture of bacteria and minimal medium were inoculated into each well of sterilized 100-well honeycomb plates, and the cell density at OD600 was monitored by using Bioscreen C automated growth curve analysis system.
Confocal laser scanning microscopy (CLSM) and image acquisition.
In LB cultures, C. testosteroni CNB-1 and other strains grew to an OD600 of 2.0, and then bacteria were statically incubated at 30°C for 48 h. The air-liquid interface biofilms (pellicles) which grew in glass test tubes were moved onto glass slides. Biofilms were stained using SYTO9 and washed with phosphate buffer three times. Double-sided tape was used around the biofilm to maintain the gap between cover glass and biofilm. All fluorescent images were acquired by a Leica SP8 (Leica Microsystems, Germany). CLSM-captured images were subjected to quantitative image analysis using COMSTAT software (60).
Genetic cloning, overexpression, and protein purification.
Overexpression and purification of proteins were performed as previously described (42). Briefly, genes were cloned into pET28a to generate an N-terminal His-tagged fusion protein. Expression of the CheA gene was induced by the addition of 0.1 mM IPTG for 5 h at 30°C, while the expression of the response regulator genes (CheY1, CheY2, FlmD, and FlmE) and their mutants was induced at 16°C for 12 h. All proteins were then purified using AKTA FPLC equipped with a HisTrap HP column. Buffer desalting and protein concentration were performed by an Amicon Ultra-15 concentrator (Merck, MA, USA).
Bacterial two-hybrid assay.
The BacterioMatch II Two-Hybrid system (Stratagene, CA, USA) was used to test the interaction between targeted proteins. Plasmid construction and screening were performed as previously described (61) and according to the manufacturer’s instructions. Briefly, overnight cultures were collected and washed by ddH2O three times. Bacteria (3 μl) were inoculated onto a selective screening medium plate containing 5 mM 3-amino-1,2,4-triazole (3-AT), 12.5 μg/ml streptomycin, 15 μg/ml tetracycline, and 25 μg/ml chloramphenicol to select positive growth cotransformants.
Phosphotransfer assay.
All reactions were performed in TGMNKD (50 mM Tris-HCl, 10% [vol/vol] glycerol, 5 mM MgCl2, 150 mM NaCl, 50 mM KCl, 1 mM dithiothreitol [pH 8.0]) buffer at 25°C (62). To initiate the phosphorylation reaction, 10 μCi [γ-32P]ATP (PerkinElmer, MA, USA) was added to 100 μl of TGMNKD buffer that was previously mixed with 5 μM CheA. After 15 min, 10 μl of sample was taken (T = 0) prior to the addition of any response regulators and quenched with 5 μl of SDS sample buffer. Then, the response regulators were added to mixtures to a final concentration of 10 μM. After specified time intervals, 10-μl samples were collected, and the reactions were stopped by the addition of 5 μl of sample buffer. The proteins were separated by 15% SDS-PAGE and exposed to a phosphorimaging screen. Quantitative analysis of the phosphotransfer efficiency was performed using Quantity One (Bio-Rad, CA, USA).
Data sources, software, and analysis.
Sequences of chemotaxis proteins and associated information from C. testosteroni CNB-2 genome (identical to CNB-1 except for the loss of pCNB plasmid) were obtained from the MiST2.2 database (30). Multiple-sequence alignments were built using the L-IN-I algorithm from the MAFFT v4.182 package (63). Complete domain architectures for chemoreceptor sequences were obtained using the CDvist server (64). Chemoreceptors were assigned to heptad classes, and CheA, CheW, CheB, and CheR were assigned to evolutionary classes using previously described hidden Markov models (31, 35) and HMMER v.2.0 package (65). Methylation sites were identified from multiple-sequence alignment of the chemoreceptor signaling domain, using the consensus sequence [ASTG]-[ASTG]-x(2)-[EQ]-[EQ]-x(2)-[ASTG]-[ASTG] (35).
