Abstract
Background-
Proper dynamics of RNA polymerase II (Pol II), such as promoter recruitment and elongation, are essential for transcription. Peroxisome proliferator-activated receptor (PPAR)-γ coactivator-1α (PGC-1α), also termed PPARGC1a, is a transcriptional coactivator that stimulates energy metabolism, and PGC-1α target genes are downregulated in the failing heart. However, whether the dysregulation of Pol II dynamics occurs in PGC-1α target genes in heart failure has not been defined.
Methods and Results-
Chromatin immunoprecipitation-sequencing (ChIP-seq) revealed that reduced promoter occupancy was a major form of Pol II dysregulation on PGC-1α target metabolic gene promoters in the pressure-overload (PO)-induced heart failure model. Cardiac-specific PGC-1α knockout (PGC-1α-cKO) mice showed phenotypic similarity to the PO-induced heart failure model in wild-type mice, such as contractile dysfunction and downregulation of PGC-1α target genes, even under basal conditions. However, the protein levels of PGC-1α was neither changed in the PO model nor in human failing hearts. ChIP assays revealed that the promoter occupancy of Pol II and PGC-1α was consistently reduced both in the PO model and PGC-1α-cKO mice. In vitro DNA binding assays using an endogenous PGC-1α target gene promoter sequence confirmed that PGC-1α recruits Pol II to the promoter.
Conclusions-
These results suggest that PGC-1α promotes the recruitment of Pol II to the PGC-1α target gene promoters. Downregulation of PGC-1α target genes in the failing heart is attributed, in part, to a reduction of the PGC-1α occupancy and the Pol II recruitment to the promoters, which might be a novel mechanism of metabolic perturbations in the failing heart.
Subject code: Metabolism
Keywords: Heart failure, metabolic remodeling, PGC-1α, RNA polymerase II
Introduction
The heart is an organ that demands high energy production and consumption in order to maintain blood flow throughout the body. To satisfy the high energy demand, cardiomyocytes harbor a many mitochondria and utilize fatty acids as a primary source of energy. Peroxisome proliferator-activated receptor (PPAR)-γ coactivator-1α (PGC-1α) is a transcriptional coactivator that broadly induces metabolic genes through co-activation of transcription factors such as nuclear receptors, including peroxisome proliferator-activated receptors (PPARs) and estrogen-related receptors (ERRs). Downregulation of PGC-1α target metabolic genes is a hallmark of heart failure, which may contribute to insufficient energy production and further progression of pathology1. Pressure-overload (PO)-induced failing heart phenotypes and downregulation of metabolic genes are exacerbated in global PGC-1α knockout mice2, 3. Notably, despite the fact that significant metabolic actions of PGC-1α are well recognized not only in the heart but also other organs such as adipose, liver, and skeletal muscle, the role of PGC-1α in the PO-induced heart failure model has not been investigated with cardiac-specific PGC-1α knockout mice. Therefore, whether PGC-1α in cardiomyocytes plays a role in cardiac function and metabolism has not been proven. To address this issue, pathological characterization of cardiac-specific PGC-1α knockout under PO conditions is important.
Although PO-induced failing heart phenotypes are exacerbated in global PGC-1α knockout mice2, this does not necessarily mean that PGC-1α is inactivated during heart failure development. It has been thought that downregulation of PGC-1α is a mechanism responsible for downregulation of the target genes in the failing heart. However, PGC-1α is downregulated neither in human heart failure patients nor in the pressure-overload-induced heart failure model in mice3, 4. Moreover, increased PGC-1α expression with the transgene in the failing heart does not rescue cardiac dysfunction and metabolic perturbations5, 6. Thus, it remains unclear whether downregulation of metabolic genes in the failing heart occurs through the dysregulation of PGC-1α.
RNA polymerase II (Pol II) is a multiple protein complex that transcribes a gene to synthesize mRNA. Pol II is recruited to promoter regions and moves to the gene body for transcription. Proper Pol II dynamics, including promoter recruitment, promoter clearance, elongation, termination and recycling, are required for transcription. Impairment of any of the processes could result in transcriptional suppression. However, it is largely unknown whether the downregulation of PGC-1α target genes in the failing heart is attributed to altered Pol II dynamics.
PGC-1α binds to specific subunits of Mediator complexes and of general transcription factors including TFIID and TFIIH7–9. Mediators and general transcription factors are essential for proper Pol II dynamics. Particularly, Mediators and TFIID play a crucial role in promoter recruitment of Pol II, also termed pre-initiation complex formation, whereas TFIIH mediates promoter clearance of Pol II. However, the extent to which PGC-1α is required for proper Pol II dynamics and whether PGC-1α regulates Pol II recruitment or clearance has not been defined.
In this study, we used cardiac-specific PGC-1α knockout mice to address the hypotheses that 1) PGC-1α has a regulatory role in Pol II dynamics and 2) Pol II is dysregulated in PO-induced heart failure model due to PGC-1α dysregulation. We demonstrate that Pol II occupancy in metabolic gene promoters was reduced in the failing heart, concurrent with reduced occupancy of PGC-1α at those promoters. Cardiac-specific deletion of PGC-1α was sufficient to reduce Pol II promoter occupancy. Conversely, overexpression of PGC-1α promoted the recruitment of Pol II to the promoters of metabolic genes in cultured cardiomyocytes and in vitro reconstitution systems. Taken altogether, these results suggest that reduced promoter occupancy of PGC-1α is a mechanism responsible for reduced Pol II recruitment leading to metabolic disturbances in the failing heart.
Methods
Data, Material, and Code Disclosure
PGC-1α-cKO mice can be distributed upon material transfer agreement from original sources of PGC-1αflox/flox and αMHC-Cre mice. Upon request to the corresponding author, plasmid and virus vectors can be distributed unless otherwise any conflicts noticed.
Animal experiments
The conditional knockout line of PGC-1αflox/flox mice with a C57BL/6 background was obtained from Jackson laboratory10. The PGC-1αflox/flox mice crossed with cardiac-specific Cre transgenic mice (αMHC-Cre). All procedures involving animals were performed in accordance with protocols approved by Rutgers Biomedical and Health Sciences.
Statistical analysis
Statistical comparisons were made using Kaplan-Meier log rank test and a nonparametric Mann-Whitney U test with GraphPad Prism 8.1. P<0.05 was defined as statistically significant. All error bars represent S.E.M.
Results
Recruitment of Pol II to metabolic gene promoters is reduced in the failing heart.
