Abstract
Macrophages are a predominant immune cell population that drive inflammatory responses and exhibit transitions in phenotype and function during tissue remodeling in disease and repair. Thus, engineering an immunomodulatory biomaterial has significant implications for resolving inflammation. In this work, we engineered a biomimetic and photoresponsive hyaluronan (HA) hydrogel nanocomposite with tunable 3-D ECM adhesion sites for dynamic macrophage immunomodulation. We exploited photo-degradative APP (alkoxylphenacyl-based polycarbonate) nanocomposites to permit user-controlled RGD (Arg-Gly-Asp) adhesive peptide release and conjugation to a HA-based extracellular matrix (ECM) for real-time integrin activation of macrophages encapsulated in 3-D HA-APP nanocomposite hydrogels. We demonstrate that photo-controlled 3-D ECM-RGD peptide conjugation can activate αvβ3 integrin of macrophages, and periodic αvβ3 integrin activation can enhance anti-inflammatory M2 macrophage polarization. Altogether, we highlight an emerging use of biomimetic, photo-responsive, and bioactive HA-APP nanocomposite hydrogel to command 3-D cell-ECM interactions for modulating macrophage polarization, which may shed light on cell-ECM interactions in innate immunity and inspire new biomaterials-based immunomodulatory therapies.
Keywords: photoresponsive biomaterials, hydrogel nanocomposites, hyaluronan, macrophages, immunomodulation
The table of contents entry
Photoresponsive HA hydrogel nanocomposite with tunable 3-D ECM adhesion sites for dynamic macrophage immunomodulation.
Graphical Abstract:
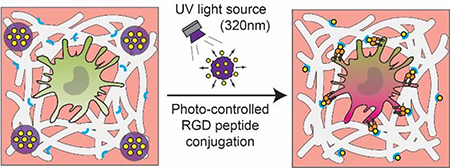
Introduction
Inflammation, predominantly facilitated by macrophages, coincides with dynamic extracellular matrix (ECM) remodeling to influence tissue plasticity in disease and repair.[1] Determining how the microenvironment triggers a phenotypic switch to either a ‘pro-inflammatory (M1)’ or an ‘anti-inflammatory (M2)’ macrophage phenotype is critical to expanding tissue destruction[2–4] or orchestrating repair.[5–7] Thus, there is a need for robust immunomodulatory strategies that can control macrophage polarization for tuning inflammatory responses that can promote tissue repair.
Although many soluble cytokine factors have been reported to be potential immunomodulation targets that dictate macrophage crosstalk,[8] the cell-ECM interactions have an underappreciated regulatory role in macrophage inflammation. Thus, it is necessary to fully explore how macrophages interact with dynamic ECM remodeling to modulate endogenous tissue repair progress. ECM adhesion receptors, specifically integrins, may dominate macrophage behaviors in tissue repair because they are an important link between ECM and intracellular cytoskeleton signaling.[9] Although there are studies that investigate the association of specific integrins to macrophage polarization, these studies lack the environmental control over dynamic integrin activation. It has been shown that the timing and extent of cell-ECM adhesion may be critical to stage-dependent cell functions.[10, 11] As a result of immune cell behaviors and functions being temporally defined,[12] a biomimetic and dynamic ECM composite is essential to studying how cell-ECM interactions modulate distinct macrophage phenotypes and developing a potent solution to modulate inflammation in a temporal manner.
In soft tissue-based organs like the brain,[13] 3-D hydrogels offer a simple, biomimetic approach to reconstruct in vivo ECM signatures. Hyaluronan (HA) is one example of a native ECM composite found in human tissue,[14] and its accumulation and turnover has been implicated to modulate inflammatory responses of macrophages in physiological and pathological conditions.[15] HA’s structural and biological properties are important to mediate cell signaling during stages of development,[16] disease,[17] or tissue repair.[18] The increase of HA deposition into sites of tissue injury and during inflammatory diseases inspired incorporating HA as a building block ECM protein in 3-D hydrogels for immunomodulation in regenerative medicine.[14] Current approaches have included patterning ligand-specific peptides such as growth factors and integrin-based ECM adhesion motifs onto 3-D scaffolds,[19] yet most are static and cannot be used as dynamic immunomodulation approaches. In recent years, ‘stimuli-responsive’ materials have attracted much attention for their use as specific and predictive drug delivery systems in response to external and internal stimuli, including physical (e.g. temperature, light, and electrical and magnetic fields), mechanical (e.g. ultrasound), and biochemical (pH, reactive oxygen species, proteases) stimuli.[20] On account of their non-invasiveness and remote temporal control, photoresponsive systems have been developed[21–23] to achieve on-demand therapeutic dosing in response to illumination at distinct wavelengths (ultraviolet (UV), visible, or near-infrared regions). However, there has been a lack of dynamic and immunomodulatory ECM-based hydrogels to control the kinetics of macrophage polarization in a biomimetic 3-D HA hydrogel scaffold. Hence, novel stimuli-responsive materials should be developed to produce a bioactive and dynamic ECM composite that can control macrophage phenotype upon on-demand stimulation.
In this work, we have engineered a biomimetic and photoresponsive HA hydrogel nanocomposite that is capable of modulating macrophage inflammatory phenotypes over time. We demonstrate that photo-controlled 3-D ECM-RGD peptide conjugation can activate αvβ3 integrin of macrophages, and periodic αvβ3 integrin activation can enhance anti-inflammatory M2 macrophage polarization. The photoresponsive HA hydrogel nanocomposite hydrogel can be applied as an in vitro 3-D culture model to probe and regulate cell-ECM interactions for real-time macrophage immunomodulation in a biomimetic ECM. Hence, this dynamic, immunomodulatory hydrogel platform can be tailored to incorporate tissue-specific cells to study diseases associated with specific organs, providing a pathway to improve current biomaterials design in regenerative hydrogel therapies.