ACKNOWLEDGMENTS
We thank Haichun Gao (Zhejiang University) and Zheng-Guo He (Huazhong Agricultural University) for providing E. coli strains and instructions on performing the bacterial two-hybrid assays. John S. Parkinson (University of Utah) provided the E. coli strains RP5232 and RP9535. Xiaolan Zhang (Institute of Microbiology, CAS) provided technical support on CLSM.
This work was supported by grants from the National Natural Science Foundation of China (31230003 to S.-J.L.) and the U.S. National Institutes of Health (GM072295 to I.B.Z).
We declare that we have no competing financial interests.
Footnotes
Citation Huang Z, Wang Y-H, Zhu H-Z, Andrianova EP, Jiang C-Y, Li D, Ma L, Feng J, Liu Z-P, Xiang H, Zhulin IB, Liu S-J. 2019. Cross talk between chemosensory pathways that modulate chemotaxis and biofilm formation. mBio 10:e02876-18. https://doi.org/10.1128/mBio.02876-18.
Contributor Information
Tarek Msadek, Institut Pasteur.
Karen Ottemann, University of California, Santa Cruz.
George O'Toole, Dartmouth.
REFERENCES
- 1.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 2.Parkinson JS, Hazelbauer GL, Falke JJ. 2015. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi S, Sourjik V. 2018. Stimulus sensing and signal processing in bacterial chemotaxis. Curr Opin Microbiol 45:22–29. doi: 10.1016/j.mib.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Ortega A, Zhulin IB, Krell T. 2017. Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev 81:e00033-17. doi: 10.1128/MMBR.00033-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welch M, Oosawa K, Aizawa S, Eisenbach M. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci U S A 90:8787–8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao R, Collins EJ, Bourret RB, Silversmith RE. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol 9:570–575. doi: 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
- 7.Springer WR, Koshland DE Jr.. 1977. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci U S A 74:533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stock JB, Koshland DE Jr.. 1978. A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci U S A 75:3659–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacal J, García-Fontana C, Muñoz-Martínez F, Ramos J-L, Krell T. 2010. Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ Microbiol 12:2873–2884. doi: 10.1111/j.1462-2920.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- 10.Bhaya D, Takahashi A, Grossman AR. 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc Natl Acad Sci U S A 98:7540–7545. doi: 10.1073/pnas.131201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, Bertrand JJ, Semmler AB, Mellick AS, Martin PR, Alm RA, Hobbs M, Beatson SA, Huang B, Nguyen L, Commolli JC, Engel JN, Darzins A, Mattick JS. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol 52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 12.Kirby JR, Zusman DR. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc Natl Acad Sci U S A 100:2008–2013. doi: 10.1073/pnas.0330944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berleman JE, Bauer CE. 2005. Involvement of a Che-like signal transduction cascade in regulating cyst cell development in Rhodospirillum centenum. Mol Microbiol 56:1457–1466. doi: 10.1111/j.1365-2958.2005.04646.x. [DOI] [PubMed] [Google Scholar]
- 14.Alexandre G. 2015. Chemotaxis control of transient cell aggregation. J Bacteriol 197:3230–3237. doi: 10.1128/JB.00121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corral-Lugo A, De la Torre J, Matilla MA, Fernández M, Morel B, Espinosa-Urgel M, Krell T. 2016. Assessment of the contribution of chemoreceptor-based signaling to biofilm formation. Environ Microbiol 18:3355–3372. doi: 10.1111/1462-2920.13170. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Sun Y, Shen R, Dang X, Liu X, Sui F, Li Y, Zhang Z, Alexandre G, Elmerich C, Xie Z. 2018. A chemotaxis-like pathway of Azorhizobium caulinodans controls flagella-driven motility, which regulates biofilm formation, exopolysaccharide biosynthesis, and competitive nodulation. Mol Plant Microbe Interact 31:737–749. doi: 10.1094/MPMI-12-17-0290-R. [DOI] [PubMed] [Google Scholar]