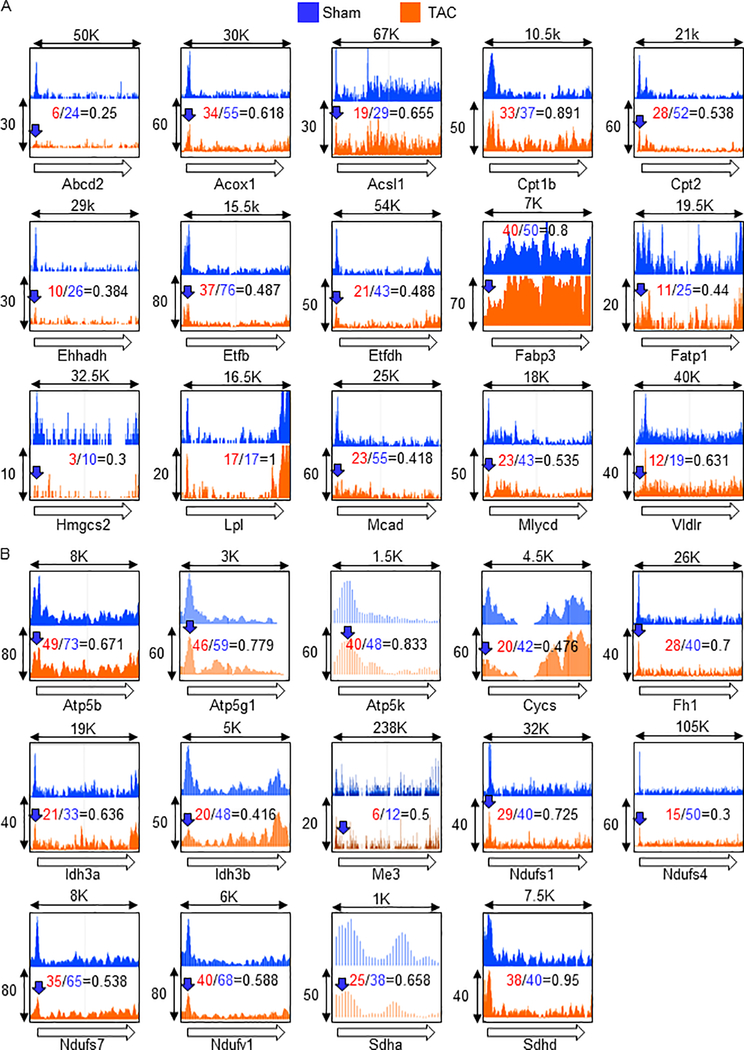
Genome-wide Pol II localization provides insight into the mechanism responsible for gene expression changes. Pressure overload (PO) induced by transverse aortic constriction (TAC) is an animal model of heart failure in which surgical constriction of the aorta induces PO in the left ventricle, leading to downregulation of metabolic genes and cardiac dysfunction in mice. To clarify how Pol II is regulated in metabolic genes under PO conditions, Pol II occupancy was examined with chromatin immunoprecipitation-sequencing (ChIP-seq). We focused on PPAR and ERR target genes, since their regulation is through co-activator PGC-1α. PPAR target genes are enriched in fatty acid metabolism including uptake, transport, and oxidation (Figure 1A), whereas ERR target genes are enriched in the mitochondrial Krebs cycle and electron transport chain (Figure 1B). The ChIP-seq analysis revealed that Pol II occupancy was reduced in 26 out of the 29 gene promoters under PO conditions (blue arrows). In addition, an increase in Pol II occupancy flanking the transcription termination site was observed in three genes: Fabp3, Cycs, and Idh3b, indicating impairment of transcriptional termination. The occupancy of Pol II on Fabp3 and Cycs also increased in the gene body, suggesting impairment of Pol II release from the transcription termination site. The specific impairment of Pol II promoter clearance was not observed in any of these genes, which was presumably evidenced by either no change or increased Pol II occupancy in the promoter but reduced Pol II occupancy in the gene body. These results suggest that inhibition of promoter recruitment is a major form of Pol II dysregulation in the PGC-1α target metabolic genes under PO conditions.
Fig. 1.
Reduced promoter occupancy is a major form of Pol II dysregulation in metabolic gene promoters under PO conditions. ChIP-seq was performed with anti-Pol II antibody after 4 days of TAC. The Pol II occupancies in known or predicted PPAR (A) and ERR (B) target gene promoters were shown. Open arrows indicate direction of gene body, and therefore, origin of the arrow indicates promoter region, whereas arrow head indicates transcription termination site. Blue arrows indicate at least 15% reduction of a peak of Pol II occupancy in the promoter region under PO conditions. X-axis is length of genomic region (Kbp). Y-axis is relative ChIP coverage provided by Integrated Genome Browser.
Cardiac-specific PGC-1α deletion induces failing heart phenotypes
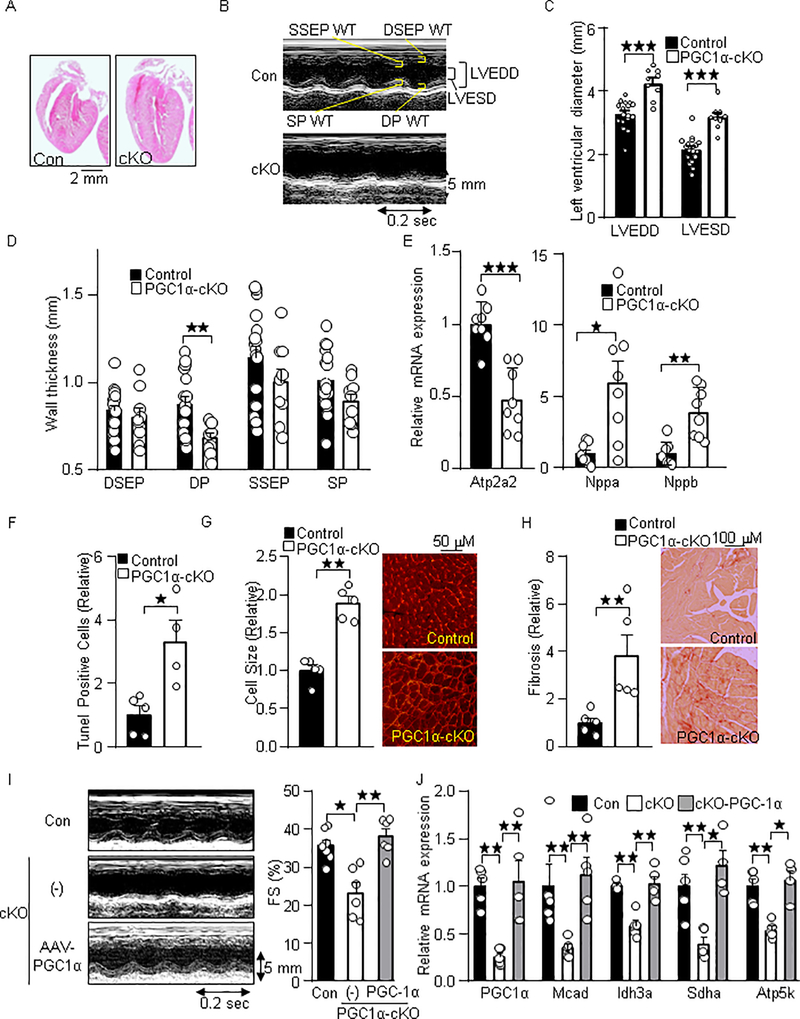
To test the role of PGC-1α in Pol II dynamics in metabolic gene promoters, we used cardiac-specific PGC-1α knockout (PGC-1α-cKO) mice generated with PGC-1αflox/flox and αMHC-Cre mice. To investigate the extent to which PGC-1α in cardiomyocytes maintains cardiac function and metabolic gene expression, we first characterized cardiac phenotypes and metabolic gene expression in the PGC-1α-cKO mice. As shown in Figure 2A, the hearts of PGC-1α-cKO mice were enlarged compared to control hearts. Echocardiographic measurements demonstrated that cardiac systolic dysfunction and increased left ventricular diameters (LVEDD (mm): control: 3.27; PGC-1α-cKO: 4.25, p<0.0001, LVESD (mm): control; 2.13, PGC-1α-cKO; 3.18, p<0.0001), and a trend of reduced wall thicknesses were also observed in PGC-1α-cKO mice, indicating a left ventricular dilation (Figure 2B–D). Gene expressional analyses showed decreased expression level of Atp2a2 (p=0.0006), and increased expression levels of Nppa (p=0.0104) and Nppb (p=0.0011), indicating cardiac hypertrophy and failure in PGC-1α-cKO mice (Figure 2E). Histological analyses showed increases in apoptotic cell death (p=0.0159), fibrosis (p=0.0079), and cellular hypertrophy (p=0.0079) in the PGC-1α-cKO mice under basal conditions (Figure 2F–H). To investigate the extent to which loss of PGC-1α negatively regulates cardiac function in the adult phase, PGC-1α was re-expressed in 6–11-week-old PGC-1α-cKO mice with an adeno-associated virus vector (AAV). Cardiac contractile dysfunction and impaired PGC-1α target gene expression observed in PGC-1α-cKO mice were normalized after 3 weeks of AAV transduction (Fractional shortening (FS) %: Control: 23.9, PGC-1α; 38.2, p=0.0043) (Figure 2I–J). In addition, cardiac dilation characterized by reduced wall thickness and increased left ventricular diameters tended to be normalized by AAV-PGC-1α (LVEDD (mm); control: 4.41, AAV-PGC-1α: 3.82; p<0.017, LVESD (mm): control: 3.37; AAV-PGC-1α: 2.37, p=0.0087) (Table 1). These results suggest that PGC-1α ablation in the adult heart resulted in mild heart failure under basal conditions.