2. Results and Discussion
2.1. Photoresponsive HA hydrogel Nanocomposite for Dynamic Macrophage Immunomodulation
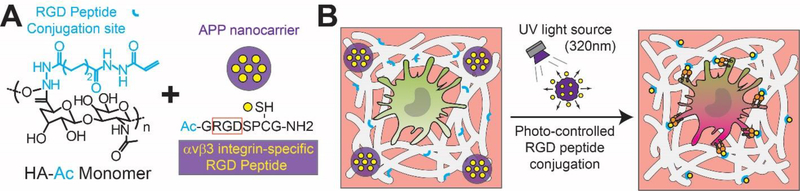
To achieve temporal control over cell-ECM interactions in a 3-D hydrogel, we synthesized a photoresponsive HA hydrogel nanocomposite (Figure 1) to modulate macrophage inflammatory responses through integrin activation. Although there are many environmental cues that drive macrophage polarization, this study dissects how photo-controlled RGD peptide release and conjugation affects macrophage phenotype and function over time. RGD (Ac-GRGDSPCG-NH2) is a tripeptide that can mimic cell adhesion ligands and bind to cell-surface receptors that promote cell survival and spreading, creating a hospitable microenvironment for cells within a 3-D crosslinked ECM polymeric network. HA (50 kDa) was modified with acrylate groups to generate an acrylated HA macromer (HA-Ac) with available binding sites for RGD peptides Ac-GRGDSPCG-NH2 specific for αvβ3 integrin. HA-Ac macromer chains were crosslinked by a cysteine-contained biodegradable MMP (matrix metalloproteinase) crosslinker peptide (GCRDVPMSMRGGDRCG) for encapsulating RAW 264.7 murine macrophages[24] in a 3-D biomimetic ECM. Previous studies have demonstrated MMP is universally expressed among all cells,[25] and hydrogels with protease-degradable crosslinkers can be rapidly cleaved by MMP-1 and MMP-2 proteases for supporting cell spreading and survival.[26] Furthermore, RGD peptide conjugation to HA chains can be achieved through a Michael addition reaction between Acrylate groups on HA and thiol groups (via cysteine residues) on RGD peptides.[27] TNBSA (trinitrobenzene sulfonic acid) assay results show that 36.7% of carboxylic acid groups were coupled with ADH (adipic dihydrazide). After reacting HA-ADH with NHS-AC at pH 7.4, the TNBSA assay did not detect free primary amines in HA-AC, yielding approximately 60 acrylates per HA chain which provides sufficient acrylation for both HA crosslinking and RGD peptide conjugation.[27] To determine if the mechanical properties of the bulk material were altered during photo-stimulation, we measured the storage (G’) and loss modulus (G”) of the photoresponsive nanocomposite hydrogel with a plate-to-plate rheometer. Results show that the G’ and G” did not statistically differ after photoresponsive nanocomposite hydrogels were stimulated with 30s UV exposure, confirming that photo-controlled RGD peptide release and conjugation is the predominant physical factor that is regulating the 3-D in vitro culture conditions (Supporting Information, Figure S1).
Figure 1.
Photoresponsive HA hydrogel nanocomposite with photo-controlled 3-D ECM adhesion sites for dynamic macrophage immunomodulation. Schematics of (A) photoresponsive HA hydrogel nanocomposites generated with HA-Ac macromers and alkoxylphenacyl-based polycarbonate (APP) nanocarrier (purple) loaded with RGD peptide (yellow), and (B), upon defined UV exposure, RGD peptides will covalently conjugate (blue “Y”) onto crosslinked HA hydrogels (white) to temporally activate αvβ3 integrin macrophage expressions for macrophage polarization in a dynamic 3-D HA ECM composite.
With the ability to pulse UV light exposure at defined times, we can achieve temporal control over 3-D cell-ECM interactions and macrophage immunomodulation with αvβ3 integrin ligand presentation. To control the kinetics of RGD peptide conjugation on HA, we developed photo-degradable alkoxylphenacyl-based polycarbonate (APP) nanocomposites to load RGD peptides and perform controlled release upon UV light with a wavelength of 302 nm. Potential toxicities associated with chromophoric or polymeric units of most photoresponsive systems have influenced their compatibility for clinical use. However, APPs are a new class of photoresponsive polymers that has been reported to possess high thermal stability, biocompatibility and biodegradability in vivo.[23, 28] Recognizing that long-term UV exposure (minutes) as an external trigger has limitations for in vivo applications due to substantial tissue penetration and detrimental photochemical reactions,[29] we shortened the extent of UV exposure (λ= 302 nm) time (seconds) of APP nancomposites in vitro to reduce cell photodamage. We validated the cytotoxic effects of UV light exposure by staining for RAW264.7 macrophage viability (Supporting Information, Figure S2) and confirmed that UV light exposure with a duration of up to 30s had no significant influence over macrophage cell viability (> 95%). A similar study investigating the potential cytotoxicity suspected with UV light degradation of APP nanocomposites also found no significant decrease in cell viability and, after 72 hrs, observed cell viabilities higher than 90% for cell culture.[28] Furthermore, we validated that the effects of the 30-second UV exposure alone had no significant effects on the macrophage phenotype and cytokine secretions. By immunostaining and quantifying the ratio of Arginase-1 (M2 macrophage marker) and iNOS (M1 macrophage marker) expressions, it was determined that pre-polarized M0, M1, and M2 macrophages exhibited no statistically significant changes to their respective markers upon 30-second UV exposure in 3-D photoresponsive nanocomposite hydrogel (Supporting Information, Figure S3). To validate that UV exposure alone had no effect on macrophage function, we screened the pro-inflammatory (TNF-α and IL-6) and anti-inflammatory cytokines (IL-10 and TGF-β) secreted by M0 macrophages in 3-D photoresponsive nanocomposite hydrogels after 30-second UV exposure, and found no statistically significant alterations to levels (Supporting Information, Figure S4). Altogether, we engineered a photoresponsive, dynamic, and immunomodulatory 3-D HA hydrogel nanocomposite for real-time and temporal control of cell-ECM interactions and macrophage polarization based on on-demand photo-stimulation.
A major advantage of a photoresponsive HA hydrogel nanocomposite is its stable controllability over user-activated RGD peptide release and conjugation, as compared to other internal stimuli systems (e.g. pH, enzyme) that may fluctuate widely at different times or disease stages and, thus, result in poor reproducibility and therapeutic efficacy. Therefore, photoresponsive hydrogel systems that apply external triggers such as light permit users to model different tissue microenvironments because photoresponsive nanocomposites are not dependent on differences in biological signatures between diseased and healthy tissue.
