Abstract
The ability to regulate gene expression in response to environmental alterations is vital for the endurance of all cells. However, unlike bacteria and unicellular organisms, cells of multicellular eukaryotes have developed this competency in a highly sophisticated manner, which ultimately allows for multiple lineages of differentiated cells. To maintain stability and generate progeny, differentiated cells must remain lineage-committed through numerous cell generations, and therefore their transcriptional modus operandi ought to be memorized and transmittable. To preserve the specialized characteristics of differentiated cells, it is crucial that transcriptional alterations that are triggered by specific external or intrinsic stimuli can last also after stimuli fading and propagate onto daughter cells. The unique composition of DNA and histones, and their ability to acquire a variety of epigenetic modifications enables eukaryotic chromatin to assimilate cellular plasticity and molecular memory. The most well studied types of epigenetic modifiers are covalently modifying DNA or histones, mostly in a reversible manner. Additional epigenetic mechanisms include histone variant replacement, energy-utilizing remodeling factors, and noncoding transcripts assembled with modifying complexes. Working with multi-functional complexes including transcription factors, epigenetic modifiers have the potential to dictate a variety of transcriptional programs underlying all cellular lineages, while utilizing in each the same source DNA as their substrates.
Keywords: epigenetics, chromatin, chromatin modifiers, DNA, histone
Introduction
One of the greatest enigmas in the field of life sciences concerns the fact that in multicellular organisms, the zygote − the very first cell formed − can give rise to strikingly diverse cell types that all share an identical genome. These specialized somatic cell types, formed by induction of pluripotent stem cells through the process of terminal differentiation, are characterized by unique transcriptional programs, manifest distinct phenotypes, respond to different cellular signals, and fulfill appropriate functions in their specialized niches. In addition, differentiated somatic cells possess heritable cellular memory that allows them to stably maintain their cell fate characteristics and correctly transmit these characteristics to their progeny during the organism’s lifespan, thus defining cell lineages and cell types (1). Remarkably, these basic concepts were all introduced in the late ‘30s as part of Conrad Waddington’s canalization model of epigenetic landscape that conceptualized how genes might interact with their surroundings and products for driving phenotypic modulation in cells (2). Waddington conceived a pluripotent stem cell as a marble placed on top of a hill, driven by gravity down through one of multiple potential trenches (representing individual developmental pathways) to reach a final stop at the bottom (symbolizing a distinct differentiated fate). Once the marble reaches its ultimate destination, it cannot travel back up hill or roll over into an alternative trench (representing a unique mature and permanent differentiation stage). Thus, this model embodies the competence of pluripotent cells carrying identical genomes to give rise to diverse cell types via their distinct epigenetic landscape. The more contemporary definition of epigenetic trait proposes that it is a stable, heritable phenotype resulting from alterations to a chromosome without changes in the DNA sequence, and it is transmittable during both meiosis and mitosis (3, 4). Other definitions of the term are broader and include transient activity states that can occur in non-dividing cells as well, thus referring essentially to molecular remodeling events that arise over the DNA fiber at any instant. In this sense the self-defining meaning of ‘epigenetics’ can be directly interpreted by its Greek prefix, ‘epi’ (over, above). Recently, an alternative term, ‘memigenetics’, was proposed for portraying more adequately the inheritance aspect of epigenetics and to accentuate its contribution to cellular memory (5). Epigenetic memory was further classified into three paradigms of heritable epigenetic memory [(i) cellular memory, (ii) transcriptional memory, and (iii) transgenerational memory] that operate over different timescales for establishing stable gene expression patterns (4). Such memigenetic mechanisms enable eukaryotic creatures to adapt to altered environmental conditions and to develop long lasting responses that can be displayed through several generations. Among the well-recognized examples are X chromosome inactivation (XCI) (6), fetal programming (7), and imprinting (8). Recent studies suggest that epigenetic memory plays a crucial role in maintaining behavioral memories in the adult brain (9). Nevertheless, other adaptive mechanisms that act transiently and reversibly to enable cells to respond to external stimuli, but do not necessarily possess the ability of post-mitotic self-propagation, have been established (1, 10). These include histone phosphorylation (11), DNA repair (12), and transcription (13), in addition to molecular complexes that are active at the kinetochore, centromeres (14), and telomeres (15). In light of this and due to the critical roles that specific epigenetic mechanisms play in gene regulation, this review will focus on the broader role of ‘epigenetics’, and designate ‘epigenetic modifiers’ as the enzymes and molecular complexes involved in modification of chromatin, its spatial conformation, and operation. Here, we review the conceptual dual-role that epigenetic modifiers play within cells as regulators of structure and function. We describe four different layers of epigenetic regulators that have been proven to function diversely within cells to often arrive at a common endpoint, and that are being continuously studied. A discussion covering some of the prominent epigenetic scenarios that are currently known allows us to step into the convoluted realm of epigenetic modifiers.
The two conceptual arms of epigenetic modifiers: governing cellular architecture and cellular behavior
In all eukaryotic cells, genomic DNA is bundled into chromosomes and confined within the nucleus. Remarkably, the accumulated length of uncoiled DNA from a single human cell is approximately 2 meters, and yet the size of the nucleus that harbors it is only 6 μm. To address this architectural challenge, eukaryotic chromosomes are not constructed of bare DNA strands, but rather consist of chromatin, a DNA-protein complex that enables condensation of DNA into a highly organized structure (16). At its basic structural unit, chromatin is assembled into nucleosomes, which are structural units consisting of 146 bp of DNA that are firmly bundled around a canonical octamer of evolutionarily conserved histone proteins that contain two pairs of histone H2A-H2B dimers and an H3-H4 histone tetramer (17). At its first level of condensation the chromatin forms a 10 nm linear array of nucleosomes, often referred to as “beads-on-a-string”, which is conducive for transcription and replication due to its lower-order DNA organization. As cells progress through the cell cycle, transitioning from interphase to metaphase, this filament of nucleosome arrays becomes further condensed by linker histones H1 and additional nonhistone chromosomal proteins, forming a higher-order chromatin structure also known as a “multinucleosome” (16). In vitro studies have suggested that the nucleosome arrays fold into fibers with approximate size of 30 nm, however, the existence of a 30 nm chromatin fiber in vivo is controversial and could be an over-simplification, as more recent studies pointed on the dynamic and polymorphic nature of chromatin (18). Primarily, the tight morphology of chromosomes is maintained by the formation of ionic bonds between the acidic sugar phosphate backbone of the DNA and the basic amino acid residues in the histones within a nucleosome. However, additional chief mechanisms that participate in regulating chromatin condensation in a spatial and temporal manner are known: (i) Major histone subtypes (such as H3, H2A) can be replaced by histone variants (H3.3, H2A.X, and H2A.Z) that affect nucleosome properties, chromatin packaging and genomic stability differently compared to canonical histones (19). (ii) ATP-dependent chromatin remodeling enzymes unwrap, slide and rewrap nucleosomes, hindering or exposing DNA sequences during the processes of DNA repair via replication factors and transcription (20). (iii) DNA methylation at the 5th carbon of cytosine residues (5mC) present in cytosine-guanine (CpG) dinucleotides by DNA methyltrasferases (DNMTs) promotes transcriptional gene silencing and establishes genome stability via suppression of transposon mobilization (21). (iv) Post-translational modification (PTM) of the N-terminal tail domains of histone proteins including methylation, acetylation, phosphorylation, and ubiquitination can facilitate or negate chromatin condensation (22).
Alongside the hierarchical compaction characteristics of DNA, chromatin can be further categorized based on large-scale morphology that manifests two gross varieties, silent heterochromatin and active euchromatin. Heterochromatin represents the portion of chromatin that remains in a condensed conformation after progressing from metaphase to interphase, and is in general found to be transcriptionally inactive, although a few transcribed genes and ncRNAs that reside in this transcriptionally repressive environment are exception to this rule (23–25). In human and Drosophila, heterochromatin comprises approximately 30% of the genome (26). Besides being constitutively silent, heterochromatic domains are distinctively characterized by low gene density, high abundance of repetitive sequences, and late-replicating DNA segments. As a basic component of the eukaryotic genome, heterochromatin is vital for the separation of chromosomes in mitosis and for protection of chromosome ends. In contrast, euchromatin is defined as open chromatin domains in which nucleosomes are less compact. Euchromatic DNA contains most of the coding genes, is transcriptionally active or permissive for transcription, and replicates early (22).
In addition to the traditional division of chromatin into heterochromatin and euchromatin, abundant evidence from multiple deep sequencing projects such as ENCODE and modENCODE suggested that a finer classification of chromatin states could be applied (27–29). For example, a recent study in Drosophila cells in which the binding profiles of 53 chromatin proteins were mapped to the genome has distinguished five major distinctive chromatin classes that substantially differ in their domains number, genome coverage, numbers of genes, as well as their transcriptional activity, biochemical properties, histone modifications, and replication timing. Two chromatin classes were characterized as known types of classic heterochromatin that marked by the presence of Heterochromatin Protein 1 (HP1), or by polycomb group (PcG) proteins and methylation of lysine 27 of histone H3 (H3K27). One chromatin class that devoid of classic heterochromatin markers was identified as repressive chromatin, which covers nearly one-half of the genome; whereas two distinct classes of transcriptionally active euchromatin were characterized by unique molecular organization and contrasting levels of H3K36me3 (28).
