SUMMARY
The discovery of microRNA (miRNA) sorting into extracellular vesicles (EVs) revealed a novel mode of intercellular communication and uncovered a link between cellular endomembrane compartments and small RNAs in EV-secreting cells. Using a two-step ultracentrifugation procedure to isolate EVs released by T cells, we found that 45% of tRNA fragments (tRFs), but fewer than 1% of miRNAs, were significantly enriched in EVs compared with the corresponding cellular RNA. T cell activation induced the EV-mediated release of a specific set of tRFs derived from the 5′ end and 3′-internal region of tRNAs without variable loops. Inhibition of EV biogenesis pathways specifically led to the accumulation of these activation-induced EV-enriched tRFs within multivesicular bodies (MVBs). Introducing antisense oligonucleotides to inhibit these tRFs enhanced T cell activation. Taken together, these results demonstrate that T cells selectively release tRFs into EVs via MVBs and suggest that this process may remove tRFs that repress immune activation.
Graphical Abstract
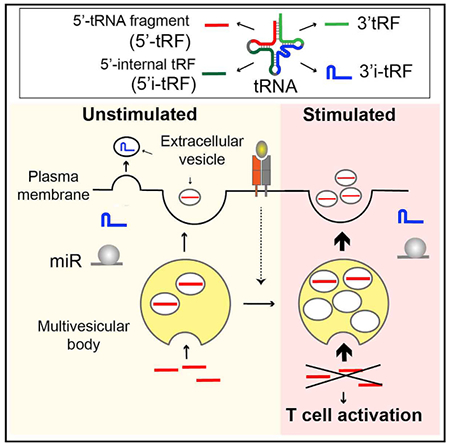
In Brief
Chiou et al. show that T cells release extracellular vesicles that carry RNA cargo enriched in tRNA fragments. Immune activating signals enhance multivesicular body formation and the secretion of vesicles containing specific tRNA fragments. Within cells, these tRNA fragments inhibit T cell activation and cytokine production.
INTRODUCTION
Extracellular vesicles (EVs) that carry extracellular RNAs (ex-RNAs) are generated from different intracellular origins. Microvesicles are assumed to be released directly from budding of the plasma membrane, whereas exosomes originate from the endosomal compartment and are released upon fusion of multivesicular bodies (MVBs) with the plasma membrane (Colombo et al., 2014). The encapsulation of exRNAs within vesicles protects them from degradation, making them stable constituents of body fluids. Moreover, exosome-associated CD47 inhibits phagocytosis, increasing retention in circulation (Kamerkar et al., 2017). These properties of exRNAs and their carriers have been exploited for biomarker discovery, and they allow exRNAs to mediate communication between exosome secreting cells and recipient cells (Tkach and Thery, 2016). In addition, the exosome biogenesis pathway modulates microRNA (miRNA) silencing activity through the association of miRNA effector proteins with MVBs (Gibbings et al., 2009).
T cells are a robust source of EVs containing small RNAs. T-cell-expressed miRNAs are associated with EVs and increase in the serum of immunized mice and humans (de Candia et al., 2013, 2014), while cellular miRNAs are globally downregulated upon T cell activation (Bronevetsky et al., 2013). Exosome secretion is important for proper immune function, as Rab27 deficiency modulates inflammatory responses and inhibits chronic inflammation in mice (Alexander et al., 2017; Okoye et al., 2014). Target cell killing by cytotoxic T cells involves the activation-induced fusion of Rab7-containing cytotoxic granules with the plasma membrane in a Rab27-dependent manner (Daniele et al., 2011; de Saint Basile et al., 2010). The fusion of MVBs with the plasma membrane in may be regulated in a similar manner to control exRNA release in exosomes. For these reasons, T cells are a good model for investigating signal-regulated mechanisms of RNA packaging into exosomes and how this process affects their biological activity in source and recipient cells.
tRNA fragments (tRFs) are generated through endonucleolytic cleavage of tRNAs (Gebetsberger and Polacek, 2013). They are among the most prevalent small RNA species detected in exRNA, and in cells they rank second in abundance only to miRNAs (Lee et al., 2009). Early studies detected tRFs in the urine of cancer patients (Borek et al., 1977; Speer et al., 1979), raising the possibility that tRFs may be oncogenic and that they may be actively released into body fluids. tRFs can be transferred from epididymosomes to sperm, epigenetically transmitting information about paternal diet and metabolism to offspring (Sharma et al., 2016). tRFs also impact a number of functions in somatic cells, including cell proliferation, cancer progression, and the activity of endogenous retroelements (Goodarzi et al., 2015; Maute et al., 2013; Schorn et al., 2017). However, their secretion and biological effects in T cells remain unexplored.
In the present study, we analyzed EVs rigorously separated from ribonucleoprotein aggregates in cell culture supernatants of activated T cells. RNA sequencing showed that compared with cellular RNAs, tRFs were enriched in EVs more than any other class of RNA, which is consistent with studies in cell lines (Baglio et al., 2015; Koppers-Lalic et al., 2014; Li et al., 2013; Liao et al., 2014; Tosar et al., 2015). We further identified specific sets of tRFs whose release via EVs is enhanced by T cell activation and showed that blocking tRF release by neutral sphingomyelinase (nSMase) inhibitor increased the cellular levels of these activation-induced EV-enriched tRFs but not other activation-independent EV-enriched tRFs. Subcellular fractionation further showed that nSMase inhibitor treatment specifically led to the accumulation of these activation-induced EV-enriched tRFs within the Rab7-containing compartments, suggesting that these tRFs were released via MVB sorting. T cells transfected with antisense oligonucleotides against these tRFs displayed enhanced T cell activation. These results indicate that activated T cells utilize EV biogenesis pathways to selectively secrete tRFs that can repress T cell activation.
RESULTS
Purified T Cell EVs Contain Intact Extracellular Small RNA Species
To establish an experimental system for investigating the effects of activating signals on exRNA biogenesis, we isolated EVs secreted by primary T cells stimulated with antibody agonists of antigen and costimulatory receptors (anti-CD3 and anti-CD28) (Figure S1A). After 3 days in these culture conditions, approximately 60% of in vitro activated T cells remain alive, and they uniformly upregulate CD44 and downregulate CD62L (Figure S1B). Since so many T cells die during activation, we expected that intracellular contents passively released from dead cells would contaminate EVs secreted by live cells. To separate EVs from contaminating particles, we used a two-stage purification procedure to isolate EVs. First, large contaminating cells and cellular debris were removed from the media by successive low and medium speed centrifugation, and EVs and other small particles were pelleted by ultracentrifugation at 100,000 × g. Second, to eliminate non-vesicle contaminants, the 100,000 × g pellet was resuspended in 80% sucrose buffer, overlaid with lower concentrations of sucrose buffer, and ultracentrifuged again to float vesicles into the less dense sucrose layers (Figure 1A).
Figure 1. EVs that Contain Intact Discrete RNA Species Are Separated from Protein Aggregates that Are Dominated by Fragmented RNAs.
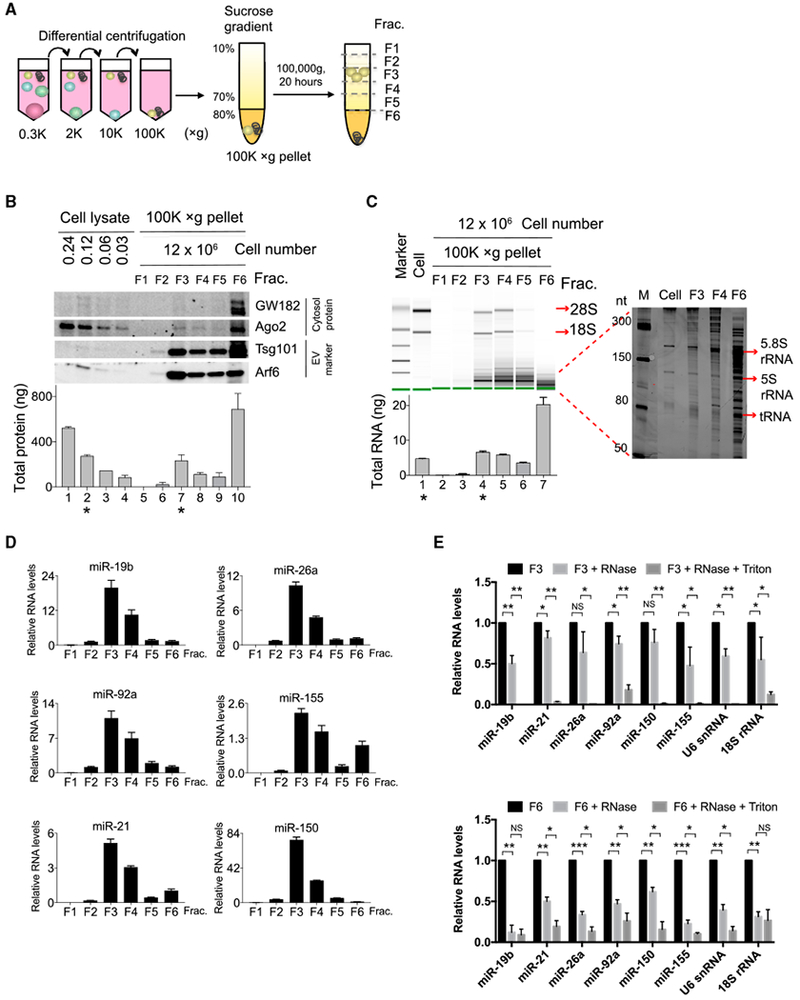
(A) Schematic of a two-step purification procedure for separation of EVs from aggregates in cell culture supernatant. The supernatant was first subjected to differential centrifugation to remove live cells, dead cells, cell debris, and, finally, EVs, and aggregates were precipitated into 100,000 × g pellets. The 100,000 × g pellets were further separated by sucrose gradient into 6 fractions.
(B) Western blot (top panel) analysis of sucrose gradient fractions of the separated 100,000 × g pellets and cell lysates prepared from the indicated numbers of cells in each lane. Bradford assay (bottom panel) determined total protein recovered in each fraction or cell lysate. * marks lanes containing similar concentrations of proteins from cell lysates (lane 2) and sucrose gradient fraction 3 (lane 7).
(C) RNA 2100 Bioanalyzer analysis of large RNA species (left panel) and PAGE analysis of small RNA species ranging from 50 to 300 bp (right panel). Bottom panel shows total RNA yield from each fraction. * marks lanes with similar RNA yield from cells (lane 1) and sucrose gradient fraction 3 (lane 4).
(D) qPCR analysis of miRNA abundance in equal volumes of RNA purified from each fraction.