- 18.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 19.Merritt JH, Ha DG, Cowles KN, Lu W, Morales DK, Rabinowitz J, Gitai Z, O'Toole GA. 2010. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1:e00183-10. doi: 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T. 2014. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ Microbiol 16:1961–1981. doi: 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- 21.Francis VI, Stevenson EC, Porter SL. 2017. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol Lett 364:fnx104. doi: 10.1093/femsle/fnx104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giacalone D, Smith TJ, Collins AJ, Sondermann H, Koziol LJ, O'Toole GA. 2018. Ligand-mediated biofilm formation via enhanced physical interaction between a diguanylate cyclase and its receptor. mBio 9:e01254-18. doi: 10.1128/mBio.01254-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. 2007. The second messenger bis-(3'-5')-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol 65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 24.Baraquet C, Harwood CS. 2013. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci U S A 110:18478–18483. doi: 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol 66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu JF, Sun CW, Jiang CY, Liu ZP, Liu SJ. 2005. A novel 2-aminophenol 1,6-dioxygenase involved in the degradation of p-chloronitrobenzene by Comamonas strain CNB-1: purification, properties, genetic cloning and expression in Escherichia coli. Arch Microbiol 183:1–8. doi: 10.1007/s00203-004-0738-5. [DOI] [PubMed] [Google Scholar]
- 27.Wu JF, Jiang CY, Wang BJ, Ma YF, Liu ZP, Liu SJ. 2006. Novel partial reductive pathway for 4-chloronitrobenzene and nitrobenzene degradation in Comamonas sp. strain CNB-1. Appl Environ Microbiol 72:1759–1765. doi: 10.1128/AEM.72.3.1759-1765.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horemans B, Breugelmans P, Hofkens J, Springael D. 2017. Carbon catabolite repression and cell dispersal affect degradation of the xenobiotic compound 3,4-dichloroaniline in Comamonas testosteroni WDL7 biofilms. FEMS Microbiol Ecol 93:fix004. doi: 10.1093/femsec/fix004. [DOI] [PubMed] [Google Scholar]
- 29.Ma YF, Zhang Y, Zhang JY, Chen DW, Zhu Y, Zheng H, Wang SY, Jiang CY, Zhao GP, Liu SJ. 2009. The complete genome of Comamonas testosteroni reveals its genetic adaptations to changing environments. Appl Environ Microbiol 75:6812–6819. doi: 10.1128/AEM.00933-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulrich LE, Zhulin IB. 2010. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res 38:D401–D407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni B, Huang Z, Fan Z, Jiang CY, Liu SJ. 2013. Comamonas testosteroni uses a chemoreceptor for tricarboxylic acid cycle intermediates to trigger chemotactic responses towards aromatic compounds. Mol Microbiol 90:813–823. doi: 10.1111/mmi.12400. [DOI] [PubMed] [Google Scholar]
- 33.Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. 2010. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol 76:889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortega DR, Fleetwood AD, Krell T, Harwood CS, Jensen GJ, Zhulin IB. 2017. Assigning chemoreceptors to chemosensory pathways in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 114:12809–12814. doi: 10.1073/pnas.1708842114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander RP, Zhulin IB. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci U S A 104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni B, Huang Z, Wu YF, Fan Z, Jiang CY, Liu SJ. 2015. A novel chemoreceptor MCP2983 from Comamonas testosteroni specifically binds to cis-aconitate and triggers chemotaxis towards diverse organic compounds. Appl Microbiol Biotechnol 99:2773–2781. doi: 10.1007/s00253-014-6216-3. [DOI] [PubMed] [Google Scholar]
- 37.Sourjik V, Schmitt R. 1996. Different roles of CheY1 and CheY2 in the chemotaxis of Rhizobium meliloti. Mol Microbiol 22:427–436. doi: 10.1046/j.1365-2958.1996.1291489.x. [DOI] [PubMed] [Google Scholar]
