Abstract
Sepsis is a rapidly progressing, life threatening immune response triggered by infection that affects millions worldwide each year. Current clinical diagnosis relies on broad physiological parameters and time consuming lab-based cell culture. If proper treatment is not provided, cases of sepsis can drastically increase in severity over the course of a few hours. Development of new point of care tools for sepsis has the potential to improve diagnostic speed and accuracy, leading to prompt administration of appropriate therapeutics, thereby reducing healthcare costs and improving patient outcomes. In this review we examine developing and commercially available technologies to assess the feasibility of rapid, accurate sepsis diagnosis, with emphasis on point of care.
Introduction
Sepsis is a life-threatening condition caused by the body’s response to microbial infection, which triggers a cascade of events that can lead to organ failure and death.1 Global estimates suggest that there were 31.5 million cases of sepsis in 2016, with 5.3 million resulting in death.2 The Center for Disease Control reports that one in three fatalities in US hospitals are related to sepsis.3 Neonatal sepsis is also a major concern, with the World Health Organization estimating an annual death incidence of over 1 million.4
The current standard for clinical sepsis diagnosis, referred to as Sequential Organ Failure Assessment (SOFA), was defined by the Third International Sepsis Consensus Task Force in 2016.5 This scoring system informs of cumulative organ dysfunction that results from a dysregulated host response to infection. The SOFA score factors in respiration (arterial oxygen partial pressure to fractional inspired oxygen), coagulation (platelet count), liver function (bilirubin levels), cardiovascular function (mean arterial pressure or intervention), central nervous system function (Glasgow Coma Scale), and renal function (creatinine and urine output).5 For each poorly functioning organ system, one point is assigned, with a score of 2 points or more representing a positive sepsis diagnosis. A positive SOFA score is associated with a hospital mortality rate of >10%.5 Quick SOFA (qSOFA) offers a rapid alternative to SOFA by using only a subset of the SOFA scoring including altered mentation, systolic blood pressure below 100 mm Hg, and a respiratory rate of 22 per minute or greater.5 qSOFA does not require laboratory testing and it can be repeated as needed. SOFA replaces the standard established in 2001 known as Systemic Inflammatory Response Syndrome (SIRS) criteria.6 SIRS scoring involves two or more of: temperature greater than 38˚C or less than 36˚C, heart rate above 90 beats per minute, respiratory rate greater than 20 per minute, or white blood cell count greater than 12,000/μL or less than 4,000/μL.7 SOFA has shown greater prognostic accuracy when compared to SIRS and qSOFA.8
Sepsis diagnosis has improved with the implementation of the SOFA scoring system; however, it still lacks specificity and requires multiple measurements. SOFA scoring leads to misdiagnoses in approximately 30% of patients, leading to unnecessary admin-istration of antibiotics, which increases hospital costs and contrib-utes to antibiotic resistance.9 Given that survival rate decreases by 7.7% for every hour delay in the administration of antimicrobial therapy, rapid detection is vital for patient survival.10 Additionally, many of the SOFA criteria require infrastructure and testing equipment, such as blood analyzers, that may be unavailable in developing healthcare communities. Due to these factors, it would be preferable to have a simpler, single test to diagnosis sepsis at the point of need that can replace SOFA scoring without decreased accuracy.
Point of care (POC) technologies based on biomolecular analysis provide rapid, actionable information to a patient or health professional at the time and site of evaluation and treatment.11 These technologies are typically low cost, easy to use, and rapid with few infrastructure requirements making them applicable in a variety of settings worldwide.12 Microfluidics,13 lateral flow,14,15 dipstick,15 and smartphone16 technologies have been used to investigate sepsis biomarkers such as proteins, nucleic acids, human cells, microbes, or pathogens.17 POC technology is capable of utilizing small sample volumes, such as a fingerprick blood sample, and compact standalone devices or only the most basic laboratory equipment, such as microscopes. These factors allow POC diagnostics the potential to reach patients in limited-resource healthcare communities.12 Additionally, use of POC testing at triage in US hospitals has been shown to reduce emergency room care times by 1 hour compared to traditional laboratory testing, decreasing hospital costs and saving lives.18 POC diagnostics are actively being explored for nutrition,19–22 infectious disease,11,13,23 cancer,24–27 and HIV/AIDS.28–30
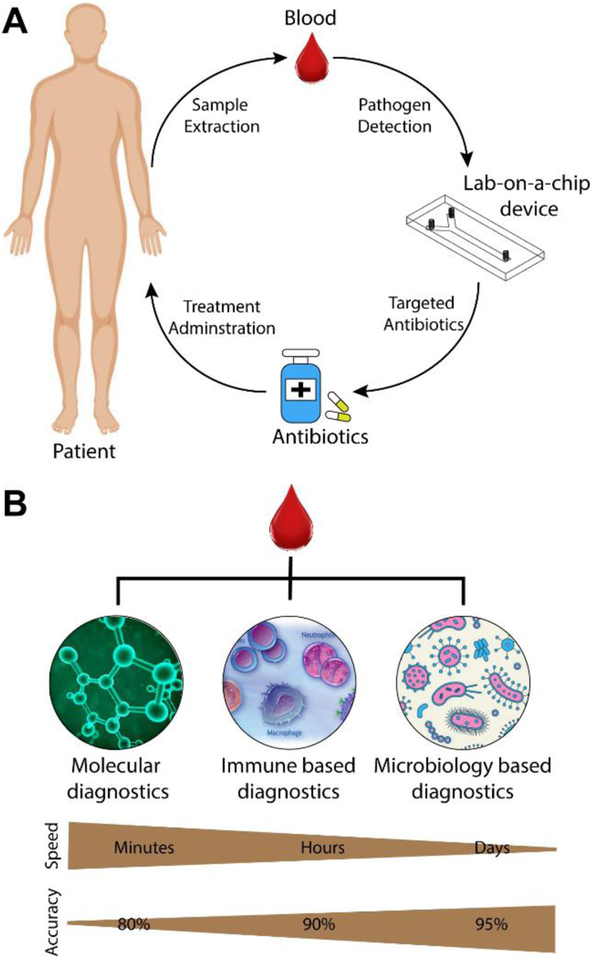
Sepsis POC diagnostics are a potential solution for risk-stratification of new patients, guiding initial treatment, monitoring progression of infection, and evaluating patients’ response to treatment (Figure 1A).31 Ideal testing times should be on the order of tens of minutes to allow antibiotic intervention to prevent spread of infection and organ failure. POC tests used in US hospitals may require high diagnostic accuracy to compete with laboratory testing while low resource settings may prioritize inexpensive production, long-term low-cost storage, and ease of use.
Figure 1.
A) Recommended use of POC sepsis chip in clinical care. Adapted with permission from Ref. 69. B) Categories of whole blood sepsis diagnostics are listed on the top and ordered by relative speed and accuracy depicted as gradients on the bottom.