Fig. 2.
Cardiac-specific loss of PGC-1α results in heart failure. (A) H&E stained images of the PGC-1α-cKO mice. (B) Representative M-mode echocardiography of the PGC-1α-cKO mice. (C) Enlarged left ventricular diameter and (D) reduced wall thickness (WT). LVEDD: LV end diastolic dimension, LVESD: LV end systolic dimension, DSEP: Diastolic septal, DP: Distolic posterior: SP: Systolic posterior, SSEP: systolic septal. (E) Heart failure marker genes expression in PGC-1α-cKO mice. The expression of indicated genes was examined by quantitative real time PCR (N=8). Atp2a2: ATPase, Ca++ transporting, cardiac muscle, slow twitch 2, Nppa: Natriuretic peptide A, and Nppb: Natriuretic peptide B. Histological analyses of myocardium in PGC-1α-cKO mice. Histochemical analyses were performed with (F) Tunel, (G) WGA, and (H) PSR staining. (I) Re-expression of PGC-1α with AAV normalizes cardiac systolic dysfunction in PGC-1α-cKO mice. Representative M-mode echocardiography (Left). Fractional shortening (Right). (J) Re-expression of PGC-1α with AAV normalizes PGC-1α target gene expression in PGC-1α-cKO mice. Echocardiographic measurements (I) and gene expression analysis (J) were performed after 3 weeks of transduction of AAV- PGC-1α and the control (GFP). The numbers of mice examined in each experimental group were: 6–17(C-D), 8 (E), 4–5 (F-H), 5–7 (I) and 4–6 (J). * p<0.05; ** p<0.01; *** p<0.001 as indicated.
Table 1.
Echocardiographic data of PGC-1α-cKO mice under PO conditions.
| Cre BL(6) | Cre PO(6) | Wild BL(17) | Wild PO(17) | cKO BL(9) | cKO PO(7) | cKO-AAV-con(5) | cKO-AAV-PGC-1α(6) | |
|---|---|---|---|---|---|---|---|---|
| HR, bpm | 429±79 | 440±36 | 456±60 | 495±52 | 438±47 | 389±38§ | 466±32 | 436±61 |
| LVEDD, mm | 3.19±0.34 | 3.74±0.53 | 3.27±0.44 | 3.96±0.43† | 4.25±0.47† | 4.55±0.49‡ | 4.41±0.46 | 3.82±0.29|| |
| LVESD, mm | 2.09±0.41 | 2.92±0.68 | 2.13±0.43 | 3.03±0.54† | 3.18±0.39† | 4.15±0.36§ | 3.37±0.59 | 2.37±0.34# |
| LVEF, % | 72.0±9.1 | 52.0±14.9 | 71.8±8.8 | 54.9±13.1† | 57.6±7.5† | 23.3±7.0§ | 54.9±12.8 | 76.0±5.6# |
| LVFS, % | 34.9±7.1 | 22.5±8.2 | 35.1±6.7 | 24.0±7.7† | 25.1±4.4† | 8.5±2.8§ | 23.9±7.2 | 38.2±4.7# |
| DSEP WT, mm | 0.71±0.12 | 0.91±0.06 | 0.83±0.13 | 0.93±0.14 | 0.80±0.15 | 0.77±0.17‡ | 0.77±0.04 | 0.81±0.20 |
| DP WT, mm | 0.77±0.22 | 0.77±0.10 | 0.88±0.17 | 0.89±0.13 | 0.68±0.09* | 0.64±0.12§ | 0.73±0.12 | 0.85±0.11|| |
| SSEP WT, mm | 0.99±0.19 | 1.06±0.16 | 1.14±0.28 | 1.17±0.26 | 1.00±0.23 | 0.83±0.17§ | 0.92±0.07 | 1.18±0.16# |
| SP WT, mm | 0.90±0.29 | 0.86±0.13 | 1.01±0.17 | 1.10±0.16 | 0.89±0.13 | 0.72±0.26§ | 0.95±0.21 | 1.18±0.20 || |
| Age Month | 2.52±0.88 | 4.17±0.60 | 3.82±0.47 | 3.09±0.97 | 3.82±0.77 | 2.68±0.81 | 3.11±0.77 | 2.80±0.74 |
HR, heart rate; LV, left ventricle; EDD, end-diastolic dimension; ESD, end-systolic dimension; EF, ejection fraction, FS, fractional shortening; DSEP, Diastolic septal; DP, Diastolic posterior; SP, Systolic posterior; SSEP, systolic septal; WT, wall thickness.
Data are mean ± SD.
p<0.01;
p<0.01vs Wild BL
p<0.05;
p<0.001 vs Wild PO
p<0.05;
p<0.01vs cKO-AAV Cont.
Parenthesis indicates N.