- 38.Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol 9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 39.Page SC, Immormino RM, Miller TH, Bourret RB. 2016. Experimental analysis of functional variation within protein families: receiver domain autodephosphorylation kinetics. J Bacteriol 198:2483–2493. doi: 10.1128/JB.00853-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertrand JJ, West JT, Engel JN. 2010. Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa. J Bacteriol 192:994–1010. doi: 10.1128/JB.01390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo M, Huang Z, Yang J. 2017. Is there any crosstalk between the chemotaxis and virulence induction signaling in Agrobacterium tumefaciens? Biotechnol Adv 35:505–511. doi: 10.1016/j.biotechadv.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 42.Huang Z, Ni B, Jiang CY, Wu YF, He YZ, Parales RE, Liu SJ. 2016. Direct sensing and signal transduction during bacterial chemotaxis toward aromatic compounds in Comamonas testosteroni. Mol Microbiol 101:224–237. doi: 10.1111/mmi.13385. [DOI] [PubMed] [Google Scholar]
- 43.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. 2015. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silversmith RE, Wang B, Fulcher NB, Wolfgang MC, Bourret RB. 2016. Phosphoryl group flow within the Pseudomonas aeruginosa Pil-Chp chemosensory system: differential function of the eight phosphotransferase and three receiver domains. J Biol Chem 291:17677–17691. doi: 10.1074/jbc.M116.737528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inclan YF, Persat A, Greninger A, Von Dollen J, Johnson J, Krogan N, Gitai Z, Engel JN. 2016. A scaffold protein connects type IV pili with the Chp chemosensory system to mediate activation of virulence signaling in Pseudomonas aeruginosa. Mol Microbiol 101:590–605. doi: 10.1111/mmi.13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenal U, Galperin MY. 2009. Single domain response regulators: molecular switches with emerging roles in cell organization and dynamics. Curr Opin Microbiol 12:152–160. doi: 10.1016/j.mib.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. 2015. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev 39:649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu Rev Genet 41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 49.Siryaporn A, Goulian M. 2008. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol Microbiol 70:494–506. doi: 10.1111/j.1365-2958.2008.06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siryaporn A, Perchuk BS, Laub MT, Goulian M. 2010. Evolving a robust signal transduction pathway from weak cross-talk. Mol Syst Biol 6:452. doi: 10.1038/msb.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Capra EJ, Perchuk BS, Skerker JM, Laub MT. 2012. Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell 150:222–232. doi: 10.1016/j.cell.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silva JC, Haldimann A, Prahalad MK, Walsh CT, Wanner BL. 1998. In vivo characterization of the type A and B vancomycin-resistant enterococci (VRE) VanRS two-component systems in Escherichia coli: a nonpathogenic model for studying the VRE signal transduction pathways. Proc Natl Acad Sci U S A 95:11951–11956. doi: 10.1073/pnas.95.20.11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A 109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrova OE, Sauer K. 2012. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol 194:5817–5828. doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt J, Musken M, Becker T, Magnowska Z, Bertinetti D, Moller S, Zimmermann B, Herberg FW, Jansch L, Haussler S. 2011. The Pseudomonas aeruginosa chemotaxis methyltransferase CheR1 impacts on bacterial surface sampling. PLoS One 6:e18184. doi: 10.1371/journal.pone.0018184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, Givskov M, Whitchurch CB, Engel JN, Tolker-Nielsen T. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol 10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 57.Armitano J, Mejean V, Jourlin-Castelli C. 2013. Aerotaxis governs floating biofilm formation in Shewanella oneidensis. Environ Microbiol 15:3108–3118. doi: 10.1111/1462-2920.12158. [DOI] [PubMed] [Google Scholar]
- 58.Gambari C, Boyeldieu A, Armitano J, Mejean V, Jourlin-Castelli C. 2019. Control of pellicle biogenesis involves the diguanylate cyclases PdgA and PdgB, the c-di-GMP binding protein MxdA and the chemotaxis response regulator CheY3 in Shewanella oneidensis. Environ Microbiol 21:81–97. doi: 10.1111/1462-2920.14424. [DOI] [PubMed] [Google Scholar]