(E) qPCR analysis of the abundance of the indicated RNA species detected in fraction 3 (top) or in fraction 6 (bottom) left untreated or treated with RNase A or RNase A and Triton X-100.
Data are representative of three independent experiments. Statistical significance is measured using a one-tailed t test: *p < 0.05, **p < 0.01, and ***p < 0.001.
Error bars indicate SD of the mean. See also Figure S1.
Fractions of these gradients were collected for further analysis. Several independent lines of evidence indicated that fraction 3 contained EVs, while fraction 6 was dominated by degraded RNA and/or protein aggregates. First, fraction 3 was composed of 34% to 52% sucrose, corresponding to a density of 1.15 to 1.20 g/ml, the known density range of exosomes (Théry et al., 2006). Second, fraction 3 was highly enriched in the microvesicle and exosome protein markers Arf6 and Tsg101 (Figure 1B). Ago2 and GW182 proteins involved in miRNA function were present but not enriched in EVs. The total protein quantity in fraction 3 was similar to that of T cell lysates made from 100-fold fewer cells (Figure 1B, bottom; compare fraction 3 of 100,000 × g pellet from 12 × 106 cells versus cell lysate from 0.12 × 106 cells), indicating that activated T cells secreted at least 1% of their cellular protein content in an EV-rich exudate. Fraction 6 contained even more total protein and was non-selectively enriched for all proteins tested by immunoblotting, suggesting that it was dominated by dense protein aggregates that remained at the bottom of the sucrose gradient.
Finally, RNA analyses showed that fraction 3 contains intact exRNA species. Bioanalyzer analysis detected intact 28S and 18S rRNA in fraction 3 but not in fraction 6, despite the latter’s higher overall RNA content (Figure 1C). Compared with cells, both fractions were enriched in smaller RNA species. PAGE separation of 50–300 nt RNAs further demonstrated that fraction 3 contained intact 5.8S and 5S rRNAs and tRNAs, whereas fraction 6 was dominated by apparently randomly fragmented RNAs. Although full-length rRNAs were detected in EV fractions, they were not enriched in EV fractions compared to cellular RNA, consistent with previous reports (Wei et al., 2017). qPCR showed that intact miRNAs were also much more abundant in fraction 3 than in fraction 6 (Figure 1D). These results suggest that the various exRNA species in fraction 3 are protected from degradation by encapsulation within EVs. To test this possibility, we treated fraction 3 or fraction 6 with RNase A in the presence or absence of the vesicle-disrupting detergent Triton X-100. For fraction 3, RNase treatment did not dramatically alter miRNA or other exRNA species detection unless Triton X-100 was present (Figure 1E, top). By contrast, RNase treatment of fraction 6 with RNase A dramatically reduced the detection of exRNAs even in the absence of Triton X-100 (Figure 1E, bottom). All together, these data show that in vitro activated T cells release EVs that contain and protect exRNAs and that our purification procedure separates these EVs from RNA and/or protein aggregates that also accumulate in the culture media.
tRFs Are Enriched in T Cell EVs
To identify specific small RNAs that were enriched in bona fide T cell EVs free from aggregate contamination, we performed sequencing of small (~15–31 nt) RNA libraries prepared from EVs (F3), aggregates (F6), and activated T cells (Figure 1B). To reduce bias in small RNA library preparation, we modified the 5′ and 3′ ligation adaptors with five random bases added to the 3′ end and 5′ end, respectively (Fuchs et al., 2015). Greater than 70% of sequence reads from cell libraries mapped to the genome, compared to 62% from EVs and 55% from aggregates (Figure S2A), consistent with lower mapping rates for EV libraries in the previous study (Cha et al., 2015).
Classifying the sequenced RNAs revealed several distinct features of the composition of small RNAs and RNA fragments in T cells, EVs, and aggregates. First, RNA fragments derived from larger RNA classes, including protein coding mRNAs and long intergenic noncoding RNAs (lincRNAs), accounted for about 50% of the total reads in all libraries, and their composition was similar among all samples (Figure 2A). Second, miRNAs were depleted compared to cellular RNA, slightly in EVs but greatly in aggregates (Figure 2A). This is consistent with qPCR results that detected much higher miRNA abundance in EVs than in aggregate fractions (Figure 1D). Third, EVs and aggregates differ greatly in their composition of other RNA classes. In particular, tRFs were specifically enriched in EVs, whereas aggregates contained more small nucleolar RNA (snoRNAs) and small nuclear RNA (snRNAs) (Figure 2B).
Figure 2. Comparing the Composition of Small RNAs in Activated T Cells, EVs, and Aggregates.
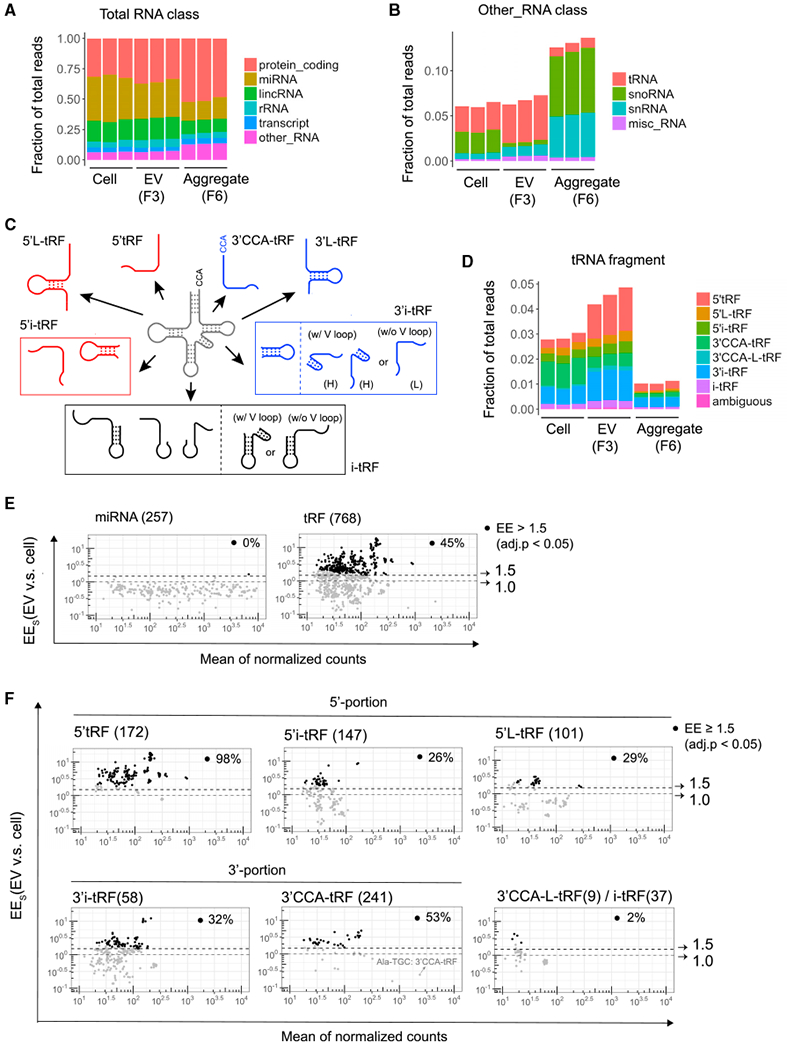
(A) Relative composition of mapped reads in small RNA libraries. RNA classification was based on the biotypes of Ensembl gene and transcript annotation. The “transcript” RNA class is the union of different biotypes, including processed transcript, antisense, and pseudogene.
(B) Sub-classification of reads shown as “other_RNA” in (A) into tRNA, snoRNA, snRNA, and misc_RNA.
(C) Pictorial summary of the 7 categories of tRFs that can arise from mature tRNAs. 3′i-tRF were further classified as hairpin (H) or linear (L) to indicate the presence or absence of tRNA variable loop-derived hairpin structures, respectively.
(D) Classification of reads in the 7 tRF classes shown in (C) and rare tRFs of ambiguous classification.
(E and F) MA plots comparing EV enrichment (EEs) (ratio of reads in EVs versus cells) and mean of normalized counts per million for all 257 detected miRNAs (E, left), all 768 detected tRFs (E, right), and subsets of tRFs in the indicated classes (F). Dotted lines and arrows indicate thresholds at 1.0 (no enrichment) and 1.5 EE. Black circles (·) mark RNA species significantly enriched in exosomes (≥ 1.5-fold; adjusted p < 0.05), and the percentage of significantly enriched RNA species among each class is indicated in each plot.
We further classified tRFs into five groups derived from different parts of full-length tRNAs: the 5′ end (5′tRF), 3′ end (3′CCA-tRF), 5′-internal region (5′i-tRF), 3′-internal regions (3′i-tRF), and central internal region (i-tRF) (Figure 2C). In each group, longer tRFs (26 to 31 nt) and shorter tRFs (14 to 25 nt) were classified as L-tRF and tRF, respectively. We found that 5′tRFs and 3′i-tFRs were enriched, but 3′CCA-tFRs were depleted in EVs compared to cellular small RNAs (Figure 2D). Analysis of tRF read length revealed that 18–21 nt 5′tRFs were dominantly enriched in EVs, while 17–18 and 22 nt 3′CCA-tRF reads were significantly depleted (Figure S2B). These data indicate that specific tRNA cleavage events generate tRFs that are preferentially released or retained in cells.
Detailed comparison of all RNA species in EVs and cells further elucidated the specificity ofTcell release of tRFs within EVs. As a fraction of all sequenced RNAs, only one miRNA (miR-150) exhibited EV enrichment (EE) as measured by the ratio of RNA reads in EVs versus cells. In contrast, 45% of tRFs showed at least 1.5-fold EE (Figure 2E; Table S1). Among the subgroups of tRFs, 5′tRFs were the most uniformly and highly enriched in EVs. 98% of all 5′tRFs displayed at least 1.5-fold EE, with some enriched as much as 10-fold (Figure 2F). In comparison, EE greater than 1.5-fold was observed for just 2%, 26%, 29%, and 36% of 3′L-tRF/i-tRF, 5′i-tRF, 5′L-tRF, and 3′i-tRF, respectively. Although 53% of 3′CCA-tRFs exhibited at least 1.5-fold EE (Figure 2F), the 3′CCA-tRF derived from tRNA-Ala-TGC that accounted for 10% of all tRF reads were depleted in EVs, resulting in fewer total reads of 3′CCA-tRFs in EVs compared to cells (Figure 2D). Taken together, our results revealed that tRFs were more significantly enriched in EVs than were miRNAs and other small RNAs and that 5′tRFs and select other tRFs were preferentially exported into EVs secreted from activated T cells.