A striking number of epigenetic modifiers operate to facilitate and maintain chromatin conformation within these genomic domains during cell cycle progression and cellular senescence. These epigenetic factors govern the dynamic state of chromatin and control genomic activation through regulation of DNA accessibility, DNA repair machinery, and transcription. Epigenetic modifiers are regarded as key regulators of proliferation that enable cells to preserve their cellular traits and phenotypic identity, while conferring sufficient plasticity to allow for adaptation to environmental alterations and developmental cues. Thus, although chromatin condensation represents a fundamental feature controlled by epigenetic mechanisms, our updated perception of the role of epigenetic modifiers has become much more penetrating and complex, and we no longer refer to them merely as mediators of DNA-compaction, but rather perceive them as direct regulators of cellular behavior and fate. From a systemic point of view, epigenetic modifiers play an extremely important role: on the one hand, they address the fundamental requirement of DNA compaction, while on the other hand they act as crucial modulators of development, differentiation, and lineage selection that enable cells to preserve their identity.
Four conceptual layers of epigenetic modifiers
During recent years, four main layers have emerged to form the basis for epigenetic modifications of DNA and chromatin. Two of these layers, namely, DNA methylation, and histone modifications, are facilitated through mechanisms that involve covalent modifications of either DNA (21) or histones (30), respectively. The third layer involves direct remodeling of nucleosomes and is mediated by multi-enzymatic complexes that employ the energy derived from ATP hydrolysis to alter DNA-histone interactions. Importantly, these enzymatic complexes often set the groundwork necessary for recruiting complementing factors that support the induction of the new chromatin states (31). The fourth layer involves noncoding RNAs (ncRNAs), which form RNA-protein complexes that can interact directly with chromatin. Importantly, chromatin dynamics facilitated by these regulatory mechanisms are generally reversible, through the recruitment of antagonistic sets of enzymatic complexes that can negate their activity, either by erasing formerly deposited modifications or by depositing new marks with nullifying effects. As part of the continuous effort to characterize proteins implicated in epigenetic activity, these complexes are widely referred to as writers, readers and erasers (32, 33). Epigenetic writers contain a catalytic site that allows them to directly modify DNA nucleotides or the histones, e.g. DNMTs, histone acetyltransferases (HATs), histone methyltransferases (HMTs), ubiquitin ligases and kinases. Readers represent proteins that harbor domains capable of recognizing and binding to previously deposited epigenetic marks. In contrast, epigenetic eraser proteins are capable of removing the epigenetic marks introduced by the writers, and include for instance, lysine demethylases (KDMs), and histone deacetylases (HDACs). Unlike writer and eraser proteins, epigenetic readers do not alter the DNA or histones but rather detect their epigenetic marks. For example, chromodomains or PHD fingers recognize histone methyl-lysine marks. Together, these epigenome constituents fulfill essential roles in establishing operative chromatin states.
DNA methylation and demethylation enzymes
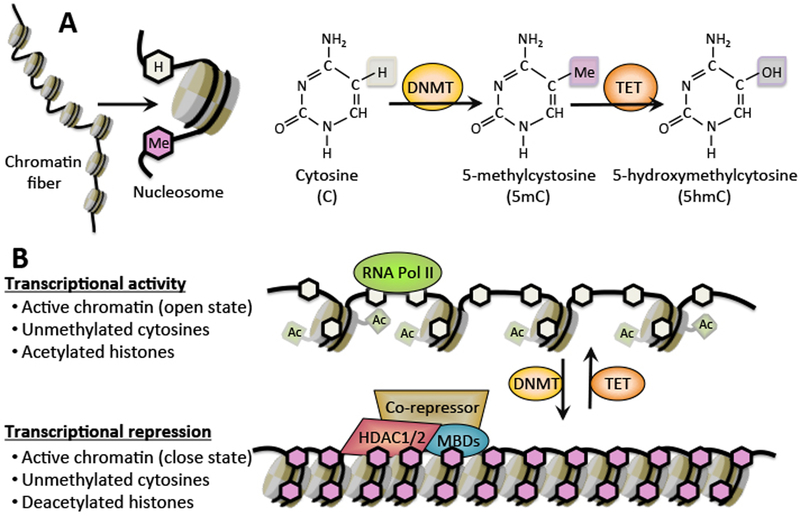
In many higher eukaryotic cells, DNA methylation, characterized by the addition of a methyl group (-CH3) to the C5 position of the cytosine pyrimidine ring thus forming 5-methylcystosine (5mC), serves as a highly stable covalent modification. This mechanism therefore allows the mammalian genome to expand its coding capability by utilizing methylated cytosine residues in addition to its four well-known nucleotide bases (Fig. 1). Interestingly, this “5th base”, generated by a post-replicative modifying mechanism catalyzed by DNMTs, primarily occurs within genomic regions rich in CpG dinucleotides. Notably, while both DNA strands are uniformly methylated at CpG dinucleotides, approximately 30–40% of these regions remain unmethylated in mammals, and the overall distribution of CpGs is not homogeneous (34). Most CpG dinucleotides are methylated and found within dense heterochromatin and repetitive regions (including centromeric repeats and satellite regions), while shorter stretches of CpG-rich DNA regions (denoted as “CpG Islands”) frequently reside within euchromatic regions, including gene promoters and first exons of housekeeping genes, and normally remain unmethylated. The overall arrangement of methylated/unmethylated genomic regions is accurately preserved during multiple cell cycle rounds, and the general concept is that DNA methylation is required for encoding heritable silencing of target genes. Nevertheless, promoters and enhancers with relatively low CpG content, especially within pluripotent and lineage-specific genes, manifest a wide spectrum of methylation levels in different tissues, and their methylation patterns further change during cell differentiation (35, 36).
Figure 1:
Basic principles of DNA methylation as an epigenetic mechanism. (A) The DNA fiber is coiled twice around histone octamers, forming nucleosomes, which are the building blocks of a chromosome. The free nucleotide cytosine is incorporated into DNA during replication. DNA methylation occurs at 5-position of cytosine residues in an enzymatic reaction catalyzed by DNA methyltransferases (DNMTs). 5-methylcystosine (5mC) can be further converted by TET1–3 enzymes to 5-hydroxymethylcytosine (5hmC), leading to DNA demethylation.
(B) Schematic of the reversible alterations in chromatin organization that impact gene expression: genes are transcribed when the chromatin is acetylated and active (open state), and they are transcriptionally repressed when the DNA is methylated and chromatin is condensed (close state). The complex, MBDs-HDAC1/2, bound to methylated, double-stranded DNA, suppresses gene transcription and converts chromatin integrity into a condensed state. White hexagons = unmethylated cytosines; pink hexagons = methylated cytosines.
DNMTs are the enzymes that catalyze the transfer of a methyl group from S-adenosyl-L-methuonin (SAM) to cytosine in a reaction that involves base flipping (37). In mammals, the DNMT family comprises five members: DNMT1, DNMT2, DNMT3A, DNMT3B and DNMT3L (DNMT3-like). DNMT1 has been implicated in heritable transmission of DNA methylation over multiple cell divisions, thus is often referred to as the “maintenance methylase”. During semi-conservative DNA replication, DNMT1 binds PCNA (proliferating cell nuclear antigen) in the replication fork and replicates the methylation pattern of parental DNA onto the newly synthesized reciprocal strand (38, 39). On the other hand, DNMT3a and DNMT3b enzymes are responsible for catalyzing de novo DNA methylation at unmethylated DNA during development (21, 40). DNMT3L lacks most of the C-terminal catalytic domain and is therefore enzymatically inactive. Nevertheless, DNMT3L interacts with DNMT3a and DNMT3b to stimulate their catalytic activity, which is crucial for methylation of retrotransposable elements in the male germ line (41), and for establishment of maternal imprints (42). Since CpG hypermethylation has been generally correlated with gene silencing, it has been proposed that DNA methylation promotes transcriptional silencing through direct inhibition of transcription factor binding to DNA. Indeed, this interpretation has been demonstrated by the identification of a number of transcription factors (including MLTF, CREB, E2F, NfkB and c-Myc) that cannot bind methylated recognition elements (43–47). Nevertheless, multiple transcription factors, such as CTF, YY1 and Sp1, were found to be insensitive to DNA methylation status (48), and it was further demonstrated that in some cases DNA methylation is capable of suppressing transcription only after chromatin has been assembled (49). In addition, relatively few transcription factors contain the dinucleotide CpG in their motif binding site (48). Thus, it was suggested that the main mechanism by which methylated DNA inhibits transcription is indirect.
DNA methylation represses transcription through an indirect mechanism by acting as a recognition site for suppressive molecules that bind to methylated CpG sites (mCpGs). These mCpG binding proteins (MBPs) further elicit transcriptional silencing by recruiting repressive chromatin modifiers and remodeling complexes (21). In mammals, three unique types of MBP proteins capable of reading the 5-methylcytosine mark have been identified to date: the methyl-CpG binding domain (MBD) family, the SRA (SET and Ring finger-associated) family (also referred to as “UHRF family”), and a family of proteins harboring zinc fingers (also denoted as “Kaiso protein family”) (50). The MBD-containing proteins (MeCP2 and MBD1–4) were the first proteins discovered that exhibit specific binding to mCpGs. With the exception of MBD3, which lacks methyl-CpG-binding activity due to sequence divergence in its MBD (51), all other MBD enzymes bind DNA in a methylation-dependent manner via their MBD. Almost all members of the MBD family are associated with transcriptional silencing activity as they form complexes with HDACs, and serve as components of the nucleosome remodeling deacetylase (NuRD) complex (51). Accordingly, nearly all MBD proteins bind DNA in highly methylated chromatin regions such as heterochromatin, endoparasitic sequences and imprinted genes, where they induce genomic stability and transcriptional silencing (52, 53).