PGC-1α-cKO mice are susceptible to pressure overload
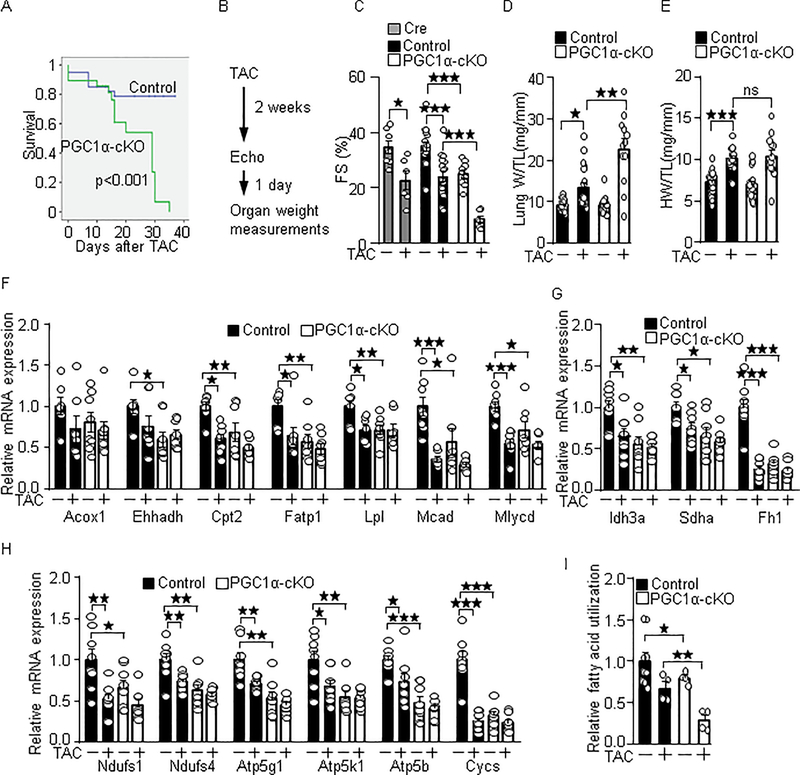
Under the PO conditions, PGC-1α-cKO mice showed a high mortality rate (Median survival: 29 days, p=0.0002), compared to the control mice (Figure 3A). Therefore, gross and echocardiographic measurements were performed after 2 weeks of PO (Figure 3B). As shown in Figure 3C, PO induced cardiac contractile dysfunction, evidenced by reduced fractional shortening in wild-type and αMHC-Cre mice. On the other hand, PGC-1α-cKO mice showed contractile dysfunction even without PO (FS (%); control: 35.1; PGC-1α-cKO: 25.1, p=0.0002), which was further exacerbated under PO conditions (FS (%); control: 24.0; PGC-1α-cKO: 8.5, p<0.0001). Echocardiographic data are summarized in Table 1. PO-induced lung congestion, an index of heart failure measured as lung weight/tibia length ratio (mg/mm) was increased in PGC-1α-cKO mice (control: 13.43; PGC-1α-cKO: 22.65, p=0.0016) (Figure 3D). PO-induced cardiac hypertrophy measured as heart weight/tibia length ratio (mg/mm) was not significantly changed in PGC-1α-cKO mice (control: 10.08; PGC-1α-cKO: 10.39, p=0.88) (Figure 3E). These results suggest that cardiac-specific PGC-1α deletion exacerbates PO-induced failing heart phenotypes. Results of organ weight measurements are summarized in Table 2. The metabolic genes involved in fatty acid metabolism (Figure 3F), Krebs cycle (Figure 3G), and mitochondrial electron transport (Figure 3H) were downregulated in the wild-type mice in response to PO and were downregulated in the PGC-1α-cKO mice even without PO. Fatty acid utilization activity was impaired in the PGC-1α-cKO mice under both basal and PO conditions (Fig. 3I). These results suggest that PGC-1α in cardiomyocytes plays an important role in cardiac energetics in both basal and PO conditions.
Fig. 3.
PGC-1α-cKO mice are susceptible to PO-induced heart failure. (A) High mortality rate in PGC-1α-cKO mice under PO conditions. PGC-1α-cKO mice were subjected to TAC. Statistical analysis was performed with the Kaplan-Meier log rank test. (B) Timeline of echocardiography and organ weight measurements. Fractional shortening (FS) (C), lung congestion (D), cardiac hypertrophy (E), PGC-1α target genes expression (F-H), and fatty acid utilization activity (I) were examined in PGC-1α-cKO mice after 2 weeks of TAC. PGC-1α target genes involved in fatty acid metabolism (F), Krebs cycle (G) and mitochondrial ATP production (H). The numbers of mice examined in each experimental group were: 39(Wt)-29(cKO) (A), 7–17(C), 12–26 (D-E), 7–8 (F-H), and 4–9 (I). * p<0.05; ** p<0.01; *** p<0.001 as indicated.
Table 2.
Organ weight data of PGC-1α-cKO mice under PO conditions.
| Wild BL(26) | Wild PO (18) | cKO BL (23) | cKO PO (12) | |
|---|---|---|---|---|
| Body weight (BW), g | 26.6±4.3 | 24.6±2.9 | 26.8±4.6 | 20.7±3.4† |
| Tibia length (TL), mm | 17.4±0.6 | 17.2±0.5 | 17.8±1.3 | 17.0±0.5 |
| Heart weight (HW), mg | 126±23 | 173±21 | 124±29 | 177±46 |
| LV weight (LVW), mg | 98.6±18.6 | 138.7±16.5 | 96.9±23.0 | 121.9±21.0† |
| Lung weight (LUW), mg | 158±19 | 230±91 | 160±34 | 386±146† |
| HW/BW | 4.75± 0.60 | 7.07±0.88 | 4.61±0.70 | 8.53±1.81* |
| LVW/BW | 3.70±0.43 | 5.68±0.77 | 3.60±0.48 | 5.91±0.86 |
| LUW/BW | 6.02±0.81 | 9.60±4.27 | 6.05±1.12 | 19.03±8.14† |
| HW/TL | 7.24±1.32 | 10.08±1.19 | 6.97±1.56 | 10.39±2.66 |
| LVW/TL | 5.66±1.06 | 8.09±0.95 | 5.44±1.12 | 7.17±1.24† |
| LUW/TL | 9.07±1.17 | 13.43±5.22 | 9.04±1.76 | 22.65±8.50† |
| Age, Month | 3.62±0.94 | 3.75±1.10 | 3.75±1.21 | 2.74±0.49 |
PO: TAC 2 weeks.
Data are mean ± SD.
p<0.05;
p<0.01 vs Wild PO.
Parenthesis indicates N.