Research on POC sepsis devices has been conducted using a variety of biomarkers (Figure 1B). Here we categorize them in three sections including blood plasma protein quantification, leukocyte monitoring, and pathogen detection. Relevant blood plasma proteins include C-reactive protein (CRP), procalcitonin (PCT), interleukins, and lactate. Leukocyte activity is measured through antibody capture or intrinsic property characterization while pathogen isolation is accomplished though many unique techniques such as magnetic separation. In this review, specific technologies are presented and evaluated for their respective advantages and disadvantages. Inclusion in this review is limited to the previous 10 years and application in a POC device or setting. A comprehensive review of all sepsis biomarkers outside of POC32–36 and the pathophysiology of sepsis37,38 can be found elsewhere.
Assays for Blood Plasma Proteins
During sepsis, various proteins are expressed or secreted into the blood as a response to infection – cytokines, interleukins, acute-phase proteins, and receptor proteins concentrations are all increased. Presently, the most frequently investigated biomarkers for POC diagnostics are interleukin-6 (IL-6), C-reactive protein (CRP), and procalcitonin (PCT).39 IL-6 is a cytokine released near the onset of infection that correlates well with severity and outcomes in septic patients, where decreasing levels are predictive of survival.40,41 A recent meta-analysis found the area under receiver operating characteristic curve (AUROC) value for IL-6 to be 0.79, indicating moderate diagnostic sensitivity.36 CRP is expressed as a response to cytokines like IL-642 and has an AUROC value of 0.77.36 PCT release is dependent on the severity of infection43 with a half-life of around 24 hours and an AUROC value of 0.8536, where decreasing levels correlate with better survival rates.39 IL-26 in combination with PCT exhibits greater diagnostic ability than either IL-26 or PCT alone.44 Other proteins tested as standalone biomarkers include lactate, sTREM-1, neopterin, TNF-α, E-selectin, S-100, presepsin, LBP, CD64, DcR3, endocan, sICAM-1, and C3a.36 Some of these markers have AUROC values greater than 0.9; however, the supporting literature is sparse so more extensive research is needed to be conclusive.
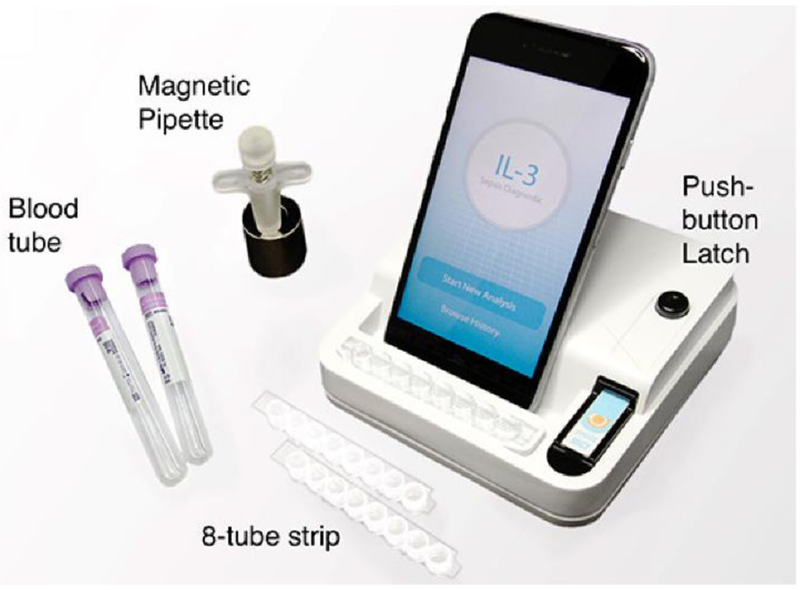
Single-analyte devices have the advantage of simplicity and are being tested for POC sepsis diagnosis. Blood lactate level has been used as a standalone sepsis biomarker for risk stratification of emergency room patients.45 Gaieski et al.46 reported that fingerprick sampling of lactate reduced the mean time from triage to result by 40% from standard laboratory analysis. Rascher et al.47 have developed a microfluidic device to detect PCT via immunofluorescence with a limit of detection (LOD) of 0.04ng/mL in less than ten minutes. Okamura et al.48 have developed an assay, called PATHFAST, to detect presepsin, showed higher specificity compared to other single biomarkers.49 A chemiluminescent enzyme immunoassay is used to run whole blood samples with results in 17 minutes. Valera et al.50 have adapted a previously designed biochip platform28,51,52 to sense IL-6 using latex beads. Measurements with this device took 5 minutes each and the limit of detection was 127 pg/mL. Min et al.53 are able to detect IL-3 levels within 1-hour using their POC device with a reported AUROC value of 0.91 (Figure 2). The smartphone functions as a touch-screen interface, as well as providing data storage and system controls. POC devices commonly integrate smartphone technology due to their imaging, computation, communication, and networking capabilities, as well as their familiarity and simplicity to almost all age groups.54
Figure 2.
Example smart phone enabled POC interleukin detection device. Reproduced with permission from Ref. 53.
Multiplexed biomarker detection has the potential to increase diagnostic power. Shapiro et al.55 suggested the most effective panel to assess patient risk would include neutrophil gelatinase-associated lipocalin, interleukin-1ra, and Protein C. Buchegger et al.56 have developed a miniaturized protein microarray chip for the detection of IL-6, IL-8, IL-10, TNF alpha, S-100, PCT, E-Selectin, CRP, and Neopterin. The device has been tailored for sepsis POC testing in neonates, building on a previous device.57 Improvements include a reduced patient sample volume of 4μL, implementation of a single-step process, reduced incubation times, and the addition of internal calibration. For the previous iteration of this device, Sauer et al.57 reported that the 2.5 hour single-step protocol suffered from lower sensitivity than the 4 hour multi-step protocol. However, the new device shows no loss of sensitivity when utilizing streptavidin conjugated magnetic particles to promote mixing and binding within the smaller sample volume. Complete sample processing was kept at 2.5 hours for the single-step process due to decreasing accuracy for analytes S-100 and CRP when incubation time was reduced. Kemmler et al.58 have developed a microarray biochip using CRP, IL-6, PCT, and NPT. The device uses an on-chip immunofluorescence assay that lasts only 25 minutes and requires between 10–75μL of sample. However, due to the high LOD of IL-6 and PCT, this device is only applicable for clinically relevant levels in the mid to high range. The clinically relevant region of interest for PCT is between 0.05 and 50ng/mL, while the PCT LOD for this device is 0.34ng/mL. Schotter et al.59 reported a magnetic lab-on-a-chip device for the detection of the sepsis-related cytokines IL-6, IL-8, IL-10, and TNF-α using embedded magneto resistive sensors.