- 59.Wu Y, Ding Y, Cohen Y, Cao B. 2015. Elevated level of the second messenger c-di-GMP in Comamonas testosteroni enhances biofilm formation and biofilm-based biodegradation of 3-chloroaniline. Appl Microbiol Biotechnol 99:1967–1976. doi: 10.1007/s00253-014-6107-7. [DOI] [PubMed] [Google Scholar]
- 60.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 61.Zhang L, Zhang L, Liu Y, Yang S, Gao C, Gong H, Feng Y, He ZG. 2009. Archaeal eukaryote-like Orc1/Cdc6 initiators physically interact with DNA polymerase B1 and regulate its functions. Proc Natl Acad Sci U S A 106:7792–7797. doi: 10.1073/pnas.0813056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Porter SL, Armitage JP. 2004. Chemotaxis in Rhodobacter sphaeroides requires an atypical histidine protein kinase. J Biol Chem 279:54573–54580. doi: 10.1074/jbc.M408855200. [DOI] [PubMed] [Google Scholar]
- 63.Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 64.Adebali O, Ortega DR, Zhulin IB. 2015. CDvist: a webserver for identification and visualization of conserved domains in protein sequences. Bioinformatics 31:1475–1477. doi: 10.1093/bioinformatics/btu836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763. [DOI] [PubMed] [Google Scholar]
- 66.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Che pathway regulates chemotaxis. (A) Diagram of the che genetic cluster. (B and C) Functional characterization of the che cluster in C. testosteroni (B) and E. coli (C) by swimming plate. Shown are the means and standard deviations from three independent experiments conducted in triplicate. Download FIG S1, TIF file, 0.4 MB (418.6KB, tif) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Orthologous relationships between components of the Flm and Chp/Pil pathways. Results of mutual best BLAST hits against corresponding genomes are shown along with their E values, percentage of identity, and length coverage. Download Table S1, PDF file, 0.01 MB (11KB, pdf) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Chemosensory apparatus of C. testosteroni CNB-2. Chemotaxis class definition for chemoreceptors is according to Alexander and Zhulin (35), where the number of helical heptads in signaling domain defines the class. Chemotaxis class definition for other proteins is according to Wuichet and Zhulin (31). Conserved methylation sites (M1 through M4) and a CheB/CheR-binding pentapeptide are deduced from the multiple sequence alignment shown in Fig. S3. Download Table S2, PDF file, 0.04 MB (38.3KB, pdf) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Domain architectures of C. testosteroni chemoreceptors revealed by using CDvist. Transmembrane regions and Pfam domains are shown. Download FIG S2, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Multiple sequence alignment of signaling domains from C. testosteroni chemoreceptors. Predicted methylation sites are shown in red, and adjucent consensus sequences are shown in gray; predicted C-terminal pentapeptides are highlighted in yellow. Download FIG S3, PDF file, 0.06 MB (61.9KB, pdf) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Single chemoreceptor deletion does not affect the biofilm formation. Biofilm formation by each single chemoreceptor mutant measured by a crystal violet assay. Download FIG S4, PDF file, 0.5 MB (543.7KB, pdf) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth rates of the receptor-free mutant (Δ20) and the wild type (CNB-1) on different carbon sources. Gen, gentisate; 4HB, 4-hydroxybenzoate; 3HB, 3-hydroxybenzoate; PCA, protocatechuate. Download FIG S5, TIF file, 2.0 MB (2MB, tif) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of chemoreceptor mutants in the presence and absence of their ligand cis-aconitate. CNB-1, wild type; Δ20/mcp2201, Δ20 mutant complemented with the corresponding chemoreceptor gene; Δ20/mcp2983, Δ20 mutant complemented with the corresponding chemoreceptor gene; Δ20, mutant lacking all chemoreceptor genes. Growth conditions are described in Materials and Methods. Download FIG S6, PDF file, 1.2 MB (1.2MB, pdf) .
Copyright © 2019 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.