PGC-1α is not significantly downregulated under pressure overload conditions
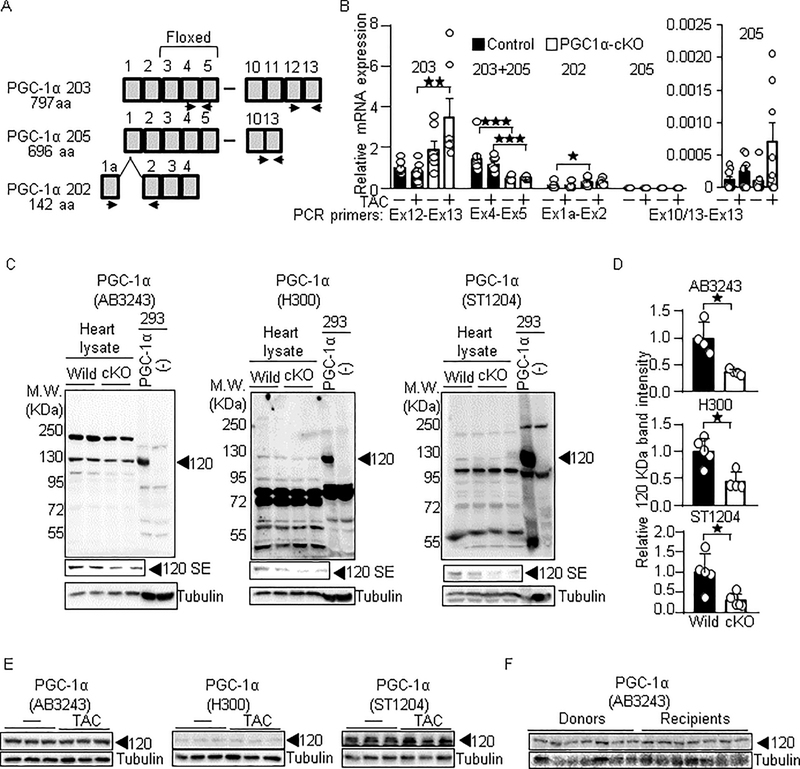
PGC-1α-cKO mice even in the absence of PO showed phenotypic similarity to the PO model in the wild-type mice, such as impaired contractile function and downregulation of PGC-1α target genes. The phenotypic similarity may be caused by downregulation of PGC-1α in the PO model and PGC-1α-cKO mice. To test this, we examined the expression levels of all three splice variants of PGC-1α mRNA that encode a protein, and excluded the variants subjected to non-sense mediated decay. As shown in Figure 4A and 4B, an authentic PGC-1α (PGC-1α 203) tended to decrease in PO conditions, but it did not attain a statistical significance (p=0.505). In addition, the other splice variants including PGC-1α 205 and PGC-1α 202 tended to increase in PO conditions. Thus, PGC-1α mRNAs do not significantly decrease in PO conditions. Notably, we found that a region of PGC-1α mRNA corresponding to the floxed exons (Exons 3 to 5) was significantly downregulated in PGC-1α-cKO mice (Basal: p=0.0007; PO: p=0.007), but the other regions were not. Thus, artificial mRNA of PGC-1α lacking floxed exons is likely expressed in PGC-1α-cKO mice. To test whether PGC-1α protein is downregulated under PO conditions, western blot analyses were performed. Because the molecular weight of PGC-1α varies from 80 to 120 KDa among the literature, we first determined the PGC-1α-specific signal in the heart lysate. While we tested several antibodies, three of them detected PGC-1α exogenously expressed in HEK293 cells, which was approximately 120 KDa (Figure 4C). The signal at 120 KDa was detected in the heart lysate, which was partly but significantly reduced in the PGC-1α-cKO mice, suggesting that the signal at 120 KDa represents PGC-1α (Figure 4D). Then, we investigated whether the PGC-1α protein at 120 KDa is downregulated under PO conditions. As shown in Figure 4E, the protein expression of PGC-1α, detected by all three antibodies, was not significantly changed in the PO model. Similarly, we did not observe significant changes in the protein expression of PGC-1α in human failing hearts (Relative band intensity: recipients: 1; donors: 1.06, p=0.62) (Figure 4F). Taken altogether, our results suggest that downregulation of PGC-1α is not necessary for the downregulation of PGC-1α target genes in the failing heart.
Fig. 4.
PGC-1α is not significantly downregulated in the failing heart. (A-B) The expression of PGC-1α mRNA in PGC-1α-cKO mice. The splice isoforms of PGC-1α are shown in Ensembl (http://useast.ensembl.org/index.html). (A) Schematic representation of splice isoforms of PGC-1α and primer sets for detecting them. PGC-1α 203, canonical full length of PGC-1α, encodes 797 amino acids (aa) of protein. PGC-1α 205, lacking Ex11 to 12, encodes 696 aa of protein. PGC-1α 202 is composed of alternative Ex1 and Ex2 to 4, which encodes 142 aa of protein. (B) Relative copy number of indicated PGC-1α splice isoforms. Relative copy number of PGC-1α 203 in wild type mice under basal conditions is defined as 1. N=6–8. (C) PGC-1α specific signal in heart lysate. Western blot analyses were performed with heart lysate derived from PGC-1α-cKO and from HEK293 cells with exogenous PGC-1α expression. Anti-PGC-1α antibodies used for these analyses were Millipore AB3243, Santa Cruz H-300, and Calbiochem ST1204. SE: Short exposure. (D) Relative signal intensities of the signal at 120 KDa/tubulin shown in right panels. N=4–6. (E-F) The proteins levels of PGC-1α are not significantly downregulated in the TAC model (E) and human failing hearts (F). Donors: Healthy hearts. Recipients: Failing hearts. * p<0.05; ** p<0.01; *** p<0.001 as indicated.
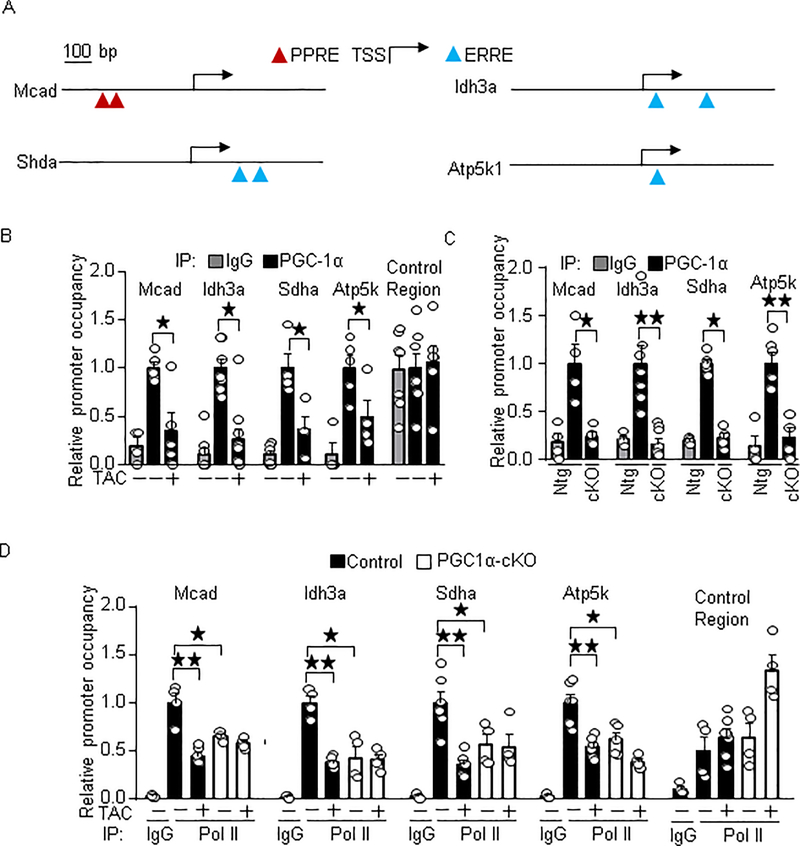
Reduced occupancy of PGC-1α in metabolic gene promoters in the PO model
It is generally thought that recruitment of PGC-1α to target gene promoters is essential for PGC-1α-induced transcriptional activation. Since PGC-1α was not significantly downregulated in the failing heart in mice and humans, we hypothesized that PGC-1α dissociates from the target gene promoters in the failing heart. To test this, we performed ChIP assays on wild-type and PGC-1α-cKO mice under PO conditions. We chose PPAR and ERR target gene promoters containing their binding sequences (PPRE and ERRE) nearby the transcription start site (TSS), such as Mcad, Idh3a, Sdha, and Atp5k (Figure. 5A). As shown in Figure 5B, PGC-1α occupancy in these promoters was significantly reduced in the PO conditions (relative promoter occupancy of PGC-1α in PGC-1α-cKO: control mice defined as 1; Mcad 0.37, p=0.037; Idh3a 0.38, p=0.032; Sdha 0.36, p=0.029; Atp5k 0.49, p=0.038). As expected, PGC-1α occupancy was significantly reduced in PGC-1α-cKO mice (Figure 5C). These results suggest that PGC-1α dissociates from target gene promoters in the failing heart. To investigate whether reduced promoter occupancy of PGC-1α is accompanied by the reduced Pol II occupancy, ChIP assays were performed with anti-Pol II antibody. Pol II occupancy in these promoters was also reduced in the PO model (relative promoter occupancy of Pol II in TAC mice: control mice defined as 1; Mcad 0.48, p=0.0095; Idh3a 0.38, p=0.0095; Sdha 0.36, p=0.0022; Atp5k 0.54, p=0.0022) and in PGC-1α-cKO mice (relative promoter occupancy of Pol II in PGC-1α-cKO mice: control mice defined as 1; Mcad 0.65, p=0.035; Idh3a 0.43, p=0.029; Sdha 0.57, p=0.038; Atp5k 0.62, p=0.0303) (Figure 5D). Taken altogether, these results suggest that PGC-1α dissociates from target gene promoters in the failing heart, which is accompanied by reduced Pol II recruitment.