Selective Signal-Regulated tRF Release by T Cells
To determine whether T cell activation modulates the export of tRFs, we isolated EVs secreted from resting T cells using the same two-step procedure (Figure S3A). Resting T cells were much smaller than stimulated T cells (Figure S3B), and 3.3-fold more resting cells were required to obtain the same yield of EVs (by protein content) as was obtained from stimulated cells (Figure 3A; compare 40 million resting versus 12 million stimulated T cells). However, the efficiency of EV production was similar under resting and stimulated conditions. Once again, approximately 1% of the protein yield from cell lysates was released into the EV fraction during culture under resting conditions (Figure 3B; compare EVs from 40 × 106 cells to cell lysate from 0.4 × 106 cells). Small RNAs under 150 nt and the exosome protein markers Tsg101 andCD81 were less enriched in EV fractions under resting conditions (Figures 3B and S3C), whereas EV fractions contained similar amounts of β-actin and total protein under resting conditions and stimulated conditions. This indicates that fewer exosomes were secreted and other types of co-purifying EVs that contained lower levels of exosomal markers and small RNAs may be more predominantly released under resting conditions. Small RNA sequencing showed that miRNAs were similar, but tRFs showed less EE under resting conditions (Figures S3D and S3E). In particular, 5′tRF and 3′i-tRF reads were less enriched, and 3′CCA-tRF reads were less depleted from EV fractions under resting conditions compared to stimulated conditions (Figure 3C).
Figure 3. T Cell Activation Induces the EV Enrichment of tRFs that Are Derived from the 5′-Portion of tRNAs and the 3′-Internal Region of tRNAs without Hairpin Structures.
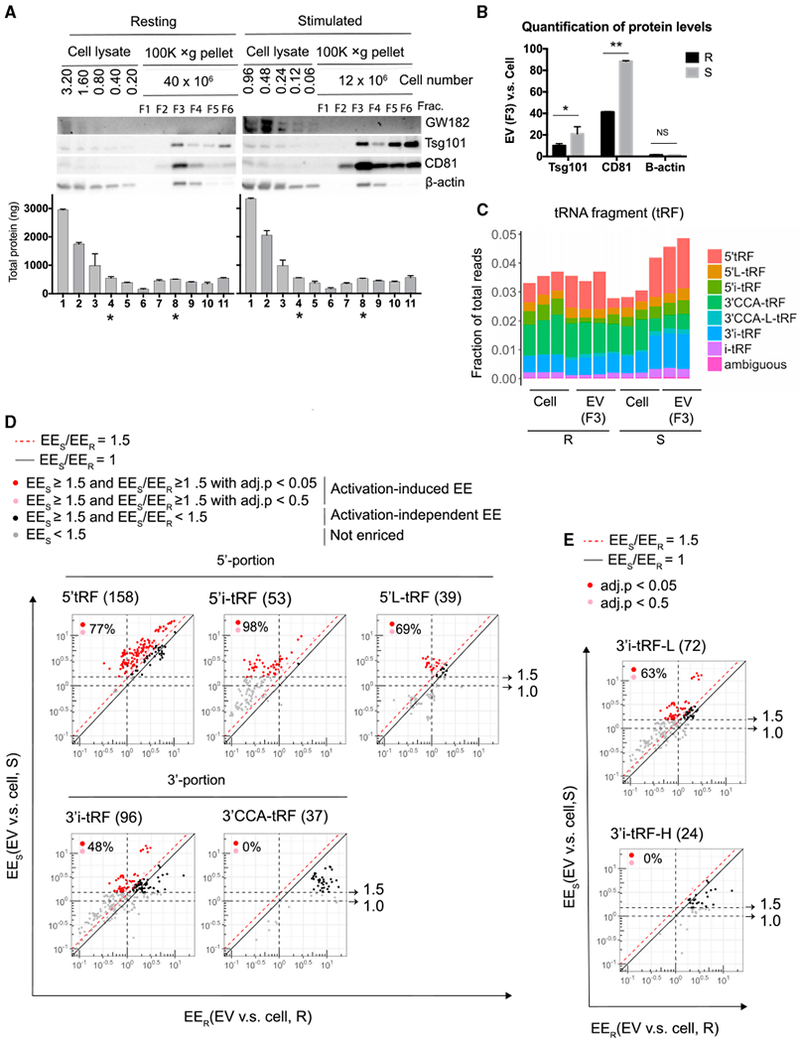
(A) Western blot (top panel) analysis of sucrose gradient fractions of the separated 100,000 × g pellets and cell lysates prepared from the indicated numbers of cells cultured under resting or stimulated conditions. Bradford assay (bottom panel) determined total protein recovered in each fraction or cell lysate. * marks lanes containing similar concentrations of proteins from cell lysates (lane 4) and EVs (sucrose gradient fraction 3; lane 8).
(B) Chemiluminescent quantification of western blot for proteins as shown in (A), expressed as the ratio of each protein in EVs versus cell lysates.
(C) tRF sequence reads from resting and stimulated cellular and EV small RNA libraries classified as in Figures 2C and 2D.
(D) EV enrichment (EE) for the indicated tRF classes from T cells cultured under resting (EER, x axis) and stimulated (EES, y axis) conditions. Dotted black lines and arrows indicate thresholds at 1.0 EES (no enrichment) and 1.5 EES. Solid black line demarcates equal EE in resting and stimulated cells (EES/EER = 1). Red dotted line indicates threshold at 1.5-fold increased EE in stimulated cells compared to resting cells (EES/EER ≥ 1.5). tRFs exhibiting activation-induced EV enrichment (EEs ≥ 1.5 and EES/EER ≥ 1.5) are marked by red circles (adjusted p < 0.05) or pink circles (adjusted p < 0.5). Black circles (·) mark tRFs with activation-independent EV enrichment in stimulated cells (EES ≥ 1.5; adjusted p < 0.05). The percentage of tRFs with activation-induced EE (union of red and pink circles) among all tRFs enriched in EVs in stimulated cells (union of black, red, and pink circles; total number indicated in parentheses after tRF class label in each panel) is indicated in each plot.
(E) 3′i-tRF-L (linear) and 3′i-tRF-H (hairpin) subsets of the total 3′i-tRF shown in (D), lower left panel.
To identify the specific tRFs that exhibit activation-induced EE, we directly compared sequencing data for EVs and cells under stimulated and resting conditions. Among the 5′tRFs, 5′L-tRFs, and 5′-i-tRFs with ≥ 1.5-fold EE in stimulated conditions, large majorities (77%, 98%, and 69%, respectively) exhibited at least 1.5-fold greater EE in stimulated T cell cultures as compared to their EE in resting T cells. In contrast, 3′CCA-tRFs displayed uniformly higher EE in resting conditions, and only 48% of 3′i-tRFs were more EV enriched in stimulated conditions (Figure 3D; Table S2). There are two structural types of 3′i-tRF, as follows: hairpin fragments (3′i-tRF-H) and linear fragments (3′i-tRF-L) that derive from tRNAs with and without variable loops, respectively (Figure 2C). 63% of the linear 3′i-tRF-L, but none of the haripin 3′i-tRF-H fragments, showed greater EE in stimulated conditions (Figure 3E). These results indicate that the positions within parent tRNAs and the secondary structures of internal tRFs correlate with selective release into EVs in response to T cell activation. tRFs derived from the 5′ end of tRNAs and from internal regions without hairpin structures exhibited EE that is enhanced upon T cell activation.
Examination of read alignments to individual representative tRNAs illustrate these regional patterns of EE and revealed that tRFs derived from the same tRNA can display different types of EE (Figures 4A and S4A). For simplicity, we classified tRFs into the following three categories: activation-induced EE, activation-independent EE, and not enriched. 5′tRF derived from the tRNAs Leu-TAA and Leu-TAG showed activation-induced EE, whereas 3′i-tRF-H from tRNA Leu-TAA exhibited activation-independent EE. Oligo(dT) qRT-PCR detected both tRFs and full-length tRNA in Poly(A) polymerase treated cellular RNA, as shown by gel electrophoresis of PCR products in the exponential phase of amplification (Figure 4B). However, since EV did not contain detectable full-length tRNA, this assay could be used to measure the relative abundance of tRFs in EVs from resting and activated T cells. Activation-induced EV-enriched tRFs were increased by 4- to 6-fold in EVs following T cell stimulation, whereas activation-independent EV-enriched tRFs were only ~2-fold more abundant in EVs from resting versus stimulated T cells (Figure 4C). RNase treatment with and without Triton X-100 indicated that these tRFs were encapsulated within EVs (Figure 4D).
Figure 4. Validation of EV Enrichment of tRFs and Their Responses to T Cell Activation.
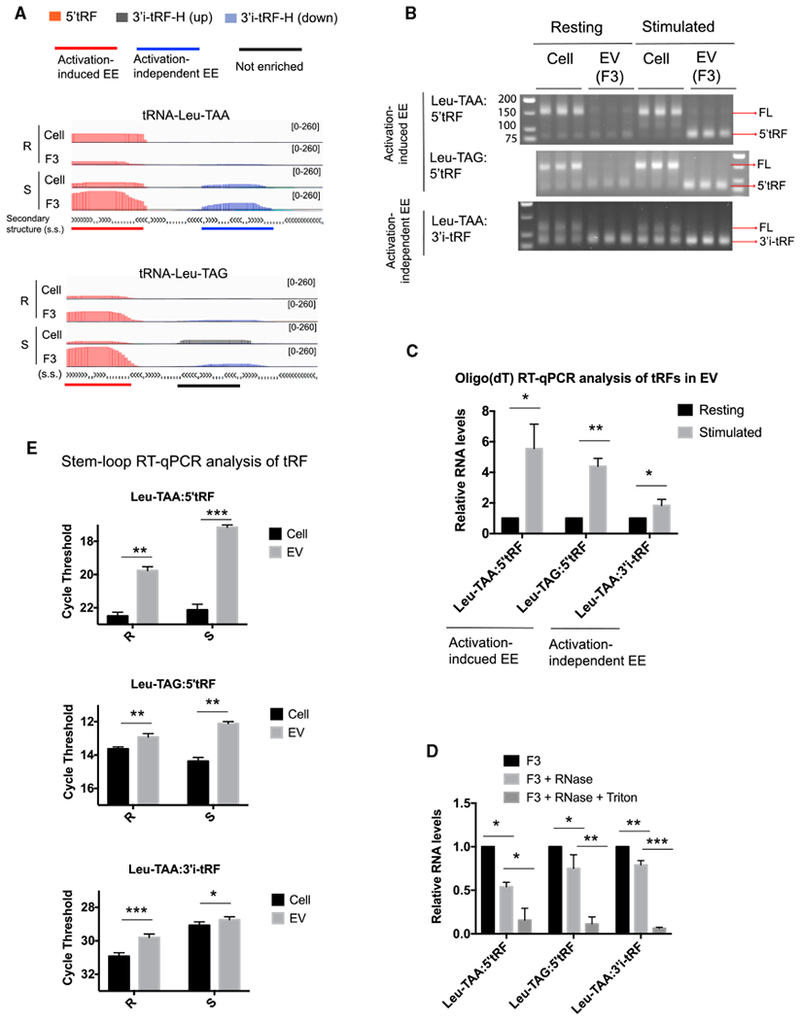
(A) IGV visualization of tRFs of the indicated classes aligned to five representative mature tRNAs.