5-Methylcytosine is considered as a major variant and a vital epigenetic modification in the genome of mammalian cells. It has been implicated in regulation of gene transcription, cell development, and disease pathogenesis (54). Nevertheless, studies from the past decade have indicated that besides DNA methylation, the methyl mark itself (5mC) can be further processed by the Ten-eleven translocation methylcytosine dioxygenase (TET) enzymes into additional cytosine variants, including 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (55). Therefore these cytosine derivatives technically represent a few more “nucleotides” that expand the coding potential of mammalians genome (56).
DNA demethylation, the reciprocal process of DNA methylation, is executed via two separate processes known as active and passive demethylation (57). Active DNA demethylation is mediated independently of DNA replication by either a base excision repair reaction catalyzed directly by DNA demethylase, which erases DNA methylation, or via an indirect reaction in which methyl-cytosine is first chemically converted to thymine through deamination prior to DNA glycosylase activity that removes 5-methylcytosine (58). Studies in plants revealed the identity of two DNA demethylases, DME (59) and ROS1 (60), and two related proteins (DML2 and DML3) as 5-methylcytosine DNA glycosylases that induce a base excision reaction for active and direct DNA demethylation (58). On the other hand, passive DNA demethylation takes place following several rounds of DNA replication when methyltransferase DNMT1 is absent or inactive, resulting in demethylation of cytosines in the newly synthesized strand. A recent study has shown that both paternal and maternal genomes undergo widespread passive and active demethylation in murine zygotes soon after fertilization and before the first mitotic division (61). In addition, replication-independent demethylase activity was demonstrated to be involved in other intracellular processes, including dendritic growth of newborn neurons in the adult hippocampus and induction of proliferation of neural progenitors (62).
Histone modifying enzymes
Evidence accumulating in recent years has demonstrated the outstanding diversity and biological significance associated with unique patterning of covalent histone marks. The rich plethora of modifications dotting the tail domains of histone combined with a variety of reader proteins and modulators, has led to the emergence of the conceptual term ‘histone language’ (also denoted as ‘histone code’). Extensive studies aimed at the elucidation of the histone code has revealed its ability to be written, read, and erased by a large collection of epigenetic modulators that can induce distinct downstream responses and cellular behaviors (63).
The canonical histones (H1, H2A, H2B, H3 and H4) and the variant histones consist of a central globular domain and COOH- and NH2- terminal tail regions. While interactions with nucleosomes are mediated through the histone core domain, interactions with chromatin modulators are largely mediated through the extended histone tails (especially via the NH2- terminal tail) (64), which can potentially adapt more than one hundred different post-translational modifications, including lysine acetylation, lysine and arginine methylation, serine, tyrosine, and threonine phosphorylation, ubiquitination, ADP-ribosylation, as well as sumoylation (65). In addition to these predominant histone tail modifications, specific residues within the histone core domain, such as H3T45, H3K56 and H4K91, can be subjected to covalent modifications as well. Due to their close proximity to the globular histone core, these residues effectively impact the positioning, mobility and stability of nucleosomes (66). In addition to the canonical histones, humans have five ubiquitous somatic variants of the linker histone H1 (H1.1, H1.2, H1.3, H1.4 and H1.5), which can carry diverse post-translational modifications (67). Adding further to the complexity, the major histone subtypes (such as H3, H2A) can be replaced by histone variants (such as H3.3, H2A.X, and H2A.Z) via a replication-independent nucleosome assembly. These histone variants possess their own specific biophysical characteristics that are suggested to affect nucleosome properties, and impact chromatin packaging and genomic stability (19), which can ultimately lead to profound effects on cellular behavior. For example, the histone variant macroH2A (mH2A) has recently been demonstrated to play a role as a tumor suppressor. A decline in mH2A expression was shown to be associated with the progression of malignant tumors in a variety of cancer types, including melanoma, colon, breast, lung, bladder, ovarian, testicular, and cervical (68). On the other hand, histone variant H2A.Z, which is constitutively expressed throughout the cell cycle and its deposition on chromatin is inversely correlated with DNA methylation, was demonstrated to act as an oncogene by promoting cell proliferation. Furthermore, several studies detected elevated levels of H2A.Z in bladder, breast, colorectal, and lung cancers (68).
Among the two central types of covalent epigenetic modifications, DNA methylation is considered highly stable and often acts to “lock in” epigenetic states, while histone modifications are regarded as more temporary (69). Interestingly, although histone modification and DNA methylation are mediated through different enzymatic reactions and catalyzed by different epigenetic factors, there seems to be a biological crosstalk between these two epigenetic mechanisms, which converge to modulate gene expression. As discussed further below, after DNA replication, DNA methylation may serve as a template for depositing specific histone marks, while on the other hand, there is data to suggest that histone modifications can impact DNA methylation patterns as well (70).
Histone acetyltransferase and deacetylases
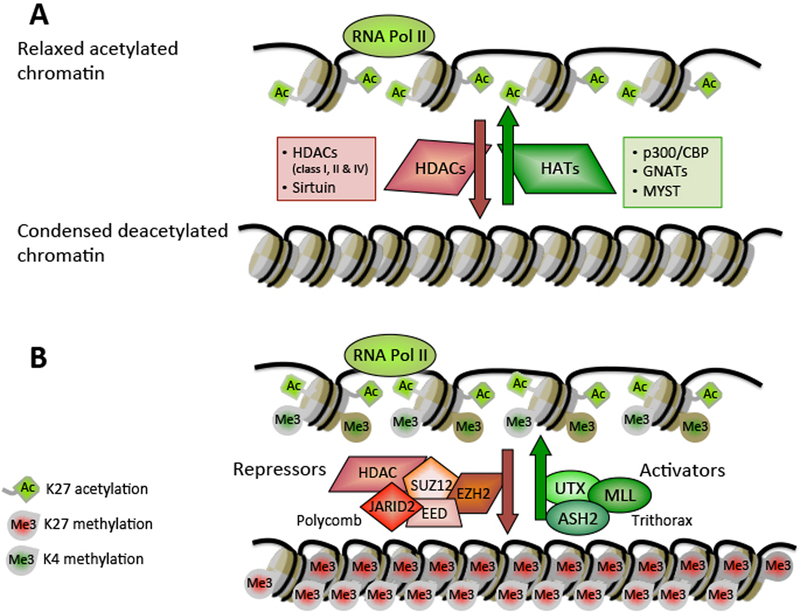
One of the most widely explored enzymatic activities is lysine acetylation, which is implicated in transcriptional activity. Histone lysine acetylation adds a negative charge to the positive lysine residues of histones and weakens the interaction between histone tails and the DNA, which leads to an overall relaxation in the chromatin structure and improves the accessibility of transcription factor binding sites within DNA (Fig. 2A). The acetylation of various residues, including K9, K14, K18, K23, and K27 on histone H3, and K5, K8, K12, and K16 on histone H4 generally promotes chromatin decompaction and transcriptional activation, whereas deacetylation of these sites is widely linked to chromatin condensation and transcriptional suppression (22). Genome-wide localization studies revealed that acetylation on distinctive histone residues is increased at different genomic regions, suggesting that individual acetylation sites are involved in different functions. For example, H3K9ac is highly enriched in promoter regions, consistent with a role in transcriptional initiation, whereas H4K12ac and H3K27ac reoccur at high levels in transcribed regions, implying a role in transcriptional elongation (71). Importantly, regulatory events that occur at distinctive genomic regions known as ‘enhancers’, are highly correlative with their activation status and their ability to induce transcription at remote promoters (72). The presence of histone H3K27ac and H3Kme1 at enhancer sites distinguishes active enhancers from inactive/poised enhancers that deposited with H3K4me1 alone, therefore conferring to this histone mark a central role in regulation of various gene expression programs (73).
Figure 2:
Schematic model for the role of methyltransferases/demethylases in transcriptional control of genes.
(A) Histone acetylation impacts chromatin structure. The dynamic state of histone acetylation/deacetylation is dictated by reciprocal reactions catalyzed by histone acetyltransferases (HATs) (e.g. p300/CBP) and histone deacetylases (HDACs) (e.g. HDAC1) enzymes. Acetylation of histones promotes chromatin deconsendation, enhances accessibility of chromatin and allows DNA binding proteins to interact with exposed sites to activate transcription of subset of genes. (B) The involvement of demethylases/methyltransferases in transcriptional control of developmental genes. Histone methyltransferases and demethylases are resided in the same complex, which methylates one mark while demethylates the antagonistic mark. The equilibrium between activities of the two opposing complexes, dictating the methylation pattern at a specific gene, is exemplified here by the activating MLL2/UTX complex and the Polycomb-repressive complexes (PRC2 complex).
Histone acetylation is a reversible enzymatic process mediated by histone acetyltransferases (HATs). By themselves, HATs do not show any sequence-specific DNA binding capability, however they can be targeted to specific DNA domains by binding other DNA binding proteins. HATs are grouped into three major families: p300/CBP (CREB-binding protein), GNATs (Gcn5-related N-acetyltransferases), and MYST proteins, all of which form multiprotein enzymatic complexes (74). The major difference between these enzymatic families stems from HAT module size, histone substrate specificity, and biological outcome.
Histone acetylation can be reversed by the activity of histone deacetylases (HDACs). HDACs are generally found in multiprotein enzymatic complexes targeted to specific genomic domains through their association with binding proteins, including hormone receptors and MBPs. HDACs are categorized grossly into two distinct families (which are comprised of four classes): the Sirtuin family, which depends on nicotinamide adenosine dinucleotide (NAD+) as a cofactor (also denoted as class III), and the classical HDAC family, which is dependent on Zn2+ and comprises classes I, II and IV. Generally speaking, HATs and HDACs have no preferential specificity towards a distinct lysine or acetyl-lysine residue (22).