Fig. 5.
PGC-1α dissociates target gene promoters in the failing heart. (A) Schematic representation of PGC-1α target gene promoters. (B-C) The promoter occupancy of PGC-1α in the target gene promoters in the PO model (B) and PGC-1α-cKO mice (C). The numbers of mice examined in each experimental group were: 4–6 (B and C) and 4–6 (D). * p<0.05; ** p<0.01 as indicated.
PGC-1α promotes the recruitment of Pol II to PGC-1α target gene promoters
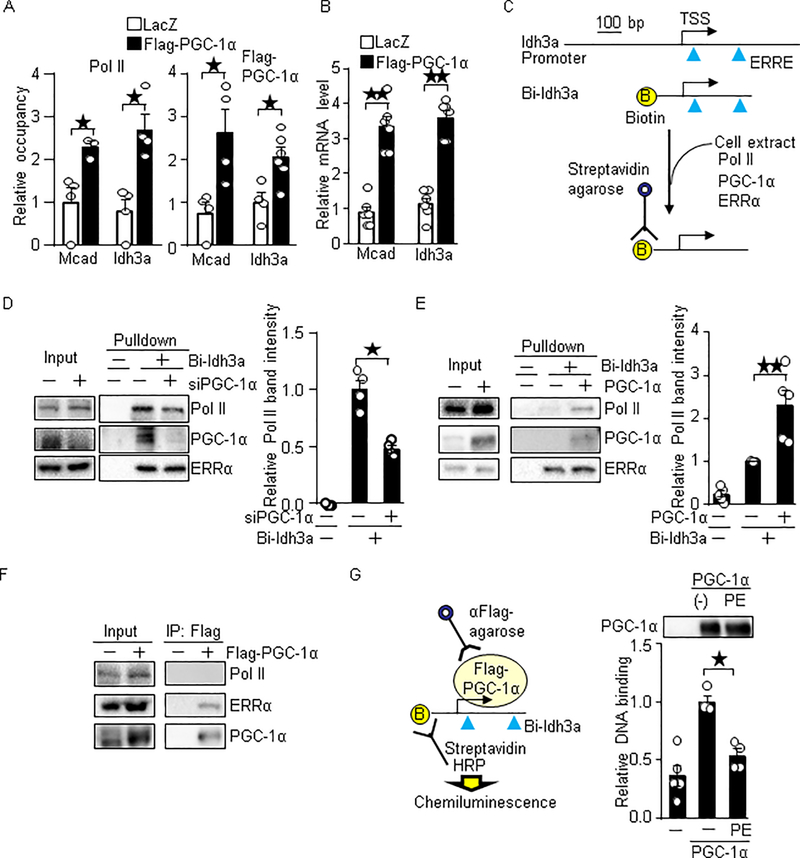
Based on our observations, we hypothesized that PGC-1α promotes Pol II recruitment, and therefore, the reduced PGC-1α promoter occupancy leads to a reduction in the Pol II recruitment in the failing heart. To test if PGC-1α promotes Pol II recruitment, we exogenously expressed PGC-1α in primary cultured cardiomyocytes. As shown in Figure 6A, overexpression of PGC-1α in cardiomyocytes promoted the recruitment of Pol II to PGC-1α target gene promoters, such as Mcad (p=0.029) and Idh3a (p=0.029). Consistent with this result, PGC-1α upregulated Mcad (p=0.0022) and Idh3a (p=0.0022) (Figure 6B). These results suggest that PGC-1α-induced Pol II recruitment is associated with the target gene induction. To verify this result, we performed in vitro DNA binding assays using biotin-labeled DNA comprising 380 bp of the Idh3a promoter containing ERREs and TSS (Figure 6C). The promoter was incubated with cell extract as a source of Pol II and general transcriptional machineries. Pol II recruitment to the Idh3a promoter was partly reduced by PGC-1α knockdown in primary cultured cardiomyocytes (relative promoter binding of Pol II: control: 1; PGC-1α knockdown: 0.48, p=0.029) (Figure 6D). Conversely, the Pol II recruitment was enhanced by overexpression of PGC-1α in HEK293 cells (relative promoter binding of Pol II: control: 1; PGC-1α overexpression: 2.3, p=0.0079) (Figure 6E). However, Flag-tagged PGC-1α (Flag-PGC-1α) did not interact with Pol II in cardiomyocytes, whereas a direct binding of Flag-PGC-1α to ERRα was detected (Figure 6F). Thus, PGC-1α does not directly bind to Pol II. To investigate whether PGC-1α’s ability to bind to promoters declines in heart failure development, we performed in vitro DNA binding assays with Flag-PGC-1α purified from cardiomyocytes treated with phenylephrine (PE) that mimics a pathological consequence of heart failure in cultured cardiomyocytes. PGC-1α from PE treated cells showed a lesser binding ability to the promoter (relative promoter binding of PGC-1α: control cells: 1; PE-treated cells: 0.53, p=0.0286). These results suggest that PGC-1α promotes Pol II recruitment and that the reduced PGC-1α occupancy of its target gene promoters results in a reduction of Pol II recruitment in the failing heart.
Fig. 6.