2.2. Photo-controlled kinetics of RGD peptide conjugation in 3-D HA-APP Hydrogel
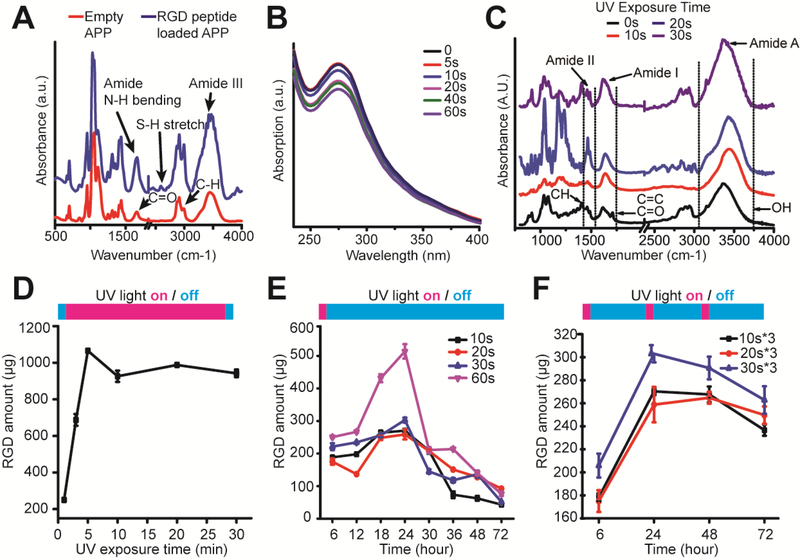
To confirm the functionality of APP nanocomposites as depots for photo-controlled RGD peptide release and conjugation, we first measured the IR spectrum of APP nanocomposites before and after RGD peptide loading (Figures 2A&C&D). IR spectras demonstrated an amide I band (1600–1700 cm−1) and S-H stretching vibrations at ~2600 cm−1, confirming a successful encapsulation of RGD peptides into APP nanocomposites. We then assessed the photosensitivity of APP nancomposites by conducting a UV spectrum analysis (Figure 2B). It was found that APP nanocomposites absorb UV light with a λmax at 280 nm. For increasing UV exposure time, there was a decrease of absorbance at λmax, indicating photodegradation of APP nanocomposites upon increasing UV light exposure. The photodegradation was further confirmed by the decreasing size of APP nanocomposites after UV exposure (Supporting Information, Figure S5). Embedded into 3-D HA hydrogels, APP nanocomposites were photodegraded, and an IR spectrum analysis was conducted to confirm HA structural changes during a two-step HA hydrogel synthesis (Supporting Information, Figure S6) and photo-controlled RGD peptide conjugation (Figure 2C). The evolution of peaks at 1640 cm−1 and 1720 cm−1 prove a successful macromer functionalization of adipic dihydrazide (HA-ADH) and acrylate (HA-AC) onto HA monomer chains. The temporally-controlled distribution and binding of RGD peptides to HA-Ac with increasing UV exposure time was confirmed by an increasing intensity at amide I region and consumption of acrylate groups.
Figure 2.
Characterization of photoresponsive RGD peptide conjugation in 3-D HA-APP hydrogel. (A) IR spectrum of alkoxylphenacyl-based polycarbonate (APP) nanocarrier before and after RGD peptide loading. (B) UV spectrum showing change in absorbance with increasing photodegradation time. (C) IR spectrum of photo-controlled RGD peptide conjugation onto HA-Ac Macromers with increasing, one-time UV exposure. (D-F) Kinetics of RGD peptide conjugation to HA-Ac macromers based on RGD peptide release profiles for (D) constant UV exposure, (E) one-time UV exposure, (F) and periodic UV exposure times. Error bars represent ± S.E.M. from 3 independent experiments. P-values were calculated using the Student’s t-test. *, P < 0.05.
We further assessed the kinetics of RGD peptide conjugation by characterizing the APP nanocomposite release profiles during controlled UV exposure times (Figures 2D-F). UV exposure times were first screened to determine the total RGD peptide release amount from APP nancomposites and optimal UV exposure times (Figure 2D). It was found that during the first 5 min. of UV exposure, RGD peptide release was elevated with increasing UV exposure time, and then levels plateaued, which indicates RGD peptide release is saturated after 10 min. We next evaluated the RGD peptide release profiles for up to 72 hrs by stimulating APP nanocomposites at different UV irradiation times at one time and then monitored changes in release amounts every 6 hrs (Figure 2E). For the first 12 hrs, it was shown that the irradiation time influences the released peptide amount, especially when increasing the irradiation time from 10 s to 60 s. Then, all released peptide amounts peaked at 24 hrs and decayed for the remaining 72 hrs. Most importantly, when selecting the appropriate UV irradiation times for controlled RGD peptide release, compared to other testing times, 60 s rapidly changes the release rate of RGD peptides within HA hydrogels at 24 hrs after a one-time UV stimulation. Thus, a single 60-second UV exposure does not fit the criteria of controllable RGD peptide release. This is an important consideration since UV light is a potent external trigger due to its low wavelength, resulting in photons of higher energy for APP photodegradation, compared to other light stimuli at higher wavelengths. To maintain a controlled release after a one-time stimulation, it can be seen that a UV light exposure duration from 10 s to 30 s can release RGD peptides without drastic release rate changes.
To show temporal control over RGD peptide conjugation while minimizing photo-damage, we repeatedly exposed HA hydrogel nanocomposites to 10 s, 20 s and 30 s UV exposure every 24 hrs (Figure 2F). Compared to a one-time exposure, periodic exposure indicates a lesser degree of decay during the remaining 72 hrs, resulting in APP nanocomposites that can maintain a constant release of RGD peptides during multiple, short rounds of UV stimulation. Similar to a one-time exposure, APP nanocomposites demonstrate controlled RGD peptide release during periodic exposure to UV stimulus with approximately equal amounts of RGD peptide added to 3-D HA hydrogels each day. This proves that the HA hydrogel nanocomposite not only has user-defined control over photodegradation time but also has defined dosage rates. Unlike other photoresponsive hydrogels that have successfully achieved temporal control with photocleavable linkers or photocage moieties, an important contribution of this study is proving high controllability over the timing and dosage of RGD peptide release and conjugation, allowing users to modulate cell-ECM interactions for studying immunity in a biomimetic and dynamic 3-D ECM composite.