Histone kinases and phosphatases
Similarly to histone acetylation, phosphorylation of amino acids influences the ionic potential of the nucleosome and has a direct impact on chromatin condensation and DNA accessibility. Histone phosphorylation is regarded as a highly reversible modification controlled by kinases and phosphatases that add and remove the phosphate group, respectively. The modification is catalyzed by histone kinases that transfer a phosphate group from an ATP molecule to the hydroxyl group of tyrosines, threonines, or serines on the tail regions of histones, resulting in the addition of a negative charge to the histone. Similar to histone acetylation, phosphorylation has been shown to play an important role in induction of transcription (75, 76), and has been implicated in chromatin dynamics in a variety of processes such as mitosis, transcription, apoptosis, and DNA repair (77, 78).
In mammals, phosphorylation of the serine at the 10th amino acid on the tails of histone H3 (H3S10ph) represents one of two phosphorylation events that have been studied the most. This phosphorylation is catalyzed by several kinases, including AKT, Msk1/2 and Aurora, whose activity is induced by cytokines, mitogens, or stress (79). H3S10ph is induced during cell division in higher eukaryotic cells and associated with chromatin condensation. After the completion of DNA replication and upon the onset of metaphase, nearly all of nucleic H3 histones become phosphorylated at this residue. Phosphorylation of H3S10 promotes the displacement of the repressive complex containing heterochromatin protein 1 (HP1) from heterochromatin and stimulates recruitment of ATP-dependent remodeling complexes, coactivators, and RNA polymerase II, which initiate transcription (80, 81). Moreover, phosphorylated H3S10 induces the recruitment of several members of the 14-3-3 phospho-binding protein family, which upon their recruitment mediate crosstalk between histone phosphorylation and acetylation during transcription elongation (82, 83). H3S10ph can be dephosphorylated by enzymatic activity of phosphatases such as protein phosphatase 1 (PP1), which interact directly with histone H3 and create enzymatic complexes with HDACs and demethylases (84).
The second well-known phosphorylation event is triggered by DNA damage, and mediated by PI3-K-like kinases, such as ATR, ATM and DNA-PKcs, which induce phosphorylation on H2AX at serine 139, commonly known as gamma-H2AX. Upon DNA repair the modification is reversed by dephosphorylation catalyzed by several phosphatases, including PP2A, PP4, PP6 and Wip1, which are required for recovery from the DNA damage checkpoint (85). In mammalian cells, deregulation of the molecular circuit that controls this modification has been reported to be associated with severe pathologies such as cancer and neuronal dysfunctions (79, 84).
Histone methyltransferases (HMTs)
Unlike histone acetylation and phosphorylation, histone methylation has no impact on the positive charge of the targeted residues − mainly, lysine or arginine − and therefore affects chromatin structure only indirectly through the recruitment of remodeling enzymes and transcription factors. In addition, in contrast to histone acetylation and phosphorylation, histone methylation can be found at both transcriptionally inactive and active chromatin. Thus, for example, H3K9 and H3K27 methylations are largely coupled with maintenance of stable heterochromatin and transcriptional suppression, while H3K4, H3K36 and H3K79 methylation is generally linked to open chromatin configurations at the promoters of active genes (22, 86). Furthermore, methylation can occur in multiple degrees, with lysine methylation occurring in mono-, di- or trimethylated forms, and arginine methylation occurring in mono- or dimethylated (symmetric or asymmetric) forms (87). Depending on the genomic region where it is deposited and the functional context, histone methylation can be displayed in any of its operative degrees. For example, while H3K4me3 is a hallmark of actively transcribed gene promoters and closely matches the distribution pattern of RNA polymerase II (RNAPII), H3K4me1 demarcates transcriptional enhancers (88, 89). As one might speculate, DNA and histones can acquire a combination of covalent marks that function synergistically or antagonistically. In ESCs for instance, nucleosomes are bivalently marked with both H3K27me3 (which is known to be coupled with silenced chromatin), and H3K4me3 (which is known to be coupled with active chromatin) methylations. In this unique combination, which has been identified primarily on silenced key developmental genes, the inducible effect of the active mark is nullified by the presence of the suppressive mark resulting in gene silencing (90). Studies have shown that this combinatorial arrangement of two “contradicting” methylation marks primes the marked promoters in a temporarily suspended state that lasts throughout early developmental stages. These bivalent genes lose their suppressive mark upon receiving differentiation signals that lead to their transcriptional induction (91). Furthermore, while H3K9 methylation and H3K9 acetylation are typically found to be mutually exclusive (92, 93), deposition of H3K9ac in human ESCs is found to be highly associated with methylation of H3K4me3 (94). Interestingly, a linkage between histone methylation and DNA methylation has also been discovered. In Arabidopsis and Neurospora, mutations that disrupt H3K9me3 sites were shown to lead to significant reductions in the levels of DNA methylation (95, 96), suggesting that DNA methylation is dependent on histone methylation. In mammalian cells, H3K9 is frequently methylated at silenced promoters enriched with hypermethylated CpG islands. Upon transcriptional induction, the H3K9 at these promoters become unmethylated and acetylated, and accordingly the CpG islands are demethylated (97).
Histone methyltransferases are substrate-specific enzymes that utilize SAM as the methyl source to catalyze the addition of methyl groups to histone tails. Although this biochemical mechanism is common to all histone methyltransferases, this enzymatic family can be split into two major categories, as follows.
Histone lysine methyltransferases (HKMT)
Methylation of lysine residues occurs on several positions on histones H3 and H4, whereas a few of these lysines, including H3K9, H3K14, H3K23, H3K27, H4K12, H4K20, and H4K79, are also substrates for acetylation. Most enzymes that catalyze histone lysine methylation share a well-conserved 140 amino acid catalytic domain, also known as SET (Su(var)3–9; Ezh2; Trithorax) domain. The first HKMT to be discovered was SUV39H1, which is the major methyltransferase that methylates H3K9 (98). Following this discovery, many additional SET domain-containing enzymes have been revealed, the majority of which were found to methylate lysines within the histone N-terminus tails. In contrast, methylation of H3K79, a lysine located within the histone globular core, is mediated by a unique HKMT, Dot1, which lacks the SET domain (99). Distinct HKMT enzymes are highly proficient in catalyzing specific methylation states. For example, SET7/9 is only capable of inducing monomethylation at H3K4 (100), while other HKMTs, such as MLL1 and DIM5, can mediate trimethylation of H3K4 (101) and H3K9 (102), respectively. The enzymatic activity of HKMTs is highly specific and substrate affinity is suggested to rise due to the assembly of these enzymes into conserved multiprotein complexes (103). Such a megacomplex, containing several histone HKMTs that possess mono-, di-, or trimethylation activities (including SUV39H1, G9a, GLP and SETDB1) was recently implicated in the catalysis of H3K9me3 at pericentric heterochromatin and silenced genes (104). Interestingly, G9a is capable of trimethylation of H3K27 as well as H3K9 (105), whereas EZH2 (enhancer of zeste homolog 2), the functional enzymatic component of the polycomb repressive complex 2 (PRC2), was found to predominantly trimethylate H3K27, and to a lesser extent H3K9 (106–108). The polycomb group (PcG) genes were first identified in Drosophila, where they were implicated in the silencing of developmental HOX genes (109), and later on as part of a central mechanism of tissue differentiation and organogenesis (110, 111). However, continuing studies have pointed out the important role that the PRC2 complex plays in additional processes such as X-chromosome inactivation (112) and tumorgenesis (113). In mammalian cells, PcG proteins are classified into two groups of protein complexes dubbed PRC1 and PRC2. The PRC2 complex, which catalyzes the di- and trimethylation of H3K27, is comprised of one catalytic subunit (EZH2), two noncatalytic subunits (zinc-finger-containing SUZ12, and WD40-repeat protein EED), a histone chaperon subunit (RbAp46/48), a cofactor subunit (AEBP2), an accessory subunit (JARID2), and a Polycomb-like (PCL) subunit (PCL1, PCL2, or PCL3, also known as PHF1, MTF2 and PHF19, respectively). Studies in cycling cells revealed that through G1 phase the PRC2 complex binds via the carboxy-terminal domain of EED to H3K27me3 that is deposited on sites of ongoing DNA replication (114, 115). This self-reinforcing loop, which enables the transmission of the H3K27me3 mark to the next generation, represents an important mechanism of epigenetic propagation across cell divisions. EZH1, the homolog protein of EZH2, forms a non-canonical PRC2 complex and catalyzes mono-, di- and trimethylation on H3K27 in the absence of EZH2 (116).
The PRC1 complexes (Polycomb repressive complex 1) contain two core components, namely RING1A/B, and BMI1 (which is composed of six members in humans: BMI/PCGF4, NSPC1/PCGF1, MEL18/PCGF2, PCGF3, PCGF5 and MBLR/PCGF6). Of these two components, RING1A/B, which is a catalytic E3 ubiquitin ligase, serves as a writer due to its ability to catalyze monoubiquitylation of histone H2A. The PRC1 repressive complex harbors additional H3K27me3-reader components and has five functional homologs (Cbx2/M33, Cbx4/Pc2, Cbx6, Cbx7 and Cbx8/Pc3) in humans that each possess well-characterized methyl-binding chromo-domains that facilitate the interaction of the PRC1 with H3K27me3. The chromatin-bound complex induces chromatin compaction and catalyzes monoubiquitylation of histone H2A, locking chromatin in a silenced state and forcing Pol-II to remain in a halted state (117).