Leukocyte Intrinsic Properties as Immune Markers
Identifying and monitoring the systemic inflammatory response is important because immune-paralysis is common in late stage sepsis, so this can be used as a marker of sepsis imitation and progression. Neutrophils have gained attention because of detectable changes in morphology, mechanics, and motility in septic conditions.1,59–62 Neutrophil CD64 expression63,64 has been shown to increase under septic conditions, making it a potential biomarker for sepsis.65–67 In response to bacterial infection, neutrophil CD64 was demonstrated to be upregulated as early as 1 hour after infection.68
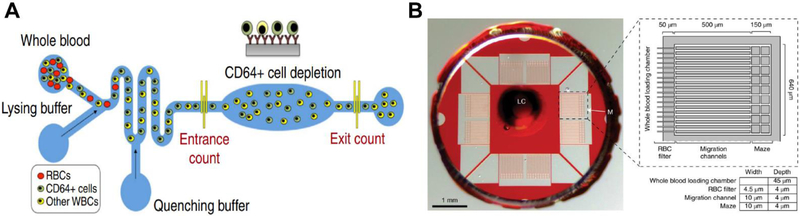
Zhang et al.69 developed a POC device with a herringbone-shaped cell capture chamber for affinity separation of CD64 cells within 2 hours. The device demonstrated an AUROC of 0.90 when compared to sepsis diagnosis based on qSOFA in a trial of 10 patient samples.69 The technology was then expanded to include CD69 and the combined panel increased the AUROC to 0.98 when tested with 12 qSOFA positive clinical samples.70 Prieto et al.71 used iso-dielectric separation to differentiate electrical properties of activated and non-activated leukocytes. Using mouse models, they demonstrated that their on-a-chip electrical cell profiling strongly correlated with flow cytometry.71 This device offers continuous monitoring of sepsis progression; however, it has not been tested clinically. Hassan et al.72 designed a microfluidic biochip to electrically quantify CD64 antigen expression on neutrophils (Figure 3A). Their device can produce test results in 30 minutes and offers an alternative to hematology analyzers and flow cytometers which may not be available in a POC setting.72 This technology builds on previous POC devices that electronically quantifies cells from whole blood samples.52,73,74 This device demonstrated an AUROC of 0.77 which is better than the AUROC of a subset of SIRS parameters (temperature, pulse, respirations and systolic blood pressure) which was found to be 0.7.72 Applications of cell capture and quantification on a chip is not limited to sepsis, and reviews of these technologies can be found elsewhere.75–77
Figure 3.
A) Microfluidic CD64 expression quantification device with lysing and quenching zones for erythrocyte removal. False-colored fluorescent image of CD64+ cells captured on anti-CD64 coated pillars in the CD64 cell depletion zone. Reproduced with permission from Ref. 74.B) Microfluidic assay for spontaneous neutrophil motility from a drop of blood. Reproduced with permission from Ref. 9.
Neutrophils undergo changes in their intrinsic properties during infection and observing them can inform on disease progression. The most frequently studied is neutrophil motility.9,60,61 Neutrophils, even once isolated from blood, exhibit a spontaneous migration signature specific to sepsis.61 Ellet et al.9 created a microfluidic assay to capitalize on this principle (Figure 3B). Using continuous fluorescent imaging and a machine learning algorithm, they quantified reverse migration, oscillation, and pauses of neutrophils isolated from a finger prick blood sample. The assay is robust, requires minimal handling, and automated imaging and analysis. It requires a four-hour time-lapse microscopy step followed by two-and-a-half-hour image processing, a longer time scale than some POC technologies but still clinically relevant. In a double blinded clinical trial using SOFA scoring, the device obtained an AUROC of 0.99 with 97% sensitivity and 98% specificity.9
While many of the blood plasma assays and leukocyte technologies hold promise, there is not currently a point of care technology that has sensitivity and specificity in the 90th percentile and can produce results in under an hour.
Pathogen Isolation as a Diagnostic and Treatment
With advances in miniaturization and development of lab-on-a-chip systems, direct isolation and detection of pathogenic bacteria from the blood for sepsis therapy and diagnosis has been explored. Microfluidic devices for pathogen isolation, lysis, and DNA extraction have been reported to identify pathogens and aid in antibiotic selection. Techniques utilizing dielectrophoresis, inertial effects, surface acoustic waves, and centrifugal microfluidics have been developed for separation of infectious agents from blood cells. Lab-on-a-chip systems with integrated bacterial separation, chemical or mechanical cell lysis, and DNA extraction for PCR or sequence specific capture have shown promise for rapid sepsis diagnosis.78–80
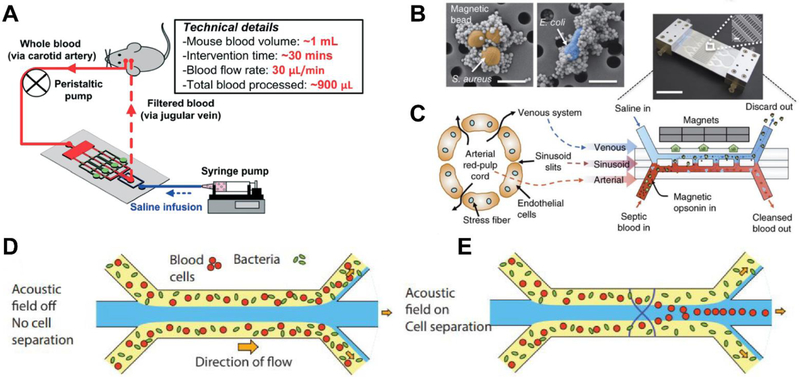
Artificial spleen type devices for the direct removal of pathogens and endotoxins from whole blood were developed for therapeutic applications (Figure 4A). Similar to dialysis, these devices process whole blood by capturing a broad range of infectious agents using ligand and antibody coated magnetic beads,81–85 porous membranes,86 biomimetic cell margination,87,88 acoustophoresis,89,90 and elasto-inertial-based migration.91 Table 1 lists the miniaturized pathogen removal devices for sepsis treatment. A comparison of pathogen separation rate and removal efficiency is shown among these devices. Lee et al.84 developed magnetic nanoparticles modified with a synthetic ligand that binds to both Gram-positive and Gram-negative bacteria. Yung et al.82 developed mannose-binding lectin (MBL) coated magnetic nanobeads that do not activate complement factors or coagulation (Figure 4B,C). They reported processing of up to 1.25 L/hr of blood and greater than 90% removal of pathogenic organisms in rats. Lopes et al.85 reported separation of six Gram-positive species, two Gram-negative species, Methicillin-susceptible Staphylococcus aureus (MSSA), and Methicillin-resistant Staphylococcus aureus (MRSA) in whole blood via mutated lysozyme coated magnetic beads. Hou et al.87 developed an extracorporeal biomimetic microfluidic device based on microcirculatory cell margination for non-specific removal of pathogens and cellular components. This method can reach up to ~1–2 L/hr throughput and when multiplexed could produce a 100-fold higher throughput. An acoustic microfluidic method (Figure 4D, E) was recently reported by Ohlsson et al.89,90 based on size separation of pathogens. This method is label-free and resulted in a greater than 80% bacterial separation from a whole blood sample. These miniaturized devices for sepsis pathogen removal have a potential for POC applications. By improving clearance rates and efficiency, these devices could be effective in aiding sepsis treatment. Separation rates similar to blood dialysis machines (~ 1–2 L/hr) would require at least 5 hours to filter all the blood from an average person. Since survival decreases by the hour, a flow rate closer to ~4–5 L/hr is more desirable for therapeutic POC applications. Such characteristics can be achieved by 1) multiplexing microfluidic devices for high throughput, 2) utilizing high affinity target binding antibodies, and 3) minimizing technological complexity for separation.