2.3. Photo-triggered immunomodulatory hydrogels upregulate αvβ3 integrin expression and control macrophage-ECM interactions
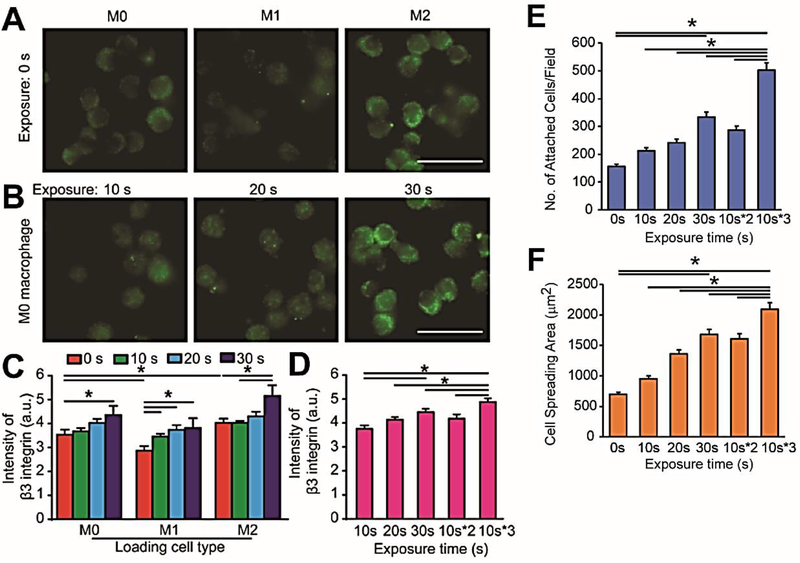
To examine the requirements of integrins for macrophage polarization, we generated M1 and M2 phenotypes with RAW 264.7 macrophages using lipopolysaccharide (LPS) for M1 phenotype induction and IL-10 for M2 phenotype induction (details in Experimental Section) with well-established protocols.[30] We first examined surface αvβ3 integrin expressions of inactivated M0 macrophages and pre-polarized M1 and M2 macrophages embedded in 3-D HA hydrogels. In HA hydrogels not conjugated with RGD, it was shown that M2 macrophages exhibited a higher fluorescent intensity of β3 integrin expression, while M1 macrophages showed a weak fluorescent intensity of β3 integrin expression, compared to M0 macrophages (Figure 3A). Next, inactivated M0 macrophages were suspended in 3-D HA-APP hydrogels and αvβ3 integrin surface expressions were quantified by immunostaining after temporal activation with increasing one-time UV exposure time from 0 s to 30 s (Figures 3B&C) or periodic exposure (Figure 3D). For one-time UV exposure, M0, M1 and M2 macrophages are steadily showing increasing αvβ3 integrin expression with increasing exposure time from 0s to 30s. However, αvβ3 integrin expression of M0 macrophages with periodic UV exposure was statistically greater than αvβ3 integrin levels of M0 macrophages stimulated with one-time UV exposure. To further prove the control over the adhesive behaviors of macrophages, we examined the cell attachment and spreading of M0 macrophages on a HA-APP hydrogel bed after 30 s of one-time UV exposure (Figure 3E&F). With increasing UV exposure time from 0 s to 30 s, macrophage attachment and spreading increased significantly, supporting that photo-controlled RGD peptide conjugation can improve macrophage ECM adhesion.
Figure 3.
Photo-triggered HA-APP hydrogels control macrophage-ECM adhesive interactions and αvβ3 integrin expression. (A) Immunofluorescent images of αvβ3 integrin expression in pre-polarized M0, M1 and M2 macrophages in unmodified 3-D HA hydrogels. Scale bar is 30 μm. (B) Immunofluorescent images of αvβ3 integrin expressions of M0 macrophages in 3-D HA-APP hydrogels with increasing one-time UV exposure. Scale bar is 30 μm. (C) Quantified αvβ3 integrin expression of inactivated M0 and pre-polarized M1 and M2 macrophages in 3-D HA-APP hydrogels with increasing one-time UV exposure. (D) Quantified αvβ3 integrin expression of M0 macrophages in 3-D HA-APP hydrogels with one-time and periodic UV exposure. (E) Quantified cell attachment of M0 macrophages on 2-D HA-APP hydrogel substrates in response to increasing one-time and periodic UV exposure. (F) Quantified cell spreading area of M0 macrophages on 2-D HA-APP hydrogel substrates in response to increasing one-time and periodic UV exposure. Error bars represent ± S.E.M. from 3 independent experiments. P-values were calculated using the Student’s t-test. *, P < 0.05.
2.4. Sustained photo-controlled αvβ3 integrin expression promotes an anti-inflammatory M2 macrophage phenotype.
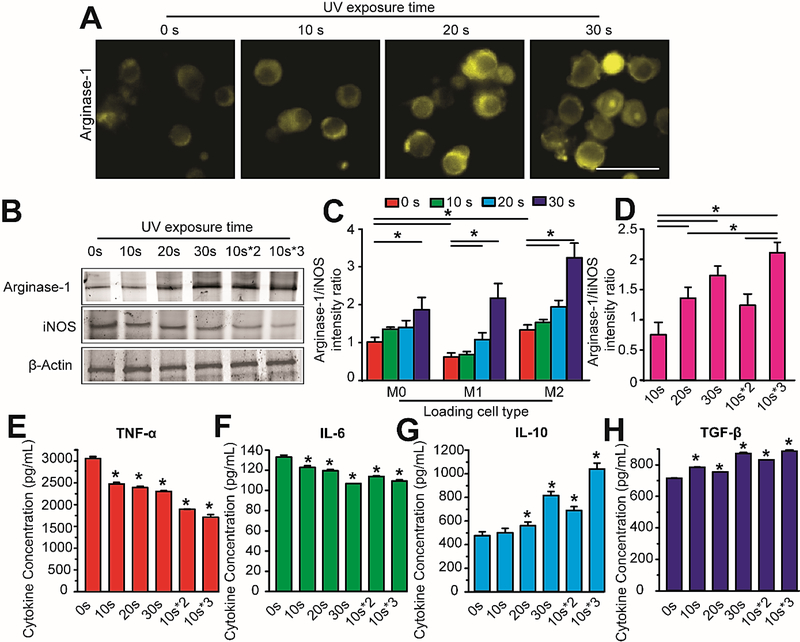
Next, we sought to determine whether photo-controlled αvβ3 integrin activation in our 3-D HA-APP hydrogel can regulate macrophage polarization to achieve immunomodulation. To this end, we classified different macrophage phenotypes by immunostaining expressions of well-established markers for iNOS (M1-specific marker) and Arginase-1 (M2-specific marker) on RAW 264.7 macrophages.[31] It was determined that M2 macrophages demonstrated elevated Arginase-1 and weak iNOS expressions, while M1 macrophages showed elevated iNOS and weak Arginase-1 levels (Figure 4). To determine if altering RGD peptide concentration in unmodified 3-D HA hydrogels induced different macrophage phenotypes, we cultured inactivated M0 and pre-polarized M1 and M2 macrophages in 1:2, 1:6, and 1:12 ratios of 3-D HA composites pre-conjugated with RGD peptides (Supporting Information, Figure S7). It was found that increasing αvβ3 integrin activation correlated to higher Arginase-1 levels compared to iNOS levels for M2 macrophages, while M1 maintained their iNOS and Arginase-1 expressions. To further investigate whether the material of the APP nanocomposite could influence macrophage phenotype, we investigated macrophage polarization with empty APP nanocomposites (Supporting Information, Figure S8). It was determined with immunostaining that empty APP nanocomposites degraded with one-time UV exposure had no significant influence over M1 or M2 macrophage polarization.