PGC-1α promotes Pol II recruitment. (A-B) Overexpression of PGC-1α promotes Pol II recruitment and target gene expression in cardiomyocytes. Flag-PGC-1α was overexpressed with adenovirus vector. (A) ChIP assays were performed with anti-Pol II and anti-Flag antibodies. N=3–8. (B) The expression levels of indicated genes were examined. (C) Schematic representation for the DNA binding assay. Biotin-labeled Idh3a promoter containing ERRE and TSS were incubated with cell lysate. The PGC-1α, ERRα and Pol II bound to the DNA was examined with Western blot analyses. (D) Knockdown of PGC-1α inhibits Pol II recruitment in vitro. N=4. (E) Overexpression of PGC-1α promotes Pol II recruitment. N=5. (F) The binding of PGC-1α to Pol II was not observed with co-immunoprecipitation assay. Flag-PGC-1α was overexpressed in cardiomyocytes with adenovirus vector. Co-immunoprecipitation assays were performed with anti-Flag antibody. (G) PGC-1α purified from phenylephrine (PE) treated cells has a lesser ability to bind to the promoter. Cardiomyocytes expressed Flag-PGC-1α were treated with 100 μM PE for 16 hours. Flag-PGC-1α was immunoprecipitated with anti-Flag-antibody. The immunocomplex was incubated with HEK293 cell lysate as a source of ERRs and general transcriptional machineries, biotin-labeled Idh3a promoter and HRP (horseradish peroxidase)-conjugated streptavidin. The binding of PGC-1α and biotin-labeled Idh3a promoter was measured by chemiluminescence. N=4–5. * p<0.05; ** p<0.01 as indicated.
Discussion
Pol II regulation during heart failure development
The failing heart is known as an energy-starved heart, in association with global downregulation of genes involved in fatty acid metabolism and mitochondrial ATP production11. Pol II plays a central role in transcription and, therefore, gene expression. Importantly, however, whether and how Pol II is dysregulated and is involved in the downregulation of metabolic genes in the failing heart has not been defined. Although impairment of any process of Pol II dynamics could result in transcriptional suppression, we here show that inhibited promoter recruitment is a major form of Pol II dysregulation in metabolic genes. Notably, however, impairment of Pol II transcriptional termination was also observed in several PGC-1α target genes. Taken altogether, although inhibited promoter recruitment is the major form of Pol II dysregulation, the other regulation may concurrently take place. Further studies are needed to clarify how Pol II dynamics are regulated in the failing heart.
Role of PGC-1α in downregulation of PGC-1α target genes in heart failure
Downregulation of PGC-1α was originally thought to be a mechanism responsible for downregulation of PGC-1α target genes in the PO-induced heart failure model, because PGC-1α mRNA is downregulated2. However, diverse outcomes of PGC-1α mRNA expressional changes have been reported, which include upregulation and no change4, 12, 13. We here observed that the mRNA was not significantly downregulated after 2 weeks of TAC in control mice. Since all control mice used for testing PGC-1α mRNA levels were PGC-1αflox/flox mice, we should be aware of the possibility in which the flox insertion may disrupt a critical element that mediates PO-induced downregulation. Concerning PGC-1α protein levels, it was reported that PGC-1α protein was significantly downregulated in the PO model4, although the same group later reported no changes in PGC-1α protein in the PO model14. No changes in PGC-1α protein was also reported by another group6. Our data also showed that there were no changes in PGC-1α protein in the PO model and in human failing hearts. Thus, it is unlikely that downregulation of PGC-1α is essentially required for downregulation of PGC-1α target genes in the failing heart. In this study, we showed that PGC-1α dissociated from the promoters in the failing heart, which could be due to a posttranslational modification or conformational changes of PGC-1α. In addition, it partially explains the downregulation of PGC-1α target genes without changing PGC-1α expression. Although the mechanism by which PGC-1α dissociates from the promoter is different between the PGC-1α-cKO and PO model, PGC-1α-cKO may mimic pathological consequences triggered by promoter dissociation of PGC-1α in the PO model. This could account for the phenotypic similarity between the PGC-1α-cKO and PO model. The mechanism by which PGC-1α dissociates from the promoters should be further investigated. In addition, ChIP-seq analysis of PGC-1α in the PO model would be an intriguing future study. Unfortunately, the H300 anti-PGC-1α antibody qualified for ChIP assay is no longer commercially available, thus we could not conduct the ChIP-seq analysis in this study.
Immunoblot analysis of PGC-1α
While the molecular weight of PGC-1α varies from 80 to 120 KDa among the literature, we showed that the signal at 120 KDa corresponded to a full length PGC-1α. The signal at 120 KDa was partly reduced in PGC-1α-cKO mice. The persistent signal in PGC-1α-cKO mice is possibly due to the following three reasons. First, Cre-induced deletion is not fully achieved. Second, there is PGC1α protein derived from non-cardiomyocytes such as fibroblasts. Indeed, persistent expression of PGC1α mRNA corresponding to even in the floxed region was detected in PGC-1α-cKO hearts (Figure 4A and 4B). Third, there may be a non-specific signal that overlaps with the specific signal. Notably, the signal at 120 KDa was a minor band detected by all three PGC-1α antibodies, whereas they detected major bands at a different molecular weight. This characteristic outcome may account for the varied PGC-1α molecular weight among the literature. Namely, investigators may interpret that a major signal is a PGC-1α specific band. Besides the band at 120 KDa, the others may be splice variants or PGC-1α homologues. The signal above the 120 KDa detected by AB3243 anti-PGC-1α antibody may be PGC-1β or PPRC1 (PGC-1 related protein 1) because of the homology with the antigen peptide and their molecular weight. In addition, several bands below the 120 KDa detected by all three antibodies may be a PGC-1α 205 splice variant which encodes approximately 100 amino acids shorter than the full length. However, the relative expression levels of PGC-1α 205 mRNA was approximately 10,000 times lower than the full length (Figure 4B). Therefore, these signals may represent a posttranslational modification such as cleavage rather than the PGC-1α 205 splice variant. Importantly, none of these signals below 120 KDa were significantly downregulated in the PO model (data not shown). Thus, even if these signals are derived from PGC-1α, they are not significantly downregulated in the PO model.
Mechanism by which PGC-1α stimulates transcription
Although PGC-1α is a well-investigated transcriptional co-activator, the mechanism by which PGC-1α stimulates transcription remains elusive. PGC-1α recruits histone acetyltransferases and chromatin remodeling complexes to the target gene promoters, thereby stimulating transcription15, 16. However, the extent to which PGC-1α is essential for their recruitment has not been investigated with a loss of PGC-1α model. We here show that PGC-1α is essential for Pol II recruitment in a subset of genes, although significant binding of PGC-1α to Pol II was not observed. Thus, it is likely that PGC-1α indirectly interacts with Pol II. PGC-1α may promote Pol II recruitment through recruitment of histone acetyltransferases and chromatin remodeling complexes. Whether recruitment of histone acetyltransferases and chromatin remodeling complexes is inhibited in the loss of PGC-1α model would be an important investigation. Notably, we show that PGC-1α promotes Pol II recruitment with in vitro DNA binding assays. Because we used naked DNA without histones, PGC-1α recruits Pol II independently of histone acetyltransferases and chromatin remodeling complexes in the in vitro DNA binding system. PGC-1α may regulate Pol II dynamics through the general transcription factors and Mediators. The mechanism by which PGC-1α promotes Pol II recruitment should be further investigated.