(B) Electrophoretic analysis of products from the exponential phase of amplification in oligo(dT) RT-PCR assays for the indicated tRFs. We detected a band of the correct size and a larger product ~60 nt longer than 5′tRF or ~30 nt longer than 3′i-tRF, corresponding to amplification from full-length (FL) tRNAs.
(C) Oligo(dT) qRT-PCR measurement of tRF abundance in EVs in resting (black bars) and stimulated (gray bars) conditions.
(D) Oligo(dT) qRT-PCR analysis of the indicated RNA species detected in fraction 3 (F3) left untreated or treated with RNase A or with RNase A and Triton X-100.
(E) tRF abundance in cells (black bars) and EVs (gray bars) under resting (R) and stimulated (S) conditions as determined by stem-loop qRT-PCR.
Data are representative of at least three independent experiments. Statistical significance is measured using a one-tailed t test: *p < 0.05, **p < 0.01, and ***p < 0.001. Error bars indicate standard deviation of the mean. See also Figure S4.
We designed stem-loop qRT-PCR assays that reliably distinguish tRFs from their corresponding full-length tRNAs (Figures S4B and S4C) and used them to further confirm that all three of these EV-enriched tRFs were enriched in EV compared to cellular RNA (Figure 4E). Moreover, the difference in tRF abundance in EV and cellular RNA was greater in stimulated conditions than resting conditions for activation-induced EV-enriched 5′tRFs Leu-TAA and Leu-TAG but not for the activation-independent EV-enriched 3′i-tRF Leu-TAA. These data validate specific abundant tRFs that were differentially enriched in EVs produced by T cells in response to activating antigen and costimulatory receptor signals.
Activation-Induced nSMase-Dependent MVB Formation and tRF Downregulation
We hypothesized that activation-induced EV release may lead to the downregulation of cellular tRFs through MVB-mediated EV biogenesis pathways. Exosomes are released when cellular MVBs fuse with the plasma membrane, releasing intraluminal vesicles and their associated cargo. This process is regulated by ESCRT (endosomal sorting complexes required for transport) complexes and the activity of nSMase (Colombo et al., 2014). The nSMase inhibitor GW4869 (Luberto et al., 2002) repressed EV production by activated T cells, as indicated by dose-dependent reductions in the recovery of EV proteins and RNA in EV fractions (Figures 5A–5C). Total EV RNA recovery was reduced to about 50% of control in GW4869 treated cells, similar to the decrease of TfR and Arf6, but less than the 95% reduction of the restricted exosomal marker Tsg101 (compare Figure 5B and Figure 5C). These data indicate that GW4869 repressed the release of exosomes more than other types of EVs produced by activated T cells.
Figure 5. nSMase Inhibitor Represses EV Enrichment of Activation-Induced and Activation-Independent EV-Enriched tRFs.
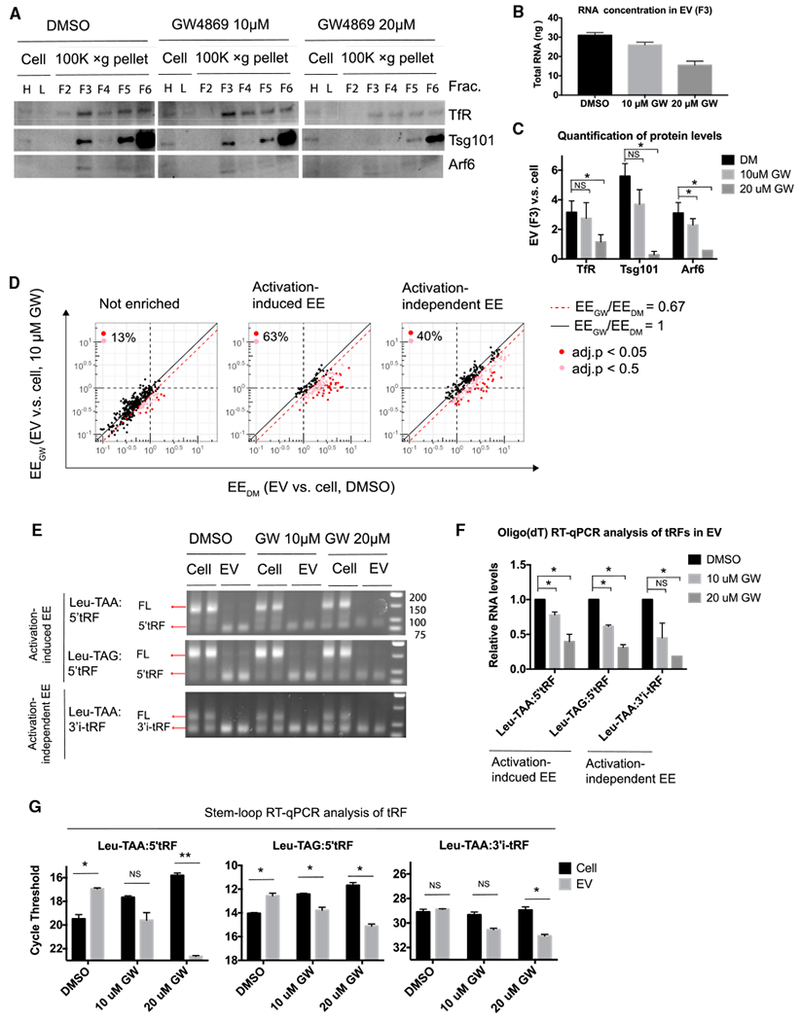
(A) Western blot analysis of sucrose gradient fractions of the separated 100,000 × g pellets and cell lysates from T cells stimulated in the presence of DMSO vehicle control or GW4869 at the indicated concentrations. Cell lysates were prepared from higher and lower amounts of cells, which are labeled as H and L, respectively.
(B) RNA yield from EV fraction (F3).
(C) Chemiluminescent quantification of western blot for proteins as shown in (A).
(D) EV enrichment (EE) for the indicated tRF classes from T cells stimulated in the presence of DMSO (EEDM, x axis) or GW4869 (EEGW, y axis). Dotted black lines indicate thresholds at 1.0 EE (no enrichment) in DMSO or GW4869 condition. Solid black line demarcates equal EE in each condition (EEDM/EEGW = 1). Red dotted line indicates threshold at 0.67-fold decreased EE in stimulated cells treated with GW4869 compared to stimulated cells treated with DMSO(EEGW/EEDM ≤ 0.67). tRFs with EE decreased by GW4869 treatment (EEGW/EEDM ≤ 0.67) are marked by red circles (adjusted p < 0.05) or pink circles (adjusted p < 0.5). The percentage of tRFs with EE decreased EE by GW4869 treatment (union of red and pink circles) among all tRFs in that class (union of black, red, and pink) is indicated in each plot.
(E) Electrophoretic analysis of products from the exponential phase of amplification in oligo(dT) RT-PCR assays for the indicated tRFs as in Figure 4B.
(F) Oligo(dT) qRT-PCR measurement of tRF abundance in EVs from cells stimulated with DMSO (black bars) and the indicated concentrations of GW4869 (gray bars).
(G) tRF abundance in cells (black bars) and in EVs (gray bars) under resting and stimulated conditions as determined by stem-loop qRT-PCR.
Statistical significance is measured using a one-tailed t test: *p < 0.05, **p < 0.01, and ***p < 0.001. Error bars indicate SD of the mean.
We compared the small RNA species in cells and EV fractions from T cells treated with GW4869 and DMSO vehicle alone by sequencing. GW4869 treatment did not significantly alter the overall distribution of broad classes of small RNAs in cells or EV fractions (Figures S5A–S5C). However, the EE of tRFs was altered by GW4869 treatment. 40% of all activation-induced EV-enriched tRFs and 63% of activation-independent EV-enriched tRFs exhibited 1.5-fold or more reduction in EE in GW4869 treated cells, compared with just 13% of non-enriched tRFs (Figure 5D; Table S2). Oligo(dT) qRT-PCR confirmed that GW4869 reduced the abundance of EV-enriched tRFs in EVs by 20%–50% at low concentration (10 μM) and by 60%–80% at high concentration (20 μM) (Figures 5E and 5F). Quantifying gel bands corresponding to tRF qRT-PCR products from cellular RNA indicated that activation-induced EV-enriched tRFs accumulated in cells upon GW4869 treatment more than activation-independent EV-enriched tRFs (Figures 5E and S5D). Stem-loop qRT-PCR confirmed that GW4869 treatment reduced both activation-induced and activation-independent EV-enriched tRF abundance in EVs while selectively increasing the cellular abundance of activation-induced EV-enriched tRFs (Figure 5G). Thus, nSMase activity is required for EV-enriched tRF enrichment in EVs and to prevent accumulation of activation-induced EV-enriched tRFs in activated T cells.
GW4869 represses not only the release of exosomes via MVB but also microvesicles that bud directly from the plasma membrane (Menck et al., 2017). To further probe the pathway of tRF release by T cells, we analyzed their subcellular localization by cellular organelle fractionation. Previous studies have reported that the 15,000 × g pellet of homogenized cells contains active MVBs that can sort proteins or RNAs into intraluminal vesicles (Shurtleff et al., 2016). We obtained 15,000 × g pellets from homogenized T cells and then further fractionated them by Opti-prep gradient centrifugation (Figure 6A). In resting T cells, the late endosome protein Rab7 was enriched in fractions (6 and 7) that contained no detectable CD81 (Figure 6B). CD81 is an exosome marker that can appear on the plasma membrane and endomembranes in lymphocytes. By contrast, in activated T cells, Rab7 was enriched in fractions (4,5, and 6) with substantial CD81 protein. These results indicate that CD81 may be more actively incorporated into Rab7-containing MVBs in activated T cells. Notably, we also observed greater enrichment of CD81 in EV fractions from activated compared with resting T cell supernatants (Figures 3A and 3B).
Figure 6. nSMase Inhibitor Induced the Accumulation of Activation-Induced EV-Enriched tRFs, but Not Activation-Independent EV-Enriched tRFs, within Rab7-Containing MVB Compartments.