Figure 4.
Temporal αvβ3 integrin activation strengthens M2 macrophage polarization in 3-D photoresponsive HA-APP hydrogels. (A) Representative immunostaining images of increased Arginase-1 (M2 macrophage-specific marker) of M0 macrophages in 3-D HA-APP hydrogels treated with increasing one-time UV exposure. Scale bar is 30 μm. (B) Western blot analysis of Arginase-1 and iNOS expressions of M0 macrophages in 3-D HA-APP hydrogels with increasing one-time UV exposure. (C) Quantified fluorescent intensity ratios of Arginase-1 and iNOS expressions for inactivated M0 macrophages and pre-polarized M1 and M2 macrophages embedded in 3-D HA-APP hydrogels treated with increasing one-time UV exposure. (D) Quantified fluorescent intensity ratios of Arginase-1 and iNOS expressions for M0 macrophages in 3-D HA-APP hydrogels treated with one-time or periodic UV exposure. (E-H) Immunoassay levels of proinflammatory cytokines, TNF-a (E) and IL-6 (F), and anti-inflammatory cytokines IL-10 (G) and TGF-β (H) secreted by M0 macrophages in 3-D HA-APP hydrogels treated with increasing one-time and periodic UV exposure. Error bars represent ± S.E.M. from 3 independent experiments. P-values were calculated compared to 0s and using the Student’s t-test. *, P < 0.05.
To examine whether temporal αvβ3 integrin activation can strengthen anti-inflammatory M2 macrophage polarization, we investigated macrophages cultured in photoresponsive 3-D HA-APP hydrogels with one-time UV exposure for 10 s, 20 s and 30 s (Figures 4A-C) or periodic exposure with 10 s (Figure 4D). In terms of increasing UV-controlled αvβ3 integrin activation, it was found that 30 s exhibited elevated Arginase-1 levels relative to decreasing UV exposure (Figure 4A). This finding was confirmed by western blotting to demonstrate that increasing or periodic UV-controlled αvβ3 integrin activation promoted Arginase-1 expressions and inhibited iNOS expressions (Figure 4B). Temporal integrin activation in 3-D HA-APP hydrogels enhanced M2 macrophage polarization based on elevated Arginase-1 expressions with 30 s of either one-time or periodic UV exposure over 72 hrs (Figures 4C&D). Yet, it was shown that tuning αvβ3 integrin expression every 10 s at each 24 hr interval for a 72 hr culture period promoted stronger Arginase-1 levels than 30 s treatment at one time (Figure 4D).
To further prove the macrophage polarization in dynamic 3-D HA-APP hydrogels, we examined cytokine levels associated with M1 and M2 macrophages. Profiles of M1, pro-inflammatory cytokines TNF-α and IL-6, and M2, anti-inflammatory cytokines, TGF-β1 and IL-10, were examined (Figures 4E-H). After 72 hrs of photo-controlled integrin activation, pro-inflammatory cytokines TNF-α and IL-6 were observed to have decreasing expressions, while the concentrations of anti-inflammatory cytokines TGF-β1 and IL-10 increased significantly. However, compared to a one-time UV exposure, a periodic 10-second UV activation promoted the weakest pro-inflammatory and strongest anti-inflammatory cytokine expressions. The increasing concentrations of TGF-β1 and IL-10 cytokine confirmed the transition of M0 to the M2 macrophage phenotype and indicated that the 3-D HA-APP hydrogel with temporal αvβ3 integrin activation can provide an efficient approach to probe the macrophage polarizations, compared to a transient, one-time exposure. Our previous study demonstrated that activating αvβ3 integrin in a 3-D brain tumor microenvironment facilitated tumor-associated macrophages to promote brain tumor angiogenesis,[30] which required both ECM and soluble cytokine factors to sustain angiogenesis. Here, we dissected how integrin-mediated macrophage-ECM interactions can modulate macrophage phenotype using an in vitro stimuli-responsive hydrogel system. This points toward a direction of designing novel ECM-based and stimuli-responsive hydrogel nanocomposites as part of an immunomodulatory approach to regulate the appropriate immune responses for efficient tissue repair.
3. Conclusion
Tissue development and repair involve dynamic, overlapping phases of macrophage-associated inflammation and ECM remodeling. Understanding the phenotypic switch of macrophages in response to biomimetic and dynamic ECM composites will provide a path towards engineering nuanced biomaterials-based immunomodulation and probing inflammation-driven tissue regeneration in damaged or diseased tissue. Here, we developed a photoresponsive 3-D HA nanocomposite hydrogel that provides users control over the activation of integrin-mediated ECM adhesion sites for macrophage immunomodulation over time. By leveraging photo-responsive nanocomposites, we provide an on-demand and dynamic 3-D hydrogel system that defines the dosage and kinetics of RGD adhesive peptide conjugation to ECM, surpassing the limitations of previous 3-D biomimetic ECM composites. In contrast to static ligand expression, our results highlighted that periodic αvβ3 integrin activation for 72 hrs is critical to sustaining an anti-inflammatory M2 phenotype. The implications of this temporal control over macrophage immunomodulation represents a novel strategy to control inflammatory responses that may accelerate endogenous tissue repair responses.
4. Experimental Section
Materials.
Ac-GRGDSPCG-NH2 (RGD) and GCRDVPMSMRGGDRCG (MMP) peptides are purchased from Genscript (Purity > 90%). Hyaluronic acid and all other chemicals are ordered from Sigma-Aldrich.