As mentioned above, deposition of methyl groups on histone residues can also be associated with active chromatin, and in mammals this biochemical reaction is known to be mediated by multiple different HKMTs, including MLL1–5, SET1A, SET1B, ASH1, and ASH2 (22). The mammalian MLL enzymes are homologous to the Drosophila Trithorax enzymes that are responsible for the transcriptional regulation of Hox genes through their counteractive action to the PcG-mediated silencing (118). SET1 and MLL1–4 are part of the multiprotein COMPASS and COMPASS-like complexes catalyzing the deposition of mono-, di- and trimethyl groups on H3K4 (119). MLL1 has been shown to be associated with target promoters and coding regions of actively transcribed genes, and to interact and co-localize with RNA Pol-II (120). Chromosomal translocations at the MLL gene locus have been reported in several forms of leukemia, and their frequency in pediatric leukemia is extremely high (121).
A few known reader proteins of histone lysine methylation have been identified, all of which harbor methyl lysine recognition domains (such as Tudor, Chromo, WD repeat, and MBT), and upon their binding to the methylated histone can recruit downstream effector proteins (122). For example, chromodomain-containing proteins within the PRC1 complex mediate the interaction with methylated H3K27 (123). Likewise, the Tudor domain within the checkpoint protein Rad9 is necessary for binding to methylated H3K79 (124) (Fig. 2B).
Similar to histone acetylation, which is modified by both acetyltransferases and deacetylases, histone methylation is a reversible process and methylation levels are balanced by the counteractive activity of methyltransferase writers and histone demethylase erasers. The first histone methyl demethylase to be discovered, LSD1 (lysine specific demethylase 1) is a well-conserved specific H3K4me1/me2 demethylase that mediates transcriptional repression via histone demethylation (125). Since then, ample additional enzymes implicated in histone demethylation have been reported, including JHDM1A, JMJD2A/JHDM3A, JMJD2B, JMJD3/UTX, and JARID1 (22, 126, 127). Based on the nature of their catalytic function, the demethylase enzymes can be split into two clades, with the oxidation-based reactions mediated by LSD1-domain enzymes, and the hydroxylation-based reactions mediated by JmjC (JumanjiC)-domain enzymes (126). Whereas JmjC-domain- harboring proteins can mediate the removal of methyl groups from mono-, di- or trimethylated lysines, LSD1 acts to remove only mono- and dimethyl residues. Lysine demethylases have been found to act as members of multiprotein complexes and the distinct outcome of their function is often affected by the enzymes they bind to and by the pattern of chromatin marks in which they recognize. Strikingly, LSD1’s binding partners, CoREST, BHC80, and the androgen receptor (AR), were shown to have a significant effect on its enzymatic activity and specificity (128–130). Thus, when associated with the AR complex, LSD1 acts as a transcriptional coactivator, demethylating H3K9me2/me1 (130) and de-repressing AR target genes, whereas when in a complex with the repressive CoREST complex, LSD1 acts as a corepressor by demethylating H3K4me2/me1 (125).
Another interesting mechanism that was recently shown to be involved in the reversal of histone methylation is H3 tail cleavage, mediated by the proteolytic activity of Cathepsin L (CTSL1) (131). The enrichment of the protease on chromatin during senescence of human fibroblasts and melanocytes was shown to be associated with the cleavage of histone H3.3 tail at two distinct sites, namely residue T22 and between K9 and K14, generating the cleaved histone products H3cs1 and H3cs2, respectively. In addition, ectopic expression of H3.3, and to an even greater extent H3.3cs1, stimulates senescence in the absence of oncogenic signals. Importantly, the senescence-associated H3.3 tail cleaved products were correlated with a significant decrease in cell cycle genes transcription, and with reduction in H3K4me3 levels during senescence. These findings therefore suggest that proteolytic processing of the H3 tail serves as a chief mechanism for reversing H3K4 methylation at cell cycle-promoting genes during senescence (131).
Histone arginine methyltransferases (RHMT)
The family of protein arginine methyltransferases (PRMTs) is comprised of 11 members, all of which share a common highly conserved catalytic region and are capable of performing mono- and dimethylation of the guanidino group of the arginine residues (132, 133). The PRMT family can catalyze the production of three different forms of methylated arginine on nitrogen atoms, namely monomethylarginine (MMA), asymmetric dimethylarginine (ADMA), and symmetric dimethylarginine (SDMA). Based on their methylated arginine products the enzymatic family can be classified into two types: Type I PRMTs (PRMT1, 2, 3, 4/CARM1, 6, and 8), which catalyze the asymmetric reaction by adding two methyl groups to the same nitrogen atom to generate ADMA, and type II PRMTs (PRMT5, 7, 9/FBXO11, 10, and 11/FBXO10), which catalyze the symmetric reaction by adding a second methyl group to the other terminal nitrogen of arginine generating SDMA. Both types of enzymes are capable of mediating the MMA reaction (132).
While the downstream cellular implications of histone arginine methylation are still poorly understood, recent findings indicate that histone arginine methylation is associated with gene activation. Methylation of arginine residues on histone H4 by PRMT1, and on histone H3 by CARM1, was shown to occur within promoters of hormone target genes during their transcriptional activation following hormone induction (134–136). In addition, chromatin immunoprecipitation studies demonstrated the existence of cycles of arginine methylation during activation of these genes. These cyclical waves of epigenetic remodeling, correlated with transcriptional induction, were postulated to act as a cellular mechanism for fine-tuning of transcription rate upon different inductive stimuli (137). To date, only a few effector proteins (‘readers’), including Tudor domain-containing protein 3 (TDRD3) (138), DNMT3a (139), p300/CBP-associated factor (PCAF) (140), and RNA polymerase-associated protein 1 (PAF1) complex (141), have been implicated in binding of methylated arginine residues in histone tails. Furthermore, a recent study that utilized an unbiased proteomic approach to unravel binding proteins of arginine-methylated histones did not identify any new targets, but instead revealed that both H4R3me2s and H4R3me2a suppressed the binding of two nuclear heterodimers that belong to the signal recognition particle (SRP), SRP68 and SRP72. Thus, arginine methylation in histones is likely to be employed in order to repel rather than to recruit effector proteins (142). Interestingly, several studies have demonstrated the dynamic interplay that exists between histone arginine methylation and other epigenetic marks. A study that utilized recombinant histones as substrates revealed that methylation of H4R3 by PRMT1 stimulates subsequent p300-induced H4 acetylation, whereas acetylation of H4 inhibits H4R3 methylation (143). Synergy between HATs and protein arginine methyltransferases was documented in several studies (144, 145), including one study that showed that depletion of PRMT1, which concomitantly led to a significant decrease in H4R3 methylation, resulted in reduction in the levels of H4ac, H3K9ac, and H3K14a, and to an increase in H3K9 and H3K27 methylation (146).
In this regard, it is interesting to note that the CRAM1 methyltransferase is also known to methylate RNAPII at its carboxyl tail domain on a single arginine residue (R1810). Upon methylation this arginine the Tudor domain–containing protein, TDRD3, is becoming recruited to RNAPII and transcriptionally active promoters (147). Although TDRD3 has no catalytic activity of its own, recent studies have suggested its function as a scaffolding protein that forms tight complexes with RNAPII and DNA topoisomerase IIIβ (TOP3B), which promote transcription efficiency (148). Importantly, inhibition of RNAPII methylation at R1810 results in genome-wide alterations of a variety of small nucleolar RNAs and small nuclear RNAs, linking arginine methylation directly to regulation of selected RNA species (147).
Currently, two biochemical reactions are known to contribute to the erasing of methyl groups from arginine residues at histone tails; deamination, catalyzed by PADI4, and demethylation, mediated by JMJD6. In humans, PADI4 belongs to a family of five PADI isoforms that are known to be involved in diverse cellular roles mediated by calcium-dependent deamination catalysis. Upon treatment with calcium ionophores histones become citrullinated by PADI4 in human granulocytes, unveiling citrullination as a new form of post-translational histone modification (149). However, since this reaction affects both non-methylated and methylated (mono- and asymmetric dimethylated) arginines, and cannot be subsequently reversed by the action of a methyltransferase, it is commonly accepted that PADI4 is not a true demethylase. In addition, while PADI4 is capable of catalyzing the deamination of monomethyl-arginines at histones H3 and H4, it is incapable of catalyzing the deamination of arginines (149, 150). Accordingly, upon hormone stimulation the estrogen-regulated pS2 promoter becomes active and through the action of the RHMTs CARM1 and PRMT1 becomes methylated on its H3R17 residues. Subsequently, and along with the downregulation of pS2 transcription, PADI4 is recruited to the pS2 promoter and mediates deaminiation of the histone H3 N-terminal tail, converting the methylated arginine into citrulline. However, since replacement of arginine by citrulline prevents further methylation of this residue, it is speculated that chromatin rearrangement can occur either via replication-dependent new histone deposition, or through the action of a yet unidentified aminotransferase enzyme that can convert citrulline back into arginine (149, 150).
Recently, a family of the Jumonji domain-containing enzymes (including JMJD2A, JMJD2B, JMJD2C, and JMJD2D) demonstrated methyl type- and site-specific demethylase activities toward a variety of lysine residues (127). Among this enzymes family, JMJD6 (PSR) is a JmjC-containing iron- and 2-oxoglutarate-dependent dioxygenase that is capable of specifically removing methyl residues from the monomethylated or dimethylated H3R2 and H4R3 (151). However, JMJD6 is incapable of demethylating H2A, H3R8, H3R17, or H3R26. Importantly, Jmjd6−knockout mice suffer numerous developmental abnormalities during embryogenesis, suggesting that arginine demethylation fulfills an essential role in cellular proliferation and differentiation during development (152, 153). Future studies would need to be performed to determine whether additional arginine-specific demethylases catalyze the reversal reaction at other sites of arginine methylation.