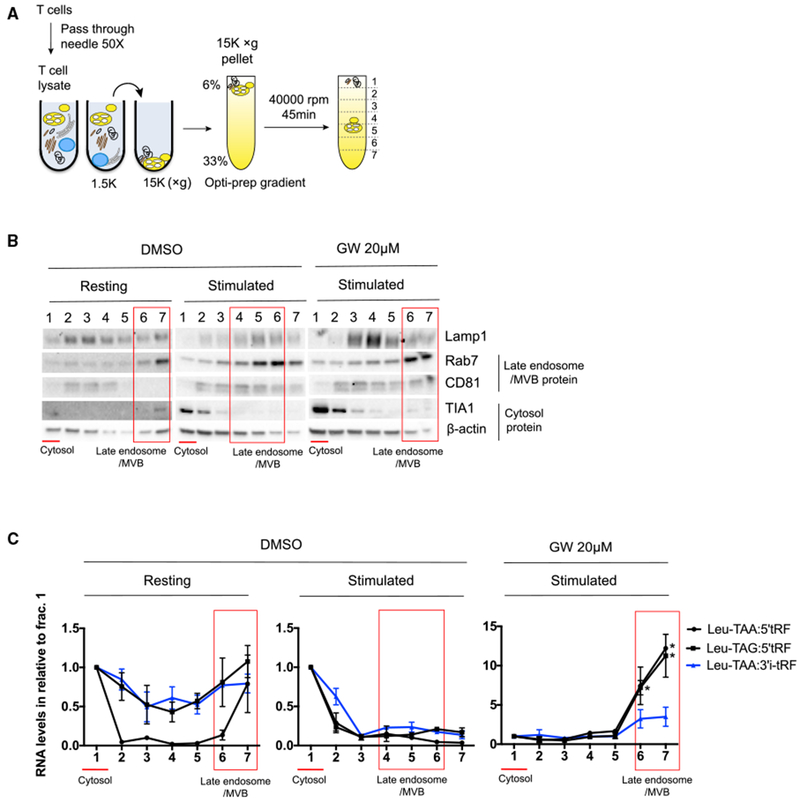
(A) Schematic of subcellular fractionation of T cells. 15,000 × g pellets obtained from differential centrifugation of T cell lysates were layered on the top of the Optiprep gradient and then centrifuged at 40,000 rpm for 45 min to separate different membrane organelles or cytosolic proteins.
(B and C) Western blot analysis of proteins (B) and stem-loop qRT-PCR quantification of tRFs in subcellular fractions obtained from resting conditions or activated conditions treated with DMSO or GW4869 (C). For each fraction, RNA concentration is normalized to fraction 1. In (B) and (C), red lines indicate cytosolic fractions, and red boxes indicate MVB fractions.
Data are representative of two independent experiments. Statistical significance is measured using a one-tailed t test: *p < 0.05, **p < 0.01, and ***p < 0.001. Error bars indicate standard deviation of the mean. See also Figure S6.
The lower-density Rab7- and CD81-containing membrane fractions may represent MVBs that are intracellular intermediates of exosome biogenesis. Consistent with this interpretation, GW4869 treatment of stimulated T cells shifted the enrichment of Rab7 back to the higher-density fractions 6 and 7. GW4869 reduces the formation of intraluminal vesicles in MVBs, which may lead to increased MVB density (Trajkovic et al., 2008). These high-density Rab7-containing fractions also contained CD81, suggesting that GW4869 blocked MVB maturation after incorporation of CD81 and potentially RNA cargo as well. Oligo(dT) RT-PCR detected both full-length tRNAs and tRFs in key cellular Optiprep gradient fractions (Figure S6). Therefore, we used stem-loop qRT-PCR to quantify tRFs (Figure 6C). Under resting conditions, both activation-independent and activation-induced EV-enriched tRFs were concentrated in cytosolic and high-density Rab7-containing fractions (Figure 6C, left panel). These tRFs were mainly restricted to the cytosolic fractions from stimulated T cells (Figure 6C, middle panel). Increased MVB formation and EV release may efficiently reduce the burden of EV-enriched tRFs in Rab7-containing endomembrane compartments in activated T cells. Consistent with this possibility, GW4869 treatment induced dramatic accumulation of activation-induced EV-enriched tRFs (and to a lesser degree, the activation-independent EV-enriched 3′i-tRF from Leu-TAA) in high-density Rab7- and CD81-containing fractions as compared to cytosolic fractions (Figure 6C, right panel). Taken together, these results indicate that T cell activating signals induce functional MVB formation and constitutive removal of activation-induced EV-enriched tRFs.
Activation-Induced EV-Enriched tRFs Inhibit T Cell Activation
We hypothesized that signal-regulated release of tRFs in EVs may serve as a means to export cellular tRFs that could alter T cell activation. To test the functional impact of these tRFs, we transfected T cells with 2′-o-methylated oligonucleotides antisense to specific abundant tRFs (Table S2) on day 1 of activation and then analyzed transfected cells on day 3. Activated T cells upregulate CD44, downregulate the lymph node homing molecule CD62L, and produce cytokines such as interleukin-2 (IL-2). Antisense oligos against the activation-independent EV-enriched 3′i-tRF derived from tRNA Leu-TAA and tRNA Ser-GCT did not affect the downregulation of CD62L or the upregulation of CD44 (Figures 7A–7C). However, antisense oligos against the activation-induced EV-enriched 5′tRFs derived from both tRNA Leu-TAA and tRNA Leu-TAG enhanced cell activation, as shown by decreased CD62Lhi cells and a corresponding increase in the frequency of fully activated CD44hiCD62Llo cells (Figures 7A–7C). We measured cytokine production by flow cytometry following restimulation with PMA (phorbol 12-myristate 13-acetate) and ionomycin (Figures 7D and 7E). IL-2 production was enhanced in T cells transfected with antisense oligo against either activation-induced EV-enriched tRF but neither of the activation-independent EV-enriched tRFs. These transfected T cells expressing more IL-2 and other activation markers also displayed lower viability in culture (Figure S7). These observations may be linked, as IL-2 sensitizes T cells to activation-induced apoptotic cell death (Marks-Konczalik et al., 2000). These results suggest that T cells engage EV biogenesis pathways to dispose of tRFs that can inhibit their activation and cytokine production.
Figure 7. Transfection of Antisense Oligos against tRFs that Are Associated with MVBs in an Activation-Induced Manner Enhances T Cell Activation.
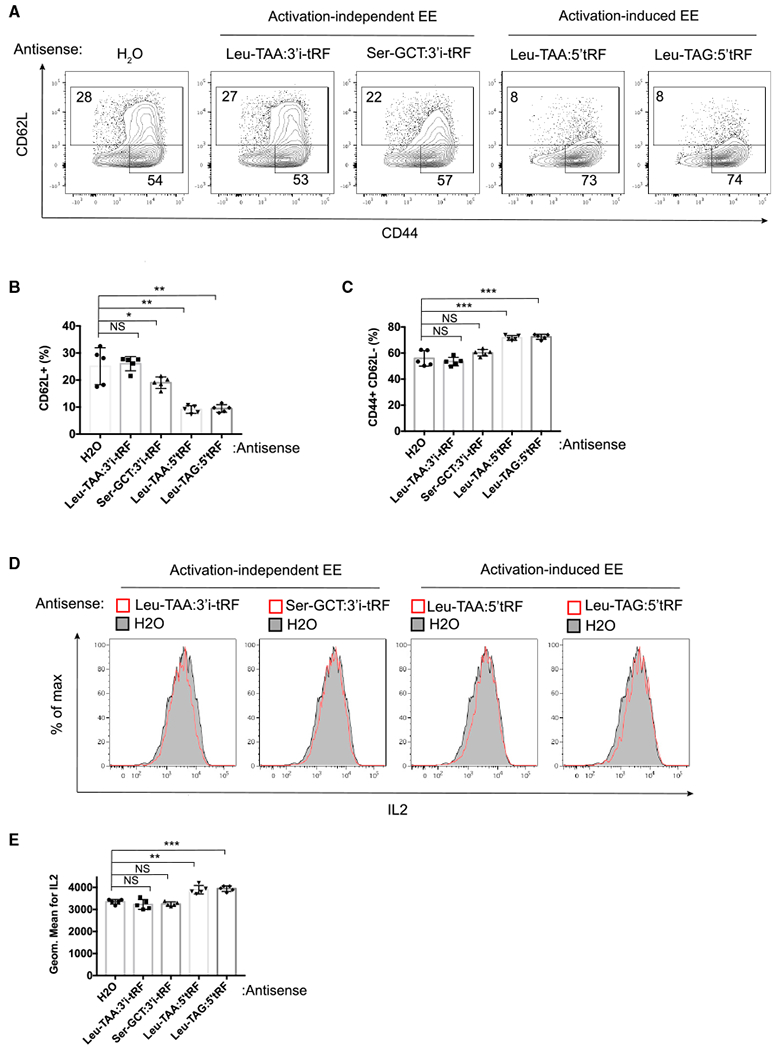
(A) Representative flow cytometric analysis of CD44 and CD62L expression on the surface of CD4+ T cells transfected with antisense oligos complementary to tRFs or water vehicle (H2O) control.
(B and C) Quantified frequency of CD62L+ (B) and CD44+ and CD62L+ (C) cells.
(D) Representative flow cytometric analysis of IL-2 intracellular staining of live CD4+ T cells restimulated with PMA and ionomycin.
(E) Geometric mean fluorescence intensity of IL-2 staining.
Data are representative of at least three independent experiments. Statistical significance is measured using a one-tailed t test: *p < 0.05, **p < 0.01, and ***p < 0.001. Error bars indicate standard deviation of the mean. See also Figure S7.
DISCUSSION
Our results show that T cells release specific sets of tRFs within EVs through their association with MVBs in a signal-regulated manner. By rigorously separating contaminating RNA and/or protein aggregates from EVs, we found that tRFs, but not miRNAs, are enriched in T-cell-derived EVs relative to all other cellular small RNA species. Specific classes of tRFs distinguished by their location in the parent tRNA and by tRNA-specific structural features were preferentially released in EVs. For some of these classes, their release was regulated during T cell activation. tRFs enriched in EVs in an activation-induced manner were mostly derived from tRNA 5′ ends, 5′-internal regions, and 3′-internal regions without variable loop hairpins. In contrast, tRFs derived from tRNA 3′ ends and 3′-internal regions with variable loop hairpins were frequently enriched in EVs from activated T cells similarly or even less than those produced by resting T cells. These patterns of EV release corresponded with differences in the accumulation of tRFs within activated T cells treated with the nSMase inhibitor GW4869, which inhibits EV biogenesis pathways. GW4869-treated activated T cells accumulated activation-induced EV-enriched tRFs, but not activation-independent EV-enriched tRFs within high-density fractions containing Rab7 and the exosome marker CD81, suggesting that these tRFs are released through an MVB-mediated EV biogenesis pathway. EVs released from activated T cells also contained more CD81 than EVs from resting T cells, further indicating that signal-regulated exosome production contributes to tRF secretion. Antisense oligos directed against activation-induced tRFs increased T cell activation. Taken together, these findings imply that secretion via exosomes and other EVs is a mechanism whereby T cells rapidly remove tRFs through endomembrane compartments to prevent them from engaging cytosolic targets that mediate T cell activation.