Discrepancy of cardiac phenotypes in PGC-1α knockout mouse lines
Previous studies investigated the role of PGC-1α in cardiac function in two independent lines of global PGC-1α knockout mice. One group used a global PGC-1α KO line to show cardiac systolic dysfunction under basal conditions, whereas the other group which used the other line, did not show any significant baseline phenotypes2. Interestingly, the PGC-1α knockout line originally showing cardiac systolic dysfunction did not show any baseline phenotypes, when these mice were characterized by the other investigators at another institute3. It is also reported that cardiac-specific PGC-1α knockout mice do not show any significant baseline phenotypes17. Notably, the cardiac-specific PGC-1α mouse line previously reported is essentially the same as the PGC-1α-cKO mice used by this study because we used the identical PGC-1αflox/flox and αMHC-Cre lines. Contrary to the previous report, we observed that the PGC-1α-cKO mouse line showed a certain degree of failing heart phenotypes under basal conditions. Despite the phenotypic variation in the PGC-1α knockout lines under basal conditions, the deteriorated cardiac dysfunction under PO conditions has been commonly observed in the two independent global PGC-1α knockout mice2, 3. We here verified the deteriorated cardiac dysfunction under PO conditions in the PGC-1α-cKO mice. Taken altogether, loss of PGC-1α exacerbates PO-induced failing heart phenotypes regardless global or cardiac specific deletion, while whether or not loss of PGC-1α spontaneously develops heart failure under basal conditions may depend upon genetic background or housing conditions.
In summary, impaired promoter recruitment is a major form of Pol II dysregulation in PGC-1α target genes in the failing heart, which is associated with downregulation of the target genes and metabolic disturbances. Additionally, we observe PGC-1α positively regulates the promoter recruitment of Pol II. Lastly, we find PGC-1α dissociates from the target gene promoters in the failing heart, which could be a mechanism responsible for the impaired promoter recruitment of Pol II.
Supplementary Material
What is new
Downregulation of genes involved in fatty acid oxidation and mitochondrial ATP production is a hallmark of heart failure, which is thought to promote the pathology due to insufficient energy production. PGC-1α is a transcriptional coactivator that induces metabolic genes. RNA polymerase II (Pol II) is an enzyme that transcribes genes, and therefore, proper Pol II dynamics is essential for transcription.
Here we show that 1) PGC-1α plays a role in the promoter recruitment of Pol II and 2) PGC-1α dissociates from the promoters in the failing heart, thereby inhibiting Pol II recruitment.
What are the clinical implications
Previous studies aim to clarify the mechanism responsible for the metabolic disturbance largely focused on expressional changes of responsible transcriptional activators such as PGC-1α. However, downregulation of PGC-1α is not uniformly observed in the failing heart. In addition, forced expression of PGC-1α does not show any therapeutic effect.
Here we show a novel mechanism responsible for the metabolic disturbance, which partly explains the downregulation of PGC-1α target genes independent of PGC-1α downregulation.
This study suggests that promoter recruitment of PGC-1α and Pol II is a novel therapeutic target to normalize PGC-1α target gene expression in the failing heart.
Acknowledgements
The authors thank Christopher D. Brady for critical reading of the manuscript. The authors declare no conflict of interest in this study.
Source of Funding
This work was supported in part by the American Heart Association (AHA) Scientist Developmental Grant 12SDG11890014 (SO), Grant in Aid 17GRNT33440031 (SO), New Jersey Health Foundation research grants (PC56–16 and PC80–17) (SO), an AHA student scholarship (KS), the Glorney-Raisbeck Medical Student Grant (KS), Ministry of Science and Technology Taiwan NSC102–2628-B075–002-MY3 (CH), Foundation Leducq Transatlantic Networks (JS), and US Public Health Service Grant HL67724, HL91469, HL102738, HL112330, and AG23039 (JS).
Footnotes
Disclosure Statement
None.
References
- 1.Sihag S, Cresci S, Li AY, Sucharov CC and Lehman JJ. PGC-1alpha and ERRalpha target gene downregulation is a signature of the failing human heart. J Mol Cell Cardiol. 2009;46:201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A and Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-gamma coactivator 1alpha. Proc Natl Acad Sci U S A. 2006;103:10086–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu Z, Xu X, Hu X, Fassett J, Zhu G, Tao Y, Li J, Huang Y, Zhang P, Zhao B and Chen Y. PGC-1 alpha regulates expression of myocardial mitochondrial antioxidants and myocardial oxidative stress after chronic systolic overload. Antioxidants & redox signaling. 2010;13:1011–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu X, Xu X, Huang Y, Fassett J, Flagg TP, Zhang Y, Nichols CG, Bache RJ and Chen Y. Disruption of sarcolemmal ATP-sensitive potassium channel activity impairs the cardiac response to systolic overload. Circ Res. 2008;103:1009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karamanlidis G, Garcia-Menendez L, Kolwicz SC Jr., Lee CF and Tian R. Promoting PGC-1alpha-driven mitochondrial biogenesis is detrimental in pressure-overloaded mouse hearts. Am J Physiol Heart Circ Physiol. 2014;307:H1307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereira RO, Wende AR, Crum A, Hunter D, Olsen CD, Rawlings T, Riehle C, Ward WF and Abel ED. Maintaining PGC-1alpha expression following pressure overload-induced cardiac hypertrophy preserves angiogenesis but not contractile or mitochondrial function. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014;28:3691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A and Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1 alpha. Cell metabolism. 2006;3:429–438. [DOI] [PubMed] [Google Scholar]
- 8.Traboulsi H, Davoli S, Catez P, Egly JM and Compe E. Dynamic partnership between TFIIH, PGC-1alpha and SIRT1 is impaired in trichothiodystrophy. PLoS genetics. 2014;10:e1004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin JW and Wang G. The Mediator complex: a master coordinator of transcription and cell lineage development. Development. 2014;141:977–87. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D and Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–35. [DOI] [PubMed] [Google Scholar]
- 11.Huss JM and Kelly DP. Mitochondrial energy metabolism in heart failure: a question of balance. J Clin Invest. 2005;115:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huss JM, Imahashi K, Dufour CR, Weinheimer CJ, Courtois M, Kovacs A, Giguere V, Murphy E and Kelly DP. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. [DOI] [PubMed] [Google Scholar]
- 13.Karamanlidis G, Nascimben L, Couper GS, Shekar PS, del Monte F and Tian R. Defective DNA replication impairs mitochondrial biogenesis in human failing hearts. Circ Res. 2010;106:1541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu X, Xu X, Lu Z, Zhang P, Fassett J, Zhang Y, Xin Y, Hall JL, Viollet B, Bache RJ, Huang Y and Chen Y. AMP activated protein kinase-alpha2 regulates expression of estrogen-related receptor-alpha, a metabolic transcription factor related to heart failure development. Hypertension. 2011;58:696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Liu C, Li N, Hao T, Han T, Hill DE, Vidal M and Lin JD. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008;8:105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B and Spiegelman BM. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–71. [DOI] [PubMed] [Google Scholar]
- 17.Patten IS, Rana S, Shahul S, Rowe GC, Jang C, Liu L, Hacker MR, Rhee JS, Mitchell J, Mahmood F, Hess P, Farrell C, Koulisis N, Khankin EV, Burke SD, Tudorache I, Bauersachs J, del Monte F, Hilfiker-Kleiner D, Karumanchi SA and Arany Z. Cardiac angiogenic imbalance leads to peripartum cardiomyopathy. Nature. 2012;485:333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.