Cell Culture and reagents.
Mouse macrophage cells (RAW264.7, ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Sigma-Aldrich) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco), and 1% penicillin/streptomycin (Gibco). All cell lines were grown no more than 20 passages in a 37 °C (5% CO2) incubator. In addition, Propidium Iodide (1 μM) and Calcein AM (2 μM) (Sigma-Aldrich) were used to examine the cell viability (Supporting Information, Figure S1) 2 hrs after UV exposure in HA hydrogel nanocomposites according to manufacturer’s protocols.
Synthesis of acrylated hyaluronic acid (HA-AC).
HA undergoes a two-step functionalization prior to the conjugation with RGD peptides.[27] Hyaluronic acid (100 mg, 0.0015 mmol, 50 kDa) is reacted with ADH (2.6 g, 8.4 mmol) in the presence of 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) (0.3 g, 0.92 mmole) at pH 4.75 overnight, and dialyzed (MWCO 6–8 kDa) in DI water for 2 days. The obtained HA-ADH is lyophilized. 36.7% of the carboxyl groups were modified with ADH based on the TNBSA (Pierce, Rockford, Illinois) assay. HA-ADH (100 mg, 0.0014 mmol) is reacted with N-Acryloxysuccinimide (NHS-AC) (108 mg, 0.47 mmol) in HEPES buffer overnight and purified through dialysis in DI water for 2 days before lyophilization.
Synthesis of alkoxylphenacyl-based polycarbonates (APP) homopolymer.
The synthesis of APP has been reported previously.[32] First, hydroxyacetophenone (10 g) is chain extended with a hydroxy terminated spacer. The product (15 g) is converted to the bromide without purification and then reacts with sodium acetate (15 g) to yield the acetate protected derivative in high yield. Following deprotection, the alkoxylphenacyl diol (0.76 g) is added to a double neck flask. The flask is vacuumed and refilled with N2 gas three times. Chloroform (7 mL) and pyridine (1.8 mL) are added. Then the solution is stirred and cooled in an ice bath. Triphosgene (0.41 g, 1.38 mmol) is dissolved in chloroform (5 mL) in a small vial, and the triphosgene solution is added dropwise to the flask. The reaction mixture is allowed to proceed for 2 hours, and the reaction temperature warms up to room temperature. Then the polymer is precipitated in cold isopropanol (200 mL). The precipitants are centrifuged and dried in vacuum oven at room temperature, and finally obtained as a white solid.
Synthesis of RGD peptide nanocomposites.
RGD peptide-loaded APP nanocomposites (RGD-APP) are prepared by nanoprecipitation. Briefly, APP homopolymer is dissolved in DMSO (2.5–40 mg/mL). Once the polymer is completely solubilized, RGD peptide solution (0–1 mg/mL) is added and mixed for 5 min at 350 rpm. This feed solution is then added dropwise into 1 mL of Pluronic solution (3% w/v) at 37 °C under vigorous stirring. The resulting RGD-APP nanocomposite is dialyzed (6–8 kDa MWCO) in DI water for 2 hrs, to remove residual DMSO from final product.
Synthesis of Hyaluronan Hydrogel Nanocomposite.
HA-AC (20 mg/mL, 50 μL) and RGD-APP (2.5 mg/mL, 50 μL) are firstly mixed and then crosslinked by the addition of MMP crosslinker (10 mg/mL, 40 μL) onto HA-AC. The mixture is injected into a PDMS microwell and the gelation reaction is carried out at pH 7.4 and 37 °C for 30 min. To encapsulate cells in 3-D HA hydrogel nanocomposites, the aforementioned mixture is mixed with 60 μL RAW264.7 macrophages (1×107 cells/mL) prior to gelation. Cell culture media is added and replaced every 24 hrs to avoid hydrogel dehydration and gels are incubated at 37 °C (5% CO2) for 72 hr culture.
PDMS Microwell Fabrication.
Poly-dimethylsiloxane (PDMS) was used to frame 5mm wells that support 3-D hyaluronan hydrogel culture. Briefly, a mixture of PDMS base and curing agent (0008952166, Sylgard 184 Silicone Elastomer, Dow Corning) with a volume-to-volume (v/v) ratio of 10:1 was prepared using a clean glass rod in a laminar flow hood to avoid dust contamination. Followed by vacuum desiccation, PDMS solution was poured into a clean, empty 100 × 15 mm petri dish (BD Falcon, 08–757-100D) to produce a thick layer (~5 mm) by curing PDMS in an 80 °C oven for 2 hrs. Once PDMS was peeled from petri dish, 5.0 mm holes were punched for micro-wells for a hydrogel loading window. PDMS layers and glass coverslips (22 × 22 mm; Thermo Fischer-Scientific) were activated with plasma (350 W, PlasmaEtch) for 2 min. and bound together and incubated in an 80°C oven overnight. To anchor 3-D HA hydrogel nanocomposites onto the PDMS-glass window, devices were sequentially activated with plasma (350 W, 2 min), coated with 0.1 mg/mL Poly-L-Lysine (P8920, Sigma-Aldrich) for 1 hr, and 1% glutaraldehyde (340855, Sigma-Aldrich) for 2 hrs. Microwell devices were rinsed completely using distilled water twice, and subsequently treated to UV-sterilization in a Type 2 class laminar flow hood for 15 min.
Macrophage polarization.
Macrophage polarization is induced in vitro as previously reported [30]. Untreated RAW264.7 cells are labeled as M0 macrophages. RAW264.7 cells (3×106 cells/mL) are seeded in 12-well plates overnight before polarization. M1 macrophages are obtained by stimulating RAW264.7 cells with lipopolysaccharide (LPS; 100 ng/mL) for 24 hrs and M2 macrophages are obtained by treating RAW264.7 cells with IL-10 (200 ng/mL).
Materials Characterization.