Figure 4.
A) Example pathogen removal apparatus. Adapted with permission from Ref. 86. B) Magnetic bead capture of S. aureus and E. coli. Reproduced with permission from Ref. 81. C)Magnetic opsonin and biospleen device for pathogen removal and blood cleansing. Reproduced with permission from Ref. 81. D-E) Acoustic separation of bacteria and blood cells. Reproduced with permission from Ref. 89.
Table 1.
State-of-the-art pathogen removal devices and techniques
| Group | Device | Targets | Separation method | Sample | Separation Rate | Reported Pathogen Clearance |
|---|---|---|---|---|---|---|
| Ingber80 | Microfluidic | Fungi (C. albicans) | Magnetic separation with antibody coated beads | Whole blood | 20 ml/hr | 80% |
| Kohane84 | Microfluidic | E. coli | MNPs modified with bis-Zn-DPA | Bovine whole blood | 60 ml/hr | 100% |
| Ingber81 | Microfluidic | S. aureus, E. coli | Magnetic beads with engineered human opsonin-MBL | Rat whole blood | 1.25 L/hr | >90% |
| Russom91 | Microfluidic | E. coli | Elasto-inertial microfluidics with viscoelastic effect | Whole blood | 60 μl/hr | 76% |
| Han87 | Microfluidic | E. coli, leukocytes, cytokines | Passive cell migration | Spiked whole blood | 150 ml/hr | 70% |
| Kreiger85 | Manual | Six Gram-positive and two Gram-negative species, MSSA, MRSA | LysE35A-coated beads | Whole blood | N/A | 85% |
| Zhan86 | Microfluidic | E. coli | Porous polycarbonate (PCTE) membrane with 2 um pores | Whole blood | 50 μl/min | 22% |
| Laurell90 | Microfluidic device with acoustic transducer | P. putida | Size based separation using acoustic impedance | Whole blood | 200 μl/min | 80% |
The blood cleansing approach is promising for the direct removal of pathogens from blood, however, this method will not remove infection localized within organs. With the blood cleansing technology in infancy, significant processing times of several hours are required to remove the circulating infection which is slower than desirable. The removal of blood pathogens has been shown to delay disease progression, providing time to identify the contamination and administer targeted antibiotics, thus improving overall survival. Combining on-chip DNA extraction and blood cleansing technology could be used to perform pathogen-specific analysis while targeted antibacterial therapies are investigated.
Commercial Devices
Most of the technologies discussed thus far are in research and development, but it is also important to consider those that are commercially available. Commercial technologies currently available for sepsis diagnosis range from simple devices like blood analysers to more complex systems for molecular diagnostics and mass spectroscopy. Table 2 provides a complete summary of commercially available diagnostic technologies for sepsis, with an evaluation of their feasibility at the POC. We have categorized them into blood analysers, microbiology based, molecular diagnostic based, and pathogen removal. Currently, most of these methods require microarrays, polymerase chain reaction (PCR), or other complex laboratory equipment that may be unavailable in low resource settings. We predict that the commercial market will also experience a shift towards POC sepsis diagnostics in the coming years.
Table 2.
Commercially Available Technologies for Sepsis Diagnosis and Treatment
| Device | Company | Technology | Sample | Turnaround time | Sepsis Diagnosis | FDA clearance | Point-of-care? | |
|---|---|---|---|---|---|---|---|---|
| Blood Analyzers | ||||||||
| EPOC® Blood Analysis System | Siemens Healthineers | Blood gas analyzer with room temperature test card | Whole blood | < 1 min | qSOFA scoring | Yes | Portable, 92 ul blood sample | |
| Microbiology Based | ||||||||
| Septi-Chek | Becton Dickinson | Blood culture with media | Whole blood | ~38 hr | Pathogen identification | Yes | Requires laboratory equipment | |
| Oxiod Signal | Thermo Fisher Scientific | Blood culture with media | Whole blood | 24hr | Pathogen identification | Yes | Requires laboratory equipment | |
| Molecular Diagnostic Based | ||||||||
| QuickFISH/PNA FISH | AdvanDx | Fluorescence in situ hybridization | Positive blood culture | ~1.5 hr | Pathogen identification | Yes | Requires microarray equipment | |
| hemoFISH | Miacom diagnostics | Fluorescence in situ hybridization | Positive blood culture | 0.5 hr | Pathogen identification | Yes | Requires microarray equipment | |
| Prove-it / Verigene | Luminex | PCR amplification and array detection | Positive blood culture | 3.5 hr | Pathogen identification | No | Requires PCR equipment | |
| FilmArray | Biofire Diagnostics | Real-time PCR | Positive blood culture | 1hr | Pathogen identification | Yes | Requires FilmArray system | |
| HYPLEX | BAG | Multiplex PCR with end point detection using an enzyme-linked sandwich assay in a microtiter plate | Positive blood culture | 3 hr | Pathogen identification | No | Requires PCR and complex instrumentation | |
| ACCU-PROBE | Gen-probe | Chemiluminescent DNA probes (rRNA) | Positive blood culture | 3 hr | Pathogen identification | No | Requires PCR and complex instrumentation | |
| PLEX-ID BAC | Abott | PCR amplification with Electrospray ionization mass spectrometric analysis | Positive blood culture | 6hr | Pathogen identification | No/CE marking | Requires PCR and complex instrumentation | |
| Staph SR | Becton Dickinson | Multiplex PCR | Positive blood culture | 3 hr | Pathogen identification | Yes | Requires PCR equipment | |
| StaphPlex | Qiagen | PCR amplification and Array detection | Positive blood culture | 5 hr | Pathogen identification | No | Requires PCR and complex instrumentation | |
| MALDI-TOF | bioMérieux | Matrix assisted laser desorption | Positive blood culture | 2hr | Pathogen identification | No | Requires complex instrumentation | |
| Magicplex | Seegene | Multiplex PCR | Whole blood | 3.5 hr | Pathogen identification | No | Requires PCR equipment | |
| SeptiFast | Roche | Multiplex PCR | Whole blood | 6 hr | Pathogen identification | Yes | Requires PCR equipment | |
| SepsiTest | Molzym, Germany | PCR and sequencing | Whole blood | 8–12 hr | Pathogen identification | No | Requires PCR and complex instrumentation | |
| VYOO | SIRS lab | Multiplex PCR and gel electrophoresis | Whole blood | 8 hr | Pathogen identification | No | Requires PCR equipment | |
| Xpert MRSA/SA | Cepheid | Real-time multiplex PCR | Whole blood | 1hr | Pathogen identification | Yes | Requires PCR equipment | |
| VAPChip | Eppendorf Array Technologies, Belgium | Multiplex PCR and hybridization | Whole blood | 5–8 hr | Pathogen identification | No | Requires PCR and complex instrumentation | |
| DiagCORE | STAT Diagnostics | 48 plex real-time PCR | Whole blood | 0.5 −1.2 hr | Pathogen identification | No/CE marking | Requires PCR equipment | |
| T2 Candida | T2 Biosystems, USA | Miniaturized magnetic resonance | Whole blood | 3–5 hr | Pathogen identification | Yes | Requires complex instrumentation | |