ATP-dependent remodeling proteins
As described above, a variety of posttranslational modifying reactions can modulate the histone N-terminal tails, leading subsequently to profound effects on recognition of nucleosomes by regulatory enzymes and its higher-order folding. Nevertheless, interconvertible chromatin structures can be modulated in a more transient and subtle manners by nucleosome remodeling enzymes that perturb DNA-histones interactions. The enzymatic catalysis often leads to the establishment of a permissive chromatin state via the sliding of nucleosomes to expose genomic sequences that were previously occluded (20). Given the large number of weak interactions that hold DNA and histones together within nucleosomes, all nucleosome-remodeling factors utilize the energy of ATP hydrolysis in order to introducing superhelical torsion into DNA. The catalytic reaction mobilizes histone proteins away from the nucleosome or mediates their replacement by histone variants in a non-covalent fashion. Nucleosome remodeling proteins are incorporated into enzymatic complexes that are composed of two to twelve subunits, that all share a common subunit from the Swi2/Snf2 ATPase family (154). Based on the resemblance of their sequence motifs that lay outside of their ATPase domains, nucleosome-remodeling enzymes are classified into a few subfamilies, of which only several possess catalytic remodeling capacity, including the Swi2/Snf2 bromodomain ATPases, the ISWI/SNF2L SANT-domain ATPases, and Mi-2/NuRD and INO80 subfamilies (154). Collectively, the action of chromatin-remodeling enzymes can lead to different outcomes, such as DNA transcription, recombination, replication, or repair (155).
Noncoding RNAs (ncRNAs)
Approximately 90% of the mammalian genome is transcribed, whereas only 1–2% of transcripts encode proteins. Accumulating evidence from recent years indicates that in addition to controlling gene expression at the transcriptional and post-transcriptional levels, ncRNAs possess essential regulatory functions in differentiation, development, and disease progression (156). Cell type specific noncoding transcripts were shown to interact with ubiquitously expressed enzymes, and to create catalytic RNA-protein complexes that further correspond with chromatin-modifying complexes, distinct coding and noncoding RNAs, as well as histones, thus stimulating the acquisition of specific chromatin configurations.
With the advent of parallel deep sequencing technology molecular scientists are now becoming exposed to the massive compendium of ncRNAs, and the accumulation of studies supports the involvement of ncRNA species, including microRNAs (miRNAs), piwi-interacting RNAs (piRNAs), and circular RNAs (circRNAs), in the regulation of gene expression. Specifically, the ability of long ncRNAs (lncRNAs) to guide and serve as a modular scaffold for different chromatin modifying complexes thrusts ncRNAs into the limelight as potent epigenetic modifiers (156, 157). Noncoding RNAs were found to not only be involved in the establishment of the patterning of various epigenetic markers, but also serve as an essential type of epigenetic marker by themselves.
Several studies have demonstrated the reciprocal dynamic interplay between DNA methylation and ncRNAs. In plants, RNA-directed DNA methylation (RdDM) is guided by small interfering RNAs (siRNAs) that target DNA methyltransferase DRM2 (homolog of DNMT3A/DNMT3B) to specific genomic regions, such as repetitive sequences or gene promoters, to mediate de novo DNA methylation (158). Furthermore, in human cells, short-segment noncoding transcripts transiently transcribed from a loci located upstream of the promoter of elongation factor 1alpha (EF1A) were found to be associated with an increase in DNA methylation within the targeted sequence (159).
The cis- or trans- function of various ncRNAs was also demonstrated to occur through the interaction with specific histone modifying enzymes that are recruited to certain genomic sites, resulting in the formation of a distinct histone modification state. For instance, the Hox antisense intergenic RNA (HOTAIR), was shown to target two specific histone modifying enzymes to act at the HOXD locus: the histone methyltransferase complex PRC2, which establishes the inhibitory H3K27me3 mark, and the LSD1 demethylase complex, which mediates enzymatic demethylation of H3K4me2. Thus, HOTAIR can serves as a modular scaffold that links both histone methylase and demethylase activity to a particular chromatin site (160).
Although the contribution of ncRNAs to the regulation and targeting of ATP-dependent chromatin remodeling enzymes remains largely unknown, there are a few examples in which ncRNAs have manifested their influence on the activity of chromatin remodeling complexes. Under one of these settings, the NoRC remodeling complex (a member of the ISWI family of ATP-dependent chromatin remodeling complexes) is targeted to the ribosomal gene promoter via an RNA-dependent mechanism, leading to the formation of heterochromatin and to transcriptional silencing of ribosomal DNA (rDNA) repeats (161). Another example was discovered in the nucleoplasmic nuclear compartment of Drosophila cells where the chromatin remodeling activity of the ATPase ISWI was found to be controlled by hsrω, a class of functionally conserved, developmentally regulated lncRNAs that form the heterogeneous nuclear ribonucleoprotein (hnRNP)-containing “omega speckles” (distinct nuclear domains localized in the nucleus in close proximity to the chromatin that are known to play an active role in storage and sequestration of diverse hnRNPs and other proteins involved in RNA processing and maturation). Observations indicated that hsrω ncRNA physically binds to the N-terminus portion of ISWI and stimulates its ATPase activity to remodel and structurally arrange omega speckles (162).
Summary and outlook
The present-day definition of epigenetics refers to the cellular phenomenon that enables phenotypic changes in gene expression without evoking alterations in the underlying genomic DNA itself. Here, we presented four fundamental mechanistic layers that are responsible for epigenetic alterations in both genomic DNA and histones. Histone proteins entwined within the DNA fiber serve as the substrates of a variety of enzymes that inflict post-translational modifications (PTMs), and represent the core target of most known epigenetic modifiers. The largest well-studied groups of PTMs are histone acetylation and methylation. Histone acetyltransferases (HATs) have been implicated in gene transcription, whereas histone methyltransferases (HMTs) function in both silencing and inducing gene expression. In general, the fastidious dynamics of histone modifications is achieved by counter processes catalyzed by dedicated enzymes (such as HDACs and KDMs), which are capable of erasing previously deposited patterns, or nullifying their biochemical outcome by depositing new epigenetic marks that reverse their outcomes. As regulators of both chromatin structural dynamics and genomic information flow, epigenetic modifiers are vital for homeostasis and cellular identity. Undoubtedly, their most essential role is in sustaining the integrity of somatic cells throughout the lifetime of an individual. An aberrant epigenetic modulation within cells leads to pathological consequences such as cancer and neurodegeneration. The advent of parallel deep sequencing techniques has greatly advanced our understanding of epigenetic regulation, however many aspects remain yet unknown. Novel regulators playing currently undefined roles are sure to be discovered and thus will join the already highly crowded enzymatic realm of epigenetic modifiers. Our knowledge of the extensive implications of epigenetic patterns on promotion of chromatin folding, looping, and other transcription enhancement activities in specific cell lineages under different pathological and physiological conditions (72) is expected to continuously grow. A more comprehensive knowledge of these molecular epigenetic mechanisms will pave the way for the discovery of targets for pharmacological treatment of diseases such as neurologic disorders and cancer.
Acknowledgements
I thank Drs. T. Granot, N. Evensen, and J. Cheng for critical reading of the manuscript. Apologies to authors who I have omitted to mention due to space constraints. The author has no conflict of interest to disclose.