Global miRNA turnover and selective downregulation also occurs during T cell activation (Bronevetsky et al., 2013). However, in contrast to tRFs, miRNAs were present but not enriched in EVs released by T cells. This finding is consistent with previous analyses of the RNA composition of exosomes secreted by B cells, stem cells, and glioblastoma, esophageal, and breast cancer cells (Baglio et al., 2015; Koppers-Lalic et al., 2014; Li et al., 2013; Liao et al., 2014; Tosar et al., 2015). Ago2 protein was also not enriched in T cell EVs compared to cellular protein, indicating that Ago2-miRNA complexes are not selected for sorting into EVs. In isogenic cell lines differing only in KRAS status, MVB association and exosomal release of Ago2 and miRNAs was decreased by an activating KRAS mutation (McKenzie et al., 2016). Since Kras is activated by T cell receptor signaling (Ebinu et al., 2000), it is possible that activated Kras represses exosomal release of miRNAs associated with Ago2 in T cells and in a variety of stem and cancer cell lines. This possibility is consistent with the report that few miRNAs are enriched in exosomes secreted by the Jurkat T cell line and that these exosomal miRNAs are not associated with Ago2 but instead interact with another RNA binding protein, hnRNPA2/B1 (Villarroya-Beltri et al., 2013). Coordinated mechanisms of cellular small RNA repertoire remodeling may be an important feature of T cell activation and immune responses.
Among small RNAs, tRFs are second only to miRNAs in their abundance, and they have been shown to regulate critical cellular functions (Lee et al., 2009; Thompson and Parker, 2009). Although tRF species have been shown to be abundant in exosomes in several studies, their biogenesis and function as exRNAs are relatively understudied. Our comprehensive analysis of tRFs in T cells and their secreted EVs showed that almost all 5′tRFs, but only a select few derived from other regions of tRNAs, are enriched in EVs. Three types of tRFs derived the 5′ terminus of tRNAs have been classified according to their length: 5′halves (31–33 nt), 5′L-tRFs (26–31 nt), and 5′tRF (less than 26 nt). Specific 5′halves repress protein translation by targeting eukaryotic translation initiation factors (Ivanov et al., 2011) and modulate the formation of stress granules to inhibit protein synthesis (Emara et al., 2010). 5′L-tRFs derived from tRNA Gly-GCC are transferred from the epididymis to sperm to regulate endogenous retroelements (Sharma et al., 2016). In contrast to 5′halves and 5′L-tRFs, 5′tRFs have no hairpin structures. Their specific and prominent activation-induced EE suggest that 5′tRFs may regulate different cellular processes that are associated with membrane compartments, allowing their rapid removal through EV-biogenesis pathways.
The positional and structural patterns apparent in tRFs that undergo activation-induced EV release indicates that mechanisms that sort specific tRFs into EVs may be coupled to tRNA processing pathways. Dicer and Angiogenin (ANG) cleave the D-loops, T-loops, and anticodon loops of mature tRNAs (Cole et al., 2009; Fu et al., 2009). ANG is a candidate mediator of tRF processing and sorting into EVs, possibly in collaboration with the RNA binding protein YBX1. ANG is a secreted protein present at high concentration in blood plasma (Barcena et al., 2015) and is itself enriched in exosomes (Wei et al., 2017). YBX1 associates with miR-223 and packages this miRNA into exosomes (Frye et al., 2009; Shurtleff et al., 2016). YBX1 also interacts with a specific set of tRFs (Goodarzi et al., 2015). Some of these tRFs could be derived from ANG cleavage. Most YBX1-associated tRFs are derived from 5′ internal regions or central regions that include the anticodon loops of tRNAs (Goodarzi et al., 2015). Both miR-223 and some of the known YBX1-associated tRFs were enriched in EVs in an activation-induced manner in T cells, making YBX1 a candidate for tRF sorting into EVs upon T cell activation. tRNA synthetases that associate with ceramide or the lysosome may also play a role in the observed selectivity of tRF processing and secretion (Bidlingmaier et al., 2016; Choi et al., 2017; Han et al., 2012).
Our findings indicate that removal of cellular tRFs via sorting into MVBs and EVs may be an important mechanism to avert inhibitory functions of tRFs during T cell activation. However, tRFs in secreted EVs may be actively transferred among cells, allowing them to mediate intercellular RNA communication. These uniquely regulated exRNA species may also have utility as biomarkers of immune activity and disease processes. Compared with miRNAs, which are ubiquitous in almost all cell types, 5′tRFs as a class appear to be selectively abundant in hematopoietic tissues (Dhahbi, 2015). In summary, our data show that tRFs are functionally active small RNAs that are dynamically regulated by differential intracellular compartmentalization and release within EVs in response to T cell activating signals.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, K. Mark Ansel (mark.ansel@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6J were purchased from Jackson Laboratory and bred in specific pathogen-free conditions in the Animal Barrier Facility at the University of California, San Francisco. Male and female mice were euthanized with CO2 at 4-7 weeks of age for spleen and lymph node removal. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco.
METHOD DETAILS
CD4+ T cell stimulation and culture
Naive CD4+ T cells from spleen and lymph nodes of mice were purified by magnetic bead selection (Invitrogen) and used in all experiments. For stimulated conditions, naive T cells were stimulated with plate-bound anti-CD3 (2.0 μg/ml) and soluble anti-CD28 (0.5 μg/ml) in the presence of 10 units/mL IL2 (National Cancer Institute) for ~60 hr at initial cell density of ~1 × 106 cells/ml. After stimulation, the cell density of live cells was similar to the initial density of the naive cells (~1 × 106 cells/ml) since not only cell proliferation but also cell death occurs during stimulation. For resting conditions, T cell cultures that were stimulated for ~48 hr were resuspended and diluted with 3 volumes of media containing 10 units/ml IL2. After expansion for ~24 hr, cells were spun down and resuspended in EV-free media containing 10 units/ml IL2 for 48 hr at initial cell density of ~2 × 106 cells/ml, which then becomes ~4 × 106 cells/ml after 48 hr-resting in IL2-containing media. Cells grown for exosome production were incubated in EV-free medium produced by ultracentrifugation at 100,000 × g (28,000 RPM) for 18 hr using an SW-28 rotor (Beckman Coulter). All T cell culture was done in DMEM high-glucose media supplemented with 10% FBS and other chemicals as previously described (Steiner et al., 2011).
Exosome/EV purification
Cell culture supernatant and T cells were harvested together by gently pipetting T cells sediment in the bottom of the plate with the conditioned medium. The collected medium containing both T cells and cell culture supernatant was centrifuged for 10 min at 300 × g. The resulting cell pellet was used for cell lysate, RNA preparation or subcellular fractionation and the resulting supernatant was further centrifuged for 20 min at 2000 × g to remove cellular debris. To remove microvesicles, the supernatant collected from 2000 × g spin was centrifuged for 30 min at 10,000 × g. The resulting supernatant containing both exosomes, other types of EVs and RNA and/or protein/membrane complexes or aggregates was further subjected to ultracentrifugation at 100,000 × g for 2 hr. To separate EVs from the contaminating aggregates, the resulting crude EV pellet was resuspended in 100 μL PBS and 1 mL 90% sucrose buffer (final sucrose concentration = 82%), which was then overlaid with 11 layers of 1 mL sucrose buffer in order from high to low concentrations, including buffers containing 70%, 64%, 58%, 46%, 40%, 34%, 28%, 22%, 16%, and 10% of sucrose. The resulting 12 mL sucrose gradient was subjected to ultracentrifugation at 100,000 × g for 18 hr. Six fractions, each of which is 2 ml, were collected from top to bottom and resuspended in 9 mL PBS, which were then further centrifuged at 100,000 × g for 1 hr. The resulting supernatants were poured out and the pellets were resuspended in 50 μL PBS. The third fraction from the top, which contains the interface of the 34% and 40% sucrose solutions, should be enriched for EVs. The sixth fraction, which contains the interface of the 70% and 82% sucrose solutions, contains protein/RNA/membrane aggregates. All sucrose buffers were made with sucrose and PBS.
Qligo(dT) RT-qPCR
Total RNA was extract from EVs by using Trizol LS (Life Technologies). For RNase protection assays, EVs were left untreated, treated with 50 μg/ml RNase A or treated with RNase A in the presence of 0.1% Triton X-100 (Sigma) for 15 min at room temperature. The relative levels of miRNAs or tRFs in EVs were quantified by miScript II RT Kit and miSYBR Green PCR Kit (QIAGEN). The miScript II RT Kit added poly (A) to the 3′ end of RNAs followed by using oligo(dT) adaptor for the RT reactions. The qPCR universal reverse primer was included in the miSYBR Green PCR Kit. The qPCR forward primers for miRNAs, U6 snRNA, and 18S rRNA were previously described (Bronevetsky et al., 2013). The qPCR forward primers for all tRFs were the full-length tRF sequences provided in Figure S4.
Stem loop RT-qPCR
The design of primers, probes and reaction conditions were as previously described with the following modifications (Chen et al., 2005; Moltzahn et al., 2011). TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems) was used for reverse transcription reactions. The 7.5 μL RT reactions contained 20 ng of total RNA, 1 × RT buffer, 1 mM dNTP, 3.33 U/ìl MultiScribe reverse transcriptase, 0.25 U/ìl RNase inhibitor and 50 nM stem-loop RT primer. RT reaction conditions: 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. The 10 μL pre-PCR reactions contained 3 μL RT-product, 1 × TaqMan Universal PCR master Mix with no AmpErase UNG (Applied Biosystems), 50 nM forward primer and 50 nM universal reverse primer. Pre-PCR conditions: 95°C for 10 min, followed by 8 cycles of 95°C for 10 s, and 65°C for 1 min. The 10 μL qPCR reactions included 2 μL pre-PCR product, 1 × TaqMan Universal PCR master Mix with no AmpErase UNG (Applied Biosystems), 0.2 ìM TaqMan probe (IDT), 1.5 ìM forward primer, and 0.7 ìM universal reverse primer. qPCR reaction conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, and 65°C for 1 min. The universal reverse primer sequence is GTGTCGTGGAGTCGGC. The stem-loop RT primer sequence is CTCAACTGGTGTCGTG GAGTCGGCAATTCAGTTGAG(X)8. The forward primer sequence is ACACTCCAGCTGGGTGGATTTATATC(X)15-16. The TaqMan probe sequence is /56-FAM/TTCAGTTGA/ZEN/G(X)8/3IABkFQ/ (PrimeTime qPCR probes, IDT). In all sequences, X indicates the tRF-specific sequence shown in Figure S4C.