| Pathogen Removal Devices | ||||||||
| Cytosorb | Cytosorb | Removes cytokines with filter | Whole blood | Up to 24 hours a day, for up to 7 consecutive days | Bacterial removal | No/CE marking | Requires extracorporeal equipment with filtration in a hospital | |
| LPS Adsorber | Alteco Medical, Sweden | Peptide binding of endotoxin with porous polyethylene matrix | Whole blood | Not available | Bacterial removal | No/CE marking | Requires extracorporeal equipment with filtration in a hospital | |
| LPS-binding Toraymyxin device | Spectral Medical | Ionic binding of Polymyxin-B and Lipid A to endotoxin | Whole blood | Recommended dosing: 2 hr hemoperfusion in 24 hr period | Bacterial removal | No/CE marking | Requires extracorporeal equipment with filtration in a hospital | |
Bacterial cultures are time consuming (24 to 48 hours), delaying the switch from broad spectrum antibiotics to more effective targeted therapies. Mature molecular diagnostic techniques such as PCR and fluorescent in-situ hybridization (FISH) have emerged as rapid diagnostic alternatives for pathogen identification.92–94 These techniques require up to 12 hours, but recently, turnaround times as small as 30 minutes are reported for the identification of 14 bacterial species by a Miacom system using FISH from positive blood culture test. Standard blood culture takes 1–3 days to increase bacterial concentration to the detectable limit for molecular diagnostic testing. By eliminating the blood culture, technologies such as Septifast (Roche) can identify 25 different common sepsis pathogens within 6 hours, DiagCORE (STAT Diagnostics) can perform a 48 plex real-time PCR test that recognizes relevant pathogens in 30 to 80 minutes, and T2 Candida panel detects fungal infections in whole blood samples using a PCR system. Although commercial systems exist, many of these technologies are not suitable for POC use. The research on sepsis POC technologies highlighted in this article could reduce the time to detection and aid in targeted antibacterial therapies at the point of need.
Commercial biomarker tests that indicate early organ dysfunction and predict septic shock have been developed. Molecular fingerprinting prior to and during treatment, including cytokine profiling95–97 and PCT tests,98 are being clinically evaluated. Many innovations are unfolding in the biomarker space such as adrenomedullin, which is a biomarker for septic shock shown to increase in blood 1–2 days before shock occurs.99 Devices for extracorporeal filtering of the infection also exist in the market. Products like Cytosorb (CE-marked) filter proinflammatory cytokines to control immune response while Alteco Medical’s LPS Adsorber and Spectral Medical’s LPS-binding Toraymyxin devices remove endotoxins released by pathogens. For POC applications, there is a need to develop and improve these commercial devices for both rapid and accurate diagnosis of sepsis.
Conclusions
In this review, we examined developing and commerically avaliable technologies to assess the feasibility of rapid, accurate sepsis diagnosis with emphasis on POC. As with many technologies, there is conflict between speed and accuracy. Some of these devices, such as the EPOC Blood Analysis System (Siemens Healthineers), are very fast but have limited use and require secondary testing and evalutation for a complete diagnosis. Others have strong monitoring potential but take hours of complex anaylsis.
Studies indicate that diagnosing sepsis using a single biomarker is challenging,100 however, biomarker panels have shown promise.56–58,101 Additional barriers to pathogen detection exist due to the breadth of infectious agents, relatively high limits of detection, and long culture times. Recent research has produced strong evidence for the use of immune system markers as indicators of sepsis over serum biomarkers. Neutrophils have been of special interest due to their critical role in innate immunity and rapid response to infection.102 We predict that markers of the immune response to sepsis will continue to be well studied in the coming years.
Some experts have called into question the homogeneity of sepsis as a disease.103–109 They argue that the term sepsis itself is too broad and that different classifications of sepsis should exist. If this is true, a single diagnostic device for all types of sepsis may not be possible. Based on a functional analysis, Sweeny et al.103 proposed three subtypes of sepsis: inflammopathic, adaptive, and coagulopathic. Davenport et al.107 suggest two phenotypes based on immune response states, prognoses, and genetic variation, whereas, Wong et al.104–106 proposed three subtypes based on genome-wide expression profiling. The heterogeneous disease progressions and immune responses associated with sepsis, may be unminding the efforts to standardize diagnostics and treatments. Additional research is required to determine the extent of heterogeneity in sepsis and the corresponding effects on diagnostics and therapies. It is possible that, rather than a single biomarker or panel to diagnosis all cases of sepsis, it will be necessary to utilize different diagnostics at different times and for different subsets of septic patients.100 We predict that research on the homogeneity of sepsis will be of increasing interest in the coming years.
The transition to SOFA scoring makes benchmarking POC diagnostic devices challenging. While some devices are tested against SOFA scoring, others are still being tested against less accurate SIRS scoring.5,8,9,110 A standardized protocol for comparison should be established to increase the power of reported results. POC devices should continue to be designed with an emphasis on low-cost and long term stability, and additional thought should be given to how diagnostic outcomes can inform treatment. Additionally, further devlopement of diagnostics could expand the ability to differentiate stages and progression of sepsis, rather than septic versus not septic.100 POC technology for the diagnosis or treatment of sepsis would be especially beneficial due to the rapid disease progression and impact on populations worldwide. A successful POC sepsis diagnostic could improve health care access, save lives, reduce healthcare costs, and minimize suffering both in countries with well-developed healthcare systems and in low-resource settings. In the last 10 years, significant progress has been made, but additional research is vital to develop a successful POC device for sepsis.
Acknowledgements
DE acknowledges support from the US National Institutes of Health through grant R01EB021331 (FeverPhone). AS acknowledges support from the US National Institutes of Health through grant R01AI132738-01A1.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Conflicts of Interest
There are no conflicts to declare.