List of abbreviations:
- XCI
chromosome X inactivation
- DNMT
DNA methyltrasferases
- PTM
post-translational modification
- ncRNA
noncoding RNA
- HAT
histone acetyltransferase
- HMT
histone methyltransferases
- KDM
lysine demethylases
- HDAC
histone deacetylases
- CpG
cytosine-guanine
- SAM
S-adenosyl-L-methuonin
- PCNA
proliferating cell nuclear antigen
- mCpGs
methylated CpG sites
- MBPs
mCpG binding proteins
- MBD
methyl-CpG binding domain
- SRA
SET and Ring finger-associated
- NuRD
nucleosome remodeling deacetylase
- mH2A
macroH2A
- CBP
CREB-binding protein
- GNAT
Gcn5-related N-acetyltransferase
- HP1
heterochromatin protein 1
- PP1
protein phosphatase 1
- ESCs
embryonic stem cells
- HMT
histone methyltransferase
- RNAPII
RNA polymerase II
- HKMT
lysine histone methyltransferase
- SET
Su(var)3–9, Ezh2, Trithorax
- EZH2
Enhancer of zeste
- PRC2
polycomb repressive complex 2
- PcG
polycomb group
- PCL
Polycomb-like
- PRC1
polycomb repressive complex 1
- LSD1
lysine specific demethylase 1
- AR
androgen receptor
- CTSL1
cathepsin L
- RHMT
histone arginine methyltransferase
- PRMT
arginine methyltransferase
- MMA
monomethylarginine
- ADMA
asymmetric dimethylarginine
- SDMA
symmetric dimethylarginine
- TDRD3
Tudor domain-containing protein 3
- PCAF
p300/CBP-associated factor
- PAF1
polymerase-associated protein 1
- SRP
signal recognition particle
- miRNAs
microRNAs
- piRNAs
piwi-interacting RNAs
- circRNAs
circular RNAs; lncRNAs, long ncRNAs
- RdDM
RNA-directed DNA methylation
- siRNAs
interfering RNAs
- EF1A
elongation factor 1alpha
- HOTAIR
Hox antisense intergenic RNA
- rDNA
ribosomal DNA
- hnRNP
heterogeneous nuclear ribonucleoproteins
References
- 1.Reik W Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–32. [DOI] [PubMed] [Google Scholar]
- 2.Waddington CH. Organisers & genes. Cambridge Eng.: The University Press; 1940. x p., 1 l., 160 p. incl. front., illus., diagrs. p. [Google Scholar]
- 3.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes & development. 2009;23(7):781–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Urso A, Brickner JH. Mechanisms of epigenetic memory. Trends in genetics : TIG. 2014;30(6):230–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mann JR. Epigenetics and memigenetics. Cellular and molecular life sciences : CMLS. 2014;71(7):1117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sado T, Brockdorff N. Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2013;368(1609):20110325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandman CA, Davis EP, Buss C, Glynn LM. Prenatal programming of human neurological function. International journal of peptides. 2011;2011:837596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabo PE, Mann JR. Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes & development. 1995;9(15):1857–68. [DOI] [PubMed] [Google Scholar]
- 9.Zovkic IB, Guzman-Karlsson MC, Sweatt JD. Epigenetic regulation of memory formation and maintenance. Learning & memory. 2013;20(2):61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katan-Khaykovich Y, Struhl K. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes & development. 2002;16(6):743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossetto D, Avvakumov N, Cote J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics : official journal of the DNA Methylation Society. 2012;7(10):1098–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossetto D, Truman AW, Kron SJ, Cote J. Epigenetic modifications in double-strand break DNA damage signaling and repair. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16(18):4543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eccleston A, Cesari F, Skipper M. Transcription and epigenetics. Nature. 2013;502(7472):461. [DOI] [PubMed] [Google Scholar]
- 14.Warburton PE. Epigenetic analysis of kinetochore assembly on variant human centromeres. Trends in genetics : TIG. 2001;17(5):243–7. [DOI] [PubMed] [Google Scholar]
- 15.Blasco MA. The epigenetic regulation of mammalian telomeres. Nature reviews Genetics. 2007;8(4):299–309. [DOI] [PubMed] [Google Scholar]
- 16.Alberts B Molecular biology of the cell. 5th ed New York: Garland Science; 2008. [Google Scholar]
- 17.Quina AS, Buschbeck M, Di Croce L. Chromatin structure and epigenetics. Biochemical pharmacology. 2006;72(11):1563–9. [DOI] [PubMed] [Google Scholar]
- 18.Grigoryev SA, Woodcock CL. Chromatin organization - the 30 nm fiber. Experimental cell research. 2012;318(12):1448–55. [DOI] [PubMed] [Google Scholar]
- 19.Kamakaka RT, Biggins S. Histone variants: deviants? Genes & development. 2005;19(3):295–310. [DOI] [PubMed] [Google Scholar]
- 20.Toto M, D’Angelo G, Corona DF. Regulation of ISWI chromatin remodelling activity. Chromosoma. 2014;123(1–2):91–102. [DOI] [PubMed] [Google Scholar]
- 21.Rottach A, Leonhardt H, Spada F. DNA methylation-mediated epigenetic control. Journal of cellular biochemistry. 2009;108(1):43–51. [DOI] [PubMed] [Google Scholar]
- 22.Kouzarides T Chromatin modifications and their function. Cell. 2007;128(4):693–705. [DOI] [PubMed] [Google Scholar]
- 23.Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nature structural & molecular biology. 2007;14(11):1041–8. [DOI] [PubMed] [Google Scholar]
- 24.Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell cycle. 2008;7(9):1161–5. [DOI] [PubMed] [Google Scholar]
- 25.Schoeftner S, Blasco MA. A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. The EMBO journal. 2009;28(16):2323–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimitri P, Caizzi R, Giordano E, Carmela Accardo M, Lattanzi G, Biamonti G. Constitutive heterochromatin: a surprising variety of expressed sequences. Chromosoma. 2009;118(4):419–35. [DOI] [PubMed] [Google Scholar]
- 27.mod EC, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330(6012):1787–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143(2):212–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Libbrecht MW, Ay F, Hoffman MM, Gilbert DM, Bilmes JA, Noble WS. Joint annotation of chromatin state and chromatin conformation reveals relationships among domain types and identifies domains of cell-type-specific expression. Genome research. 2015;25(4):544–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J, Grunstein M. 25 years after the nucleosome model: chromatin modifications. Trends in biochemical sciences. 2000;25(12):619–23. [DOI] [PubMed] [Google Scholar]
- 31.Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & development. 1999;13(18):2339–52. [DOI] [PubMed] [Google Scholar]
- 32.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell research. 2011;21(4):564–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor perspectives in biology. 2014;6(4):a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic acids research. 1982;10(8):2709–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell stem cell. 2008;2(2):160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts RJ, Cheng X. Base flipping. Annual review of biochemistry. 1998;67:181–98. [DOI] [PubMed] [Google Scholar]
- 38.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–12. [DOI] [PubMed] [Google Scholar]
- 39.Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends in cell biology. 2009;19(1):29–41. [DOI] [PubMed] [Google Scholar]
- 40.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–3. [DOI] [PubMed] [Google Scholar]
- 41.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431(7004):96–9. [DOI] [PubMed] [Google Scholar]
- 42.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129(8):1983–93. [DOI] [PubMed] [Google Scholar]
- 43.Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes & development. 1988;2(9):1136–43. [DOI] [PubMed] [Google Scholar]
- 44.Kovesdi I, Reichel R, Nevins JR. Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(8):2180–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251(4990):186–9. [DOI] [PubMed] [Google Scholar]
- 46.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes & development. 1989;3(5):612–9. [DOI] [PubMed] [Google Scholar]
- 47.Bednarik DP, Duckett C, Kim SU, Perez VL, Griffis K, Guenthner PC, et al. DNA CpG methylation inhibits binding of NF-kappa B proteins to the HIV-1 long terminal repeat cognate DNA motifs. The New biologist. 1991;3(10):969–76. [PubMed] [Google Scholar]
- 48.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64(6):1123–34. [DOI] [PubMed] [Google Scholar]
- 49.Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Current biology : CB. 1997;7(3):157–65. [DOI] [PubMed] [Google Scholar]
- 50.Sasai N, Defossez PA. Many paths to one goal? The proteins that recognize methylated DNA in eukaryotes. The International journal of developmental biology. 2009;53(2–3):323–34. [DOI] [PubMed] [Google Scholar]
- 51.Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends in genetics : TIG. 2003;19(5):269–77. [DOI] [PubMed] [Google Scholar]
- 52.Dhasarathy A, Wade PA. The MBD protein family-reading an epigenetic mark? Mutation research. 2008;647(1–2):39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: reading the wrong words. British journal of cancer. 2008;98(12):1881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends in biochemical sciences. 2006;31(2):89–97. [DOI] [PubMed] [Google Scholar]
- 55.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song CX, Yi C, He C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nature biotechnology. 2012;30(11):1107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133(7):1145–8. [DOI] [PubMed] [Google Scholar]
- 58.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annual review of genetics. 2009;43:143–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124(3):495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111(6):803–14. [DOI] [PubMed] [Google Scholar]
- 61.Guo F, Li X, Liang D, Li T, Zhu P, Guo H, et al. Active and passive demethylation of male and female pronuclear DNA in the Mammalian zygote. Cell stem cell. 2014;15(4):447–58. [DOI] [PubMed] [Google Scholar]
- 62.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323(5917):1074–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. [DOI] [PubMed] [Google Scholar]
- 64.Luger K, Richmond TJ. The histone tails of the nucleosome. Current opinion in genetics & development. 1998;8(2):140–6. [DOI] [PubMed] [Google Scholar]
- 65.Turner BM. Epigenetic responses to environmental change and their evolutionary implications. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2009;364(1534):3403–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ye J, Ai X, Eugeni EE, Zhang L, Carpenter LR, Jelinek MA, et al. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Molecular cell. 2005;18(1):123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harshman SW, Young NL, Parthun MR, Freitas MA. H1 histones: current perspectives and challenges. Nucleic acids research. 2013;41(21):9593–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vardabasso C, Hasson D, Ratnakumar K, Chung CY, Duarte LF, Bernstein E. Histone variants: emerging players in cancer biology. Cellular and molecular life sciences : CMLS. 2014;71(3):379–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emery AEH, Korf BR, Pyeritz RE, Rimoin DL. Emery and Rimoin’s essential medical genetics. Boston: Academic Press,; 2013. Available from: http://catalog.himmelfarb.gwu.edu/iii/encore/record/C__Rb1627009. [Google Scholar]
- 70.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature reviews Genetics. 2009;10(5):295–304. [DOI] [PubMed] [Google Scholar]
- 71.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics. 2008;40(7):897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blum R Activation of muscle enhancers by MyoD and epigenetic modifiers. Journal of cellular biochemistry. 2014;115(11):1855–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(50):21931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nature reviews Molecular cell biology. 2007;8(4):284–95. [DOI] [PubMed] [Google Scholar]
- 75.Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, et al. Snf1--a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293(5532):1142–6. [DOI] [PubMed] [Google Scholar]