Structural modifications of hydrogel composite were studied by attenuated total reflectance Fourier transform Infrared spectroscopy (ATR-FTIR, ThermoScientific Nicolet-6700). The spectra were recorded in the wavelength region of 350–4000 cm−1 with 256 scans and a 4 cm−1 resolution. A background was collected before sample measurement and was updated every 20 min. The size of nanocarrier was measured by dynamic light scattering (DLS, Zetasizer NanoS, Malvern Instruments). The identification of APP nanocomposite structure was carried out by using scanning electron microscope (Merlin FESEM, Carl Zeiss). The UV absorbance spectrum of APP nanocomposites after different exposure times was measured by the microplate reader (SpectraMax M2, Molecular Devices). The storage (G’) and loss (G”) modulus were measured with a plate-to-plate rheometer (ARES LS-1, TA Instruments) using an 8mm plate under a constant strain of 0.05 and frequency range of 0.1 to 10 rad/s. HA-APP hydrogels were synthesized as described above, treated with UV stimulation for 30s, cut to a size of 8.0mm in diameter to fit the plate. A humid hood was used to prevent the hydrogel from drying and the temperature was maintained at 25°C.
Studying RGD release kinetics.
The UV light exposure was conducted by a compact and handheld UV lamp (UVP, λ= 302 nm, 6 W). RGD-encapsulated APP nanocomposites (500 μL) treated by different UV exposure time are loaded into centrifugal filter unit (Amicon, MW 3000), after centrifuged at 3000 rpm for 10min, the free peptides are transferred through the membrane of concentrator to the centrifuge tube bottom, then collected into filtered solution for quantification analysis. The RGD release amount was quantified by manual instructions provide by the Pierce Quantification Fluorometric Peptide Assay (Thermo Fisher-Scientific, USA). Briefly, the released peptides are specifically labeled with an amine-reactive fluorescent reagent, and the fluorescently labeled peptides are detected using Ex390/Em475. Then, the quantitative information is determined by comparing release profiles with a standard curve. For multiple days of UV exposure, after each day the residuals in concentrator are collected and DI water is added to make a total volume (500 μL). Then the solution is treated with specific UV time and loaded back into centrifugal filter. The released free peptides are collected as previously described.
Immunofluorescence staining and analysis.
To investigate Arginase-1 (Arg-1), iNOS antibody and integrin β3 expressions, RAW 264.7 macrophages in different conditions, macrophage cells (M0, M1, M2) were cultured in HA-APP gel composites in different conditions for 72 hrs. Cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, USA) for 30 min, permeabilized with 0.3% Triton X-100 (Sigma-Aldrich, USA) for 10 min, and then blocked with 3% donkey serum for 1 hr on ice to eliminate unspecified binding. Cells were incubated with mixture of Arginase-1 primary antibodies (0.5 mg/mL, 1:100, Thermo Fisher-Scientific) and iNOS primary antibody (4 mg/mL, 1:50, Thermo Fisher-Scientific) for 1 hr, and then visualized with Alexa Fluor 647 conjugated donkey anti-goat IgG secondary antibodies (2 mg/mL, 1:200, Thermo Fisher-Scientific) and 488 conjugated donkey anti-rabbit IgG secondary antibodies (0.5 mg/mL, 1:250, Thermo Fisher-Scientific). Integrin β3 was visualized by anti-integrin β3 antibody (5 mg/mL, 1:500, Abcam) and Alexa Fluor 555 conjugated goat anti-rabbit IgG secondary antibodies (5 mg/mL, 1:200, Thermo Fisher-Scientific). Cells were incubated for 30 min with Hoechst 33342 to counterstain nuclei. Fluorescent images were obtained using an inverted microscope (Zeiss Axio Observer.Z1) equipped with a digital CMOS camera (ORCA-Flash4.0 LT Digital CMOS camera, Hamamatsu Photonics) and a 40X objective. Arginase-1, iNOS expressions, and Integrin β3 expressions are quantified by the mean intensity of fluorescent regions in grayscale images for each cell.
ELISA immunoassays.
After macrophage polarization, cytokine secretions were examined by using Mouse IL-10 (ELISA Max, BioLegend, USA), IL-6 (ELISA Max, BioLegend, USA), TNF-α (ELISA Max, BioLegend, USA), and TGF-β1 (Abcam) Cytokine ELISA kit, respectively according to manufacturer’s protocols. Briefly, supernatants were collected every 24 hrs of polarization and centrifuged at 2000 g for 10 min at 4 °C to remove cellular debris. The cytokines were determined by the ELISA kit. Results were normalized to the total amount of protein.
Western blot analysis.
Cells were dissociated with an ice-cold lysis buffer of Halt Protease and Phosphatase Inhibitor Cocktail (1:100; 78440, Thermo Fisher-Scientific) in RIPA buffer (89900, Thermo Fisher-Scientific) before undergoing a bicinchoninic acid (BCA, Sigma-Aldrich) assay to assess total protein content, with each sample run in triplicate. Equivalent amounts of protein were resolved via SDS-PAGE using polyacrylamide gel (Mini-PROTEAN TGX Precast Gels, Bio-rad) and transferred to PVDF membranes (Millipore, USA). Furthermore, membranes were blocked with 5% non-fat dry milk in Tris-buffered saline for 2 hrs and then probed overnight incubation at 4 °C with Arginase-1 or iNOS primary antibodies (1:1000, Abcam). The membranes were then incubated with a secondary antibody, horse radish peroxidase (HRP)-conjugated anti-rabbit, anti-goat IgG (1:5000, Bio-rad or Thermo scientific) in blocking buffer for 1 hr and then detected with ECL substrates (1705060S, Clarity, Bio-rad), respectively. Chemiluminescence was imaged on a ChemiDoc Imaging Systems (Bio-rad).
Cell Attachment and Cell Spreading.
HA-APP hydrogels were made as described above and loaded into a 24-well plate. 5000 RAW264.7 cells were loaded in 500 uL complete cell culture media in each well. After 1 hr of cell loading, each well was washed three times with fresh cell culture media to remove the unattached cells. Cell attachment was quantified by counting cell numbers per field under a 20x objective. Cell spreading was quantified by tracing cell boundaries per field under a 40x objective.
Data analysis.
Samples were tested at least three times and data are expressed at means ± S.E.M. unless otherwise specified. The statistical analysis is carried out using Student’s t-test. Significance was assessed at a P-value of less than 0.05.
Supplementary Material
Acknowledgements
We acknowledge financial support from the National Science Foundation (CBET 1701322), National Institute of Health (NIH/NIBIB 1R21EB025406-01A1), and the American Heart Association Scientist Development Grant (16SDG31020038), and the New York University Whitehead Fellowship. The authors would like to acknowledge the Chemistry Department Shared Instrumentation Facility (SIF) at New York University for the permission of using SEM and ATR-FTIR. We would like to thank Dr. Ayaskanta Sahu, Dr. Jin Kim Montclare, and Dr. David Pine in the Department of Chemical and Biomolecular Engineering at New York University for permitting use of the Schlenk line, lyophilizer, DLS, microplate reader and ARES Rheometer for this study.