References
- 1.Hotchkiss RS and Karl IE, N. Engl. J. Med, 2003, 348, 138–150. [DOI] [PubMed] [Google Scholar]
- 2.Fleischmann C, Scherag A, Adhikari NKJ, Hartog CS, Tsaganos T, Schlattmann P, Angus DC and Reinhart K, Am. J. Respir. Crit. Care Med, 2016, 193, 259–272. [DOI] [PubMed] [Google Scholar]
- 3.CDC: Sepsis Statistics, https://www.cdc.gov/sepsis/datareports/index.html, (accessed 27 March 2018). [Google Scholar]
- 4.Edmond K and Zaidi A, PLoS Med, , DOI: 10.1371/journal.pmed.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer M, Deutschman CS, Seymour C, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, Der Poll T, Vincent JL and Angus DC, JAMA - J. Am. Med. Assoc, 2016, 315, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, Cohen J, Opal SM, Vincent JL and Ramsay G, in Intensive Care Medicine, 2003. [DOI] [PubMed] [Google Scholar]
- 7.Bone RC, Balk RA, Cerra F and Al E., Crit. Care Med, 1992, 20, 864–874.1597042 [Google Scholar]
- 8.Raith EP, Udy AA, Bailey M, McGloughlin S, MacIsaac C, Bellomo R and Pilcher DV, JAMA - J. Am. Med. Assoc, 2017, 317, 290–300. [DOI] [PubMed] [Google Scholar]
- 9.Ellett F, Jorgensen J, Marand AL, Liu YM, Martinez MM, Sein V, Butler KL, Lee J and Irimia D, Nat. Biomed. Eng, 2018, 2, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A and Cheang M, Crit. Care Med, 2006, 34, 1589–1596. [DOI] [PubMed] [Google Scholar]
- 11.Kozel TR and Burnham-marusich AR, J. Clin. Microbiol, 2017, 55, 2313–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yager P, Domingo GJ and Gerdes J, Annu. Rev. Biomed. Eng, 2008, 10, 107–144. [DOI] [PubMed] [Google Scholar]
- 13.Foudeh AM, Fatanat Didar T, Veres T and Tabrizian M, Lab Chip, 2012, 12, 3249–3266. [DOI] [PubMed] [Google Scholar]
- 14.Hu J, Wang SQ, Wang L, Li F, Pingguan-Murphy B, Lu TJ and Xu F, Biosens. Bioelectron, 2014, 54, 585–597. [DOI] [PubMed] [Google Scholar]
- 15.Yetisen AK, Akram MS and Lowe CR, Lab Chip, 2013, 13, 2210–2251. [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Akay A, Wei H, Wang S, Pingguan-Murphy B, Erlandsson BE, Li X, Lee W, Hu J, Wang L and Xu F, Proc. IEEE, 2015, 103, 236–247. [Google Scholar]
- 17.Gubala V, Harris LF, Ricco AJ, Tan MX and Williams DE, Anal. Chem, 2012, 84, 487–515. [DOI] [PubMed] [Google Scholar]
- 18.Singer AJ, Taylor M, LeBlanc D, Meyers K, Perez K, Thode HC and Pines JM, J. Emerg. Med, 2018, 55, 172–178. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, O’Dell D, Hohenstein J, Colt S, Mehta S and Erickson D, Sci. Rep, 2016, 6, e28237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srinivasan B, O’Dell D, Finkelstein JL, Lee S, Erickson D and Mehta S, Biosens. Bioelectron, 2018, 99, 115–121. [DOI] [PubMed] [Google Scholar]
- 21.Lu Z, O’Dell D, Srinivasan B, Rey E, Wang R, Vemulapati S, Mehta S and Erickson D, Proc. Natl. Acad. Sci, 2017, 114, 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vemulapati S, Rey E, O’Dell D, Mehta S and Erickson D, Sci. Rep, 2017, 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Mehta S and Erickson D, Anal. Chem, 2016, 88, 8359–8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Biosens. Bioelectron, 2006, 21, 1887–1892. [DOI] [PubMed] [Google Scholar]
- 25.Soper SA, Brown K, Ellington A, Frazier B, Garcia-Manero G, Gau V, Gutman SI, Hayes DF, Korte B, Landers JL, Larson D, Ligler F, Majumdar A, Mascini M, Nolte D, Rosenzweig Z, Wang J and Wilson D, Biosens. Bioelectron, 2006, 21, 1932–1942. [DOI] [PubMed] [Google Scholar]
- 26.Rusling JF, Kumar CV, Gutkind JS and Patel V, Analyst, 2010, 135, 2496–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciatto S, JAMA, 2005, 293, 810–816.15713770 [Google Scholar]
- 28.Watkins NN, Hassan U, Damhorst G, Ni HK, Vaid A, Rodriguez W and Bashir R, Sci. Transl. Med, 2013, 5, 214ra170. [DOI] [PubMed] [Google Scholar]
- 29.Wesolowski LG, MacKellar DA, Facente SN, Dowling T, Ethridge SF, Zhu JH and Sullivan PS, Aids, 2006, 20, 1661–1666. [DOI] [PubMed] [Google Scholar]
- 30.Cheng X, Gupta A, Chen C, Tompkins RG, Rodriguez W and Toner M, Lab Chip, 2009, 9, 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umlauf VN, Dreschers S and Orlikowsky TW, Int. J. Pediatr, 2013, e63191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierrakos C and Vincent JL, Crit. Care, 2010, 14, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kibe S, Adams K and Barlow G, J. Antimicrob. Chemother, 2011, 66, 33–40. [DOI] [PubMed] [Google Scholar]
- 34.Marshall JC and Reinhart K, Crit. Care Med, 2009, 37, 2290–2298. [DOI] [PubMed] [Google Scholar]
- 35.Bloos F and Reinhart K, Virulence, 2014, 5, 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Hou J, Li Q, Chen K, Wang S-N and Wang J, Springerplus, 2016, 5, 2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hotchkiss RS, Moldawer LL, Opal SM, Reinhart K, Turnbull IR and Vincent JL, Nat. Rev. Dis. Prim, 2016, 2, 16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angus DC and van der Poll T, N. Engl. J. Med, 2013, 369, 840–851. [DOI] [PubMed] [Google Scholar]
- 39.Bloos F and Reinhart K, Virulence, 2014, 5, 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettilä V, Hynninen M, Takkunen O, Kuusela P and Valtonen M, Intensive Care Med, 2002, 28, 1220–1225. [DOI] [PubMed] [Google Scholar]
- 41.Casey LC, Balk RA and Bone RC, Ann. Intern. Med, 1993, 119, 771–778. [DOI] [PubMed] [Google Scholar]
- 42.Gabay C and Kushner I, N. Engl. J. Med, 1999, 340, 448–454. [DOI] [PubMed] [Google Scholar]