Subcellular fractionation
107 cells T cells were washed 3 times in PBS and resuspended in 2 volumes of homogenization buffer (400 mM sucrose, 20 mM HEPES, pH 7.5, 1mM EDTA) with Complete Mini protease inhibitor (Roche) and 1U RNasin (Promega). The activated T cells in homogenization buffer were passed ~50 times through a 22 gauge needle, and resting T cells were passed ~80 times through a 25 gauge needle until 50%–80% of cells were disrupted, as assessed by trypan blue staining. The homogenized cells were then centrifuged at 1,500 × g for 5 min to remove nuclei and the resulting supernatant was centrifuged at 15,000 × g for 20 min to spin down large cellular membranes, which were then subjected to OptiPrep gradient centrifugation. The OptiPrep gradient was made by overlaying 10 layers of 0.4 mL buffer in order from high to low concentration (33%, 30%, 27%, 24%, 21%, 18%, 15%, 12%, 9% and 6%) of OptiPrep. All OptiPrep solutions were made in buffer containing 50 mM HEPES, 78 mM NaCl, 4 mM MgCl2 and 8.4 mM CaCl2 and 10 mM EDTA. The 15,000 × g pellet fraction was resuspended in 100 μL PBS and added on the top of the OptiPrep gradients, and centrifuged at 40,000 rpm for 45 min using a SW55 rotor (Beckman Coulter). Seven fractions of 600 μL each were collected from top to bottom. 200 μL of each fraction was extracted in 700 μL Trizol (Invitrogen) for RNA purification and 400 μL of each fraction was further concentrated by StrataClean Resin (Agilent Genomics) for western blot analysis. Briefly, 10 μL of StrataClean Resin was added to each fraction followed by incubation at 4°C for 1 hr on a rotator mixer. After spinning down resin and removing supernatant, 30 μL of 1.5 × Laemmli buffer was added to each sample, which was then frozen for at −80°C until use.
Small RNA cDNA library preparation
Barcoded small RNA cDNA libraries were prepared as previously described with modifications (Williams et al., 2013). Briefly, to reduce ligation bias, 5 random nucleotides were added into the 3′ end of 5′-adaptor and the 5′end of 3′-adaptors during the oligo synthesis. Barcoded 3′-adaptors were adenylated by 5′DNA adenylation kit (NEB). 19-nt and 24-nt synthetic oligoribonucleotides that were used as size markers were radiolabeled at the 5′ end by T4 PNK (NEB). 10 ng RNA from each EV or cell sample and the radiolabeled size markers were ligated separately to 10 μM adenylated 3′-adaptor using T4 RNA ligase 2, truncated K227Q (NEB) at 4°C for overnight. The 3′-ligation reactions containing EV or cell samples were pooled, ethanol-precipitated and loaded into a single lane of a 15% denaturing polyacrylamide gel. The ligation reactions containing radiolabeled 19-nt and 24-nt oligos were loaded into the nearby lane to be the size markers for cutting the band containing 3′-ligated products of pooled EV and cell samples from gels. The RNAs within the cut gel pieces were eluted at 4°C for overnight followed by ethanol precipitation. The purified 3′-ligated products were further ligated to 10 μM 5′-adaptor using T4 RNA ligase 1 (NEB) at 37°C for 1 hr. Ligated small RNAs were again cut and purified from a 12% polyacrylamide gel based on the radiolabeled size markers. The purified ligated small RNAs were reverse-transcribed using SuperScript III (Invitrogen), and amplified by PCR. The PCR products that were in the exponential phase of amplification, which is 8-12 cycles in this study, were purified from 2.5% agarose gel (Semkem LE agarose, Lonza) using a gel extraction kit (QIAGEN). The gel purification step was performed twice to increase the removal of adaptor dimers from ligated small RNAs. Except for the five random nucleotides within the 3′ end of 5′-adaptor and the 5′ end of 3′-adaptors, all the sequences including 5′-adaptors, barcoded 3′-adaptors, PCR primers were the same as those published in the previous studies (Farazi et al., 2012).
Annotation of tRFs from RNA-seq data
The obtained sequence files were trimmed for the constant regions of the adaptor sequences by FASTQ/A Trimmer. The resulting files were split into the separate samples according to the barcode sequences in the 3′ end of the read by FASTQ/A Barcode splitter. The reads in the resulting files contain 5 random nucleotides in the 5′ end and barcoded sequences followed by 5 random nucleotides in the 3′ end, which were both trimmed by FASTQ/A Trimmer. Extracted reads were mapped by Bowtie2 v2.2.4 (default settings) to mouse genome reference sequence (GRCm38/mm10). The mapped reads were then intersected with gene annotation (Ensemble GRCm38 release 91) supplemented with tRNA annotation from the Genomic tRNA Database (Lowe and Eddy, 1997). The numbers of mapped reads that overlap with each annotations were counted by bedtools coverage. For annotating tRNA fragments, a bed file that contains the coordination of different tRF groups, as described in Figure 2C, within 472 annotated tRNAs was created. The coverage of tRNA reads in EV or cell samples for different tRF groups annotated in this bed file was calculated by bedtools coverage. The tRNA read was assigned to a specific tRF group that has the largest coverage by the read. More then 90% of tRNA reads were assigned to specific tRFs with more then 70% coverage. tRNA reads were annotated as ambigious when their coverage for all tRF groups was less than 70%.
tRF antisense oligonucleotides transfection
Antisense 2′-o-methyl oligonucleotides (IDT) were transfected using a Neon transfection kit and device (Invitrogen). A total of 5 × 105 T cells that have been stimulated for 1 day were washed two times with PBS before suspension in 10 μL of buffer T (Neon kit, Invitrogen). Antisense oligos (1 μL of 125 μM) were added to the cell suspension to a final volume of 11 μL and mixed. 10 μL of the suspension was electroporated with a Neon electroporation device (Invitrogen; 1,600 V, 10 ms, three pulses). The transfected T cells were then cultured in 0.5 mL media so that the final concentration of tRF antisense oligos is 250 nM. The 0.5 mL media for culturing transfected T cells were in 24-well plates with plate-bound anti-CD3 (2.0 μg/ml), soluble anti-CD28 (0.5 μg/ml) and 10 units/mL IL2. After 2 days of culture, the transfected T cells were harvested and analyzed by flow cytometry for surface CD44 and CD62L expression. The sequence of antisense oligo for Leu-TAA:5′tRF was CCACUCGGCCAUUCUG, for Leu-TAG:5′tRF was CCGCUCGGCCACGCUA, for Ser-GCT:3′i-tRF was CCCUCGCGUGCAAAGCACA, and for Leu-TAA:3′i-tRF was ACGCGGAUAUAAAUCC.
Intracellular cytokine staining
After 2 days of culture, T cells transfected with antisense oligos were restimulated for 3 hours with 10 nM PMA and 1 μM Ionomycin in the presence of 5 μg/ml brefeldin A. The harvested cells were stained with cell viability dye (eFluor 780, Invitrogen) followed by fixation, permeabilization and IL-2 staining as previously described (Steiner et al., 2011).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical information pertaining to the number of replicates and significance can be found in the figure legends. All experiments were performed at least three times independently where possible. In instances where the experiments were performed two times due to limited availability of primary cells or reagents, independent techniques were used to validate the findings.
RNA-seq data analysis
The ‘Deseq2′ package (Love et al., 2014) was used for analysis of differential expression. ‘ggplot2′ was used to produce scatterplots. EV enrichment (EE) in Figures 2, 3, and 5 was the fold-change of read counts in EVs versus cells. Deseq2 was used for analyzing the fold-changes of all annotated genes in EVs versus in cells and then the fold-changes of miRNAs and tRFs were subset from all genes. Thus, the EE of miRNAs and tRFs is a relative value that is compared to small RNAs derived from all genes. Deseq2 was also used for analyzing the fold-changes of the EE of all annotated genes in stimulated versus resting conditions or in stimulated conditions with DMSO versus with GW4869. The fold-changes of the EE of tRFs were subset from all genes. Thus, the fold-change of the EE of tRFs is also a relative value that is compared to small RNAs derived all genes.
DATA AND SOFTWARE AVAILABILITY
RNA sequencing data are deposited at GEO under the accession number GEO: GSE121724.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| GW182 | Bethyl Laboratories | Cat# A302-329A; RRID: AB_1850240 |
| Ago2 (C34C6) | Cell Signaling | Cat# 2897S; RRID: AB_2054446 |
| Tsg101 (C-2) | Santa Cruz Biotechnology | Cat# sc-7964; RRID: AB_671392 |
| CD81 (D5O2Q) | Cell Signaling | Cat# 10037S; RRID: AB_2714207 |
| Arf6 (3A-1) | Santa Cruz Biotechnology | Cat# sc-7971; RRID: AB_2289810 |
| Transferrin Receptor | Abcam | Cat# ab84036; RRID: AB_10673794 |
| Beta-actin (AC-74) | Sigma | Cat# A5316; RRID: AB_476743 |
| Lamp1 | Abcam | Cat# ab24170; RRID: AB_775978 |
| Rab7 (D95F2) | Cell Signaling | Cat# 9367S; RRID: AB_1904103 |
| TIA1 (D-9) | Santa Cruz Biotechnology | Cat# sc-48371; RRID: AB_628358 |
| Rabbit IgG (H+L) | Bio-Rad | Cat# 170-6515; RRID: AB_11125142 |
| Mouse IgG (H+L) | Bio-Rad | Cat# 170-6516; RRID: AB_11125547 |
| CD62L-BV605 (MEL-14) | Biolegend | Cat# 563252; RRID: AB_11125577 |
| CD44-A700 (IM7) | eBioscience | Cat# 56-0441-80; RRID: AB_494012 |
| IL2-PerCP-Cy5.5 (JES6-5H4) | eBioscience | Cat# 503821; RRID: AB_961403 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| GW4869 | Sigma | Cat# D1692 |
| StrataClean Resin | Agilent Technologies | Cat# 400714 |
| Deposited Data | ||
| Raw and processed small RNA sequencing data | This paper | GEO: GSE121724 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 | Jackson Laboratory | JAX:000664 |
| Oligonucleotides | ||
| Leu-TAA-5RF: ACCAGAATGGCCGAGTGGTTA | This paper | N/A |
| Leu-TAG-5RF: GGTAGCGTGGCCGAGCGGTC | This paper | N/A |
| Leu-TAA-3RF: GGATTTATATCCGCGTGGG | This paper | N/A |
| LeuTAA5_FP_2: ACACTCCAGCTGGGACCAGAATGGCCGA | This paper | N/A |
| LeuTAA3_FP_2: ACACTCCAGCTGGGTGGATTTATATCCG | This paper | N/A |
| LeuTAG5_FP_2: ACACTCCAGCTGGGGGTAGCGTGGCCGA | This paper | N/A |
| LeuTAA5_RP_2: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCACTCG | This paper | N/A |
| LeuTAA3_RP_2: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCACGCGG | This paper | N/A |
| LeuTAG5_RP_2: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCGCTCG | This paper | N/A |
| LeuTAA3_Pr_2: 5′- /56-FAM/TTCAGTTGA/ZEN/GCCACGCGG/3IBkFQ/ –3′ | This paper | N/A |
| LeuTAG5_Pr_2: 5′- /56-FAM/TTCAGTTGA/ZEN/GACCGCTCG/3IBkFQ/ –3′ | This paper | N/A |
| LeuTAA5_Pr_2: 5′- /56-FAM/TTCAGTTGA/ZEN/GACCACTCG/3IBkFQ/ –3′ | This paper | N/A |
| Universal RP: GTGTCGTGGAGTCGGC | This paper | N/A |
| Software and Algorithms | ||
| Bowtie2 v2.2.4 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2 |
| Samtools v1.7 | Li et al., 2009 | https://github.com/samtools/samtools |
| Bedtools v2.27.1 | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2 |
| DEseq2 v1.18.1 | Love et al., 2014 | https://bioconductor.org/biocLite.R |
| FASTX-Toolkit v0.0.14 | Gregory Hannon lab at cold spring harbor laboratory | http://hannonlab.cshl.edu/fastx_toolkit |
Highlights.