Contributor Information
Dr. Haoyu Wang, Department of Mechanical and Aerospace Engineering, New York University, Brooklyn, NY 11201, USA
Renee-Tyler Tan Morales, Department of Mechanical and Aerospace Engineering, New York University, Brooklyn, NY 11201, USA.
Dr. Xin Cui, Department of Mechanical and Aerospace Engineering, New York University, Brooklyn, NY 11201, USA
Jiongxian Huang, Department of Mechanical and Aerospace Engineering, New York University, Brooklyn, NY 11201, USA.
Weiyi Qian, Department of Mechanical and Aerospace Engineering, New York University, Brooklyn, NY 11201, USA.
Dr Jie Tong, Department of Mechanical and Aerospace Engineering, New York University, Brooklyn, NY 11201, USA.
Prof. Weiqiang Chen, Department of Mechanical and Aerospace Engineering, New York University, Brooklyn, NY 11201, USA Department of Biomedical Engineering, New York University, Brooklyn, NY 11201, USA.
References
- [1].Hu X, Leak RK, Shi Y, Suenaga J, Gao Y, Zheng P, Chen J, Nature Reviews Neurology 2014, 11, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ekdahl CT, Claasen J-H, Bonde S, Kokaia Z, Lindvall O, Proceedings of the National Academy of Sciences of the United States of America 2003, 100, 13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liu Z, Fan Y, Won SJ, Neumann M, Hu D, Zhou L, Weinstein PR, Liu J, Stroke 2007, 38, 146. [DOI] [PubMed] [Google Scholar]
- [4].Wang Q, Tang XN, Yenari MA, Journal of neuroimmunology 2007, 184, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang Y, Zhao L, Wang X, Ma W, Lazere A, Qian H.-h., Zhang J, Abu-Asab M, Fariss RN, Roger JE, Wong WT, Science Advances 2018, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Thored P, Heldmann U, Gomes-Leal W, Gisler R, Darsalia V, Taneera J, Nygren JM, Jacobsen SE, Ekdahl CT, Kokaia Z, Lindvall O, Glia 2009, 57, 835. [DOI] [PubMed] [Google Scholar]
- [7].Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, Lafaille JJ, Hempstead BL, Littman DR, Gan W-B, Cell 2013, 155, 1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Martinez FO, Gordon S, F1000Prime Reports 2014, 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu X, Reddy DS, Pharmacology & therapeutics 2012, 134, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cha BH, Shin SR, Leijten J, Li YC, Singh S, Liu JC, Annabi N, Abdi R, Dokmeci MR, Vrana NE, Ghaemmaghami AM, Khademhosseini A, Advanced healthcare materials 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kwon MY, Vega SL, Gramlich WM, Kim M, Mauck RL, Burdick JA, Advanced healthcare materials 2018, 7, e1701199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gelderblom M, Leypoldt F, Steinbach K, Behrens D, Choe C-U, Siler DA, Arumugam TV, Orthey E, Gerloff C, Tolosa E, Magnus T, Stroke 2009, 40, 1849. [DOI] [PubMed] [Google Scholar]
- [13].Barnes JM, Przybyla L, Weaver VM, Journal of cell science 2017, 130, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Burdick JA, Prestwich GD, Advanced materials (Deerfield Beach, Fla.) 2011, 23, H41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Petrey AC, de la Motte CA, Frontiers in Immunology 2014, 5, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fowke TM, Karunasinghe RN, Bai J-Z, Jordan S, Gunn AJ, Dean JM, Scientific Reports 2017, 7, 44135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim Y, Kumar S, Molecular cancer research : MCR 2014, 12, 1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Moshayedi P, Carmichael ST, Biomatter 2013, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Moshayedi P, Nih LR, Llorente IL, Berg AR, Cinkornpumin J, Lowry WE, Segura T, Carmichael ST, Biomaterials 2016, 105, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mura S, Nicolas J, Couvreur P, Nature Materials 2013, 12, 991. [DOI] [PubMed] [Google Scholar]
- [21].Basuki JS, Qie F, Mulet X, Suryadinata R, Vashi AV, Peng YY, Li L, Hao X, Tan T, Hughes TC, Angewandte Chemie (International ed. in English) 2017, 56, 966. [DOI] [PubMed] [Google Scholar]
- [22].Kang H, Trondoli AC, Zhu G, Chen Y, Chang Y-J, Liu H, Huang Y-F, Zhang X, Tan W, ACS Nano 2011, 5, 5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wehrung D, Chamsaz EA, Joy A, Oyewumi MO, European Journal of Pharmaceutics and Biopharmaceutics 2014, 88, 962. [DOI] [PubMed] [Google Scholar]
- [24].Berghaus LJ, Moore JN, Hurley DJ, Vandenplas ML, Fortes BP, Wolfert MA, Boons G-J, Comparative immunology, microbiology and infectious diseases 2010, 33, 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sternlicht MD, Werb Z, Annual review of cell and developmental biology 2001, 17, 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA, Proceedings of the National Academy of Sciences 2003, 100, 5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lei Y, Gojgini S, Lam J, Segura T, Biomaterials 2011, 32, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wehrung D, Sun S, Chamsaz EA, Joy A, Oyewumi MO, Journal of pharmaceutical sciences 2013, 102, 1650. [DOI] [PubMed] [Google Scholar]
- [29].Olejniczak J, Carling CJ, Almutairi A, Journal of controlled release : official journal of the Controlled Release Society 2015, 219, 18. [DOI] [PubMed] [Google Scholar]
- [30].Cui X, Morales RT, Qian W, Wang H, Gagner JP, Dolgalev I, Placantonakis D, Zagzag D, Cimmino L, Snuderl M, Lam RHW, Chen W, Biomaterials 2018, 161, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lisi L, Ciotti GM, Braun D, Kalinin S, Curro D, Dello Russo C, Coli A, Mangiola A, Anile C, Feinstein DL, Navarra P, Neuroscience letters 2017, 645, 106. [DOI] [PubMed] [Google Scholar]
- [32].Sun S, Chamsaz EA, Joy A, ACS Macro Letters 2012, 1, 1184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.