- 43.Bloos F, Marshall JC, Dellinger RP, Vincent J-L, Gutierrez G, Rivers E, Balk RA, Laterre P-F, Angus DC, Reinhart K and Brunkhorst FM, Crit. Care, 2011, 15, R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong HR, Liu KD, Kangelaris KN, Lahni P and Calfee CS, J. Crit. Care, 2014, 40, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikkelsen ME, Miltiades AN, Gaieski DF, Goyal M, Fuchs BD, Shah CV, Bellamy SL and Christie JD, Crit. Care Med, 2009, 37, 1670–1677. [DOI] [PubMed] [Google Scholar]
- 46.Gaieski D, Drumheller B, Goyal M, Fuchs B, Shofer F and Zogby K, West. J. Emerg. Med, 2013, 14, 58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rascher D, Geerlof A, Kremmer E, Krämer P, Michael S, Hartmann A and Rieger M, Biosens. Bioelectron, 2014, 59, 251–258. [DOI] [PubMed] [Google Scholar]
- 48.Okamura Y and Yokoi H, Clin. Chim. Acta, 2011, 412, 2157–2161. [DOI] [PubMed] [Google Scholar]
- 49.Yaegashi Y, Sato N, Suzuki Y, Kojika M, Imai S, Takahashi G, Miyata M, Endo S, Shirakawa K and Furusako S, J. Infect. Chemother, 2005, 11, 234–238. [DOI] [PubMed] [Google Scholar]
- 50.Valera E, Berger J, Hassan U, Ghonge T, Liu J, Rappleye M, Winter J, Abboud D, Haidry Z, Healey R, Hung NT, Leung N, Mansury N, Hasnain A, Lannon C, Price Z, White K and Bashir R, Lab Chip, 2018, 18, 1461–1470. [DOI] [PubMed] [Google Scholar]
- 51.Watkins NN, Sridhar S, Cheng X, Chen GD, Toner M, Rodriguez W and Bashir R, Lab Chip, 2011, 11, 1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hassan U, Watkins NN, Reddy B, Damhorst G and Bashir R, Nat. Protoc, 2016, 11, 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Min J, Nothing M, Coble B, Zheng H, Park J, Im H, Weber GF, Castro CM, Swirski FK, Weissleder R and Lee H, ACS Nano, 2018, 12, 3378–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erickson D, O’Dell D, Jiang L, Oncescu V, Gumus A, Lee S, Mancuso M and Mehta S, Lab Chip, 2014, 14, 3159–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shapiro NI, Trzeciak S, Hollander JE, Birkhahn R, Otero R, Osborn TM, Moretti E, Nguyen HB, Gunnerson KJ, Milzman D, Gaieski DF, Goyal M, Cairns CB, Ngo L and Rivers EP, Crit. Care Med, 2009, 37, 96–104. [DOI] [PubMed] [Google Scholar]
- 56.Buchegger P, Sauer U, Toth-Székély H and Preininger C, Sensors, 2012, 12, 1494–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauer U, Domnanich P and Preininger C, Anal. Biochem, 2011, 419, 46–52. [DOI] [PubMed] [Google Scholar]
- 58.Kemmler M, Sauer U, Schleicher E, Preininger C and Brandenburg A, Sensors Actuators, B Chem, 2014, 192, 205–215. [Google Scholar]
- 59.Schotter J, Shoshi A and Brueckl H, J. Magn. Magn. Mater, 2009, 321, 1671–1675. [Google Scholar]
- 60.Zonneveld R, Molema G and Plötz FB, Crit. Care Med, 2016, 44, 218–228. [DOI] [PubMed] [Google Scholar]
- 61.Jones CN, Moore M, Dimisko L, Alexander A, Ibrahim A, Hassell BA, Warren HS, Tompkins RG, Fagan SP and Irimia D, PLoS One, 2014, 9, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Celik IH, Demirel G, Aksoy HT, Erdeve O, Tuncer E, Biyikli Z and Dilmen U, Pediatr. Res, 2012, 71, 121–125. [DOI] [PubMed] [Google Scholar]
- 63.Cid J, Aguinaco R, Sánchez R, García-Pardo G and Llorente A, J. Infect, 2010, 60, 313–319. [DOI] [PubMed] [Google Scholar]
- 64.Gros A, Roussel M, Sauvadet E, Gacouin A, Marqué S, Chimot L, Lavoué S, Camus C, Fest T and Le Tulzo Y, Intensive Care Med, 2012, 38, 445–452. [DOI] [PubMed] [Google Scholar]
- 65.Ajmani S, Agarwal V and Gurjar M, K Glob Infect Dis, 2018, 10, 33–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhandari V, Wang C, Rinder C and Rinder H, Pediatrics, 2008, 121, 129–134. [DOI] [PubMed] [Google Scholar]
- 67.Hassan U, Zhu R and Bashir R, Lab Chip, 2018, 18, 1231–1240. [DOI] [PubMed] [Google Scholar]
- 68.van der Meer W, Pickkers Peter P, Scott CS, van der Hoeven JG and Gunnewiek JK, J. Endotoxin Res, 2007, 13, 94–100. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y, Li W, Zhou Y, Johnson A, Venable A, Hassan A, Griswold J and Pappas D, Analyst, 2018, 143, 241–249. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Zhou Y, Li W, Lyons V, Johnson A, Venable A, Griswold J and Pappas D, Anal. Chem, 2018, 12, 7204–7211. [DOI] [PubMed] [Google Scholar]
- 71.Prieto JL, Su H-W, Hou HW, Vera MP, Levy BD, Baron RM, Han J and Voldman J, Lab Chip, 2016, 16, 4333–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hassan U, Ghonge T, Reddy B, Patel M, Rappleye M, Taneja I, Tanna A, Healey R, Manusry N, Price Z, Jensen T, Berger J, Hasnain A, Flaugher E, Liu S, Davis B, Kumar J, White K and Bashir R, Nat. Commun, 2017, 8, e15949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hassan U, Reddy B, Damhorst G, Sonoiki O, Ghonge T, Yang C and Bashir R, TECHNOLOGY, 2015, 3, 201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hassan U, Watkins NN, Edwards C and Bashir R, Lab Chip, 2014, 14, 1469–1476. [DOI] [PubMed] [Google Scholar]
- 75.Wyatt Shields C Iv, Reyes CD and López GP, Lab Chip, 2015, 15, 1230–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun T and Morgan H, Microfluid. Nanofluidics, 2010, 8, 423–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nilsson J, Evander M, Hammarström B and Laurell T, Anal. Chim. Acta, 2009, 649, 141–157. [DOI] [PubMed] [Google Scholar]
- 78.Zelenin S, Hansson J, Ardabili S, Ramachandraiah H, Brismar H and Russom A, Biotechnol. Lett, 2015, 37, 825–830. [DOI] [PubMed] [Google Scholar]
- 79.Mahalanabis M, Al-Muayad H, Kulinski MD, Altman D and Klapperich CM, Lab Chip, 2009, 9, 2811–2817. [DOI] [PubMed] [Google Scholar]