- 76.Banerjee T, Chakravarti D. A peek into the complex realm of histone phosphorylation. Molecular and cellular biology. 2011;31(24):4858–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hurd PJ, Bannister AJ, Halls K, Dawson MA, Vermeulen M, Olsen JV, et al. Phosphorylation of histone H3 Thr-45 is linked to apoptosis. The Journal of biological chemistry. 2009;284(24):16575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461(7265):819–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keppler BR, Archer TK. Chromatin-modifying enzymes as therapeutic targets--Part 2. Expert opinion on therapeutic targets. 2008;12(11):1457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eissenberg JC, Elgin SC. Molecular biology: antagonizing the neighbours. Nature. 2005;438(7071):1090–1. [DOI] [PubMed] [Google Scholar]
- 81.Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, et al. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Molecular cell. 2006;24(3):367–81. [DOI] [PubMed] [Google Scholar]
- 82.Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138(6):1122–36. [DOI] [PubMed] [Google Scholar]
- 83.Karam CS, Kellner WA, Takenaka N, Clemmons AW, Corces VG. 14–3-3 mediates histone cross-talk during transcription elongation in Drosophila. PLoS genetics. 2010;6(6):e1000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Franklin TB, Mansuy IM. The prevalence of epigenetic mechanisms in the regulation of cognitive functions and behaviour. Current opinion in neurobiology. 2010;20(4):441–9. [DOI] [PubMed] [Google Scholar]
- 85.Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: to be or not to be. FEBS letters. 2010;584(17):3717–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–81. [DOI] [PubMed] [Google Scholar]
- 87.Shilatifard A Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annual review of biochemistry. 2006;75:243–69. [DOI] [PubMed] [Google Scholar]
- 88.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. [DOI] [PubMed] [Google Scholar]
- 89.Orford K, Kharchenko P, Lai W, Dao MC, Worhunsky DJ, Ferro A, et al. Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Developmental cell. 2008;14(5):798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26. [DOI] [PubMed] [Google Scholar]
- 91.Hublitz P, Albert M, Peters AH. Mechanisms of transcriptional repression by histone lysine methylation. The International journal of developmental biology. 2009;53(2–3):335–54. [DOI] [PubMed] [Google Scholar]
- 92.Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, Alako BT, et al. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS pathogens. 2009;5(9):e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Current opinion in cell biology. 2001;13(3):263–73. [DOI] [PubMed] [Google Scholar]
- 94.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130(1):77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414(6861):277–83. [DOI] [PubMed] [Google Scholar]
- 96.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416(6880):556–60. [DOI] [PubMed] [Google Scholar]
- 97.Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer research. 2002;62(24):7213–8. [PubMed] [Google Scholar]
- 98.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–9. [DOI] [PubMed] [Google Scholar]
- 99.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes & development. 2002;16(12):1518–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421(6923):652–6. [DOI] [PubMed] [Google Scholar]
- 101.Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Molecular and cellular biology. 2009;29(22):6074–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, et al. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nature genetics. 2003;34(1):75–9. [DOI] [PubMed] [Google Scholar]
- 103.Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Charge-based interaction conserved within histone H3 lysine 4 (H3K4) methyltransferase complexes is needed for protein stability, histone methylation, and gene expression. The Journal of biological chemistry. 2012;287(4):2652–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Molecular cell. 2010;37(1):46–56. [DOI] [PubMed] [Google Scholar]
- 105.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. The Journal of biological chemistry. 2001;276(27):25309–17. [DOI] [PubMed] [Google Scholar]
- 106.de la Cruz CC, Kirmizis A, Simon MD, Isono K, Koseki H, Panning B. The polycomb group protein SUZ12 regulates histone H3 lysine 9 methylation and HP1 alpha distribution. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2007;15(3):299–314. [DOI] [PubMed] [Google Scholar]
- 107.Sun F, Chan E, Wu Z, Yang X, Marquez VE, Yu Q. Combinatorial pharmacologic approaches target EZH2-mediated gene repression in breast cancer cells. Molecular cancer therapeutics. 2009;8(12):3191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & development. 2002;16(22):2893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Beuchle D, Struhl G, Muller J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development. 2001;128(6):993–1004. [DOI] [PubMed] [Google Scholar]
- 110.Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(22):E149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aldiri I, Vetter ML. PRC2 during vertebrate organogenesis: a complex in transition. Developmental biology. 2012;367(2):91–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Muyrers-Chen I, Hernandez-Munoz I, Lund AH, Valk-Lingbeek ME, van der Stoop P, Boutsma E, et al. Emerging roles of Polycomb silencing in X-inactivation and stem cell maintenance. Cold Spring Harbor symposia on quantitative biology. 2004;69:319–26. [DOI] [PubMed] [Google Scholar]
- 113.Hock H A complex Polycomb issue: the two faces of EZH2 in cancer. Genes & development. 2012;26(8):751–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, et al. A model for transmission of the H3K27me3 epigenetic mark. Nature cell biology. 2008;10(11):1291–300. [DOI] [PubMed] [Google Scholar]
- 115.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461(7265):762–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Molecular cell. 2008;32(4):491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nature reviews Molecular cell biology. 2009;10(10):697–708. [DOI] [PubMed] [Google Scholar]
- 118.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134(2):223–32. [DOI] [PubMed] [Google Scholar]
- 119.Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nature reviews Molecular cell biology. 2012;13(9):543–7. [DOI] [PubMed] [Google Scholar]
- 120.Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(41):14765–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nature genetics. 2002;30(1):41–7. [DOI] [PubMed] [Google Scholar]
- 122.Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cellular and molecular life sciences : CMLS. 2006;63(23):2755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes & development. 2003;17(15):1823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Grenon M, Costelloe T, Jimeno S, O’Shaughnessy A, Fitzgerald J, Zgheib O, et al. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast. 2007;24(2):105–19. [DOI] [PubMed] [Google Scholar]
- 125.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–53. [DOI] [PubMed] [Google Scholar]
- 126.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes & development. 2008;22(9):1115–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shi Y Histone lysine demethylases: emerging roles in development, physiology and disease. Nature reviews Genetics. 2007;8(11):829–33. [DOI] [PubMed] [Google Scholar]
- 128.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437(7057):432–5. [DOI] [PubMed] [Google Scholar]
- 129.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Molecular cell. 2005;19(6):857–64. [DOI] [PubMed] [Google Scholar]
- 130.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437(7057):436–9. [DOI] [PubMed] [Google Scholar]
- 131.Duarte LF, Young AR, Wang Z, Wu HA, Panda T, Kou Y, et al. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nature communications. 2014;5:5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cha B, Jho EH. Protein arginine methyltransferases (PRMTs) as therapeutic targets. Expert opinion on therapeutic targets. 2012;16(7):651–64. [DOI] [PubMed] [Google Scholar]
- 133.Litt M, Qiu Y, Huang S. Histone arginine methylations: their roles in chromatin dynamics and transcriptional regulation. Bioscience reports. 2009;29(2):131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO reports. 2002;3(1):39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, et al. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Current biology : CB. 2001;11(24):1981–5. [DOI] [PubMed] [Google Scholar]
- 136.Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Current biology : CB. 2001;11(12):996–1000. [DOI] [PubMed] [Google Scholar]
- 137.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115(6):751–63. [DOI] [PubMed] [Google Scholar]
- 138.Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, et al. TDRD3 is an effector molecule for arginine-methylated histone marks. Molecular cell. 2010;40(6):1016–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nature structural & molecular biology. 2009;16(3):304–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li X, Hu X, Patel B, Zhou Z, Liang S, Ybarra R, et al. H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood. 2010;115(10):2028–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu J, Xu W. Histone H3R17me2a mark recruits human RNA polymerase-associated factor 1 complex to activate transcription. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(15):5675–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li J, Zhou F, Zhan D, Gao Q, Cui N, Li J, et al. A novel histone H4 arginine 3 methylation-sensitive histone H4 binding activity and transcriptional regulatory function for signal recognition particle subunits SRP68 and SRP72. The Journal of biological chemistry. 2012;287(48):40641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293(5531):853–7. [DOI] [PubMed] [Google Scholar]
- 144.Koh SS, Chen D, Lee YH, Stallcup MR. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. The Journal of biological chemistry. 2001;276(2):1089–98. [DOI] [PubMed] [Google Scholar]
- 145.Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Current biology : CB. 2002;12(24):2090–7. [DOI] [PubMed] [Google Scholar]
- 146.Huang S, Litt M, Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes & development. 2005;19(16):1885–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sims RJ 3rd, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ 3rd, et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332(6025):99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Molecular cell. 2014;53(3):484–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306(5694):279–83. [DOI] [PubMed] [Google Scholar]
- 150.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118(5):545–53. [DOI] [PubMed] [Google Scholar]
- 151.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318(5849):444–7. [DOI] [PubMed] [Google Scholar]
- 152.Kunisaki Y, Masuko S, Noda M, Inayoshi A, Sanui T, Harada M, et al. Defective fetal liver erythropoiesis and T lymphopoiesis in mice lacking the phosphatidylserine receptor. Blood. 2004;103(9):3362–4. [DOI] [PubMed] [Google Scholar]
- 153.Li MO, Sarkisian MR, Mehal WZ, Rakic P, Flavell RA. Phosphatidylserine receptor is required for clearance of apoptotic cells. Science. 2003;302(5650):1560–3. [DOI] [PubMed] [Google Scholar]
- 154.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annual review of biochemistry. 2002;71:247–73. [DOI] [PubMed] [Google Scholar]
- 155.Santenard A, Torres-Padilla ME. Epigenetic reprogramming in mammalian reproduction: contribution from histone variants. Epigenetics : official journal of the DNA Methylation Society. 2009;4(2):80–4. [DOI] [PubMed] [Google Scholar]
- 156.Huang B, Jiang C, Zhang R. Epigenetics: the language of the cell? Epigenomics. 2014;6(1):73–88. [DOI] [PubMed] [Google Scholar]
- 157.Ma H, Hao Y, Dong X, Gong Q, Chen J, Zhang J, et al. Molecular mechanisms and function prediction of long noncoding RNA. TheScientificWorldJournal. 2012;2012:541786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Huang CF, Zhu JK. RNA Splicing Factors and RNA-Directed DNA Methylation. Biology. 2014;3(2):243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305(5688):1289–92. [DOI] [PubMed] [Google Scholar]
- 160.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Molecular cell. 2006;22(3):351–61. [DOI] [PubMed] [Google Scholar]
- 162.Onorati MC, Lazzaro S, Mallik M, Ingrassia AM, Carreca AP, Singh AK, et al. The ISWI chromatin remodeler organizes the hsromega ncRNA-containing omega speckle nuclear compartments. PLoS genetics. 2011;7(5):e1002096. [DOI] [PMC free article] [PubMed] [Google Scholar]