Specific tRFs are enriched in extracellular vesicles released by T cells
T cell activating signals regulate tRF secretion
Activation enhances multivesicular body formation and cellular tRF downregulation
Activation-induced, extracellular-vesicle-enriched tRFs inhibit T cell activation
ACKNOWLEDGMENTS
This publication is part of the NIH Extracellular RNA Communication Consortium paper package and was supported by the NIH Common Fund’s exRNA Communication Program. We thank Darryl Mar, Priti Singh, and Annie Wang for help with mouse colony maintenance. This work was supported by the NIH (U19CA179512) and the Sandler Asthma Basic Research Center.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.11.073.
REFERENCES
- Alexander M, Ramstead AG, Bauer KM, Lee SH, Runtsch MC, Wallace J, Huffaker TB, Larsen DK, Tolmachova T, Seabra MC, et al. (2017). Rab27-Dependent Exosome Production Inhibits Chronic Inflammation and Enables Acute Responses to Inflammatory Stimuli. J. Immunol 199, 3559–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglio SR, Rooijers K, Koppers-Lalic D, Verweij FJ, Pérez Lanzón M, Zini N, Naaijkens B, Perut F, Niessen HW, Baldini N, and Pegtel DM (2015). Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther 6, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcena C, Stefanovic M, Tutusaus A, Martinez-Nieto GA, Martinez L, Garcia-Ruiz C, de Mingo A, Caballeria J, Fernandez-Checa JC, Mari M, and Morales A (2015). Angiogenin secretion from hepatoma cells activates hepatic stellate cells to amplify a self-sustained cycle promoting liver cancer. Sci. Rep 5, 7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidlingmaier S, Ha K, Lee NK, Su Y, and Liu B (2016). Proteome-wide Identification of Novel Ceramide-binding Proteins by Yeast Surface cDNA Display and Deep Sequencing. Mol. Cell. Proteomics 15, 1232–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, and Waalkes TP (1977). High turnover rate of transfer RNA in tumor tissue. Cancer Res 37, 3362–3366. [PubMed] [Google Scholar]
- Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, Kremmer E, Heissmeyer V, McManus MT, Erle DJ, et al. (2013). T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med 210, 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S, et al. (2015). KRAS-dependent sorting of miRNA to exosomes. eLife 4, e07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Son JB, Kang J, Kwon J, Kim JH, Jung M, Kim SK, Kim S, and Mun JY (2017). Leucine-induced localization of Leucyl-tRNA synthetase in lysosome membrane. Biochem. Biophys. Res. Commun 493, 1129–1135. [DOI] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, and Hutvagner G (2009). Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15, 2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Raposo G, and Théry C (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol 30, 255–289. [DOI] [PubMed] [Google Scholar]
- Daniele T, Hackmann Y, Ritter AT, Wenham M, Booth S, Bossi G, Schintler M, Auer-Grumbach M, and Griffiths GM (2011). A role for Rab7 in the movement of secretory granules in cytotoxic T lymphocytes. Traffic 12, 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Candia P, Torri A, Gorletta T, Fedeli M, Bulgheroni E, Cheroni C, Marabita F, Crosti M, Moro M, Pariani E, et al. (2013). Intracellular modulation, extracellular disposal and serum increase of MiR-150 mark lymphocyte activation. PLoS ONE 8, e75348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Candia P, Torri A, Pagani M, and Abrignani S (2014). Serum micro-RNAs as Biomarkers of Human Lymphocyte Activation in Health and Disease. Front. Immunol 5, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Basile G, Ménasché G, and Fischer A (2010). Molecular mechanisms of biogenesis and exocytosis of cytotoxic granules. Nat. Rev. Immunol 10, 568–579. [DOI] [PubMed] [Google Scholar]
- Dhahbi JM (2015). 5′ tRNA Halves: The Next Generation of Immune Signaling Molecules. Front. Immunol 6, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebinu JO, Stang SL, Teixeira C, Bottorff DA, Hooton J, Blumberg PM, Barry M, Bleakley RC, Ostergaard HL, and Stone JC (2000). RasGRP links T-cell receptor signaling to Ras. Blood 95, 3199–3203. [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N,Tisdale S, Kedersha N, Hu GF, and Anderson P (2010). Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem 285, 10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Brown M, Morozov P, Ten Hoeve JJ, Ben-Dov IZ, Hovestadt V, Hafner M, Renwick N, Mihailović A, Wessels LF, and Tuschl T (2012). Bioinformatic analysis of barcoded cDNA libraries for small RNA profiling by next-generation sequencing. Methods 58, 171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye BC, Halfter S, Djudjaj S, Muehlenberg P, Weber S, Raffetseder U, En-Nia A, Knott H, Baron JM, Dooley S, et al. (2009). Y-box protein-1 is actively secreted through a non-classical pathway and acts as an extracellular mitogen. EMBO Rep 10, 783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, and Zheng X (2009). Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583, 437–442. [DOI] [PubMed] [Google Scholar]
- Fuchs RT, Sun Z, Zhuang F, and Robb GB (2015). Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. PLoS ONE 10, e0126049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J, and Polacek N (2013). Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 10, 1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, and Voinnet O (2009). Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol 11, 1143–1149. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, and Tavazoie SF (2015). Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 161,790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, and Kim S (2012). Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149, 410–424. [DOI] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, and Anderson P (2011). Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, and Kalluri R (2017). Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 546, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX, et al. (2014). Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep 8, 1649–1658. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, and Dutta A (2009).Anovel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CC, Eaton SA, Young PE, Lee M, Shuttleworth R, Humphreys DT, Grau GE, Combes V, Bebawy M, Gong J, et al. (2013). Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol 10, 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Liu R, Yin L, and Pu Y (2014). Expression profiling of exosomal miRNAs derived from human esophageal cancer cells by Solexa high-throughput sequencing. Int. J. Mol. Sci 15, 15530–15551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, and Eddy SR (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luberto C, Hassler DF, Signorelli P, Okamoto Y, Sawai H, Boros E, Hazen-Martin DJ, Obeid LM, Hannun YA, and Smith GK (2002). Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J. Biol. Chem 277, 41128–41139. [DOI] [PubMed] [Google Scholar]
- Marks-Konczalik J, Dubois S, Losi JM, Sabzevari H, Yamada N, Feigenbaum L, Waldmann TA, and Tagaya Y (2000). IL-2-induced activation-induced cell death is inhibited in IL-15 transgenic mice. Proc. Natl. Acad. Sci. USA 97, 11445–11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, and Dalla-Favera R (2013). tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 110, 1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie AJ, Hoshino D, Hong NH, Cha DJ, Franklin JL, Coffey RJ, Patton JG, and Weaver AM (2016). KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep 15, 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menck K, Sönmezer C, Worst TS, Schulz M, Dihazi GH, Streit F, Erdmann G, Kling S, Boutros M, Binder C, and Gross JC (2017). Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. J. Extracell. Vesicles 6, 1378056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moltzahn F, Olshen AB, Baehner L, Peek A, Fong L, Stöppler H, Simko J, Hilton JF, Carroll P, and Blelloch R (2011). Microfluidic-based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients. Cancer Res 71, 550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye IS, Coomes SM, Pelly VS, Czieso S, Papayannopoulos V, Tolmachova T, Seabra MC, and Wilson MS (2014). MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity 41, 89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn AJ, Gutbrod MJ, LeBlanc C, and Martienssen R (2017). LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 170, 61–71.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, and Schekman R (2016). Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 5, e19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer J, Gehrke CW, Kuo KC, Waalkes TP, and Borek E (1979). tRNA breakdown products as markers for cancer. Cancer 44, 2120–2123. [DOI] [PubMed] [Google Scholar]
- Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, and Ansel KM (2011). MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 35, 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Amigorena S, Raposo G, and Clayton A (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol 30, 3.22.1–3.22.29. [DOI] [PubMed] [Google Scholar]
- Thompson DM, and Parker R (2009). Stressing out over tRNA cleavage. Cell 138, 215–219. [DOI] [PubMed] [Google Scholar]
- Tkach M, and Théry C (2016). Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 164, 1226–1232. [DOI] [PubMed] [Google Scholar]
- Tosar JP, Gámbaro F, Sanguinetti J, Bonilla B, Witwer KW, and Cayota A (2015). Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res 43, 5601–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, and Simons M (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319,1244–1247. [DOI] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, and Sánchez-Madrid F (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun 4, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Batagov AO, Schinelli S, Wang J, Wang Y, El Fatimy R, Rabinovsky R, Balaj L, Chen CC, Hochberg F, et al. (2017). Coding and non-coding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun 8, 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, and Tuschl T (2013). Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. USA 110, 4255–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data are deposited at GEO under the accession number GEO: GSE121724.