- 80.Knob R, Hanson RL, Tateoka OB, Wood RL, Guerrero-Arguero I, Robison RA, Pitt WG and Woolley AT, J. Chromatogr. A, 2018, 1562, 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yung CW, Fiering J, Mueller AJ and Ingber DE, Lab Chip, 2009, 9, 1171–1177. [DOI] [PubMed] [Google Scholar]
- 82.Kang JH, Super M, Yung CW, Cooper RM, Domansky K, Graveline AR, Mammoto T, Berthet JB, Tobin H, Cartwright MJ, Watters AL, Rottman M, Waterhouse A, Mammoto A, Gamini N, Rodas MJ, Kole A, Jiang A, Valentin TM, Diaz A, Takahashi K and Ingber DE, Nat. Med, 2014, 20, 1211–1216. [DOI] [PubMed] [Google Scholar]
- 83.Cooper RM, Leslie DC, Domansky K, Jain A, Yung C, Cho M, Workman S, Super M and Ingber DE, Lab Chip, 2014, 14, 182–188. [DOI] [PubMed] [Google Scholar]
- 84.Lee JJ, Jeong KJ, Hashimoto M, Kwon AH, Rwei A, Shankarappa SA, Tsui JH and Kohane DS, Nano Lett, 2014, 14, 1–5. [DOI] [PubMed] [Google Scholar]
- 85.Lopes ALK, Cardoso J, dos Santos FRCC, Silva ACG, Stets MI, Zanchin NIT, Soares MJ and Krieger MA, J. Microbiol. Methods, 2016, 128, 96–101. [DOI] [PubMed] [Google Scholar]
- 86.Aran K, Morales M, a Sasso L, Lo J, Zheng M, Johnston I, Kamdar N, Ündar A and Zahn JD, 15th Int. Conf. Miniaturized Syst. Chem. Life Sci, 2011, 1, 497–499. [Google Scholar]
- 87.Hou HW, Wu L, Amador-Munoz DP, Vera MP, Coronata A, Englert JA, Levy BD, Baron RM and Han J, Lab Chip, 2016, 16, 688–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hou HW, Gan HY, Bhagat AA, Li LD, Lim CT and Han J, Biomicrofluidics, 2012, 6, 024115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ohlsson P, Evander M, Petersson K, Mellhammar L, Lehmusvuori A, Karhunen U, Soikkeli M, Seppä T, Tuunainen E, Spangar A, Von Lode P, Rantakokko-Jalava K, Otto G, Scheding S, Soukka T, Wittfooth S and Laurell T, Anal. Chem, 2016, 88, 9403–9411. [DOI] [PubMed] [Google Scholar]
- 90.Ohlsson P, Petersson K, Augustsson P and Laurell T, Sci. Rep, 2018, 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Faridi MA, Ramachandraiah H, Banerjee I, Ardabili S, Zelenin S and Russom A, J. Nanobiotechnology, 2017, 15, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chun K, Syndergaard C, Damas C, Trubey R, Mukindaraj A, Qian S, Jin X, Breslow S and Niemz A, J. Lab. Autom, 2015, 20, 539–561. [DOI] [PubMed] [Google Scholar]
- 93.Afshari A, Schrenzel J, Ieven M and Harbarth S, Crit. Care, 2012, 16, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lebovitz EE and Burbelo PD, Mol. Diagnosis Ther, 2013, 17, 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ulloa L and Tracey KJ, Trends Mol. Med, 2005, 11, 56–63. [DOI] [PubMed] [Google Scholar]
- 96.Bozza FA, Salluh JI, Japiassu AM, Soares M, Assis EF, Gomes RN, Bozza MT, Castro-Faria-Neto HC and Bozza PT, Crit. Care, 2007, 11, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mera S, Tatulescu D, Cismaru C, Bondor C, Slavcovici A, Zanc V, Carstina D and Oltean M, Apmis, 2011, 119, 155–163. [DOI] [PubMed] [Google Scholar]
- 98.Kibe S, Adams K and Barlow G, J. Antimicrob. Chemother, 2011, 66, 34–40. [DOI] [PubMed] [Google Scholar]
- 99.Gabrielczyk T, Eur. Biotechnol, 2016, 15–22. [Google Scholar]
- 100.Reddy B, Hassan U, Seymour C, Angus DC, Isbell TS, White K, Weir W, Yeh L, Vincent A and Bashir R, Nat. Biomed. Eng, 2018, 2, 640–648. [DOI] [PubMed] [Google Scholar]
- 101.Harbarth S, Holeckova K, Pittet D, Ricou B, Grau GE and Vadas L, Am. J. Respir. Crit. Care Med, 2001, 164, 396–402. [DOI] [PubMed] [Google Scholar]
- 102.Kolaczkowska E and Kubes P, Nat. Rev. Immunol, 2013, 13, 159–175. [DOI] [PubMed] [Google Scholar]
- 103.Sweeney TE, Azad TD, Donato M, Haynes WA, Perumal TM, Henao R, Bermejo-Martin JF, Almansa R, Tamayo E, Howrylak JA, Choi A, Parnell GP, Tang B, Nichols M, Woods CW, Ginsburg GS, Kingsmore SF, Omberg L, Mangravite LM, Wong HR, Tsalik EL, Langley RJ and Khatri P, Crit. Care Med, 2018, 46, 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wong HR, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Willson DF, Freishtat RJ, Anas N, Meyer K, Checchia PA, Monaco M, Odom K and Shanley TP, BMC Med, 2009, 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wong HR, Cvijanovich NZ, Allen GL, Thomas NJ, Robert J, Anas N, Meyer K, Checchia PA, Lin R, Shanley TP, Bigham MT, Wheeler DS, Doughty LA, Tegtmeyer K, Poynter SE, Kaplan JM, Chima RS, Stalets E, Basu RK, Varisco BM and Barr FE, 2012, 39, 2511–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wong HR, Cvijanovich NZ, Anas N, Allen GL, Thomas NJ, Bigham MT, Weiss SL, Fitzgerald J, Checchia PA, Meyer K, Shanley TP, Quasney M, Hall M, Gedeit R, Freishtat RJ, Nowak J, Shekhar RS, Gertz S, Dawson E, Howard K, Harmon K, Beckman E, Frank E and Lindsell CJ, Am. J. Respir. Crit. Care Med, 2015, 191, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Davenport EE, Burnham KL, Radhakrishnan J, Humburg P, Hutton P, Mills TC, Rautanen A, Gordon AC, Garrard C, Hill AVS, Hinds CJ and Knight JC, Lancet Respir. Med, 2016, 4, 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Burnham KL, Davenport EE, Radhakrishnan J, Humburg P, Gordon AC, Hutton P, Svoren-Jabalera E, Garrard C, Hill AVS, Hinds CJ and Knight JC, Am. J. Respir. Crit. Care Med, 2017, 196, 328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prescott HC, Calfee CS, Taylor Thompson B, Angus DC and Liu VX, Am. J. Respir. Crit. Care Med, 2016, 194, 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sinto R, Chandra AT, Lie KC and Suwarto S, IOP Conf. Ser. Earth Environ. Sci, 2018, 125, 12022. [Google Scholar]